Abstract
An epidemiological survey for the monitoring of bovine tuberculosis transmission was carried out in western Liguria, a region in northern Italy. Fifteen Mycobacterium bovis strains were isolated from 63 wild boar samples (62 from mandibular lymph nodes and 1 from a liver specimen). Sixteen mediastinal lymph nodes of 16 head of cattle were collected, and 15 Mycobacterium bovis strains were subsequently cultured. All M. bovis strains isolated from cattle and wild boars were genotyped by spoligotyping and by restriction fragment length polymorphism (RFLP) analysis with the IS6110 and IS1081 probes. All M. bovis strains showed the typical spoligotype characterized by the absence of the 39 to 43 spacers in comparison with the number in M. tuberculosis. A total of nine different clusters were identified by spoligotyping. The largest cluster included 9 strains isolated from wild boars and 11 strains isolated from cattle, thus confirming the possibility of transmission between the two animal species. Fingerprinting by RFLP analysis with the IS6110 probe showed an identical single-band pattern for 29 of 30 strains analyzed, and only 1 strain presented a five-band pattern. The use of IS1081 as a second probe was useful for differentiation of M. bovis from M. bovis BCG but not for differentiation among M. bovis strains, which presented the same undifferentiated genomic profile. In relation to the epidemiological investigation, we hypothesized that the feeding in pastures contaminated by cattle discharges could represent the most probable route of transmission of M. bovis between the two animal species. In conclusion, our results confirmed the higher discriminatory power of spoligotyping in relation to that of RFLP analysis for the differentiation of M. bovis genomic profiles. Our data showed the presence of a common M. bovis genotype in both cattle and wild boars, confirming the possible interspecies transmission of M. bovis.
Mycobacterium bovis, the causative agent of bovine tuberculosis, has also been implicated as the etiologic agent of disease in a variety of domestic and wild animals (3–5, 7, 20, 28, 31).
In Italy, control of bovine tuberculosis in cattle is based primarily upon a surveillance program that uses the single intradermal comparative skin test followed by compulsory slaughtering and postmortem meat inspection.
Recently, in western Liguria, a region in northern Italy, several cases of tuberculosis-like lesions have been reported following anatomopathological examinations of wild boars which had been killed during the hunting season (1). M. bovis infection has been described in wild boars worldwide (2, 5, 9, 13, 19). The possible presence of the disease in a population of wild animals which is not easily controllable raises problems of public health and wildlife management and may interfere with plans for the eradication of bovine tuberculosis. Moreover, in such a context, the identification of possible alternative sources of infection is important in the effort to control the disease (11, 16, 19, 27–29).
Molecular typing is a valuable tool in epidemiological investigations and for identification of potential sources of infection and has recently been recommended as a means of drawing conclusions as to whether and to what extent transmission from badgers to cattle takes place (6, 9–11, 32–36, 39).
Spoligotyping is a PCR-based method designed to detect the presence or the absence of a unique spacer within the direct repeat (DR) locus of the M. bovis genome (22, 24). Spoligotyping analysis has recently been performed with M. bovis strains obtained from different animals to show the interaction between species (2, 34, 39).
Because of this observation and in consideration of the ongoing cattle tuberculosis issue in the area, we decided to undertake a study aimed at verifying the etiology of the lesions observed in wild boars and characterizing the epidemiology of the infection. In particular, our goal was to verify the presence of mycobacteria in the lesions, to identify the various species of mycobacteria involved, and to better study the sources of infection and the relative epidemic routes to investigate the possibility of interspecies M. bovis transmission.
MATERIALS AND METHODS
Geographic area of study.
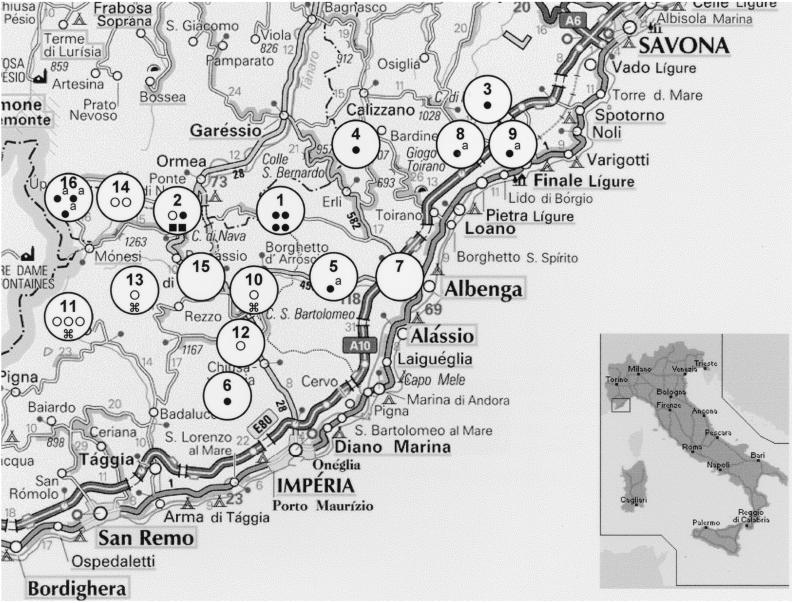
Our research was carried out in the provinces of Imperia and Savona, which are located in western Liguria, a region in northwestern Italy that borders France (Fig. 1). The cattle examined came from farms at seven different locations in the province of Imperia and from two towns located in the province of Savona. This area is quite mountainous, thus making it difficult to record accurately the population of wild animals that inhabits the zone. It is a common practice to pasture the cows there in summer and sometimes year-round. The area where the wild boars were killed had a radius of 20 km, and wild boars were killed at 12 locations in the province of Imperia. Generally, wild boars tend to shift 10 to 15 km in the zones that they inhabit; during the hunting season they may even cover greater distances. In Fig. 1 the numbers 1 to 16 indicate the origins of the wild boars and cattle.
FIG. 1.
Geographical origins of M. bovis strains isolated from cattle and wild boars aligned with the corresponding spoligotype. ●, spoligotype b strains from cattle; ●a, spoligotype b strains from cattle that pastured in zone 16 during the summer; ○, spoligotype b strains from wild boars; ■, spoligotype a strains from cattle;  , spoligotype c strains from wild boars.
, spoligotype c strains from wild boars.
Specimen collection. (i) Wild boars.
During the 1993 to 1995 hunting seasons, mandibular lymph nodes and one liver sample were taken from 63 wild boars which had been killed and which showed tuberculosis-like lesions. Samples were retrieved from 8 boars hunted in 1993, 9 boars hunted in 1994, and 46 boars hunted in 1995. The tissue samples showed purulent, sarcomatous, caseous necrotic, and calcified lesions. At the time that the tissue samples were collected a record was compiled for each animal examined. The record described the necroscopic lesions seen and the place where the animal was killed. The age was determined by means of a dental analysis, and the animals examined were grouped into three classes: age ≤1 year, age from 1 to 2 years, and age >2 years. In order to avoid cross-contamination, different sets of sterile instruments were used to examine each animal.
(ii) Cattle.
The mediastinal lymph nodes of 16 head of cattle were examined. These cattle had been subjected to compulsory slaughter after testing positive by intradermal tuberculization carried out as indicated by the national program for the eradication of bovine tuberculosis. The cattle came from nine farms (identified by the letters A to I, respectively). Seven of these were situated in mountainous areas (identified by the numbers 1 to 7, respectively) in the province of Imperia within a radius of 10 km. The other two farms were located in two different zones of the province of Savona and are identified by the numbers 8 and 9, respectively (Fig. 1). The origins of the 16 cattle were as follows: five were from farm A, three were from farm B, two were from farm D, and the remaining six were from farms C, E, F, G, H, and I respectively. The cattle from the two farms in the province of Savona (farms D and F) and the cattle from farm E, situated in zone 5, were pastured during the summer in the mountains in zone 16, located in the province of Imperia (Fig. 1).
Storage of specimens and decontamination procedure.
The frozen specimens were delivered to the laboratory, stored at −20°C, and examined within 6 months of the time that they were collected. The specimen to be cultured was, on average, of small dimensions, sometimes less than 5 g, and was so strongly contaminated as to not allow decontamination with cetylpyridine. Therefore, the samples were homogenized in a mortar and underwent decontamination with 2% NaOH (23).
The upper part of the pellet was used for both the cultural and microscopic examinations.
Microscopic examination.
The microscopic examination was performed by smearing a loop of 10 μl on a glass slide. The sample on the slide was then stained with Ziehl-Neelsen stain and was observed by the standard method (23).
Cultural examination.
For cultural examination one loop of 10 μl was cultured on two 7H10 (Difco, Detroit, Mich.) tubes modified by means of substituting sodium pyruvate (12 g/liter) for glycerol on two Lowenstein-Jensen tubes and on two Stonebrink self-prepared tubes. The tubes were incubated at 37°C for 6 months and were checked weekly for growth. In the tubes that showed growth during the 6-month period, confirmation of the suspected colonies was made by Ziehl-Neelsen staining, followed by biochemical identification (23).
DNA extraction.
Mycobacteria were harvested from Stonebrink slopes, transferred into a microcentrifuge tube containing 400 μl of 1× Tris-EDTA (TE) buffer, and incubated at 80°C for 20 min. Fifty microliters of 10 mg of lysozyme per liter was added, and the mixture was incubated at 37°C for at least 1 h. Seventy-five microliters of a mixture containing 150 μl of 10 mg of proteinase K (Sigma Aldrich) per ml and 2.1 ml of 10% sodium dodecyl sulfate (SDS) was added to the lysozyme-treated samples. After vortexing and incubation for 10 min at 65°C, 100 μl of 5 M NaCl was added, and subsequently, 100 μl of N-cetyl-N,N,N,-trimethyl ammonium bromide–NaCl solution was added with the aim of binding cell wall debris, denatured proteins, and polysaccharides. After vortexing until the liquid content became white (“milky”), the mixture was incubated for 10 min at 65°C. DNA was then extracted by the standard phenol-chloroform extraction method (15).
Spoligotyping.
Spoligotyping was performed as described previously by Kamerbeek et al. (22). This technique is based on the in vitro amplification of the DNA of the highly polymorphic DR genomic locus present in the M. tuberculosis complex chromosome. In detail, DNA amplification was performed with 50-μl volumes of reaction mixtures containing 5 μl of 10× reaction buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl), 3 μl of 25 mM MgCl2, 20 pmol of each oligonucleotide primer, 1.2 U of Taq polymerase enzyme (Ampli Taq Gold; Perkin-Elmer, Norwalk, Conn.), and approximately 10 ng of template DNA. The oligonucleotide primers DRa (5′-CCGAGAGGGGACGGAAAC-3′) and DRb (5′-GGTTTTGGGTCTGACGAC-3′) amplify the spacer sequences between the DRs. The reverse primer (DRb) was biotinylated at the 5′ end. The conditions used for PCR were 10 min at 95°C, followed by 20 cycles of 1 min at 95°C, 1 min at 55°C, and 30 s at 72°C. M. tuberculosis H37Rv, M. bovis BCG P3, M. avium, and purified water were included as controls with every batch of tests. A set of 43 oligonucleotides (22), each corresponding to one of the unique spacer DNA sequences within the DR locus, were covalently bound to a Byodine C membrane (Pall Biosupport, Portsmouth, United Kingdom). Briefly, the Byodine C membrane was activated by 10 min of incubation in freshly prepared 16% 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide. The membrane was rinsed with water, and it was immediately placed in a miniblotter system (Immunetics, Cambridge, Mass.). The slots of the miniblotter were filled in parallel lines with 150 μl of a 0.125 μM amino-linked oligonucleotide solution in 500 mM NaHCO3 (pH 8.4). The membrane was then removed from the miniblotter and was inactivated with 100 mM NaOH for 10 min. After washing in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.4]) supplemented with 0.1% SDS, the membrane was stored in 20 mM EDTA at 4°C in sealed plastic bags until it was used.
The membrane was washed for 5 min at 60°C in 2× SSPE supplemented with 0.1% SDS and was then placed in the miniblotter. For hybridization, 20 μl of the PCR product was diluted in 150 μl of 2× SSPE–0.1% SDS and was then heat denatured. Almost 150 μl of the diluted samples was pipetted into the parallel miniblotter channels in such a way that they were perpendicular to the rows of oligonucleotides that had been loaded previously. The hybridization was performed for 60 min at 60°C. The membrane was then removed, washed with 2× SSPE supplemented with 0.5% SDS for 10 min at 60°C, and incubated with 1:4,000-diluted streptavidin-peroxidase conjugate (Boehringer Mannheim Biochemicals, East Sussex, United Kingdom) for 60 min at 42°C. The membrane was washed twice for 10 min each time in 250 ml of 2× SSPE–0.5% SDS at 42°C and was rinsed with 250 ml of 2× SSPE for 5 min at room temperature. The hybridized DNA was detected with enhanced chemiluminescence detection liquid (Amersham International plc, Buckinghamshire, United Kingdom) and by exposure of the membrane to X-ray film (Hyperfilm; Amersham International plc) for 5 min.
The membrane was stripped by washing twice for 30 min each time in 1% SDS at 80°C, followed by 15 min of incubation in 20 mM EDTA (pH 8) at room temperature.
RFLP DNA fingerprinting analysis.
Restriction fragment length polymorphism (RFLP) analysis with IS6110 and IS1081 as genetic probes was performed with all isolates as described previously by van Embden and colleagues (37–40).
PCR control procedures.
Each PCR was performed with positive and negative controls by means of sterile procedures and by use of contamination-free guidelines to prevent false-positive results. The chance of PCR contamination was minimized by physical separation of the amplified products from the starting materials, and all pre-PCR handling of materials took place in a room separate from the site where PCR was performed (which had a circulation-free, sterile bench and UV lighting). Another room was dedicated to the processing and analysis of all of the amplification products.
Computer-assisted analysis of the patterns.
Gel Compar Software, version 4.1 (Applied Maths, Kortrijk, Belgium), was used to compare the hybridization patterns obtained by spoligotyping and RFLP DNA fingerprinting. In the spoligotyping analysis, each positive spot was defined as a band. The software clustered strains with the same spoligotyping pattern, and defined-similarity dendrograms were used to join the clusters that were obtained. The pattern for each strain was checked by visual inspection.
RESULTS
Wild boar specimens.
The grouping into classes of the 63 wild boars by age indicated that 38 animals were 1 year of age, 19 animals were between 1 and 2 years of age, and 6 animals were older than 2 years of age. Of the 63 samples collected, 21 were negative by Ziehl-Neelsen staining and cultural examination and 42 (66.6%) were positive by Ziehl-Neelsen staining, while 23 (37%) were positive by cultural examination, and so all of the latter samples were included in the 42 Ziehl-Neelsen staining-positive samples. Biochemical tests with the 23 culture-positive samples allowed the identification of 15 strains of M. bovis, 5 strains of M. avium, 1 strain of M. tuberculosis, and 2 strains of M. fortuitum. The average time for isolation of M. avium was 64 days (range, 40 to 124 days), and that for M. bovis was 80 days (range, 28 to 124 days). Fourteen of the 15 strains of M. bovis were isolated from mandibular lymph nodes and 1 was isolated from the liver of a wild boar with the generalized form of M. bovis infection. M. bovis was frequently isolated from young animals (12 of 15 wild boars less than 1 year old). All M. bovis isolates were collected in seven mountainous zones in the province of Imperia, as indicated by the numbers 2 and 10 to 15 (Fig. 1), and were located within an area with a radius of 20 km.
Cattle specimens.
Fifteen of the 16 cattle specimens (94%) were positive by microscopic examination; 15 strains of M. bovis were isolated from these samples. The average isolation time was 27 days (range, 6 to 63 days).
Genotypic analysis.
Spoligotyping was performed for all the mycobacteria cultured (23 from wild boars and 15 from cattle). All the experiments were performed three times, with concordant results obtained each time. The control strains M. tuberculosis H37Rv and M. bovis BCG yielded consistent patterns in all tests. A blank pattern was obtained when M. avium and water were analyzed by spoligotyping, with no contamination being found. The five strains of M. avium and the two strains of M. fortuitum were constantly negative by genotyping analysis by spoligotyping. The other 30 strains analyzed, 15 from cattle and 15 from wild boars, showed patterns characterized by the absence of 39 to 43 spacers, a property which allows M. bovis to be differentiated from M. tuberculosis and which confirms the biochemical test results (Fig. 2). One strain isolated from a wild boar identified as M. tuberculosis by the biochemical tests had a spoligotype compatible with that of M. tuberculosis (Fig. 2).
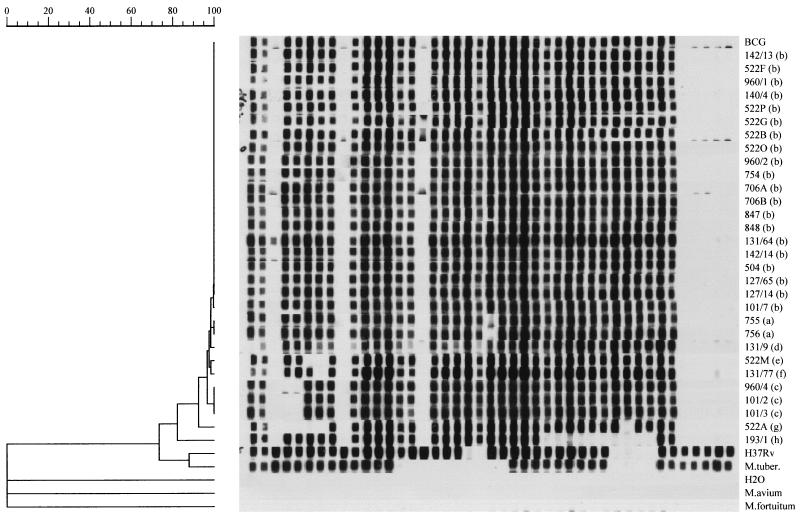
FIG. 2.
Dendrogram drawn with the GelCompar program showing the relationship of eight representative spoligotypes of 30 M. bovis isolates from cattle and wild boars.
According to the genomic differentiation, spoligotyping analysis led to the identification of nine different spoligotypes, which were designated spoligotypes a to h.
The M. bovis strains isolated from cattle belonged to four different spoligotypes (spoligotypes a, b, e, and g). Of 15 strains analyzed, spoligotype b appeared 11 times, spoligotype a appeared twice, and spoligotype e and g appeared one time each.
The M. bovis isolates collected from wild boars belonged to five different spoligotypes (spoligotypes b, c, d, f, and h). Among the strains examined, spoligotype b appeared nine times, spoligotype c appeared three times, and spoligotypes d, f, and h appeared one time each. The results are reported in Table 1 and Fig. 2.
TABLE 1.
DNA fingerprinting results for 30 M. bovis strains with individual RFLP types by RFLP analysis with two probes and individual spoligotypes
| Strain | Animal species | Farm | Geographic origin | Spoligotype | Type by RFLP analysis
|
|
|---|---|---|---|---|---|---|
| IS6110 | IS1081 | |||||
| 755 | Bovine | B | 2 | a | a | α |
| 756 | Bovine | B | 2 | a | a | α |
| 847 | Bovine | B | 2 | b | a | α |
| 522-O | Bovine | H | 4 | b | a | α |
| 960/2 | Wild boar | 12 | b | a | α | |
| 102/7 | Wild boar | 2 | b | a | α | |
| 754 | Bovine | A | 1 | b | a | α |
| 706 A | Bovine | A | 1 | b | a | α |
| 706 B | Bovine | A | 1 | b | a | α |
| 504 | Bovine | A | 1 | b | a | α |
| 848 | Bovine | C | 3 | b | a | α |
| 131/64 | Wild boar | 14 | b | a | α | |
| 142/14 | Wild boar | 13 | b | a | α | |
| 127/65 | Wild boar | 10 | b | a | α | |
| 127/14 | Wild boar | 11 | b | a | α | |
| 522 G | Bovine | F | 9, 16 | b | a | α |
| 142/13 | Wild boar | 14 | b | a | α | |
| 522 F | Bovine | E | 5, 16 | b | a | α |
| 522 B | Bovine | D | 8, 16 | b | a | α |
| 140/4 | Wild boar | 11 | b | a | α | |
| 522 P | Bovine | I | 6 | b | a | α |
| 960/1 | Wild boar | 11 | b | b | α | |
| 960/4 | Wild boar | 13 | c | a | α | |
| 101/2 | Wild boar | 10 | c | a | α | |
| 101/3 | Wild boar | 11 | c | a | α | |
| 131/9 | Wild boar | 13 | d | a | α | |
| 522 M | Bovine | G | 7 | e | a | α |
| 131/77 | Wild boar | 13 | f | a | α | |
| 522 A | Bovine | D | 8, 16 | g | a | α |
| 193/1 | WIld boar | 15 | h | a | α | |
The strains were from wild boars and cattle from different geographical areas in northern Italy.
Data processing with GelCompar software, version 4.1, identified three clusters. One cluster (spoligotype a) was made up of two strains collected from two cattle from the same farm (farm B), one cluster (spoligotype c) included three strains from wild boars killed in three bordering zones (zones 10, 11, and 13) in different years (strain 960/4 in 1993 and strains 101/2 and 101/3 in 1994). Finally, one cluster (spoligotype b) consisted of 20 strains, 11 of which were from cattle and nine of which were from wild boars. In the latter cluster, four head of cattle came from farm A located in zone 1; seven head of cattle came from seven different farms located in seven different zones. Of the nine wild boars whose strains belonged to this cluster, two were killed in 1994 and seven were killed in 1995; three were killed in zone 11, two were killed in zone 14, and the remaining four were killed in zones 2, 10, 12, and 13, respectively (all size zones are located in an area with a radius of about 5 km). The six M. bovis strains not included in this cluster belonged to four different spoligotypes; strains of spoligotype c appeared three times; strains of spoligotypes d, f, and h appeared once each. Only strains of spoligotype b were identified among strains collected from both wild boars and cattle, and spoligotype b was the prevalent spoligotype within both species (Table 1).
With regard to the bovine strains, it is worthy of note that for farm A, the four strains belonged to a single spoligotype (spoligotype b). For farm B, of the three strains analyzed, two belonged to spoligotype a and one belonged to spoligotype b; the investigations carried out by the local health authorities indicated that the latter farm had not recently purchased animals. It must be pointed out that farm B pastures the animals permanently in zone 2 and that a strain collected from a wild boar killed in the same zone belonged to spoligotype b. For farm D, located in the province of Savona, the two isolates examined were genotypically different from each other: one belonged to spoligotype b, while the other belonged to spoligotype g; the two head of cattle from which these strains were isolated had recently been bought from a farm which pastures its animals in the province of Imperia in zone 16. Only a single strain belonging to spoligotype b was isolated from cattle on farms C, E, F, H, and I. Farm E is located in zone 5 but practices summer alpine pasturing in zone 16. A unique strain (spoligotype e) was isolated from an animal on farm G (Table 1).
RFLP DNA fingerprinting analysis was performed for 30 M. bovis strains. By using IS6110 probe, 29 of 30 strains (97%) showed a single-band RFLP pattern characterized by a 1.9-kb PvuII restriction fragment. The remaining strain, isolated from a cow, showed a five-band pattern. However, when typed by spoligotyping the latter strain (strain 960/1) belonged to spoligotype b (Fig. 2). IS1081-based typing was subsequently performed and showed that all strains had unique patterns that were different from the M. bovis BCG genomic profile due to the absence of the specific band at the 8.07-kb position that is typically present in the genotypic profiles of all BCG strains.
DISCUSSION
M. bovis, the causative agent of bovine tuberculosis, has the widest host range of any other member of the M. tuberculosis complex. Several reports have recently focused attention on the diffusion of tuberculosis caused by M. bovis in wild animals and on the role played by different feral animals (deer, badgers, and feral pigs) in the diffusion and persistence of M. bovis disease in cattle (2–5, 7–9, 15, 26, 29–31). In areas where there is close contact between feral and farm animals, it has been described that programs aimed at eradication of bovine tuberculosis are still unable to prevent the occurrence of sporadic outbreaks (2, 8, 13, 19). In this context, it has been suggested that wild animals may play a role in the maintenance of bovine tuberculosis. Moreover, in countries where control of M. bovis diffusion has proven to be difficult, the identification of alternative sources of infection in other farm animals, such as goats and sheep, and wild animals, such as deer and badgers, has been helpful in the effort to control the disease (7–9, 12, 20, 25, 26, 30, 31). In particular, feral pigs and wild boars have been postulated to play an important role in the maintenance of M. bovis infection in cattle (2, 13, 19, 28).
Eastern Liguria, the geographical area that was the focus of our study, is not officially free of bovine tuberculosis. In the years of our study (1993 to 1995), the Italian Ministry of Health reported that the prevalence of skin test-positive animals ranged from 0.5 to 1.1%. The overall proportion of farms with positive cattle reaches 1.1 to 1.6% (21).
Before our study, no evidence of M. bovis isolates in wild boars had been shown in any region of Italy outside Liguria. A previous study, conducted in a region that borders Liguria, showed that neither macroscopic nor microscopic Mycobacterium lesions were ever observed in 20 wild boars killed during the 1996-1997 hunting season (1).
During the 3-year study period, we have isolated Mycobacterium spp. from 37% of the specimens from wild boars with mandibular lymph node pathological lesions, which are indicative of tuberculosis. M. bovis was the most frequently isolated species of mycobacteria, being detected in 68% (15 of 22) of the specimens. Similarly, all strains isolated from cattle were M. bovis. These data are comparable to those reported by Wakelin and Churchman (41) and Corner et al. (13) in two studies carried out in New Zealand, where mycobacteria were isolated from about 40% of the acid fast-positive pathological specimens analyzed. However, the percentage of isolation is substantially smaller than that reported by Ekdahl et al. (19) in New Zealand (100%).
In agreement with what has already been stated in the literature (2, 14, 16, 17, 34, 39), in our study spoligotyping was confirmed to be more efficient than RFLP DNA fingerprinting for discrimination of M. bovis strains. The typing of the 30 available isolates by RFLP DNA fingerprinting with IS6110 identified only two different genomic patterns. Again, the application of RFLP analysis with a second probe (IS1081) successfully identified the BCG genomic profile in strains with the M. bovis genotype, but it still did not provide any additional information on the genomic relatedness of the different M. bovis strains isolated. On the contrary, spoligotyping allowed the identification of eight different genomic patterns, with a large cluster of patterns found for strains from the two different animal species. Therefore, in our hands, the spoligotyping method was confirmed to be a more discriminatory technique, allowing us to disclose several epidemiological links that the RFLP fingerprinting technique failed to identify. These observations support the hypothesis that spoligotyping analysis is a sensitive and specific method for typing of M. bovis strains.
The typing of the 38 isolates collected during the 3 years of the study by spoligotyping revealed the predominance of strains of a single genotype that were present both in cattle (73%) and in wild boars (60%), while strains of all the other genomic patterns identified were unique both in cattle and in wild boars. Strains of some spoligotypes were restricted to small areas, forming small groups of related endemic strains, while strains of the prevalent genotype were widespread within the area, likely because of the movement of cattle among the pastures.
The identification of strains of the same spoligotype in both animal species favors the hypothesis of interspecies transmission of the infection. Although no definitive epidemiological connection could be established, the transmission from wild animals to cattle or vice versa might have taken place when the animals shared the same pasture (18, 27). Therefore, the observation that the M. bovis strains from both cattle and wild boars had spoligotypes that clustered in a main group seems to indicate a behavior somewhat different from that for M. bovis isolates from other animal reservoirs, which may contain different M. bovis types, depending on the host (9, 11, 32).
It has been described previously that possible routes of transmission of M. bovis to wild boars could be the ingestion of offal from infected cattle or the spread of aerosols during close contact of the different animal species (9, 11, 32). However, neither of these hypotheses seems to fit the situation in our study because in the area studied, close contacts between cattle and wild boars were virtually impossible. The feeding of wild boars in pastures contaminated by cattle discharges represents a more reasonable route of transmission. Moreover, as far as the spread of cattle is concerned, the transmission of M. bovis might have occurred when cattle ate or came into close contact with the carcasses of infected wild boars. Instead, the transmission of M. bovis from wild boars to cattle seems more difficult in our survey because of both the lack of close contacts and the low prevalence of pulmonary and generalized disease in infected wild boars, which reduce the probability of transmission. In fact, the main clinical manifestation of tuberculosis in wild boars is mandibular lymph node lymphadenitis. The localization of the pathological lesions, which are limited to the lymph nodes of the head, that we observed in 62 of 63 boars examined and the low number of mycobacteria detected in such lesions could be explained by the high degree of resistance of wild boars to M. bovis infection, as has already been hypothesized by Corner et al. (13). This finding limits the possibility of disease spread between different species and renders unlikely the possibility that wild boars may play an important role in the diffusion of M. bovis infection. This was also proven by the lack of any documented case of endemic M. bovis infection in wild boars in the absence of other infected hosts (13, 28). Therefore, we can hypothesize that wild boars are the end host for M. bovis infection.
In conclusion, spoligotyping has been confirmed to be a more appropriate molecular epidemiological typing method for genetic analysis of M. bovis strains, mainly because of its capability, in comparison with that of RFLP analysis, to differentiate isolates by use of a single insertion sequence element, IS6110. Our results point out the presence of a single M. bovis genotype in both cattle and wild boars, possibly indicating interspecies M. bovis transmission. However, our study suggests that it is unlikely that wild boars act as a reservoir for bovine tuberculosis. We are confident that more exhaustive epidemiological conclusions regarding the dynamics of interspecies transmission of M. bovis between wild boars and cattle will be achieved by further observations after the completion of the bovine tuberculosis eradication program.
ACKNOWLEDGMENT
This work is supported by a grant from the Italian National Institute of Health, “2nd National Tuberculosis Project.”
REFERENCES
- 1.Antonini, M. Unpublished data.
- 2.Aranaz A, Liebana E, Mateos A, Dominguez L, Vidal D, Domingo M, Gonzales M, Rodriguez-Ferri E F, Buschoten A E, van Embden J D A, Cousins D. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J Clin Microbiol. 1996;34:2734–2740. doi: 10.1128/jcm.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernabè A, Gòmez M A, Navarro J A, Gòmez S, Sànchez J, Sidrach J, Menchén V. Pathological changes of spontaneous dual infection of tuberculosis and paratuberculosis in goats. Small Ruminant Res. 1991;5:377–390. [Google Scholar]
- 4.Bernabè A, Gòmez M A, Navarro J A, Gòmez S, Sànchez J, Sidrach J, Menchén V, Vera A, Sierra M A. Morphopathology of caprine tuberculosis. J Pulmonary Tuberc An Vet Murcia. 1991;6–7:9–20. [Google Scholar]
- 5.Bolske G, Englund L, Wahlstrom H, de Lisle G W, Collins D M, Croston P S. Bovine tuberculosis in Swedish deer farms: epidemiological investigations and tracing using restriction fragment analysis. Vet Rec. 1995;136:414–417. doi: 10.1136/vr.136.16.414. [DOI] [PubMed] [Google Scholar]
- 6.Cave M D, Eisenach K D, Salfinger M, Bates J H, Crawford J T. Usefulness of IS6110 in fingerprinting DNA of Mycobacterium bovis. Med Microbiol Lett. 1992;1:96–102. [Google Scholar]
- 7.Clifton-Hadley R S, Wilesmith J W. Tuberculosis in deer: a review. Vet Rec. 1991;129:5–12. doi: 10.1136/vr.129.1.5. [DOI] [PubMed] [Google Scholar]
- 8.Clifton-Hadley R S, Wilesmith J W, Stuart F A. Mycobacterium bovis in the European badger (Meles meles): epidemiological finding in tuberculous badger from a naturally infected population. Epidemiol Infect. 1993;111:9–19. doi: 10.1017/s0950268800056624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins D M, Gabric D M, De Lisle G W. Typing of Mycobacterium bovis isolates from cattle and other animals in the same locality. N Z Vet J. 1987;36:45–46. doi: 10.1080/00480169.1988.35476. [DOI] [PubMed] [Google Scholar]
- 10.Collins D M, Erasmuson S K, Stephens D M, Yates G F, De Lisle G W. DNA fingerprinting of Mycobacterium bovis strains by restriction fragment analysis and hybridization with insertion elements IS1081 and IS6110. J Clin Microbiol. 1993;31:1143–1147. doi: 10.1128/jcm.31.5.1143-1147.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins D M, Radford A J, de Lisle G W, Billman-Jacobe H. Diagnosis and epidemiology of bovine tuberculosis using molecular biological approaches. Vet Microbiol. 1994;40:83–94. doi: 10.1016/0378-1135(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 12.Cordes D O, Bullians J A, Lake D E, Carter M E. Observations on tuberculosis caused by Mycobacterium bovis in sheep. N Z Vet J. 1981;29:60–62. doi: 10.1080/00480169.1981.34798. [DOI] [PubMed] [Google Scholar]
- 13.Corner L A, Barrett R H, Lepper A W D, Lewis V, Pearson C W. A survey of mycobacteriosis of feral pigs in the Northern Territory. Aust Vet J. 1981;57:537–542. doi: 10.1111/j.1751-0813.1981.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 14.Cousin D V, Williams S N, Ross B C, Ellis T M. Use of a repetitive element isolated from Mycobacterium tuberculosis in hybridization studies with Mycobacterium bovis. A new tool for epidemiological studies of bovine tuberculosis. Vet Microbiol. 1993;37:1–17. doi: 10.1016/0378-1135(93)90178-a. [DOI] [PubMed] [Google Scholar]
- 15.Cousin D V, Francis B R, Casey R, Mayberry C. Mycobacterium bovis infection in a goat. Aust Vet J. 1993;70:262–263. doi: 10.1111/j.1751-0813.1993.tb08045.x. [DOI] [PubMed] [Google Scholar]
- 16.Cousins D, Williams S, Liebana E, Aranaz A, Bunschoten A E, van Embden J D, Ellis T. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J Clin Microbiol. 1998;36:168–178. doi: 10.1128/jcm.36.1.168-178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cousins D V, Skuce R A, Kazwala R R, van Embden J D A. Towards a standardized approach to DNA fingerprinting of Mycobacterium bovis. Int J Tuberc Lung Dis. 1998;2:471–478. [PubMed] [Google Scholar]
- 18.Duffield B J, Young D A. Survival of Mycobacterium bovis in defined environmental conditions. Vet Microbiol. 1985;10:193–197. doi: 10.1016/0378-1135(85)90021-5. [DOI] [PubMed] [Google Scholar]
- 19.Ekdahl M O, Smith B L, Money D F L. Tuberculosis in some wild and feral animals in New Zealand. N Z Vet J. 1970;18:44–45. doi: 10.1080/00480169.1970.33860. [DOI] [PubMed] [Google Scholar]
- 20.Gutiérrez M, Samper S, Gavigan J A, Garcia Marin J F, Martin C. Differentiation by molecular typing of Mycobacterium bovis strains causing tuberculosis in cattle and goats. J Clin Microbiol. 1995;33:2953–2956. doi: 10.1128/jcm.33.11.2953-2956.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Italian Ministry of Health. Unpublished data.
- 22.Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J D A. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent P T, Kubica G P. Public health mycobacteriology. A guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 24.Liébana E, Aranaz A, Francis B, Cousins D. Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J Clin Microbiol. 1996;34:933–938. doi: 10.1128/jcm.34.4.933-938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liébana E, Aranaz A, Dominguez L, Mateos A, Gonzàles-Llamazares O, Rodriguez-Ferri E F, Domingo M, Vidal D, Cousin D. The insertion element IS6110 is a useful tool for DNA fingerprinting of Mycobacterium bovis isolates from cattle and goats in Spain. Vet Microbiol. 1997;54:223–233. doi: 10.1016/s0378-1135(96)01282-5. [DOI] [PubMed] [Google Scholar]
- 26.Little T W A, Naylor P F, Wilesmith J W. Laboratory study of Mycobacterium bovis infection in badger and calves. Vet Rec. 1982;111:550–557. [PubMed] [Google Scholar]
- 27.McIlroy S G, Neill S D, McCracken R M. Pulmonary lesions and Mycobacterium bovis excretion from respiratory tract of tuberculin reacting cattle. Vet Rec. 1986;118:718–721. doi: 10.1136/vr.118.26.718. [DOI] [PubMed] [Google Scholar]
- 28.McInerney J, Small K J, Caley P. Prevalence of Mycobacterium bovis infection in feral pigs in the northern territory. Aust Vet J. 1995;72:448–451. doi: 10.1111/j.1751-0813.1995.tb03486.x. [DOI] [PubMed] [Google Scholar]
- 29.Neill S D, Hanna J, O’Brien J J, McCracken R M. Transmission of tuberculosis from experimentally infected cattle to in-contact calves. Vet Rec. 1989;124:269–271. doi: 10.1136/vr.124.11.269. [DOI] [PubMed] [Google Scholar]
- 30.Nolan A, Wilesmith J W. Tuberculosis in badger (Meles meles) Vet Microbiol. 1994;40:179–191. doi: 10.1016/0378-1135(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 31.Rogers L M, Cheeseman C L, Mallison P J, Clifton-Hadley R S. The demography of a high-density badger population in the west of England. J Zool. 1997;242:705–728. [Google Scholar]
- 32.Romano M I, Alito A, Fisanotti J C, Bigi F, Kantor I, Cicuta M E, Cataldi A. Comparison of different genetic markers for molecular epidemiology of bovine tuberculosis. Vet Microbiol. 1996;50:59–71. doi: 10.1016/0378-1135(95)00197-2. [DOI] [PubMed] [Google Scholar]
- 33.Roring S, Hughes M S, Beck L A, Skuce R A, Neill S D. Rapid diagnosis and strain differentiation of Mycobacterium bovis in radiometric culture by spoligotyping. Vet Microbiol. 1998;61:71–80. doi: 10.1016/s0378-1135(98)00167-9. [DOI] [PubMed] [Google Scholar]
- 34.Roring S, Brittain D, Bunschoten A E, Hughes M S, Skuce R A, van Embden J D, Neill S D. Spacer oligotyping of Mycobacterium bovis isolates compared to typing by restriction fragment length polymorphism using PGRS, DR and IS6110 probes. Vet Microbiol. 1998;61:111–120. doi: 10.1016/s0378-1135(98)00178-3. [DOI] [PubMed] [Google Scholar]
- 35.Skuce R A, Brittain D, Hughes M S, Neill S D. Differentiation of Mycobacterium bovis isolates from animals by DNA typing. J Clin Microbiol. 1996;34:2469–2474. doi: 10.1128/jcm.34.10.2469-2474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szewzyk R, Svenson S B, Hoffner S E, Bölske G, Wahlström H, Englund L, Engvall A, Källenius G. Molecular epidemiological studies of Mycobacterium bovis infections in human and animals in Sweden. J Clin Microbiol. 1995;33:3183–3185. doi: 10.1128/jcm.33.12.3183-3185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Embden J D A, Cave M D, Crawford J T, Dale J D, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Soolingen D, De Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Soolingen D, Dehaas P E W, Haagsma J, Eger T, Hermans P W M, Ritacco V, Alito A, van Embden J D. Use of various genetic markers in differentiation of Mycobacterium bovis strains from animals and humans and for studying epidemiology of bovine tuberculosis. J Clin Microbiol. 1994;32:2425–2433. doi: 10.1128/jcm.32.10.2425-2433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Soolingen D, Hermans P W M, de Haas P E W, van Embden J D A. Insertion element IS1081-associated restriction fragment length polymorphism in Mycobacterium tuberculosis complex species: a reliable tool for recognizing Mycobacterium bovis BCG. J Clin Microbiol. 1992;30:1772–1777. doi: 10.1128/jcm.30.7.1772-1777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakelin C A, Churchman O T. Prevalence of bovine tuberculosis in feral pigs in Central Otago. Surveillance. 1991;18:19–20. [Google Scholar]