Abstract
Around 30% of the patients that undergo bariatric surgery (BS) do not reach an appropriate weight loss. The OBEGEN study aimed to assess the added value of genetic testing to clinical variables in predicting weight loss after BS. A multicenter, retrospective, longitudinal, and observational study including 416 patients who underwent BS was conducted (Clinical.Trials.gov- NCT02405949). 50 single nucleotide polymorphisms (SNPs) from 39 genes were examined. Receiver Operating Characteristic (ROC) curve analysis were used to calculate sensitivity and specificity. Satisfactory response to BS was defined as at nadir excess weight loss >50%. A good predictive model of response [area under ROC of 0.845 (95% CI 0.805–0.880), p < 0.001; sensitivity 90.1%, specificity 65.5%] was obtained by combining three clinical variables (age, type of surgery, presence diabetes) and nine SNPs located in ADIPOQ, MC4R, IL6, PPARG, INSIG2, CNR1, ELOVL6, PLIN1 and BDNF genes. This predictive model showed a significant higher area under ROC than the clinical score (p = 0.0186). The OBEGEN study shows the key role of combining clinical variables with genetic testing to increase the predictability of the weight loss response after BS. This finding will permit us to implement a personalized medicine which will be associated with a more cost-effective clinical practice.
Keywords: obesity, bariatric surgery, weight loss, genetics, polygenic risk
1. Introduction
Obesity is a multifactorial and complex disease, caused by the contribution and interaction of environmental and genetic factors [1,2]. Its prevalence has increased dramatically in recent decades, and up to one-fifth of the whole world’s population is expected to live with obesity by 2025 [3]. The discouraging results provided by the conventional treatment have led to the progressive use of the bariatric surgery (BS) as a main treatment for morbidly obesity in Western countries [4].
However, around 30% of the patients that undergo BS do not reach an appropriate weight loss and/or do not resolve the comorbidities associated with obesity [5,6,7]. This fail is associated with a decline in health-related quality of life and patients report feelings of frustration, anger, and even depression [8,9]. Therefore, the identification of new predictive factors of response to BS seems mandatory [10,11,12,13]. This strategy will permit us to identify the best candidates to BS and even to select the type of BS technique, thus optimizing the health care resources.
Inheritance is responsible for 40–75% of all the causes of obesity, a percentage modulated by the epigenetic influence [2,14]. Through genome-wide association studies (GWAS), a series of gene variants and single nucleotide polymorphisms (SNPs) in more than 120 genes have been linked with eating behavior, energy expenditure, response to diet, or lifestyle interventions [15,16]. As each variant alone has little effect on body weight, genetic predisposition is conditioned by the simultaneous presence of SNPs in multiple genes [11]. The possibility of elucidating the best combination of SNPs responsible for the variability of the response to BS in terms of weight loss offers the opportunity to design individualized therapy strategies. Our group has recently showed that an algorithm based on the selection of SNPs associated with predisposition to obesity, appetite regulation, and weight loss was able to predict the percentage of excess body weight loss (%EWL) after BS with high sensitivity and specificity [17]. It should be noted that this was a pilot study including only women who undergo a Roux-en-Y gastric bypass (RYGB), precluding the extrapolation of the results to the whole population.
The objective of the OBEGEN study is to confirm and extend our previous data regarding the added value of genetic testing to clinical variables in predicting the weight loss after BS. For this purpose, a total of 416 subjects (women and men) were evaluated, both RYGB and sleeve gastrectomy (SG) were included, and 50 SNPs were assessed.
2. Materials and Methods
2.1. Statement on Ethics
The OBEGEN study was approved by the human ethics committee of the University Arnau de Vilanova Hospital of Lleida (CEIC-1743). All potential participants gave written informed consent to join the study, which was conducted according to the Helsinki Declaration and the Good Clinical Practice Guidelines. The study was registered in Clinical.Trials.gov- NCT02405949 (accessed on 16 October 2021).
2.2. Study Design and Description of the Study Population
The OBEGEN project was a multicenter, retrospective, longitudinal, and observational study investigating the role of some genetic variants added to clinical variables to predict weight loss after BS. A total of 416 patients who underwent BS between January 2017 and August 2018 at the Obesity Units of four University Hospitals in Catalonia (Spain) were included.
Eligible patients were men and women ≥18 years old, who underwent BS at least 18 months prior to the study. Among the 449 patients who met these criteria, 33 were excluded because of the following reasons: current pregnancy (n = 2), development of drug or alcohol abuse or eating disorders after bariatric surgery (n = 5), markedly mobility problems (n = 4), a different bariatric surgery technique apart from RYGB or SG (n = 9), use of weight lowering pharmacotherapy (n = 7) or a second surgical intervention required during follow-up (n = 6). The study flow chart is displayed in Figure 1.
Figure 1.
Flow chart of the OBEGEN study population.
All the patients agreed to participate in the study and signed the informed consent underwent complete medical history, anthropometric measurement, physical examination, and DNA sampling.
2.3. Outcome Weight Measures
The primary endpoint was the %EWL at nadir. Excess body weight (EBW) was defined as the amount of weight that was in excess from the ideal body weight (IBW). IBW was estimated according to the 1983 Metropolitan Life Insurance Tables (use the midpoint for medium frame) [18]. The %EWL was calculated according to the formula: [total preoperative weight (kg) − weight after bariatric surgery (kg)/EBW (kg)] × 100. Those with a reduction in their %EWL >50% at nadir were considered “good responders” [19].
2.4. Genotyping
DNA was extracted from saliva samples and processed by GoldenGate® Genotyping Assay for VeraCode. The genetic predisposition was assessed using the 50 SNPS in 39 genes included in a commercial nutrigenomic product, the Nutri inCode (NiC) (Ferrer inCode, Barcelona, Spain). This product includes SNPs that had previously been associated with susceptibility to weight loss, both in response to lifestyle intervention and BS [8,20,21]. In addition, Nutri inCode also includes selected variants of published GWAS studies or replication studies related to genetic susceptibility to regulate appetite and develop type 2 diabetes and obesity [22,23]. Finally, in the present study, the panel has been enriched with new 11 SNPs compared to our pilot study (Table 1) [17].
Table 1.
Selected genes and single nucleotide polymorphisms evaluated in the OBEGEN study.
| Gene | Name | Chromosome Allocation of Human Ortholog | SNP |
|---|---|---|---|
| ACE | angiotensin I converting enzyme | 17q23.3 | rs4343 |
| ADIPOQ | adiponectin, C1Q and collagen domain containing | 3q27.3. | rs16861209 rs2241766 |
| ADRB3 | adrenoceptor beta 3 | 8p11.23. | rs4994 rs9693898 |
| AGRP | Agouti related neuropeptide | 16q22.1 | rs11575892 * |
| AGTR1 | angiotensin II receptor type 1 | 3q24. | rs5186 |
| APOA2 | apolipoprotein A2 | 11q23.3. | rs5082 |
| APOA5 | apolipoprotein A5 | 11q23.3. | rs651821 |
| APOC3 | apolipoprotein C3 | 11q23.3. | CD010 |
| BDNF | brain derived neurotrophic factor | 11p14.1. | rs6265 rs925946 |
| CCDC93 | coiled-coil domain containing 93 | 2q14.1 | rs10490628 * |
| CDKAL1 | CDK5 regulatory subunit associated protein 1 like 1 | 6p22.3. | rs7754840 |
| CDKN2B | cyclin dependent kinase inhibitor 2B | 9p21.3. | rs10811661 |
| CLOCK | clock circadian regulator | 4q12. | rs4580704 rs4864548 |
| CNR1 | cannabinoid receptor 1 | 6q15 | rs6454674 * |
| ELOVL6 | ELOVL fatty acid elongase 6 | 4q25. | rs682447 |
| ESR1 | estrogen receptor 1 | 6q25.1 | rs3778099 * |
| FTO | fat mass and obesity associated | 16q12.2. | rs9939609 |
| GHRL | ghrelin and obestatin prepropeptide | 3p25.3. | rs696217 |
| IGF2 | insulin like growth factor 2 | 11p15.5 | rs680 * |
| INSIG2 | insulin induced gene 2 | 2q14.1. | rs7566605 rs3771942 * |
| IL-1B | interleukin 1 beta | 2q14.1 | rs1143643 * |
| IL6 | interleukin 6 | 7p15.3. | rs1800795 |
| LEP | leptin | 7q32.1. | rs12535708 |
| LEPR | leptin receptor | 1p31.3. | rs1137100 |
| LPL | lipoprotein lipase | 8p.22. | rs328 |
| MC4R | melanocortin 4 receptor | 18q21.32. | rs12970134 rs52820871 rs17700633 rs2229616 rs17782313 |
| MTCH2 | mitochondrial carrier 2 | 11p11.2. | rs10838738 |
| NEGR1 | neuronal growth regulator 1 | 1p31.1 | rs2568958 * |
| PLIN1 | perilipin 1 | 15q26.1. | rs1052700 rs894160 |
| PPARA | peroxisome proliferator activated receptor alpha | 22q13.31. | rs1800206 |
| PPARG | peroxisome proliferator activated receptor gamma | 3p25.2. | rs1801282 |
| PCSK1 | proprotein convertase subtilisin/kexin type 1 | 5q15. | rs6235 |
| PON1 | paraoxonase 1 | 7q21.3. | CD014 |
| SIRT1 | sirtuin 1 | 10q21.3 | rs7069102 * |
| TCF7L2 | transcription factor 7 like 2 | 10q25.2. | rs7903146 |
| TMEM18 | transmembrane protein 18 | 2p25.3 | rs2867125 * |
| UCP1 | uncoupling protein 1 | 4q31.2 | rs45539933 * |
| UCP2 | uncoupling protein 2 | 11q13.4. | rs659366 |
| UCP3 | uncoupling protein 3 | 11q13.4. | rs1800849 |
| WFS1 | wolframin ER transmembrane glycoprotein | 4p16.1. | rs10010131 |
SNP: single nucleotide polymorphism. *: Genetic variants that have been added from the pilot study [17].
2.5. Statistical Methods
A normal distribution of the variables was established using the Kolmogorov–Smirnov test, and data are expressed as the mean ± SD, median (interquartile range), or as a percentage. Comparisons between groups were made using the Student’s t-test and the Mann–Whitney U test for quantitative variables, and the Pearson’s chi-squared for categorical variables. The relationship between continuous variables was examined by the Pearson linear correlation test.
To assess the best predictive clinical–genetic risk score, patients were distributed into two groups according to the BS response: (i) %EWL ≤ 50% (n = 113); (ii) %EWL > 50% n = 301). On one side, the different SNPs were coded as 0, 1, or 2 according to the number of risk alleles associated with a favorable weight response. Clinical variables analyzed included gender, age at surgery, preoperative weight and BMI, type of surgery (RYGB or SG), EBW, and presence of type 2 diabetes. Univariate and multivariate logistic regressions were used to establish associations between the genetic and/or the clinical variables and the loss of weight. In this way, we generated three risk scores: (i) a clinical risk score, (ii) a genetic risk score, and (iii) the OBEGEN clinical-genetic risk score (OBEGEN-CGRS), which includes both the selected clinical and genetic variants in the multivariate logistic regression.
Akaike Information Criterion (AIC)-based backward selection was used to remove not significant variables, from an initial model containing all the candidate predictors. The calibration of the logistic model’s adequacy was determined using the test of fit by the Hosmer–Lemeshow. The accuracy of different scores/models in discriminating those who obtained the objective weight loss (%EWL > 50%) from those who did not achieve the objective weight loss (for evaluating the prediction performance of the models) was evaluated using a Receiver Operating Characteristic (ROC) curve analysis. The cut-offs to calculate the sensitivity and specificity of the developed algorithms were selected as the point which maximizes the Youden index. An odds ratio with its 95% confidence interval was finally calculated. The total area under the ROC (AUROC) curve was interpreted following guidelines: 0.9–1.0 excellent, 0.8–0.9, good; 0.7–0.8, fair; 0.6–0.7, poor; and 0.5–0.6, not useful. Comparisons between the obtained AUROC were compared using the method of Hanley and McNeil.
All the contrasts were bilateral with a significance level of 0.05. The data were analyzed with the Statistical Package for the Social Sciences software (IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY, USA).
3. Results
3.1. Baseline Characteristics of Patients
The main baseline characteristics of patients included in the OBEGEN study are shown in Table 2. After a follow-up period of 14.6 ± 0.8 months, 301 (72.3%) patients achieved a %EWL higher than 50%. Patients with a favorable weight response were younger, mainly women, and underwent RYGB.
Table 2.
Main clinical characteristics, metabolic, and anthropometry data of patients included in the study and according to the weight response to bariatric surgery.
| Total | %EWL > 50% | %EWL ≤ 50% | p | |
|---|---|---|---|---|
| Patients, n (%) | 416 | 301 (72.3) | 115 (27.6) | <0.001 |
| Female, n (%) | 348 (83.6) | 260 (86.3) | 88 (76.5) | <0.001 |
| Age (yrs) | 48.3 ± 10.3 | 49.0 ± 10.4 | 51.5 ± 9.2 | 0.003 |
| SG, n (%) | 137 (32.9) | 105 (34.8) | 32 (27.8) | <0.001 |
| RYGB, n (%) | 280 (67.3) | 218 (72.4) | 62 (53.9) | <0.001 |
| Initial BMI (Kg/m2) | 44.3 ± 7.9 | 44.5 ± 9.6 | 44.2 ± 7.3 | 0.554 |
| Initial weight (Kg) | 113.0 ± 18.4 | 112.3 ± 20.3 | 113.4 ± 13.8 | 0.361 |
| Excess weight (Kg) | 49.3 ± 17.3 | 48.7 ± 19.2 | 51.3 ± 12.2 | 0.020 |
| Nadir BMI (Kg/m2) | 29.9 ± 5.8 | 28.5 ± 4.5 | 37.0 ± 6.8 | <0.001 |
| Type 2 diabetes, n (%) | 173 (41.5) | 128 (42.5) | 45 (39.1) | <0.001 |
| Hypertension (%) | 286 (68.7) | 204 (67.7) | 82 (71.3) | <0.001 |
| Dyslipidemia (%) | 306 (73.5) | 212 (70.4) | 94 (81.7) | 0.010 |
| Sleep apnea (%) | 116 (27.8) | 88 (29.2) | 28 (24.3) | <0.001 |
Data are mean ± SD, median (range) or n (percentage). SG: sleeve gastrectomy; RYGB: Roux-en-Y gastric bypass; BMI: body mass index. EWL: excess of weight loss. Hypertension was defined by increased systolic (≥140 mmHg) or increased diastolic (≥90 mmHg) blood pressure or using antihypertensive drugs, according to current guidelines. Dyslipidemia was defined using lipid-lowering drugs, decreased values of HDL cholesterol (men< 0.9 mmol/L, women <1.0 mmol/L) or by at least one increased value of total cholesterol (>5.2 mmol/L), LDL cholesterol or triglycerides (>1.7 mmol/L).
3.2. Construction of a Clinical Risk Score
When only available clinical data were evaluated, the multivariable logistic regression model showed than age at BS, type of BS and presence of type 2 diabetes were independent risk factors for predicting a favorable weight loss in the entire population (Table 3). Therefore, a clinical risk score was developed including these three variables that showed an AUROC for predicting a good response to BS of 0.775 [95% confidence interval (CI) 0.731 to 0.814, p < 0.0001], with a sensitivity of 93.0% and a specificity of 50.4%. The calibration of the adequacy of the model determined by the Hosmer–Lemeshow test was 0.522.
Table 3.
Clinical and genetic variables that significantly predicts favorable weight loss after BS in the entire population of the OBEGEN project.
| Clinical Variables | Genetic Variants | All selected Variants | |||
|---|---|---|---|---|---|
| Coefficient | Coefficient † | Coefficient † | |||
| Age | −0.03458 | rs16861209 | −0.30388 | CRS | 1.13897 |
| Type of surgery | 0.69588 | rs17782313 | 0.32234 | GRS | 1.30048 |
| Type 2 Diabetes | 3.05077 | rs1800795 | −0.33407 | Constant | −1.34401 |
| Constant | −3.75577 | rs1801282 | 0.33407 | ||
| rs3771942 | −0.17997 | ||||
| rs6454674 | 0.24788 | ||||
| rs682447 | 0.41113 | ||||
| rs894160 | 0.28848 | ||||
| rs925946 | 0.28604 | ||||
| Constant | −0.30768 | ||||
†: Coefficients in multiple logistic regression model. CRS: clinical risk score; GRS: genetic risk score.
3.3. Construction of a Genetic Risk Score
Additionally, when genetic data were analyzed alone, the multivariate logistic regression equation for predicting a favorable weight loss response after the BS included nine insSNPs located in ADIPOQ, MC4R, IL-6, PPARG, INSIG2, CNR1, ELOVL6, PLIN1, and BDNF (Table 3). This genetic risk score showed an AUROC of 0.648 (95% CI 0.597 to 0.696, p < 0.0001), with a sensitivity of 48.7% and a specificity of 75.0%. The calibration of the adequacy of the model determined by the Hosmer–Lemeshow test was 0.922.
3.4. Construction of the OBEGEN Clinical-Genetic Risk Score
Based on the clinical and genetic data from our population, we created the OBEGEN-CGRS, including age at surgery, type of surgery, presence of type 2 diabetes, and the nine SNPs associated with weight loss in response to BS (Table 3). The OBEGEN-CGRS score ranges from −4 to +4 points, with a cut-off point to define a good responder of 0.662. This predictive model showed an AUROC of 0.845 (95% CI 0.805 to 0.880, p < 0.001), with a sensitivity of 90.1% and a specificity of 65.5%. The calibration of the model’s adequacy determined by the Hosmer–Lemeshow test was 0.927. An internal validation of this clinical-genetic algorithm was performed using a Bootstrap method to quantify the uncertainty associated with the AUROC. The result was an AUC of 0.845 (95% CI: 0.800 to 0.888).
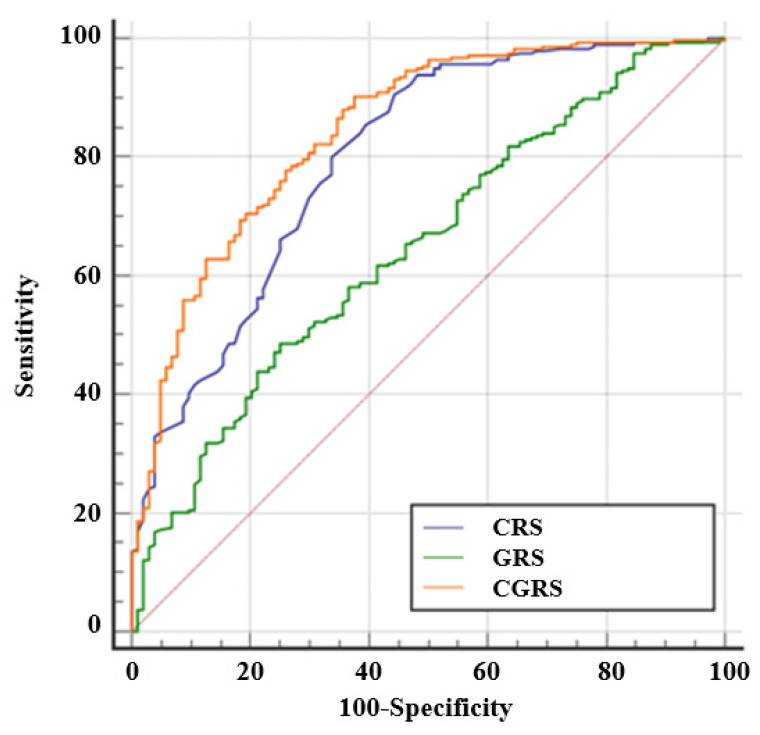
The OBEGEN-CGRS score showed a significant higher AUROC than either the clinical score (p = 0.0186) or the genetic score (p < 0.0001) (Figure 2).
Figure 2.
The predictive capacity of favorable weight loss (%EWL > 50%) obtained by the clinical risk score (CRS), the genetic risk score (GRS), and the clinical plus genetic risk score (CGRS) in our study population in the OBEGEN project.
4. Discussion
In this study we provide evidence that the combination of clinical plus genetic data is a reliable method to predict the weight response after BS. The OBEGEN-CGRS permits us to progress towards the personalization in the management of patients with severe obesity, seeking maximum efficiency with the least surgical damage.
Although BS provides successful weight loss in most of the cases, 25–30% of patients who undergo BS may not achieve the desired weight reduction [5,6,7]. This failure is considered multifactorial, with some preoperative factors associated with the hospital center (i.e., surgeons’ experience, bariatric procedure, preoperative education, and recommended weight loss before BS), the patient (age, gender, ethnicity, preoperative BMI, and comorbidities associated with obesity), and psychosocial features (economic resources, household type, and personality disorders) [11,12,13,24,25,26]. The real factors that predict weight loss following BS are still far to be determined. This is due to the inconsistency in reporting and the methodological weaknesses in analysis, which include a little if any consideration of genetic factors [21,27].
The development of new predictive tools for BS, based on some of the previous predictive factors, but at the same time fueled by new components, is a real need for clinicians who treat obesity worldwide. These instruments should help physicians to identify the best candidates who must undergo BS and to surgeons to optimize the surgical procedure.
Few studies have addressed the influence of genetics on long-term dynamic changes in body weight with ambiguous results [16,28]. The Swedish Obesity Study analyzed the impact of various SNPs from 11 genes in 1443 BS cases [29]. After the evaluation of 20 gene SNPs in 249 morbidly obese subjects undergoing RYGB, Velázquez-Fernández et al. showed that POMC rs1042571 was the only associated with favorable weight loss [30]. In another group of 1011 subjects, an increasing number of SNPs alleles in or near the FTO, INSIG2, MC4R, and PCSK1 genes negatively influenced weight loss trajectories after RYGB in those with an initial BMI >50 kg/m2 [10]. More recently, in a group of 146 individuals, carriers of another variant of the FTO gene (rs9939609) showed a lower success rate, as well as a greater and faster weight recovery beyond 2 years after BS [31]. Nevertheless, the same SNP was not associated with different weight loss after 6 months of SG in 74 morbidly obese patients [32]. Regarding the MC4R gene, among 1433 subjects with a follow-up period of 12 months after RYGB, carriers of the I251L variant lost 9% more weight compared with the noncarriers [20]. Finally, a prospective observational study with 105 patients evaluated SNPs in the leptin receptor, FTO and FABP2 genes [33]. This study showed that carriers of the LEP223 (rs1137101) experienced close to 25% lower excess weight at 12 and 24 months after bariatric surgery.
Beyond the isolated study of one or the other gene, genetic risk scores composed by adiposity-related SNPs have been related with weight loss after RYGB or SG in Swiss, Danish, and Greek populations [34,35,36]. It should be noted that only three of the nine genes included in the OBEGEN-CGRS appeared previously reported in association with the weight loss response after BS [10,20,36]. This fact highlights the complexity of the genetic basis associated with the development of obesity, but also that the genes related to the therapeutic response may be different from those proposed so far. In the OBEGEN study, including both genders and different bariatric procedures, the combination of clinic and genetic data enhanced the predictive capacity of the genetic risk score, with improved sensitivity and specificity. Altogether, our findings support the use of genetic testing in clinical practice.
Among the multiple variants of genes that were evaluated, nine of them interact with clinical variables to modify the weight response to BS in the OBEGEN study. While low serum adiponectin levels have been associated with central obesity, insulin resistance, metabolic syndrome, and type 2 diabetes, ADIPOQ (rs16861209) has been significantly associated with elevated fasting serum adiponectin levels [37,38]. Similarly, although IL-6 significantly increases the risk of obesity, in the PREDIMED trial carriers of the rs1800795 showed greater weight loss with the Mediterranean diet with supplements of olive oil compared to a Mediterranean diet low-fat diet than heterozygous and non-carrier carriers after 3 years of intervention [39,40]. In addition, PPARG (rs1801282) was associated not only with short-term (6-month) and long-term (2-year) weight loss but also with weight regain in the Diabetes Prevention Program [41]. PLIN1, a circadian lipid stabilizing protein in the adipocyte, has been associated with body weight regulation and PLIN1 (rs894160) with variability in weight loss [42,43]. Elovl6, a microsomal enzyme involved in the elongation of saturated and monounsaturated fatty acids with 12, 14, and 16 carbons, regulates mitochondrial function and thermogenic capacity in brown adipose tissue [44]. The BDNF (rs925946), INSIG2 (rs3771942) and CNR1 (rs6454674) variants are well stablished genetic determinants of obesity [45,46,47]. Similarly, MC4R (rs17782313) is associated with high dietary intake and different obesity-related phenotypic traits, such as insulin resistance, type 2 diabetes, and hypertriglyceridemia [48]. However, so far there are no data on the response to weight loss treatments regarding these last four variants.
The main differences between our CGRS and previous studies is located in the methodology assessment. Previous studies have been interested in analyzing the association between the number of genetic risk variables and the greater or lower weight loss following BS. However, we have focused our interest on the elaboration of predictive equations mixing clinical and genetic variables and the identification of predictive cut-off in those predictive equations. We have also calculated the prognostic capability of our score in identifying the “good” and “bad” responders to BS. We believe that this is a concept of great interest and more reliable in daily clinical practice. In this way, a patient who requires BS is exposed to an intervention that is difficult to reverse, so it is vital to be able to predict success or failure with a high degree of certainty. Currently, the choice of surgical technique is based on the baseline BMI and the presence of comorbidities. The introduction of a genetic predisposition score in the decision-making algorithms will bring us closer to the best selection of the patient, to choose the most convenient surgery, and to improve current health outcomes in our population.
There are some potential limitations that need to be considered when contemplating the results of our study. First, we evaluated a selected population of patients who underwent bariatric surgery, excluding those who underwent to other surgical techniques other than RYGB and SG, as well as those who had used weight lowering pharmacotherapy or had required a second surgical intervention. However, we believe that the inclusion of these more extreme cases might increase rather than reduce the reliability of our score. Second, the OBEGEN project is a retrospective one, meaning that no irrefutable clinical consequences can still be inferred to general population. Prospective long-term studies testing the genetic basis of patients with severe obesity before the recommendation of bariatric procedures are needed. Third, the characterization of favorable weight loss after BS is controversial [19]. In the OBEGEN study we have used a classic definition (%EWL>50) to better confirm our previous pilot study, so the results could be different if the chosen definition had been another. Finally, our model needs to be validated in an untrained dataset to fully demonstrate its applicability in the real clinical practice.
5. Conclusions
In conclusion, the OBEGEN project shows how the addition of genetic testing to the currently used clinical variables significantly improve our capability to identify patients with obesity who will be “good” or “bad” responders to BS in terms of weight loss. This information should help us both to personalize the therapeutic approach in severe obesity (e.g., select beforehand surgical techniques with a higher degree of malabsorption in those patients identified as “bad” responders), thus optimizing the limited health resources.
Author Contributions
Conceptualization, A.C. (Andreea Ciudin), C.H., E.S. (Eduardo Salas), R.S., and A.L. (Albert Lecube); methodology A.C. (Andreea Ciudin), R.S., and A.L. (Albert Lecube); validation, E.F., L.G.-C., A.C. (Andreea Ciudin), A.C. (Assumpta Caixàs), N.V., S.P., and A.S.-S.; formal analysis, A.C. (Andreea Ciudin), E.S. (Eduardo Salas), R.S., and A.L. (Albert Lecube); investigation, E.F., L.G.-C., A.C. (Andreea Ciudin), A.C. (Assumpta Caixàs), N.V., S.P., A.S.-S., A.L. (Alexis Luna), E.S. (Enric Sánchez), M.R., C.H., R.V., A.R., M.d.l.F., and A.L. (Albert Lecube); resources, A.L. (Albert Lecube); data curation, E.F., L.G.-C., A.C. (Andreea Ciudin), A.C. (Assumpta Caixàs), N.V., S.P., A.S.-S., E.S. (Enric Sánchez), and M.R.; writing—original draft preparation, A.C. (Andreea Ciudin) and A.L. (Albert Lecube); writing—review and editing, A.C. (Andreea Ciudin), E.S. (Eduardo Salas), R.S., and A.L. (Albert Lecube); supervision, A.C. (Andreea Ciudin) and A.L. (Albert Lecube); funding acquisition, A.L. (Albert Lecube). All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the “Pla Estratègic de Recerca i Innovació en Salut” (PERIS) 2016–2020 (SLT002/16/00497), the Instituto de Salud Carlos III (PI PI18/00964), Fondos FEDER “Una manera de hacer Europa”), and Menarini España. CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM) is an initiative of the Instituto Carlos III.
Institutional Review Board Statement
The OBEGEN study was approved by the human ethics committee of the University Arnau de Vilanova Hospital of Lleida (CEIC-1743). Written informed consent was obtained from all subjects involved in the study, which was conducted according to the Helsinki Declaration and the Good Clinical Practice Guidelines. The study was registered in Clinical.Trials.gov- NCT02405949.
Informed Consent Statement
Written informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Swinburn B.A., Sacks G., Hall K.D., McPherson K., Finegood D.T., Moodie M., Gortmaker S.L. The global obesity pandemic: Shaped by global drivers and local environments. Lancet. 2011;378:804–814. doi: 10.1016/S0140-6736(11)60813-1. [DOI] [PubMed] [Google Scholar]
- 2.Locke A., Kahali B., Berndt S.I., Justice A.E., Pers T., Day F., Powell C., Vedantam S., Buchkovich M.L., Yang J., et al. Genetic studies of body mass index yield new insights for obesity biology. Nat. Cell Biol. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Null N. Trends in adult body-mass index in 200 countries from 1975 to 2014: A pooled analysis of 1698 population-based measurement studies with 19·2 million participants. Lancet. 2016;387:1377–1396. doi: 10.1016/s0140-6736(16)30054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angrisani L., Santonicola A., Iovino P., Vitiello A., Higa K., Himpens J., Buchwald H., Scopinaro N. IFSO Worldwide Survey 2016: Primary, Endoluminal, and Revisional Procedures. Obes. Surg. 2018;28:3783–3794. doi: 10.1007/s11695-018-3450-2. [DOI] [PubMed] [Google Scholar]
- 5.Sarabu N. Weight and Metabolic Outcomes 12 Years after Gastric Bypass. N. Engl. J. Med. 2018;378:93–96. doi: 10.1056/nejmc1714001. [DOI] [PubMed] [Google Scholar]
- 6.Kowalewski P.K., Olszewski R., Walędziak M., Janik M., Kwiatkowski A., Gałązka-Świderek N., Cichoń K., Brągoszewski J., Paśnik K. Long-Term Outcomes of Laparoscopic Sleeve Gastrectomy—A Single-Center, Retrospective Study. Obes. Surg. 2017;28:130–134. doi: 10.1007/s11695-017-2795-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth A.E., Thornley C.J., Blackstone R.P. Outcomes in Bariatric and Metabolic Surgery: An Updated 5-Year Review. Curr. Obes. Rep. 2020;9:380–389. doi: 10.1007/s13679-020-00389-8. [DOI] [PubMed] [Google Scholar]
- 8.Karlsson J., Taft C., Rydén A., Sjöström L., Sullivan M. Ten-year trends in health-related quality of life after surgical and conventional treatment for severe obesity: The SOS intervention study. Int. J. Obes. 2007;31:1248–1261. doi: 10.1038/sj.ijo.0803573. [DOI] [PubMed] [Google Scholar]
- 9.Kubik J.F., Gill R.S., Laffin M., Karmali S. The Impact of Bariatric Surgery on Psychological Health. J. Obes. 2013;2013:1–5. doi: 10.1155/2013/837989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Still C.D., Wood G.C., Chu X., Erdman R., Manney C.H., Benotti P.N., Petrick A.T., Strodel W.E., Mirshahi U.L., Mirshahi T., et al. High Allelic Burden of Four Obesity SNPs Is Associated With Poorer Weight Loss Outcomes Following Gastric Bypass Surgery. Obesity. 2011;19:1676–1683. doi: 10.1038/oby.2011.3. [DOI] [PubMed] [Google Scholar]
- 11.Livhits M., Mercado C., Yermilov I., Parikh J.A., Dutson E., Mehran A., Ko C.Y., Gibbons M.M. Preoperative Predictors of Weight Loss Following Bariatric Surgery: Systematic Review. Obes. Surg. 2011;22:70–89. doi: 10.1007/s11695-011-0472-4. [DOI] [PubMed] [Google Scholar]
- 12.Aliakbarian H., Bhutta H.Y., Heshmati K., Kunju S.U., Sheu E.G., Tavakkoli A. Pre-operative Predictors of Weight Loss and Weight Regain Following Roux-en-Y Gastric Bypass Surgery: A Prospective Human Study. Obes. Surg. 2020;30:4852–4859. doi: 10.1007/s11695-020-04877-7. [DOI] [PubMed] [Google Scholar]
- 13.Kitamura R., Chen R., Trickey A., Eisenberg D. Positive and Negative Independent Predictive Factors of Weight Loss After Bariatric Surgery in a Veteran Population. Obes. Surg. 2020;30:2124–2130. doi: 10.1007/s11695-020-04428-0. [DOI] [PubMed] [Google Scholar]
- 14.Moustafa J.S.E.-S., Froguel P. From obesity genetics to the future of personalized obesity therapy. Nat. Rev. Endocrinol. 2013;9:402–413. doi: 10.1038/nrendo.2013.57. [DOI] [PubMed] [Google Scholar]
- 15.Pigeyre M., Yazdi F.T., Kaur Y., Meyre D. Recent progress in genetics, epigenetics and metagenomics unveils the pathophysiology of human obesity. Clin. Sci. 2016;130:943–986. doi: 10.1042/CS20160136. [DOI] [PubMed] [Google Scholar]
- 16.Bray M.S., Loos R., McCaffery J., Ling C., Franks P., Weinstock G., Snyder M.P., Vassy J., Agurs-Collins T., The Conference Working Group NIH working group report—Using genomic information to guide weight management: From universal to precision treatment. Obesity. 2015;24:14–22. doi: 10.1002/oby.21381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciudin A., Fidilio E., Ortiz A., Pich S., Salas E., Mesa J., Hernández C., Simó-Servat O., Lecube A., Simó R. Genetic Testing to Predict Weight Loss and Diabetes Remission and Long-Term Sustainability after Bariatric Surgery: A Pilot Study. J. Clin. Med. 2019;8:964. doi: 10.3390/jcm8070964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Listed N. 1983 metropolitan height and weight tables. Stat. Bull. Metrop. Life Found. 1983;64:3–9. [PubMed] [Google Scholar]
- 19.Grover B.T., Morell M.C., Kothari S.N., Borgert A.J., Kallies K.J., Baker M.T. Defining Weight Loss After Bariatric Surgery: A Call for Standardization. Obes. Surg. 2019;29:3493–3499. doi: 10.1007/s11695-019-04022-z. [DOI] [PubMed] [Google Scholar]
- 20.Mirshahi U.L., Still C.D., Masker K.K., Gerhard G.S., Carey D.J., Mirshahi T. TheMC4R(I251L) Allele Is Associated with Better Metabolic Status and More Weight Loss after Gastric Bypass Surgery. J. Clin. Endocrinol. Metab. 2011;96:E2088–E2096. doi: 10.1210/jc.2011-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moleres A., Rendo-Urteaga T., Zulet M.A., Marcos A., Campoy C., Garagorri J.M., Martínez J.A., Azcona-Sanjulián M.C., Marti A. Obesity Susceptibility Loci on Body Mass Index and Weight Loss in Spanish Adolescents after a Lifestyle Intervention. J. Pediatr. 2012;161:466–470.e2. doi: 10.1016/j.jpeds.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Blakemore A.I.F., Froguel P. Investigation of Mendelian forms of obesity holds out the prospect of personalized medicine. Ann. N. Y. Acad. Sci. 2010;1214:180–189. doi: 10.1111/j.1749-6632.2010.05880.x. [DOI] [PubMed] [Google Scholar]
- 23.Welter D., MacArthur J., Morales J., Burdett A., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2013;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parri A., Benaiges D., Schröder H., Izquierdo-Pulido M., Ramón J., Villatoro M., Roux J.F.-L., Goday A. Preoperative Predictors of Weight Loss at 4 Years Following Bariatric Surgery. Nutr. Clin. Pr. 2015;30:420–424. doi: 10.1177/0884533614568154. [DOI] [PubMed] [Google Scholar]
- 25.Gerken A.L.H., Rohr-Kräutle K.-K., Weiss C., Seyfried S., Reissfelder C., Vassilev G., Otto M. Handgrip Strength and Phase Angle Predict Outcome After Bariatric Surgery. Obes. Surg. 2020;31:200–206. doi: 10.1007/s11695-020-04869-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen M.S., Christensen B.J., Schmidt J.B., Tækker L., Holm L., Lunn S., Ritz C., Albrechtsen N.J.W., Holst J.J., Schnurr T.M., et al. Predictors of weight loss after bariatric surgery—A cross-disciplinary approach combining physiological, social, and psychological measures. Int. J. Obes. 2020;44:2291–2302. doi: 10.1038/s41366-020-0576-9. [DOI] [PubMed] [Google Scholar]
- 27.Cottam S., Cottam D., Cottam A. Sleeve Gastrectomy Weight Loss and the Preoperative and Postoperative Predictors: A Systematic Review. Obes. Surg. 2019;29:1388–1396. doi: 10.1007/s11695-018-03666-7. [DOI] [PubMed] [Google Scholar]
- 28.McCaffery J.M. Precision behavioral medicine: Implications of genetic and genomic discoveries for behavioral weight loss treatment. Am. Psychol. 2018;73:1045–1055. doi: 10.1037/amp0000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarzynski M.A., Jacobson P., Rankinen T., Carlsson B., Sjöström L., Bouchard C., Carlsson L.M.S. Associations of markers in 11 obesity candidate genes with maximal weight loss and weight regain in the SOS bariatric surgery cases. Int. J. Obes. 2010;35:676–683. doi: 10.1038/ijo.2010.166. [DOI] [PubMed] [Google Scholar]
- 30.Velázquez-Fernández D., Mercado-Celis G., Flores-Morales J., Clavellina-Gaytán D., Vidrio R., Vidrio E., Mosti M., Sánchez-Aguilar H., Rodriguez D., León P., et al. Analysis of Gene Candidate SNP and Ancestral Origin Associated to Obesity and Postoperative Weight Loss in a Cohort of Obese Patients Undergoing RYGB. Obes. Surg. 2016;27:1481–1492. doi: 10.1007/s11695-016-2501-9. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues G.K., Resende C.M., Durso D.F., Rodrigues L.A., Silva J.L.P., Reis R.C., Pereira S.S., Ferreira D.C., Franco G.R., Alvarez-Leite J. A single FTO gene variant rs9939609 is associated with body weight evolution in a multiethnic extremely obese population that underwent bariatric surgery. Nutrition. 2015;31:1344–1350. doi: 10.1016/j.nut.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 32.Balasar Ö., Çakır T., Erkal Ö., Aslaner A., Cekic B., Uyar M., Bülbüller N., Oruç M.T. The effect of rs9939609 FTO gene polymorphism on weight loss after laparoscopic sleeve gastrectomy. Surg. Endosc. 2015;30:121–125. doi: 10.1007/s00464-015-4169-y. [DOI] [PubMed] [Google Scholar]
- 33.Kops N.L., Vivan M.A., Horvath J.D.C., de Castro M.L.D., Friedman R. FABP2, LEPR223, LEP656, and FTO Polymorphisms: Effect on Weight Loss 2 Years After Bariatric Surgery. Obes. Surg. 2018;28:2705–2711. doi: 10.1007/s11695-018-3213-0. [DOI] [PubMed] [Google Scholar]
- 34.Bandstein M., Voisin S., Nilsson E.K., Schultes B., Ernst B., Thurnheer M., Benedict C., Mwinyi J., Schiöth H.B. A Genetic Risk Score is Associated with Weight Loss Following Roux-en Y Gastric Bypass Surgery. Obes. Surg. 2016;26:2183–2189. doi: 10.1007/s11695-016-2072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aasbrenn M., Schnurr T., Have C.T., Svendstrup M., Hansen D.L., Worm D., Balslev-Harder M., Hollensted M., Grarup N., Burgdorf K.S., et al. Genetic Determinants of Weight Loss after Bariatric Surgery. Obes. Surg. 2019;29:2554–2561. doi: 10.1007/s11695-019-03878-5. [DOI] [PubMed] [Google Scholar]
- 36.Katsareli E.A., Amerikanou C., Rouskas K., Dimopoulos A., Diamantis T., Alexandrou A., Griniatsos J., Bourgeois S., Dermitzakis E., Ragoussis J., et al. A Genetic Risk Score for the Estimation of Weight Loss After Bariatric Surgery. Obes. Surg. 2020;30:1482–1490. doi: 10.1007/s11695-019-04320-6. [DOI] [PubMed] [Google Scholar]
- 37.Peters K., Beilby J., Cadby G., Warrington N.M., Bruce D.G., Davis W.A., Davis T.M., Wiltshire S., Knuiman M., McQuillan B.M., et al. A comprehensive investigation of variants in genes encoding adiponectin (ADIPOQ) and its receptors (ADIPOR1/R2), and their association with serum adiponectin, type 2 diabetes, insulin resistance and the metabolic syndrome. BMC Med. Genet. 2013;14:15. doi: 10.1186/1471-2350-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyriakou T., Collins L.J., Spencer-Jones N.J., Malcolm C., Wang X., Snieder H., Swaminathan R., Burling K.A., Hart D.J., Spector T.D., et al. Adiponectin gene ADIPOQ SNP associations with serum adiponectin in two female populations and effects of SNPs on promoter activity. J. Hum. Genet. 2008;53:718–727. doi: 10.1007/s10038-008-0303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gholami M., Sharifi F., Shahriari S., Khoshnevisan K., Larijani B., Amoli M.M. Association of interleukin-6 polymorphisms with obesity: A systematic review and meta-analysis. Cytokine. 2019;123:154769. doi: 10.1016/j.cyto.2019.154769. [DOI] [PubMed] [Google Scholar]
- 40.Razquin C., Martinez J.A., Martinez-Gonzalez M.A., Fernández-Crehuet J., Santos J.M., Marti A. A Mediterranean diet rich in virgin olive oil may reverse the effects of the −174G/C IL6 gene variant on 3-year body weight change. Mol. Nutr. Food Res. 2010;54:S75–S82. doi: 10.1002/mnfr.200900257. [DOI] [PubMed] [Google Scholar]
- 41.Delahanty L.M., Pan Q., Jablonski K.A., Watson K.E., McCaffery J., Shuldiner A., Kahn S.E., Knowler W.C., Florez J.C., Franks P.W., et al. Genetic Predictors of Weight Loss and Weight Regain After Intensive Lifestyle Modification, Metformin Treatment, or Standard Care in the Diabetes Prevention Program. Diabetes Care. 2011;35:363–366. doi: 10.2337/dc11-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soenen S., Mariman E.C., Vogels N., Bouwman F.G., Hoed M.D., Brown L., Westerterp-Plantenga M.S. Relationship between perilipin gene polymorphisms and body weight and body composition during weight loss and weight maintenance. Physiol. Behav. 2009;96:723–728. doi: 10.1016/j.physbeh.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 43.Garaulet M., Vera B., Bonnet-Rubio G., Gómez-Abellán P., Lee Y.-C., Ordovás J.M. Lunch eating predicts weight-loss effectiveness in carriers of the common allele at PERILIPIN1: The ONTIME (Obesity, Nutrigenetics, Timing, Mediterranean) study. Am. J. Clin. Nutr. 2016;104:1160–1166. doi: 10.3945/ajcn.116.134528. [DOI] [PubMed] [Google Scholar]
- 44.Tan C.Y., Virtue S., Bidault G., Dale M., Hagen R., Griffin J.L., Vidal-Puig A. Brown Adipose Tissue Thermogenic Capacity Is Regulated by Elovl6. Cell Rep. 2015;13:2039–2047. doi: 10.1016/j.celrep.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akbarian S.-A., Salehi-Abargouei A., Pourmasoumi M., Kelishadi R., Nikpour P., Heidari-Beni M. Association of Brain-derived neurotrophic factor gene polymorphisms with body mass index: A systematic review and meta-analysis. Adv. Med. Sci. 2018;63:43–56. doi: 10.1016/j.advms.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Fox C.S., Heard-Costa N., Cupples L.A., Dupuis J., Vasan R.S., Atwood L.D. Genome-wide association to body mass index and waist circumference: The Framingham Heart Study 100K project. BMC Med. Genet. 2007;8:S18. doi: 10.1186/1471-2350-8-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benzinou M., Chèvre J.-C., Ward K.J., Lecoeur C., Dina C., Lobbens S., Durand E., Delplanque J., Horber F.F., Heude B., et al. Endocannabinoid receptor 1 gene variations increase risk for obesity and modulate body mass index in European populations. Hum. Mol. Genet. 2008;17:1916–1921. doi: 10.1093/hmg/ddn089. [DOI] [PubMed] [Google Scholar]
- 48.Yu K., Li L., Zhang L., Guo L., Wang C. Association between MC4R rs17782313 genotype and obesity: A meta-analysis. Gene. 2020;733:144372. doi: 10.1016/j.gene.2020.144372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.