Abstract
The genus Apiospora is known as a cosmopolitan genus, found across various substrates. In this study, four Apiospora taxa were obtained from the decaying stems of bamboo and maize in northern Thailand. Apiospora collections were compared with known species based on the morphological characteristics and the DNA sequence data of internal transcribed spacer (ITS), the partial large subunit nuclear rDNA (LSU), the translation elongation factor 1-alpha gene (TEF1-α) and beta-tubulins (TUB2). Apiospora chiangraiense sp. nov. and two new host records (Ap. intestini and Ap. rasikravindra) are introduced here based on the morphological characteristics and multi-locus analyses. Additionally, thirteen species previously identified as Arthrinium are introduced as new combinations in Apiospora, viz., Ap. acutiapica, Ap. bambusicola, Ap. biserialis, Ap. cordylines, Ap. cyclobalanopsidis, Ap. euphorbiae, Ap. gelatinosa, Ap. locuta-pollinis, Ap. minutispora, Ap. pseudorasikravindrae, Ap. septate, Ap. setariae and Ap. sorghi.
Keywords: one new species, new combinations, new host records, phylogeny, taxonomy
1. Introduction
Apiospora was introduced by Saccardo with Ap. montagnei as the type species [1]. The genus was reported in both sexual and asexual morphs. The sexual morphs are characterized by multi-locular perithecial stromata with hyaline ascospores surrounded by a thick gelatinous sheath [2,3,4]. The asexual morph of Apiospora was characterized by basauxic conidiogenesis, with globose to subglobose conidia, which are usually lenticular in the side view, obovoid and pale brown to brown [2,5,6]. Species of Apiospora are similar in morphology, thus it is difficult to distinguish them without molecular phylogenetic data. The size, color and shape of conidia and the morphology of conidiophores (e.g., size, shape and septation) should be used together to better identify them. For example, conidiophores of some species reduce to conidiogenous cells (e.g., Ap. bambusae, Ap. acutiapicum), while some species have semi-micronematous to macronematous conidiophores (e.g., Ap. bambusicola, Ap. intestini).
Apiospora species have a worldwide distribution and can be found from various hosts [3,7,8,9]. Most Apiospora species are associated with plants as endophytes, pathogens or saprobes, especially on bamboo [2,3,10,11]. To date, more than 25 species have been found from bamboo [2,3,10,11]. Apiospora species can cause leaf necrosis and twig dieback in the olive tree (Olea europaea), leaf edge spot of the peach (Prunus persica), blight disease of bamboo (Schizostachyum), leaf spot of rosemary (Salvia rosmarinus), kernel blight of barley (Hordeum vulgare) and brown culm streak of Phyllostachys praecox [11,12,13,14,15,16,17]. Some species have also been isolated from lichens, air, soil and animal tissues, and a few species are human pathogens which can cause cutaneous infections in humans [9,18,19,20,21,22,23].
The morphological relationships between Arthrinium and Apiospora have long been debated after Ellis [24], as the morphological characteristics of these two genera are similar and difficult to distinguish based on morphology alone. Apiospora was synonymized under Arthrinium by Crous et al. [3] as they found that Apiospora is the sexual morph of Arthrinium and phylogenetic analyses showed that the two genera formed a monophyletic clade. Meanwhile, the phylogenic analyses results from Pintos et al. [25] showed Arthrinium forms a monophyletic clade that separates from all other sequences of Apiospora and suggested that Arthrinium s. str. could actually be phylogenetically different from Apiospora, but this is in need of clarification using the phylogeny of additional species before making a conclusive taxonomic decision on the issue. Recently, Pintos and Alvarado [4] showed that Apiospora and Arthrinium present independent lineages, thus they separate well into two genera.
Morphologically, the conidia of Apiospora are more or less rounded in the face view and lenticular in the side view and conidiophores sometimes develop forming acervuli. Whereas the conidia of Arthrinium are variously shaped (angular, curved, fusiform, globose, polygonal, navicular) and the conidiophores of some species have thick blackish septa [14]. Ecologically, Apiospora species are mostly reported on Poaceae, while Arthrinium species commonly occur on Cyperaceae and Juncaceae. Moreover, Apiospora has a worldwide distribution, and species in the genus can be found from tropical and subtropical areas to the Mediterranean, temperate and cold regions, while Arthrinium species are rarely found from tropical and subtropical habitats. Hence, Pintos and Alvarado [4] considered that genetic, morphological and ecological differences are sufficient to support the taxonomic separation of the two genera, and accordingly, 55 Arthrinium species were transferred to Apiospora based on the phylogenetic analyses. Presently, 117 records of Apiospora are listed in the Index Fungorum [26].
The aims of this study are to determine the phylogenetic placement of the genus Apiospora and describe the three taxa that were isolated from maize and bamboo in Chiang Rai province, Thailand. Based on the morphological characteristics and phylogenetic analyses of a combined dataset of the internal transcribed spacer (ITS), the partial large subunit nuclear rDNA (LSU), the translation elongation factor 1-alpha gene (TEF1-α) and beta-tubulins (TUB2), a new species, Ap. chiangraiense, as well as two new host records, Ap. rasikravindrae and Ap. intestini, are introduced. In addition, thirteen species of Arthrinium were synonymized under Apiospora.
2. Materials and Methods
2.1. Sample Collection, Isolation and Morphological Characteristic Examination
Fresh specimens of bamboo and maize culms with fungal fruiting bodies were collected from Chiang Rai, Thailand from September–October 2020. Specimens were brought to the laboratory in plastic Ziploc bags for observation. Senanayake et al. [27] were followed for the morphological observations and single-spore isolation. The morphological characteristics were examined under a stereomicroscope (Motic SMZ-171, Wetzlar, Germany). The conidiomata were observed and photographed using a Nikon ECLIPSE Ni-U compound microscope connected to a Nikon camera series DS-Ri2 (New York, United States). The germinating ascospores were transferred aseptically to fresh potato dextrose agar (PDA) media and incubated at room temperature (25 °C) for 2–4 weeks. The morphological characteristics of cultures were checked and recorded after 30–60 days.
The herbarium specimens have been deposited at the herbarium of Mae Fah Luang University (MFLU) and Kunming Institute of Botany (HKAS), while the living cultures have been deposited at Mae Fah Luang University Culture Collection (MFLUCC). The Faces of Fungi and the Index Fungorum numbers are registered as outlined in Jayasiri et al. [28], and the Index Fungorum [26].
2.2. DNA Extraction, PCR Amplification and Sequencing
The genomic DNA was extracted from living pure cultures using the Biospin Fungus Genomic DNA extraction Kit (BioFlux, P.R. China) following the manufacturer’s protocol. The internal transcribed spacer (ITS) with the primer pair of ITS4/ITS5 [29], the partial large subunit nuclear rDNA (LSU) with the primer pair of LR0R/LR5 [30], the translation elongation factor 1-alpha gene (TEF1-α) with the primers of EF1-728F/EF-2 [31,32] and the TUB2 with primers of bt2a/bt2b [33] were used to amplify the genes ITS, LSU, TEF1-α and TUB2. The polymerase chain reaction (PCR) was carried out under the following protocol: the final volume of 25 μL consisting of 2 μL of DNA template, 1 μL of each forward and reverse primers, 12.5 μL of 2× FastTaq Premix (mixture of Taq DNA polymerase, dNTPs, and a buffer) and 9.5 μL of deionized water. The PCR thermal cycle program was as follows: for ITS and LSU: initial denaturation at 95 °C for 5 min, then 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 1 min and final extension at 72 °C for 10 min; for TEF1-α: initial denaturation at 94 °C for 5 min, then 35 cycles of denaturation at 94 °C for 1 min, annealing at 56 °C for 1 min and extension at 72 °C for 90 s and final extension at 72 °C for 10 min; for TUB2: initial denaturation at 95 °C for 5 min, then 35 cycles of denaturation at 94 °C for 45 s, annealing at 55 °C for 45 s and extension at 72 °C for 1 min and final extension at 72 °C for 10 min. The PCR products were checked in 1% agarose gels and sent to Tsing Ke Biological Technology (Kunming) Co., China for sequencing. The sequence quality was checked, and the sequences were condensed with SeqMan. The sequences derived in this study were deposited in the GenBank, and the accession numbers were obtained (Table 1).
Table 1.
Taxa names, strain numbers, host, countries and corresponding GenBank accession numbers of the taxa used in the phylogenetic analyses of this study.
| Taxa Names | Strain Numbers | Substrates | Countries | GenBank Accession Numbers | |||
|---|---|---|---|---|---|---|---|
| ITS | LSU | TUB2 | TEF 1-α | ||||
| Apiospora acutiapica | KUMCC 20-0210 | Bambusa bambos | China | MT946343 | MT946339 | MT947366 | MT947360 |
| Apiospora aquaticum | S-642 | Submerged wood | China | MK828608 | MK835806 | - | - |
| Apiospora arundinis | CBS 133509 | Aspergillus flavus sclerotium Buried in sandy field | USA | KF144886 | KF144930 | KF144976 | KF145018 |
| Apiospora arundinis | CBS 449.92 | Bamboo | Canada | KF144887 | KF144931 | KF144977 | KF145019 |
| Apiospora aurea | CBS 244.83 | - | Japan | AB220251 | KF144935 | KF144981 | KF145023 |
| Apiospora balearica | CBS 145129 | Undetermined poaceae | Spain | MK014869 | MK014836 | MK017975 | MK017946 |
| Apiospora bambusae | ICPM 6889 | Bamboo | New Zealand | MK014874 | MK014841 | MK017980 | MK017951 |
| Apiospora bambusae | CBS 145133 | Phyllostachys aurea | Spain | MK014875 | MK014842 | MK017981 | MK017952 |
| Apiospora bambusicola | MFLUCC20-0144 | Schizostachyum brachycladum | Thailand | MW173030 | MW173087 | - | MW183262 |
| Apiospora biserialis | CGMCC 3.20135 | Bamboo | China | MW481708 | MW478885 | MW522955 | MW522938 |
| Apiospora biserialis | GZCC 20_0099 | Bamboo | China | MW481709 | MW478886 | MW522956 | MW522939 |
| Apiospora biserialis | GZCC 20_0100 | Bamboo | China | MW481710 | MW478887 | MW522957 | MW522940 |
| Apiospora camelliae-sinensis | LC 5007 | Camellia sinensis | China | KY494704 | KY494780 | KY705173 | KY705103 |
| Apiospora camelliae-sinensis | LC 8181 | Brassica rapa | China | KY494761 | KY494837 | KY705229 | KY705157 |
| Apiospora chromolaenae | MFLUCC17-1505 | Chromolaena odorata | Thailand | MT214342 | MT214436 | - | MT235802 |
| Apiospora chiangraiense ▲ | MFLUCC21-0053 | Dead culms of bamboo | Thailand | MZ542520 | MZ542524 | MZ546409 | - |
| Apiospora cordylinae | GUCC 10026 | Cordyline fruticosa | China | MT040105 | - | MT040147 | MT040126 |
| Apiospora cyclobalanopsidis | CGMCC 3.20136 | Cyclobalanopsidis glauca | China | MW481713 | MW478892 | MW522962 | MW522945 |
| Apiospora cyclobalanopsidis | GZCC 20_0103 | Cyclobalanopsidis glauca | China | MW481714 | MW478893 | MW522963 | MW522946 |
| Apiospora descalsii | CBS 145130 | Ampelodesmos mauritanicus | Spain | MK014870 | MK014837 | MK017976 | MK017947 |
| Apiospora dichotomanthi | LC 4950 | Dichotomanthes tristaniicarpa | China | KY494697 | KY494773 | KY705167 | KY705096 |
| Apiospora dichotomanthi | LC 8175 | Dichotomanthes tristaniicarpa | China | KY494755 | KY494831 | KY705223 | KY705151 |
| Apiospora esporlensis | CBS 145136 | Phyllostachys aurea | Spain | MK014878 | MK014845 | MK017983 | MK017954 |
| Apiospora euphorbiae | IMI 285638b | Bambusa sp. | Bangladesh | AB220241 | AB220335 | AB220288 | - |
| Apiospora gaoyouensis | CFCC 52301 | Phragmites australis | China | MH197124 | - | MH236789 | MH236793 |
| Apiospora gaoyouensis | CFCC 52302 | Phragmites australis | China | MH197125 | - | MH236790 | MH236794 |
| Apiospora garethjonesii | KUMCC 16-0202 | Dead culms of bamboo | China | KY356086 | KY356091 | - | - |
| Apiospora gelatinosa | KHAS 11962 | Bamboo | China | MW481706 | MW478888 | MW522958 | MW522941 |
| Apiospora gelatinosa | GZAAS 20-0107 | Bamboo | China | MW481707 | MW478889 | MW522959 | MW522942 |
| Apiospora guizhouensis | LC 5318 | Air in karst cave | China | KY494708 | KY494784 | KY705177 | KY705107 |
| Apiospora guizhouensis | LC 5322 | Air in karst cave | China | KY494709 | KY494785 | KY705178 | KY705108 |
| Apiospora hispanica | IMI 326877 | Beach sand | Spain | AB220242 | AB220336 | AB220289 | - |
| Apiospora hydei | CBS 114990 | Bambusa tuldoides | China | KF144890 | KF144936 | KF144982 | KF145024 |
| Apiospora hydei | KUMCC 16-0204 | Dead culms of bamboo | China | KY356087 | KY356092 | - | - |
| Apiospora hyphopodii | MFLUCC15-0003 | Bambusa tuldoides | China | KR069110 | - | - | - |
| Apiospora hyphopodii | KUMCC 16-0201 | Culms of bamboo | China | KY356088 | KY356093 | - | - |
| Apiospora iberica | CBS 145137 | Arundo donax | Portugal | MK014879 | MK014846 | MK017984 | MK017955 |
| Apiospora intestini | CBS 135835 | Gut of a grasshopper | India | KR011352 | MH877577 | KR011350 | KR011351 |
| Apiospora intestini ▲ | MFLUCC 21-0052 | Dead culms of bamboo | Thailand | MZ542521 | MZ542525 | MZ546410 | MZ546406 |
| Apiospora italica | CBS 145138 | Arundo donax | Italy | MK014880 | MK014847 | MK017985 | MK017956 |
| Apiospora italica | CBS 145139 | Phragmites australis | Spain | MK014881 | MK014848 | MK017986 | - |
| Apiospora jatrophae | AMH-9557 | Jatropha podagrica | India | JQ246355 | - | - | - |
| Apiospora jatrophae | AMH-9556 | Jatropha podagrica | India | HE981191 | - | - | - |
| Apiospora jiangxiensis | LC 4494 | Phyllostachys sp. | China | KY494690 | KY494766 | KY705160 | KY705089 |
| Apiospora jiangxiensis | LC 4577 | Maesa sp. | China | KY494693 | KY494769 | KY705163 | KY705092 |
| Apiospora kogelbergensis | CBS 113332 | Cannomois virgata | South Africa | KF144891 | KF144937 | KF144983 | KF145025 |
| Apiospora kogelbergensis | CBS 113333 | Dead culms of Restionaceae | South Africa | KF144892 | KF144938 | KF144984 | KF145026 |
| Apiospora locuta -pollinis | LC 11688 | Bee bread | China | MF939596 | - | MF939623 | MF939618 |
| Apiospora locuta -pollinis | LC 11683 | Brassica campestris | China | MF939595 | - | MF939622 | MF939616 |
| Apiospora longistroma | MFLUCC 11-0479 | Dead culms of bamboo | Thailand | KU940142 | KU863130 | - | - |
| Apiospora longistroma | MFLUCC11-0481 | Dead culms of bamboo | Thailand | KU940141 | KU863129 | - | - |
| Apiospora malaysiana | CBS 102053 | Macaranga hullettii | Malaysia | KF144896 | KF144942 | KF144988 | KF145030 |
| Apiospora marii | CBS 497.90 | Beach sands | Spain | AB220252 | KF144947 | KF144993 | KF145035 |
| Apiospora marii | DiSSPA_A1 | Oleaeuropaea | Italy | MK602320 | - | MK614695 | MK645472 |
| Apiospora mediterranea | IMI 326875 | Air | Spain | AB220243 | AB220337 | AB220290 | - |
| Apiospora minutispora | 1.70E-41 | Mountain soil | Korea | LC517882 | - | LC518888 | LC518889 |
| Apiospora mytilomorpha | DAOM 214595 | Andropogon sp. | India | KY494685 | - | - | - |
| Apiospora neobambusae | LC 7106 | Leaves of bamboo | China | KY494718 | KY494794 | KY705186 | KY806204 |
| Apiospora neobambusae | LC 7124 | Leaves of bamboo | China | KY494727 | KY494803 | KY705195 | KY806206 |
| Apiospora neochinensis | CFCC 53036 | Fargesia qinlingensis | China | MK819291 | - | MK818547 | MK818545 |
| Apiospora neochinensis | CFCC 53037 | Fargesia qinlingensis | China | MK819292 | - | MK818548 | MK818546 |
| Apiospora neogarethjonesii | DQD 2019a | Bamboo | China | MK070897 | MK070898 | - | - |
| Apiospora neosubglobosa | JHB 006 | Bamboo | China | KY356089 | KY356094 | - | - |
| Apiospora neosubglobosa | KUMCC 16-0203 | Bamboo | China | KY356090 | KY356095 | - | - |
| Apiospora obovata | LC 4940 | Lithocarpus sp. | China | KY494696 | KY494772 | KY705166 | KY705095 |
| Apiospora obovata | LC 8177 | Lithocarpus sp. | China | KY494757 | KY494833 | KY705225 | KY705153 |
| Apiospora ovata | CBS 115042 | Arundinaria hindsii | China | KF144903 | KF144950 | KF144995 | KF145037 |
| Apiospora paraphaeosperma | MFLUCC13-0644 | Dead culms of bamboo | Thailand | KX822128 | KX822124 | - | - |
| Apiospora phragmitis | CPC 18900 | Phragmites australis | Italy | KF144909 | KF144956 | KF145001 | KF145043 |
| Apiospora phyllostachydis | MFLUCC18-1101 | Phyllostachys heteroclada | China | MK351842 | MH368077 | MK291949 | MK340918 |
| Apiospora piptatheri | CBS 145149 | Piptatherum miliaceum | Spain | MK014893 | MK014860 | - | MK017969 |
| Apiospora pseudomarii | GUCC 10228 | Aristolochia debilis | China | MT040124 | - | MT040166 | MT040145 |
| Apiospora pseudoparenchymatica | LC 7234 | Leaves of bamboo | China | KY494743 | KY494819 | KY705211 | KY705139 |
| Apiospora pseudoparenchymatica | LC 8173 | Leaves of bamboo | China | KY494753 | KY494829 | KY705221 | KY705149 |
| Apiospora pseudorasikravindrae | KUMCC 20-0208 | Bambusa dolichoclada | China | MT946344 | - | MT947367 | MT947361 |
| Apiospora pseudosinensis | CPC 21546 | Leaves of bamboo | Netherlands | KF144910 | KF144957 | - | KF145044 |
| Apiospora pseudospegazzinii | CBS 102052 | Macaranga hullettii | Malaysia | KF144911 | KF144958 | KF145002 | KF145045 |
| Apiospora pterosperma | CBS 123185 | Machaerina sinclairii | New Zealand | KF144912 | KF144959 | KF145003 | - |
| Apiospora pterosperma | CPC 20193 | Lepidosperma gladiatum | Australia | KF144913 | KF144960 | KF145004 | KF145046 |
| Apiospora qinlingensis | CFCC 52303 | Fargesiaqinlingensis | China | MH197120 | - | MH236791 | MH236795 |
| Apiospora qinlingensis | CFCC 52304 | Fargesia qinlingensis | China | MH197121 | - | MH236792 | MH236796 |
| Apiospora rasikravindrae | LC 8179 | Brassica rapa | China | KY494759 | KY494835 | KY705227 | KY705155 |
| Apiospora rasikravindrae | NFCCI 2144 | Soil | Norway | JF326454 | - | - | - |
| Apiospora rasikravindrae ▲ | MFLUCC 21-0051 | Dead culms of bamboo | Thailand | MZ542523 | MZ542527 | MZ546412 | MZ546408 |
| Apiospora rasikravindrae ▲ | MFLUCC 21-0054 | Dead culms of Maize | Thailand | MZ542522 | MZ542526 | MZ546411 | MZ546407 |
| Apiospora sacchari | CBS 372.67 | Air | - | KF144918 | KF144964 | KF145007 | KF145049 |
| Apiospora sacchari | CBS 664.74 | Soil under Calluna vulgaris | Netherlands | KF144919 | KF144965 | KF145008 | KF145050 |
| Apiospora saccharicola | CBS 191.73 | Air | Netherlands | KF144920 | KF144966 | KF145009 | KF145051 |
| Apiospora saccharicola | CBS 831.71 | - | Netherlands | KF144922 | KF144969 | KF145012 | KF145054 |
| Apiospora septata | CGMCC 3.20134 | bamboo | China | MW481711 | MW478890 | MW522960 | MW522943 |
| Apiospora septata | GZCC 20_0109 | bamboo | China | MW481712 | MW478891 | MW522961 | MW522944 |
| Apiospora serenensis | IMI 326869 | Food, pharmaceutical excipients, atmosphere and home dust | Spain | AB220250 | AB220344 | AB220297 | - |
| Apiospora setariae | MT492005 | Setaria viridis | China | MT492005 | - | MT497467 | MW118457 |
| Apiospora setostroma | KUMCC 19-0217 | Dead branches of bamboo | China | MN528012 | MN528011 | - | MN527357 |
| Apiospora sorghi | URM 93000 | Sorghum bicolor | Brazil | MK371706 | - | MK348526 | - |
| Apiospora subglobosa | MFLUCC11-0397 | Dead culms of bamboo | Thailand | KR069112 | KR069113 | - | - |
| Apiospora subrosea | LC 7291 | Leaves of bamboo | China | KY494751 | KY494827 | KY705219 | KY705147 |
| Apiospora subrosea | LC 7292 | Leaves of bamboo | China | KY494752 | KY494828 | KY705220 | KY705148 |
| Apiospora thailandica | MFLUCC 15-0199 | Dead culms of bamboo | Thailand | KU940146 | KU863134 | - | - |
| Apiospora thailandica | MFLUCC15-0202 | Dead culms of bamboo | Thailand | KU940145 | KU863133 | - | - |
| Apiospora vietnamensis | IMI 99670 | Citrus sinensis | Vietnam | KX986096 | KX986111 | KY019466 | - |
| Apiospora xenocordella | CBS 478.86 | Soil from roadway | Zimbabwe | KF144925 | KF144970 | KF145013 | KF145055 |
| Apiospora xenocordella | CBS 595.66 | Soil | Austria | KF144926 | KF144971 | - | - |
| Apiospora yunnana | DDQ 00281 | Phyllostachys nigra | China | KU940148 | KU863136 | - | - |
| Apiospora yunnana | MFLUCC15-1002 | Phyllostachys nigra | China | KU940147 | KU863135 | - | - |
| Arthrinium austriacum | GZU 345004 | Carex pendula | Austria | MW208928 | - | - | - |
| Arthrinium austriacum | GZU 345006 | Carex pendula | Austria | MW208929 | MW208860 | - | - |
| Arthrinium cf. sporophleoides | GZU 345102 | Carex firma | Austria | MW208944 | MW208866 | - | - |
| Arthrinium caricicola | CBS 145127 | Carex ericetorum | China | MK014871 | MK014838 | MK017977 | MK017948 |
| Arthrinium crenatum | AG 19066 | Poaceae, Carex | France | MW208931 | MW208861 | - | - |
| Arthrinium curvatum | AP 25418 | Leaves of Carex sp. | China | MK014872 | MK014839 | MK017978 | MK017949 |
| Arthrinium japonicum | IFO 30500 | - | Japan | AB220262 | AB220356 | AB220309 | - |
| Arthrinium japonicum | IFO 31098 | Leaves of Carex despalata | Japan | AB220264 | AB220358 | AB220311 | - |
| Arthrinium luzulae | AP7619-3 | Luzula sylvatica | Spain | MW208937 | MW208863 | - | - |
| Arthrinium morthieri | GZU 345043 | Cyperaceae carex | Austria | MW208938 | MW208864 | - | - |
| Arthrinium phaeospermum | CBS 114317 | Leaves of Hordeum vulgare | Iran | KF144906 | KF144953 | KF144998 | KF145040 |
| Arthrinium phaeospermum | CBS 114318 | Leaves of Hordeum vulgare | Iran | KF144907 | KF144954 | KF144999 | KF145041 |
| Arthrinium puccinioides | CBS 549.86 | Lepidosperma gladiatum | Germany | AB220253 | AB220347 | AB220300 | - |
| Arthrinium sphaerospermum | AP25619/CBS 146355 | Poaceae | Norway | MW208943 | MW208865 | - | - |
| Arthrinium sporophleum | CBS 145154 | Dead leaves of Juncus sp. | Spain | MK014898 | MK014865 | MK018001 | MK017973 |
| Arthrinium trachycarpum | CFCC 53038 | Trachycarpus fortune | China | MK301098 | - | MK303394 | MK303396 |
| Arthrinium urticae | IMI 326344 | - | - | AB220245 | AB220339 | AB220292 | - |
| Arthrinium trachycarpum | CFCC 53039 | Trachycarpus fortune | China | MK301099 | - | MK303395 | MK303397 |
| Nigrospora aurantiaca | CGMCC 3.18130 | Nelumbo sp. | China | KX986064 | KX986098 | KY019465 | KY019295 |
| Nigrospora camelliae -sinensis | CGMCC 3.18125 | Camellia sinensis | China | KX985986 | KX986103 | KY019460 | KY019293 |
| Nigrospora chinensis | CGMCC 3.18127 | Machilus breviflora | China | KX986023 | KX986107 | KY019462 | KY019422 |
| Nigrospora gorlenkoana | CBS 480.73 | Vitis vinifera | Kazakhstan | KX986048 | KX986109 | KY019456 | KY019420 |
| Nigrospora guilinensis | CGMCC 3.18124 | Camellia sinensis | China | KX985983 | KX986113 | KY019459 | KY019292 |
| Nigrospora hainanensis | CGMCC 3.18129 | Musa paradisiaca | China | KX986091 | KX986112 | KY019464 | KY019415 |
| Nigrospora lacticolonia | CGMCC 3.18123 | Camellia sinensis | China | KX985978 | KX986105 | KY019458 | KY019291 |
| Nigrospora musae | CBS 319.34 | Musa sp. | Australia | MH855545 | KX986110 | KY019455 | KY019419 |
| Nigrospora oryzae | LC2693 | Neolitsea sp. | China | KX985944 | KX986101 | KY019471 | KY019299 |
| Nigrospora osmanthi | CGMCC 3.18126 | Hedera nepalensis | China | KX986010 | KX986106 | KY019461 | KY019421 |
| Nigrospora pyriformis | CGMCC 3.18122 | Citrus sinensis | China | KX985940 | KX986100 | KY019457 | KY019290 |
| Nigrospora rubi | LC2698 | Rubus sp. | China | KX985948 | KX986102 | KY019475 | KY019302 |
| Nigrospora sphaerica | LC7298 | Nelumbo sp. | China | KX985937 | KX986097 | KY019606 | KY019401 |
| Nigrospora vesicularis | CGMCC 3.18128 | Musa paradisiaca | China | KX986088 | KX986099 | KY019463 | KY019294 |
| Sporocadus trimorphus | CBS 114203 | Rosa canina | Sweden | MH553977 | MH554196 | MH554636 | MH554395 |
Notes: Newly generated sequences are indicated by ▲ after the species name. Ex-type strains are in bold. - = information not available. Abbreviations: AMH: Ajrekar Mycological Herbarium, Pune, Maharashtra, India; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands; CFCC: China Forestry Culture Collection Center, Beijing, China; CPC: Culture collection of Pedro Crous, housed at the Westerdijk Fungal Biodiversity Institute; DAOM: Canadian Collection of Fungal Cultures, Ottawa, Canada; DDQ: D.Q. Dai; GUCC: Guizhou University Culture Collection, Guizhou, China; ICMP: International Collection of Microorganisms from Plants, New Zealand; IFO: Institute for Fermentation, Osaka, Japan; IMI: Culture collection of CABI Europe UK Centre, Egham, UK; JHB: H.B. Jiang; KUMCC: Culture collection of Kunming Institute of Botany, Yunnan, China; LC: Personal culture collection of Lei Cai, housed in the Institute of Microbiology, Chinese Academy of Sciences, China; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; NFCCI: National Fungal Culture Collection of India.
2.3. Phylogenetic Analyses
The sequences generated in this study were subjected to a basic local alignment search tool (BLAST) search in the GenBank to identify closely related Apiospora taxa to the taxa obtained in this study. The sequences of Apiospora were also obtained from recently published data [4,18,34,35,36,37,38,39]. Consensus sequences were assembled and aligned using BioEdit and MAFFT v.7.110 online program, respectively (http://mafft.cbrc.jp/alignment/server, accessed on 12 August 2021) [40], and manually edited using BioEdit v7.2.3 [41].
The construction of the combined phylogenetic trees was completed using maximum likelihood (ML) and Bayesian inference posterior probabilities (BYPP), with Sporocadus trimorphus (CBS 114203) as the outgroup taxon. The models were selected as GTRGAMMA for maximum likelihood, while the best-fit models were selected as GTR + I + G for ITS, LSU and HKY + I + G for TUB2, and TEF1-α for the Bayesian posterior probability analysis. The maximum likelihood (ML) analysis was performed using RAxML-HPC v.8 [42,43] on the XSEDE TeraGrid of the CIPRES Science Gateway (https://www.phylo.org, accessed on 12 August 2021) [44] with a rapid bootstrap analysis, followed by 1000 bootstrap replicates. The final tree was selected amongst the suboptimal trees from each run by comparing the likelihood scores under the GTRGAMMA substitution model. The Bayesian analyses were performed by MrBayes v. 3.2 [45]. Markov chain Monte Carlo (MCMC) was run for 5,000,000 generations, and the trees were sampled every 100th generation. The first 10% of the trees that represented the burn-in phase were discarded, and only the remaining 90% of the trees were used for calculating the posterior probabilities (PP) for the majority rule consensus tree. Phylogenetic trees were visualized with FigTree v. 1.4.2 [46] and modified in Adobe Illustrator CS5 software (Adobe Systems, USA). The newly obtained sequences in this study were deposited in the GenBank.
3. Results
3.1. Phylogeny
The combined ITS, LSU, TEF1-α and TUB2 dataset comprised 138 strains, including four newly sequenced strains, with Sporocadus trimorphus (CBS 114203) as the outgroup taxon. Multi-locus sequences were concatenated, which comprised 2820 nucleotide characters, including gaps (ITS: 1–637, LSU: 638–1518, TEF1-α: 1519–1971 and TUB2: 1972–2799). The phylogenic tree from the RAxML analysis had similar topology to the Bayesian analysis. The RAxML analysis of the combined dataset yielded the best scoring tree (Figure 1) with a final ML optimization likelihood value of −27840.652840. The matrix had 1446 distinct alignment patterns, with 27.45% undetermined characters or gaps. the estimated base frequencies were as follows: A = 0.238477, C = 0.253732, G = 0.254209, T = 0.253582; substitution rates AC = 1.244445, AG = 3.021293, AT = 1.211434, CG = 1.060781, CT = 4.719948, GT = 1.000000; gamma distribution shape parameter α = 0.298987.
Figure 1.
The best-scoring RAxML tree constructed from a concatenated ITS, LSU, TEF1-α, and TUB2 dataset. The tree is rooted with Sporocadus trimorphus (CBS 114203). Nodes were annotated if bootstrap supported value ≥ 70% maximum likelyhood bootstrap proportion (ML, left) or ≥0.95 Bayesian posterior probability (PP, right). The newly described species are in red and new combination species are in blue.
The phylogenetic trees generated by maximum likelihood and Bayesian show the taxonomic placements of our total strains belong to Apiospora. The strains MFLUCC 21-0051 and MFLUCC 21-0054 clustered together with members of Apiospora and grouped with Ap. rasikravindrae (NFCCL 2144 and LC 8179). The strain MFLUCC 21-0052 presented as a distinct lineage and sister to Ap. intestine (CBS 135835) with significant statistical support (ML/BI = 100/1.00). The strain MFLUCC 21-0053 clustered with Ap. intestine (CBS 135835), but in a distinct clade with high support (ML/BI = 100/1.00).
3.2. Taxon Treatment
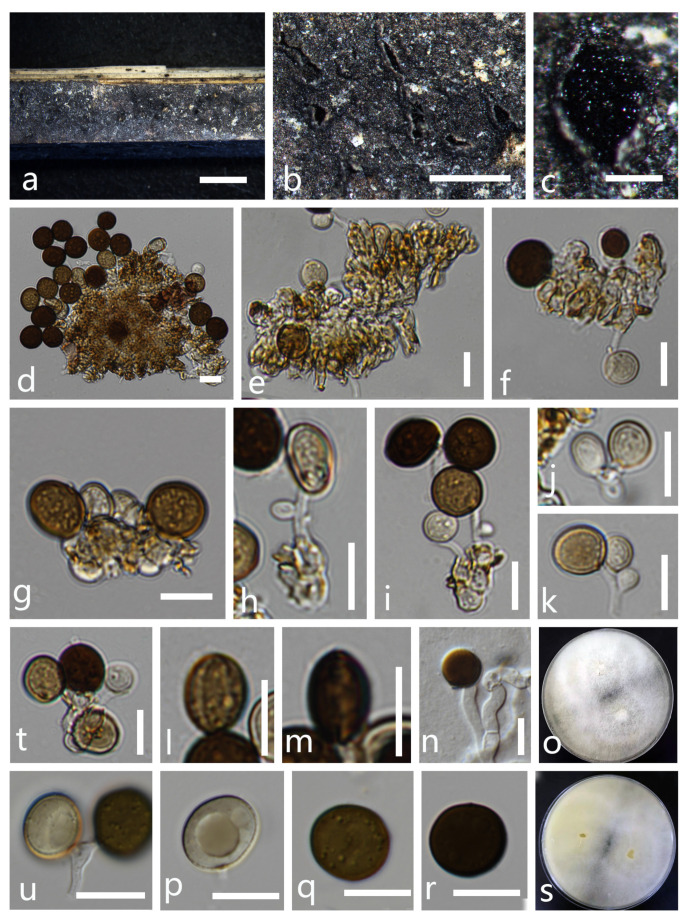
Apiospora chiangraiense X.G. Tian and Tibpromma S., sp. nov. (Figure 2).
Figure 2.
Apiospora chiangraiense (MFLU 21-0046, holotype). (a–c) Appearance of the fungus on dead culms of bamboo. (d–i) Conidia with conidiophores. (j–l) Conidiogenous cells bearing conidia. (n,o,q) Conidia. (p) Conidia with germ-slit. (m) Germinated conidium. (r,s) Colonies on PDA media (r forward, s reversed). Scale bars: a = 4000 μm, b = 1000 μm, c = 200 μm and d–q = 10 µm.
Index Fungorum number: IF558492; Faces of Fungi number: FoF 09905.
Etymology: Referring to Chiang Rai Province, Thailand, where the fungus was collected.
Saprobic on dead culms of bamboo. Sexual morph: undetermined. Asexual morph: Colonies on natural substrate are dry, dark brown to black, velvety, dull, consisting of a sterile mycelial outer zone and a round, glistening, abundantly sporulating center, with conidia readily liberated when disturbed. Mycelium is superficial, branched, hyaline to dark brown, septate, smooth-walled and hyphae. Conidiophores are reduced to conidiogenous cells, grouped together to form sporodochia. Conidiogenous cells are 4–7.5 µm high × 3–4 µm diam. ( = 6 × 3.5 µm2, n = 30), monoblastic or polyblastic, aggregated in clusters on hyphae, hyaline to light brown, smooth and cylindrical to subcylindrical. Conidia are aseptate, pale brown to dark brown, in the surface view 6.5–8 × 6–8 μm2 ( = 7.5 × 7 μm2, n = 50), in the lateral view 6–7.5 × 4–5.5 μm2 ( = 7 × 5 μm2, n = 50), with a central scar, globose in the surface view, a lenticular inside view, with straight germ slit spore length.
Culture characteristics: Conidia germinating on PDA within 24 h at room temperature (25 °C). On the PDA, the colonies’ surfaces are white, lightly yellow, wooly, flat, spreading, filiform, with abundant aerial mycelia and reverse off-white to yellow.
Material examined: THAILAND, Chiang Rai Province, Muang District, on dead culms of bamboo, 23 October 2020, X.G. Tian, bb-4-5, (MFLU 21-0046, holotype); ex-type culture, MFLUCC 21-0053.
Notes: In the phylogenetic analyses, Apiospora chiangraiense formed a distinct clade sister to Ap. intestini with strong bootstrap support values (ML/BI = 100/1.00). Morphologically, Ap. chiangraiense is distinct from Ap. intestine by conidiogenous cells, conidiophores and conidia. The conidiophores of Ap. intestini are usually hyaline, macronematous, mononematous and transversely septate, while Ap. chiangraiense has reduced conidiophores and conidiogenous cells grouped together to form sporodochia. Additionally, Ap. chiangraiense has larger conidia compared to Ap. intestini (surface view 6.5–8 × 6–8 μm2, lateral view 6–7.5 × 4–5.5 μm versus surface view (4.5–) 5.5 (–6) μm diam, side view (2–) 4 (–6) μm diam). Based on pairwise nucleotide comparisons, Ap. chiangraiense is different from Ap. intestini (CBS 135835) in 27/583 bp (4.63/) of the ITS, 9/814 (1.1%) of the LSU and 61/696 bp (8.76%) of TUB2, but we were unable to compare TEF-α pairwise nucleotides as the amplification of TEF-α was not successful for this species. However, both the phylogenetic analyses and morphological characteristics supported our species as a distinct new species.
Apiospora intestini (Kajale, Sonawane and Rohit Sharma) Pintos and P. Alvarado, Fungal Systematics and Evolution 7: 206 (2021) (Figure 3).
Figure 3.
Apiospora intestini (MFLU 21-0045). (a–c) Appearance of the fungus on dead culms of bamboo. (d–g) Conidia with conidiophores. (h–j,l,m) Conidiogenous cells bearing conidia. (n–s) Conidia. (k) Germinated conidium. (t,u) Colonies on PDA media (t forward, u reversed). Scale bars: a = 4000 μm, b = 1000 μm, c = 200 μm, d–g = 20 µm, h–m = 10 μm and n–s = 5 μm.
Index Fungorum number: IF 837744.
Saprobic on dead culms of bamboo. Sexual morph: undetermined. Asexual morph: Colonies are on natural substrate surface, gregarious, powdery, dark brown to black, dull with conidia readily liberated when disturbed. Conidiophores are 1.5–2 µm wide ( = 2 µm, n = 40) hyaline, macronematous, mononematous, transversely septate, thick-walled, brown. Conidiogenous cells are 6–9.5 × 1.5–2 µm2 ( = 7.5 × 2 µm2, n = 30), intercalary, cylindrical, hyaline. Conidia are 6.5–5 × 6–10 µm2 ( = 7 × 5.5 µm2, n = 50), borne as bunches on conidiophores, lateral, pale brown to brown, smooth-walled, globose to subglobose or irregularly round, aseptate, with a central scar and without germ slit.
Culture characteristics: Conidia germinating on PDA within 24 h at room temperature. The colonies’ surfaces are white, cottony, flat, spreading, filiform, mycelia not tightly attached to the surface and the reverse lightly pigmented.
Material examined: THAILAND, Chiang Rai Province, Muang District, on dead culms of bamboo, 23 October 2020, X. G. Tian bb-4-2 (MFLU 21-0045), living culture, MFLUCC 21-0052.
Notes: Apiospora intestini was introduced by Crous et al. [19] based on the morphology of asexual morph and the phylogenetic analyses. In this paper, our new isolate (MFLUCC 21-0052) clustered with the ex-type strain of Ap. intestini with relatively high support (ML/BI = 100/1.00). Morphologically, the conidia of the new isolate (MFLUCC 21-0052) are similar to the holotype Ap. intestini (CBS 135835) in having similar size of conidiophores that are borne as bunches, intercalary and terminal, brown, smooth, aseptate and globose to subglobose. Based on nucleotide comparisons, Ap. intestini (MFLUCC 21-0052) is slightly different from Ap. intestini in 12/580 bp (2.07%) of the ITS, 2/814 (0.24%) of the LSU, 2/684 bp (0.29%) of TUB2 and 2/610 bp (0.32%) of TEF1-α. Based on both phylogeny and morphology, the new isolate (MFLUCC 21-0052) is identified as Ap. intestini. This is the first report of Ap. intestini (MFLUCC 21-0052) isolated from dead culms of bamboo in Thailand, which was originally isolated from a grasshopper’s gut in India.
Apiospora rasikravindrae (Shiv M. Singh, L.S. Yadav, P.N. Singh, Rahul Sharma and S.K. Singh) Pintos and P. Alvarado, Fungal Systematics and Evolution 7: 207 (2021) (Figure 4).
Figure 4.
Apiospora rasikravindrae (MFLU 21-0045). (a–c) Appearance of the fungus on dead culms of bamboo. (d–i) Conidia with conidiophores. (j,k,t,u) Conidiogenous cells bearing conidia. (l,m) Conidia with germ-slit. (p–r) Conidia. (n) Germinated conidium. (o,s) Colonies on PDA media (o forward, s reversed). Scale bars: a = 4000 μm, b = 2000 μm, c = 200 μm and d–s = 10 μm.
Index Fungorum number: IF 837716; Faces of Fungi number: FoF 01994.
Saprobic on dead culms of bamboo. Colonies appear as spotty patches on natural substrate surface. Conidiomata are immersed, pycnidial, scattered, globose to slightly conical, ostiolate, black, coriaceous. Conidiophores are 9–26 × 1–2.5 μm2 ( = 17.5 × 2 μm2, n = 15), arising mostly from swollen basal cells, micro to semi-macronematous, mononematous, unbranched, straight or flexuous, smooth and thin-walled, hyaline. Conidiogenous cells are basauxic, discrete, hyaline, smooth-walled. Conidia in surface view are 9–11 × 9–10.5 μm2 ( = 10 × 10 μm2, n = 50), in lateral view 10–11 × 6.5–8 μm2 ( = 10.5 × 7.5 μm2, n = 20), lenticular, globose to ovoid, occasionally elongated to ellipsoidal, brown to dark brown, smooth-walled, with a longitudinal, thin-walled, with a pale equatorial slit.
Material examined: THAILAND, Chiang Rai Province, Muang District, isolated as Saprobic on dead culms of bamboo, 23 October 2020, X. G. Tian, bb-4-1 (MFLU 21-0044), living culture, MFLUCC 21-0051; ibid decaying maize culms, 11 November 2020, X. G. Tian, corn-1-1 (HKAS 115764), living culture, MFLUCC 21-0054
Notes: The National Center for Biotechnology Information (NCBI) BLAST results of ITS, LSU, TUB2 and TEF1-α sequences of our new isolate (MFLUCC 21-0054) showed high similarities with Apiospora rasikravindrae (LC 8179) (100%, 100%, 98.90% and 98.97%, respectively), while the new isolate (MFLUCC 21-0051) also showed high similarities with Apiospora rasikravindrae (LC 8179) (99.83%, 100%, 99.61% and 99.51%, respectively). Our phylogenetic analyses showed that the two new isolates clustered with the ex-type strain of Ap. rasikravindrae (NFCCI 2144) and Ap. rasikravindrae (LC 8179). Morphologically, our new isolate is closely related to the holotype of Ap. rasikravindrae in having lenticular, globose to ovoid, occasionally elongated to ellipsoidal, brown to dark brown, smooth-walled, germ-slit conidia and micro-semi-macronematous, mononematous, unbranched, straight or flexuous, smooth and thin-walled and hyaline conidiophores. Hence, the two new isolates are identified as Ap. rasikravindrae.
Apiospora rasikravindrae was originally isolated from soil in Norway [47]. Apiospora rasikravindrae occurred on Capsicum, Kappaphycus alvarezii, Pinus, Platanus acerifolia, rice, Sargassum thunbergia and Triticum aestivum from Brazil, China, India, Japan, Netherlands, Svalbard and Thailand [3,48]. Dai et al. [3] describe and illustrate both sexual and asexual morphs for Ap. rasikravindrae and it was collected on the stems of bamboo. In this study, the isolate MFLUCC 21-0051 was newly collected from bamboo, while the isolate MFLUCC 21-0054 was newly recorded from maize.
3.3. New Combinations
Apiospora acutiapica (Senan. and Cheew) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558499.
Basionym: Arthrinium acutiapicum Senan. and Cheew, Frontiers in Microbiology 11. 2020.
Notes: Arthrinium acutiapicum was introduced by Senanayake et al. [34] and was collected on dead twigs of Bambusa bambos in China. Senanayake et al. [34] mentioned that this species is distinct from Ar. pseudorasikravindrae, which is a sister to Ar. acutiapicum, by the reduction of conidiophores to conidiogenous cells, cylindrical to ampulliform, pale brown conidiogenous cells with pointed, hyaline apex and brown to dark brown and smooth-walled conidia with a dark equatorial slit [34].
In our phylogenetic analysis based on combined LSU, ITS, TEF1-α and TUB2 sequence data, Arthrinium acutiapicum clustered with Apiospora pseudorasikravindrae (=Ar. pseudorasikravindrae) with high support (ML/BI = 95/-). Thus, we propose the transfer of Ar. acutiapicum under the new combination Ap. acutiapica, based on the morphological and phylogenetic analyses.
Apiospora bambusicola (X. Tang, K.D. Hyde and J.C. Kang) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558493; Faces of Fungi number: FoF 09162.
Basioym: Arthrinium bambusicola X. Tang, K.D. Hyde and J.C. Kang, Biodiversity Data Journal 8 (e58755): 11 2020.
Notes: Arthrinium bambusicola was introduced by Tang et al. [18] and was collected on dead culms of Schizostachyum brachycladum in Thailand. Tang et al. [18] mentioned that Ar. bambusicola were retrieved as a sister taxon of Ar. gutiae with high support (ML/BI = 83/0.99), but differs from Ar. gutiae in having larger conidia and irregularly rounded, guttulate to roughened conidia. Pintos and Alvarado [4] transferred Ar. gutiae to Apiospora based on the phylogenetic analyses and morphological characters.
In our phylogenetic analyses based on combined LSU, ITS, TEF1-α and TUB2 sequence data, Arthrinium bambusicola is a sister to the newly introduced species Ap. chiangraiense with high support (ML/BI = 80/0.99). Thus, we propose the transfer of Ar. bambusicola under the new combination Ap. bambusicola, based on the morphological and phylogenetic analyses.
Apiospora biserialis (Y. Feng and Z.Y. Liu) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558502; Faces of Fungi number: FoF 09569.
Basioym: Arthrinium biseriale Y. Feng, J.K. Liu, C.G. Lin, Y.Y. Chen, M.M. Xiang and Z.Y. Liu, Frontiers in Microbiology 12. 2021.
Notes: Arthrinium biseriale was introduced by Feng et al. [49] from dead culms of bamboo in China. The phylogenetic analysis of Feng et al. [49] showed that Ar. biseriale is closely related to Ar. gelatinosum, but they are phylogenetically distinct and can be recognized as two different species. Morphologically, Ar. biseriale has smaller stromata and the spores of Ar. biseriale are more curved than those of Ar. gelatinosum [49].
In our phylogenetic analyses based on combined LSU, ITS, TEF1-α and TUB2 sequence data, Arthrinium biseriale clustered with Apiospora gelatinosa with high support (ML/BI = 90/0.99). Thus, we propose the transfer of Ar. biseriale under the new combination Ap. biserialis, based on the morphological and phylogenetic analyses.
Apiospora cordylines (T.Z. Chen, Yong Wang bis and K.D. Hyde) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558494.
Basionym: Arthrinium cordylines T.Z. Chen, Yong Wang bis and K.D. Hyde, Mycotaxon 136(1): 189 2021.
Notes: Arthrinium cordylines was introduced by Chen et al. [39] from the leaves of Cordyline fruticosa in China. Chen et al. [39] mentioned that Ar. cordylinae formed a well-supported branch with type strains of Ar. aureum (CBS 244.83) and Ar. hydei (CBS 114990). Meanwhile, a base difference comparison also confirmed Ar. cordylinae is a distinct species.
In our phylogenetic analyses, Arthrinium cordylines is a sister to Ap. hydei with high support (ML/BI = 96/0.99). Thus, we propose the transfer of Ar. cordylines under the new combination Ap. cordylines.
Apiospora cyclobalanopsidis (Y. Feng and Z.Y. Liu) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558503; Faces of Fungi number: FoF 09572.
Basioym: Arthrinium cyclobalanopsidis Y. Feng, J.K. Liu, C.G. Lin, Y.Y. Chen, M.M. Xiang and Z.Y. Liu, Frontiers in Microbiology 12. 2021.
Notes: Arthrinium cyclobalanopsidis was introduced by Feng et al. [49] from a leaf of Cyclobalanopsidis glauca Oerst in China. Feng et al. [49] showed that Ar. cyclobalanopsidis clustered with Ar. camelliae-sinensis, but can be distinguished from Ar. camelliae-sinensis by conidiogenous cells. Pintos and Alvarado [4] transferred Ar. camelliae-sinensis to Apiospora camelliae-sinensis, based on the phylogenetic analyses and morphological characteristics.
In our phylogenetic analyses based on combined LSU, ITS, TEF1-α and TUB2 sequence data, Arthrinium cyclobalanopsidis clustered with Ap. camelliae-sinensis with high support (ML/BI = 78/1.00). Thus, we propose the transfer of Ar. cyclobalanopsidis under the new combination Ap. cyclobalanopsidis, based on the morphological and phylogenetic analyses.
Apiospora euphorbiae (M.B. Ellis) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558495.
Basioym: Arthrinium euphorbiae M.B. Ellis, Mycol. Pap. 103: 6 1965.
Notes: Arthrinium euphorbiae was introduced by Ellis et al. [24] from the dead stems of Euphorbia in Zambia. Tang et al. [18] showed that Ar. euphorbiae is phylogenetically closely related to Ar. malaysianum, Ar. vietnamensis, and Ar. chromolaenae [4,18].
In our phylogenetic analyses, Ar. euphorbiae is a sister to Ap. malaysiana (=Ar. malaysianum) with strong bootstrap support values (ML/PP = 94/0.99). Thus, we propose the transfer of Ar. euphorbiae under the new combination Ap. euphorbiae.
Apiospora gelatinosa (Y. Feng and Z.Y. Liu) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558504; Faces of Fungi number: FoF 09570.
Basioym: Arthrinium gelatinosum Y. Feng, J.K. Liu, C.G. Lin, Y.Y. Chen, M.M. Xiang and Z.Y. Liu, Frontiers in Microbiology 12. 2021.
Notes: Arthrinium gelatinosum was introduced by Feng et al. [49] from dead culms of bamboo in China. Feng et al. [49] mentioned that Ar. gelatinosum is a sister to Ar. biseriale with a well-supported lineage (ML/MP/BI = 93/98/1.00) [49].
In our phylogenetic analyses, Arthrinium gelatinosum clustered with Apiospora biserialis with high support (ML/BI = 90/0.99). Thus, we propose the transfer of Ar. gelatinosum under the new combination Ap. gelatinosa.
Apiospora locuta-pollinis (F. Liu and L. Cai) Pintos and P. Alvarado), X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: 834523; Faces of Fungi number: FoF05221.
Synonyms: Arthrinium pseudomarii T.Z. Chen, Yong Wang bis and K.D. Hyde, Mycotaxon 136(1): 189. 2021.
Basionym: Arthrinium locutum-pollinis F. Liu and L. Cai (as ‘locuta- pollinis’), Mycosphere 9: 1094. 2018.
Notes: Arthrinium pseudomarii was introduced by Chen et al. [39] from the leaves of Aristolochia debilis in China. Chen et al. [39] mentioned that Ar. pseudomarii differs from Ar. hispanicum, Ar. marii and Ar. mediterranei by larger, subglobose to ellipsoid conidia and showed a close relationship with three species with high bootstrap support values (ML/MP = 95/93) [39].
In our phylogenetic analyses, Ar. pseudomarii (GUCC 10228) is a sister to Ap. locuta-pollinis (=Ar. locuta-pollinis) with high support of 95% ML. Based on the nucleotide comparisons, Ar. pseudomarii is slightly different from Ap. locuta-pollinis in 10/582 bp (1.72%) of ITS, but no base pair differences were observed in TUB2 and TEF1-α. Morphologically, the conidia of Ar. pseudomarii are similar to the holotype Ap. locuta-pollinis (LC 11683) in having similar size, brown with a hyaline equatorial rim, smooth, subglobose to ellipsoid condia and hyaline to pale brown, smooth, ampulliform to doliiform conidiogenous cells aggregated into clusters on the hyphae. Thus, we identified that they are the same species, and we synonymize Ar. pseudomarii under the Ap. locuta-pollinis, based on the morphological and phylogenetic analyses.
Apiospora minutispora (K. Das, S.Y. Lee and H.Y. Jung) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558497.
Basionym: Arthrinium minutisporum K. Das, S.Y. Lee and H.Y. Jung, Mycobiology 48(6): 453 2020.
Notes: Arthrinium minutisporum was introduced by Das et al. [37] from mountain soil in Korea. Morphologically, Ar. minutisporum is quite similar to Ar. phragmites, Ar. aureum and Ar. Hydei. However, the conidia of Ar. minutisporum are smaller than those of Ar. phragmites, Ar. aureum and Ar. Hydei, and Ar. minutisporum produce smaller conidiogenous cells than Ar. phragmites [39]. Pintos and Alvarado [4] transferred Ar. phragmites, Ar. aureum and Ar. hydei to Apiospora phragmites, Ap. aureum and Ap. hydei, based on the phylogenetic analyses and morphological characteristics. Whereas Ar. minutisporum was maintained in Arthrinium.
In our phylogenetic analyses, Arthrinium minutisporum forms a distinct subclade and is close to Apiospora aurea, Ap. balearica and Ap. descalsii with strong bootstrap support values (ML/PP = 99/1.00) within Apiospora. Thus, we propose the transfer of Ar. minutisporum under the new combination Ap. minutispora.
Apiospora pseudorasikravindrae (Senan. and Cheew) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF 558505.
Basionym: Arthrinium pseudorasikravindrae Senan. and Cheew, Frontiers in Microbiology 11. 2020.
Notes: Arthrinium pseudorasikravindrae was introduced by Senanayake et al. [34] from dead twigs of Bambusa bambos Voss. in China. Arthrinium pseudorasikravindrae is morphologically distinct from Ar. chinense, Ar. paraphaeospermum and Ar. rasikravindrae by its thick-walled, finely roughened conidia with a pale, equatorial slit and ampulliform, cylindrical or doliiform, basauxic conidiogenous cells [34]. Pintos and Alvarado [4] transferred Ar. chinense, Ar. paraphaeospermum and Ar. rasikravindrae to Apiospora and synonymized them under Apiospora chinense, Ap. paraphaeospermum and Ap rasikravindrae, respectively, based on the phylogenetic analyses and morphological characteristics.
Our phylogenetic analyses based on combined LSU, ITS, TEF1-α and TUB2 sequence data show Ar. pseudorasikravindrae is a sister to the new combinations Ap. acutiapica (=Ar. acutiapicum) with high support (ML/BI = 77/0.99). Thus, we propose the transfer of Ar. pseudorasikravindrae under the new combination Ap. pseudorasikravindrae.
Apiospora septata (Y. Feng and Jian K. Liu) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558506; Faces of Fungi number: FoF 09571.
Basioym: Arthrinium septatum Y. Feng, J.K. Liu, C.G. Lin, Y.Y. Chen, M.M. Xiang and Z.Y. Liu, Frontiers in Microbiology 12. 2021.
Notes: Arthrinium septatum was introduced by Feng et al. [49] from dead bamboo culms in China. Feng et al. [49] showed that Arthrinium septatum forms a well-supported clade and appears to be distinct from other Arthrinium species. Arthrinium septatum resembles Ar. biseriale in having a biseriate, broad fusiform to cylindrical ascospores and cylindrical, clavate asci. However, Ar. septatum differs from Ar. biseriale by having smaller stromata [49].
In our phylogenetic analyses, Arthrinium septatum groups in a well-supported clade with Ap. pseudospegazzinii and Ap. gelatinosa. Thus, we propose the transfer of Ar. septatum under the new combination Ap. septata, based on the morphological and phylogenetic analyses.
Apiosporasetariae (Jiang, N.; Tian, C.M.) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF835609.
Basioym: Arthrinium setariae JIANG, N.; TIAN, C.M. Phytotaxa 483, 149-159. 2021.
Notes: Arthrinium setariae was introduced by Jing et al. [38] from Setaria viridis in China. Jing et al. [38] mentioned that this species has larger conidia and is phylogenetically closely related to Ar. jiangxiense. Pintos and Alvarado [4] transferred Ar. jiangxiense to Apiospora and synonymized Ap. jiangxiens based on the phylogenetic analyses and morphological characteristics.
In our phylogenetic analyses based on combined LSU, ITS, TEF1-α and TUB2 sequence data, Arthrinium setariae clustered with Apiospora jiangxiense with high support (ML/BI = 87/1.00). Thus, we propose the transfer of Ar. setariae under the new combination Ap. setariae, based on the morphological and phylogenetic analyses.
Apiospora sorghi (J.D.P. Bezerra, C.M Gonçalves and C.M. Souza-Motta) X.G. Tian and Tibpromma S., comb. nov.
Index Fungorum number: IF558498; Faces of Fungi number: FoF 05762.
Basioym: Arthrinium sorghi J.D.P. Bezerra, C.M Gonçalves and C.M. Souza-Motta, Fungal Diversity: 10.1007, 73 2020.
Notes: Arthrinium sorghi was introduced as an endophyte by Bezerra et al. [36] from the leaves of Sorghum bicolor in Brazil. Bezerra et al. [36] mentioned that Ar. sorghi is treated as a unique lineage within Arthrinium based on ITS phylogenetic analysis. Morphologically, Ar. sorghi resembles Ar. pseudosinense, Ar. ovatum and Ar. phaeospermum, but differs from them by the culture characteristics, conidiophores and conidia size [36]. Pintos and Alvarado [4] transferred Ar. pseudosinense, Ar. ovatum and Ar. phaeospermum to Apiospora pseudosinensis, Ap. ovata and Ap. phaeospermum based on the phylogenetic analyses and morphological characteristics.
In our phylogenetic analyses based on combined LSU, ITS, TEF1-α and TUB2 sequence data, Arthrinium sorghi clustered with Apiospora bambusucila with high support (ML/BI = 78/0.99). Thus, we propose the transfer of Ar. sorghi under the new combination Ap. sorghi, based on the morphological and phylogenetic analyses.
4. Discussion
In this study, the new taxon Apiospora chiangraiense and two new host records, viz., Ap. intestini and Ap. rasikravindrae, are introduced based on the morphological and phylogenetic analyses. In addition, thirteen new combinations are proposed based on the phylogenetic analyses.
Apiospora was previously synonymized under Arthrinium, but Pintos et al. [14] re-evaluated the placements of these two genera and transferred 55 species to Apiospora based on a phylogenetic analysis. Currently, 117 species of Apiospora are listed in the Index Fungorum [33]. Among these 117 species, 55 species have been confirmed in Apiospora by phylogenetic analyses [4]; however, the remaining 62 species need to be confirmed, as the sequence data of these species are not available. The morphology of Apiospora and Arthrinium are quite similar, so it is difficult to distinguish Apiospora and Arthrinium based only on morphology.
In the phylogenetic analyses, two Arthrinium species, viz., Arthrinium trachycarpum and Ar. urticae, formed a distinct clade out of Arthrinium, and this result is consistent with previous studies [18]. However, the morphologies of these two species are not significantly different from Arthrinium; thus, more collections are required to clarify the placement of these two species [24,50]. In addition, our phylogenetic analyses showed that Apiospora sorghi, Ap. bambusucila, Ap. chiangraiense and Ap. intesini are not clustered together in Apiospora major clades (Figure 1). We also compared the LSU sequence of these four species with other Apiospora species, but a few base pair differences were found. Moreover, their morphologies fit well within the species concept of Apiospora. Thus, further phylogenetic analyses are necessary to confirm whether Apiospora is a species complex or not.
Author Contributions
Data curation, X.T.; funding acquisition, S.T.; methodology, X.T.; supervision, S.C.K., A.M. and S.T.; writing—original draft, X.T. and D.B.; Writing—review and editing, X.T., S.C.K., A.M., I.P., J.X., D.B. and S.T. All authors have read and agreed to the published version of the manuscript.
Funding
Saowaluck Tibpromma would like to thank the International Postdoctoral Exchange Fellowship Program (number Y9180822S1), CAS President’s International Fellowship Initiative (PIFI) (number 2020PC0009), China Postdoctoral Science Foundation and the Yunnan Human Resources and Social Security Department Foundation for funding her postdoctoral research. Samantha C. Karunarathna thanks CAS President’s International Fellowship Initiative (PIFI) for funding his postdoctoral research (number 2018PC0006) and the National Science Foundation of China (NSFC) for funding this research work under project code 31750110478. Itthayakorn Promputtha is grateful to Chiang Mai University for partial support of this research. Austin Smith at World Agroforestry (ICRAF), Kunming Institute of Botany, China, is thanked for English editing.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Saccardo P. Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novis vel criticis, systemate carpologico dispositorum. Atti Soc. Veneziana-Trent.-Istriana Sci. Nat. 1875;4:77–100. [Google Scholar]
- 2.Dai D.Q., Jiang H.B., Tang L.Z., Bhat D.J. Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere. 2016;7:1332–1345. doi: 10.5943/mycosphere/7/9/7. [DOI] [Google Scholar]
- 3.Dai D.Q., Phookamsak R., Wijayawardene N.N., Li W.J., Bhat D.J., Xu J.C., Taylor J.E., Hyde K.D., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2017;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 4.Pintos Á., Alvarado P. Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Syst. Evol. 2020;4:197–221. doi: 10.3114/fuse.2021.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunze G. Zehn neue Pilzgattungen. Mykol. Hefte. 1817;1:1–18. [Google Scholar]
- 6.Hyde K., Fröhlich J., Taylor J. Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia. 1998;50:21–80. [Google Scholar]
- 7.Hyde K., Norphanphoun C., Maharachchikumbura S., Bhat D., Jones E., Bundhun D., Chen Y., Bao D., Boonmee S., Calabon M. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 8.Ramos H.P., Braun G.H., Pupo M.T., Said S. Antimicrobial activity from endophytic fungi Arthrinium state of Apiospora montagnei Sacc. and Papulaspora immersa. Braz. Arch. Biol. Technol. 2010;53:629–632. doi: 10.1590/S1516-89132010000300017. [DOI] [Google Scholar]
- 9.Wang M., Tan X.M., Liu F., Cai L. Eight new Arthrinium species from China. MycoKeys. 2018;1:1–24. doi: 10.3897/mycokeys.39.27014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuels G., McKenzie E., Buchanan D.E. Ascomycetes of New Zealand 3. Two new species of Apiospora and their Arthrinium anamorphs on bamboo. N. Z. J. Bot. 1981;19:137–149. doi: 10.1080/0028825X.1981.10425113. [DOI] [Google Scholar]
- 11.Yin C., Luo F., Zhang H., Fang X., Zhu T., Li S. First Report of Arthrinium kogelbergense Causing Blight Disease of Bambusa intermedia in Sichuan Province, China. Plant Dis. 2021;105:214. doi: 10.1094/PDIS-06-20-1159-PDN. [DOI] [Google Scholar]
- 12.Zhao Y., Deng C., Chen X. Arthrinium phaeospermum causing dermatomycosis, a new record of China. Acta Mycol. Sin. 1990;9:232–235. [Google Scholar]
- 13.Piccolo S.L., Mondello V., Giambra S., Conigliaro G., Torta L., Burruano S. Arthrinium phaeospermum, Phoma cladoniicola and Ulocladium consortiale, New Olive Pathogens in Italy. J. Phytopathol. 2014;162:258–263. doi: 10.1111/jph.12179. [DOI] [Google Scholar]
- 14.LI S.j., ZHU T.h. Binding site of toxic protein from Arthrinium phaeospermum on plasmalemma of hybrid bamboo. J. Zhejiang Univ. (Agric. Life Sci.) 2012;38:355–361. [Google Scholar]
- 15.Ji Z.L., Zhang S., Zhu F., Wan B., Liang R. First Report of Arthrinium arundinis Causing Leaf Edge Spot of Peach in China. Plant Dis. 2020;104:3077. doi: 10.1094/PDIS-12-19-2666-PDN. [DOI] [Google Scholar]
- 16.Chen K., Wu X.Q., Huang M.X., Han Y.Y. First report of brown culm streak of Phyllostachys praecox caused by Arthrinium arundinis in Nanjing, China. Plant Dis. 2014;98:1274. doi: 10.1094/PDIS-02-14-0165-PDN. [DOI] [PubMed] [Google Scholar]
- 17.Bagherabadi S., Zafari D., Anvar F.G. First report of leaf spot caused by Arthrinium arundinis on rosemary in Iran. J. Plant Pathol. 2014;96:4–126. [Google Scholar]
- 18.Tang X., Goonasekara I.D., Jayawardena R.S., Jiang H.B., Li J.F., Hyde K.D., Kang J.C. Arthrinium bambusicola (Fungi, Sordariomycetes), a new species from Schizostachyum brachycladum in northern Thailand. Biodivers. Data J. 2020;8:e58755. doi: 10.3897/BDJ.8.e58755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crous P.W., Wingfield M.J., Le Roux J.J., Richardson D.M., Strasberg D., Shivas R.G., Alvarado P., Edwards J., Moreno G., Sharma R. Fungal Planet description sheets: 371–399. Pers. Mol. Phylogeny Evol. Fungi. 2015;35:264. doi: 10.3767/003158515X690269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma R., Kulkarni G., Sonawane M.S., Shouche Y.S. A new endophytic species of Arthrinium (Apiosporaceae) from Jatropha podagrica. Mycoscience. 2014;55:118–123. doi: 10.1016/j.myc.2013.06.004. [DOI] [Google Scholar]
- 21.Elissawy A.M., Ebada S.S., Ashour M.L., Özkaya F.C., Ebrahim W., Singab A.B., Proksch P. Spiroarthrinols A and B, two novel meroterpenoids isolated from the sponge-derived fungus Arthrinium sp. Phytochem. Lett. 2017;20:246–251. doi: 10.1016/j.phytol.2017.05.008. [DOI] [Google Scholar]
- 22.Goodenough A.E., Stallwood B., Dandy S., Nicholson T.E., Stubbs H., Coker D.G. Like mother like nest: Similarity in microbial communities of adult female Pied Flycatchers and their nests. J. Ornithol. 2017;158:233–244. doi: 10.1007/s10336-016-1371-1. [DOI] [Google Scholar]
- 23.Wang H., Umeokoli B.O., Eze P., Heering C., Janiak C., Müller W.E., Orfali R.S., Hartmann R., Dai H., Lin W. Secondary metabolites of the lichen-associated fungus Apiospora montagnei. Tetrahedron Lett. 2017;58:1702–1705. doi: 10.1016/j.tetlet.2017.03.052. [DOI] [Google Scholar]
- 24.Ellis M.B. Dematiaceous Hyphomycetes: IV. Mycol. Pap. 1963;29:1–33. [Google Scholar]
- 25.Pintos Á., Alvarado P., Planas J., Jarling R. Six new species of Arthrinium from Europe and notes about A. caricicola and other species found in Carex spp. hosts. MycoKeys. 2019;49:15. doi: 10.3897/mycokeys.49.32115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Index Fungorum (2021) [(accessed on 12 August 2021)]. Available online: http://www.indexfungorum.org/Names/Names.asp.
- 27.Senanayake I., Rathnayaka A., Marasinghe D., Calabon M., Gentekaki E., Lee H., Hurdeal V., Pem D., Dissanayake L., Wijesinghe S. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere. 2020;11:2678–2754. doi: 10.5943/mycosphere/11/1/20. [DOI] [Google Scholar]
- 28.Jayasiri S.C., Hyde K.D., Ariyawansa H.A., Bhat J., Buyck B., Cai L., Dai Y.C., Abd-Elsalam K.A., Ertz D., Hidayat I. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015;74:3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- 29.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Volume 18. Academic Press; Cambridge, MA, USA: 1990. p. 7. [Google Scholar]
- 30.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- 32.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senanayake I.C., Bhat J.D., Cheewangkoon R., Xie N. Bambusicolous Arthrinium Species in Guangdong Province, China. Front. Microbiol. 2020;11:602773. doi: 10.3389/fmicb.2020.602773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerin D., Nigro F., Faretra F., Pollastro S. Identification of Arthrinium marii as Causal Agent of Olive Tree Dieback in Apulia (Southern Italy) Plant Dis. 2020;104:694–701. doi: 10.1094/PDIS-03-19-0569-RE. [DOI] [PubMed] [Google Scholar]
- 36.Yuan H.S., Lu X., Dai Y.C., Hyde K.D., Kan Y.H., Kušan I., He S.H., Liu N.G., Sarma V.V., Zhao C.L. Fungal diversity notes 1277–1386: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2020;104:1–266. doi: 10.1007/s13225-020-00461-7. [DOI] [Google Scholar]
- 37.Das K., Lee S.Y., Choi H.W., Eom A.H., Cho Y.J., Jung H.Y. Taxonomy of Arthrinium minutisporum sp. nov., Pezicula neosporulosa, and Acrocalymma pterocarpi: New Records from Soil in Korea. Mycobiology. 2020;48:450–463. doi: 10.1080/12298093.2020.1830741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.JIANG N., TIAN C.-M. The holomorph of Arthrinium setariae sp. nov.(Apiosporaceae, Xylariales) from China. Phytotaxa. 2021;483:149–159. doi: 10.11646/phytotaxa.483.2.7. [DOI] [Google Scholar]
- 39.Chen T.Z., Zhang Y., Ming X.B., Zhang Q., Long H., Hyde K.D., Li Y., Wang Y. Morphological and phylogenetic resolution of Arthrinium from medicinal plants in Yunnan, including A. cordylines and A. pseudomarii spp. nov. Mycotaxon. 2021;136:183–199. doi: 10.5248/136.183. [DOI] [Google Scholar]
- 40.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; Proceedings of the Nucleic Acids Symposium Series; London, UK. 2–6 September 1999; pp. 95–98. [Google Scholar]
- 42.Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 43.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 44.Miller M., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the Gateway Computing Environments Workshop; New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 45.Nylander J.A.A. MrModeltest v2 Program Distributed by the Autho. Evol. Biol. Cent. Upps. Univ. Upps. Sweden. 2004 [Google Scholar]
- 46.Rambaut A. FigTree v1.4: Tree Figure Drawing Tool. [(accessed on 12 August 2021)]. Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 47.Singh S.M., Yadav L.S., Singh P.N., Hepat R., Sharma R., Singh S.K. Arthrinium rasikravindrii sp. nov. from Svalbard, Norway. Mycotaxon. 2013;122:449–460. doi: 10.5248/122.449. [DOI] [Google Scholar]
- 48.Rana S., Singh P.N., Gaikwad S.B., Singh S.K. Morphology, phylogeny and ex situ conservation of Arthrinium rasikravindrae (Apiosporaceae: Xylariales): A new record from India. Kavaka. 2017;49:1–5. [Google Scholar]
- 49.Feng Y., Liu J.K.J., Lin C.G., Chen Y.Y., Xiang M.M., Liu Z.Y. Additions to the genus Arthrinium (Apiosporaceae) from bamboos in China. Front. Microbiol. 2021;12:661281. doi: 10.3389/fmicb.2021.661281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan H., Jiang N., Liang L.Y., Yang Q., Tian C.M. Arthrinium trachycarpum sp. nov. from Trachycarpus fortunei in China. Phytotaxa. 2019;400:203–210. doi: 10.11646/phytotaxa.400.3.7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.