Abstract
We compared two commercial molecular assays (the Murex Hybrid Capture CMV DNA assay [HCA], version 2, and the Roche Amplicor plasma PCR assay) with a standard shell vial assay in detecting and predicting cytomegalovirus (CMV) disease in a group of renal transplant patients and assessed the role of viral load measurements (using the HCA) in their management. The sensitivity of the HCA and Amplicor assay in terms of disease detection was 100%, compared to 71% for the shell vial assay. Both the HCA and the PCR assay detected all cases of disease, at medians of 11 and 12.5 days before the onset of symptoms, respectively. Significantly higher viral loads were detected in those patients with symptoms (7.9 × 105 copies/ml) than in patients without symptoms (7.9 × 104 copies/ml; P < 0.0001). There was also a trend towards higher viral loads in those patients with primary infections (7.8 × 105 copies/ml) than in those patients with reactivations of CMV disease or reinfections. Successful treatment with ganciclovir was associated with a >90% reduction in viral load. Both of these new assays are sensitive and easy to use. A comparison of accurate quantitation is also useful in monitoring responses to antiviral therapy.
Infection with cytomegalovirus (CMV) is a significant cause of morbidity in renal transplant recipients, with up to 60% of primary infections (i.e., in a seronegative recipient of a seropositive organ) resulting in symptomatic CMV disease (12). With the development of effective antiviral agents, such as ganciclovir, the emphasis has switched to the early identification and treatment of those patients at greatest risk of disease. Effective preemptive therapy following the detection of CMV in the blood is dependent on the development of laboratory assays which are sensitive, specific, and rapid so that all patients at risk of disease are identified quickly and the number of patients treated inappropriately (i.e., who were not destined to develop disease) is kept to a minimum.
The rapid detection of CMV was first achieved with the shell vial assay (SVA). While this assay was an improvement on standard culture techniques, a significant proportion of patients with CMV viremia were still missed (11). The newer techniques, such as PCR (with leukocytes, plasma, or whole blood), are more sensitive (1, 2, 6, 10); however, as a variety of in-house methods and sample types are used, comparison of results between centers is difficult (3). The detection of CMV pp65 antigen in polymorphonuclear leukocytes is another, more sensitive, alternative to the SVA. However, the rapid processing of samples and the subjective interpretation of fluorescence in the cytospin preparations make this test unsuitable for use in some laboratories (7). A recently developed DNA hybridization test, the hybrid capture CMV DNA assay (HCA), offers the advantage of objective CMV DNA detection and quantification without the hazards of PCR, such as contamination or inhibition.
Quantitative techniques based on PCR- and DNA-based assays have demonstrated a relationship between the quantity of virus (viral load) and the likelihood of CMV disease (2, 5) and can also be used to monitor responses to therapy (9).
In this study, we compared the performance of the HCA (version 2; Murex Ltd., Dartford, United Kingdom) and the Amplicor CMV DNA plasma PCR assay (Roche Diagnostic Systems, Branchburg, N.J.) with a 24-h SVA in detecting and predicting CMV disease in a group of adult renal transplant patients. The clinical significance of CMV viral load measurements and the changes which occurred following the introduction of anti-CMV therapy were also assessed with the HCA results.
MATERIALS AND METHODS
Patient population.
Between January and November 1997, 52 adult (32 male and 20 female) patients underwent renal transplantation and were included in the study. Paired heparinized (10-ml) and EDTA-anticoagulated (2 × 4.5-ml) blood samples were collected at weekly intervals for 12 weeks, starting the week after transplantation. The heparinized sample was used for the SVA; the SVA results alone were used in patient management. Both the HCA and the plasma PCR assay were performed retrospectively; plasma from one EDTA sample was used for the PCR assay and the leukocyte fraction from the other sample was used in the HCA (storage details are described below). Additional samples were taken if CMV was suspected and previous SVA results had been negative.
Immunosuppression with azathioprine (1.5 mg/kg of body weight daily), cyclosporine (5 mg/kg twice daily for 3 days, reduced to 4 mg/kg twice daily according to levels in serum), and prednisolone (20 mg/kg daily for the first month, reduced to 10 mg/kg daily by week 12) was started immediately posttransplantation. Rejection was treated with either methylprednisolone (0.5 g daily for 3 days) or anti-thymocyte globulin (2.5 mg/kg/day for 10 days) if rejection symptoms were severe. Episodes of CMV disease were treated with ganciclovir (10 mg/kg daily or less depending on renal function) for 10 to 14 days.
Pretransplant sera from both recipients and donors were tested for CMV antibodies (total) by the Serodia passive particle agglutination assay (Fujero, Tokyo, Japan).
Definitions.
The diagnosis of CMV disease was based on the criteria defined at the International CMV Workshop (8), in which the detection of CMV viremia (using the SVA) is associated with one or more of the following symptoms and signs: (i) pyrexia with or without general malaise and with or without leukopenia (defined as fever of ≥38°C for ≥48 h in the absence of transplant rejection or intercurrent infection), (ii) pneumonitis (defined as symptoms of hypoxia with or without interstitial changes on chest X ray and the detection of CMV in bronchoalveolar lavage fluid), and (iii) hepatitis (defined as a alanine aminotransferase level twice the upper limit of normal).
SVA.
The polymorphonuclear leukocyte fraction was inoculated onto a confluent human fibroblast monolayer (human embryonic cells) for up to 24 h before being stained with a fluorescein isothiocyanate-labelled monoclonal antibody directed against the 72-kDa immediate-early protein of CMV. Infected cells exhibited apple-green kidney-shaped fluorescence.
HCA.
The leukocyte fraction was separated and stored at −20°C for future use, as described in the manufacturer’s protocol. Briefly, this involved two rounds of selective erythrocyte lysis with lysis buffer, followed by centrifugation and pelleting of the leukocyte fraction. Following cell lysis and denaturation of the leukocyte pellet, the CMV DNA was hybridized to a CMV-specific RNA probe; the resultant RNA-DNA hybrids were then immobilized onto capture tubes and detected following the addition of alkaline phosphatase-conjugated antibodies. Following the addition of a chemiluminescent substrate, light was emitted and measured on a luminometer as relative light units. The intensity of the light was proportional to the quantity of CMV DNA present in the original sample. Positive samples were defined as those giving twice the mean value of the negative control. The quantity of CMV DNA, expressed as either picograms per milliliter or genome copies per milliliter, was determined by reference to the three positive standards in the assay.
Amplicor CMV PCR test.
All plasma samples were separated and stored at −70°C until the assay was performed. The manufacturer’s instructions were followed for all stages of the process, including sample preparation, amplification, and detection. The risk of contamination was reduced by the inclusion of Amperase (uracyl-N-glycosylase) in the master mixture and the substitution of dUTP for dTTP. Briefly, DNA was extracted from the patients’ plasma samples following a 30-min incubation at 100°C in the extraction reagent. Amplification, after the addition of the master mixture reagents, consisted of 40 cycles of denaturation (94°C, 30 s), annealing (65°C, 30 s), and extension (72°C, 1 min). The denatured amplicons were then transferred to a microwell plate and incubated at 37°C for 1.5 h in the presence of hybridization buffer. Following the sequential addition of avidin-horseradish peroxidase conjugate and substrate to each well, the reaction was stopped and the optical density at 450 nm was measured. Values of ≥0.25 indicate the presence of CMV DNA in the original sample.
Statistical methods.
The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for each of the assays with respect to the development of CMV disease. Differences between peak viral loads in patients with and without symptoms and differences among the three donor-recipient subgroups were assessed with the Mann-Whitney U test and the Kruskal-Wallis test, respectively. Using logistic regression, the probability of developing disease at a given viral load was also determined. A receiver operator curve (ROC) analysis was used on the HCA quantitative results to further analyze the usefulness of viral load measurements. By defining an optimum viral load cutoff level, those patients at greatest risk of developing CMV disease could be identified.
RESULTS
Clinical features.
A total of 14 of the 52 patients (27%) developed signs of CMV disease (8, persistent spiking fever; 4, pneumonitis; and 2, esophagitis), and 13 of these patients were treated with ganciclovir. The fever in the remaining patient resolved before ganciclovir was started. Of those patients with CMV disease, 6 had primary infections (seropositive donor organ and seronegative recipient [D+/R−]), 3 had reactivations (D−/R+), and 5 had either reactivations or reinfections (D+/R+). All 14 patients had CMV disease detected by the HCA or the PCR assay, whereas only 10 patients had samples which were positive with the SVA. The ability to predict CMV disease was based on samples from 13 patients who had a complete set of weekly samples taken immediately before the onset of symptoms. Of the 38 patients who remained symptom free, 28, 13, and 20 had negative results for all sequential samples tested by the SVA, HCA, and PCR assay, respectively.
CMV was first detected by the SVA at a median of 6 weeks (range, 2 to 11 weeks) posttransplant, compared to a median of 5 weeks (range, 2 to 10 weeks) for the HCA or PCR assay (range, 1 to 10 weeks). The interval from transplantation to the earliest detection of CMV (viremia or DNAemia) was significantly lower with all the assays for those patients with symptoms than for those who remained symptom free: SVA, 5 and 7 weeks, respectively (P = 0.027); HCA, 4 and 6 weeks (P = 0.001); and PCR assay, 4 and 6 weeks (P = 0.046).
Comparison of the assays.
Four hundred twenty-seven samples were tested by all three assays. The sensitivity, specificity, PPV, and NPV for detection of CMV disease by each of the assays are shown in Table 1. Both the HCA and the PCR assay were superior to the SVA and were able to detect all the cases of CMV disease. Sequential samples were consistently found to be positive during an episode of viremia over successive weeks when the HCA (median number of positives, 6; range, 2 to 10) or the PCR assay (median, 4; range, 1 to 10) was used, whereas with the SVA a positive result usually occurred in isolation. The median times, in days, between a positive result and the onset of symptoms were 1.5 (range, −30 to +10) for the SVA, 11 (range, −51 to 0) for the PCR assay, and 12.5 (range, −51 to 0) for the HCA.
TABLE 1.
Comparison of two new commercial assays with the SVA in the detection of CMV disease
| Assay and cutoff | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| SVA | 0.71 | 0.74 | 0.50 | 0.88 |
| PCR | 1.0 | 0.52 | 0.44 | 1.0 |
| HCAa | 1.0 | 0.34 | 0.36 | 1.0 |
| HCA, ≥7.9 × 104 c/mlb | 0.93 | 0.68 | 0.52 | 0.96 |
| HCA, ≥1.3 × 105 c/mlb | 0.93 | 0.84 | 0.68 | 0.97 |
| HCA, ≥2.0 × 105 c/mlb | 0.79 | 0.87 | 0.69 | 0.92 |
Qualitative results (i.e., positive or negative).
Quantitative results. The optimum cutoff viral load (determined by ROC analysis) is given. c/ml, copies per milliliter.
The specificities of the three tests (SVA, PCR assay, and HCA) were 0.74, 0.52, and 0.34, respectively; the PPVs were 0.5, 0.44, and 0.36. By applying a ROC analysis to the quantitative values of the HCA, an optimum cutoff viral load of 1.3 × 105 copies/ml was determined. The sensitivity and specificity at this level were 0.93 and 0.84, respectively.
Role of CMV viral load.
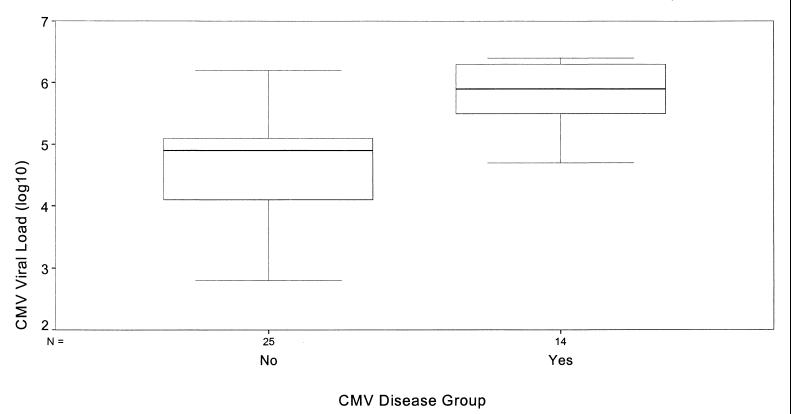
For the 39 patients with positive HCA results, the relationship between viral load and the presence or absence of posttransplant CMV disease is shown in Fig. 1. Thirteen patients had undetectable CMV loads; all of these remained symptom free. The median peak viral load in the blood of symptomatic patients was 7.9 × 105 copies/ml (range, 5.0 × 104 to 2.5 × 106), which was significantly higher than that observed in the asymptomatic group (median, 7.9 × 104 copies/ml; range, 6.2 × 102 to 1.6 × 106; P < 0.0001). There was no significant difference in the median peak viral loads in the three donor-recipient subgroups (P = 0.15). There was a trend toward higher viral loads in those patients with primary infections (D+/R−), with a median peak viral load of 7.8 × 105 copies/ml (range, 4.8 × 104 to 2.2 × 106), followed by those in the D+/R+ group, with 2.0 × 105 copies/ml (range, 1.6 × 103 to 2.3 × 106). The lowest median viral load was in those patients experiencing reactivations of latent virus (D−/R+), with 8.7 × 104 copies/ml (range, 1 × 103 to 1.4 × 106).
FIG. 1.
Box-and-whisker plot to show the relationship between virus load and CMV disease. Yes, 14 patients with CMV disease; No, 25 patients without CMV disease.
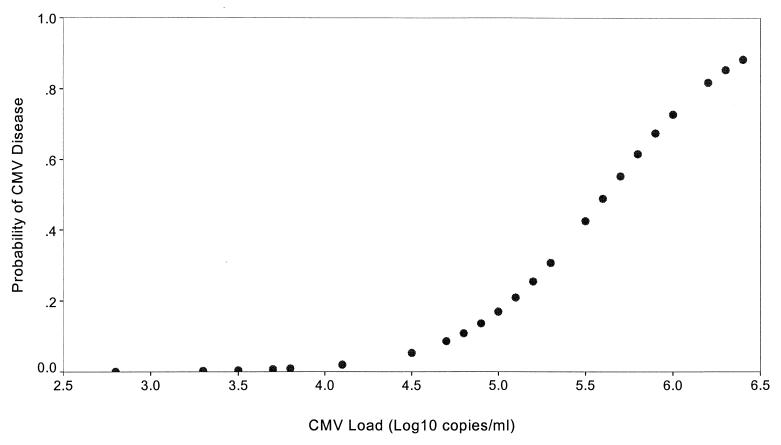
Using logistic regression analysis, we identified viral load as a significant risk factor for CMV disease (P < 0.002). The logistic regression coefficient for viral load was 2.57 (standard error, 0.83), indicating an increased-odds ratio of 13.0 (95% confidence interval, 2.56 to 66.7) for each log10 increase in viral load. For example, the risk of disease increased from 1.5% at a virus load of 104.0 copies/ml to 73% at 106.0 copies/ml (Fig. 2).
FIG. 2.
Probability of acquiring CMV disease with increasing viral load.
Effect of ganciclovir therapy.
The response to ganciclovir was monitored in seven patients. The remaining six patients were not included, either because of death within 1 week of starting therapy (two patients) or because there were insufficient samples taken (four patients). In four of the seven patients, a clinical response was mirrored by a >90% reduction in viral load within 14 days of starting ganciclovir. Of the remaining three patients, two developed recurrent CMV disease. In both cases the reduction in viral load was less than 90% from baseline: 66% (from 2.2 × 106 to 7.4 × 105 copies/ml) for one and 84% (from 5.8 × 105 to 8.9 × 104 copies/ml) for the other. In the remaining patient, disseminated herpes zoster developed 2 days after ganciclovir was started for CMV disease. By day 14, the CMV load had fallen from 1.4 × 106 to 7.9 × 105 copies/ml (a 44% reduction).
DISCUSSION
Although the mortality from established CMV disease following renal transplantation has fallen since the introduction of ganciclovir, the virus is still associated with significant morbidity (12). While preemptive therapy reduces the incidence and severity of CMV disease, it is dependent on early laboratory identification of those at greatest risk of disease.
To date, the newer techniques, such as PCR and the pp65 assay, have either relied on in-house reagents or required subjective interpretation of immunofluorescence, and there has been little agreement as to which is the most appropriate test. With the development of commercial formats of these assays, some of these problems may now be addressed. We compared CMV disease detection by two commercial assays and an SVA and assessed the role of viral load in the management of CMV disease.
Both the HCA and the plasma PCR assay were highly sensitive, detecting CMV disease in all 14 patients tested, whereas the SVA remained negative for 4 patients with CMV disease. Although the calculated sensitivity of the SVA was more than 70%, the test was positive only once during an episode of CMV viremia. Subsequent samples were either negative or toxic. The molecular methods, by contrast, were repeatedly positive with sequential samples. In a busy diagnostic laboratory, the confirmation of positive results is required and is usually achieved by testing another sample. This is particularly important in the absence of clinical symptoms. Therefore, despite a positive SVA result being a strong predictor of CMV disease (especially in the D+/R− subgroup), an isolated positive result may be difficult to interpret, especially when the assay is dependent on the subjective interpretation of immunofluorescence and other variables, such as the quality of the cell culture used to prepare the shell vials.
As has been shown in other studies of CMV infection following renal transplantation (5, 9, 13), we observed significantly higher levels of CMV DNA (with the HCA) in those patients with CMV disease than in those with asymptomatic infections. Within the three donor-recipient subgroups, there was a trend towards higher viral loads in those patients with primary infections (D+/R−) than in those with either reactivations or reinfections (D+/R+) or reactivations (D−/R+).
Using logistic regression analysis, we have shown that the risk of disease increases as the viral load increases (e.g., a 50% probability of disease was reached at a virus load of 4.0 × 105 copies/ml and an 80% risk was reached at 1.6 × 106 copies/ml). By using a ROC analysis, we were able to determine a cutoff CMV viral load of 1.3 × 105 copies/ml (Table 1), so only those values greater than or equal to this value are regarded as clinically significant (i.e., highly predictive of CMV disease). This results in sensitivity and specificity values of 0.93 and 0.84, respectively. Identification of a cutoff level would therefore reduce the number of patients treated with preemptive therapy who were not destined to develop CMV disease; however, the selection of such a cutoff value needs confirmation by testing a larger number of patients.
These newer techniques allow the earlier introduction of preemptive therapy, which should reduce the number of patients developing significant CMV disease. In view of the toxic side effects of the currently available anti-CMV agents, preemptive therapy should be aimed at those patients at greatest risk of developing disease. Our preliminary results suggest that the accurate measurement of viral load may help to define further the group at greatest risk.
The detection of CMV DNA in plasma suggests active viral replication with spread of the virus from the leukocytes into the plasma fraction (14). Despite this, the specificity of the plasma PCR was still only 50%. Our HCA data suggest that accurate quantification may further define those at greatest risk of developing disease. We are currently evaluating a quantitative format of the Roche PCR assay in patients with CMV DNA detectable in plasma.
Clinical response to ganciclovir is associated with a reduction in viral load (1), and it is possible that viral load may be used to guide the duration of therapy. Although we were able to include only seven treated patients in this study, our preliminary results suggest that effective treatment should be continued until the viral load is reduced by at least 90%. In most patients, such a reduction should occur after 10 to 14 days; however, some may require a longer course of treatment to prevent recurrent disease.
In summary, the two commercial assays are superior to the SVA. Both are objective methods of detection not dependent on the subjective interpretation of immunofluorescence (as with SVA or the antigenemia assay) or the local preparation of reagents (as with in-house PCR). The assays were equivalent in terms of disease detection, but the plasma PCR assay was easier to perform. The HCA offered the advantage of being an easy method of measuring the viral load, allowing the accurate prediction of disease risk and monitoring of response to therapy.
REFERENCES
- 1.Cope A V, Sabin C, Burroughs K, Rolles K, Griffiths P D, Emery V C. Inter-relationships among quantity of human cytomegalovirus (HCMV) DNA in blood, donor-recipient serostatus, and administration of methylprednisolone as risk factors for HCMV disease following liver transplantation. J Infect Dis. 1997;176:1484–1490. doi: 10.1086/514145. [DOI] [PubMed] [Google Scholar]
- 2.Fox J C, Kidd I M, Griffiths P D, Sweny P, Emery V C. Longitudinal analysis of cytomegalovirus load in renal transplant recipients using a quantitative polymerase chain reaction: correlation with disease. J Gen Virol. 1995;76:309–319. doi: 10.1099/0022-1317-76-2-309. [DOI] [PubMed] [Google Scholar]
- 3.Grundy J E, Ehrnst A, Einsele H, Emery V C, Hebart H, Prentice H G, Ljungman P. A three-center European external quality control study of PCR for detection of cytomegalovirus DNA in blood. J Clin Microbiol. 1996;34:1166–1170. doi: 10.1128/jcm.34.5.1166-1170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho M. Advances in understanding cytomegalovirus infection after transplantation. Transplant Proc. 1994;26:7. [PubMed] [Google Scholar]
- 5.Imbert-Marcille B M, Cantarovich D, Ferre-Aubineau V, Richet B, Soulilou J P, Billaudel S. Usefulness of DNA viral load quantification for cytomegalovirus disease monitoring in renal and pancreas/renal transplant recipients. Transplantation. 1997;63:1476–1481. doi: 10.1097/00007890-199705270-00018. [DOI] [PubMed] [Google Scholar]
- 6.Jiwa N M, Van Gemert G, Raap A, et al. Rapid detection of human cytomegalovirus DNA in peripheral leucocytes of viraemic transplant recipients by the polymerase chain reaction. Transplantation. 1989;48:72–76. doi: 10.1097/00007890-198907000-00017. [DOI] [PubMed] [Google Scholar]
- 7.Landry M L, Ferguson D, Cohen S, Huber K, Wetherill P. Effect of delayed specimen processing on cytomegalovirus antigenemia test results. J Clin Microbiol. 1995;33:257–259. doi: 10.1128/jcm.33.1.257-259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ljungman P, Plotkin S. Workshop on CMV disease—definitions, clinical severity scores and new syndromes. Scand J Infect Dis Suppl. 1995;99:87–89. [Google Scholar]
- 9.Macartney M, Gane E J, Portman B, Williams R. Comparison of a new quantitative cytomegalovirus DNA assay with other detection methods. Transplantation. 1997;63:1803–1806. doi: 10.1097/00007890-199706270-00017. [DOI] [PubMed] [Google Scholar]
- 10.Patel R, Smith T F, Espy M, Wiesner R H, Kron R A F, Portela D, Paya C V. Detection of cytomegalovirus DNA in sera of liver transplant recipients. J Clin Microbiol. 1994;32:1431–1434. doi: 10.1128/jcm.32.6.1431-1434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillay D, Charman H, Lok K, Griffiths P D. Detection of cytomegalovirus by a rapid culture system: a comparison of monoclonal antibodies in a clinical setting. J Virol Methods. 1992;40:219–224. doi: 10.1016/0166-0934(92)90070-t. [DOI] [PubMed] [Google Scholar]
- 12.Smiley L, Wlodaver C, Grossman R, et al. The role of pre-transplant immunity in protection from cytomegalovirus disease following renal transplantation. Transplantation. 1985;40:157. doi: 10.1097/00007890-198508000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Van den Berg A P, Van der Bij W, Van Son W J, et al. Cytomegalovirus antigenaemia as a useful marker of symptomatic cytomegalovirus infection after renal transplant—a report of 130 consecutive patients. Transplantation. 1989;48:991–995. doi: 10.1097/00007890-198912000-00019. [DOI] [PubMed] [Google Scholar]
- 14.Wolf D, Spector S. Early diagnosis of human CMV disease in transplant recipients by DNA amplification in plasma. Transplantation. 1993;56:330–334. doi: 10.1097/00007890-199308000-00014. [DOI] [PubMed] [Google Scholar]