Abstract
Lactic acid bacteria (LAB) are widely used as probiotics in the food industry owing to their beneficial effects on human health. However, numerous antibiotic resistance genes have been found in LAB strains, especially tetracycline resistance genes. Notably, the potential transferability of these genes poses safety risks. To comprehensively evaluate tetracycline resistance in LAB, we determined the tetracycline susceptibility patterns of 478 LAB strains belonging to four genera and eight species. By comparing phenotypes with genotypes based on genome-wide annotations, five tetracycline resistance genes, tet(M), tet(W/N/W), tet(L), tet(S), and tet(45), were detected in LAB. Multiple LAB strains without tetracycline resistance genes were found to be resistant to tetracycline at the currently recommended cutoff values. Thus, based on the minimum inhibitory concentrations of tetracycline for these LAB strains, the species-specific microbiological cutoff values for Lactobacillus (para)gasseri, Lactobacillus johnsonii, and Lactobacillus crispatus to tetracycline were first developed using the Turnidge, Kronvall, and eyeball methods. The cutoff values for Lactiplantibacillus plantarum were re-established and could be used to better distinguish susceptible strains from strains with acquired resistance. Finally, we verified that these five genes play a role in tetracycline resistance and found that tet(M) and tet(W/N/W) are the most widely distributed tetracycline resistance genes in LAB.
Keywords: lactic acid bacteria, tetracycline resistance, minimum inhibitory concentration, tetracycline resistance gene, microbiological cutoff value
1. Introduction
Tetracyclines are widely used antibiotics in human medicine and animal husbandry owing to their broad-spectrum antibacterial activity, low production cost, and lack of serious adverse reactions [1]. However, with the extensive and unreasonable use of tetracyclines, bacterial tetracycline resistance has become a serious concern [2], and the acquisition of tetracycline resistance genes has been identified as the main cause of bacterial tetracycline resistance [3]. Most tetracycline resistance genes are dependent of the bacteria. So, usually the most frequent genes for Gram-negative bacteria are tet(A) and tet(B), which are relatively highly distributed [4,5]. Most tetracycline resistance genes are linked to transmissible plasmids, transposons, and conjugative transposons, which can quickly spread among bacteria in humans, animals, and the environment [6,7]. Accordingly, these genes pose a great threat to human and animal health [8].
Lactic acid bacteria (LAB) have been consumed for thousands of years and are “generally recognized as safe” microorganisms [9,10]. Several species of LAB have been granted the qualified presumption of safety (QPS) status [11]. However, in recent years, owing to the improper use of antibiotics (overuse and misuse), many LAB have developed drug resistance [12]. Several studies have revealed that LAB isolated from food harbor a variety of antibiotic resistance genes (ARGs) [13,14], which are located on the mobile genetic elements and have a potential risk of horizontal gene transfer [15,16]. When these strains enter the human intestinal tract with food, they may transmit their resistance genes to the intestinal pathogenic bacteria and opportunistic pathogenic bacteria through the food chain and confer drug resistance to hosts, thereby threatening human health. Some studies have also confirmed that the tetracycline resistance gene in LAB can be transferred to other bacteria through horizontal gene transfer [17]. In 2002, the Food and Agriculture Organization and the World Health Organization proposed that probiotics used for food consumption should be used to evaluate the safety of antibiotic resistance in commercial applications [18]. The European Food Safety Agency (EFSA) document highlighted the need to determine whether there is no acquired or transferable resistance factor in a candidate probiotic or starter culture to declare it safe for human and animal consumption and to obtain QPS status [19]. Moreover, the document proposes that antibiotic susceptibility testing should be conducted according to international standards, such as those of the International Standard Organization (ISO) and Clinical and Laboratory Standards Institute [20].
Antimicrobial susceptibility testing is a traditional method of drug resistance testing. These testing methods include K-B disk diffusion, broth macrodilution, broth microdilution, agar dilution, and E-test [21,22]. These culture-based tests determine the growth of bacteria in the presence of antibiotics and evaluate bacterial resistance based on the drug resistance phenotype [23]. Microbiological cutoff values (MCOFFs) are usually used as the interpretation criteria to identify antibiotic resistance and to differentiate strains with acquired resistance from susceptible strains [24]. However, the cutoff values of some LAB species have not yet been determined. The EFSA guidelines classify these LAB species according to their fermentation type and determine the cutoff values based on their fermentation type [20].
The drug resistance of bacteria is usually determined by sequencing of coding genes. With the breakthrough of genome-wide sequencing technology, researchers have developed a sequence alignment method to identify antimicrobial resistance genes through sequence similarity [25]. By comparing the nucleic acid sequence or protein sequence of a strain with the sequence in the antimicrobial resistance database, the ARGs in the genome of LAB strains can be quickly identified and characterized to evaluate their drug resistance and risk of transfer [26]. Furthermore, the transformation of drug resistance evaluation from phenotype-based to genotype-based was promoted by such comparisons.
This study aimed to determine the tetracycline sensitivity of eight species of LAB from different geographical locations and different sources and to establish a new sensitive-resistance cutoff value at the species level to distinguish sensitive strains without resistance genes from strains that have acquired resistance, using the sensitivity analysis and genotype association results. The epidemiology and species distribution of tetracycline resistance genes in LAB were determined through genotype-phenotype association analysis.
2. Materials and Methods
2.1. Strains and Cultural Conditions
Details of the 478 strains belonging to Lacticaseibacillus paracasei (n = 116), Lacticaseibacillus rhamnosus (n = 68), Limosilactobacillus reuteri (n = 47), Lactiplantibacillus plantarum (n = 99), Lactobacillus (para)gasseri (n = 100), Lactobacillus johnsonii (n = 18), or Lactobacillus crispatus (n = 30) are presented in Table S1. All strains were identified at the species level based on 16S rRNA sequencing and were held in cryotubes with 15%–30% (w/v) glycerol and deposited in the Culture Collection of Food Microorganisms (CCFM) of Jiangnan University. All LAB strains were grown in de Man, Rogosa, and Sharpe (MRS) liquid medium at 37 °C (L. plantarum at 28 °C) for 16–24 h. Before susceptibility testing, all strains were propagated for three generations under the specified culture conditions, as mentioned above.
2.2. Antibiotic Susceptibility Testing
The minimum inhibitory concentration (MIC) was determined by the microdilution broth method using hand-made 96-well plates, and the specific operation followed the international standard method ISO 10932 (IDF 223:2010) [27]. Briefly, 100 μL of serial two-fold dilutions of tetracycline were distributed into each well of the 96-well plates. The bacterial suspensions were diluted until the optical density (OD) was between 0.16 and 0.2 at 625 nm (Shimadzu UV-1800 spectrophotometer, Kyoto, Japan), with a corresponding concentration of 3 × 108 CFU/mL. The suspensions were diluted again 1000 times, and then 100 μL was added to each well of the 96-well plates. The 96-well plates were incubated under anaerobic conditions at 37 °C (L. plantarum incubated at 28 °C) for 48 h. Tetracycline was purchased from Sangon Biotech (Shanghai, China). The MIC for each LAB strain was defined as the lowest antibiotic concentration without visible growth. MICs were measured in triplicate. Lacticaseibacillus paracasei ATCC 334 and L. plantarum ATCC 14917 served as quality control strains. The interpretation criteria used to differentiate wild-type strains from non-wild-type strains were defined as the MCOFFs by EFSA [20,28].
2.3. Identification of Tetracycline Resistance Genes
The genome sequences of the tested strains were aligned with the Comprehensive Antibiotic Resistance Database (CARD, http://arpcard.Mcmaster.ca (accessed on 22 August 2019)) through the Resistance Gene Identifier (RGI, version 5.1.0) to identify all known ARGs [29]. A gene was recognized as a putative ARG if the identity value at the amino acid level was not lower than 70%. Putative ARGs related to phenotypic resistance were plotted as a heatmap using TBtools v1.09852 [30]. The results of protein sequence alignment were visualized using ESPript 3.0.
2.4. Statistical Analysis and Determination of Tentative Microbiological Cutoff Values (TMCOFFs)
The TMCOFFs were determined using two classical statistical methods, as described by Turnidge et al. [31] and Kronvall [32]. The MIC cumulative frequency distribution table was obtained by statistical analysis of the MIC values. The MIC frequency distribution data were imported into ECOFFinder_XL_2010_v2.0 and Automatic_NRI-MIC_Win_V01beta data tables, respectively, according to the corresponding method instructions. The cutoff values containing 99% wild-type strains were rounded off to the adjacent twofold dilution antibiotic concentration and were defined as TMCOFFs in ECOFFinder Excel sheets. A visual method was also employed to formulate TMCOFFs. The visual method defined TMCOFFs as the MIC at the second twofold dilution concentration higher than the model MIC and that contained at least 95% of wild-type strains [33]. The final TMCOFFs obtained in this study were the median values of the cutoff values obtained from the three methods.
2.5. Sample Collection and RT-PCR
Lactic acid bacteria strains containing tetracycline resistance genes were inoculated into MRS liquid medium and incubated at 37 °C (L. plantarum incubated at 28 °C) to achieve logarithmic growth phase. Tetracycline was added at a final concentration of 1/2 × MIC (Table S1) of tetracycline. The bacterial culture without tetracycline was used as the control, and three independent biological repeats were employed in each group. The bacterial cells was collected by centrifugation at 2500× g for 10 min at 4 °C (Eppendorf 5424R centrifuge, Hamburg, Germany) after continuous culture for 1 h, and three biologically repetitive bacterial cells were mixed for subsequent RNA extraction [34].
Total RNA was extracted using the Bacteria RNA Extraction Kit (R403-01, Vazyme Biotech Co., Ltd., Nanjing, China) according to the manufacturer’s protocol. Three independent biological replicates were employed for each treatment, and the extracted RNA samples were mixed for each treatment. RNA purity and concentration were determined using an ultramicrospectrophotometer (Implen, Munich, Germany). RNA integrity was detected through agarose gel electrophoresis. Total RNA extracted from the bacteria was reverse transcribed into cDNA using HiScript III-RT SuperMix for qPCR (R323-01, Vazyme Biotech Co., Ltd., Nanjing, China) according to the manufacturer’s instructions.
By using the CFX connect real-time qPCR system (Bio-Rad, Hercules, CA, USA), the 16S rRNA gene as an internal reference gene, and the relative quantitative method, expression of the drug resistance gene was determined. All amino acid sequences of the same tetracycline resistance gene detected in this study were compared using the MEGA X v10.2.4 software to find the conserved sites of the sequence. The Primer-BLAST program (http://www.ncbi.nlm.nih.gov/tools/Primer-Blast/) (accessed on 10 October 2020) was used to design primers in the conserved region and check the specificity of primers [35]. The primers were synthesized by Shanghai Sunni Biotechnology Co., Ltd. The primer sequences were as follows: tet(M)-F, TTACTGTATCACCCGCTTCC; tet(M)-R, CAGTCGTCACATTCCAACC [36]; tet(W/N/W)-F, TGGAAAGACGACCTTGACGG; tet (W/N/W)-R, ACATCTGTGCCACTGGAAGG; tet(L)-F, CATTTGGTCTTATTGGATCG; tet (L)-R, ATTACACTTCCGATTTCGG; tet(S)-F, ACGCTATGGGTGTGAACAAGG; tet (S)-R, CAATAGGCGCAAGCATTCGG; 16S rRNA-F, AGAGTTTGATCCTGGCTCAG; 16S rRNA-R, CTACGGCTACCTTGTTACGA [37]; tet(45)-F, ACCTGCGAGTACAAACTGGG; and tet(45)-R, AACCCAATTACCGACCCGAA. The final reaction volume for RT-PCR was 10 μL, and the mixture comprised the following: 2 × iTaqTM Universal SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA), 5 μL; forward primers (500 nM), 0.5 μL; reverse primers (500 nM), 0.5 μL; cDNA (100 ng), 1 μL; and ddH2O, 3 μL. The reaction conditions were 95 °C for 30 s; 95 °C, 5 s; 60 °C, 30 s; and 72 °C, 20 s. In the subsequent three steps, 39 cycles were performed, and the fluorescence was measured at 60 °C. The dissociation curve was generated under the following conditions: 65 °C, 5 s; 95 °C, 0.5 °C. Three biological replicates were set for each sample, and the experiment was repeated three times. A heatmap of the gene expression data was plotted using TBtools v1.09852 [38], and the expression level was log2 transformed [39].
3. Results
3.1. Determination of the MICs and Identification of the Resistance Phenotype
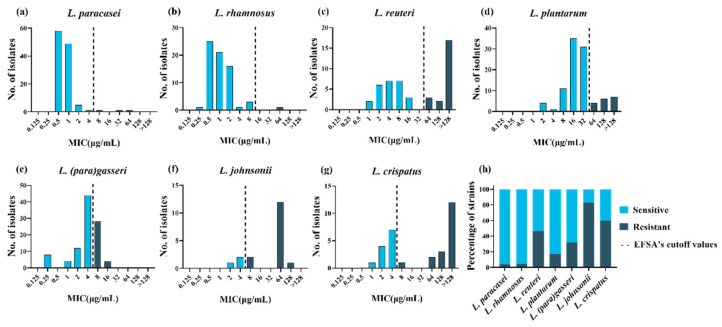
To explore the tolerance level of different species of LAB to tetracycline, the MICs of tetracycline were tested using the broth microdilution method for 478 LAB strains. The MICs of tetracycline for the two quality control strains in this study were within the quality control range, and the MICs for all strains are presented in Table S1. Differences were observed in the intra- and interspecies levels of tetracycline susceptibility. Lactiplantibacillus plantarum and L. reuteri had a wide MIC range that covered 8 twofold dilutions compared to the remaining species. Furthermore, L. paracasei, L. rhamnosus and L. crispatus covered 7 twofold dilutions, while L. johnsonii and L. (para)gasseri covered only 5–6 twofold dilutions (Figure 1a–g). The MICs for L. paracasei and L. rhamnosus were mainly between 0.5 and 2 μg/mL, while those for L. reuteri, L. (para)gasseri, L. johnsonii, and L. crispatus were mainly between 2 and 8 μg/mL. In particular, compared to other species, L. plantarum showed higher MICs, ranging from 8 to 32 μg/mL. The MICs for L. johnsonii, L. crispatus, and L. reuteri showed an obvious bimodal distribution, suggesting that these species may contain acquired tetracycline resistance genes.
Figure 1.
Minimum inhibitory concentration (MIC) distributions and drug resistance rate within 478 lactic acid bacteria strains. (a–g): Distribution of the MICs of tetracycline for eight lactic acid bacterial species. The black dotted lines represent epidemiological cutoff values reported by the European Food Safety Agency (EFSA). Sky blue represents phenotypically sensitive strains, and blue-black represents phenotypically resistant strains. (h) Distribution of tetracycline-resistant (black) and -susceptible (gray) strains according to the EFSA epidemiological cutoff values.
Phenotypic resistance was interpreted based on the MCOFFs reported by EFSA, which served as the interpretation criteria to distinguish susceptible strains free of phenotypically discoverable acquired resistance mechanisms from resistant strains. Generally, a strain was classified as susceptible when the MIC of a given antibiotic was not more than the established MCOFF. Herein, 23% (108/478) of the LAB strains were resistant to tetracycline; most L. johnsonii and L. crispatus strains showed tetracycline resistance, with resistance levels of 83% (15/18) and 60% (18/30), respectively. Lacticaseibacillus rhamnosus and L. paracasei showed the least common phenotypic resistance, with tetracycline resistance observed in 1% (1/68) and 3% (3/116) of the strains examined, respectively. Lactiplantibacillus plantarum, L. (para)gasseri, and L. reuteri exhibited tetracycline resistance levels of 17% (17/99), 32% (32/100), and 47% (22/47), respectively (Figure 1h).
3.2. Identification of ARGs and Their Correlation with Phenotype
The MCOFF is often used as an interpretation standard for the separation of sensitive strains from acquired resistance strains. If the MIC of one or more antibiotics for one strain is greater than the cutoff value, it is necessary to further explore its resistance mechanism at the genetic level [18]. Therefore, the genome sequences of 478 strains of LAB, which were tested for phenotypic resistance, were subjected to sequence alignment with CARD. Based on the selection standards, among the 478 strains, five tetracycline resistance-related genes encoded tetracycline target protection proteins (tet(W/N/W), tet(M), and tet(S)) and efflux pumps (tet(45) and tet(L)).
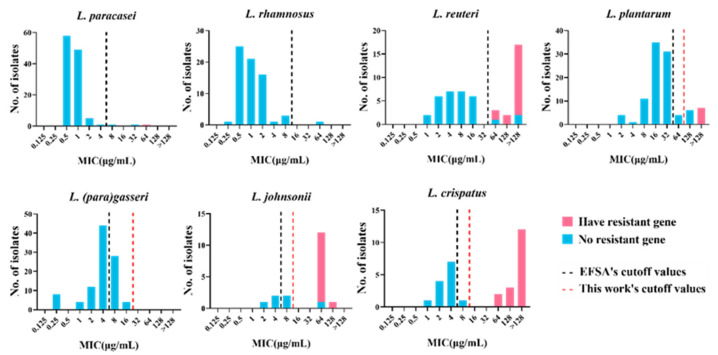
To identify the resistance determinants of tetracycline-resistant strains, we analyzed the association between phenotypically resistant strains and their genotypes (Table 1). Among the three tetracycline-resistant strains of L. paracasei, the strain FCQHC12L3 was characterized by the presence of tet(M). However, no tetracycline resistance gene was found in the remaining two phenotypically resistant strains. Among the 17 tetracycline-resistant strains of L. plantarum, six were characterized by the presence of tet(M), while QHLJZD13-L6 was characterized by the presence of tet(S), and no related resistance genes were detected in 10 strains. Among the 22 tetracycline-resistant strains of L. reuteri, two strains were characterized by the presence of tet (M) and tet(L), whereas two strains were characterized by the presence of tet (W/N/W) and tet(L). Three strains and eleven strains harbored only tet(M) or tet(W/N/W), respectively. Furthermore, strain FYNLJ83L8 was characterized by the presence of tet(45), and three strains were not associated with resistance genes. Across the 15 tetracycline-resistant strains of L. johnsonii, the gene tet(W/N/W) was found in 11 resistant strains. In addition, FHNXY70M2 was characterized by the presence of tet(W/N/W) and tet (L), and the remaining three strains did not display resistance-related genes. Of the 18 resistant strains of L. crispatus, tet (M) was found in two resistant strains, and tet(W/N/W) was found in four resistant strains. Ten strains were characterized by the presence of tet(W/N/W) and tet(L), and the strain FHNXY70M14 was characterized by the presence of tet(M), tet(W/N/W), and tet(L). However, no resistance-related genes were detected in strain FHNXY56M7. No tetracycline resistance genes were found in any of the 32 tetracycline-resistant strains of L. (para)gasseri and one tetracycline-resistant strain of L. rhamnosus. In particular, two strains (L. plantarum RS41-7 and L. gasseri FHNFQ34_L1) harbored a tet(M) gene but were sensitive to tetracycline. Furthermore, the tet(M) sequence of the two strains had obvious deletions in the functional sites, resulting in the loss of resistance function (Figure 2). Therefore, in this study, we did not classify these two strains as carriers of the gene tet(M). In brief, through genotypic and phenotypic association analysis, these five tetracycline resistance genes could explain the resistance phenotypes of 33% (1/3) of L. paracasei, 41% (7/17) of L. plantarum, 80% (12/15) of L. johnsonii, 94% (17/18) of L. crispatus, and 86% (19/22) of L. reuteri strains. However, resistance phenotypes of 48% (52/108) of the resistant strains could not be explained based on their genotypes (Figure 3).
Table 1.
Association between tetracycline-resistant strains and tetracycline resistance genes in eight lactic acid bacterial species.
| Tetracycline Resistance Genes | Number of Tetracycline-Resistant Strains |
|---|---|
| tet(M) | L. paracasei (1), L. plantarum (6), L. reuteri (3), L.crispatus (2) |
| tet(W/N/W) | L. reuteri (11), L.johnsonii (11), L.crispatus (4) |
| tet(S) | L. plantarum (1) |
| tet(45) | L. reuteri (1) |
| tet(M) and tet(L) | L. reuteri (2), L.crispatus (10) |
| tet(W/N/W) and tet(L) | L. reuteri (2), L.johnsonii (1) |
| tet(M), tet(W/N/W), and tet(L) | L.crispatus (1) |
| No tetracycline resistance genes | L. paracasei (2), L. rhamnosus (1), L. plantarum (10), L. reuteri (3), L.johnsonii (3), L.crispatus (1), L. (para)gasseri (32) |
Number in parentheses represents the number of strains with tetracycline resistance genes detected in the corresponding lactic acid bacterial species.
Figure 2.
Multi-sequence alignment of tet(M) sequences of Lactiplantibacillus plantarum RS41-7 and Lactobacillus gasseri FHNFQ34_L1 (the bottom two sequences) with the tet(M) sequences of other strains in this study.
Figure 3.
MIC distribution of 478 lactic acid bacteria strains with or without the tetracycline resistance gene. Blue: strains without resistance gene. Pink: strains with the tetracycline resistance gene. Black dotted line: cutoff value established by EFSA. Red dotted line: the new cutoff value established by this work.
3.3. Definition of New Susceptibility–Resistance Cutoff Values
The results of genotype-phenotype association analysis revealed that the genetic basis for the resistance of 48% (52/108) of the strains with tetracycline resistance phenotype could not be determined, including that of two strains of L. paracasei, one strain of L. rhamnosus, 10 strains of L. plantarum, one strain of L. crispatus, three strains of L. reuteri, three strains of L. johnsonii, and 32 strains of L. (para)gasseri. Lactobacillus (para)gasseri has a high drug resistance rate; however, the resistance determinants of all phenotypically resistant strains could not be identified. We speculate that the cutoff value based on the fermentation type may not be applicable to all species of LAB. Thus, it is recommended to develop MCOFFs at the species level. Lactobacillus (para)gasseri, L. crispatus, and L. johnsonii did not have species-specific cutoff values; therefore, we statistically analyzed the MIC frequency distribution of these species of Lactobacillus to establish species-specific TMCOFFs, which could better distinguish the resistant strains from the sensitive strains without acquired ARGs. Lacticaseibacillus paracasei, L. rhamnosus, L. plantarum, and L. reuteri had MCOFFs at the species level; however, 10% of L. plantarum had resistance phenotype but no resistance determinants. Therefore, we reformulated the cutoff value of L. plantarum to determine whether the strains containing resistance genes could be better distinguished from sensitive strains. Based on the MIC distribution of tetracycline of L. (para)gasseri, L. crispatus, L. johnsonii, and L. plantarum in this study, two different statistical approaches (Turnidge and Kronvall) and a “visual estimation” approach (eyeball method) were used to determine the new susceptibility–resistance cutoff values (Table 2).
Table 2.
Comparison of tentative microbiological cutoff values (TMCOFFs) for tetracycline calculated using two statistical methods and the eyeball method.
| TMCOFFs Obtained Using the Indicated Method (%) a | |||||
|---|---|---|---|---|---|
| Species | EFSA Cut Off | Method of Turnidge et al. b |
Method of Kronvall |
Eyeball Method | Median for the Method |
| L. (para)gasseri | 4 (68%) | 16 (100%) | 256 (100%) | 16 (100%) | 16 (100%) |
| L. johnsonii | 4 (17%) | 32 (38%) | 16 (38%) | 16 (38%) | 16 (38%) |
| L. crispatus | 4 (40%) | 8 (50%) | 16 (50%) | 16 (50%) | 16 (50%) |
| L. plantarum | 32 (83%) | 64 (87%) | 64 (87%) | 64 (87%) | 64 (87%) |
a Values in percentages denote the proportion of isolates with an MIC that is not greater than the TMCOFFs. b Calculated TMCOFFs including 99% of the strains in the wild-type population.
Based on the new cutoff values, all L. (para)gasseri strains were classified as sensitive, and the strains of L. crispatus containing resistance genes were distinguished from the sensitive strains without acquired ARGs. One strain belonging to L. johnsonii was not associated with the tetracycline resistance gene and was classified as phenotypically resistant. However, the MIC for this strain was found to be equivalent to that for another L. johnsonii strain containing the tetracycline resistance gene. Accordingly, we speculated that it contained potential tetracycline resistance genes. Therefore, the new cutoff value could completely distinguish the strains containing resistance genes from sensitive strains in L. johnsonii. The cutoff value of tetracycline for L. plantarum was newly formulated as 64 μg/mL, which is the same as that formulated by Flórez et al. [40]. However, the resistance determinants of six strains of L. plantarum still need to be further explored.
3.4. Prevalence and Distribution of Tetracycline Resistance Genes in LAB
To explore the distribution and prevalence of these five tetracycline resistance genes in LAB, we performed statistical analysis on the detection of tetracycline resistance genes in eight species of LAB (Table 3). The most widely distributed tetracycline resistance gene in LAB was tet(M), which was detected in four LAB species, including L. paracasei, L. plantarum, L. reuteri, and L. crispatus. The genes tet(W/N/W) and tet(L) were detected in three species of LAB, including L. reuteri, L. johnsonii, and L. crispatus. The tet(S) gene was detected in only one strain of L. plantarum, and the tet (45) gene was found in only one strain of L. reuteri. The tet(W/N/W) gene was the most frequently detected tetracycline resistance gene in LAB; it was detected in 30 strains of LAB in this study. The genes tet(M), tet(L), tet(S), and tet(45) had detection frequencies of 26, 16, 1, and 1, respectively. Most types of the tetracycline resistance genes were detected in L. reuteri, including tet(M), tet(W/N/W), tet(L), and tet(45). Three of the tetracycline resistance genes were detected in L. crispatus, namely tet(M), tet(W/N/W), and tet(L). Two genes, tet(W/N/W) and tet(L), were detected in L. johnsonii. Two tetracycline resistance genes, tet(M) and tet(S), were found in L. plantarum. Among L. paracasei strains, only one harbored the tet(M) gene. No tetracycline resistance genes were found in any of the L. rhamnosus and L. (para)gasseri strains in this study. Herein, the tetracycline resistance gene was detected in 12% (56/478) of the strains. A total of 26 LAB strains contained only the tet(W/N/W) gene, while 12 LAB strains contained only the tet(M) gene. Interestingly, the gene tet(L) did not appear alone in LAB but was always detected together with other tetracycline resistance genes. The genes tet(M) and tet(L) were detected together in 12 strains of LAB. The genes tet(W/N/W) and tet(L) were detected concurrently in three strains of LAB. Three tetracycline resistance genes, tet(M), tet(W/N/W), and tet(L), were detected simultaneously in one L. crispatus strain. One strain of L. plantarum contained only the tet(S) gene, and one L. reuteri strain contained only the tet(45) gene.
Table 3.
The detailed distribution of the detected tetracycline resistance genes in different lactic acid bacterial species.
| Species | Total Strain Number | TETR | tet(M) | tet(W/N/W) | tet(L) | tet(S) | tet(45) |
|---|---|---|---|---|---|---|---|
| L. paracasei | 116 | 3 | 1 | 0 | 0 | 0 | 0 |
| L. rhamnosus | 68 | 1 | 0 | 0 | 0 | 0 | 0 |
| L. plantarum | 99 | 17 | 6 | 0 | 0 | 1 | 0 |
| L. reuteri | 47 | 22 | 5 | 13 | 4 | 0 | 1 |
| L. johnsonii | 18 | 15 | 0 | 12 | 1 | 0 | 0 |
| L. crispatus | 30 | 18 | 13 | 5 | 11 | 0 | 0 |
| L. (para)gasseri | 100 | 32 | 0 | 0 | 0 | 0 | 0 |
| Total | 478 | 108 | 25 | 30 | 16 | 1 | 1 |
TETR represents the number of tetracycline-resistant strains based on the EFSA breakpoint value.
The genes of tet(M) and tet(W/N/W) are the most widely distributed tetracycline resistance genes in LAB. In order to further explore their phylogenetic relationship, multiple sequence alignments of a total of 25 tet(M) and 30 tet(W/N/W) gene sequences identified in this paper together with the same genotype sequences retrieved from the NCBI database were performed by ClustalW, and the phylogenetic tree was constructed using the Neighbor-Joining method by MEGA X (Figures S1 and S2). Most of the same species were in the same branch, suggesting that the genes had adaptive mutations when transferred to different species, and the tet(M) and tet(W/N/W) genes of LAB of the same species may have come from the same host.
3.5. Expression of Tetracycline Resistance Gene Based on RT-PCR
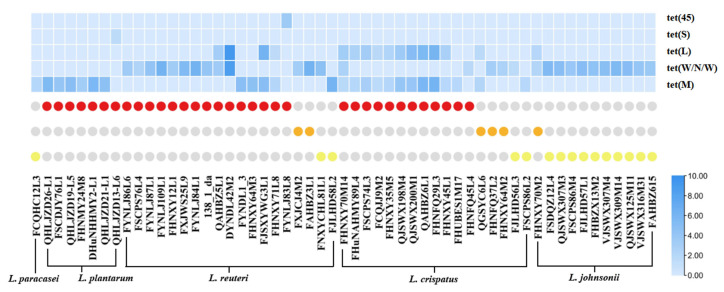
To verify that the five tetracycline resistance genes play a role in the resistance to tetracycline, the expression of drug resistance genes carried by drug-resistant strains induced by tetracycline at a concentration of 1/2 × MIC was determined using RT-PCR. After being induced by tetracycline, the expression of these five drug-resistant genes in the drug-resistant strains was upregulated, indicating that these genes are indeed involved in the resistance to tetracycline. In the same species of LAB, some strains with the same resistance determinants had different MICs; this result may be attributed to the differences in gene expression. However, assessing the relative expression of genes alone cannot completely explain this phenomenon. We also attempted to directly compare the expression of the same resistance genes in different strains; however, the strains with the same resistance determinants had different MICs, which could not be explained; we thus speculated that it may be mainly related to gene structure [41]. Interestingly, the expression level of the tet (L) gene was similar to that of other tetracycline resistance genes in the same strain, suggesting that these genes may be located in the same gene cluster (Figure 4).
Figure 4.
The expression of resistance genes in phenotypically resistant strains induced by tetracycline at the concentration of 1/2 × MIC. The heatmap above represents the level of gene expression. The circle chart represents the MIC for the strain, the red circle indicates that the MIC for the corresponding strain is greater than 128 μg/mL, the orange circle indicates that the MIC for the corresponding strain is equal to 128 μg/mL, and the yellow circle indicates that the MIC for the corresponding strain is equal to 64 μg/mL.
4. Discussion
At present, there are many LAB species with QPS status for which MCOFFs at the species level have not been determined. However, many researchers have found that different LAB species have different tolerances to the same antibiotic [42]. Species-specific cutoff values must be set when determining phenotypic resistance. Studies have shown, based on the cutoff values recommended by the EFSA at the species level, that some LAB species have high resistance levels [43], suggesting that these cutoff values should be reexamined according to the genetic basis for resistance. Therefore, we formulated a species-specific cutoff value of tetracycline for L. (para)gasseri, L. crispatus, and L. johnsonii. The drug resistance rate of L. plantarum was relatively high. Furthermore, the determinants of tetracycline resistance could not be found for several phenotypically resistant strains. The cutoff values formulated by EFSA for L. plantarum have only been defined for strains used in animal feed [44]. The strains used in the present study were of different origins. Based on the MIC distribution data for the abovementioned four species, we developed a new cutoff value. Similar results for both the absolute MIC and MIC ranges for these species have been reported by other authors [45].
The newly established cutoff value for L. plantarum is greater than that established by EFSA but equivalent to that formulated by Flórez et al. [40]. The newly established cutoff values for L. (para)gasseri, L. crispatus, and L. johnsonii were also greater than those established by EFSA according to the fermentation metabolism category. The new cutoff values can completely distinguish the sensitive strains of L. (para)gasseri, L. crispatus, L. johnsonii, and L. plantarum from the strains with acquired resistance genes, except one strain of L. johnsonii (MIC = 64 μg/mL) and six strains of L. plantarum (MIC = 128 μg/mL), which were suspected to possess acquired resistance to this antibiotic; we are currently exploring their resistance using the transcriptional technology [46]. Among the remaining three species of LAB, six phenotypically resistant strains that were not associated with known tetracycline resistance genes were present. These LAB strains may carry other genes that have not been found to play a role in the function of tetracycline resistance before. We also used transcriptome analysis to explore whether their resistance phenotypes are mediated by unknown tetracycline resistance genes, evolution, or other internal factors [47], which made it difficult to predict the resistance phenotype by relying on the database, to determine whether the existing cutoff value is reasonable.
To explore the resistance determinants of phenotypically resistant strains, we compared the protein sequences of all strains with the CARD. Sequences with similarity greater than 30% are believed to be homologous; however, when an identity >30% was used as the threshold to screen resistance genes, all phenotypically sensitive strains had multiple tetracycline resistance genes (Figure S3). Thus, we gradually increased the threshold with a gradient of 10% and found that when identity was > 70%, all phenotypically sensitive strains did not have tetracycline resistance genes. As a result, the threshold was set at 70%. Among the phenotypically resistant strains with tetracycline resistance genes, five key genes, tet(M), tet(W/N/W), tet(L), tet(S), and tet(45), related to tetracycline resistance phenotype were identified in their genomes. In particular, among all LAB strains in this study, the genes tet(M) and tet(W/N/W) were the most widely distributed tetracycline resistance genes [48] and were usually located on mobile genetic elements. The genes tet(Q), tet(K), and tet(O) were reported to be detected in LAB strains [49,50]; however, they were not detected in this study; this result may be related to the species, region, and source of LAB [13]. The common mechanisms of tetracycline resistance include ribosome protection, antibiotic efflux, and antibiotic inactivation [51]. Among all tetracycline-resistant strains in this study, the resistance mechanism of encoding ribosomal protective protein was dominant, followed by the efflux pump protein, indicating that tetracycline resistance is mainly mediated by the acquisition of tetracycline resistance genes encoded by the two resistance mechanisms, and that the two mechanisms work together to enhance the resistance of bacteria.
5. Conclusions
Based on the MIC distribution data of 478 strains of LAB, LAB showed moderate resistance to tetracycline. Further, formulating the breakpoint value at the species level was found to be necessary. Therefore, the species-specific microbiological cutoff values for L. (para)gasseri, L. crispatus, and L. johnsonii against tetracycline were formulated, and new susceptibility-resistance cutoff values for L. plantarum were defined. The genes tet(M), tet(W/N/W), tet(L), tet(S), and tet(45) were the key resistance genes for the tetracycline resistance phenotype and were found to widely exist in LAB. The determination of antibiotic resistance in probiotic strains is related to food safety issues. The findings of this study provide certain guiding significance and reference values at the phenotype and genotype levels for the safe application of LAB in the food industry and the formulation of probiotic resistance evaluation standards.
Acknowledgments
Thanks for the help from the Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province. We thank Yutao Chen and Zihuan Wu for technical assistance; Zhen Jin, Xinyi Wang, Li Qian, and Qianwen Wang for their helpful advice.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9102128/s1, Table S1. Details of 478 strains belonging to 8 lactic acid bacterial species, including their origin, MIC values, resistance phenotype, and accession number. Figure S1. Phylogenetic analysis of tet(M) belonging to 25 strains of lactic acid bacteria identified in this paper together with the same genotype sequences retrieved from NCBI database. Figure S2. Phylogenetic analysis of tet(W/N/W) belonging to 30 strains of lactic acid bacteria identified in this paper together with the same genotype sequences retrieved from NCBI database. Figure S3. The resistance genes of 478 strains of lactic acid bacteria were detected with an identity greater than 30% as the threshold.
Author Contributions
Conceptualization, H.Z., J.Z. (Jianxin Zhao), W.C. and W.L.; Data curation, Q.M., Z.P. and Z.F.; Formal analysis, Q.M.; Funding acquisition, H.Z., J.Z. (Jianxin Zhao), W.C. and W.L.; Investigation, Q.M.; Methodology, Q.M., Z.P. and Z.F.; Project administration, Q.M.; Resources, Q.M., Y.-k.L., H.W. and J.Z. (Jinlin Zhu); Software, Q.M. and Z.P.; Supervision, H.W., J.Z. (Jinlin Zhu) and W.L.; Validation, Q.M.; Visualization, Q.M., Z.P. and Z.F.; Writing—original draft, Q.M.; Writing—review and editing, Q.M.,Z.P., Z.F., H.W., J.Z. (Jinlin Zhu), Y.-k.L.,H.Z., J.Z. (Jianxin Zhao), W.C. and W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31820103010), the National Key Research and Development Program of China (No. 2019YFF0217601).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Of the 478 LAB strains deposited in the NCBI GenBank database, 439 were released as part of our previous studies [52,53,54,55], and the remaining 39 genomes were deposited under project accession no. PRJNA658852. The accession numbers for all individual genomes used in this study are presented in Table S1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jamal M., Shareef M., Sajid S. Lincomycin and tetracycline resistance in poultry. Review. Matrix Sci. Pharma. 2017;1:33–38. doi: 10.26480/msp.01.2017.33.38. [DOI] [Google Scholar]
- 2.Rudra P., Hurst-Hess K., Lappierre P., Ghosh P. High levels of intrinsic tetracycline resistance in Mycobacterium abscessus are conferred by a tetracycline-modifying monooxygenase. Antimicrob. Agents Chemothe. 2018;62:e00119-18. doi: 10.1128/AAC.00119-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tao R., Ying G.-G., Su H.-C., Zhou H.-W., Sidhu J.P. Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl rivers in South China. Environ. Pollut. 2010;158:2101–2109. doi: 10.1016/j.envpol.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Guarddon M., Miranda J.M., Rodríguez J.A., Vázquez B.I., Cepeda A., Franco C.M. Real-time polymerase chain reaction for the quantitative detection of tetA and tetB bacterial tetracycline resistance genes in food. Int. J. Food Microbiol. 2011;146:284–289. doi: 10.1016/j.ijfoodmicro.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 5.Arredondo A., Àlvarez G., Nart J., Mor C., Blanc V., León R. Detection and expression analysis of tet(B) in Streptococcus oralis. J. Oral Microbiol. 2019;11:1643204. doi: 10.1080/20002297.2019.1643204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speer B.S., Shoemaker N.B., Salyers A.A. Bacterial resistance to tetracycline: Mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 1992;5:387–399. doi: 10.1128/CMR.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He T., Wang R., Liu D., Walsh T.R., Zhang R., Lv Y., Ke Y., Ji Q., Wei R., Liu Z., et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019;4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 8.Phillips I., Casewell M., Cox T., De Groot B., Friis C., Jones R., Nightingale C., Preston R., Waddell J. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 2004;53:28–52. doi: 10.1093/jac/dkg483. [DOI] [PubMed] [Google Scholar]
- 9.Liu S.-n., Han Y., Zhou Z.-j. Lactic acid bacteria in traditional fermented Chinese foods. Food Res. Int. 2011;44:643–651. doi: 10.1016/j.foodres.2010.12.034. [DOI] [Google Scholar]
- 10.Damania P., Patel R., Shaw R., Kataria R.P., Wadia A. Development of antimicrobial packaging materials for food preservation using bacteriocin from Lactobacillus casei. Microbiol. Res. 2016;7:19–22. doi: 10.4081/mr.2016.6622. [DOI] [Google Scholar]
- 11.Leuschner R.G., Robinson T.P., Hugas M., Cocconcelli P.S., Richard-Forget F., Klein G., Licht T.R., Nguyen-The C., Querol A., Richardson M. Qualified presumption of safety (QPS): A generic risk assessment approach for biological agents notified to the European Food Safety Authority (EFSA) Trends Food Sci. Technol. 2010;21:425–435. doi: 10.1016/j.tifs.2010.07.003. [DOI] [Google Scholar]
- 12.Shazali N., Foo H.L., Loh T.C., Choe D.W., Rahim R.A. Prevalence of antibiotic resistance in lactic acid bacteria isolated from the faeces of broiler chicken in Malaysia. Gut Pathog. 2014;6:1–7. doi: 10.1186/1757-4749-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Çataloluk O., Gogebakan B. Presence of drug resistance in intestinal lactobacilli of dairy and human origin in Turkey. FEMS Microbiol. Lett. 2004;236:7–12. doi: 10.1111/j.1574-6968.2004.tb09620.x. [DOI] [PubMed] [Google Scholar]
- 14.Abriouel H., Muñoz M.d.C.C., Lerma L.L., Montoro B.P., Bockelmann W., Pichner R., Kabisch J., Cho G.-S., Franz C.M., Galvez A., et al. New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res. Int. 2015;78:465–481. doi: 10.1016/j.foodres.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 15.Broaders E., Gahan C.G., Marchesi J.R. Mobile genetic elements of the human gastrointestinal tract: Potential for spread of antibiotic resistance genes. Gut microbes. 2013;4:271–280. doi: 10.4161/gmic.24627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei Z., Sadiq F.A., Han X., Zhao J., Zhang H., Ross R.P., Lu W., Chen W. Identification, characterization, and phylogenetic analysis of eight new inducible prophages in Lactobacillus. Virus Res. 2020;286:198003. doi: 10.1016/j.virusres.2020.198003. [DOI] [PubMed] [Google Scholar]
- 17.Das D.J., Shankar A., Johnson J.B., Thomas S. Critical insights into antibiotic resistance transferability in probiotic Lactobacillus. Nutrition. 2020;69:110567. doi: 10.1016/j.nut.2019.110567. [DOI] [PubMed] [Google Scholar]
- 18.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastro. Hepat. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 19.Hazards E.P.o.B. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update) EFSA J. 2013;11:3449. doi: 10.2903/j.efsa.2013.3449. [DOI] [Google Scholar]
- 20.Additives E.P.o., Feed P.o.S.u.i.A. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012;10:2740. doi: 10.2903/j.efsa.2012.2740. [DOI] [Google Scholar]
- 21.Balouiri M., Sadiki M., Ibnsouda S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016;6:71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kushiro A., Chervaux C., Cools-Portier S., Perony A., Legrain-Raspaud S., Obis D., Onoue M., van de Moer A. Antimicrobial susceptibility testing of lactic acid bacteria and bifidobacteria by broth microdilution method and Etest. Int. J. Food Microbiol. 2009;132:54–58. doi: 10.1016/j.ijfoodmicro.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 23.McLain J.E., Cytryn E., Durso L.M., Young S. Culture-based methods for detection of antibiotic resistance in agroecosystems: Advantages, challenges, and gaps in knowledge. J. Environ. Qual. 2016;45:432–440. doi: 10.2134/jeq2015.06.0317. [DOI] [PubMed] [Google Scholar]
- 24.Kahlmeter G., Giske C.G., Kirn T.J., Sharp S.E. Point-counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute recommendations for reporting antimicrobial susceptibility results. J. Clin. Microbiol. 2019;57:e01129-19. doi: 10.1128/JCM.01129-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boolchandani M., D’Souza A.W., Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019;20:356–370. doi: 10.1038/s41576-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su M., Satola S.W., Read T.D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 2019;57:e01405-18. doi: 10.1128/JCM.01405-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International Organization for Standardization (ISO) Milk and Milk Products—Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB) ISO; Geneva, Switzerland: 2010. [(accessed on 12 September 2019)]. ISO 10932:2010 (IDF 223:2010) Available online: https://www.iso.org/standard/46434.html. [Google Scholar]
- 28.Additives E.P.o., Feed P.o.S.u.i.A., Rychen G., Aquilina G., Azimonti G., Bampidis V., Bastos M.d.L., Bories G., Chesson A., Cocconcelli P.S., et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018;16:e05206. doi: 10.2903/j.efsa.2018.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu M., Zhao Y. Comparative resistomic analyses of Lysobacter species with high intrinsic multidrug resistance. J. Glob. Antimicrob. Resist. 2019;19:320–327. doi: 10.1016/j.jgar.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Chen C., Chen H., He Y., Xia R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv. 2018;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [Google Scholar]
- 31.Turnidge J., Kahlmeter G., Kronvall G. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin. Microbiol. Infect. 2006;12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- 32.Kronvall G.r. Normalized resistance interpretation as a tool for establishing epidemiological MIC susceptibility breakpoints. J. Clin. Microbiol. 2010;48:4445–4452. doi: 10.1128/JCM.01101-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cafarchia C., Iatta R., Immediato D., Puttilli M.R., Otranto D. Azole susceptibility of Malassezia pachydermatis and Malassezia furfur and tentative epidemiological cut-off values. Med. Mycol. 2015;53:743–748. doi: 10.1093/mmy/myv049. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y.-j., Singh R.P., Lan X., Zhang C.-s., Sheng D.-h., Li Y.-q. Whole transcriptome analysis and gene deletion to understand the chloramphenicol resistance mechanism and develop a screening method for homologous recombination in Myxococcus xanthus. Microb. Cell Factories. 2019;18:1–15. doi: 10.1186/s12934-019-1172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelm-Nelson C.A., Stevenson S.A., Ciucci M.R. Data in support of qPCR primer design and verification in a Pink1−/− rat model of Parkinson disease. Data Brief. 2016;8:360. doi: 10.1016/j.dib.2016.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X.J., Tian A.J., Lv S.M., Du A.D., Chen B.L., Zhang S.X. Detection of resistance and resistance genes of swine Escherichia coli in Guizhou to tetracyclines. China Anim. Husb. Vet. Med. 2018;45:1367–1373. doi: 10.16431/j.cnki.1671-7236.2018.05.030. [DOI] [Google Scholar]
- 37.Uchida M., Amakasu H., Satoh Y., Murata M. Combinations of lactic acid bacteria and yeast suitable for preparation of marine silage. Fish. Sci. 2004;70:507–517. doi: 10.1111/j.1444-2906.2004.00832.x. [DOI] [Google Scholar]
- 38.Wang Q., Xiong Y., Zhang S., Sui Y., Yu C., Liu P., Li H., Guo W., Gao Y., Przepiorski A. The Dynamics of Metabolic Characterization in iPSC-Derived Kidney Organoid Differentiation via a Comparative Omics Approach. Front. Genet. 2021;12:132. doi: 10.3389/fgene.2021.632810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua S., Kittler R., White K.P. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flórez A.B., Egervärn M., Danielsen M., Tosi L., Morelli L., Lindgren S., Mayo B. Susceptibility of Lactobacillus plantarum strains to six antibiotics and definition of new susceptibility–resistance cutoff values. Microb. Drug Resist. 2006;12:252–256. doi: 10.1089/mdr.2006.12.252. [DOI] [PubMed] [Google Scholar]
- 41.Cui C.-Y., He Q., Jia Q.-L., Li C., Chen C., Wu X.-T., Zhang X.-J., Lin Z.-Y., Zheng Z.-J., Liao X.-P. Evolutionary Trajectory of the Tet (X) Family: Critical Residue Changes towards High-Level Tigecycline Resistance. Msystems. 2021;6:e00050-21. doi: 10.1128/mSystems.00050-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao Y., Zhang W., Guo H., Pan L., Zhang H., Sun T. Comparative studies on antibiotic resistance in Lactobacillus casei and Lactobacillus plantarum. Food Control. 2015;50:250–258. doi: 10.1016/j.foodcont.2014.09.003. [DOI] [Google Scholar]
- 43.Campedelli I., Mathur H., Salvetti E., Clarke S., Rea M.C., Torriani S., Ross R.P., Hill C., O’Toole P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019;85:e01738-18. doi: 10.1128/AEM.01738-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Authority E.F.S. Opinion of the Scientific Panel on additives and products or substances used in animal feed (FEEDAP) on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance. EFSA J. 2005;3:223. doi: 10.2903/j.efsa.2005.223. [DOI] [Google Scholar]
- 45.Danielsen M., Wind A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003;82:1–11. doi: 10.1016/S0168-1605(02)00254-4. [DOI] [PubMed] [Google Scholar]
- 46.Matzrafi M., Shaar-Moshe L., Rubin B., Peleg Z. Unraveling the transcriptional basis of temperature-dependent Pinoxaden resistance in Brachypodium hybridum. Front. Plant Sci. 2017;8:1064. doi: 10.3389/fpls.2017.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersson D.I. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 2003;6:452–456. doi: 10.1016/j.mib.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Comunian R., Daga E., Dupré I., Paba A., Devirgiliis C., Piccioni V., Perozzi G., Zonenschain D., Rebecchi A., Morelli L., et al. Susceptibility to tetracycline and erythromycin of Lactobacillus paracasei strains isolated from traditional Italian fermented foods. Int. J. Food Microbiol. 2010;138:151–156. doi: 10.1016/j.ijfoodmicro.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 49.Egervärn M., Roos S., Lindmark H. Identification and characterization of antibiotic resistance genes in Lactobacillus reuteri and Lactobacillus plantarum. J. Appl. Microbiol. 2009;107:1658–1668. doi: 10.1111/j.1365-2672.2009.04352.x. [DOI] [PubMed] [Google Scholar]
- 50.Huys G., D’HAENE K., Danielsen M., Maettoe J., Egervaern M., Vandamme P. Phenotypic and molecular assessment of antimicrobial resistance in Lactobacillus paracasei strains of food origin. J. Food Prot. 2008;71:339–344. doi: 10.4315/0362-028X-71.2.339. [DOI] [PubMed] [Google Scholar]
- 51.Thaker M., Spanogiannopoulos P., Wright G.D. The tetracycline resistome. Cell. Mol. Life Sci. 2010;67:419–431. doi: 10.1007/s00018-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou X., Yang B., Stanton C., Ross P., Zhao J., Zhang H., Chen W. Comparative analysis of Lactobacillus gasseri from Chinese subjects reveals a new species-level taxa. BMC Genom. 2020;21:2–16. doi: 10.1186/s12864-020-6527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Q., Zhang L., Ross P., Zhao J., Zhang H., Chen W. Comparative genomics of Lactobacillus crispatus from the gut and vagina reveals genetic diversity and lifestyle adaptation. Genes. 2020;11:360. doi: 10.3390/genes11040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang D., Yang B., Chen Y., Stanton C., Ross R.P., Zhao J., Zhang H., Chen W. Comparative genomic analyses of Lactobacillus rhamnosus isolated from Chinese subjects. Food Biosci. 2020;36:100659. doi: 10.1016/j.fbio.2020.100659. [DOI] [Google Scholar]
- 55.Pei Z., Sadiq F.A., Han X., Zhao J., Zhang H., Ross R.P., Lu W., Chen W. Comprehensive Scanning of Prophages in Lactobacillus: Distribution, Diversity, Antibiotic Resistance Genes, and Linkages with CRISPR-Cas Systems. Msystems. 2021;6:e01211-20. doi: 10.1128/mSystems.01211-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Of the 478 LAB strains deposited in the NCBI GenBank database, 439 were released as part of our previous studies [52,53,54,55], and the remaining 39 genomes were deposited under project accession no. PRJNA658852. The accession numbers for all individual genomes used in this study are presented in Table S1.