Abstract
Passion fruit (Passiflora edulis Sims.) is an ever-increasing interest crop in Italy because it is mainly cultivated for its edible fruit and, secondly, as an ornamental evergreen climber. During the summer of 2020, two-year-old plants of purple passion fruit in one of the most important expanding production areas of Sicily (southern Italy) showed symptoms of yellowing, wilting, and vascular discoloration. Fusarium-like fungal colonies were consistently yielded from symptomatic crown and stem tissues. Five representative isolates were characterized by a morphological and molecular analysis based on a multilocus phylogeny using RNA polymerase’s second largest subunit (RPB2) and translation elongation factor 1-alpha (EF-1α) genes, as Fusarium nirenbergiae (Fusarium oxysporum species complex). Pathogenicity tests conducted on healthy 1-year-old passion fruit cuttings revealed symptoms similar to those observed in the field. To our knowledge, this is the first report of Fusarium wilt on passion fruit caused by Fusarium nirenbergiae. This report focuses on the phytopathological implications of this fungal pathogen, which may represent a future significant threat for the expanding passion fruit production in Italy and Europe.
Keywords: wilt, passion fruit, Fusarium oxysporum species complex
1. Introduction
In recent years, tropical fruit production increased worldwide due to the increasing demand of global markets and more efficient transportation and storage techniques [1,2]. Most of the tropical fruit is destined for fresh consumption or industrial transformation. Among these, passion fruit (Passiflora edulis Sims.) is one of the most exported and consumed fruit commodities. It originated in tropical and subtropical America [3], and it is now extensively cultivated worldwide, including Australia, New Zealand, India, Africa, and South America [4,5]. Passion fruit is mainly cultivated for its edible fruit but secondarily also for its attractive flowers on ornamental evergreen vines.
In Italy, the cultivation of P. edulis (also known as purple passion fruit) as a fruit crop in some regions characterized by a Mediterranean climate (e.g., Sicily and Calabria) is gaining growing interest by local farmers, and it is carried out under greenhouse and, to a lower extent, open field conditions. Indeed, although the crop is well adapted to a wide rainfall range (1000–2500 mm for crop season), minimum temperatures below 5 °C should be avoided because they seriously compromise the plant growth and nutrient uptake [6,7,8]. In this regard, it should be noted that a process of reconversion of protected tomato and vegetable crops into tropical fruit plantations is currently taking place in southern Italy and Sicily.
Unfortunately, this species is affected by many diseases during its different growth stages, and this reduces the yield and the farmers’ income [9]. One of the most widely reported fungal pathogens affecting passion fruit is Fusarium oxysporum f. sp. passiflorae, which causes the Fusarium wilt. It was first reported in Australia [10] but is nowadays spread worldwide [11,12,13]. Among Fusarium diseases, Neocosmospora solani (=Fusarium solani) is responsible for the basal stem rot [14,15,16]. According to Viana & Costa [17], the species F. oxysporum f. sp. passiflorae and N. solani are the most damaging ones to passion fruit crops. Minor diseases have been reported on passion fruit, such as the damping-off of seedlings and collar and root rot in adult plants caused by Rhizoctonia solani [18] and collar rot caused by Phytophthora spp. [19]. During a recent survey performed in Sicily, young passion fruit plants showing symptoms of general yellowing and wilting were observed in some of the most representative production areas. Given the increasing interest of local growers in expanding passion fruit cultivation, the aim of this study was to characterize the fungal species associated with those symptoms and test their pathogenicity, in order to better understand the syndrome’s aetiology.
2. Results
The symptoms observed in the greenhouse consisted of leaf yellowing and wilting (Figure 1a,b), external crown and root rot and wood discoloration moving upward to the canopy (Figure 1c). The disease incidence reached 10% of the cultivated plants. Colonies with white or light purple aerial mycelia and violet pigmentation on the underside of the cultures developed after 14 days on PDA, being firstly identified as Fusarium-like. Sporodochial macroconidia with 2 to 5 septa, grown on OA, measured (23.09–) 28.76 ± 3.06 (–35.48) μm × (1.99–) 3.84 ± 0.58 (–4.75) μm (Figure 1f,g). Oval, unicellular microconidia developed on short monophialides, grown on OA, measured (3.1–) 5.17 ± 1.35 (–9.17) μm × (1.3–) 1.98 ± 0.37 (–2.9) μm (Figure 1i).
Figure 1.
Disease symptoms and Fusarium nirenbergiae features: (a,b), yellowing and wilting of passion fruit plants in greenhouse; (c), vascular discoloration on a collar portion; (d,e), F. nirenbergiae (Di3A-Pef1 isolate) grown on 7 day-old (up) and 14 day-old (down) OA; (f), sporodochia on OA; (g), sporodochial conidia (macroconidia); (h), chlamydospores on SNA; (i), aerial conidia (microconidia). Scale bars, (f): 2 mm; (g–i): 50 μm.
PCR edit amplicons resulted in 528 bp for the partial ITS region, 287 bp for EF-1α and 953 bp for RPB2. The sequences were registered in GenBank as follows: MZ398141, MZ398142, MZ398143, MZ398144, MZ398145 for ITS, MZ408109, MZ408110, MZ408111, MZ408112, MZ408113 for RPB2 and MZ408114, MZ408115, MZ408116, MZ408117, MZ408118 for EF1-α. A GenBank BLASTn analysis and a pairwise sequence alignment on the MLST database indicated that all the isolates from passion fruit belonged to the Fusarium oxysporum species complex (FOSC). In particular, the MLST search resulted in high identity values (96–100%) (Acc. number MH582354) for the EF1-α gene and 98% (Acc. number MH582140) for the RBP2 gene with a F. oxysporum species complex (FOSC). The MP heuristic search resulted in 83 parsimony-informative characters, while 109 were variable and parsimony-uninformative and 1412 were constant. A maximum of 320 equally most parsimonious trees were retained (Tree length = 249, CI = 0.851, RI = 0.898 and RC = 0.765).
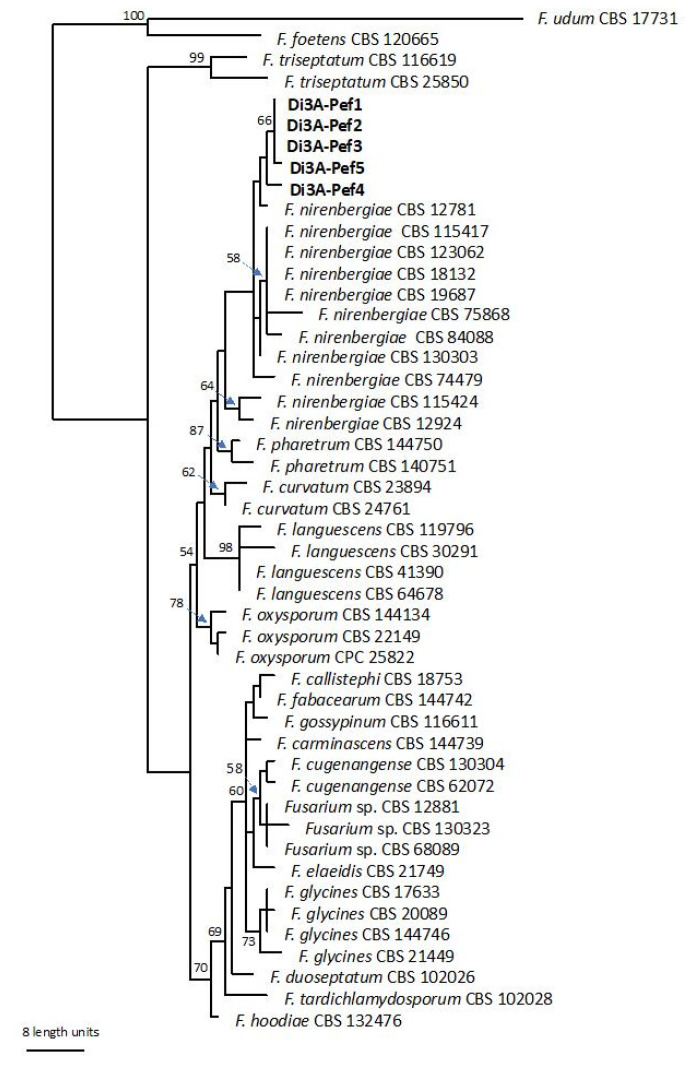
The bootstrap support values from the parsimony analysis are shown close to the branch node. The group of representative isolates Di3A-Pef1-5 clustered with the reference strain of F. nirenbergiae, as shown in Figure 2, and were clearly separated by the other sequences provided in the study by Lombard et al. [20]. The isolates were then identified as Fusarium nirenbergiae L. Lombard & Crous.
Figure 2.
Single most parsimonious phylogenetic tree resulting from the MP analysis of combined EF1-α and rbp2 sequence data. The isolates in bold were sequenced in this study. The numbers represent MP bootstrap values.
The inoculated isolate after five months caused symptoms similar to those observed under greenhouse conditions in all inoculated plants. The symptoms consisted of leaf yellowing and wilting. After 7 months all plants died. A longitudinal section of the inoculated plants reveals the internal discolorations moving upward to the canopy. The control remains symptomless. From the symptomatic tissues, F. nirembergiae was always re-isolated, and it was characterized as previously described.
3. Discussion and Conclusions
To the best of our knowledge, this paper represents the first report of F. nirembergiae, belonging to the FOSC complex, as a causal agent of Fusarium wilt of passion fruit. In this regard, both the morphological characterization and the analysis of the ITS, EF1-α and RBP2 sequences allowed us to correctly allocate a representative number of detected strains within the F. nirenbergiae group, being distinctly separated by the other taxa, as recently shown by Lombard et al. [20] and Crous et al. [21]. Based on the present findings, F. nirenbergiae was strongly grouped in a separated subclade of FOSC, phylogenetically close to F. curvatum. Although little information regarding F. nirembergiae’s pathogenicity and host range is currently available, except for the study by Zhao et al. [22] on Acer negundo, this species (belonging to FOSC) is able to colonize and infect host vascular tissues; for this reason, it is reported worldwide as responsible for Fusarium wilt [14]. The first symptoms consist of leaf yellowing and wilt, followed by the plant’s collapse. This disease is observed in adult and young plants under favorable conditions for the infection development, such as high temperature and humidity and a high potential inoculum in the soil [5,23]. Once this fungal pathogen is established in the field, its control is very difficult, since fungicide application does not result in a significant reduction of the disease amount, and the pathogen can persist in the soil for many years in the absence of the host [14]. Hence, the incidence data are very worrying as regards the nature of the fungal pathogen and dissemination ability of F. nirenbergiae under greenhouse conditions. If, on the one hand, protected systems could facilitate the cultivation of purple passion fruit, on the other hand they could aggravate the consequences of this phytopathological issue. Indeed, this could represent a future threat for the expansion of this tropical crop, which is replacing protected tomato cultivation in different areas of southern Italy. Therefore, disease management should be focused mainly on preventative and pathogen exclusion measures, avoiding plantation in areas with a severe history of Fusarium wilt infections or selecting healthy propagation material in combination with adequate agronomic practices. Additionally, other sustainable strategies should include the use of resistant cultivars, as recommended by several authors [24,25]. Comprehensively, the increasing trend of tropical plantations in Italy leads us to focus more on fungal diseases that could represent limiting factors for future production. According to presented data combined with recent findings [20,21], it cannot be excluded that some past reports of F. oxysporum f. sp. passiflorae could confirm that F. nirembergiae is a causal agent of Fusarium wilt. However, further surveys should be performed on P. edulis orchards in Italy and worldwide to confirm the new aetiology of the Fusarium wilt of passion fruit and its real diffusion.
4. Materials and Methods
4.1. Field Survey, Isolations and Morphological Characterization
In July of 2020, 50 two-year-old ‘purple’ passion fruit plants cultivated in a greenhouse in the Syracuse province (Sicily, Italy) appeared stunted, defoliated and severely wilted. Diseased vascular tissues (0.5 cm2) were surface-disinfected for 1 min in a 1.2% sodium hypochlorite (NaOCl) solution, rinsed in sterile water, placed on a potato dextrose agar (PDA, Lickson, Vicari, Italy) amended with 0.1 g/L of streptomycin sulphate (Sigma-Aldrich, St. Louis, MO, USA), to prevent bacterial growth, and then incubated at 25 ± 1 °C until fungal colonies were observed. Single-spore isolates were obtained from pure cultures grown on APDA. To induce sporulation, five representative single-spore isolates (named Di3A-Pef1, Di3A-Pef2, Di3A-Pef3, Di3A-Pef4 and Di3A-Pef5) were selected and transferred on a synthetic nutrient-poor agar (SNA) [26], Oatmeal Agar (OA, Difco, Detroit, MI, USA) and PDA for morphological characterization. A total of 50 macro- and micro-conidia were measured (length and width size) using a fluorescence microscope (Olympus-BX61) coupled to an Olympus DP70 digital camera; images and measurements were captured using the software analySIS Image Processing. Conidia sizes are reported as the minimum and maximum in parentheses, and the average is reported with the standard deviation.
4.2. Molecular Characterization and Phylogeny
Genomic DNA of the selected isolates (Di3A-Pef 1-2-3-4-5) was extracted using the Gentra Puregene Yeast/Bact kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The internal transcribed spacer of the ribosomal DNA (rDNA-ITS), partial translation elongation factor alpha gene (EF-1α) and RNA polymerase II gene (RPB2) were targeted for PCR amplification and sequencing. The primers used for these regions were: ITS5 and ITS4 for ITS [27], EF1-728F and EF1-986R for EF-1α [28] and 5f2 and 7cr for RPB2 [29]. The PCR products were purified and sequenced in both directions by Macrogen Inc. (Seoul, Korea). The sequences were edited using MEGAX: Molecular Evolutionary Genetics Analysis across computing platforms [30], manual adjustments of alignments were made when necessary and submitted to GenBank. Moreover, the sequences were blasted in the NCBIs GenBank nucleotide database and on the Fusarium MLST database of the Westerdijk Fungal Biodiversity Institute (http://www.westerdijkinstitute.nl/fusarium/, accessed on 21 May 2021). For comparison, 44 additional sequences were selected according to the recent literature [20] (Table 1). The phylogenetic analysis was based on the Maximum Parsimony (MP). The MP analysis was done using PAUP v. 4.0a165 [31]. Phylogenetic relationships were estimated by heuristic searches with 100 random addition sequences. A tree bisection-reconnection was used, with the branch swapping option set to ‘best trees’ only, with all characters weighted equally and alignment gaps treated as the fifth state. The tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency index (RC) were calculated for parsimony and the bootstrap analyses were based on 1000 replicates [32]. Fusarium foetens (CBS 120665) and F. udum (CBS 12881) served as outgroups.
Table 1.
Characteristics of Fusarium isolates included in the phylogenetic analysis.
| Species | Culture Accession | Host/Substrates | Special form | Origin | GeneBank Accession | |
|---|---|---|---|---|---|---|
| rpb2 | EF1-α | |||||
| Fusarium callistephi | CBS 187.53 | Callistephus chinensis | callistephi | The Netherlands | MH484875 | MH484966 |
| F. carminascens | CBS 144739 | Zea mays | South Africa | MH484934 | MH485025 | |
| F. cugenengense | CBS 620.72 | Crocus sp. | gladioli | Germany | MH484879 | MH484970 |
| F. cugenengense | CBS 130304 | Gossypium barbadense | vasinfectum | China | MH484921 | MH485012 |
| F.curvatum | CBS 247.61 | Matthiola incana | matthiolae | Germany | MH484876 | MH484967 |
| F.curvatum | CBS 238.94 | Beaucarnia sp. | meniscoideum | The Netherlands | MH484893 | MH484984 |
| F. duoseptatum | CBS 102026 | Musa sapientum | cubense | Malaysia | MH484896 | MH484987 |
| F. elaeidis | CBS 217.49 | Elaeis sp. | elaeidis | Zaire | MH484870 | MH484961 |
| F. fabacearum | CBS 144742 | Zea mays | South Africa | MH484938 | MH485029 | |
| F. foetens | CBS 120665 | Nicotiana tabacum | Iran | MH484918 | MH485009 | |
| F. glycines | CBS 144746 | Glycine max | South Africa | MH484942 | MH485033 | |
| F. glycines | CBS 20089 | Ocimum basilicum | basilici | Italy | MH484888 | MH484979 |
| F. glycines | CBS 17633 | Linum usitatissium | lini | Unknown | MH484868 | MH484959 |
| F. glycines | CBS 21449 | Unknown | Argentina | MH484869 | MH484960 | |
| F. gossypinum | CBS 116611 | Gossypium hirsutum | vasinfectum | Ivory Coast | MH484907 | MH484998 |
| F. hoodiae | CBS 132474 | Hoodia gordonii | hoodiae | South Africa | MH484929 | MH485020 |
| F. languescens | CBS 41390 | Solanum lycopersicum | lycopersici | Israel | MH484890 | MH484981 |
| F. languescens | CBS 119796 | Zea mays | South Africa | MH484917 | MH485008 | |
| F. languescens | CBS 30,291 | Solanum lycopersicum | lycopersici | The Netherlands | MH484892 | MH484983 |
| F. languescens | CBS 646.78 | Solanum lycopersicum | lycopersici | Morocco | MH484881 | MH484972 |
| F. nirembergiae | CBS 744.79 | Passiflora edulis | passiflorae | Brazil | MH484882 | MH484973 |
| F. nirembergiae | CBS 115424 | Agothosma betulina | South Africa | MH484906 | MH484997 | |
| F. nirembergiae | CBS 12924 | Secale cereale | Unknown | MH484864 | MH484955 | |
| F. nirembergiae | CBS 12781 | Chrysanthemum sp. | chrysanthemi | USA | MH484883 | MH484974 |
| F. nirembergiae | CBS 130303 | Solanum lycopersicum | radicis-lycopersici | USA | MH484923 | MH485014 |
| F. nirembergiae | CBS 115417 | Agothosma betulina | South Africa | MH484903 | MH484994 | |
| F. nirembergiae | CBS 19687 | Bouvardia longiflora | bouvardiae | Italy | MH484886 | MH484977 |
| F. nirembergiae | CBS 123062 | Tulip roots | USA | MH484919 | MH485010 | |
| F. nirembergiae | CBS 18132 | Solanum tuberosum | USA | MH484867 | MH484958 | |
| F. nirembergiae | CBS 75868 | Solanum lycopersicum | lycopersici | The Netherlands | MH484877 | MH484968 |
| F. nirembergiae | CBS 840.88 | Dianthus caryophyllus | dianthi | The Netherlands | MH484887 | MH484978 |
| F. oxysporum | CBS 221.49 | Camellia sinensis | medicaginis | South East Asia | MH484872 | MH484963 |
| F. oxysporum | CPC 25822 | Protea sp. | South Africa | MH484943 | MH485034 | |
| F. oxysporum | CBS 144134 | Solanum tuberosum | Germany | MH484953 | MH485044 | |
| F. pharetrum | CBS 144751 | Aliodendron dichotomum | South Africa | MH484952 | MH485043 | |
| F. pharetrum | CBS 144750 | Aliodendron dichotomum | South Africa | MH484951 | MH485042 | |
| F. trachichlamydosporum | CBS 102028 | Musa sapientum | cubense | Malaysia | MH484897 | MH484988 |
| F. triseptatum | CBS 258.50 | Ipomea batatas | batatas | USA | MH484873 | MH484964 |
| F. triseptatum | CBS 116619 | Gossypium hirsutum | vasinfectum | Ivory Coast | MH484910 | MH485001 |
| F. udum | CBS 177.31 | Digitaria ariantha | South Africa | MH484866 | MH484957 | |
| Fusarium sp. | CBS 12881 | Chrysanthemum sp. | chrysanthemi | USA | MH484884 | MH484975 |
| Fusarium sp. | CBS 130323 | Human nail | Australia | MH484927 | MH485018 | |
| Fusarium sp. | CBS 68089 | Cucumis sativus | cucurbitacearum | The Netherlands | MH484889 | MH484980 |
4.3. Pathogenicity Tests
In order to fulfil Koch’s postulates, pathogenicity tests were conducted on one-year-old potted cuttings using the mycelial plug technique. In detail, 18 healthy cuttings were inoculated, removing a piece of bark of the crown root with a scalpel blade and applying a mycelial plug (0.3 cm2), taken from a 14-day-old Di3A-Pef 1 isolate, upside down on the wound and subsequently covered with soil to prevent desiccation. The controls consisted of sterile PDA plugs applied as described above to the same number of healthy young plants. Re-isolation attempts were performed from representative inoculated plants.
Acknowledgments
All authors are grateful to all technicians and growers who supported this research.
Author Contributions
Conceptualization, D.A., A.V. and G.P.; methodology, D.A., A.F. and G.R.L.; software, A.F.; validation, D.A., A.V. and G.P.; formal analysis, A.V.; investigation, A.F. and G.R.L.; resources, A.V. and G.P.; data curation, D.A., A.F. and A.V.; writing—original draft preparation, D.A., A.F. and A.V.; writing—review and editing, A.V. and G.P.; visualization, A.V.; supervision, G.P.; project administration, G.P.; funding acquisition, A.V. and G.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the following grants: Programma Ricerca di Ateneo MEDIT-ECO UNICT 2020–2022 Linea 2-University of Catania (Italy); Starting Grant 2020, University of Catania (Italy); Fondi di Ateneo 2020–2022, University of Catania (Italy), Linea Open Access. Research Project 2016–2018, University of Catania 5A722192134.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ding P. Tropical fruits. In: Thomas B., Murphy D.J., Murray B.G., editors. Encyclopedia of Applied Plant Sciences. 2nd ed. Academic Press; Oxford, UK: 2017. pp. 431–434. [Google Scholar]
- 2.Underhill S. Fruits of Tropical Climates: Commercial and Dietary Importance. In: Caballero B., editor. Encyclopedia of Food Sciences and Nutrition. 2nd ed. Academic Press; Oxford, UK: 2003. pp. 2780–2785. [Google Scholar]
- 3.Cervi A.C. O Gênero Passiflora (Passifloraceae) No Brasil, Es- Pécies Descritas Após o Ano de 1950. Real Jardin Botánico; Madrid, Spain: 2006. [Google Scholar]
- 4.Manicom B., Ruggiero C., Ploetz R.C., de Goes A. Diseases of passion fruit. In: Ploetz R.C., editor. Diseases of Tropical Fruit Crops. CAB International; Wallingford, UK: 2003. pp. 413–441. [Google Scholar]
- 5.Vanderplank J. Passion Flowers. 2nd ed. MIT Press; London, UK: 1996. p. 224. [Google Scholar]
- 6.Das M.R., Hossain T., Mia M.B., Ahmed J., Kariman A.S., Hossain M.M. Fruit setting behavior of passion fruit. Am. J. Plant Sci. 2013;4:1066–1073. doi: 10.4236/ajps.2013.45132. [DOI] [Google Scholar]
- 7.NDA-ARC . Growing Granadillas. Institute of Tropical and Subtropical Crops; Pretoria, South Africa: 1999. pp. 1–9. [Google Scholar]
- 8.Rao B.N., Jha A.K., Deo C., Kumar S., Roy S.S., Ngachan S.V. Effect of irrigation and mulching on growth, yield and quality of passion fruit (Passiflora edulis Sims.) J. Crop Weed. 2013;9:94–98. [Google Scholar]
- 9.Fischer I.H., Lourenco S.A., Martins M.C., Kimati H., Amorim L. Seleção de plantas resistentes e de fungicidas para o controle da podridão do colo do maracujazeiro causada por Nectria hematococca. Fitopatol. Bras. 2005;30:250–258. doi: 10.1590/S0100-41582005000300006. [DOI] [Google Scholar]
- 10.McKnight T. A wilt disease of the passion vines (Passiflora edulis) caused by a species of Fusarium. Queensl. J. Agric. Sci. 1951;8:1–4. [Google Scholar]
- 11.Garcia E., Paiva D., Costa J., Portugal A., Ares A. First report of Fusarium wilt caused by Fusarium oxysporum f. sp. passiflorae on Passion Fruit in Portugal. Plant Dis. 2019;103:2680. [Google Scholar]
- 12.Liberato J.R., Costa H. Maracujá: Tecnologia de produção, Pós-Colheita, Agroindústria, Mercado. Cinco Continentes; Porto Alegre, Brazil: 2001. Doenças fúngicas, bacterianas e fitonematóides; pp. 243–276. [Google Scholar]
- 13.Rooney-Latham S., Blomquist C.L., Scheck H.J. First report of Fusarium wilt caused by Fusarium oxysporum f. sp. passiflorae on Passion fruit in north America. Plant Dis. 2011;95:1478. doi: 10.1094/PDIS-03-11-0261. [DOI] [PubMed] [Google Scholar]
- 14.Fischer I.H., Rezende J.A.M. Diseases of passion flower (Passiflora spp.) Pest Technol. 2008;2:1–19. [Google Scholar]
- 15.Li D.F., Yang J.Q., Zhang X.Y., Sun L.F. Identification of the pathogen causing collar rot of passion fruit in Fujian. Acta Phytopathol. Sin. 1993;23:372. [Google Scholar]
- 16.Ploetz R.C. Sudden wilt of passionfruit in southern Florida caused by Nectria haematococca. Plant Dis. 1991;75:1071–1073. doi: 10.1094/PD-75-1071. [DOI] [Google Scholar]
- 17.Viana F.M.P., Costa A.F. Doenças do maracujazeiro. In: Freire F.C.O., Cardoso J.E., Viana F.M.P., editors. Doenças de Fruteiras Tropicais de Interesse Agroindustrial. Embrapa informação Tecnológica; Brasília, Brazil: 2003. pp. 276–285. [Google Scholar]
- 18.Bezerra J.L., De Oliveira M.L. Damping-off of passion fruit caused by Rhizoctonia sp. [Passiflora edulis] Fitopatol. Brasil. 1984;9:273–276. [Google Scholar]
- 19.Young B.R. Root rot of passionfruit vine (Passiflora edulis Sims.) in the Auckland area, New Zealand. J Agric. Res. 1970;13:119–125. [Google Scholar]
- 20.Lombard L., Sandoval-Denis M., Lamprecht S.C., Crous P.W. Epitypification of Fusarium oxysporum clearing the taxonomic chaos. Persoonia. 2019;43:1–47. doi: 10.3767/persoonia.2019.43.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crous P., Lombard L., Sandoval-Denis M., Seifert K., Schroers H.-J., Chaverri P., Gené J., Guarro J., Hirooka Y., Bensch K., et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021;98:100116. doi: 10.1016/j.simyco.2021.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X., Li H., Zhou L., Chen F., Chen F. Wilt of Acer negundo L. caused by Fusarium nirenbergiae in China. J. For. Res. 2020;31:2013–2022. doi: 10.1007/s11676-019-00996-9. [DOI] [Google Scholar]
- 23.Bennett R.S., Davis R.M. Method for rapid production of Fusarium oxysporum f. sp. vasinfectum chlamydospores. J. Cotton Sci. 2013;17:52–59. [Google Scholar]
- 24.De Carvalho J.A., de Jesus J.G., Araujo K.L., Serafim M.E., Gilio T.A.S., Neves L.G. Passion fruit (Passiflora spp.) species as sources of resistance to soil phytopathogens Fusarium solani and Fusarium oxysporum f. sp. passiflorae complex. Rev. Bras. Frutic. 2021;43 doi: 10.1590/0100-29452021427. [DOI] [Google Scholar]
- 25.Silva A.S., Oliveira E.J., Haddad F., Jesus O.N., Oliveira S.A.S., Costa M.A.P. Variação genética em isolados de Fusarium oxysporum f. sp. passiflorae com marcadores AFLP. Sci. Agric. 2013;70:108–115. [Google Scholar]
- 26.Nirenberg H.I. A simplified method for identifying Fusarium spp. occurring on wheat. Can. J. Bot. 1981;59:1599–1609. doi: 10.1139/b81-217. [DOI] [Google Scholar]
- 27.White T.J., Bruns T., Lee S., Taylor J.L. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innes M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; New York, NY, USA: 1990. pp. 315–322. [Google Scholar]
- 28.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- 29.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 30.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swofford D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), v. 4.0b10. Sinauer Associates; Sunderland, MA, USA: 2003. [Google Scholar]
- 32.Hillis D.M., Bull J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993;42:182–192. doi: 10.1093/sysbio/42.2.182. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.