Abstract
Interleukin-32 (IL-32) is well known as a proinflammatory cytokine that is expressed in various immune cells and cancers. However, the clinical relevance of IL-32 expression in cutaneous melanoma has not been comprehensively studied. Here, we identified the prognostic value of IL32 expression using various systematic multiomic analyses. The IL32 expressions were significantly higher in cutaneous melanoma than in normal tissue, and Kaplan–Meier survival analysis showed a correlation between IL32 expression and good prognosis in cutaneous melanoma patients. In addition, we analyzed the correlation between IL32 expression and the infiltration of natural killer (NK) cells to identify a relevant mechanism between IL32 expression and prognosis in cutaneous melanoma (p = 0.00031). In the relationship between IL32 expression and the infiltration of NK cells, a negative correlation was found in resting NK cells (rho = −0.38, p = 3.95 × 10−17) whereas a strong positive correlation was observed only in active NK cells (rho = 0.374, p = 1.23 × 10−16). Moreover, IL32 expression was markedly positively correlated with the cytolytic molecules, such as granzyme and perforin. These data suggest that IL32 expression may increase patient survival through the infiltration and activation of NK cells, representative anticancer effector cells, in cutaneous melanoma. Collectively, this study provides the prognostic value of IL32 expression and its potential role as an effective predictive biomarker for NK cell infiltration in cutaneous melanoma.
Keywords: interleukin-32 (IL-32), cutaneous melanoma, multiomic analysis, immune cell infiltration, natural killer (NK) cells
1. Introduction
Despite its relatively low incidence rate, melanoma is the most critical type of cancer due to its high levels of malignancy and invasive activity [1,2]. Cutaneous melanoma, a type of melanoma, is characterized by high metastasis and poor prognosis; therefore, it accounts for 75% of skin-related deaths [3]. In recent decades, the incidence of cutaneous melanoma has continued to increase worldwide, with approximately 96,000 new cases in 2019 [4]. Various methods have been used to treat melanoma, but achieving significant therapeutic effects is difficult due to its high resistance to classical therapies, such as chemotherapy and radiotherapy [5,6,7]. Recently, various immunotherapies, such as vaccines and immuno-checkpoint inhibitors, have attempted to improve the side effects and to overcome resistance. To date, monoclonal antibodies targeting the checkpoint molecules cytotoxic T lymphocyte-associated protein 4 (CTLA-4) and programmed cell death protein 1 (PD-1), and the ligand PD-L1 are well known as the most effective immunotherapies [8,9]. These tumor immunotherapies have been established as key treatments for the clinical treatment of cancer [10]. The composition and immune contexture of the tumor microenvironment (TME) are closely related to the successful response of immunotherapy, and an increase in specific cells in the TME is associated with increased survival of patients with various types of cancer [11,12,13]. Thus, the interaction of melanoma cells with immune cells within the TME significantly influences tumor proliferation, differentiation, and progression [14,15]. Melanoma is a major immunogenic tumor, and the number of related studies using immunotherapy to control immune responses is increasing. In particular, melanoma cells are known to regulate their close association with the TME to promote tumor progression, invasion, and metastasis [16,17]. Composed of a complex network of immune cells, various growth factors, and cytokines, the TME is closely associated with melanoma, which affects the treatment effect [18].
The presence, localization, and phenotype of tumor-infiltrating lymphocytes (TILs) within the TME have been predicted to determine immunotherapy and the key regulators of disease progression [19,20,21]. TILs contain not only effector cells but also immunosuppressive cells such as regulatory T lymphocytes, tumor associated macrophage (TAM), and myeloid-derived suppressor cells (MDSC) as contributors to anticancer effects [22,23]. Therefore, an infiltration of effector cells including natural killer (NK) cells and CD8+ T cells in the tumor is highly correlated with a good prognosis in cancer patients [24,25]. The presence and activation of NK cells in the TME is associated with tumor suppression [26,27,28,29]. NK cells, a major effector cell of the innate immune system, are the first line of defense in the anticancer immune system [30]. When NK cells are activated, cytolytic molecules are released to induce apoptosis of tumor cells, and cytokines such as interferon-gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α) are produced to regulate adaptive immune T cell-mediated immune responses [26,27,28,29]. NK cell receptors induced by proinflammatory cytokines are associated with NK cell activity in patients with melanoma and are known to improve cell toxicity in tumors [31]. To predict the prognosis of cutaneous melanoma, understanding the role of TILs in the tumor environment is necessary.
Interleukin-32 (IL-32) was first demonstrated as a natural killer cell transcript (NK4), which was detected in activated NK cells and T cells [32]. IL-32 contains eight small exons and is located on human chromosome16p13.3 [33]. IL-32, a proinflammatory cytokine differentially expressed in IL-18-responsive cells, induces the expressions of TNF-α, IL-1β, and C–X–C chemokine family members and activates the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and p38 mitogen-activated protein kinase (MAPK) pathways [34,35,36]. IL-32 is more highly expressed in immune cells than in nonimmune cells and is closely related to anticancer effects in various types of cancer [37,38]. IL-32 has been known to affect tumor death by regulating immune cells including NK cells. NK cells release TNF-related apoptosis-inducing ligand (TRAIL), TNF, Fas ligand (FASL), and death receptor 3 (DR3) ligand to induce cancer cell death. Moreover, IL-32 expression increased the number of NK cells and CD8+ T cells in blood and recruited NK cells and CD8+ T cells in tumor tissues. Recently, a novel role of IL-32 for immunotherapy has been suggested by identifying the mechanism by which IL-32 primes CD8+ T cells and recruits activating intratumoral DCs and macrophages [39]. In contrast, several reports show the procancer effects of IL-32. The overexpression of IL-32 increases the tumor size and lymph node metastasis in breast cancer, and IL-32 expression is also associated with tumor metastasis and cancer cell migration in gastric and lung cancers [40,41,42].
As such, while studies with conflicting results have reported contradictory effects of IL-32 in various tumors, a comprehensive analysis of the clinical relevance of IL32 expression has yet to be performed [37,38,43]. Based on various databases, we investigated the correlation between IL32 expression levels and cancer patient outcomes. In addition, the correlation between IL32 expression and NK cells, which are effector cells, was analyzed to confirm the impact on cutaneous melanoma survival rate. The results of this study identified the important role of IL32 in cutaneous melanoma and provided its interaction and function with TILs.
2. Materials and Methods
2.1. IL32 mRNA Expression in Various Tumors
Gene Expression Profiling Interactive Analysis (GEPIA2; http://gepia.cancer-pku.cn/, accessed on 21 March 2021) was used to compare the IL32 mRNA expression between various tumors and their normal tissues. GEPIA2 provides customizable functionalities based on data from The Cancer Genome Atlas (TCGA) and the Genotype Tissue Expression project (GTEx) [44]. GEPIA2 also offers IL32 mRNA levels in cutaneous melanoma (SKCM; skin cutaneous melanoma) and normal tissues. The transcription of IL32 expression levels between SKCM and normal tissues was shown with box plots.
2.2. Analysis of the Correlation between IL32 Expression and Prognostic Value
To estimate the correlation between IL32 mRNA expression and patient survival in SKCM, various web tools were used. The prognostic value of IL32 expression was analyzed in TCGA datasets using the OncoLnc (http://www.oncolnc.org/, accessed on 28 March 2021) database and GEPIA2. We compared the survival of two groups of patients with low and high IL32 mRNA expression levels and provided hazard ratios (HR) with 95% confidence intervals and Kaplan–Meier (KM) survival curves, and the log-rank p-values were analyzed using GEPIA2. We used the TIMER databases to evaluate the clinical relevance of IL32 expression. TIMER v.1.0 provides KM plots to visualize the survival differences and p-values of the log-rank test to compare the survival curves (log-rank p < 0.05). HR indicates the hazard ratio, and its lower and upper 95% confidential intervals (CIs) were shown in each plot [45]. TIMER v.2.0 also offers Cox regression results including Z-score and statistical significance p-value. A KM plot was performed using a Cox proportional hazard model with clinical factors including race, gender, and tumor stage. Z-scores were compared to assess whether the outcome of the gene expression modulated by clinical factors was significant (increased risk: p < 0.05, Z > 0; decreased risk: p < 0.05, Z < 0; and not significant: p > 0.05) [46]. The Cox regression results of the clinical factors including gender, tumor stage, race, and tumor purity are presented in Supplementary Table S1.
2.3. Analysis of IL32 Gene Mutations in Skin Cutaneous Melanoma (SKCM)
A comprehensive open-source platform, cBioportal (http://www.cbioportal.org/, accessed on 2 April 2021) provides various cancer genomic datasets. To investigate the IL32 gene mutation in SKCM, we used the cBioportal database version 3.2.14, which provides various cancer genomic datasets [47,48]. The mutation diagram of the IL32 gene was generated using default parameter settings. The statistical analysis was performed using an unpaired t-test of GraphPad 7 software. The genomic alterations of IL32 include copy number amplification (CNA), deletion, and nonsense and missense mutations with unknown significance. In addition, we analyzed the promoter methylation of IL32 using the UALCAN for the TCGA-SKCM dataset. The statistical method of UALCAN was used to analyze changes in the expression levels between normal and other tumor grades, and p < 0.05 was considered statistically significantly different.
2.4. Correlation between IL32 Expression and Infiltration of Various Immune Cells
Tumor immune system interactions (TISIDB, http:// cis.hku.hk./TISIDB/index.php, accessed on 25 May 2021) is a database used to analysis the relative abundance of tumor immune systems and TILs. Here, it was used to determine the interactions between IL32 and TILs in SKCM. TIMER is an online web tool used to systematically analyze the correlation of immune infiltrates with various factors such as gene expression and prognostic values. TIMER v.1.0 was used to analyze the comprehensive correlation between IL32 mRNA and tumor-infiltrating immune cell signatures. The correlation between IL32 expression levels and the abundances of immune infiltrates including tumor purity, B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells, was visualized using scatter plots. In addition, we analyzed the correlation between IL32 expression and the gene markers of infiltrating NKs using the GEPIA2. We used TIMER v.2.0 (http://timer.cistrome.org/, accessed on 5 April 2021) to confirm the significance of IL32 expression with activated NK cells [49]. In addition, we compared the activated NK cell infiltration levels and resting NK cell infiltration levels using CIBERSORT in TIMER v.2.0 to determine the interactions between IL32 and TILs in SKCM.
2.5. Correlation between Coexpressed Genes and IL32
We used the TCGA-SKCM dataset in cBioportal to analyze the coexpression genes of IL32 expression. Then, we identified the 25 strongest correlated genes with the highest Spearman correlation value and the lowest p-value. We used the UCSC Xena browser (https://xena.ucsc.edu/, accessed on 13 May 2021) to analyze the correlation between IL32 and the gene with the highest positive correlation using a heatmap and scatter plot with TCGA-SKCM. The R2 database was used to visualize the correlation of IL32 with the highest positive correlation using the Tumor melanoma metastasis Bhardwaj-44 dataset via scatter plot. We also analyzed the correlation between IL32 and its altered genes to identify gene ontology terms using Enricher (http://amp.pharm.mssm.edu/Enrichr/, accessed on 25 April 2021).
3. Results
3.1. The Expression Analysis of IL32 in Various Types of Cancers
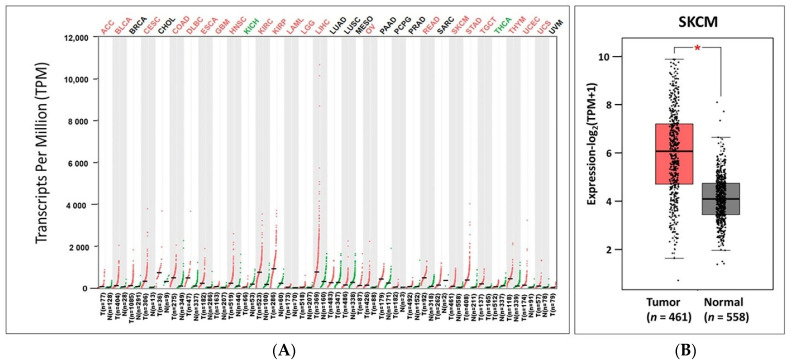
To analyze IL32 mRNA expression in tumors and normal tissues, we identified IL32 mRNA levels using various databases. GEPIA2 showed that the IL32 mRNA expression levels were significantly elevated in most types of tumors, including cutaneous melanoma (SKCM; skin cutaneous melanoma), as indicated in red (Figure 1A). However, the level of IL32 mRNA expression was higher in normal tissues than in tumor tissues of kidney chromophobe (KICH) and thyroid carcinoma (THCA), as indicated in green. Detailed findings of particular tumor types are compiled in Supplementary Table S2. We further compared the mRNA levels of IL32 between SKCM (461 samples) and normal (558 samples) tissues based on data from TCGA and GTEx. Figure 1B shows that the IL32 expression was significantly higher in SKCM tissues than in normal tissues (p < 0.05). Collectively, the data from all databases showed that IL32 mRNA expression in SKCM was markedly higher than that in normal tissues.
Figure 1.
IL32 mRNA expression levels in various types of cancers and their normal tissues. (A) Difference in IL32 expressions between tumor (red) and normal tissues (green) using the GEPIA2 database. Supplementary Table S2 presents the abbreviations of various types of cancers. (B) The box plots represent the IL32 mRNA expressions in SKCM (n = 461) and normal tissues (n = 558) using the GEPIA2. * p < 0.05.
3.2. Correlation between IL32 Expression and Patient Survival in Various Types of Cancers
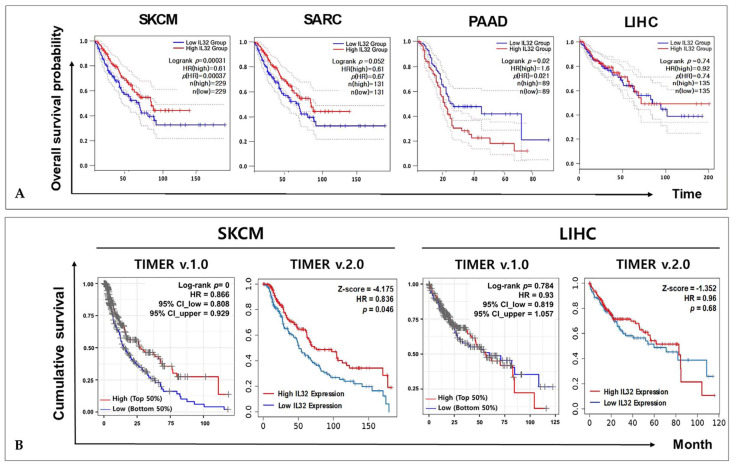
To investigate the correlation between IL32 mRNA expression and patient survival rate in various types of cancers, the overall survival probabilities were compared using the Cox regression model and the OncoLnc online tool. The Cox regression results for IL32 mRNA in various cancer types are shown in Supplementary Table S3. A Cox regression analysis for IL32 expression was performed for four types of cancers: SKCM, sarcoma (SARC), pancreatic adenocarcinoma (PAAD), and liver hepatocellular carcinoma (LIHC) (p < 0.01). As shown in Figure 2A, we found that a higher IL32 expression level was correlated with a better overall survival of SKCM (log-rank p = 0.00031) and PAAD (log-rank p = 0.02). No significant correlation was found between patient survival in SARC (log-rank p = 0.052) and LIHC (log-rank p = 0.74). The correlations between IL32 expression and disease free survival (DFS) of various types of cancers by GEPIA2 are shown in Supplementary Figure S4. Interestingly, the IL32 mRNA expression in LIHC was significantly elevated, as in SKCM (Figure 1A); therefore, LIHC was used as a control in subsequent analyses. To evaluate the clinical relevance of IL32 expression, we further identified the cumulative survival of patients with SKCM and LIHC using TIMER web tools. The KM plots were performed on a Cox proportional hazard model to determine the significance of IL32 expression on outcome, and the Cox regression results including log-rank p-value and Z-score were provided. As shown in Figure 2B, a higher IL32 expression was associated with a better prognosis in SKCM (log-rank p = 0, HR = 0.866) in the TIMER v.1.0 database. Moreover, the TIMER v.2.0 database also shows that an IL32 mRNA expression was associated with good prognosis for SKCM patients (Z-score = –4.175). The Cox regression results adjusted by clinical factors such as race, age, gender, and tumor stages are shown in Supplementary Table S1. In contrast, no significant correlation was found between IL32 expression and patient survival in LIHC (log-rank p = 0.784, HR = 0.93, Z-score = −1.352). These findings suggest that IL32 mRNA expression influences the prognosis of patients with SKCM.
Figure 2.
Correlation between IL32 expression and the prognosis of various types of cancers. Kaplan–Meier survival curves generated using the GEPIA2 website indicate that IL32 expression is higher (red) and lower (blue) than the median value of the TCGA data (p < 0.05): (A) SKCM, SARC, PAAD, and LIHC; (B) cumulative survival curves demonstrating the survival rates of patients with high (blue) or low (red) IL32 expression using the TIMER web tools (log-rank p < 0.05, Z < 0).
3.3. Genome Change of IL32 Expression in Melanoma
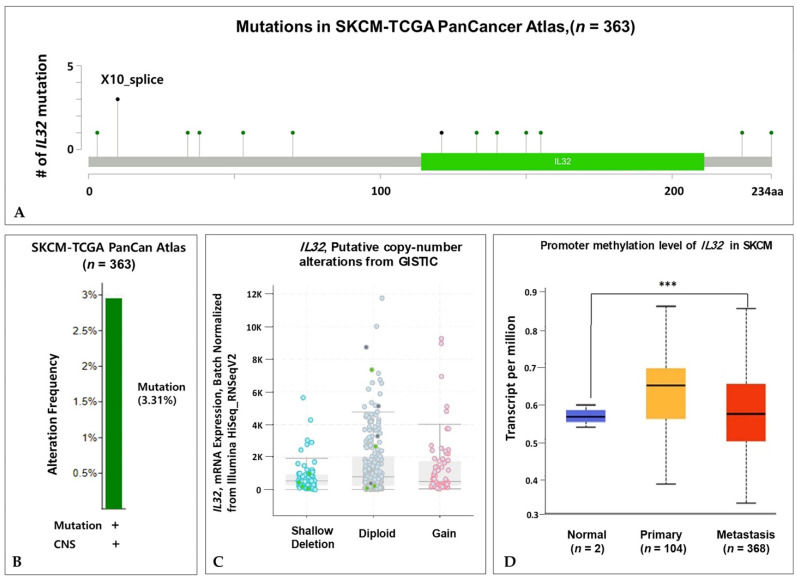
We explored the status of IL32 gene alteration in SKCM using cBioportal. A total of 363 patient samples were included from the TCGA database, and mutation types including 11 missense, 3 splice, and 1 nonsense mutation(s) were observed in the IL32 coding region (Figure 3A). The X10 splice mutation was confirmed three times more often than the other mutations. Fifteen mutations in the IL32 gene are shown in Supplementary Table S5. Figure 3B shows that the alteration frequency of the IL32 gene was 3.3% in the TCGA PanCan Atlas dataset. Additionally, we analyzed the correlation between BRAF mutation status and IL32 expression to identify other mutations in SKCM. BRAF mutations have been observed in several types of cancer, such as melanoma and colorectal cancer, and are associated with cancer cell growth and proliferation [50]. As shown in Supplementary Figure S6, no significant difference was found between mutated BRAF and wild-type BRAF with IL32 expression in melanoma (p = 0.064, p < 0.05). To further examine whether the IL32 CNA status was associated with mRNA expression, we analyzed IL32 mRNA expression for each CNA status. The IL32 expression was markedly lower in the shallow deletion samples than in the diploid and gain samples (Figure 3C). However, we found no significant differences in the IL32 expression between the diploid and gain samples. These data suggest that a shallow deletion of the CNA status could contribute to the high expression of IL32 in SKCM. To further analyze the methylation status of the IL32 gene in SKCM, we investigated the TCGA-SKCM dataset using the UALCAN database. As shown in Figure 3D, promoter methylation was significantly increased in metastatic melanoma (p = 5.95 × 10−5) compared with that in primary tissues (p = 4.23 × 10−1). Promoter methylation is an epigenetic regulator, and increased methylation is indicative of tumors [51]. Taken together, these results suggest that a positive correlation is found between DNA methylation and the mRNA expression of IL32.
Figure 3.
Genome alterations and IL32 expression in SKCM. (A) The lollipop plot shows the type and location of each mutation frequency in the TCGA-SKCM PanCan Atlas dataset using the cBioPortal database (n = 363). (B) The mutations of the IL32 gene expression are presented in SKCM. (C) The correlation between IL32 expression and CNA status of shallow deletion (SD), diploid (D), and gain (G). (D) The box plots reveal the promoter methylation levels of IL32 in the TCGA-SKCM dataset using the UALCAN database. Normal vs. Primary (p = 4.23 × 10−1), Normal vs. Metastasis (p = 5.95 × 10−5), and Primary vs. Metastasis (p = 9.15 × 10−1); *** p < 0.001.
3.4. Correlation of IL32 Expression with Immune Infiltrates
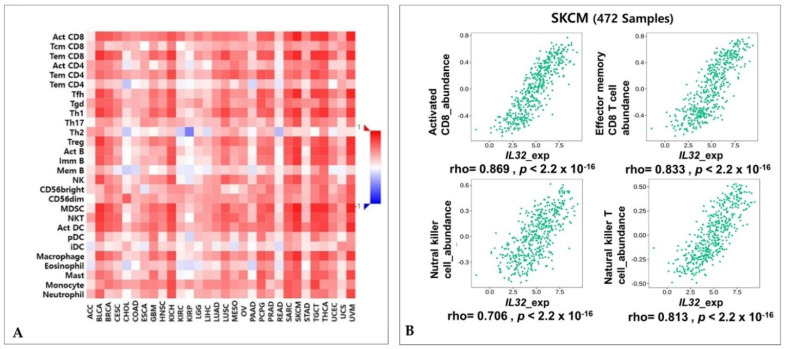
The fact that IL32 expression is involved in various cancer malignancies, including breast cancer (BRCA) and colon adenocarcinoma (COAD), is well known. However, our data showed that a high IL32 expression is associated with high survival rates. To determine the mechanism associated with clinical relevance, we investigated the correlation between IL32 expression levels and immune cell infiltration in SKCM. Supplementary Figure S7 shows that an analysis of the data using TIMER v.1.0 shows that a positive correlation is found between IL32 expression and immune cell infiltration in SKCM. In contrast, the IL32 expression levels were not significantly correlated with tumor purity and immune infiltrates in LIHC. These results demonstrate that IL32 in SKCM may be expressed by infiltrated immune cells. We further identified significant correlations of IL32 with 28 types of TILs among various cancers using the web portal TISIDB (Figure 4A). In Figure 4A, the IL32 expression has a positive correlation with various immune cells in many types of cancer. In the correlation between IL32 and NK cell infiltration, only five types of cancer including SKCM show significant positive correlations (Supplementary Figure S8). IL32 expression was markedly correlated with the abundance of NK cells (rho = 0.706, p < 2.2 × 10−16), natural killer T (NKT) cells (rho = 0.813, p < 2.2 × 10−16), activated CD8+ T cells (rho = 0.869, p < 2.2 × 10−16), and effector memory CD8+ T cells in SKCM (rho = 0.833, p < 2.2 × 10−16) (Figure 4B). Overall, these data suggest that the expression of higher IL32 is significantly involved in activated NK cell and CD8+ T cell infiltration and suggest that this leads to the antitumor activity of the effector cell.
Figure 4.
Correlations between IL32 expression with lymphocytes. (A) Correlation between the expressions of IL32 and lymphocytes in various cancer types. (B) The expression of IL32 was significantly correlated with the immune cells in SKCM: activated CD8+ T cell, effector CD8+ T cell, natural killer (NK) cell, and NKT T cell (rho > 0.4, p < 0.001).
3.5. Correlation between IL32 Expression and Various Subsets of Immune Cells in Melanoma
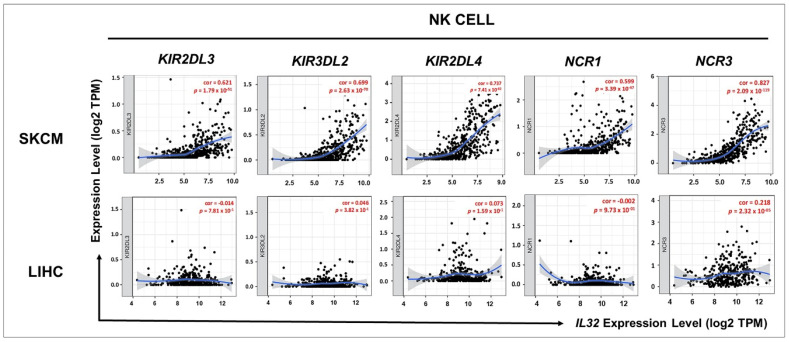
To further investigate the correlation between IL32 expression and various subsets of immune cell infiltrations in SKCM, we analyzed the correlations between IL32 and immune cell markers, including subsets of each immune cell in SKCM (Supplementary Table S9 and Figure 5). Supplementary Table S9 shows the immune cell markers including subsets of T cells (general T cells, CD8+ T cells, CD4+ T cells, regulatory T cells, and T cell exhaustion), B cells, monocytes, NK cells, TAM, M1 and M2 macrophages, and neutrophils in SKCM. These data reveal that IL32 expression is correlated significantly with most of the immune marker genes of NK cells in SKCM and that IL32 expression was significantly correlated with NK cells in SKCM but not in LIHC. Therefore, to examine the association between IL32 expression and the infiltration of NK cell subsets, the correlation between IL32 expression and gene marker expression in each immune cell was examined in SKCM. As shown in Figure 5, IL32 expression was positively correlated with KIR2DL3 (cor = 0.621, p = 1.79 × 10−51), KIR3DL2 (cor = 0.699, p = 2.63 × 10−70), KIR2DL4 (cor = 0.737, p = 7.41 × 10−82), NCR1 (cor = 0.599, p = 3.39 × 10−47), and NCR3 (cor = 0.827, p = 2.09 × 10−119) gene expressions in SKCM, whereas IL32 expression was not significantly correlated with the expression of gene markers in LIHC (KIR2DL3, cor = −0.014, p = 7.81 × 10−1; KIR3DL2, cor = 0.046, p = 3.82 × 10−1; KIR2DL4, cor = 0.073, p = 1.59 × 10−1; NCR1, cor = −0.002, p = 9.73 × 10−1; and NCR3, cor = 0.218, p = 2.32 × 10−5). Moreover, Figure 5 and Table 1 show that the expression of IL32 and the expression of NK cell markers are not correlated in LIHC but are highly correlated in SKCM, which is also shown in data from the TIMER database. Taken together, these data suggest that IL32 expression is markedly correlated with infiltrated NK cells in SKCM.
Figure 5.
Correlation of IL32 expression with NK markers. The correlation between IL32 expression and various gene markers of NK was examined using the TIMER v.1.0 web tool. The KIR2DL3, KIR3DL2, KIR2DL4, NCR1, and NCR3 genes were used as gene markers for NK cells. IL32 expression is positively related to the expression of gene markers for NK cells in SKCM. IL32 expression is not significantly correlated with the expression of most gene markers in LIHC. Supplementary Table S10 provides the p-values and correlation constants.
Table 1.
Correlation between IL32 and NK cell markers in GEPIA2.
| Gene Markers | SKCM | LIHC | |||
|---|---|---|---|---|---|
| R | p | R | p | ||
| NK cell | KIR2DL1 | 0.33 | *** | −0.0046 | 0.93 |
| KIR2DL3 | 0.35 | *** | −0.055 | 0.29 | |
| KIR2DL4 | 0.6 | 0 | 0.056 | 0.28 | |
| KIR3DL1 | 0.37 | 0 | 0.018 | 0.72 | |
| KIR3DL2 | 0.58 | 0 | −0.024 | 0.64 | |
| KIR3DL3 | 0.14 | * | 0.029 | 0.57 | |
| KIR2DS4 | 0.21 | *** | −0.052 | 0.32 | |
| KLRK1 (NKG2D) | 0.69 | 0 | 0.065 | 0.21 | |
| NCR1 (NKp46) | 0.33 | *** | −0.039 | 0.46 | |
| NCR2 (NKp44) | 0.11 | 0.019 | −0.025 | 0.63 | |
| NCR3 (NKp30) | 0.47 | 0 | 0.09 | 0.085 | |
* p < 0.01 and *** p < 0.0001.
3.6. Correlation between IL32 Expression and Activation of NK Cells
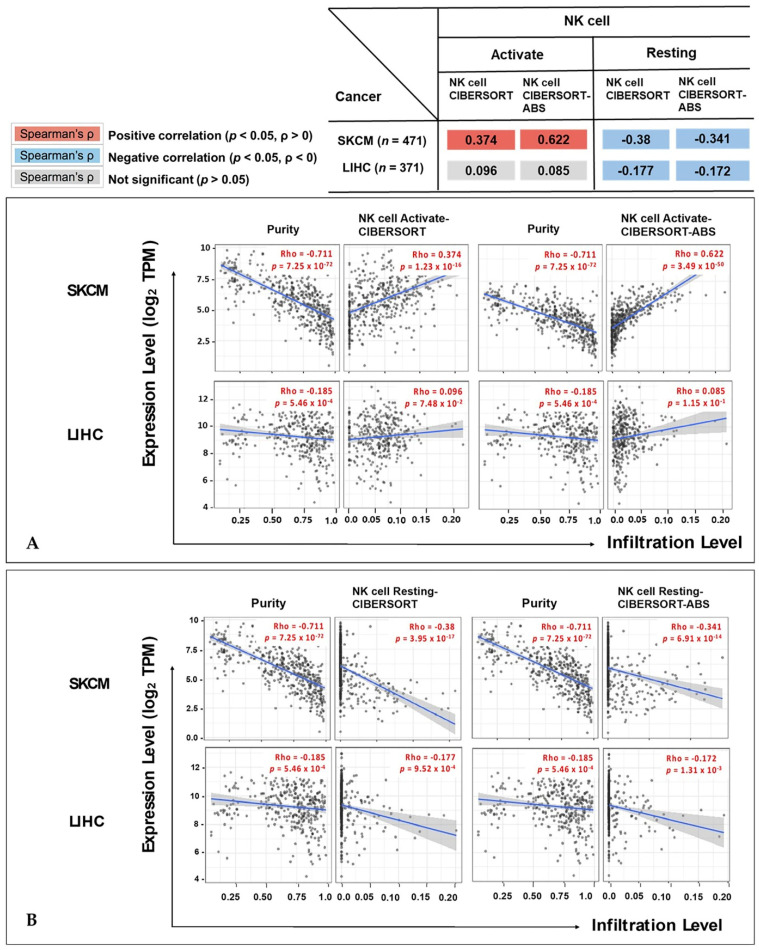
To confirm the difference between activating and resting NK cell infiltration by IL32 expression, we analyzed the expression of activated NK cells from CIBERSORT using TIMER v.2.0. As shown in Figure 6A, a high IL32 expression was positively correlated with the infiltration of activated NK cells (rho = 0.374, p = 1.23 × 10−16) in SKCM, but no correlation was found between IL32 expression in LIHC (activated NK cells; rho = 0.096, p = 7.48 × 10−2). Interestingly, the infiltration of resting NK cells showed a significant negative correlation with IL32 expression in SKCM (rho = −0.38, p = 3.95 × 10−17) and LIHC (rho = −0.177, p = 9.52 × 10−4) (Figure 6B). We further analyzed NK cell infiltration according to SKCM status to prove the correlation between IL32 expression and survival. These results suggest that IL32 expression in SKCM tissue induces the infiltration of specially activated NK cells. Overall, in this study, IL32 expression controlled activated NK cell infiltration in SKCM and improved the prognosis of melanoma patients.
Figure 6.
Correlation between IL32 expression and the infiltration levels of NK cells in SKCM and LIHC. The relationship between expression levels of IL32 and (A) activated NK cells, and (B) resting NK cell was investigated by the online tool TIMER v.2.0. (p < 0.05).
3.7. Correlation between IL32 Expression and Cytolytic Cell of NK Cells
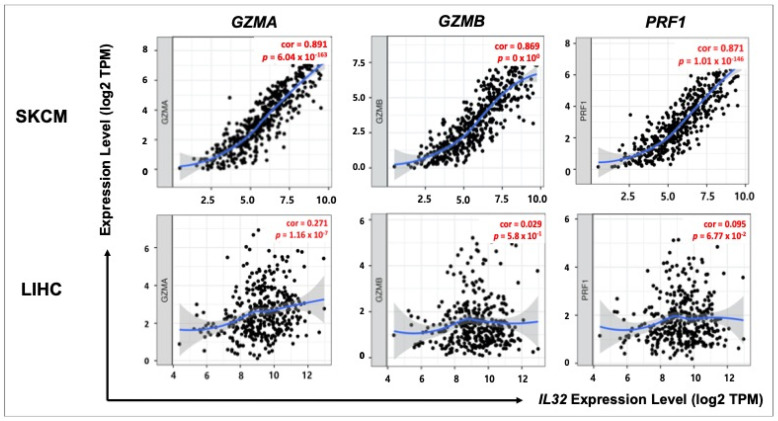
NK cells are well-known representative cytolytic effector cells that release cytotoxic molecules, such as granzyme and perforin [52]. Therefore, we analyzed the correlation between IL32 expression and gene markers of the cytolytic molecules granzyme A (GZMA), granzyme B (GZMB), and perforin (PRF1) using TIMER v.1.0. As shown in Figure 7, IL32 expression was significantly positively correlated with GZMA (cor = 0.891, p = 6.04 × 10−163), GZMB (cor = 0.869, p = 0 × 100), and RPF1 (cor = 0.871, p = 1.01 × 10−146) gene expressions in SKCM (n = 103), but no correlation was found between IL32 expression and gene markers of cytolytic molecules in LIHC (n = 371) (GZMA, cor = 0.271, p = 1.16 × 10−7; GZMB, cor = 0.029, p = 5.8 × 10−1; and RPF1, cor = 0.095, p = 6.77 × 10−2). Collectively, these results suggest that infiltrating NK cells improve SKCM patient survival by impeding tumor progression via the release of granzyme and perforin.
Figure 7.
Correlation between IL32 expression and cytolytic effector cells in SKCM and LIHC. Scatter plots were generated using TIMER v.1.0. IL32 expression and gene markers of the cytolytic effector were positively correlated in SKCM. IL32 expression was not significantly correlated with either gene in LIHC. The correlation constants and p-values are listed in Supplementary Table S11. (p < 0.05).
3.8. Coexpressed Genes and IL32 in Melanoma
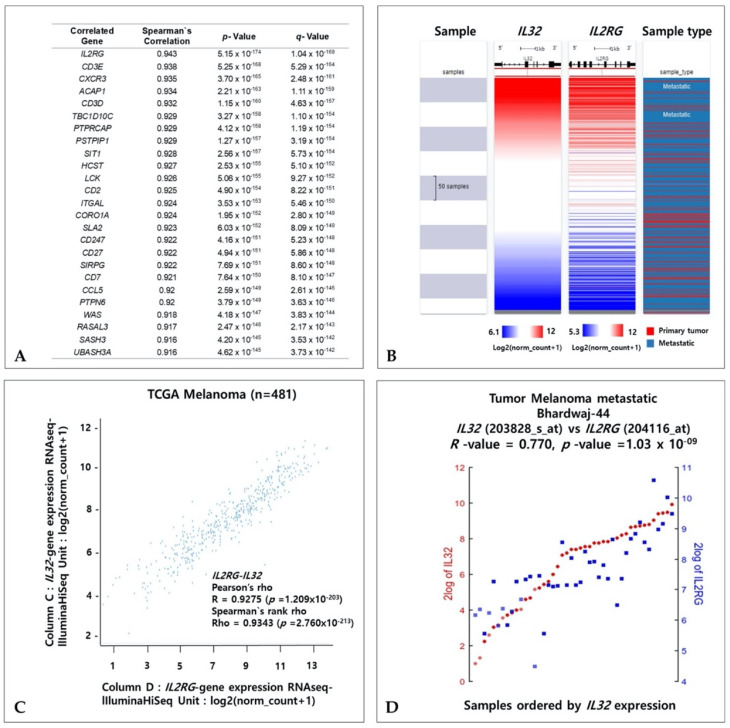
To investigate the coexpressed genes with IL32 in SKCM, we identified genes that exhibit correlated expressions with IL32 in SKCM using the TCGA dataset of cBioPortal. Figure 8A shows that 25 genes are the most positively coexpressed with IL32 in SKCM. In the TCGA-SKCM dataset, the gene expression of the interleukin 2 receptor group (IL2RG) showed the strongest positive correlation with IL32. The coexpressed patterns of IL32 and IL2RG were analyzed via a heatmap and a dot plot using the UCSC Xena web tool (Figure 8B,C). The coexpression patterns of IL32 and IL2RG in primary and metastatic melanomas were visually expressed through a heatmap (Figure 8B). In Figure 8C, the strong positive correlation between IL32 and IL2RG expression was confirmed using Pearson’s (R = 0.9275, p = 1.209 × 10−203) and Spearman’s (R = 0.9343, p = 2.76 × 10−213) correlation analyses in primary and metastatic SKCM. Additionally, the correlations with IL32 and IL2RG were confirmed using the R2 platform (Tumor melanoma metastatic Bhardwaj-44, R = 0.770, p = 1.03 × 10−9) (Figure 8D). Taken together, these data suggest that IL32 and coaltered IL2RG with biological processes related to IL-32 may be involved in melanoma progression.
Figure 8.
Coexpression genes of the IL32 gene in SKCM. (A) Twenty-five genes most positively coexpressed with IL32 in SKCM using the TCGA dataset of cBioPortal. (B) IL32 and IL2RG mRNA expressions in a heatmap using the UCSC Xena Browser. (C) Dot plot of IL32 and IL2RG mRNA expressions in the TCGA-SKCM dataset. (D) Correlation between IL32 and IL2RG expressions in the Tumor Melanoma Metastatic Bhardwaj-44 dataset using the R2 web server 3. (p < 0.05).
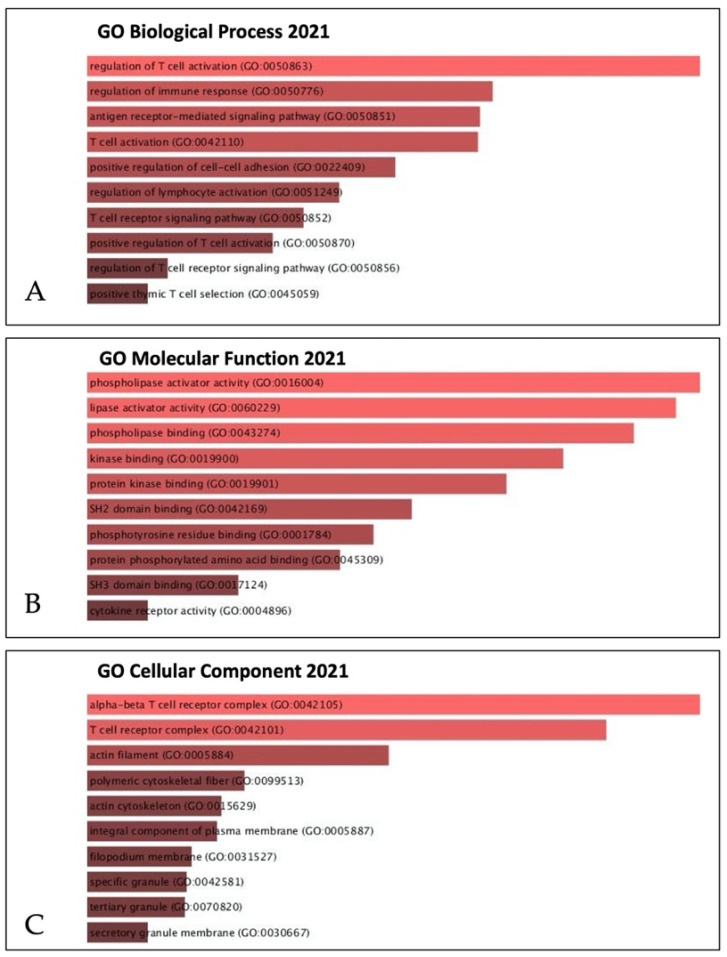
In addition, to identify biological processes and functions, we further analyzed the correlation between IL32 and coaltered genes in SKCM using the gene ontology (GO) analysis (Figure 9). In the GO biological process analysis, IL32 and IL32 coaltered genes were mainly associated with regulated immne response (Figure 9A). In the GO molecular function analysis, IL32 coaltered genes were most significantly enriched in phospholipase cativator activity, and lipase activator and phospholipase binding (Figure 9B). Figure 9C showed the most abundant results in the GO cell components associated with the alpha–beta T cell receptor complex (Figure 9C). Overall, a gene enrichment analysis of IL32 and IL32 coaltered genes suggests that IL32 may be associated with the regulation of lymphocyte activation.
Figure 9.
Correlation between IL32 gene and coexpression genes in the SKCM signaling pathway. IL32 gene and its top 25 coexpressed genes were analyzed by the Enricher web tool (https://amp.pharm.mssm.edu/Enrichr/, accessed on 25 April 2021). (A) GO biological process (2021), (B) GO molecular function (2021), and (C) GO cellular component (2021).
4. Discussion
IL-32 is a proinflammatory cytokine involved in various isoforms and is known to have both anticancer and procancer properties [53]. To date, nine isoforms have been identified, and the four most studied isoforms, IL-32α, IL-32β, IL-32γ, and IL-32δ, were first described in NK cells [38,54]. All of these isoforms in IL-32 have different sizes and secondary structures, which can lead to differences in protein function and efficacy due to changes in the tertiary protein structure [55]. Indeed, various isoforms of IL-32 show differences in efficacy to elicit a specific effect and to induce different reactions in malignant tumors [56,57]. Exogenous treatment with IL-32α inhibited proliferation and increased apoptosis in HTB-72 human melanoma cell lines in relation to the upregulation of p21, p53, and TRAIL receptor 1 (TRAILR1) [58]. However, the high expression of IL-32β was increased in cancer tissues and serum in patients with hepatocellular carcinoma, and the inhibition of IL-32α expression using siRNA resulted in decreased expression of the antiapoptotic protein bcl-2, inhibiting cell growth and apoptosis [59]. In addition, Oh et al. showed that IL-32β and IL-32γ inhibit melanoma and colon cancer tumor growth via inhibition of the activated NF-κB and STAT3 in a transgenic mouse model, while the expression of these isoforms is associated with increased infiltration and migration of breast cancer cells [43,60,61]. Although many studies have revealed the mechanism of the role of IL-32 isoforms in various types of cancers, the role of IL-32 is controversial and still unclear.
Recently, IL32 expression and cancer-related immune cells have been reported to be highly correlated in various types of cancers [62,63]. IL32 is produced by representative antitumor immune cells such as T cells, NK cells, monocytes, and macrophages [34,38]. In colorectal and prostate cancer cells, IL-32β has been shown to enhance NK cytotoxicity against cancer cells in vitro via the activation of caspase-3 [64]. In particular, IL-32β expression in metastatic mice increases the level of IL-10, an immunosuppressive cytokine, and induces infiltration of cytotoxic T cells and NK cells in tumors, resulting in the suppression of tumor growth [61]. These results suggest that IL-32 may stimulate antitumor immune responses in tumor microenvironments by inducing cytolytic activity and the infiltration of NK cells and T cells. Although various biological activities of IL-32 in tumor progression have been reported, a comprehensive analysis is required in clinical studies along with in vitro and in vivo studies because IL-32 has dual effects in tumor biology, such as procancer effects and anticancer effects. Therefore, a systematic analysis of the correlation between IL32 expression and patient survival is essential to comprehensively understand the role of IL-32 in melanoma patients.
Here, we found that IL32 mRNA expression was higher in cutaneous melanoma (SKCM; skin cutaneous melanoma) tissue than in normal tissue and that a higher expression of IL32 was significantly correlated with patient survival (Figure 1 and Figure 2), suggesting that a higher IL32 expression leads to better clinical outcomes in SKCM patients. Our analysis also showed that IL32 expression levels were positively correlated with the levels of infiltration of various immune cells, especially NK cells (Figure 4). The fact that high infiltration levels of immune cells result in better prognosis in various types of cancers is well known [65]. As shown in Figure 5, a strong negative correlation was found between IL32 expression and tumor purity, indicating that IL32 mRNA expression in SKCM tissues is likely to have been derived from infiltrated immune cells. Therefore, the reason why the patient’s survival rate increases with the increase in IL32 mRNA expression in SKCM tissues can be presumed to be due to the infiltration of activated NK cells expressing IL32. In fact, IL32 was mainly detected in activated NK cells and dendritic cells (DCs), which induce tumor cell apoptosis by producing cytolytic molecules such as perforin and granzyme from activated NK cells in tumor environments [66,67]. Nevertheless, the fact that the activities and functions of effector cells in the TME are suppressed is known [68,69]. Tumor cells secrete immunosuppressive cytokines such as IL-10, or immune cells, such as regulated T cells and M2 macrophages in the TME, inhibit the activity of antitumor effector cells due to their immune inhibition function [52,70]. Therefore, in immunotherapy, maintaining the activity of immune cells and increasing the infiltration of effector cells in tumors are strongly related to better outcomes in patients. In this regard, this study attempted to determine whether the increase in patient survival rate following IL32 mRNA expression was due to the infiltration of activated effector cells. Figure 5 and Supplementary Table S10 show a strong positive correlation between IL32 expression and the expression of specific markers for NK cells. Interestingly, the expression of IL32 and resting NK cell infiltration were negatively correlated while activated NK cell infiltration was positively correlated (Figure 6). These results indicate that activated NK cells were infiltrated within the tumor of SKCM and that tumor suppression was induced through the activation of NK cell cytotoxicity. In fact, this study identified a positive correlation between IL32 expression and cytolytic molecule genes such as GZMA, GZMB, and PRF1 (Figure 7). This suggests that IL32 could increase patient survival by enhancing the infiltration and cytolytic activities of NK cells in effector cells such as granzyme and perforin. Therefore, our study found that IL32 expression levels are correlated with NK cell infiltration in SKCM.
In addition, we investigated the co-expressed genes with biological processes related to IL32 in SKCM. Of the 25 genes that were positively correlated, IL-2 receptor γ (IL2RG) showed the strongest positive correlation with IL32 expression as shown in Figure 8. IL2RG is well known as a common subunit for the signaling of various interleukins, including IL-2, -4, -7, -9, and -15. It is essential for affinity binding and signaling of cytokines and plays an important role in the development and survival of immune cell subgroups, such as NK cells and T cells [71]. In addition, Figure 9 shows that IL32 is involved in the regulation of lymphocyte activation by GO analysis. Therefore, these data suggest that IL32 and IL32-coaltered genes are involved in antitumor immune regulation by immune cells, including NK cells, and that IL32 expression may be a novel biomarker for predicting immune cell activation in SKCM.
5. Conclusions
In conclusion, this study shows that the increased IL32 mRNA expression is significantly related to the infiltration of NK cells in cutaneous melanoma tissues, resulting in a good prognosis in cutaneous melanoma patients. The main finding of this study is that the correlation between IL32 mRNA expression and activated NK cell infiltration is significant but that the correlation between IL32 mRNA expression and a resting NK cell is not. In addition, the correlation between IL32 and various genes of cytolytic molecules, such as GZMA, GZMB, and PRF1, is positive, suggesting that IL32 mRNA expression may increase patient survival through the infiltration and activation of anticancer effector cells in cutaneous melanoma. This systematic analysis provides evidence suggesting the potential role of IL32 as an effective biomarker for patient survival in the tumor microenvironment.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jcm10204691/s1, Supplementary Table S1. The Cox regression results of IL32 expression in SKCM and LIHC; Table S2. Tumor abbreviations; Figure S3. Cox regression results for IL32 with TCGA data in various types of cancers; Table S4. Correlation between IL32 expression and DFS of various types of cancers; Table S5. Fifteen mutations in the IL32 gene; Figure S6. Correlation IL32 expression and BRAF mutation status in SKCM; Figure S7. Correlation of IL32 expression with immune cell infiltration level in SKCM and LIHC; Figure S8. Correlations between IL32 expression and NK cells of various types of cancers; Table S9. Correlation analysis between IL32 and gene markers of immune cells; Table S10. Correlation constants and p-values in Figure 5; Table S11. Correlation constants and p-values in Figure 7.
Author Contributions
Conceptualization, K.E.K.; methodology, J.Y.K. and K.E.K.; validation, K.E.K.; formal analysis, J.Y.K.; investigation, J.Y.K.; data curation, K.E.K.; writing—original draft preparation, J.Y.K.; writing—review and editing, J.Y.K. and K.E.K.; supervision, K.E.K.; project administration, K.E.K.; funding acquisition, K.E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT) (NRF-2020R1A2C1101129).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found in the link mentioned in the section of Materials and Methods.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schadendorf D., van Akkooi A.C.J., Berking C., Griewank K.G., Gutzmer R., Hauschild A., Stang A., Roesch A., Ugurel S. Melanoma. Lancet. 2018;392:971–984. doi: 10.1016/S0140-6736(18)31559-9. [DOI] [PubMed] [Google Scholar]
- 2.Li Y., Li M., Shats I., Krahn J.M., Flake G.P., Umbach D.M., Li X., Li L. Glypican 6 is a putative biomarker for metastatic progression of cutaneous melanoma. PLoS ONE. 2019;14:e0218067. doi: 10.1371/journal.pone.0218067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olszanski A.J. Current and Future Roles of Targeted Therapy and Immunotherapy in Advanced Melanoma. J. Manag. Care Pharm. 2014;20:346–356. doi: 10.18553/jmcp.2014.20.4.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naik P.P. Cutaneous Malignant Melanoma: A Review of Early Diagnosis and Management. World J. Oncol. 2021;12:7–19. doi: 10.14740/wjon1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fecher L.A., Cummings S.D., Keefe M.J., Alani R.M. Toward a Molecular Classification of Melanoma. J. Clin. Oncol. 2007;25:1606–1620. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- 6.Chin L., Garraway L.A., Fisher D.E. Malignant melanoma: Genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 7.Soengas M.S., Lowe S.W. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138–3151. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 8.Topalian S.L., Drake C.G., Pardoll D.M. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribas A., Wolchok J.D. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maibach F., Sadozai H., Jafari S.M.S., Hunger R.E., Schenk M. Tumor-Infiltrating Lymphocytes and Their Prognostic Value in Cutaneous Melanoma. Front. Immunol. 2020;11:2105. doi: 10.3389/fimmu.2020.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fridman W.H., Zitvogel L., Sautes-Fridman C., Kroemer G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 12.Park Y.-J., Kuen D.-S., Chung Y. Future prospects of immune checkpoint blockade in cancer: From response prediction to overcoming resistance. Exp. Mol. Med. 2018;50:1–13. doi: 10.1038/s12276-018-0130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes T.A., Amir E. HYPE or HOPE: The prognostic value of infiltrating immune cells in cancer. Br. J. Cancer. 2018;118:e5. doi: 10.1038/bjc.2017.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtin J.A., Fridlyand J., Kageshita T., Patel H.N., Busam K.J., Kutzner H., Cho K.-H., Aiba S., Bröcker E.-B., LeBoit P.E., et al. Distinct Sets of Genetic Alterations in Melanoma. N. Engl. J. Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 15.Passarelli A., Mannavola F., Stucci L.S., Tucci M., Silvestris F. Immune system and melanoma biology: A balance between immunosurveillance and immune escape. Oncotarget. 2017;8:106132–106142. doi: 10.18632/oncotarget.22190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falletta P., del Campo L.S., Chauhan J., Effern M., Kenyon A., Kershaw C., Siddaway R., Lisle R.J., Freter R., Daniels M., et al. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 2017;31:18–33. doi: 10.1101/gad.290940.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim I.S., Heilmann S., Kansler E.R., Zhang Y., Zimmer M., Ratnakumar K., Bowman R.L., Simon-Vermot T., Fennell M., Garippa R., et al. Microenvironment-derived factors driving metastatic plasticity in melanoma. Nat. Commun. 2017;8:14343. doi: 10.1038/ncomms14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falcone I., Conciatori F., Bazzichetto C., Ferretti G., Cognetti F., Ciuffreda L., Milella M. Tumor Microenvironment: Implications in Melanoma Resistance to Targeted Therapy and Immunotherapy. Cancers. 2020;12:2870. doi: 10.3390/cancers12102870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letca A.F., Ungureanu L., Șenilă S.C., Grigore L.E., Pop S., Fechete O., Vesa Ş.C., Cosgarea R. Regression and Sentinel Lymph Node Status in Melanoma Progression. Med. Sci. Monit. 2018;24:1359–1365. doi: 10.12659/MSM.905862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paijens S.T., Vledder A., de Bruyn M., Nijman H.W. Tumor-infiltrating lymphocytes in the immunotherapy era. Cell. Mol. Immunol. 2021;18:842–859. doi: 10.1038/s41423-020-00565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruosso T., Gigoux M., Manem V.S.K., Bertos N., Zuo D., Perlitch I., Saleh S.M.I., Zhao H., Souleimanova M., Johnson R.M., et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Investig. 2019;129:1785–1800. doi: 10.1172/JCI96313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gata V.A., Lisencu C.I., Vlad C.I., Piciu D., Irimie A., Achimas-Cadariu P. Tumor infiltrating lymphocytes as a prognostic factor in malignant melanoma. Review of the literature. J. BUON. 2017;22:592–598. [PubMed] [Google Scholar]
- 23.Gata V.A., Kubelac P.M., Buiga R., Vlad I.C., Valean D., Muntean M.V., Morariu D.S., Bonci E.A., Irimie A., Dina C., et al. The value of tumor infiltrating lymphocytes as prognostic factor for lymph node status and survival amongst patients with cutaneous malignant melanoma. J. BUON. 2021;25:2700–2707. [PubMed] [Google Scholar]
- 24.Maimela N.R., Liu S., Zhang Y. Fates of CD8+ T cells in Tumor Microenvironment. Comput. Struct. Biotechnol. J. 2019;17:1–13. doi: 10.1016/j.csbj.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S.-Y., Fu T., Jiang Y.-Z., Shao Z.-M. Natural killer cells in cancer biology and therapy. Mol. Cancer. 2020;19:120. doi: 10.1186/s12943-020-01238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gajewski T.F., Schreiber H., Fu Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saddawi-Konefka R., Seelige R., Gross E.T., Levy E., Searles S.C., Washington A., Santosa E.K., Liu B., O’Sullivan T., Harismendy O., et al. Nrf2 Induces IL-17D to Mediate Tumor and Virus Surveillance. Cell Rep. 2016;16:2348–2358. doi: 10.1016/j.celrep.2016.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seelige R., Searles S., Bui J.D. Mechanisms regulating immune surveillance of cellular stress in cancer. Cell. Mol. Life Sci. 2017;75:225–240. doi: 10.1007/s00018-017-2597-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kmiecik J., Poli A., Brons N.H., Waha A., Eide G.E., Enger P.Ø., Zimmer J., Chekenya M. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J. Neuroimmunol. 2013;264:71–83. doi: 10.1016/j.jneuroim.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Konjević G., Martinović K.M., Vuletić A., Jović V., Jurisic V., Babović N., Spužić I. Low expression of CD161 and NKG2D activating NK receptor is associated with impaired NK cell cytotoxicity in metastatic melanoma patients. Clin. Exp. Metastasis. 2007;24:1–11. doi: 10.1007/s10585-006-9043-9. [DOI] [PubMed] [Google Scholar]
- 31.Konjević G., Martinović K.M., Vuletić A., Babović N. In-vitro IL-2 or IFN-α-induced NKG2D and CD161 NK cell receptor expression indicates novel aspects of NK cell activation in metastatic melanoma patients. Melanoma Res. 2010;20:459–467. doi: 10.1097/CMR.0b013e32833e3286. [DOI] [PubMed] [Google Scholar]
- 32.Dahl C.A., Schall R.P., He H.L., Cairns J.S. Identification of a novel gene expressed in activated natural killer cells and T cells. J. Immunol. 1992;148:597–603. [PubMed] [Google Scholar]
- 33.Yan H., He D., Huang X., Zhang E., Chen Q., Xu R., Liu X., Zi F., Cai Z. Role of interleukin-32 in cancer biology (Review) Oncol. Lett. 2018;16:41–47. doi: 10.3892/ol.2018.8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paz H., Tsoi J., Kalbasi A., Grasso C.S., McBride W.H., Schaue D., Butterfield L.H., Maurer D.M., Ribas A., Graeber T.G., et al. Interleukin 32 expression in human melanoma. J. Transl. Med. 2019;17:113. doi: 10.1186/s12967-019-1862-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinarello C.A., Kim S.-H. IL-32, a novel cytokine with a possible role in disease. Ann. Rheum. Dis. 2006;65:iii61–iii64. doi: 10.1136/ard.2006.058511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi J.-D., Bae S.-Y., Hong J.-W., Azam T., Dinarello C.A., Her E., Choi W.-S., Kim B.-K., Lee C.-K., Yoon D.-Y., et al. Identification of the most active interleukin-32 isoform. Immunology. 2009;126:535–542. doi: 10.1111/j.1365-2567.2008.02917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yun H.-M., Park K.-R., Kim E.-C., Han S.B., Yoon D.Y., Hong J.T. IL-32α suppresses colorectal cancer development via TNFR1-mediated death signaling. Oncotarget. 2015;6:9061–9072. doi: 10.18632/oncotarget.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloot Y.J.E., Smit J.W., Joosten L.A.B., Netea-Maier R.T. Insights into the role of IL-32 in cancer. Semin. Immunol. 2018;38:24–32. doi: 10.1016/j.smim.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Gruber T., Kremenovic M., Sadozai H., Rombini N., Baeriswyl L., Maibach F., Modlin R.L., Gilliet M., von Werdt D., Hunger R.E., et al. IL-32γ potentiates tumor immunity in melanoma. JCI Insight. 2020;5:5. doi: 10.1172/jci.insight.138772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng Q., Li S., Zhou Y., Ou W., Cai X., Zhang L., Huang W., Huang L., Wang Q. Interleukin-32 contributes to invasion and metastasis of primary lung adenocarcinoma via NF-kappaB induced matrix metalloproteinases 2 and 9 expression. Cytokine. 2014;65:24–32. doi: 10.1016/j.cyto.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Tsai C.-Y., Wang C.-S., Tsai M.-M., Chi H.-C., Cheng W.-L., Tseng Y.-H., Chen C.-Y., Lin C.D., Wu J.-I., Wang L.-H., et al. Interleukin-32 Increases Human Gastric Cancer Cell Invasion Associated with Tumor Progression and Metastasis. Clin. Cancer Res. 2014;20:2276–2288. doi: 10.1158/1078-0432.CCR-13-1221. [DOI] [PubMed] [Google Scholar]
- 42.Wang S., Chen F., Tang L. IL-32 promotes breast cancer cell growth and invasiveness. Oncol. Lett. 2015;9:305–307. doi: 10.3892/ol.2014.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J.S., Choi S.Y., Lee J.-H., Lee M., Nam E.S., Jeong A.L., Lee S., Han S., Lee M.-S., Lim J.-S., et al. Interleukin-32β stimulates migration of MDA-MB-231 and MCF-7cells via the VEGF-STAT3 signaling pathway. Cell. Oncol. 2013;36:493–503. doi: 10.1007/s13402-013-0154-4. [DOI] [PubMed] [Google Scholar]
- 44.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B., Liu X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Rodriguez I.P., Chakravarthi B.V.S.K., Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., Li B., Liu X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel H., Yacoub N., Mishra R., White A., Yuan L., Alanazi S., Garrett J.T. Current Advances in the Treatment of BRAF-Mutant Melanoma. Cancers. 2020;12:482. doi: 10.3390/cancers12020482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holčáková J. Effect of DNA Methylation on the Development of Cancer. Klin. Onkol. Cas. Ceske Slov. Onkol. Spol. 2018;31:41–45. doi: 10.14735/amko20182S41. [DOI] [PubMed] [Google Scholar]
- 52.Gil M., Kim K.E. Systematic Multiomic Analysis of Ly75 Gene Expression and Its Prognostic Value through the Infiltration of Natural Killer (NK) Cells in Skin Cutaneous Melanoma. J. Clin. Med. 2020;9:1383. doi: 10.3390/jcm9051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong J.T., Son D.J., Lee C.K., Yoon D.-Y., Lee D.H., Park M.H. Interleukin 32, inflammation and cancer. Pharmacol. Ther. 2017;174:127–137. doi: 10.1016/j.pharmthera.2017.02.025. [DOI] [PubMed] [Google Scholar]
- 54.Kim S.-H., Han S.-Y., Azam T., Yoon D.-Y., Dinarello C.A. Interleukin-32: A Cytokine and Inducer of TNFα. Immunity. 2005;22:131–142. doi: 10.1016/j.immuni.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Heinhuis B., Koenders M., Berg W.B.V.D., Netea M.G., Dinarello C.A., Joosten L.A.B. Interleukin 32 (IL-32) Contains a Typical α-Helix Bundle Structure That Resembles Focal Adhesion Targeting Region of Focal Adhesion Kinase-1. J. Biol. Chem. 2012;287:5733–5743. doi: 10.1074/jbc.M111.288290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joosten L.A.B., Heinhuis B., Netea M.G., Dinarello C.A. Novel insights into the biology of interleukin-32. Cell. Mol. Life Sci. 2013;70:3883–3892. doi: 10.1007/s00018-013-1301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinhuis B., Koenders M., van de Loo F.A., Netea M.G., Berg W.B.V.D., Joosten L.A.B. Inflammation-dependent secretion and splicing of IL-32 in rheumatoid arthritis. Proc. Natl. Acad. Sci. USA. 2011;108:4962–4967. doi: 10.1073/pnas.1016005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicholl M.B., Chen X., Qin C., Bai Q., Zhu Z., Bs M.R.D., Fang Y. IL-32α has differential effects on proliferation and apoptosis of human melanoma cell lines. J. Surg. Oncol. 2016;113:364–369. doi: 10.1002/jso.24142. [DOI] [PubMed] [Google Scholar]
- 59.Kang Y.H., Park M.-Y., Yoon D.-Y., Han S.R., Lee C.I., Ji N.Y., Myung P.-K., Lee H.G., Kim J.W., Yeom Y.I., et al. Dysregulation of overexpressed IL-32α in hepatocellular carcinoma suppresses cell growth and induces apoptosis through inactivation of NF-κB and Bcl-2. Cancer Lett. 2012;318:226–233. doi: 10.1016/j.canlet.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 60.Oh J.H., Cho M.-C., Kim J.M., Lee S.Y., Kim H.J., Park E.S., Ban J.O., Kang J.-W., Lee D.-H., Shim J.-H., et al. IL-32γ inhibits cancer cell growth through inactivation of NF-κB and STAT3 signals. Oncogene. 2011;30:3345–3359. doi: 10.1038/onc.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yun H.-M., Oh J.H., Shim J.-H., Ban J.O., Park K.-R., Kim J.-H., Lee D.H., Kang J.-W., Park Y.H., Yu D., et al. Antitumor activity of IL-32β through the activation of lymphocytes, and the inactivation of NF-κB and STAT3 signals. Cell Death Dis. 2013;4:e640. doi: 10.1038/cddis.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung M.Y., Son M.H., Kim S.H., Cho D., Kim T.S. IL-32γ Induces the Maturation of Dendritic Cells with Th1- and Th17-Polarizing Ability through Enhanced IL-12 and IL-6 Production. J. Immunol. 2011;186:6848–6859. doi: 10.4049/jimmunol.1003996. [DOI] [PubMed] [Google Scholar]
- 63.Daltroy L.H., Larson M.G., Eaton H.M., Partridge A.J., Pless I.B., Rogers M.P., Liang M.H., Walker L.S., Van Slyke D.A., Newbrough J.R. Psychosocial Adjustment in Juvenile Arthritis. J. Pediatr. Psychol. 1992;17:277–289. doi: 10.1093/jpepsy/17.3.277. [DOI] [PubMed] [Google Scholar]
- 64.Park M.H., Song M.J., Cho M.-C., Moon D.C., Yoon D.Y., Han S.B., Hong J.T. Interleukin-32 enhances cytotoxic effect of natural killer cells to cancer cells via activation of death receptor 3. Immunology. 2012;135:63–72. doi: 10.1111/j.1365-2567.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pagès F., Galon J., Dieu-Nosjean M.-C., Tartour E., Sautes-Fridman C., Fridman W.H. Immune infiltration in human tumors: A prognostic factor that should not be ignored. Oncogene. 2009;29:1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 66.Gil M., Kim K.E. Interleukin-18 Is a Prognostic Biomarker Correlated with CD8+ T Cell and Natural Killer Cell Infiltration in Skin Cutaneous Melanoma. J. Clin. Med. 2019;8:1993. doi: 10.3390/jcm8111993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han S., Yang Y. Interleukin-32: Frenemy in cancer? BMB Rep. 2019;52:165–174. doi: 10.5483/BMBRep.2019.52.3.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rossi G.R., Trindade E.S., Souza-Fonseca-Guimaraes F. Tumor Microenvironment-Associated Extracellular Matrix Components Regulate NK Cell Function. Front. Immunol. 2020;11:73. doi: 10.3389/fimmu.2020.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vitale M., Cantoni C., Pietra G., Mingari M.C., Moretta L. Effect of tumor cells and tumor microenvironment on NK-cell function. Eur. J. Immunol. 2014;44:1582–1592. doi: 10.1002/eji.201344272. [DOI] [PubMed] [Google Scholar]
- 70.Li C., Jiang P., Wei S., Xu X., Wang J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer. 2020;19:116. doi: 10.1186/s12943-020-01234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ren J., Yu D., Fu R., An P., Sun R., Wang Z., Guo R., Li H., Zhang Y., Li Z., et al. IL2RG -deficient minipigs generated via CRISPR/Cas9 technology support the growth of human melanoma-derived tumours. Cell Prolif. 2020;53:e12863. doi: 10.1111/cpr.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found in the link mentioned in the section of Materials and Methods.