Abstract
SNAI1, a zinc finger transcription factor, not only acts as the master regulator of epithelial-mesenchymal transition (EMT) but also functions as a driver of cancer progression, including cell invasion, survival, immune regulation, stem cell properties, and metabolic regulation. The regulation of SNAI1 occurs at the transcriptional, translational, and predominant post-translational levels including phosphorylation, acetylation, and ubiquitination. Here, we discuss the regulation and role of SNAI1 in cancer metastasis, with a particular emphasis on epigenetic regulation and post-translational modifications. Understanding how signaling networks integrate with SNAI1 in cancer progression will shed new light on the mechanism of tumor metastasis and help develop novel therapeutic strategies against cancer metastasis.
Keywords: SNAI1, metastasis, post-translational modifications, epigenetic, EMT
1. Introduction
Tumor metastasis, the spreading of cancer cells from original tumor sites to distant organs followed by development of secondary tumors, is the foremost cause of cancer-related deaths [1]. Initiation of the metastatic program is often followed by exploitation of an embryonic development process referred to as epithelial-mesenchymal transition (EMT) [2]. During EMT, epithelial cells attain mesenchymal phenotypes such as increased motility and invasiveness by dissolving cell–cell junctions and rebuilding cell–matrix connections, accompanied by loss of epithelial markers and a gain of mesenchymal markers [3]. EMT is activated by a plethora of EMT-activating transcription factors (EMT-TFs), such as those from the SNAIL, zinc finger E-box binding homeobox (ZEB), and TWIST families [4].
SNAI1 was the first discovered and most intensively studied transcription repressor of E-cadherin, a hallmark of EMT encoded by the epithelial gene CDH1. SNAI1 directly binds to E-boxes present in the CDH1 promoter to transcriptionally repress its expression. On the other hand, SNAI1 also acts as a transcriptional activator. SNAI1 not only enhances mesenchymal markers including fibronectin, collagens, and the matrix degradation enzyme matrix metalloproteinases 2 and 9 (MMP2 and MMP9), it also increases other EMT transcription factors such as TWIST and ZEB1 [5,6]. In addition, SNAI1 positively regulates transcriptional activation of target genes involved in Drosophila development through direct binding to the promoters [7]. In collaboration with early growth response 1(EGR1) and SP1, SNAI1 may directly activate transcription of p15INK4b, lymphoid enhancer-binding factor (LEF), and cyclooxygenase 2 (COX2) by directly binding on a consensus motif in HepG2 cells stimulated by the phorbol ester tumor promoter 12-O-tetradecanoyl-phorbol 13-acetate (TPA) [5,8,9,10]. SNAI1 induces resistance to apoptosis, confers tumor recurrence and drug resistance, generates breast cancer stem cell (CSC)-like properties, and induces aerobic glycolysis [11,12,13,14]. Interestingly, SNAI1 is tightly controlled at both transcriptional and protein levels. Many growth factors and cytokines can transcriptionally regulate SNAI1 expression [15]. In addition, SNAI1 protein levels are regulated by post-translational modifications (PTMs). These PTMs have diverse effects on the function of SNAI1.
Because of the reversible plasticity of EMT, epigenetic alternations are required in the EMT process. In eukaryotic cells, genomic DNA interacts with histone proteins and RNA to form chromatin, which holds epigenetic information independent of the DNA genetic data [16]. Alteration of chromatin occurs through regulators responsible for DNA methylation, post-translational modifications of nucleosomal histone tails, and/or non-coding RNA modulation; these epigenetic modifications play a key role in regulating gene expression by defining whether chromatins at a given genomic locus will be transcriptionally active or inactive [17]. For EMT, a variety of epigenetic regulators are critical requirements that interpret signals passed from stimulators to transcription factors [18]. Indeed, the expression of CDH1 is regulated by multiple enzymes involving epigenetic modification. SNAI1 collaborates with multiple epigenetic enzyme complexes, such as DNA methyltransferases, histone deacetylases, and histone methyltransferase and demethylase, in the transcriptional regulation of CDH1. Recent studies suggest a crucial role of epigenetic alterations in the regulation of SNAI1 and EMT markers.
Here, we summarize the regulation of SNAI1 with an emphasis on PTMs. Moreover, we describe recent insights into the epigenetic mechanisms of SNAI1-induced cancer metastasis, focusing on the cooperation of SNAI1 with epigenetic regulators.
2. Regulation of SNAI1
Expression of SNAI1 is governed at multiple levels from gene transcription, post-transcriptional regulation, and translation to PTMs such as phosphorylation, ubiquitination, acetylation, and sumoylation.
2.1. Structure of SNAI1
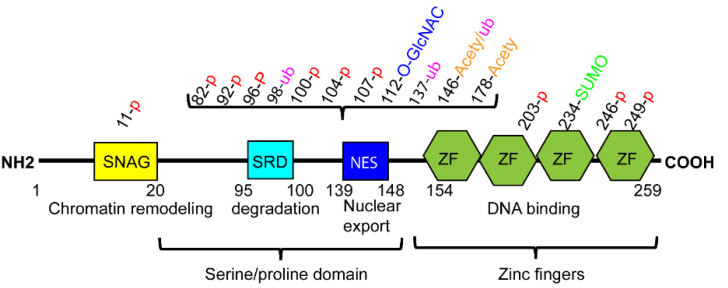
SNAI1 belongs to the SNAIL family which consists of SNAI1 (Snail), SNAI2 (Slug), and SNAI3 (Smuc) [19]. The amino termini of SNAI1 contains the evolutionarily conserved SNAI1/Gfi (SNAG) domain, which interacts with several co-repressor complexes or epigenetic remodeling complexes (Figure 1). Drosophila SNAIL lacks the SNAG domain but has a consensus PxDLSx motif and exerts their repressive function through the interaction with the co-repressor c-terminal binding protein (CtBP) [19]. In the central region, a serine-rich domain (SRD) is adjacent to the nuclear export sequence (NES) (Scheme 1). SRD controls ubiquitination and proteasome degradation while NES is involved in the regulation of its protein stability and subcellular translocation. The c-terminal zinc finger domain with four C2H2-type zinc fingers is highly conserved. This domain mediates sequence-specific interactions with their target DNA promoters containing an E-box sequence (CAGGTG).
Figure 1.
The structure of SNAI1 and potential post-translational modification sites. SNAI1 contains four main domains: Scheme 1. Gfi-1 (SNAG) domain, serine-rich domain (SRD), nuclear export sequence (NES) and zinc-finger (ZF) domain. P: phosphorylation. Acety: Acetylation. Ub: Ubiquitination. SUMO: Sumoylation.
2.2. Transcriptional and Post-Transcriptional Regulation
A diverse repertoire of molecular mechanisms regulating SNAI1 at the transcriptional level have been documented in a variety of organisms. Cytokines, chemokines, and growth factors, such as tumor necrosis factor (TNFα), transforming growth factor (TGFβ), interleukin-6 (IL-6), fibroblast growth factors (FGF), epidermal growth factor (EGF), and hepatocyte growth factor (HGF) [20], trigger an intracellular signaling cascade that leads to the binding of a transcription factor to the SNAI1 promoter to regulate its expression. Extensive evaluation of this regulation has been covered in other excellent reviews [21,22]. Interestingly, the expression of SNAI1 can also be regulated by other EMT-TFs. For example, both SNAI1 and SNAI2 were upregulated under TGFβ stimulation [23]. Depletion of SNAI2 increases SNAI1 expression and vice versa; this compensatory regulation could be indispensable for EMT and cancer progression [24]. In addition, TWIST induces SNAI1 and the Twist-SNAI1 axis is critically involved in EMT and tumor metastasis [25].
Post-transcriptional control provides a fundamental regulatory mechanism for gene expression. Besides regulation by microRNAs [26], SNAI1 transcript stability is also regulated extensively. For example, recent work showed that upon activation of EGF receptor, UDP-glucose 6-dehydrogenase (UGDH) is phosphorylated in human lung cancer cells. Phosphorylated UGDH not only converts UDP-glucose to UDP-glucuronic acid but also interacts with Hu antigen R (an RNA-binding protein that binds to short-lived mRNAs to increase their stability). This interaction attenuates the UDP-glucose-mediated inhibition and therefore enhances the stability of SNAI1 mRNA [27]. In addition, the mRNA of SNAI1 can be modified with N6-Methyladenosine (m6A) by the methyltransferase-like 3 (METTL3) and YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) (m6A readers). m6A in the coding sequence of SNAI1 triggers polysome-mediated translation of SNAI1 mRNA in cancer cells [28]. The stability of SNAI1 mRNA is also enhanced by heterogeneous nuclear ribonucleoprotein, which thus promotes invasion, metastasis, and EMT in breast cancer [29].
2.3. Post-Translational Regulation
Because of their critical roles in cancer metastasis, much attention has focused on the PTMs of SNAI1. PTMs function in the regulating protein stability, transcriptional activity, and intracellular localization of SNAI1. Among the number of modifications, phosphorylation and ubiquitination represent the best characterized and control a variety of biological activities, such as apoptosis, transcription, metabolism, and stem cell properties. Therefore, gaining deeper insight into the PTMs may help elucidate important steps in cancer metastasis.
2.3.1. Phosphorylation Regulation
SNAI1 stability is extensively regulated by phosphorylation (Table 1). On one hand, phosphorylation of SNAI1 promotes its proteasomal-mediated ubiquitination degradation. Both casein kinase 1(CK1) and dual specificity tyrosine phosphorylation regulated kinase 2 (DYRK2)-mediated SNAI1 phosphorylation at serine (Ser) 104 act to prime phosphorylations that allow glycogen synthase kinase 3 β (GSK3β)-mediated phosphorylation at Ser96 and Ser100, leading to β-TRCP-induced poly-ubiquitination and degradation [30,31]. Protein kinase D1 (PKD1)-mediated phosphorylation at Ser11 of SNAI1 facilitates F-box protein 11 (FBXO11)-mediated SNAI1 degradation [32]. Under intact apical-basal polarity, α protein kinase C (PKC) kinases promote degradation through phosphorylation of SNAI1 S249 [33]. On the other hand, some SNAI1 phosphorylations prevent its degradation. Most commonly, the main mechanism that regulates SNAI1 stability is phosphorylation at specific sites that reduce its affinity for GSK3β, thus blocking ubiquitination. For example, phosphorylation of SNAI1 at Ser100 by ataxia-telangiectasia mutated (ATM) and DNA-PKCs inhibits SNAI1 ubiquitination by reducing interaction with GSK3β [34,35]. Recently, it was shown that p38 stabilizes SNAI1 through phosphorylation at Ser107, which suppresses DYRK2-mediated Ser104 phosphorylation and subsequent GSK3β-mediated SNAI1 degradation [36]. However, stabilization of SNAI1 also occurs independent of GSK3β. Protein kinase A (PKA) and CK2 have been characterized as the main kinases responsible for in vitro SNAI1 phosphorylation at Ser11 and 92, respectively [37]. Phosphorylation of these two sites control SNAI1 stability and positively regulate SNAI1 repressive function and its interaction with the mSin3A corepressor. Alternatively, confinement of SNAI1 to the nucleus prevents degradation. ERK2-mediated Ser82/Ser104 phosphorylation of SNAI1 leads to nuclear SNAI1 accumulation [38]. P21 (RAC1) activated kinase 1 (PAK1) and GRO-α phosphorylate SNAI1 on Ser246 and increase SNAI1′s accumulation in the nucleus, which thus promotes transcriptional activity of SNAI1 [39,40,41]. Large tumor suppressor kinase 2 (Lats2) phosphorylates SNAI1 at threonine (Thr)203 in the nucleus, which prevents nuclear export, thereby supporting stabilization [42]. Recently, we also found that serine/threonine kinase 39 (STK39) enhances SNAI1 stability by phosphorylation at Thr203 [43]. Notably, phosphorylation can be reversed by phosphatases. We identified c-terminal domain phosphatase (SCP) as a specific phosphatase for SNAI1 [44]. SCP physically interacts with and stabilizes SNAI1 by direct dephosphorylation [44,45].
Table 1.
Phosphorylation regulators involved in SNAI1.
| Function | Regulation Factor | Phosphorylation Sites |
|---|---|---|
| Degradation | PKD1 | Ser11 |
| αPKC | Ser249 | |
| CK1 | Ser104/Ser107 | |
| GSK3β | Ser96/Ser100/S104/Ser107 | |
| DYRK2 | Ser104 | |
| Stabilization | PTK6 | Tyr342 |
| ATM | Ser100 | |
| CK2 | Ser92 | |
| p38 | Ser107 | |
| Nuclear accumulation and stabilization | ERK | Ser82/Ser104 |
| PAK1 | Ser246 | |
| GROα | ||
| LATS2 | Thr203 | |
| STK39 |
2.3.2. Ubiquitination and Deubiquitination
SNAI1′s ubiquitination and degradation are controlled by a number of F-box ligases, including β-TRCP1/FBXW1, FBXL14, FBXL5, FBXO11, and FBXO45 [46]. Recently, more E3 ligases have been discovered (Figure 2). F-box E3 ubiquitin ligase FBXO22 elicits antimetastatic effects by targeting SNAI1 ubiquitin-mediated proteasomal degradation in a GSK3β phosphorylation-dependent manner [47]. Through a luciferase-based genome-wide screening using small interfering RNA library against ~200 of E3 ligases and ubiquitin-related genes, SOCS box protein SplA/ryanodine receptor domain and SOCS box containing 3 (SPSB3) was identified as a novel E3 ligase component [48]. SPSB3 targets SNAI1 to promote polyubiquitination and degradation in response to GSK3β phosphorylation of SNAI1. Through yeast two-hybrid screening, the carboxyl terminus of Hsc70-interacting protein (CHIP) was identified as a novel SNAI1 ubiquitin ligase that interacts with SNAI1 to induce ubiquitin-mediated proteasomal degradation [49]. Recently, it was reported that SNAI1 was monoubiquitinated by the ubiquitin-editing enzyme A20. This monoubiquitylation of SNAI1 reduces the affinity of SNAI1 for GSK3β, and thus SNAI1 is stabilized in the nucleus [50].
Figure 2.
The ubiquitination and de-ubiquitination of SNAI1. SNAI1 is degraded by multiple E3 ligases. By contrast, de-ubiquitinases counteract E3 ligase activity and prevent SNAI1 degradation. USP: ubiquitin-specific protease; OTUB1: OTU deubiquitinase, ubiquitin aldehyde binding 1; PSMD14: proteasome 26S subunit, Non-ATPase 14; EIF3H: eukaryotic translation initiation factor 3 subunit H; DUB: deubiquitinase; UPS: ubiquitin/proteasome system; FBXW1: F-box/WD repeat-containing protein 1; FBXL: F-box and leucine rich repeat protein; FBXO: F-boxes other; SPSB3: SplA/ryanodine receptor domain and SOCS box containing 3; CHIP: carboxy-terminus of Hsc70 interacting protein.
Deubiquitinases (DUBs) counteract the SNAI1 degradation process to maintain a high level of SNAI1 protein in cancer cells. We recently identified DUB3 as a SNAI1 deubiquitinase that interacts with and stabilizes SNAI1 [51]. Independent research indicated that DUB3 is a target of cyclin-dependent kinase (CDK)4/6, and CDK4/6-mediated activation of DUB3 is essential to deubiquitinate and stabilize SNAI1 [52]. Resistance to platinum-based chemotherapy is a common event associated with tumor dissemination and metastasis in cancer patients. Upon platinum treatment, the ubiquitin-specific protease 1 (USP1) is phosphorylated by ATM and RAD3-related (ATR) and binds to SNAI1. Then, USP1 de-ubiquitinates and stabilizes SNAI1 expression, conferring resistance to platinum, increased stem cell-like features, and metastatic ability [53]. USP29 can be induced by major EMT and metastatic-inducing factors such as TGFβ, TNFα, and hypoxia. This protease enhances the interaction of SNAI1 and SCP1, and results in simultaneous dephosphorylation and de-ubiquitination of SNAI1 and thereafter cooperative prevention of SNAI1 degradation [54]. TGFβ also induces USP27X expression, which increases SNAI1 stability by deubiquitination [55]. Recently, more deubiquitinases have been identified. Eukaryotic translation initiation factor 3 subunit H (EIF3H), OTU deubiquitinase, ubiquitin aldehyde binding 1(OTUB1), USP3, proteasome 26S subunit, Non-ATPase 14 (PSMD14), USP26, USP36, USP37 also target SNAI1 for de-ubiquitination and stabilization (Figure 2) [56,57,58,59,60,61].
2.3.3. Other Post-Translational Regulation
Beyond the well-characterized PTMs of phosphorylation and ubiquitination, at least three other PTMs are involved in regulating SNAI1 protein abundance and activity. First, the sumoylation pathway is very similar to its biochemical analog, ubiquitylation, and regulates diverse cellular processes including transcription and protein stability, chromosome organization, DNA repair, and other cellular processes. TGFβ induces sumoylation of SNAI1 at its lysine (K) 234 residue, which is critical for the EMT-activating function of SNAI1 [62]. Second, the O-linked β-N-acetylglucosamine (O-GlcNAc) modification is a monosaccharide addition. SNAI1 is subject to O-GlcNAc at Ser112 under hyperglycemic conditions [63]. This modification leads to stabilization of SNAI1 by inhibition of GSK3β-mediated phosphorylation. Consequently, the O-GlcNAc SNAI1 promotes EMT. Finally, SNAI1 is also acetylated by the histone acetyltransferase adenovirus E1A-associated protein (p300) and CREB binding protein (CBP), two key transcriptional coactivators implicated in a multitude of cellular processes including cancer progression. CBP and p300 interact with SNAI1 to acetylate SNAI1 at K146 and K187, which consequently reduces SNAI1 ubiquitination and thus enhances its protein stability [64] (Figure 3).
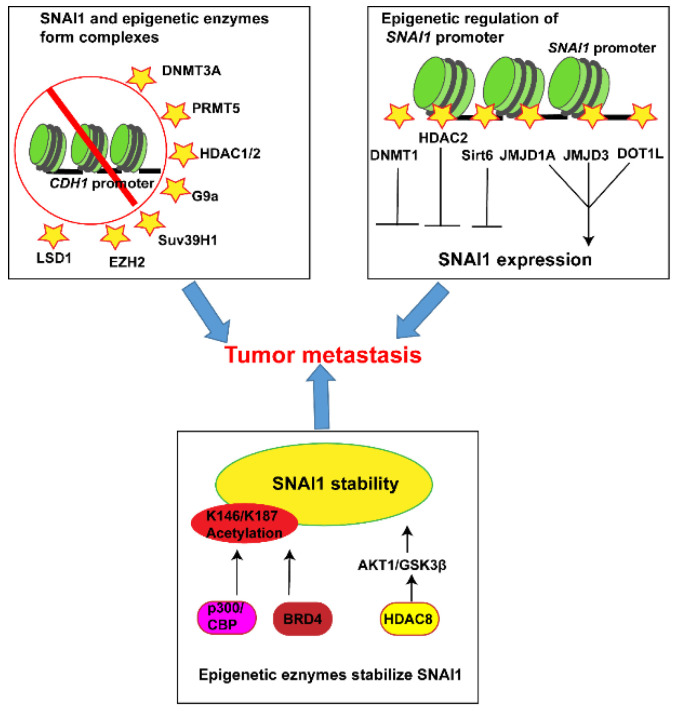
Figure 3.
The interplay between SNAI1 and epigenetic regulators. SNAI1 collaborates with epigenetic regulators to repress CDH1 expression. Epigenetic regulators are recruited to the SNAI1 promoter, leading to transcriptional activation or repression of SNAI1. In addition, epigenetic regulators regulate SNAI1 stability by post-translational modifications. The increase in SNAI1 expression via multiple epigenetic mechanisms leads to the cancer metastasis that is accompanied by the loss of CDH1. DNMT: DNA methyltransferases; PRMT: protein arginine methyltransferases; HDAC: histone deacetylases; Suv39H1: suppressor of variegation 3-9 homolog 1; EZH2: enhancer of zeste 2 polycomb repressive complex 2 subunit; LSD1: lysine-specific demethylase 1; JMJD: Jumonji C domain-containing; DOT1L: DOT1 like histone lysine methyltransferase; BRD4: bromodomain-containing protein 4; CBP: CREB binding protein.
3. The Interplay between SNAI1 and Epigenetic Regulators in Tumor Metastasis
Because EMT is a reversible and transient process, as well as having reversibility of the epigenetic marks and the enzymatic nature of the regulators, EMT-TFs and chromatin-remodeling enzymes are intimately connected (Figure 3). During tumor metastasis, SNAI1 recruits epigenetic regulators to the CDH1 promoter, thus repressing its expression. Epigenetic alterations also play a crucial role in SNAI1 expression. The interplay between SNAI1 and epigenetic regulators indicate the complexity of epigenetic mechanisms and the potentially crucial role of histone modifications for regulating SNAI1.
3.1. SNAI1 and DNA Methylation
DNA methylation involves a covalent attachment of a methyl group to cytosine residues at CpG-rich dinucleotide sequences through DNA methyltransferases (DNMTs). Upon induction of EMT, hypermethylation of the CDH1 promoter through DNMTs, which are recruited by EMT-TFs, is constantly observed in a wide variety of cancer cells. For example, SNAI1 interacts with DNMT3A to repress CDH1 expression via DNA hypermethylation and histone modifications of H3K9me2 and H3K27me3 in gastric cancer [65]. Previous research also indicated that DNMT1 was implicated in cell metastasis, such that downregulation or inhibition of DNMT1 could facilitate the metastasis of cancer cells [66]. DNMT1 can decrease the expression of CDH1 by increasing promoter methylation. Interestingly, DNMT1 can also act on CDH1 expression independent of its catalytic activity [67]. DNMT1 interacts with SNAI1 to prevent its interaction with the CDH1 promoter; this interaction leads to full CDH1 expression. Furthermore, DNMT1 is recruited to the SNAI1 promoter by AT-rich interactive domain-containing protein 2 (ARID2), a subunit of SWI/SNF chromatin remodeling complex. This complex increases the DNA methylation and suppresses SNAI1 transcription, leading to a repression of EMT. During hepatocellular carcinoma progression, loss/mutation of ARID2 impairs recruitment of DNMT1 to the SNAI1 promoter. As a result of decreased methylation at the SNAI1 promoter, there is an upregulation of SNAI1 expression that ultimately promotes EMT [68]. These results suggest that DNMT1 plays a cellular context-dependent role in tumor metastasis. Protein arginine methyltransferase (PRMT) 5 is a type II protein arginine methyltransferase. PRMT5 physically associates with SNAI1 and the NuRD (MAT1) complex to form a transcriptionally repressive complex that catalyzes a simultaneous histone demethylation and deacetylation. In addition, this complex also inhibits tet methylcytosine dioxygenase 1 (TET1) and contributes to DNA hypermethylation [69].
3.2. SNAI1 and Histone Modification
3.2.1. Acetylation
A variety of transcriptional co-activating complexes, which contain lysine acetyltransferase, catalyze lysine acetylation of histone tails. Because acetylation masks the positive charge on lysine residues and weakens the DNA–histone association and relaxes the chromatin structure, histone acetylation is often associated with gene activation. SNAI1 recruits the p300 activator complex to the VEGF and Sox2 promoters to stimulate their expression, leading to endothelium generation and tumor growth [70].
3.2.2. Deacetylation
Histone deacetylation by histone deacetylase (HDAC) is believed to restrict gene transcription because it reveals the positive charge of lysine and permits the DNA–histone interaction. HDACs, in particular HDAC1 and HDAC2, are often recruited by EMT-TFs to gene promoter regions and form protein complexes to deacetylate histones and silence expression of epithelial gene factors. For instance, SNAI1 mediates recruitment of the HDAC1/2 that contain Sin3A or NuRD repressor complexes to inhibit CDH1 expression by deacetylation of histones H3 and H4. This effect was abolished by treatment with the HDAC inhibitor Trichostatin A (TSA) [71,72]. Interestingly, HDAC2 can also be recruited by the HOP homeobox to epigenetically inhibit SNAI1 transcription, leading to the enhanced histone H3K9 deacetylation, which subsequently suppresses tumor progression [73]. Similarly, HDAC1 can be recruited by SATB homeobox 2 (SATB2) to the SNAI1 promoter, repressing SNAI1 transcription and inhibiting EMT [74]. Recently, it has been reported that HDAC8 increases the protein stability of SNAI1 via AKT/GSK3β signals [75]. HDAC8 interacts with AKT1 to decrease acetylation while increasing its phosphorylation, which further increases Ser9-phosphorylation of GSK3β. Sirt6, the class III histone deacetylates, functions as an NAD+-dependent histone deacetylase. Sirt6 interacts with p65 and attenuates NF-kB regulated SNAI1 expression by removing acetyl residues of histone H3K9 and H3K56 in the promoter regions of SNAI1 [76].
3.2.3. Acetylation Readers
The bromodomain-containing proteins (BRDs) are acetylation readers that bind to ε-N-aminoacetyl groups of nucleosomal histone lysines and recruit histone modifiers and transcriptional/remodeling factors to gene promoters; these processes promote upregulation or repression of gene expression. Ever increasing studies in different cancer cells have demonstrated the contribution of BRDs to cancer progression [77]. For instance, BRD4 interacts with SNAI1 if certain K146 and K187 are acetylated. This interaction prevents recognition of SNAI1 by its E3 ubiquitin ligases FBXL14 and β-TrCP1, thereby inhibiting SNAI1 polyubiquitination and proteasomal degradation [78]. In addition, BRD4 increases SNAI1 expression by diminishing the PKD1-mediated proteasome degradation pathway. BRD4 inhibition suppresses the expression of Gli1, which is required for transcriptional activation of SNAI1, indicating that BRD4 controls malignancy of breast cancer cells via both transcriptional and post-translational regulation of SNAI1 [79]. Therefore, inhibition of BRD4 is a promising therapeutic approach for cancer patients with metastatic lesions.
3.2.4. Methylation
Histone lysine methylation is catalyzed by lysine methyltransferases, which directly recruit or inhibit the recruitment of histone-binding proteins. Usually, H3K9 and H3K27 methylation is associated with transcriptional repression, while H3K79 is often linked with gene activation. G9a is responsible for the transcriptionally repressive modification of H3K9. In aggressive lung cancer cells, G9a is preferentially expressed, and its elevated expression correlates with poor prognosis. G9a represses a cell adhesion molecule EPCAM, which stimulates EMT and cancer metastasis by catalyzing H3K9me2 on its promoter [80]. In breast cancer cells, SNAI1 recruits G9a to the CDH1 promoter for transcription silencing. Therefore, inhibition of G9a reduces promoter H3K9me2 as well as DNA methylation which abrogates EMT and tumor metastasis [81]. Meanwhile, SNAI1 also interacts with Suv39H1, a histone methyltransferase for the trimethylation of histone H3 at lysine K9 (H3K9me3) to the CDH1 promoter to repress its transcription. EZH2, the catalytic subunit of the polycomb repressive complex 2 (PRC2), promotes transcriptional silencing of CDH1 by H3K27me3 [82,83]. EZH2 can interact with HDAC1/HDAC2 in association with SNAI1 to form a complex that represses CDH1 expression [84]. DOT1L catalyzes the methylation of an active transcription mark histone H3K79, which is crucial for tumor development [85]. In breast cancers, DOT1L forms a transcriptionally active complex with c-Myc and p300 to facilitate H3K79 methylation and acetylation in the promoter regions of SNAI1 that enhances SNAI1 de-repression, consequently promoting EMT [86].
3.2.5. Demethylation
Histone lysine-specific demethylase 1 (LSD1) functions as an epigenetic regulator by removing methyl groups on the transcription-activating H3K4 or repressing H3K9 residues through an amine oxidase reaction [87,88]. LSD1 takes part in a variety of chromatin-remodeling protein complexes to regulate tumor progression. We found that the amine oxidase domain of LSD1 interacts with the SNAG domain of SNAI1 [89]. SNAI1 recruits LSD1 and forms the SNAI1-LSD1-CoREST complex to repress CDH1 expression and enhance cell migration [89]. Another study indicated that SNAI1 recruits LSD1 on epithelial gene promoters for H3K4me2 demethylation, thereby silencing their expression and promoting EMT [90]. The chromatin remodeling factor Jumonji C (JmjC) domain-containing protein 3 (JMJD3, also known as KDM6B) is a α-ketoglutarate-dependent demethylase which is responsible for the demethylation of di- and trimethyllysine 27 (H3K27m2/3) on histone H3. JMJD3 demethylates H3K27m3 at the SNAI1 promoter to activate the transcription of SNAI1 during TGFβ-induced EMT [91]. In addition, JMJD1A also transcriptionally activates SNAI1 expression via H3K9me1 and H3K9me2 demethylation at its promoter [92].
4. Potential Pharmacological Inhibitors of SNAI1
Given the important role of SNAI1 in driving cancer progression, targeting SNAI1 would be an attractive anticancer therapeutic approach. However, the development of small molecules to inhibit SNAI1′s functions is hindered as there is no clear “ligand-binding domain” for targeting SNAI1. However, other strategies have been successfully attempted. First, the E-box, a SNAI1-binding site, was chosen as a target. A Co(III) complex conjugated to a CAGGTG hexanucleotide was synthesized. This complex binds to SNAI1 and prevents any interaction with DNA, thus reducing the invasive potential of tumor cells [93]. Second, the SNAI1-p53 complex acts as a target. Two leader compounds, GN25 and GN29, increase the expression of p53 and uncouple it from SNAI1. These two compounds selectively inhibit K-ras mutated cells [94]. Third, the LSD1-SNAI1 complex was chosen as a target. Inhibiting its interactions blocks cancer cell invasion [95,96]. Fourth, CYD19, a small-molecule compound, binds to SNAI1 and disrupts the SNAI1 interaction with p300, leading to SNAI1 degradation [97]. CYD19 impairs EMT-associated tumor invasion and metastasis by reversing SNAI1-driven EMT; this finding provides evidence that pharmacologic interference with SNAI1 acetylation may exert potent therapeutic effects in patients with cancer. Finally, chemical classes of synthetic and natural compounds affecting the transcriptional activity and expression of SNAI1 have already been characterized. For example, disulfiram inhibits cell migration, invasion, and growth of tumor grafts through the ERK/NF-κB/SNAI1 signaling pathway [98]. The proteasome inhibitor, NPI-0052, also inhibits SNAI1 expression via inhibition of NF-kB [99]. In all, targeting the SNAI1 complex or suppression of SNAI1 expression is one major approach to specifically inhibit SNAI1 activity.
Proteolysis-targeting chimeras (PROTACs) that hijack the ubiquitin-proteasome system for targeted protein degradation have expanded significantly in years [100]. This technology circumvents some of the limitations associated with traditional small-molecule therapeutics. PROTAC consists of a ligand for an E3 ligase and a ligand for a protein of interest (POI) connected by a chemical linker to form a ternary complex. In 2021, TRAnscription Factor Targeting Chimeras (TRAFTACs) technology was developed. The TRAFTAC system is composed of a HaloTag-fused dCas9 protein and a chimeric oligonucleotide that can bind transcription factor of interest (TOI) and dCas9 simultaneously [101]. This system labels the TOI with ubiquitin which then degrades the TOI by proteasomal machinery. This strategy was applied to target several transcription factors including E2F1 and NF-kB [102]. It will be attractive to design a TRAFTAC targeting SNAI1.
5. Conclusions and Perspective
SNAI1 as the key EMT regulator plays important roles in invasion and metastasis. The molecular events mediated by SNAI1 are of interest as therapeutic targets, in particular for resistant metastatic tumors. Although direct targeting of SNAI1 is unsuccessful, identifying inhibitors for PTMs of SNAI1 hold significant potential, and thus are a high priority in the development of future cancer treatments. Indeed, many pharmacological approaches, including chemical inhibitors and monoclonal antibodies that target these modification enzymes including deubiquitinase and kinase, have been devised and show promise for the treatment of tumor metastasis [103]. Furthermore, identification of SNAI1′s post-transcriptional and PTMs is crucial given that these changes could be identified in the primary tumor before metastasis occurs. Such knowledge would facilitate better prediction of patients who have genotypes that are more likely to follow an aggressive clinical course and who are prone to development of metastases.
In addition, because of the intimate connection between SNAI1 and chromatin-remodeling enzymes, targeting the epigenetic enzymes to reverse the EMT process is also an efficient and promising approach [104]. Indeed, abundant pre-clinical and clinical studies examining the effects of these epigenetic enzyme inhibitors alone or in combination with other anti-cancer agents are under development [105]. However, the impact of these epigenetic alternations on tumor metastasis differs greatly in various types of cancers. Therefore, it is urgent to comprehensively understand the mechanisms of action and roles of epigenetic modulations on EMT in different cancer types. These detailed mechanisms of epigenetic regulation in tumor metastasis will provide a bright future for the use of an efficient and specific “epigendrug” as one of the important therapeutic strategies in the fight against tumor metastasis.
Acknowledgments
We sincerely apologize to colleagues whose studies have been omitted from this review owing to space limitations. We thank Cathy Anthony for the critical editing of this manuscript.
Author Contributions
All authors contributed to the manuscript content and editing for this review. B.D. wrote the manuscript. Y.W. wrote the manuscript, provided supervision and financial support. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Shared Resources of the University of Kentucky Markey Cancer Center (P30CA177558). This research was also supported by grants from American Cancer Society Research Scholar Award (RSG13187) and NIH (P20GM121327 and CA230758) to Y.W.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors have declared that no conflict of interest exists.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Anderson R.L., Balasas T., Callaghan J., Coombes R.C., Evans J., Hall J.A., Kinrade S., Jones D., Jones P.S., Jones R., et al. A framework for the development of effective anti-metastatic agents. Nat. Rev. Clin. Oncol. 2019;16:185–204. doi: 10.1038/s41571-018-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 3.Yuan S., Norgard R.J., Stanger B.Z. Cellular Plasticity in Cancer. Cancer Discov. 2019;9:837–851. doi: 10.1158/2159-8290.CD-19-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puisieux A., Brabletz T., Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 5.Hu C.-T., Chang T.-Y., Cheng C.-C., Liu C.-S., Wu J.-R., Li M.-C., Wu W.-S. Snail associates with EGR-1 and SP-1 to upregulate transcriptional activation of p15INK4b. FEBS J. 2010;277:1202–1218. doi: 10.1111/j.1742-4658.2009.07553.x. [DOI] [PubMed] [Google Scholar]
- 6.Guaita S., Puig I., Francí C., Garrido M., Domínguez D., Batlle E., Sancho E., Dedhar S., de Herreros A.G., Baulida J. Snail Induction of Epithelial to Mesenchymal Transition in Tumor Cells Is Accompanied by MUC1 Repression andZEB1 Expression. J. Biol. Chem. 2002;277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- 7.Rembold M., Ciglar L., Yáñez-Cuna J.O., Zinzen R., Girardot C., Jain A., Welte M., Stark A., Leptin M., Furlong E.E. A conserved role for Snail as a potentiator of active transcription. Genes Dev. 2014;28:167–181. doi: 10.1101/gad.230953.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu C.-T., Wu J.-R., Chang T.Y., Cheng C.-C., Wu W.-S. The transcriptional factor Snail simultaneously triggers cell cycle arrest and migration of human hepatoma HepG2. J. Biomed. Sci. 2008;15:343–355. doi: 10.1007/s11373-007-9230-y. [DOI] [PubMed] [Google Scholar]
- 9.Ly T.M., Chen Y.-C., Lee M.-C., Hu C.-T., Cheng C.-C., Chang H.-H., You R.-I., Wu W.-S. Snail Upregulates Transcription of FN, LEF, COX2, and COL1A1 in Hepatocellular Carcinoma: A General Model Established for Snail to Transactivate Mesenchymal Genes. Cells. 2021;10:2202. doi: 10.3390/cells10092202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu W.-S., You R.-I., Cheng C.-C., Lee M.-C., Lin T.-Y., Hu C.-T. Snail collaborates with EGR-1 and SP-1 to directly activate transcription of MMP 9 and ZEB1. Sci. Rep. 2017;7:17753. doi: 10.1038/s41598-017-18101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kajita M., McClinic K.N., Wade P.A. Aberrant Expression of the Transcription Factors Snail and Slug Alters the Response to Genotoxic Stress. Mol. Cell. Biol. 2004;24:7559–7566. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani S.A., Guo W., Liao M.-J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong C., Yuan T., Wu Y., Wang Y., Fan T.W., Miriyala S., Lin Y., Yao J., Shi J., Kang T., et al. Loss of FBP1 by Snail-Mediated Repression Provides Metabolic Advantages in Basal-like Breast Cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y., Zhou B.P. Snail: More than Emt. Cell Adhes. Migr. 2010;4:199–203. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrallo-Gimeno A., Nieto M.A. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 16.Greally J.M. A user’s guide to the ambiguous word ‘epigenetics’. Nature reviews. Mol. Cell Biol. 2018;19:207–208. doi: 10.1038/nrm.2017.135. [DOI] [PubMed] [Google Scholar]
- 17.Wen B., Wu H., Shinkai Y., Irizarry R.A., Feinberg A.P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam W.L., Weinberg R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nieto M.A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 20.Joyce J.A., Pollard J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 2008;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Shi J., Chai K., Ying X., Zhou B.P. The Role of Snail in EMT and Tumorigenesis. Curr. Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufhold S., Bonavida B. Central role of snail1 in the regulation of emt and resistance in cancer: A target for therapeutic intervention. J. Exp. Clin. Cancer Res. 2014;33:1–19. doi: 10.1186/s13046-014-0062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito D., Kyakumoto S., Chosa N., Ibi M., Takahashi N., Okubo N., Sawada S., Ishisaki A., Kamo M. Transforming growth factor-beta1 induces epithelial-mesenchymal transition and integrin alpha3beta1-mediated cell migration of hsc-4 human squamous cell carcinoma cells through slug. J. Biochem. 2013;153:303–315. doi: 10.1093/jb/mvs144. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura R., Ishii H., Endo K., Hotta A., Fujii E., Miyazawa K., Saitoh M. Reciprocal expression of Slug and Snail in human oral cancer cells. PLoS ONE. 2018;13:e0199442. doi: 10.1371/journal.pone.0199442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smit M.A., Geiger T.R., Song J.Y., Gitelman I., Peeper D.S. A twist-snail axis critical for trkb-induced epithelial-mesenchymal transition-like transformation, anoikis resistance, and metastasis. Mol. Cell Biol. 2009;29:3722–3737. doi: 10.1128/MCB.01164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skrzypek K., Majka M. Interplay among SNAIL Transcription Factor, MicroRNAs, Long Non-Coding RNAs, and Circular RNAs in the Regulation of Tumor Growth and Metastasis. Cancers. 2020;12:209. doi: 10.3390/cancers12010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Liu R., Zhu W., Chu H., Yu H., Wei P., Wu X., Zhu H., Gao H., Liang J., et al. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature. 2019;571:127–131. doi: 10.1038/s41586-019-1340-y. [DOI] [PubMed] [Google Scholar]
- 28.Lin X., Chai G., Wu Y., Li J., Chen F., Liu J., Luo G., Tauler J., Du J., Lin S., et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019;10:2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Li F., Zhao H., Su M., Xie W., Fang Y., Du Y., Yu Z., Hou L., Tan W. Hnrnp-f regulates emt in bladder cancer by mediating the stabilization of snail1 mrna by binding to its 3’ UTR. EBioMedicine. 2019;45:208–219. doi: 10.1016/j.ebiom.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou B.P., Deng J., Xia W., Xu J., Li Y.M., Gunduz M., Hung M.C. Dual regulation of snail by gsk-3beta-mediated phos-phorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 31.Mimoto R., Taira N., Takahashi H., Yamaguchi T., Okabe M., Uchida K., Miki Y., Yoshida K. Dyrk2 controls the epithe-lial-mesenchymal transition in breast cancer by degrading snail. Cancer Lett. 2013;339:214–225. doi: 10.1016/j.canlet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Zheng H., Shen M., Zha Y.L., Li W., Wei Y., Blanco M.A., Ren G., Zhou T., Storz P., Wang H.Y., et al. Pkd1 phosphor-ylation-dependent degradation of snail by scf-fbxo11 regulates epithelial-mesenchymal transition and metastasis. Cancer Cell. 2014;26:358–373. doi: 10.1016/j.ccr.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung H.Y., Fattet L., Tsai J.H., Kajimoto T., Chang Q., Newton A.C., Yang J. Apical-basal polarity inhibits epitheli-al-mesenchymal transition and tumour metastasis by par-complex-mediated snai1 degradation. Nat. Cell Biol. 2019;21:359–371. doi: 10.1038/s41556-019-0291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun M., Guo X., Qian X., Wang H., Yang C., Brinkman K.L., Serrano-Gonzalez M., Jope R.S., Zhou B., Engler D.A., et al. Activation of the ATM-Snail pathway promotes breast cancer metastasis. J. Mol. Cell Biol. 2012;4:304–315. doi: 10.1093/jmcb/mjs048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyun B.-J., Seo H.R., Lee H.-J., Jin Y.B., Kim E.-J., Kim N.H., Kim H.S., Nam H.W., Yook J.I., Lee Y.-S. Mutual regulation between DNA-PKcs and snail1 leads to increased genomic instability and aggressive tumor characteristics. Cell Death Dis. 2013;4:e517. doi: 10.1038/cddis.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryu K.J., Park S.M., Park S.H., Kim I.K., Han H., Kim H.J., Kim S.H., Hong K.S., Kim H., Kim M., et al. P38 stabilizes snail by suppressing dyrk2-mediated phosphorylation that is required for gsk3beta-betatrcp-induced snail degradation. Cancer Res. 2019;79:4135–4148. doi: 10.1158/0008-5472.CAN-19-0049. [DOI] [PubMed] [Google Scholar]
- 37.MacPherson M.R., Molina P., Souchelnytskyi S., Wernstedt C., Martin-Pérez J., Portillo F., Cano A. Phosphorylation of serine 11 and serine 92 as new positive regulators of human snail1 function: Potential involvement of casein kinase-2 and the camp-activated kinase protein kinase a. Mol. Biol. Cell. 2010;21:244–253. doi: 10.1091/mbc.e09-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K., Corsa C., Ponik S., Prior J.L., Piwnica-Worms D., Eliceiri K., Keely P.J., Longmore G.D. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat. Cell Biol. 2013;15:677–687. doi: 10.1038/ncb2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z., Rayala S., Nguyen D., Vadlamudi R.K., Chen S., Kumar R. Pak1 Phosphorylation of Snail, a Master Regulator of Epithelial-to-Mesenchyme Transition, Modulates Snail’s Subcellular Localization and Functions. Cancer Res. 2005;65:3179–3184. doi: 10.1158/0008-5472.CAN-04-3480. [DOI] [PubMed] [Google Scholar]
- 40.Chen L., Pan X.-W., Huang H., Gao Y., Yang Q.-W., Wang L.-H., Cui X.-G., Xu D.-F. Epithelial-mesenchymal transition induced by GRO-α-CXCR2 promotes bladder cancer recurrence after intravesical chemotherapy. Oncotarget. 2017;8:45274–45285. doi: 10.18632/oncotarget.16786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Escriva M., Peiro S., Herranz N., Villagrasa P., Dave N., Montserrat-Sentis B., Murray S.A., Franci C., Gridley T., Virtanen I., et al. Repression of pten phosphatase by snail1 transcriptional factor during gamma radiation-induced apoptosis. Mol. Cell Biol. 2008;28:1528–1540. doi: 10.1128/MCB.02061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K., Rodriguez-Aznar E., Yabuta N., Owen R.J., Mingot J.M., Nojima H., Nieto M.A., Longmore G.D. Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. EMBO J. 2012;31:29–43. doi: 10.1038/emboj.2011.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu Z., Dong B., Guo W., Piotr R., Longmore G., Yang X., Yu Z., Deng J., Evers B.M., Wu Y. STK39 promotes breast cancer invasion and metastasis by increasing SNAI1 activity upon phosphorylation. Theranostics. 2021;11:7658–7670. doi: 10.7150/thno.62406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y., Evers B.M., Zhou B.P. Small C-terminal Domain Phosphatase Enhances Snail Activity through Dephosphorylation. J. Biol. Chem. 2009;284:640–648. doi: 10.1074/jbc.M806916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y., Liu J., Chen F., Feng X.H. C-terminal domain small phosphatase-like 2 promotes epithelial-to-mesenchymal tran-sition via snail dephosphorylation and stabilization. Open Biol. 2018;8:170274. doi: 10.1098/rsob.170274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Q., Zhou B.P., Wu Y. The regulation of snail: On the ubiquitin edge. Cancer Cell Microenviron. 2017;4:e1567. doi: 10.14800/ccm.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun R., Xie H.-Y., Qian J.-X., Huang Y.-N., Yang F., Zhang F.-L., Shao Z.-M., Li D.-Q. Fbxo22 possesses both protumor-igenic and antimetastatic roles in breast cancer progression. Cancer Res. 2018;78:5274–5286. doi: 10.1158/0008-5472.CAN-17-3647. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y., Zhou H., Zhu R., Ding F., Li Y., Cao X., Liu Z. Spsb3 targets snail for degradation in gsk-3beta phosphoryla-tion-dependent manner and regulates metastasis. Oncogene. 2018;37:768–776. doi: 10.1038/onc.2017.370. [DOI] [PubMed] [Google Scholar]
- 49.Park S., Park S., Ryu K., Kim I.-K., Han H., Kim H., Kim S., Hong K., Kim H., Kim M., et al. Downregulation of CHIP promotes ovarian cancer metastasis by inducing Snail-mediated epithelial–mesenchymal transition. Mol. Oncol. 2019;13:1280–1295. doi: 10.1002/1878-0261.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J.-H., Jung S.M., Yang K.-M., Bae E., Ahn S.G., Park J.S., Seo D., Kim M., Ha J., Lee J., et al. A20 promotes metastasis of aggressive basal-like breast cancers through multi-monoubiquitylation of Snail1. Nat. Cell Biol. 2017;19:1260–1273. doi: 10.1038/ncb3609. [DOI] [PubMed] [Google Scholar]
- 51.Wu Y., Wang Y., Lin Y., Liu Y., Wang Y., Jia J., Singh P., Chi Y.-I., Wang C., Dong C., et al. Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting Snail1 degradation. Nat. Commun. 2017;8:14228. doi: 10.1038/ncomms14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu T., Yu J., Deng M., Yin Y., Zhang H., Luo K., Qin B., Li Y., Wu C., Ren T. Cdk4/6-dependent activation of dub3 regulates cancer metastasis through snail1. Nat. Commun. 2017;8:1–12. doi: 10.1038/ncomms13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonego M., Pellarin I., Costa A., Vinciguerra G.L.R., Coan M., Kraut A., D’Andrea S., Dall’Acqua A., Castillo-Tong D.C., Califano D., et al. USP1 links platinum resistance to cancer cell dissemination by regulating Snail stability. Sci. Adv. 2019;5:eaav3235. doi: 10.1126/sciadv.aav3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian W., Li Q., Wu X., Li W., Li Q., Zhang J., Li M., Zhang D., Zhao H., Zou X., et al. Deubiquitinase USP29 promotes gastric cancer cell migration by cooperating with phosphatase SCP1 to stabilize Snail protein. Oncogene. 2020;39:6802–6815. doi: 10.1038/s41388-020-01471-0. [DOI] [PubMed] [Google Scholar]
- 55.Lambies G., Miceli M., Martinez-Guillamon C., Olivera-Salguero R., Pena R., Frias C.P., Calderon I., Atanassov B.S., Dent S.Y.R., Arribas J., et al. Tgfbeta-activated usp27x deubiquitinase regulates cell migration and chemoresistance via stabilization of snail1. Cancer Res. 2019;79:33–46. doi: 10.1158/0008-5472.CAN-18-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan L., Chen Z., Wu X., Cai X., Feng S., Lu J., Wang H., Liu N. Ubiquitin-Specific Protease 3 Promotes Glioblastoma Cell Invasion and Epithelial–Mesenchymal Transition via Stabilizing Snail. Mol. Cancer Res. 2019;17:1975–1984. doi: 10.1158/1541-7786.MCR-19-0197. [DOI] [PubMed] [Google Scholar]
- 57.Zhou H., Liu Y., Zhu R., Ding F., Cao X., Lin D., Liu Z. OTUB1 promotes esophageal squamous cell carcinoma metastasis through modulating Snail stability. Oncogene. 2018;37:3356–3368. doi: 10.1038/s41388-018-0224-1. [DOI] [PubMed] [Google Scholar]
- 58.Zhu R., Liu Y., Zhou H., Li L., Li Y., Ding F., Cao X., Liu Z. Deubiquitinating enzyme PSMD14 promotes tumor metastasis through stabilizing SNAIL in human esophageal squamous cell carcinoma. Cancer Lett. 2018;418:125–134. doi: 10.1016/j.canlet.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 59.Guo X., Zhu R., Luo A., Zhou H., Ding F., Yang H., Liu Z. EIF3H promotes aggressiveness of esophageal squamous cell carcinoma by modulating Snail stability. J. Exp. Clin. Cancer Res. 2020;39:175. doi: 10.1186/s13046-020-01678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai J., Li M., Wang X., Li L., Li Q., Hou Z., Jia H., Liu S. USP37 Promotes Lung Cancer Cell Migration by Stabilizing Snail Protein via Deubiquitination. Front. Genet. 2020;10:1324. doi: 10.3389/fgene.2019.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L., Zhou H., Zhu R., Liu Z. USP26 promotes esophageal squamous cell carcinoma metastasis through stabilizing Snail. Cancer Lett. 2019;448:52–60. doi: 10.1016/j.canlet.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 62.Gudey S.K., Sundar R., Heldin C.H., Bergh A., Landstrom M. Pro-invasive properties of snail1 are regulated by sumoylation in response to tgfbeta stimulation in cancer. Oncotarget. 2017;8:97703–97726. doi: 10.18632/oncotarget.20097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park S.Y., Kim H.S., Kim N.H., Ji S., Cha S.Y., Kang J.G., Ota I., Shimada K., Konishi N., Nam H.W., et al. Snail1 is stabilized by o-glcnac modification in hyperglycaemic condition. EMBO J. 2010;29:3787–3796. doi: 10.1038/emboj.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsu D.S.-S., Wang H.-J., Tai S.-K., Chou C.-H., Hsieh C.-H., Chiu P.-H., Chen N.-J., Yang M.-H. Acetylation of Snail Modulates the Cytokinome of Cancer Cells to Enhance the Recruitment of Macrophages. Cancer Cell. 2014;26:534–548. doi: 10.1016/j.ccell.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 65.Cui H., Hu Y., Guo D., Zhang A., Gu Y., Zhang S., Zhao C., Gong P., Shen X., Li Y., et al. DNA methyltransferase 3A isoform b contributes to repressing E-cadherin through cooperation of DNA methylation and H3K27/H3K9 methylation in EMT-related metastasis of gastric cancer. Oncogene. 2018;37:4358–4371. doi: 10.1038/s41388-018-0285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee E., Wang J., Yumoto K., Jung Y., Cackowski F.C., Decker A.M., Li Y., Franceschi R.T., Pienta K.J., Taichman R.S. DNMT1 Regulates Epithelial-Mesenchymal Transition and Cancer Stem Cells, Which Promotes Prostate Cancer Metastasis. Neoplasia. 2016;18:553–566. doi: 10.1016/j.neo.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Espada J., Peinado H., Lopez-Serra L., Setién F., Lopez-Serra P., Portela A., Renart J., Carrasco E., Calvo M., Juarranz A., et al. Regulation of SNAIL1 and E-cadherin function by DNMT1 in a DNA methylation-independent context. Nucleic Acids Res. 2011;39:9194–9205. doi: 10.1093/nar/gkr658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang H., Cao H.-J., Ma N., Bao W.-D., Wang J.-J., Chen T.-W., Zhang E.-B., Yuan Y.-M., Ni Q.-Z., Zhang F.-K., et al. Chromatin remodeling factor ARID2 suppresses hepatocellular carcinoma metastasis via DNMT1-Snail axis. Proc. Natl. Acad. Sci. USA. 2020;117:4770–4780. doi: 10.1073/pnas.1914937117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao J., Liu R., Feng D., Huang W., Huo M., Zhang J., Leng S., Yang Y., Yang T., Yin X., et al. Snail/PRMT5/NuRD complex contributes to DNA hypermethylation in cervical cancer by TET1 inhibition. Cell Death Differ. 2021;28:2818–2836. doi: 10.1038/s41418-021-00786-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang Z., Zhang Y., Liu J., Zheng Y., Li H., Kong Y., Li P., Peng H., Shi Y., Cao B., et al. Snail promotes the generation of vascular endothelium by breast cancer cells. Cell Death Dis. 2020;11:457. doi: 10.1038/s41419-020-2651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peinado H., Ballestar E., Esteller M., Cano A. Snail Mediates E-Cadherin Repression by the Recruitment of the Sin3A/Histone Deacetylase 1 (HDAC1)/HDAC2 Complex. Mol. Cell. Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujita N., Jaye D., Kajita M., Geigerman C., Moreno C.S., Wade P.A. MTA3, a Mi-2/NuRD Complex Subunit, Regulates an Invasive Growth Pathway in Breast Cancer. Cell. 2003;113:207–219. doi: 10.1016/S0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 73.Ren X., Yang X., Cheng B., Chen X., Zhang T., He Q., Li B., Li Y., Tang X., Wen X., et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat. Commun. 2017;8:14053. doi: 10.1038/ncomms14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y.-Q., Jiang D.-M., Hu S.-S., Zhao L., Wang L., Yang M.-H., Ai M.-L., Jiang H.-J., Han Y., Ding Y.-Q., et al. SATB2-AS1 Suppresses Colorectal Carcinoma Aggressiveness by Inhibiting SATB2-Dependent Snail Transcription and Epithelial–Mesenchymal Transition. Cancer Res. 2019;79:3542–3556. doi: 10.1158/0008-5472.CAN-18-2900. [DOI] [PubMed] [Google Scholar]
- 75.An P., Chen F., Li Z., Ling Y., Peng Y., Zhang H., Li J., Chen Z., Wang H. Hdac8 promotes the dissemination of breast cancer cells via akt/gsk-3beta/snail signals. Oncogene. 2020;39:4956–4969. doi: 10.1038/s41388-020-1337-x. [DOI] [PubMed] [Google Scholar]
- 76.Chen F., Ma X., Liu Y., Ma D., Gao X., Qian X. SIRT6 inhibits metastasis by suppressing SNAIL expression in nasopharyngeal carcinoma cells. Int. J. Clin. Exp. Pathol. 2021;14:63–74. [PMC free article] [PubMed] [Google Scholar]
- 77.Belkina A., Denis G.V. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qin Z.-Y., Wang T., Su S., Shen L.-T., Zhu G.-X., Liu Q., Zhang L., Liu K.-W., Zhang Y., Zhou Z.-H., et al. BRD4 promotes gastric cancer progression and metastasis through acetylation-dependent stabilization of Snail. Cancer Res. 2019;79:4869–4881. doi: 10.1158/0008-5472.CAN-19-0442. [DOI] [PubMed] [Google Scholar]
- 79.Lu L., Chen Z., Lin X., Tian L., Su Q., An P., Li W., Wu Y., Du J., Shan H., et al. Inhibition of brd4 suppresses the ma-lignancy of breast cancer cells via regulation of snail. Cell Death Differ. 2020;27:255–268. doi: 10.1038/s41418-019-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen M.-W., Hua K.-T., Kao H.-J., Chi C.-C., Wei L.-H., Johansson G., Shiah S.-G., Chen P.-S., Jeng Y.-M., Cheng T.-Y., et al. H3K9 Histone Methyltransferase G9a Promotes Lung Cancer Invasion and Metastasis by Silencing the Cell Adhesion Molecule Ep-CAM. Cancer Res. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 81.Dong C., Wu Y., Yao J., Wang Y., Yu Y., Rychahou P., Evers B.M., Zhou B.P. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Investig. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herranz N., Pasini D., Díaz V.M., Francí C., Gutierrez A., Dave N., Escrivà M., Hernandez-Muñoz I., Di Croce L., Helin K., et al. Polycomb Complex 2 Is Required for E-cadherin Repression by the Snail1 Transcription Factor. Mol. Cell. Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao Q., Yu J., Dhanasekaran S.M., Kim J.H., Mani R., Tomlins S., Mehra R., Laxman B., Cao X., Kleer C.G., et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tong Z.-T., Cai M.-Y., Wang X.-G., Kong L.-L., Mai S.-J., Liu Y.-H., Zhang H.-B., Liao Y.-J., Zheng F., Zhu W.-G., et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2011;31:583–594. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- 85.Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V.M., Su L., Xu G., Zhang Y. hDOT1L Links Histone Methylation to Leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 86.Cho M.H., Park J.-H., Choi H.-J., Park M.-K., Won H.-Y., Park Y.-J., Lee C.H., Oh S.-H., Song Y.-S., Kim H.S., et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat. Commun. 2015;6:7821. doi: 10.1038/ncomms8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Metzger E., Wissmann M., Yin N., Müller J.M., Schneider R., Peters A.H., Günther T., Buettner R., Schüle R. Lsd1 de-methylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 88.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R., Shi Y. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 89.Lin Y., Wu Y., Li J., Dong C., Ye X., Chi Y.-I., Evers B.M., Zhou B.P. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin T., Ponn A., Hu X., Law B.K., Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramadoss S., Chen X., Wang C.-Y. Histone Demethylase KDM6B Promotes Epithelial-Mesenchymal Transition. J. Biol. Chem. 2012;287:44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tang D.E., Dai Y., Fan L.L., Geng X.Y., Fu D.X., Jiang H.W., Xu S.H. Histone demethylase jmjd1a promotes tumor pro-gression via activating snail in prostate cancer. Mol. Cancer Res. 2020;18:698–708. doi: 10.1158/1541-7786.MCR-19-0889. [DOI] [PubMed] [Google Scholar]
- 93.Vistain L.F., Yamamoto N., Rathore R., Cha P., Meade T.J. Targeted Inhibition of Snail Activity in Breast Cancer Cells by Using a CoIII-Ebox Conjugate. ChemBioChem. 2015;16:2065–2072. doi: 10.1002/cbic.201500289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee S.-H., Shen G.-N., Jung Y.S., Lee S.-J., Chung J.-Y., Kim H.-S., Xu Y., Choi Y., Lee J.-W., Ha N.-C., et al. Antitumor effect of novel small chemical inhibitors of Snail-p53 binding in K-Ras-mutated cancer cells. Oncogene. 2010;29:4576–4587. doi: 10.1038/onc.2010.208. [DOI] [PubMed] [Google Scholar]
- 95.Baron R., Binda C., Tortorici M., McCammon J.A., Mattevi A. Molecular Mimicry and Ligand Recognition in Binding and Catalysis by the Histone Demethylase LSD1-CoREST Complex. Structure. 2011;19:212–220. doi: 10.1016/j.str.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ferrari-Amorotti G., Fragliasso V., Esteki R., Prudente Z., Soliera A.R., Cattelani S., Manzotti G., Grisendi G., Dominici M., Pieraccioli M., et al. Inhibiting Interactions of Lysine Demethylase LSD1 with Snail/Slug Blocks Cancer Cell Invasion. Cancer Res. 2013;73:235–245. doi: 10.1158/0008-5472.CAN-12-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li H.M., Bi Y.R., Li Y., Fu R., Lv W.C., Jiang N., Xu Y., Ren B.X., Chen Y.D., Xie H., et al. A potent cbp/p300-snail in-teraction inhibitor suppresses tumor growth and metastasis in wild-type p53-expressing cancer. Sci. Adv. 2020;6:eaaw8500. doi: 10.1126/sciadv.aaw8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han D., Wu G., Chang C., Zhu F., Xiao Y., Li Q., Zhang T., Zhang L. Disulfiram inhibits tgf-β-induced epitheli-al-mesenchymal transition and stem-like features in breast cancer via erk/nf-κb/snail pathway. Oncotarget. 2015;6:40907–40919. doi: 10.18632/oncotarget.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baritaki S., Chapman A., Yeung K., Spandidos D., Palladino M., Bonavida B. Inhibition of epithelial to mesenchymal transition in metastatic prostate cancer cells by the novel proteasome inhibitor, NPI-0052: Pivotal roles of Snail repression and RKIP induction. Oncogene. 2009;28:3573–3585. doi: 10.1038/onc.2009.214. [DOI] [PubMed] [Google Scholar]
- 100.Burslem G., Crews C.M. Proteolysis-Targeting Chimeras as Therapeutics and Tools for Biological Discovery. Cell. 2020;181:102–114. doi: 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Samarasinghe K.T., Jaime-Figueroa S., Burgess M., Nalawansha D.A., Dai K., Hu Z., Bebenek A., Holley S.A., Crews C.M. Targeted degradation of transcription factors by TRAFTACs: TRAnscription Factor TArgeting Chimeras. Cell Chem. Biol. 2021;28:648–661.e5. doi: 10.1016/j.chembiol.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu Z., Crews C.M. Recent Developments in PROTAC-Mediated Protein Degradation: From Bench to Clinic. ChemBioChem. 2021 doi: 10.1002/cbic.202100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lai K.P., Chen J., Tse W.K.F. Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy. Int. J. Mol. Sci. 2020;21:2548. doi: 10.3390/ijms21072548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dong B., Qiu Z., Wu Y. Tackle Epithelial-Mesenchymal Transition with Epigenetic Drugs in Cancer. Front. Pharmacol. 2020;11:596239. doi: 10.3389/fphar.2020.596239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park J.W., Han J.-W. Targeting epigenetics for cancer therapy. Arch. Pharmacal Res. 2019;42:159–170. doi: 10.1007/s12272-019-01126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.