Abstract
Computer-assisted DNA fingerprinting with the complex probe Ca3 has been used to analyze the relatedness of isolates collected from individuals with nosocomial bloodstream infections (BSIs) and hospital care workers (HCWs) in the surgical and neonatal intensive care units (ICUs) of four hospitals. The results demonstrate that for the majority of patients (90%), isolates collected from commensal sites before and after collection of a BSI isolate were highly similar or identical to the BSI isolate. In addition, the average similarity coefficient for BSI isolates was similar to that for unrelated control isolates. However, the cluster characteristics of BSI isolates in dendrograms generated for each hospital compared to those of unrelated control isolates in a dendrogram demonstrated a higher degree of clustering of the former. In addition, a higher degree of clustering was observed in mixed dendrograms for HCV isolates and BSI isolates for each of the four test hospitals. In most cases, HCW isolates from an ICU were collected after the related BSI isolate, but in a few cases, the reverse was true. Although the results demonstrate that single, dominant endemic strains are not responsible for nosocomial BSIs in neonatal ICUs and surgical ICUs, they suggest that multiple endemic strains may be responsible for a significant number of cases. The results also suggest that cross-contamination occurs between patients and HCWs and between HCWs in the same ICU and in different ICUs. The temporal sequence of isolation also suggests that in the majority of cases HCWs are contaminated by isolates from colonized patients, but in a significant minority, the reverse is true. The results of this study provide the framework for a strategy for more definitive testing of the origins of Candida albicans strains responsible for nosocomial infections.
Immunosuppression and other compromising conditions can result in life-threatening fungal bloodstream infections (BSIs). When such infections arise during hospitalization, they are referred to as nosocomial infections (31). There is an inclination to assume that because these infections arise in a hospital setting, the origin of the infecting strain is the hospital staff or environment. However, because Candida albicans and related species, which are responsible for the majority of nosocomial fungal infections (31), reside in the natural microflora of a majority of immunocompetent individuals as relatively benign commensal organisms (55), a nosocomial infection may also originate from the commensal strain carried into the hospital by the patient. If a nosocomial infection arises from an endogenous commensal strain, prior mucosal colonization has been implicated as an independent risk factor (9, 18, 41, 63) and the gastrointestinal tract has been implicated as the most likely reservoir (1, 28, 30, 38, 41, 60, 61, 63). In the case of exogenous transmission, contaminated infusates, biomedical devices, and the hands of health care workers (HCWs) represent documented sources (11, 12, 17, 26, 39, 46, 47, 51). In cases of nosocomial infections in newborns, the infection must originate in the hospital setting (40) since we can assume that the fetus is sterile in utero.
The epidemiology of nosocomial infections of Candida spp. has been investigated by a variety of DNA fingerprinting methods, including restriction fragment length polymorphism analysis (3, 6, 19, 39), electrophoretic karyotyping (8, 14, 59), randomly amplified polymorphic DNA analysis (13, 21, 42), and Southern blot hybridization with discriminating probes (27, 34, 44, 48, 56). Although the majority of these methods hold the potential for use in strain discrimination and valid cluster analyses, in most studies that have used them there has been no attempt to validate the methods used, no attempt to quantitate the levels of similarity or dissimilarity of isolates, and no attempt to perform cluster analyses of moderately related isolates. Instead, there has been complete reliance on subjective interpretations. Even more disturbing is the absence in most studies of a collection of unrelated isolates analyzed by the same fingerprinting method for comparison. Straightforward methods have been developed to assess the efficacy of a DNA fingerprinting method and to test whether the method possesses the necessary attributes for broad epidemiological studies (36, 53, 58). These attributes include the capacity to (i) identify the same strain in different isolates, (ii) distinguish between completely unrelated strains, (iii) cluster moderately related isolates, and (iv) distinguish microevolution in highly similar but nonidentical isolates. Recently, the use of Southern blot hybridization with the complex species-specific probe Ca3 was validated for DNA fingerprinting of C. albicans by demonstrating parity between it and both the method of randomly amplified polymorphic DNA analysis and the method of multilocus enzyme electrophoresis (36). This characterization of Ca3 fingerprinting provided quantitative measures of (i) identicalness, (ii) microevolution and high levels of relatedness, (iii) thresholds for clustering of moderately related isolates, and (iv) unrelatedness. It also provided cluster characteristics for a set of unrelated isolates that can be used to assess the relatedness of other sets of C. albicans isolates, such as collections of nosocomial isolates (36).
In the present study, we have used this validated DNA fingerprinting method to examine the relatedness of isolates of C. albicans from candidemia patients in the neonatal and surgical intensive care units (NICUs and SICUs, respectively) of four medical centers participating in the National Epidemiology of Mycoses Survey (NEMIS) (29, 35). The collection included 35 isolates primarily from the blood of 30 patients with candidemia and 75 isolates from stool, urine, respiratory, and/or gastric specimens from 28 of these patients collected before, during, and/or after collection of the isolates that caused candidemia. Infections did not occur in close temporal association in any of the four hospitals. In addition, 42 isolates were obtained from the hands of HCWs in the same intensive care units (ICUs) at the time of or very close to the time of infection.
MATERIALS AND METHODS
Collection of isolates.
The NEMIS study was established under the auspices of Pfizer Inc. (New York, N.Y.) to define the spectrum of pathogens in seven hospitals causing nosocomial fungal infections in SICUs and NICUs, to characterize the organisms with respect to their susceptibilities to commonly used antifungal agents, and to define the molecular epidemiology of these infections with respect to endogenous sources and cross-infection (29, 35). The four hospitals in the study reported here were located in New York, N.Y. (hospital A), Iowa City, Iowa (hospital B), San Antonio, Tex. (hospital C), and Atlanta, Ga. (hospital D). Prospective surveillance was conducted over a 2-year period (1993 to 1995) for all patients who were hospitalized for at least 72 h in the SICU and NICU of each study site. ICU-acquired candidemia was defined as the occurrence of a new episode after a minimum of 72 h of hospitalization in the respective ICU. Microbiologic studies included weekly surveillance cultures of stool and urine for Candida spp. (rectal swab only for NICU patients), as described previously (28, 35). Thirty-five isolates were obtained from 30 ICU patients with infections, and for 28 of these patients, 75 additional isolates were collected from other body locations that can normally be colonized by commensal strains. Because 31 of the 35 isolates from ICU patients with infections were derived from patients with BSIs, all such isolates will be referred to as BSI isolates for convenience. The remaining four isolates from ICU patients with infections were obtained from peritoneal fluid, ascitic fluid, a tissue biopsy specimen, and an abscess. Specimens were also obtained from the hands of HCWs by the broth-bag method (57). Specimens from hands were obtained on a monthly basis and whenever an episode of candidemia was recognized in an ICU. Forty-two C. albicans isolates were obtained. Because of privacy rules at the respective institutions, HCWs could be identified only by their professional role, and therefore, specific HCWs could not be tracked over time, although in most cases HCWs were distinguishable from one another by professional role, ICU, and time of sampling. Isolates from patients were initially labeled according to patient, day of isolation, and body location. For example, an isolate obtained from the stool of patient 4 on day 106 was labeled P4(106)st. In dendrograms developed exclusively for BSI isolates, the isolates were also labeled according to ICU and date of isolation. For example, an isolate from patient 2 in the NICU collected on 29 March 1995 was labeled P2 N 3/29/95. HCW isolates were labeled according to HCW title, ICU, and date of collection. For example, an isolate from HCW5, a registered nurse in the NICU, collected on 20 March 1995 was labeled HW5RN N 3/20/95. The following HCW titles and abbreviations are used: registered nurse, RN; supervisor, SU; medical doctor, MD; X-ray technician, XT; nurse’s assistant, NA; technician, TE; respiratory therapist, RT; nurse’s orderly, NU; clerk, CL; and other, OT.
Organism identification.
All isolates of Candida spp. were initially identified to the species level by routine procedures established at each participating institution and were then sent to the University of Iowa Hospitals and Clinics for banking and further analysis (35). Upon receipt at the University of Iowa, isolates were subcultured onto potato dextrose agar (Remel, Lenexa, Kan.) and CHROMagar (Hardy Diagnostics, Santa Maria, Calif.) to assess viability and species homogeneity. Species were then identified with Vitek and API products (bio Merieux, St. Louis, Mo.) and by other conventional methods as required (62). All Candida isolates were stored as water suspensions or on agar slants at ambient temperature.
DNA fingerprinting.
The complex DNA probe Ca3 (2, 22, 45) was used to assess the genetic relatedness and microevolution of the C. albicans isolates (22, 23, 55). The methods for DNA preparation and electrophoresis have been presented in detail elsewhere (24). DNA from reference strain 3153A was run in the outer two lanes of each gel in order to normalize the gel image.
DNA fingerprint analysis.
To compare the fingerprints of isolates, the DENDRON software package (version 2.0; Solltech Inc., Oakdale, Iowa), based in a Macintosh computer, was used. The methods for analysis of fingerprint patterns have been described in detail elsewhere (53). Autoradiogram images were digitized and processed for distortions. Lanes and bands were automatically identified, and the similarity coefficient (SAB) between patterns for every pair of isolates A and B was computed by the formula SAB = 2E/(2E + a + b), where E is the number of common bands in the patterns of A and B, a is the number of bands in pattern A with no correlates in pattern B, and b is the number of bands in pattern B with no correlates in pattern A. Dendrograms based on SAB values were generated by the unweighted pair-group method with arithmetic averages (UPGMA) (52). To test the stability of clusters generated by UPGMA, the Test Dendrogram Stability option of the DENDRON, version 2.0, software package was used. In this assessment, the order of data entry was randomized 20 times, and members of the major clusters at an SAB threshold of 0.80 were assessed.
Statistical tests.
A two-sample t test for independent samples with unequal variances was used to compare the average SABs between defined collections (43). The distributions of SABs were verified to be normal enough for the t test. A chi-square test was used to compare proportions of isolates in clusters generated at a particular SAB threshold (43).
RESULTS
DNA fingerprinting with the Ca3 probe.
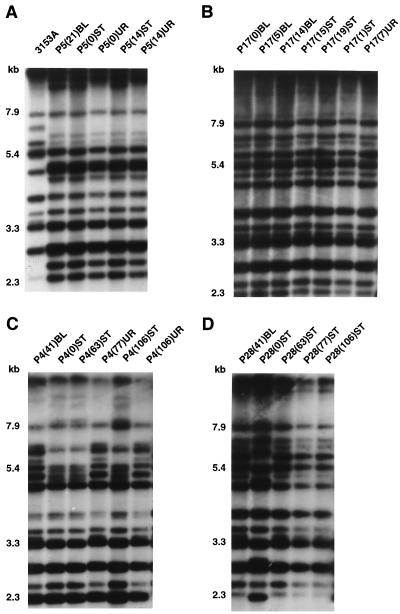
Representative Ca3 Southern blot hybridization patterns obtained with probe Ca3 are presented in Fig. 1A through D for collections from patients P5, P17, P4, and P28, respectively. Ca3 hybridized to between 15 and 20 bands in each Southern blot under the conditions used, but only the 10 to 15 bands above 2.3 kb were used to compute SABs (48). With the Ca3 probe, EcoRI-digested genomic DNA, and an SAB based on band position alone, it has been empirically demonstrated that (i) an SAB of 1.00 is achieved with multiple samples of the same clone; (ii) SABs ranging between 0.90 and 0.99 represent highly similar but nonidentical patterns and usually reflect microevolution of a single strain when isolates are obtained from the same patient; (iii) SABs ranging between 0.80 and 0.89 represent patterns for less related isolates that can still be clustered in a reproducible fashion; and (iv) SABs below 0.75 represent patterns for unrelated isolates (22, 23, 36, 37, 53).
FIG. 1.
Examples of the Southern blot hybridization patterns obtained with the complex probe Ca3. (A) Isolates from patient P5 and the reference strain 3153A; (B) isolates from patient P17; (C) isolates from patient P4; (D) isolates from patient P28. Isolate labels are explained in Materials and Methods. Molecular weights (in kilobases) are noted to the left of each Southern blot hybridization pattern.
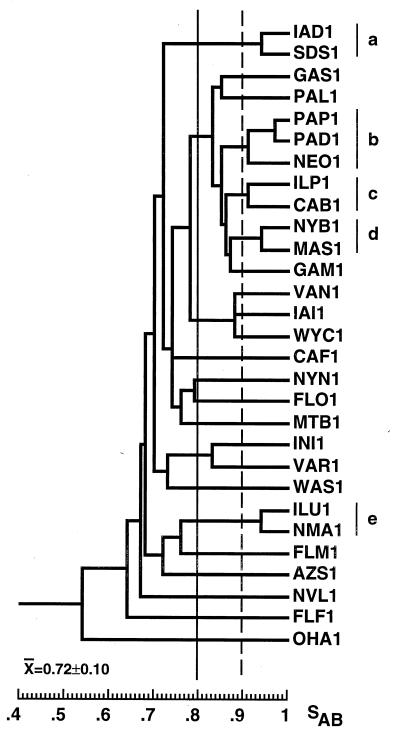
In Fig. 2, a dendrogram has been generated from the SABs computed for 29 unrelated BSI isolates, each collected in a different hospital in the continental United States (34). The average SAB for this control collection is 0.72 ± 0.10, which represents an estimate of unrelatedness for BSI isolates that will be used in this study. The dendrogram generated for this collection also provides a measure of clustering among unrelated isolates at selected thresholds (53).
FIG. 2.
Dendrogram of a control collection of 29 unrelated BSI isolates each collected from a different hospital across the continental United States. a through e, clusters of two or more isolates with SABs of ≥ 0.90. SAB thresholds for cluster analysis are drawn at 0.80 (straight line) and 0.90 (dashed line).
Comparison of BSI and commensal isolates from the same patients.
In Table 1, the sequence of isolates, their anatomical origins, and the times of isolation are presented for isolates from each of the 30 infected patients. In addition, the average SAB, the SAB between the BSI isolate and the commensal isolate obtained immediately preceding collection of the BSI isolate, and the SAB between the BSI isolate and the commensal isolate obtained immediately succeeding collection of the BSI isolate are presented for each patient. For 19 patients, an isolate was obtained from a site of commensal carriage prior to collection of the first BSI isolate (Table 1). In 17 of these patients (89%), the average SAB between the commensal isolates and the subsequent BSI isolate ranged between 0.91 and 1.00, a range of values considered to reflect high levels of relatedness (22, 23, 34, 53). The patterns of the isolates from patient P5 obtained by hybridization with the Ca3 probe (Fig. 1A) provide an example of the high level of relatedness observed between the BSI and preceding isolates in a majority of patients. Isolate P5(21)BL and the isolates from two prior urine samples, isolates P5(0)UR and P5(14)UR, differed by only one high-molecular-mass band, while isolate P5(21)BL and the two isolates from prior stool samples, isolates P5(0)ST and P5(14)ST, were identical. For only 2 of the 17 patients, patients P4 and P28, did the initial commensal isolates differ markedly from the subsequent BSI isolates (Fig. 1C and D, respectively). Interestingly, the times between collection of the commensal isolates and the subsequent BSI isolates in these two patients were the most extensive in the collection: 41 days for each patient (Table 1).
TABLE 1.
Description of collections of C. albicans isolates from individuals with BSIs or other candidemiasa
| Hospital | ICU | Patient no. | No. of isolates | Anatomic origins of isolatesa | Time of isolation (days)a | Time (days) between first isolate and Candidemia | Average SAB | SAB for ST and UR isolates and subsequent blood isolates | SAB for blood isolate and subsequent ST and UR isolates |
|---|---|---|---|---|---|---|---|---|---|
| A | SICU | P1 | 5 | ST-ST-UR, UR-BL | 0-7-15, 15-17 | 17 | 0.99 | 1.00 | |
| NICU | P2 | 4 | UR-BL-UR-ST | 0-4-6-12 | 4 | 1.00 | 1.00 | 1.00 | |
| NICU | P3 | 3 | ST-ST-BL | 0-7-13 | 13 | 0.94 | 0.91 | ||
| SICU | P16 | 4 | ST, TR-AB-ST | 0, 0-7-8 | 7 | 1.00 | 1.00 | 1.00 | |
| NICU | P17 | 7 | BL-ST-BL-UR-BL-ST-ST | 0-1-5-7-14-15-19 | 0 | 1.00 | 1.00 | ||
| NICU | P18 | 5 | BL-GA-ST-BL-ST | 0-1-2-3-21 | 0 | 0.96 | 0.93 | ||
| B | SICU | P10 | 6 | ST, UR-ST, UR-BL-ST | 0, 0-4, 4-5-7 | 5 | 1.00 | 1.00 | 1.00 |
| SICU | P11 | 3 | ST-BL-UR | 0-9-10 | 9 | 0.98 | 0.97 | 0.97 | |
| NICU | P12 | 2 | ST-BL | 0-2 | 2 | 0.97 | 0.97 | ||
| NICU | P13 | 2 | ST-BL | 0-21 | 21 | 1.00 | 1.00 | ||
| SICU | P14 | 4 | ST-ST-BL-PR | 0-20-27-28 | 27 | 1.00 | 1.00 | 1.00 | |
| SICU | P21 | 5 | TB-ST-AS-ST, UR | 0-6-10-11, 11 | 10 | 0.93 | 0.91 | 0.88 | |
| SICU | P22 | 2 | BL-UR | 0-2 | 0 | 1.00 | 1.00 | ||
| SICU | P23 | 5 | TR-UR-BL-TR, UR | 0-8-10-11, 11 | 10 | 1.00 | 1.00 | 1.00 | |
| NICU | P24 | 2 | BL-ST | 0-2 | 0 | 1.00 | 1.00 | ||
| C | SICU | P15 | 4 | UR-UR, SP, BL | 0-12, 12, 12 | 12 | 1.00 | 1.00 | |
| SICU | P27 | 2 | BL-ST | 0-1 | 0 | 1.00 | 1.00 | ||
| NICU | P31 | 1 | BL | 0 | |||||
| D | SICU | P4 | 6 | ST-BL-ST-LR-ST, UR | 0-41-63-77-106, 106 | 41 | 0.83 | 0.73 | 0.73 |
| SICU | P5 | 5 | ST, UR-ST, UR-BL | 0, 0-14, 14-21 | 21 | 0.98 | 0.97 | ||
| NICU | P6 | 4 | ST, BL, ST, ST | 0-2-7-14 | 2 | 1.00 | 1.00 | 1.00 | |
| NICU | P7 | 4 | ST, BL-ST-ST | 0-6-7-14 | 6 | 1.00 | 1.00 | 1.00 | |
| NICU | P8 | 4 | ST-ST-ST, BL | 0-7-13, 13 | 13 | 1.00 | 1.00 | ||
| NICU | P9 | 4 | ST-ST-ST-BL | 0-7-14-17 | 17 | 1.00 | 1.00 | ||
| NICU | P19 | 4 | ST-ST-BL-ST | 0-8-11-15 | 11 | 1.00 | 1.00 | 1.00 | |
| NICU | P20 | 3 | BL-ST-ST | 0-5-20 | 0 | 1.00 | 1.00 | ||
| NICU | P26 | 2 | BL-ST | 0-60 | 0 | 1.00 | 1.00 | ||
| SICU | P28 | 6 | ST-BL-ST-ST-ST, UR | 0-41-63-77-106, 106 | 41 | 0.92 | 0.79 | 1.00 | |
| NICU | P29 | 1 | BL | 0 | |||||
| NICU | P30 | 1 | BL | 0 |
Abbreviations: ST, stool; UR, urine; BL, blood; TR, tracheal lavage; AB, abscess; GA, gastric aspirate; PR, peritoneal fluid; TB, tissue biopsy specimen; AS, ascitic fluid; SP, sputum. For simplicity, isolates from blood, peritoneal fluid, tissue biopsy specimens, abscesses, and ascitic fluid are referred to as BSI isolates in the text, since 28 of the 30 infected patients (93%) had BSIs. Sequences of anatomic origin and dates of collection are noted.
For 18 patients an isolate was collected from a site of commensal carriage after the BSI isolate was collected (Table 1). For 16 of these patients (89%), the SAB between the BSI and subsequent commensal isolates ranged between 0.91 and 1.00 (Table 1). In two patients (11%), patients P4 and P28, a subsequent commensal isolate was unrelated to the BSI isolate (Table 1; Fig. 1C and D, respectively). In the case of patient P4, isolate P4(106)ST was unrelated to P4(41)BL (Fig. 1C). Interestingly, the stool isolate, isolate P4(0)ST, was also unrelated to the BSI isolate but was identical to subsequent stool isolates P4(63)ST and P4(106)ST (Fig. 1C). These results suggest that patient P4 was initially colonized by two unrelated strains, one in the stool and one in the urine, and that the strain in the urine emerged as the cause of the BSI. In the case of patient P28, preceding stool isolate P28(0)ST was unrelated to P28(41)BL, but subsequent stool isolates P28(63)ST, P28(77)ST, and P28(106)ST were identical to P28(41)BL (Fig. 1D; Table 1), suggesting strain replacement. In a third scenario, the BSI isolate was similar but nonidentical to the commensal isolates, suggesting significant microevolution. In patient P21, the BSI isolate and both the succeeding commensal isolates were similar but nonidentical, with SABs of 0.91 and 0.88, respectively (Table 1).
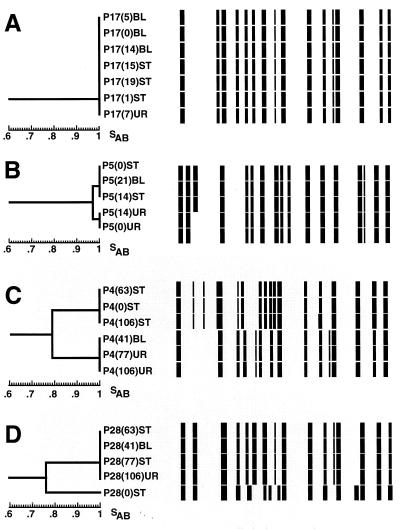
For the 18 collections of isolates with an average SAB of 1.00 (Table 1), the dendrogram that was generated was composed of a single cluster at an SAB of 1.00. The collection of isolates from patient P17 provides an example of such a dendrogram (Fig. 3A). For dendrograms of collections with average SABs below 1.00, the complexity (degree of branching) of the dendrogram increased. For example, the dendrogram generated for the collection of isolates from patient P5, which had an average SAB of 0.98 (Table 1), contained two clusters separated by a node at an SAB of 0.97. One cluster contained two identical stool isolates collected at days 0 and 14, and the second cluster contained three identical isolates, two from urine collected on days 0 and 14, suggesting that substrains resulting from microevolution had established themselves in alternative body locations. For this patient the blood isolate clustered with the stool isolates (Figure 3B). In Fig. 3C, a dendrogram is presented for the collection of isolates from patient P4; the average SAB for these isolates was 0.83 (Table 1). The dendrogram contained two clusters with a node at an SAB of 0.79. Just as in the case of the dendrogram for the collection of isolates from patient P5, the isolates from stool and urine samples separated into respective clusters, but in this case the separated clusters appeared to represent two unrelated strains. The blood isolate from this patient clustered with the urine isolates, not the stool isolates (Fig. 3C). In Fig. 3D, a dendrogram is presented for the collection of isolates from patient P28; the average SAB for these isolates was 0.92 (Table 1). A node at an SAB of 0.76 separated the first stool isolate from a cluster containing the subsequent blood, stool, and urine isolates. The combined results summarized in Table 1 demonstrate that for the majority of patients, commensal isolates and the subsequent BSI isolate from the same patient are highly similar, and for the majority of patients, a BSI isolate and subsequent commensal isolates from the same patient are highly similar. The results also suggest that in one-third of the patients, microevolution occurs in the colonizing strain.
FIG. 3.
Examples of dendrograms for collections of isolates from patients in which all isolates are identical (A), isolates show some variability reflecting microevolution (B), and isolates separate into unrelated clusters of isolates (C and D). Horizontal models of the Ca3 hybridization patterns are displayed next to the respective isolates in the dendrograms.
Genetic relatedness of BSI isolates obtained from the same hospital.
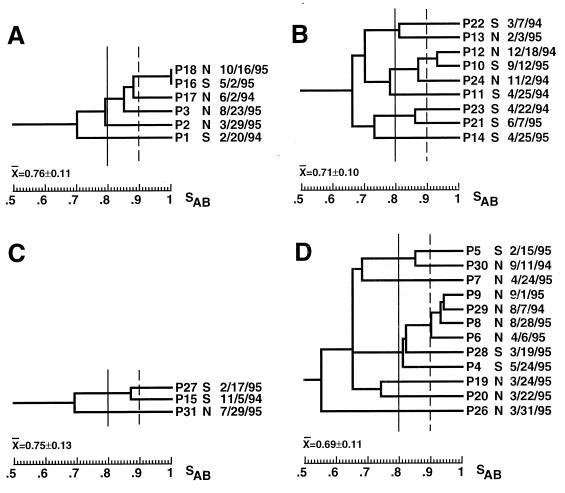
To obtain a measure of the genetic diversity of BSI isolates in each test hospital, dendrograms that included only one BSI isolate from each patient were generated for each hospital (Fig. 4). The average SABs for these restricted collections from hospitals A, B, C, and D were 0.76 ± 0.10 (n = 6), 0.71 ± 0.10 (n = 9), 0.75 ± 0.13 (n = 3), and 0.69 ± 0.11 (n = 12), respectively (Table 2). The average SAB for the combined collection of isolates from patients with candidemia from the four hospitals was 0.72 ± 0.10 (n = 30), which was identical to the value obtained for the 29 unrelated BSI isolates described previously (34) (Table 2). While the average SABs for the collections from hospitals B and D were lower than that for the random control collection of BSI isolates (P = 0.358 and 0.616, respectively), the average SABs for the collections from hospitals A and C were slightly higher (P = 0.265 and 0.615, respectively).
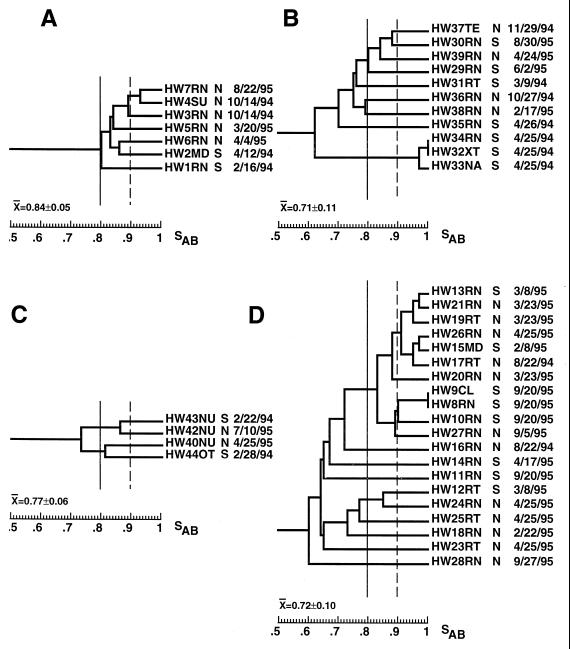
FIG. 4.
Dendrograms of the BSI isolates from the patients in hospitals A (A), B (B), C (C), and D (D). Only one BSI isolate from each patient was incorporated. The type of ICU and the date of collection are noted to the right of each isolate. Arbitrary SAB thresholds are drawn at 0.80 (straight line) and 0.90 (dashed line).
TABLE 2.
Average SABs for patients and staff of select hospitals compared with that of control isolates
| Hospital(s) | Group | No. of isolates | Avg SAB |
|---|---|---|---|
| A, B, C, D | PATa | 30 | 0.72 ± 0.10 |
| HCW | 42 | 0.73 ± 0.11 | |
| A | PAT, HCW | 13 | 0.83 ± 0.08 |
| PAT | 6 | 0.76 ± 0.11 | |
| HCW | 7 | 0.84 ± 0.05 | |
| HCW (NICU) | 5 | 0.85 ± 0.05 | |
| HCW (SICU) | 2 | 0.80 ± 0.00 | |
| B | PAT, HCW | 19 | 0.71 ± 0.10 |
| PAT | 8 | 0.71 ± 0.10 | |
| HCW | 11 | 0.71 ± 0.11 | |
| HCW (NICU) | 3 | 0.79 ± 0.01 | |
| HCW (SICU) | 8 | 0.71 ± 0.12 | |
| C | PAT, HCW | 8 | 0.73 ± 0.11 |
| PAT | 3 | 0.75 ± 0.13 | |
| HCW | 6 | 0.77 ± 0.06 | |
| HCW (NICU) | 4 | 0.65 ± 0.10 | |
| HCW (SICU) | 2 | 0.79 ± 0.00 | |
| D | PAT, HCW | 31 | 0.73 ± 0.12 |
| PAT | 11 | 0.69 ± 0.11 | |
| HCW | 20 | 0.72 ± 0.10 | |
| HCW (NICU) | 12 | 0.71 ± 0.11 | |
| HCW (SICU) | 8 | 0.74 ± 0.11 | |
| 29 hospitals (control) | PAT | 29 | 0.72 ± 0.10 |
PAT, patients.
The average SABs computed for each hospital collection suggested that in each hospital the BSI isolates were approximately as diverse as the control collection of unrelated isolates (Table 2). However, a simple comparison of the average SABs can be misleading, since one very unrelated isolate in a small collection can have an inordinately strong influence (52). The cluster characteristics of the dendrogram for BSI isolates from each of the four hospitals (Fig. 4A to D) were therefore individually compared to the cluster characteristics of the dendrogram generated for the control collection of unrelated BSI isolates (Fig. 2). The dendrogram for the collection of six BSI isolates from hospital A (Fig. 4A) included two (from patients P18 and P16) with identical patterns and four (from patients P18, P16, P17, P3), which represented 67% of the collection, in a cluster defined at an SAB threshold of 0.86. In the dendrogram for the control collection (Fig. 2), no isolates exhibited identical patterns and only 52% of the isolates in the control collection were members of clusters defined at an SAB threshold of 0.86 (Fig. 2) (P = 0.03). These results suggest a significantly higher than expected level of clustering of highly and moderately related isolates in hospital A (P = 0.03). The dendrogram for the collection of nine BSI isolates from hospital B (Fig. 4B) included five in clusters defined at an SAB threshold of 0.86, which represented 56% of the collection, compared to 52% for the control collection. The dendrogram for the three BSI isolates from hospital C (Fig. 4C) included two in a cluster defined at an SAB threshold of 0.86, which represented 67% of the collection, compared to 52% for the control collection. Finally, the dendrogram for 12 BSI isolates from hospital D (Fig. 4D) contained 6 isolates in clusters defined at an SAB threshold of 0.86, which represented 50% of the collection, which is roughly the same proportion as that for the control collection (Fig. 2). However, the hospital D collection contained a cluster of four isolates that were defined at an SAB threshold of 0.90 and that were collected from patients in the same NICU (Fig. 4D). This cluster represented 33% of the hospital D collection. In the control collection of unrelated BSI isolates (Fig. 2), the largest cluster defined at a threshold of 0.90 contained three isolates, which represented only 10% of the collection. The difference was significant, with a P value of 0.0002.
In the dendrograms generated for BSI isolates in each of the four respective hospitals (Fig. 4), there was no indication of temporal associations of highly related BSI isolates. In the hospital A collection, none of the isolates in the major cluster were collected within 2 months of one another, and even the identical pair of isolates, from patients P18 and P6, were collected 5 months apart and from different ICUs (Fig. 4A). In the hospital B collection, the two most closely related isolates, from patients P12 and P10, were collected 9 months apart, and two isolates, from patients P11 and P23, were collected within 3 days of one another in the same ICU and were unrelated (Fig. 4B). In the hospital D collection, two of the three most related isolates, from patients P8 and P9, were collected from the same ICU within 4 days of each other (Fig. 4D). However, four isolates, collected from patients P28, P19, P20, and P26 within 12 days of each other, were not highly related (Fig. 4D). These results together demonstrate the absence of single dominant endemic strains responsible for BSIs that occur in close temporal proximity in both the NICUs and the SICUs of the four hospitals in this study. However, even though related isolates did not exhibit temporal clustering, the proportion of clusters in the dendrogram for each hospital suggested a greater number of groups of related isolates than would be expected on the basis of comparisons with the control collection of unrelated BSI isolates.
Genetic relatedness of isolates from HCWs in individual hospitals.
The average SABs for isolates collected from the hands of HCWs from hospitals A, B, C, and D were 0.84 ± 0.05 (n = 7), 0.71 ± 0.11 (n = 11), 0.77 ± 0.06 (n = 4), and 0.72 ± 0.10 (n = 20), respectively (Table 2). The average SABs for HCW isolates from hospitals A and C were higher than the SAB for the 29 unrelated BSI isolates in the control collection, as was the case for the SABs for BSI isolates from the same hospitals. Only in the case of the HCW isolates in hospital A was the difference with the control collection significant (P = 0.004). Dendrograms for the HCW isolates from each hospital (Fig. 5) had some of the same characteristics as those generated for BSI isolates from the respective hospitals (Fig. 4). For instance, the dendrogram for the HCW isolate collection from hospital A was dominated by a cluster defined by an SAB threshold of 0.83 that contained six of the seven isolates (86%) (Fig. 5A). A similar cluster of four of the six BSI isolates (67%) from hospital A defined by an SAB threshold of 0.86 dominated the dendrogram for isolates from that hospital (Fig. 4D). Several additional characteristics of the dendrograms for HCW isolates are noteworthy. At an SAB threshold of 0.89, 38% of the control collection of BSI isolates formed clusters (Fig. 2). Except for one cluster of three isolates, which represented 10% of the control collection, all other clusters contained two isolates (Fig. 2). In addition, there were no clusters in the control collection defined by an SAB threshold of 0.96 (Fig. 2). In contrast, the proportion of HCW isolates in clusters that were defined at an SAB threshold of 0.89 and that contained three or more isolates for hospitals A, B, and D were 43, 27, and 50%, respectively; all of these values are higher than the proportion of 10% for the control BSI isolate collection (P = 0.0001, 0.0005 and 0.0001, respectively). In addition, the dendrograms for HCW isolates from hospitals B and D each contained a pair of identical isolates (Fig. 5B and D). Finally, at a threshold of 0.91 the dendrogram for HCW isolates from hospital D contained a cluster of six isolates (Fig. 5D), which was twice as large as the largest cluster in the dendrogram for control BSI isolates at that threshold (Fig. 2).
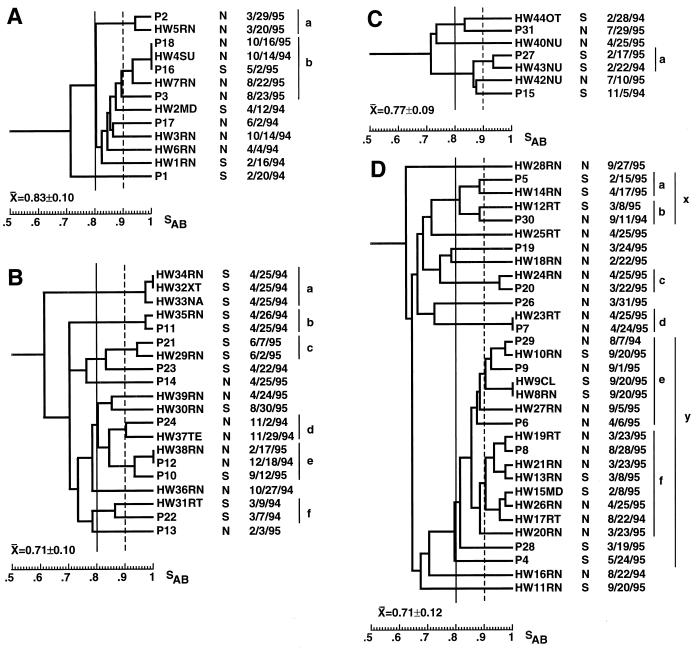
FIG. 5.
Dendrograms of the isolates from HCWs of hospitals A (A), B (B), C (C), and D (D). The type of HCW is noted immediately to the right of health care worker number. The type of ICU and the date of collection are noted to the right of each isolate. Arbitrary SAB thresholds are drawn at 0.80 (straight line) and 0.90 (dashed line). Abbreviations for HCWs are provided in Materials and Methods.
In contrast to the relative lack of temporal associations observed among BSI isolates from individual hospitals, there were several examples of this among HCW isolates. In the hospital A collection, two of the three isolates (isolates HW4SU and HW3RN) in the most highly related cluster were collected from different individuals in the NICU on the same day (Fig. 5A). In the hospital B collection, three isolates (isolates HW34RN, HW32XT, and HW33NA) in the most related cluster were collected from three different individuals in the SICU on the same day (Fig. 5B). Finally, in the hospital D collection, three of the seven isolates (isolates HW21RN, HW19RT, and HW20RN) in the largest cluster were collected from different individuals in the NICU on the same day, and an additional isolate in the cluster (isolate HW13RN) had been collected 15 days earlier (Fig. 5D). Three of the four isolates (isolates HW9CL, HW8RN, and HW10RN) in the second largest cluster were collected from individuals in the SICU on the same day, and the fourth isolate (isolate HW27RN) in this cluster had been collected 15 days earlier from an individual in the NICU (Fig. 5D). These results demonstrate transfer of strains among HCWs or from patients to several HCWs in the same ICUs.
Genetic relatedness of BSI and HCW isolates from the same hospitals.
To assess the relatedness between BSI and HCW isolates collected in the same hospitals, mixed dendrograms were generated (Fig. 6). One BSI isolate from each patient and from each HCW was included. The average SAB for the combined collection of hospital A isolates was 0.83 ± 0.10, which was significantly higher than that for the control collection of unrelated BSI isolates (Table 2) (P = 0.002). The proportion of isolates in clusters defined at an SAB of 0.85 was 85% (Fig. 6A), which was higher than the 59% value for the control BSI isolate population (P = 0.07). The combined collection of isolates from hospital A formed two clusters at an SAB threshold of 0.89 (Fig. 6A). Cluster a included two isolates, one from patient P2 and one from HW5RN; these were collected 9 days apart. The HCW isolate was collected 9 days before collection of the BSI isolate (Fig. 6A) and 5 days before collection of the first commensal isolate from this patient. Cluster b included five isolates. Three of the five (from P18, P16, and HW4SU) were identical (SABs = 1.00). Interestingly, the three isolates were collected over a 1-year period. None were collected within a month of each other. This observation is one of the strongest supporting the establishment of an endemic strain in this study. Again, the HCW isolate was collected before the BSI isolates, 1 year prior to collection of the isolate from patient P18, and 7 months before collection of the isolate from patient P16. The remaining two isolates in cluster b included one from P3 and one from HW7RN, and these were collected 1 day apart.
FIG. 6.
Dendrograms for combined BSI isolates and HCW isolates from hospitals A (A), B (B), C (C), and D (D). Only one BSI isolate from each patient was used. Isolate labels are explained in Materials and Methods. Arbitrary SAB thresholds are drawn at 0.80 (straight line) and 0.90 (dashed line). Clusters determined by a threshold of 0.90 are delineated to the right of each dendrogram.
The average SAB for the combined collection of isolates from hospital B was similar to that for the control collection of BSI isolates (Table 2). However, while 59% of the control collection of unrelated BSI isolates formed clusters at an SAB threshold of 0.85, 80% of the combined collection from hospital B formed clusters at this threshold (Fig. 6B) (P = 0.002). The combined collection from hospital B formed five clusters of two to three isolates each at an SAB threshold of 0.90 (Fig. 6B). The first cluster, cluster a, contained three HCW isolates (from HW34RN, HW32XT, and HW33NA), all collected on the same day. The next three clusters, clusters b, c, and d, each contained one patient isolate and one HCW isolate (from P11 and HW35RN, P21, and HW29RN, and P24 and HW37TE, respectively) collected 1, 5, and 27 days apart, respectively. The fifth cluster, cluster e, contained two patient isolates and one HCW isolate (from P10, P12, and HW38RN). None of the latter three isolates were collected within a month of each other, even though the three were highly related or identical. The HCW isolate in this cluster was collected 2 months after collection of the first BSI isolate but 7 months prior to collection of the second BSI isolate. The final cluster, cluster f, defined at an SAB threshold of 0.86, included isolates from P22 and HW31RT, collected within 2 days of each other.
The average SAB for the combined collection from hospital C was 0.77 ± 0.09, which was somewhat higher than that for the control collection (Table 2). The dendrogram for the hospital C collection contained a cluster of two highly related isolates, one from HW43NU and one from P27 (Fig. 6C). In this case, the HCW isolate was collected a year prior to the collection of the patient isolate.
The average SAB for the combined collection from hospital D was also similar to that for the control collection of unrelated BSI isolates (Table 2). However, the proportion of isolates in clusters defined at an SAB threshold of 0.85 was 74% (Fig. 6D), compared to 59% for the control collection (P = 0.214). The combined collection from hospital D contained six clusters defined at an SAB threshold of 0.88 (Fig. 6D). Clusters a, b, c, and d each contained a patient isolate and an HCW isolate (from P5 and HW14RN, P30 and HW12RT, P20 and HW24RN, and P7 and HW23RT, respectively) collected 2 months, 6 months, 1 month, and 1 day apart, respectively. In each of these instances, the patient isolate was collected before collection of the HCW isolate. Cluster e contained isolates from three patients and four HCWs. Five of the seven isolates in this cluster (from HW10RN, P9, HW9CL, HW8RN, and HW27RN) were collected within 19 days of each other. Finally, cluster f contained eight isolates, one from a patient and seven from HCWs. Seven of the eight isolates were collected in a 6-month period. Among the isolates in the latter cluster, the isolates from P8 and HW19RT were connected at an SAB node of 0.98. In this case, the HCW isolate was collected more than 1 year prior to collection of the patient isolate.
In 11 cases, a BSI and an HCW isolate collected from the same hospitals within a 2-month period were highly related (SAB ≥ 0.88) (Table 3). In all but one of the cases, the BSI isolate was collected in the same ICU as the HCW isolate. In the one exceptional case, isolates from three HCWs in the SICU of hospital D were highly similar to the isolate from patient P9 in the NICU (Table 3). The isolates from the SICU in this case were collected 19 days after collection of the BSI isolate from patient P9 in the NICU and 15 days after collection of a related isolate from an HCW in the NICU. Although in the preceding analysis, cases were noted of HCW isolates collected prior to related BSI isolates, in the majority of cases, BSI isolates were collected prior to related HCW isolates from the same hospital (Table 3). In all but one of these cases, isolates were also obtained from HCWs in the same ICU prior to collection of the BSI isolate that were unrelated to the BSI isolates (Fig. 6).
TABLE 3.
Highly related BSI and HCW isolates collected from the same hospitals within 2 months of each other
| Hospital | BSI patient | HCW | Type of ICU
|
SAB | Time (no. of days before or after) isolation from HCW | |
|---|---|---|---|---|---|---|
| BSI patient | HCW | |||||
| A | P2 | HW5RN | N | N | 0.94 | −9 |
| P3 | HW7RN | N | N | 0.89 | −1 | |
| B | P11 | HW35RN | S | S | 0.97 | +1 |
| P21 | HW29RN | S | S | 0.94 | +5 | |
| P24 | HW37TE | N | N | 0.90 | +27 | |
| P12 | HW38RN | N | N | 1.00 | +62 | |
| P22 | HW31RT | S | S | 0.87 | +2 | |
| D | P5 | HW14RN | S | S | 0.88 | +62 |
| P20 | HW24RN | N | N | 0.96 | +33 | |
| P7 | HW23RT | N | N | 1.00 | +1 | |
| P9 | HW27RN | N | N | 0.88 | +4 | |
| HW8RN | N | S | 0.91 | +19 | ||
| HW10RN | N | S | 0.93 | +19 | ||
| HW9CL | N | S | 0.91 | +19 | ||
Time before (−) isolation of the BSI isolate or time after (+) isolation of the BSI isolate.
Stability of clusters.
In this analysis, we have considered unrelatedness to be reflected by an SAB of 0.72, on average, and a high degree of relatedness to be reflected by SABs above 0.90. In analyzing the cluster characteristics of collections, we used thresholds of 0.85 or 0.86 to define clusters. Since the order of data input by the UPGMA method can affect branching and, thus, the stability of clusters in a dendrogram (5), we randomized data input for the largest dendrogram in the study, the combination dendrogram for patients and HCWs from hospital D (Fig. 6D). The input was randomized 20 times. At the SAB threshold of 0.80, 100% of the isolates that separated into the two major clusters x and y remained in those two clusters, demonstrating that the intermediately rooted branches that defined those clusters were stable. In addition, all clusters above the SAB threshold of 0.90 remained intact, demonstrating that the highly related clusters were also stable.
DISCUSSION
The National Nosocomial Infections Surveillance System conducted by the Centers for Disease Control and Prevention reported an increase from 2.0 nosocomial fungal infections per 1,000 discharges in 1980 to 3.8 per 1,000 discharges in 1990, an approximately twofold increase (7). In that survey, significant increases were observed in medical, surgical, and newborn services, as well as in subspecialty services such as burn and trauma, cardiac surgery, and high-risk nursery services, in a 10-year period (7). The rates of nosocomial fungal infections, therefore, have increased in all types of hospitals, for all types of specialty services, and at all sites of infection (32).
Candida spp. account for approximately 8% of all nosocomial BSIs (31–33), and of these, C. albicans accounts for the majority (50 to 70%) (7, 30, 31–33). Because Candida spp. can be carried as commensal organisms, several possible origins of nosocomial infections must be considered. First, it has been demonstrated that at least two-thirds of healthy individuals carry a Candida sp. in their natural microflora (55). In a significant number of these cases of Candida carriage, individuals carry Candida spp. in at least two anatomical niches, most notably the vaginal canal and the oral cavity. In approximately two-thirds of such individuals, unrelated C. albicans strains or different species colonize the alternative anatomical locales, and in the remaining third, substrains that are highly related but nonidentical colonize the alternative locales (55). Since there is growing genetic evidence suggesting that in the majority of patients commensal organisms are the source of subsequent infection (38, 60), commensal organisms established in the patient at the time of hospitalization should represent the major source of nosocomial yeast infections. However, just as the majority of patients carry commensal organisms prior to infection, so do the HCWs who interact with patients and so do individuals who visit patients. Therefore, there is also the possibility that infectious yeasts can be transferred from the latter individuals to susceptible patients (4, 16, 41, 43, 44, 55). In addition, there is growing concern, especially in the case of aspergillosis, that the physical environment of the hospital can harbor endemic strains of infectious fungi that may be responsible for a portion of nosocomial infections (10, 20, 25). A recent analysis of the genetic diversity of BSI isolates by the same fingerprinting methods used here suggested that particular BSI strains are more highly concentrated in particular geographical locales, that established BSI strains may be endemic in some hospitals, and that these endemic strains may adapt through microevolution to those hospital settings (34). Molecular genetic studies have also demonstrated that single strains have been responsible for a number of temporally associated outbreaks of candidemia in the same hospital or ICU (15, 26, 39, 46, 47, 51). In some cases, the isolates cultured from the hands of HCWs have been found to be genetically similar or identical to nosocomial strains (11, 12, 15, 35), although the direction of transfer in these cases was usually not apparent.
Isolates from the same patient.
There is compelling genetic evidence from a variety of studies that have used a variety of DNA fingerprinting methods that individuals usually harbor the same commensal or infecting strain of C. albicans over extended periods of time (23, 38, 42, 46, 47, 50, 54, 56, 60) and that over time colonizing strains undergo microevolution that can be monitored through reorganization of the hypervariable regions identified by the C1 fragment of the Ca3 probe, which contains a cluster of the repeat element RPS (23, 55). Here, we have compared isolates from urine, stool, and other sites of infection with commensal organisms with BSI isolates from the same individuals. Stool and urine isolates collected prior to and after collection of the first BSI isolate were similar or identical to the BSI isolates in approximately 90% of the patients. However, because all isolates were obtained from each patient after the patient entered the SICU, we cannot be certain that in all patients the infecting strain originated from the commensal strain carried into the hospital by the patient. Indeed, a variety of strains rather than a single C. albicans strain may have become endemic in a particular hospital setting, leading to a variety of nosocomial isolate genotypes similar in diversity to the variety of genotypes of isolates carried as commensal organisms in healthy individuals. Therefore, similar levels of diversity (e.g., equal SABs) do not exclude the possibility that a variety of endemic hospital strains are responsible. A second study is therefore planned. In that study high-risk patients will be sampled prior to and after entering the hospital, and isolates from a control group of healthy individuals from the same geographical locale will be used for comparison. In the case of infants who acquire nosocomial infections in NICUs, Candida colonization must originate from either the mother or the hospital setting.
The proportion of patient isolate collections that included isolates with highly related but nonidentical patterns and that were therefore undergoing microevolution was 33%. This value is below the values of 66 and 55% previously observed for collections of commensal isolates and isolates that caused vaginitis, respectively (22). The difference may be due to the time frame of the study. Carriage of the same commensal strain usually continues for very long periods of time, and the same established strain is responsible for recurrent vaginal infections over periods of up to several years (22). Therefore, in both patients who carry commensal organisms and patients with recurrent yeast vaginitis, the colonizing strain has ample chance to diversify through microevolution. The lower figure for hospitalized patients suggests that the strains that colonize patients in the respective ICUs have not had ample time to diversify, supporting the idea either that they recently colonized their present hosts or that one commensal substrain recently dominated the colonizing population.
Possibility of endemic BSI strains in ICUs.
The average SAB for BSI isolates in each of the four test hospitals was similar to that for the unrelated control collection and could therefore be interpreted to support the conclusion that the isolates from patients with candidemia in each hospital collection were unrelated and therefore did not emanate from the hospital environment. However, there were more isolates in clusters in three of the four hospital collections than in the control collection of BSI isolates. Therefore, while no single strain was responsible for the majority of nosocomial BSIs in any of the ICUs of the four hospitals in this study, BSI isolates showed more group relationships, on average. The latter point is reinforced by two additional observations. First, the collections of BSI isolates from hospitals A and D each contained a pair of identical isolates from different patients, while no identical isolates emerged in the collection of 29 unrelated control BSI isolates. Second, hospital D contained a cluster of 4 BSI isolates defined at an SAB threshold of 0.90 that represented 33% of isolates from that hospital, while the largest cluster in the control BSI collection defined at that threshold contained 3 of 29 isolates, which represented only 10% of the collection. Schmid et al. (49) obtained similar results in an analysis of surveillance isolates from 32 patients in different wards of a hospital in New Zealand. Using Ca3 fingerprinting to analyze relatedness, they found that isolates in each ward were, on average, more highly related than isolates in general.
Strong relationships exist between BSI isolates and isolates obtained from the hands of HCWs.
The isolates from the hands of HCWs in each of the four hospitals exhibited cluster characteristics similar to those of BSI isolates from the respective hospitals. The same two hospitals, hospitals A and C, exhibited the highest average SABs for both BSI and HCW isolates, and hospital A had the highest proportion of both BSI and HCW isolates in clusters at an SAB threshold of 0.86, 67 and 71%, respectively, compared to 52% for the control collection of unrelated BSI isolates. There were several additional cluster characteristics that suggested that the BSI isolates and HCW isolates were related. In the HCW isolate collection from hospital A, six of the seven isolates grouped in one cluster at an SAB threshold of 0.85, suggesting that an endemic strain had cross-contaminated the hands of hospital coworkers over a period of approximately 1 year and that this strain had undergone significant microevolution. In a combined dendrogram of patient and HCW isolates from hospital A, the four BSI isolates that formed a major cluster in the dendrogram for the BSI isolates mixed with the isolates in the HCW cluster. Isolates in the mixed cluster were distributed between the SICU and NICU of hospital A, suggesting a general endemic strain. Isolates in this mixed cluster represented 69% of isolates in the combined collection of isolates from hospital A. An additional cluster of isolates from one patient and one HCW isolate with a node at an SAB of 0.95 also emerged in the dendrogram for the mixed collection of isolates, raising the proportion of isolates from hospital A in mixed clusters defined at an SAB threshold of 0.85 to 85%. This value was significantly higher than the value of 59% obtained for the control collection of BSI isolates at the same SAB threshold. Similar results were obtained in the dendrograms for the mixture of patient and HCW isolates from hospitals B and D. Mixed clusters defined at an SAB threshold of 0.85 dominated each mixed dendrogram. The proportions of isolates in clusters defined at an SAB threshold of 0.85 for collections of isolates from hospitals B and D were 80 and 74%, respectively; again, both values were significantly higher than the value of 59% obtained for the control collection.
For 11 BSI patients from the four hospitals, isolates that were collected from HCWs within a 2-month period were related to the BSI isolates at an SAB threshold of 0.86. In all but one of these cases, the BSI and HCW isolates were obtained in the same ICU. For example, isolates from HW38RN and patient P12 had an SAB of 1.00 and were collected 2 months apart in the same NICU, and isolates from HW4SU and P18 had an SAB of 1.00 and were collected 1 year apart in the same NICU. In the case of patient P9 in the NICU of hospital D, related isolates with SABs of ≥ 0.90 were obtained 19 days later from HCWs in the SICU of the same hospitals. Fifteen days earlier, a related isolate had been collected from an HCW in the NICU, suggesting cross-contamination between HCWs in the alternative ICUs. The times of isolation of identical isolates from patients and HCWs in the same ICUs were sometimes separated by several months. For example, an isolate from HW4SU in the NICU and an identical isolate from patient P16 in the SICU of hospital A were collected 7 months apart. Clustering in the mixed dendrograms for isolates from each hospital demonstrated cross-contamination between the hands of HCWs and patients in the same hospitals. In 12 of the 14 cases in which related BSI and HCW isolates were collected within 2 months in the same hospital, the HCW isolate was collected after collection of the BSI isolate. These results suggest that in the majority of cases, transmission is from a BSI patient to an HCW, but in a minority of documented cases, HCW isolates were collected up to several months prior to collection of highly related BSI isolates, suggesting transmission from an HCW to a patient.
Conclusion.
By computing average SABs and comparing the clustering characteristics of BSI isolates in the test collections and a control collection of unrelated BSI isolates, we can conclude that in none of the four test hospitals was a single endemic strain responsible for the majority of BSIs in an ICU. However, the characteristics of clustering in dendrograms suggest that endemic strains may account for some infections, and comparisons between BSI and HCW isolates demonstrate a high degree of relatedness in many cases that can only be interpreted as cross-contamination. Our evidence suggests that cross-contamination occurs between patients and HCWs, between HCWs in the same ICU, and between HCWs in different ICUs of the same hospital. The temporal sequence of isolation in most cases supports the conclusion that HCWs are contaminated by isolates from infected or colonized patients, but in a minority of cases transmission appears to be from HCW to patient. Our results demonstrate that DNA fingerprinting with the complex probe Ca3 provides the resolution necessary for such studies and provides the framework necessary for development of a surveillance study that will address, first, the origins of nosocomial infections in NICUs, second, the relationship between commensal organisms carried by healthy individuals prior to hospitalization and subsequent infecting strains in SICUs, third, the impact of transfer from HCW to patient, and, fourth, the microevolution of endemic strains in hospital settings.
ACKNOWLEDGMENTS
This research was supported by a grant from Pfizer Inc., and by Public Health Service grants AI39735 and DE10758 from the National Institutes of Health (awarded to D.R.S.). S.R.L. was supported by training grant AG00214 from the National Institutes of Health. Francesc Marco was partially supported by a grant from Fondo de Investigaciones Sanitatarias (FIS 97/5144) and a Permiso de Ampliación de Estudios from Hospital Clínic, Barcelona, Spain.
REFERENCES
- 1.Akova M, Akalin H E, Uzun Ö, Hayran M, Tekuzman G, Kansu E, Aslan S, Telatar H. Efficacy of fluconazole in the treatment of upper gastrointestinal candidiasis in neutropenic patients with cancer: factors influencing the outcome. Clin Infect Dis. 1994;18:298–304. doi: 10.1093/clinids/18.3.298. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J, Srikantha T, Morrow B, Miyasaki S H, White T C, Agabian N, Schmid J, Soll D R. Characterization and partial sequence of the fingerprinting probe Ca3 of Candida albicans. J Clin Microbiol. 1993;31:1472–1480. doi: 10.1128/jcm.31.6.1472-1480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arif S, Barkham T, Power E G, Howell S A. Techniques for investigation of an apparent outbreak of infections with Candida glabrata. J Clin Microbiol. 1996;34:2205–2209. doi: 10.1128/jcm.34.9.2205-2209.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnaviehle A, Blanchard A, Mallie M, Quilci M, Bastide J M. Multilocus enzyme electrophoresis analysis of Candida albicans isolates from three intensive care units. An epidemiological study. Mycoses. 1997;40:159–167. doi: 10.1111/j.1439-0507.1997.tb00208.x. [DOI] [PubMed] [Google Scholar]
- 5.Backeljau T, Debruyn L, DeWolf H, Jordaens K, Van Dongen S, Winnepenninckx B. Multiple UPGMA and neighbor-joining trees and the performance of some computer packages. Mol Biol Evol. 1996;13:309–313. [Google Scholar]
- 6.Bart-Delabesse E, van Deventer H, Goessens W, Poirot J L, Lioret N, van Belkum A, Dromer F. Contribution of molecular typing methods and antifungal susceptibility testing to the study of a candidemia cluster in a burn care unit. J Clin Microbiol. 1995;33:3278–3283. doi: 10.1128/jcm.33.12.3278-3283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck-Sague C M, Jarvis W R. National Nosocomial Infections Surveillance System. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 8.Branchini M L, Geiger D C, Fischman O, Pignatari A C. Molecular typing of Candida albicans strains isolated from nosocomial candidemia. Rev Instit Med Trop Sao Paulo. 1995;37:483–487. doi: 10.1590/s0036-46651995000600002. [DOI] [PubMed] [Google Scholar]
- 9.Bross J, Talbot G H, Maislin G, Hurwitz S, Strom B L. Risk factors for nosocomial candidemia: a case-control study in adults without leukemia. Am J Med. 1989;87:614–620. doi: 10.1016/s0002-9343(89)80392-4. [DOI] [PubMed] [Google Scholar]
- 10.Buffington J, Reporter R, Lasker B A, McNeil M M, Lanson J M, Ross L A, Mascola L, Jarvis W R. Investigation of an epidemic of invasive aspergillosis: utility of molecular typing with the use of random amplified polymorphic DNA probes. Pediatr Infect Dis J. 1994;13:386–393. [PubMed] [Google Scholar]
- 11.Burnie J P. Candida and hands. J Hosp Infect. 1986;8:1–4. doi: 10.1016/0195-6701(86)90099-x. [DOI] [PubMed] [Google Scholar]
- 12.Burnie J P, Matthews R, Lee W, Philpott-Howard J, Brown R, Damani N, Breuer J, Honeywell K, Jordans Z. Four outbreaks of nosocomial systemic candidiasis. Epidemiol Infect. 1987;99:201–211. doi: 10.1017/s0950268800067030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Antonio D, Violante B, Mazzoni A, Bonfini T, Capuani M A, D’Aloia F, Iacone A, Schioppa F, Romano F. A nosocomial cluster of Candida inconspicua infections in patients with hematological malignancies. J Clin Microbiol. 1998;36:792–795. doi: 10.1128/jcm.36.3.792-795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diekema D J, Messer S A, Hollis R J, Wenzel R P, Pfaller M A. An outbreak of Candida parapsilosis prosthetic valve endocarditis. Diagn Microbiol Infect Dis. 1997;29:147–153. doi: 10.1016/s0732-8893(97)81804-4. [DOI] [PubMed] [Google Scholar]
- 15.Doebbling B N, Hollis R J, Isenberg H D, Wenzel R P, Pfaller M A. Restriction fragment analysis of a Candida tropicalis outbreak of sternal wound infections. J Clin Microbiol. 1991;29:1268–1270. doi: 10.1128/jcm.29.6.1268-1270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doi M, Homma M, Iwaguchi S, Horiba K, Tanaka K. Strain relatedness of Candida albicans strains isolated from children with leukemia and their bedside parents. J Clin Microbiol. 1994;32:2253–2259. doi: 10.1128/jcm.32.9.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isenberg H D, Tucci V, Cintron F, Singer C, Weinstein G S, Tyras D H. Single-source outbreak of Candida tropicalis complicating coronary bypass surgery. J Clin Microbiol. 1989;27:2426–2428. doi: 10.1128/jcm.27.11.2426-2428.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karabinis A, Hill C, Leclercq B, Tancrede C, Baume D, Andremont A. Risk factors for candidemia in cancer patients: a case-control study. J Clin Microbiol. 1988;26:429–432. doi: 10.1128/jcm.26.3.429-432.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatib R, Thirumoorthi M C, Riederer K M, Sturm L, Oney L A, Baran J., Jr Clustering of Candida infections in the neonatal intensive care unit: concurrent emergence of multiple strains simulating intermittent outbreaks. Pediatr Infect Dis J. 1998;17:130–134. doi: 10.1097/00006454-199802000-00010. [DOI] [PubMed] [Google Scholar]
- 20.Leenders A, van Belkum A, Janssen S, de Marie S, Kluytmans J, Wielenga J, Lowenberg B, Verbrugh H. Molecular epidemiology of apparent outbreak of invasive aspergillosis in a hematology ward. J Clin Microbiol. 1996;34:345–351. doi: 10.1128/jcm.34.2.345-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin D, Lehmann P F. Random amplified polymorphic DNA for strain delineation within Candida tropicalis. J Med Vet Mycol. 1995;33:241–246. doi: 10.1080/02681219580000491. [DOI] [PubMed] [Google Scholar]
- 22.Lockhart S, Fritch J J, Meier A S, Schröppel K, Srikantha T, Galask R, Soll D R. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol. 1995;33:1501–1509. doi: 10.1128/jcm.33.6.1501-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lockhart S, Reed B, Pierson C, Soll D R. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J Clin Microbiol. 1996;34:767–777. doi: 10.1128/jcm.34.4.767-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockhart, S. R., and D. R. Soll. Generation of species-specific DNA probes and their applications for analysis of fungal populations. In D. Coleman and K. Haynes (ed.), Methods in molecular medicine series: medical mycology protocols, in press. Humana Press, Totowa, N.J.
- 25.Loudon K W, Coke A P, Burnie J P. “Pseudoclusters” and typing by random amplification of polymorphic DNA of Aspergillus fumigatus. J Clin Pathol. 1995;48:183–184. doi: 10.1136/jcp.48.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moro M L, Maffei C, Manso E, Morace G, Polonelli L, Biavasco F. Nosocomial outbreak of systemic candidiasis associated with parenteral nutrition. Infect Control Hosp Epidemiol. 1990;11:27–35. doi: 10.1086/646075. [DOI] [PubMed] [Google Scholar]
- 27.O’Connell B, Coleman D C, Bennett D, Sullivan D, McCann S R, Keane C T. An epidemiological study of Candida species infection in cancer patients using genetic fingerprinting and morphotyping. J Hosp Infect. 1995;31:211–217. doi: 10.1016/0195-6701(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 28.Pfaller M, Cabezudo L, Koontz F, Bale M, Gingrich R. Predictive value of surveillance cultures for systemic infection due to Candida species. Eur J Clin Microbiol. 1987;6:628–633. doi: 10.1007/BF02013057. [DOI] [PubMed] [Google Scholar]
- 29.Pfaller M A. Epidemiology of fungal infections: current perspectives and future directions. Clin Infect Dis. 1995;20:1525. doi: 10.1093/clinids/20.6.1525. [DOI] [PubMed] [Google Scholar]
- 30.Pfaller M A. Epidemiology of fungal infections: the promise of molecular typing. Clin Infect Dis. 1995;20:1535–1539. doi: 10.1093/clinids/20.6.1535. [DOI] [PubMed] [Google Scholar]
- 31.Pfaller M A. Nosocomial candidiasis: emerging species, reservoirs and modes of transmission. Clin Infect Dis. 1996;22:S89–S94. doi: 10.1093/clinids/22.supplement_2.s89. [DOI] [PubMed] [Google Scholar]
- 32.Pfaller M A, Jones R N, Doern G V, Sader H S, Hollis R J, Messer S A. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and antifungal susceptibility of isolates collected in 1997 in the United States, Canada and South America for the SENTRY Program. J Clin Microbiol. 1998;36:1886–1889. doi: 10.1128/jcm.36.7.1886-1889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaller M A, Jones R N, Messer S A, Edmond M B, Wenzel R P the SCOPE Participant Group. National surveillance of nosocomial blood stream infections due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn Microbiol Infect Dis. 1998;31:327–332. doi: 10.1016/s0732-8893(97)00240-x. [DOI] [PubMed] [Google Scholar]
- 34.Pfaller M A, Lockhart S R, Pujol C A, Swails-Wenger J A, Messer S A, Edmund M B, Jones R N, Wenzel R P, Soll D R. Hospital specificity, regional specificity and fluconozole resistance of Candida albicans bloodstream isolates. J Clin Microbiol. 1998;36:1518–1529. doi: 10.1128/jcm.36.6.1518-1529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaller M A, Messer S A, Houston A, Rangel-Frausto M S, Wilson T, Blumberg H M, Edwards J E, Jarvis W, Martin M A, Neu H C, Saiman L, Patterson J E, Dibb J C, Roldan C M, Rinaldi M G, Wenzel R P. National Epidemiology of Mycoses Survey: a multicenter study of strain variation and antifungal susceptibility among isolates of Candida species. Diagn Microbiol Infect Dis. 1998;31:289–296. doi: 10.1016/s0732-8893(97)00245-9. [DOI] [PubMed] [Google Scholar]
- 36.Pujol C, Joly S, Lockhart S, Noel S, Tibayrenc M, Soll D R. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive probe Ca3 for fingerprinting Candida albicans. J Clin Microbiol. 1997;35:2348–2358. doi: 10.1128/jcm.35.9.2348-2358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pujol, C., S. Joly, B. Nolan, T. Srikantha, S. Lockhart, and D. R. Soll. Unpublished observations.
- 38.Reagan D R, Pfaller M A, Hollis R J, Wenzel R P. Characterization of the sequence of colonization and nosocomial candidemia using DNA fingerprinting and a DNA probe. J Clin Microbiol. 1990;28:2733–2738. doi: 10.1128/jcm.28.12.2733-2738.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reagan D R, Pfaller M A, Hollis R J, Wenzel R P. Evidence of nosocomial spread of Candida albicans causing blood stream infection in a neonatal intensive care unit. Diagn Microbiol Dis. 1995;21:191–194. doi: 10.1016/0732-8893(95)00048-f. [DOI] [PubMed] [Google Scholar]
- 40.Reef S A, Lasker B A, Butcher D S, McNeil M M, Pruitt R, Keyserling H, Jarvis W R. Nonperinatal nosocomial transmission of Candida albicans in a neonatal intensive care unit: prospective study. J Clin Microbiol. 1998;36:1255–1259. doi: 10.1128/jcm.36.5.1255-1259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richet H M, Andremont A, Tancrede C, Pico J L, Jarvis W R. Risk factors for candidemia in patients with acute lymphocytic leukemia. Rev Infect Dis. 1991;13:211–215. doi: 10.1093/clinids/13.2.211. [DOI] [PubMed] [Google Scholar]
- 42.Robert F, Lebreton F, Bougnoux M E, Paugam A, Wassermann D, Schlotterer M, Tourte-Schaefer C, Dupouy-Camet J. Use of random amplified polymorphic DNA as a typing method for Candida albicans in epidemiological surveillance of a burn unit. J Clin Microbiol. 1995;33:2366–2371. doi: 10.1128/jcm.33.9.2366-2371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosner B. Fundamentals of biostatistics. Belmont, Calif: Duxbury Press; 1995. [Google Scholar]
- 44.Ruiz-Diez B, Martinez V, Alvarez M, Rodriguez-Tudela J L, Martinez-Suarez J V. Molecular tracking of Candida albicans in a neonatal intensive care unit: long-term colonizations versus catheter-related infections. J Clin Microbiol. 1997;35:3032–3036. doi: 10.1128/jcm.35.12.3032-3036.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadhu C, McEachern M J, Rustchenko-Bulgac E P, Schmid J, Soll D R, Hicks J. Telomeric and dispersed repeat sequences in Candida yeasts and their use in strain identification. J Bacteriol. 1991;173:842–850. doi: 10.1128/jb.173.2.842-850.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez V, Vazquez J A, Barth-Jones D, Dembry L, Sobel J D, Zervos M J. Epidemiology of nosocomial acquisition of Candida lusitaniae. J Clin Microbiol. 1992;30:3005–3008. doi: 10.1128/jcm.30.11.3005-3008.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez V, Vazquez J A, Barth-Jones D, Dembry L, Sobel J D, Zervos M J. Nosocomial acquisition of Candida parapsilosis: an epidemiologic study. Am J Med. 1993;94:577–582. doi: 10.1016/0002-9343(93)90207-6. [DOI] [PubMed] [Google Scholar]
- 48.Schmid J, Voss E, Soll D R. Computer-assisted methods for assessing Candida albicans strain relatedness by Southern blot hybridization with repetitive sequence Ca3. J Clin Microbiol. 1990;28:1236–1243. doi: 10.1128/jcm.28.6.1236-1243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmid J, Tay Y P, Wan L, Carr M, Parr D, McKinney W. Evidence for nosocomial transmission of Candida albicans obtained by Ca3 fingerprinting. J Clin Microbiol. 1995;33:1223–1230. doi: 10.1128/jcm.33.5.1223-1230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroeppel K, Rotman M, Galask R, Mac K, Soll D R. The evolution and replacement of C. albicans strains during recurrent vaginitis demonstrated by DNA fingerprinting. J Clin Microbiol. 1994;32:2646–2654. doi: 10.1128/jcm.32.11.2646-2654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherer R J, Gledhill K S, Hampton K D, Pfaller M A, Givner L B, Abramson J S, Dillard R G. Outbreak of Candida blood stream infections associated with retrograde medication administration in a neonatal intensive care unit. J Pediatr. 1992;120:455–461. doi: 10.1016/s0022-3476(05)80920-5. [DOI] [PubMed] [Google Scholar]
- 52.Sneath P, Sokal R. Numerical Taxonomy. The principles and practice of numeric classification. San Francisco, Calif: W. H. Freeman & Co.; 1973. [Google Scholar]
- 53.Soll, D. R. The “ins and outs” of DNA fingerprinting the infectious fungi. Unpublished data. [DOI] [PMC free article] [PubMed]
- 54.Soll D R, Galask R, Isley S, Rao T V G, Stone D, Hicks J, Schmid J, Mac K, Hanna C. “Switching” of Candida albicans during successive episodes of recurrent vaginitis. J Clin Microbiol. 1989;27:681–690. doi: 10.1128/jcm.27.4.681-690.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soll D R, Galask R, Schmid J, Hanna C, Mac K, Morrow B. Genetic dissimilarity of commensal strains carried in different anatomical locations of the same healthy women. J Clin Microbiol. 1991;29:1702–1710. doi: 10.1128/jcm.29.8.1702-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soll D R, Staebell M, Langtimm C J, Pfaller M A, Hicks J, Rao T V G. Multiple Candida strains in the course of a single systemic infection. J Clin Microbiol. 1988;26:1448–1459. doi: 10.1128/jcm.26.8.1448-1459.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strausbaugh L J, Sewell D L, Tjoelker R C, Heitzman T, Webster T, Ward T T, Pfaller M A. Comparison of three methods for recovery of yeasts from hands of health care workers. J Clin Microbiol. 1996;34:471–473. doi: 10.1128/jcm.34.2.471-473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tibayrenc M, Neubauer K, Barnabé C, Guerrini F, Skaresky D, Ayala F J. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vazquez J A, Boikov D, Boikov S G, Dajani A S. Use of electrophoretic karyotyping in the evaluation of Candida infections in a neonatal intensive-care unit. Infect Control Hosp Epidemiol. 1997;18:32–37. doi: 10.1086/647498. [DOI] [PubMed] [Google Scholar]
- 60.Voss A, Hollis R J, Pfaller M A, Wenzel R P, Doebbling B N. Investigation of the sequence of colonization and candidemia in non-neutropenic patients. J Clin Microbiol. 1994;32:975–980. doi: 10.1128/jcm.32.4.975-980.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walsh T J, Merz W G. Pathologic features in the human alimentary tract associated with invasiveness of Candida tropicalis. Am J Clin Pathol. 1986;85:498–502. doi: 10.1093/ajcp/85.4.498. [DOI] [PubMed] [Google Scholar]
- 62.Warren N G, Hazen K C. Candida, Cryptococcus, and other yeasts of medical importance. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington D.C: American Society for Microbiology; 1995. pp. 723–737. [Google Scholar]
- 63.Wey S B, Mori M, Pfaller M A, Woolson R F, Wenzel R P. Risk factors for hospital-acquired candidemia: a matched case-control study. Arch Intern Med. 1989;149:2349–2353. [PubMed] [Google Scholar]