Abstract
Human-made hydrocarbon-rich environments are important reservoirs of microorganisms with specific degrading abilities and pathogenic potential. In particular, black fungi are of great interest, but their presence in the environment is frequently underestimated because they are difficult to isolate. In the frame of a biodiversity study from fuel-contaminated sites involving 30 diesel car tanks and 112 fuel pump dispensers (52 diesel and 60 gasoline, respectively), a total of 181 black fungal strains were isolated. The long cold incubation (LCI) of water-suspended samples, followed by plating on Dichloran Rose Bengal Chloramphenicol Agar (DRBC), gave isolation yields up to six times (6.6) higher than those of direct plating on DRBC, and those of enrichment with a phenolic mix. The sequencing of ITS and LSU-rDNA confirmed the dominance of potentially pathogenic fungi from the family Herpotrichiellaceae and Exophiala xenobiotica. Moreover, other opportunistic species were found, including E. opportunistica, E. oligosperma, E. phaeomuriformis, and Rhinocladiella similis. The recurrent presence of E. crusticola, Knufia epidermidis, Aureobasidium melanogenum, Cladosporium spp., and Scolecobasidium spp. was also recorded. Interestingly, 12% of total isolates, corresponding to 50% of taxa found (16/32), represent new species. All the novel taxa in this study were isolated by LCI. These findings suggest that black fungal diversity in hydrocarbon-rich niches remains largely unexplored and that LCI can be an efficient tool for further investigations.
Keywords: black yeasts, diesel car tank, diesel gasoline fuel pump dispenser, Knufia, opportunistic Exophiala species, extreme environments, hydrocarbon bioremediation, fuel degraders, Scolecobasidium, volatile organic compounds (VOCs)
1. Introduction
Extreme environments are reservoirs for specialized microorganisms able to maintain their metabolic activity despite physical and chemical restrictive conditions. Black fungi is the epithet used since the 1990s to identify a group of polyextremotolerant melanized organisms adapted to hostile environments where they cope with several types of stresses including rapid changes in key environmental parameters [1,2]. For their outstanding morpho-ecological features, black fungi are also called black yeasts, rock-inhabiting fungi (RIF), black meristematic fungi, and microcolonial fungi (MCF) [3]. These fungi should not be confused with the emerging colloquially homonymous ‘black fungus’ from the order Mucorales, responsible for opportunistic infections associated with the COVID-19 pandemic [4]. Indeed, the poikilo-tolerant black fungi belong to two main lineages, namely Dothideomycetes and Eurotiomycetes [5,6]. The former are overrepresented in cold natural habitats, while the latter, with a few exceptions, thrive in hot, semi-arid climates [5]. Chaetothyrialean fungi are well known for their dualism, where the ability to assimilate alkylbenzenes (hydrocarbonoclastic activity) and the tendency toward being virulent represent the two sides of the same coin. This feature is particularly marked within the family Herpotrichiellaceae, including the causative agents of severe infections in humans, not limited to immunosuppressed hosts, and in cold-blooded animals with a preference towards the central nervous system [7,8,9,10]. Currently, a clear delimitation between hydrocarbonoclastic and pathogenic species is missing, since the known toluene-growing species from the genus Exophiala, such as E. oligosperma, E. xenobiotica, and E. mesophila, are recognized as biosafety level 2 (BSL2) microorganisms associated with opportunistic infections in humans [6,10].
Chaetothyrealean fungi have been isolated from several man-made environments that are generally grouped into two categories. The first category includes wet oligotrophic environments subjected to extreme temperatures and biocidal products (e.g., detergents) such as dishwashers, kitchen sinks [11,12,13,14,15,16], moist surfaces and wet cells in bathrooms [17,18,19,20,21], laundry machines [22,23,24], hospital environments [25], and public bathing facilities [26,27]. The second consists of niches related to petroleum and coal products, including gasoline car tanks, creosote-treated wood (e.g., railway sleepers), surfaces containing machinery oil, dirt, and hydrocarbon-polluted soils [14,24,28,29,30,31,32]. Due to their oligotrophic nature and low competitiveness under standard laboratory conditions, these fungi typically require specific isolation methods that control fast-growing microorganisms. Several protocols have been developed to isolate black fungi in relation to the type of sample and/or the desired fungal traits: (1) pre-incubation in acidic medium at high temperature to favor thermophilic species [33]; (2) extraction via mineral oil to select hydrophobic black fungal propagules [30,31]; and (3) enrichment on volatile aromatic hydrocarbons including toluene, styrene, or a mixture of phenolic compounds to isolate fungi with aromatic-degrading abilities [24,32,34,35,36].
Hydrocarbon-contaminated sites may harbor microorganisms with biodegradative potential and/or opportunistic/pathogenic traits [6,8,11,19,24,37]. As indicated by its epithet, Exophiala xenobiotica is a species frequently found in habitats rich in monoaromatic hydrocarbons and alkanes which can also cause opportunistic mycoses [37]. To increase our knowledge on hydrocarbonoclastic fungi and their associated potential biohazards during the handling of fuels, we focused on their presence in car fuel tank caps and fuel pump dispensers. Previous studies have been carried out with gasoline [24] but, to the best of our knowledge, the fungal colonization of diesel car tanks and fuel dispensers has never been investigated. The efficiency of the isolation methods can change with the sample type and/or the environmental matrix investigated. Therefore, the development of additional protocols under different conditions is of great significance. In this work, we tested the efficacy of three isolation protocols to deepen our knowledge on the biodiversity of black fungi able to thrive on fuel pump dispensers and diesel car tanks. The collected samples were processed using three different protocols: (1) direct plating, (2) enrichment with a phenolic mix, and (3) long incubation (six to eight months) of water suspended samples at a low temperature before plating. The last is a new protocol inspired by direct observation of low-temperature (1 ± 1 °C), long-stored sample suspension vials showing clear signs of black fungal growth on their walls [38].
The obtained isolates were identified by ITS and LSU rDNA sequencing, and black fungal diversity results were analyzed and discussed in terms of their ecophysiology and phylogeny.
2. Materials and Methods
2.1. Sampling
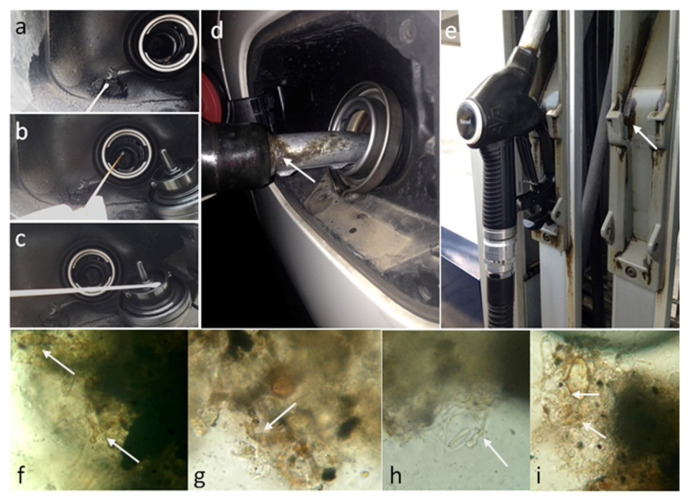
Samples were collected between Jan and Mar 2014 using sterile cotton swabs. The investigated sites were 30 diesel car tanks (DCT), 52 diesel pump dispensers (DPD), and 60 gasoline pump dispensers (GPD). Only diesel cars ten years or older were considered for this study, since spore landing may primarily occur during refueling procedures, and colonies may take considerable time to develop [24]. Three samples were taken from each diesel car: (1) from the inner part, (2) from the external part of the tank fuel pouring hole, and (3) from the internal lid surface (Figure 1a–c). Only one sample was taken from each fuel pump dispenser where dripped fuel and dark deposits were visible (Figure 1d,e). Samples were collected from different geographical areas in central Italy.
Figure 1.
Sampled surfaces: (a–c) diesel car tanks; (d,e) fuel pumps dispenser. (f–i) Magnification of biofilms taken from dripping fuels; white arrows indicate thick dark hyphae.
2.2. Isolation Protocols
Sampling swabs were washed in 1.5 mL centrifuge tubes containing 1 mL of sterile distilled water (dH2O) and stored at 1 ± 1 °C until use. Sample suspensions were used for (1) direct plating (DP, 50 μL) on Dichloran Rose Bengal Chloramphenicol Agar (DRBC, Laboratorios Conda, Madrid, Spain); (2) inoculating an enrichment liquid medium containing a phenolic mix of 4-hydroxybenzoic acid, protocatechuic acid, phenylacetic acid, and catechol (0.125 g/L each) and plated in the same solid medium as described by Isola et al. [24] (phenolic enrichment, PE); and (3) a six to eight months long storing/incubation (long cold incubation, LCI) at low temperature (1 ± 1 °C) and plated on DRBC (50 μL). Plates were then incubated at room temperature (18 ± 1 °C) and inspected daily for two to three weeks. All morphologically distinct black colonies were transferred onto malt agar (MA, malt extract 30 g/L, agar 15 g/L) and processed for molecular identification. Isolated strains were deposited in the Culture Collection of Fungi from Extreme Environments (CCFEE) of the Tuscia University (Viterbo, Italy).
2.3. Molecular Identification
Genomic DNA was extracted using a Nucleospin Plant kit (Macherey-Nagel, Düren, Germany) following the protocol optimized for fungi. ITS amplifications were performed using BioMix (BioLine, Luckenwalde, Germany) in a total volume of 25 μL. In each reaction solution, 5 pmol of each primer (ITS4 and ITS5) and about 40 ng of template DNA were added. Amplifications were carried out using MyCycler™ Thermal Cycler (Bio-Rad Laboratories, Munich, Germany), applying the following protocol: initial denaturation step for 3 min at 95 °C, 35 cycles of 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 32 s, followed by a final extension at 72 °C for 5 min [39]. When ITS matching was unfruitful, additional LSU amplifications (LR0R-LR7) were generated and analyzed to better define taxa boundaries. The applied amplification protocol was: initial denaturation step for 3 min at 95 °C, 35 cycles of denaturation at 95 °C for 45 s, annealing at 52 °C for 30 s, and extension at 72 °C for 2 min, with a 5 min final extension at 72 °C [39]. Sequences obtained from Macrogen Inc. (Madrid, Spain) were analyzed using ChromasPro v. 1.41 (Technelysium, Southport, Queensland, Australia).
Similarity searches were performed using BLASTn (NCBI, National Center for Biotechnological Information) excluding “uncultured/environmental sample sequences” from the comparison and referring mainly to CBS collection strains, preferably to ex-type strains. The obtained sequences were deposited in GenBank. To define the phylogenetic position of the isolates within the families Herpotrichiellaceae, Cyphellophoraceae, Trichomeriaceae (order Chaetothyriales), Mycosphaerellaceae (order Mycosphaerellales), and Sympoventuriaceae (order Venturiales), four ITS datasets were generated. Sequences were aligned iteratively with MUSCLE in MEGA6 [40], and the final alignments were improved manually. Maximum likelihood (ML) trees were generated using the Tamura-Nei model, and the robustness of the ML phylogenetic inference was estimated using the bootstrap method [41], with 1000 pseudoreplicates. Cyphellophora, Cladophialophora, Cladosporium, and Verruconis were used as outgroups for the Herpotrichiellaceae, Trichomeriaceae, Mycosphaerellaceae, and Sympoventuriaceae trees, respectively.
2.4. Black Fungal Diversity
The re-isolation of the same species can occur when samples are processed using different isolation protocols, and/or when multiple samples are collected from the same diesel car tank. Therefore, to assess the black fungal populations in the three studied sites (namely DCT, DPD, and GPD), the general isolation results were analyzed as species presence–absence for each sampling site regardless of the isolation method used or the surface sampled (i.e., inner, external or lid sample as in DCT). A similar presence–absence evaluation scheme was performed to evaluate the different isolation methods. The species isolation frequencies were used to define the black fungal populations in the three studied sites and to calculate the Shannon and Simpson biodiversity indices and the Chao-1 species richness.
2.5. Statistical Analysis
Confidence intervals (95% CIs) calculated with the Wald method modified by Agresti–Coull [42] and Chi-square Fisher exact test were used to statistically evaluate differences in isolation frequencies (e.g., by site: DCT, GPD, DPD). PAST software (PAleontological STatistics, ver. 4.06b, [43]) was used to calculate the biodiversity indices and to perform a hierarchical clustering analysis of the black fungal populations using the UPGMA algorithm and the Bray–Curtis similarity index with 100,000 bootstrap replicates.
3. Results
3.1. General Isolation and Molecular Identification Results
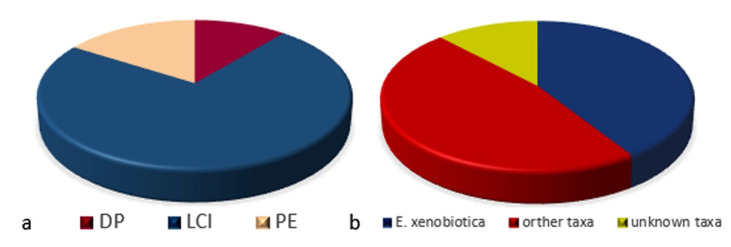
One hundred and eighty-one black fungal isolates were obtained (Figure 2, Figure S1). No significant differences on sample positivity were recorded in the three studied sites (Figure S2a). A total of 87 black fungi were isolated from DCT, 37 from DPD, and 59 from GPD (Figure S2b). Significant differences in isolation yields were recorded when LCI was compared to DP and PE (p < 0.00001). The total number of isolates obtained using LCI was up to 6.6 times greater than DP, and 4.6 times greater than PE (Figure 3a). No significant differences were recorded comparing DP and PE results (p = 0.3221). Similarly, LCI sample positivity (41.6%, 84/202) was statistically significant (p < 0.00001) when compared to PE (10.4%, 21/202) and DP (8.9%, 18/202).
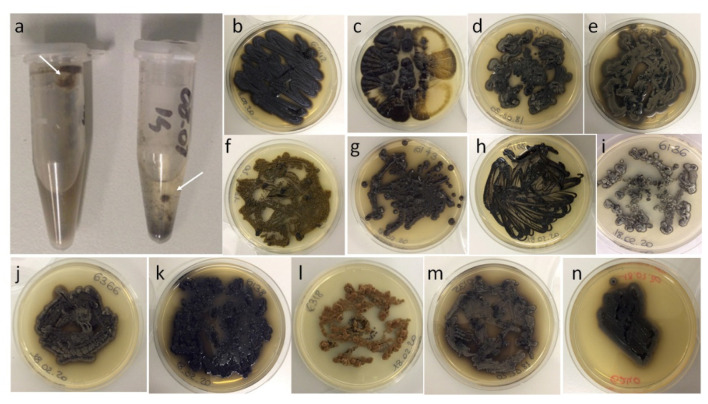
Figure 2.
(a) Example of sample suspension vials after a long cold incubation (white arrows indicate fungal growth) and pure cultures of selected isolates: (b) E. phaeomuriformis CCFEE 6242; (c) Aureobasidium melanogenum CCFEE6236; (d) E. xenobiotica CCFEE 6142; (e) Exophiala sp. 2 CCFEE 6334; (f) Aulographina pinorum CCFEE 6222; (g) E. xenobiotica CCFEE 6143; (h) E. crusticola CCFEE 6188; (i) Neodevriesia sp. CCFEE 6136; (j) K. epidermidis CCFEE 6366; (k) K. epidermidis CCFEE 6138; (l) Scolecobasidium sp. 1 CCFEE 6318; (m) Exophiala sp. 1 CCFEE 6135; (n) E. heteromorpha CCFEE 6240.
Figure 3.
General results based on the 181 isolated strains: (a) isolation yields by method: LCI: long cold incubation, DP: direct plating, and PE: phenolic enrichment; (b) prevalence of E. xenobiotica compared to other known and unknown taxa.
Most of the 181 black fungal isolates were identified at the species level (Table 1). Exophiala xenobiotica was the most frequent isolated species (Figure 3b), while 12% of the isolates represented novel fungal taxa. The isolation frequencies in the three niches under investigation were used for the subsequent biodiversity analysis.
Table 1.
List of the 181 isolated strains. The identifications were performed by BLASTn comparison and ML ITS-based trees (Figures S3–S6). The taxonomic novelties found were identified at the genus or family level.
| Species | CCFEE Collection No | Origin | GenBank Accession No | |
|---|---|---|---|---|
| ITS | LSU | |||
| Aulographina pinorum | 6220, 6222, 6230 | DPD, Italy | MZ573423 | |
| Aureobasidium melanogenum group | 6141, 6145, 6146, 6226, 6234, 6235, 6213, 6216, 6227, 6236, 6407, 6244, 6245, 6246, 6403, 6408 | DCT, DPD, GPD, Italy | MZ573420, MZ573421, MZ573422 | |
| Cladophialophora sp. | 6390 | DCT, Italy | MZ573430 | MZ956973 |
| Cladosporium herbarum group | 6184, 6199, 6016, 6192, 6193, 6027, 6029, 6030, 6031, 6054, 6148, 6191, 6378 | DCT, DPD, GPD, Italy | MZ573425, MZ573426, MZ573427 | |
| Cladosporium cladosporioides group | 6197, 6225, 6018, 6231, 6183, 6218, 6412 | DCT, DPD, GPD, Italy | MZ573428, MZ573429 | |
| Coniosporium cft. uncinatum | 6149 | GPD, Italy | MZ573424 | |
| Cyphellophora reptans | 6373, 6007, 6413, 6379, 6398 | DCT, DPD, GPD, Italy | MZ573432, MZ573434, MZ573433 | |
| Cyphellophora sp. | 6028 | DPD, Italy | MZ573431 | MZ956974 |
| Exophiala bonariae | 6041 | DPD, Italy | MZ573435 | |
| Exophiala crusticola | 6039, 6051, 6058, 6004, 6005, 6006, 6010, 6014, 6015, 6019, 6020, 6022, 6023, 6024, 6025, 6026, 6178, 6188, 6224, 6232 | DCT, DPD, GPD, Italy | MZ573436, MZ573437, MZ573438 | |
| Exophiala heteromorpha | 6150, 6181, 6339, 6343, 6240, 6241, 6243 | DCT, DPD, GPD, Italy | MZ573439, MZ573440 | |
| Exophiala oligosperma | 6139, 6345, 6360 | DCT, Italy | MZ573441 | |
| Exophiala xenobiotica | 6140, 6142, 6143, 6179, 6180, 6185, 6186, 6187, 6190, 6196, 6219, 6223, 6229, 6333, 6335, 6340, 6341, 6342, 6344, 6346, 6351, 6352, 6353, 6362, 6364, 6365, 6367, 6368, 6369, 6371, 6372, 6374, 6375, 6376, 6381, 6382, 6383, 6384, 6385, 6386, 6389, 6393, 6394, 6395, 6396, 6397, 6399, 6404, 6157, 6189, 6194, 6195, 6221, 6233, 6237, 6336, 6337, 6338, 6377, 6410, 6008, 6182, 6217, 6228, 6238, 6239, 6350, 6354, 6355, 6356, 6357, 6400, 6405, 6406 | DCT, DPD, GPD, Italy | MZ573442, MZ573443, MZ573444 | |
| Exophiala phaeomuriformis | 6242, 6358, 6359 | DCT, DPD, Italy | MZ573445, MZ573446 | |
| Exophiala sp. 1 | 6135 | DPD, Italy | MZ573447 | OK178849 |
| Exophiala sp. 2 | 6334, 6402 | DCT, Italy | MZ573448 | MZ956975 |
| Exophiala sp. 3 | 6348 | GPD, Italy | MZ573449 | MZ956976 |
| Exophiala sp. 4 | 6370 | DCT, Italy | MZ573450 | MZ956977 |
| Exophiala sp. 5 | 6387 | DCT, Italy | MZ573451 | MZ956978 |
| Extremus cft antarcticus | 6349 | DCT, Italy | MZ573453 | |
| Herpotrichiellaceae sp. | 6392 | DCT, Italy | MZ573459 | MZ956980 |
| Knufia epidermidis | 6138, 6198, 6366 | DCT, Italy | MZ573455 | |
| Knufia tsunedae | 6411 | GPD, Italy | MZ573456 | |
| Knufia sp. | 6034 | DPD, Italy | MZ573457 | MZ956979 |
| Lithohypha sp. | 6069 | GPD, Italy | MZ573458 | OK178850 |
| Neodevriesia sp. | 6136 | GPD, Italy | MZ573461 | OK178852 |
| Rhinocladiella similis | 6361 | DCT, Italy | MZ573467 | |
| Scolecobasidium sp. 1 | 6154, 6155, 6156, 6152, 6153, 6318 | DCT, GPD, Italy | MZ573462, MZ573463 | MZ956981 |
| Scolecobasidium sp. 2 | 6363 | DCT, Italy | MZ573464 | MZ956982 |
| Scolecobasidium sp. 3 | 6391 | DCT, Italy | MZ573465 | MZ956983 |
| Scolecobasidium sp. 4 | 6388 | DCT, Italy | MZ573466 | MZ956984 |
| Teratosphaeriaceae sp. | 6137 | GPD, Italy | MZ573460 | OK178851 |
Black CCFEE numbers represent the strains isolated from diesel car tanks (DCT); red, those from diesel pump dispensers (DPD); and green, from gasoline pump dispensers (GPD). Bold character indicates strains for which sequences have been deposited in GenBank.
3.2. Black Fungal Diversity
3.2.1. Black Fungal Diversity in the Three Fuel-Contaminated Sites
Diesel car tanks revealed the highest species richness (21); the difference between the two fuel pump dispensers was slight, being characterized by 13 (DPD) and 14 (GPD) taxa (Table 2).
Table 2.
Isolate data and diversity indices summary for the three investigated sites: diesel car tank (DCT), diesel pump dispensers (DPD), and gasoline pump dispensers (GPD).
| DCT | DPD | GPD | Total | |
|---|---|---|---|---|
| No. of isolates | 52 | 30 | 42 | 124 |
| No. of positive sampling sites | 25 | 21 | 29 | 75 |
| No. of taxa | 21 | 13 | 14 | 32 |
| Shannon index | 2.522 | 2.434 | 2.367 | 2.687 |
| Simpson index | 0.8145 | 0.8828 | 0.8722 | 0.8558 |
| Chao-1 index | 43.31 | 18.08 | 19.13 | 101.4 |
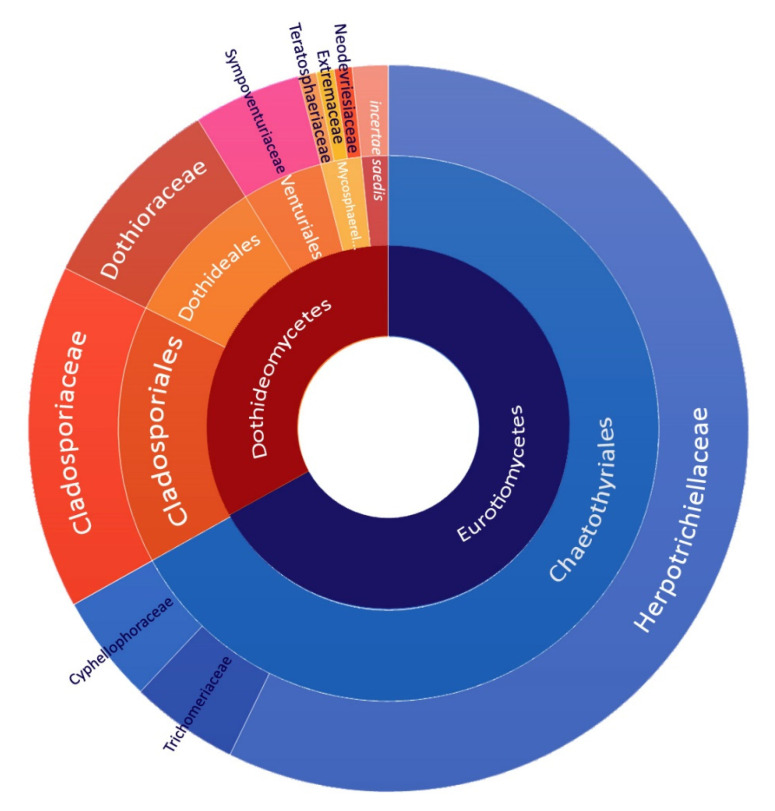
The order Eurotiomycetes, class Chaetothyriales, and family Herpotrichielaceae were the dominant taxa (Figure 4). The ratio between the classes Eurotiomycetes and Dothideomycetes was about 2:1 (67% vs. 33%). At the order level, Chaetothyriales dominated (67%), followed by Cladosporiales (15%) and Dothideales (8.7%). More than half of all isolates belonged to the family Herpotrichiellaceae (57%), followed by Cladosporiaceae (15%), and Dothioraceae (8.7%). Trichomeriaceae, Cyphellophoraceae (Eurotiomycetes), and Sympoventuariaceae (Dothideomycetes) accounted for less than 5% of the isolates each.
Figure 4.
High taxonomic rank of 124 black fungi isolated from DCT (52), DPD (32) and GPD (40). Family (outer ring), order (central ring), and class (internal ring). Different shades of the same color in the chart indicate different families within each order and class.
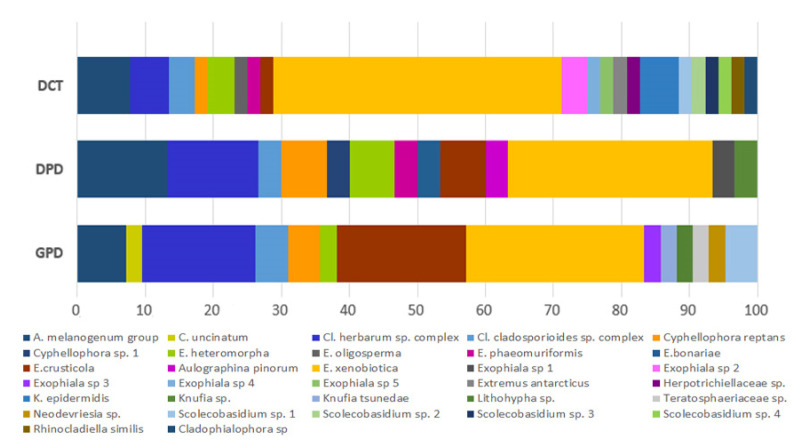
The diversity at species level is shown in Figure 5. A total of 32 taxa were assigned. In all investigated sites, two groups of cladosporiacean fungi (namely Cl. herbarum and Cl. cladosporioides groups) were found, as well as fungi very close to the type strain of Aureobasidium melanogenonum, from 100% to 98.2% in sequence similarity. A more precise species definition within these groups was not possible by using only the ITS sequences. Additionally, E. crusticola and E. xenobiotica were present in all sites, but their ratio among sites changed from 8:11 in gasoline pump dispensers to 1:22 in diesel car tanks.
Figure 5.
Relative abundance of the cultivable black fungal diversity recorded in diesel car tanks (DCT), diesel pump dispensers (DPD), and gasoline pump dispensers (GPD). The relative abundance is expressed as percentage (x axis).
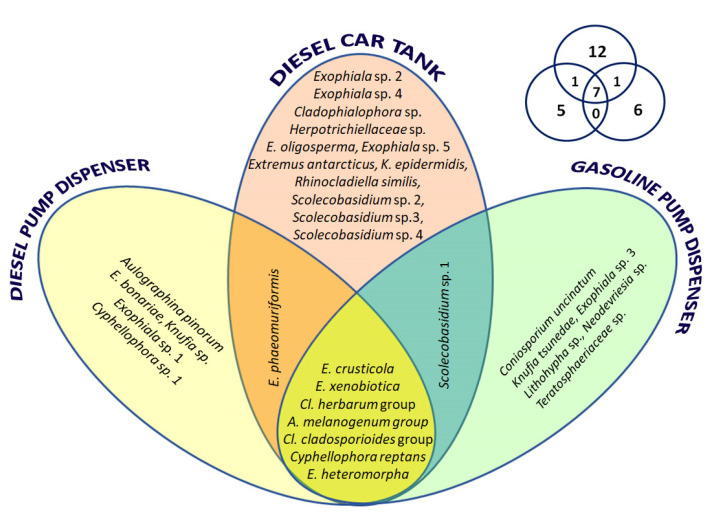
Of the 32 taxa found, seven taxa were common to all sampling sites, while some were exclusive of one site (Figure 5 and Figure 6): 12 for DCT, five for DPD, and six for GPD. Figure 6 shows the distribution of the new taxa within the studied sites. In particular, three were isolated from DPD (3 out 30 isolates counted, 10%), eight from DCT (9/52, 17%), and five from GPD (5/42, 11%). Hierarchical clustering analysis demonstrated, instead, that populations found on both fuel pump dispensers (namely from GPD and DPD) were more similar than those from DCT (Figure S7).
Figure 6.
Venn diagram showing the presence of culturable black fungal species detected in diesel car tanks (DCT), diesel pump dispensers (DPD), and gasoline pump dispensers (GPD). On the upper right side, the Venn diagram of species richness is shown.
3.2.2. Black Fungal Diversity with Regard to the Isolation Methods
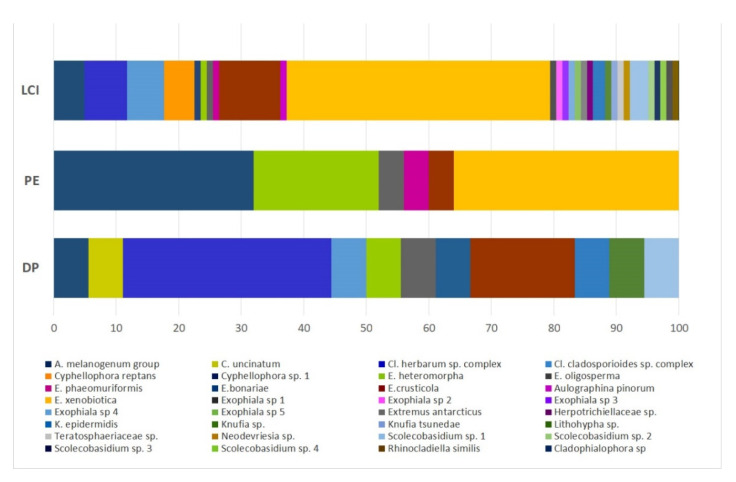
Filtered raw data (counting a single species per site and each isolation method) were used to assess the influence of the protocol used on the isolated species. A total of 145 strains were counted, 102 of which were obtained using LCI. The remaining isolates were isolated using PE (25) and DP (18). As shown in Figure 7, about 91% of taxa recorded in this study (29/32) were isolated using LCI. Regarding the 16 new taxa, they were all isolated by LCI (17 strains); while only one was isolated by DP (2 strains). The twenty-five isolates obtained using PE fall into six different taxa. Exophiala xenobiotica (36%, yellow), A. melanogenum (32%, blue), and E. heteromorpha (20%, green) were the most frequent species; no cladosporian species were recorded. The strains isolated by DP fall into 11 taxa, which highlights the absence of E. xenobiotica and the prevalence of the ubiquitous Cladosporium species complex representing 38.9% (7/18, cobalt blue and light blue) of isolates. Significant differences in PE isolation frequencies were found for A. melanogenum (p = 0.0005) and E. heteromorpha (p = 0.001) when compared to LCI, and for Cladosporium spp. when the DP results were compared to the other methods (DP vs. LCI p = 0113; DP vs. PE p = 0.001). When compared to DP, LCI and PE significantly increased the E. xenobiotica isolation frequencies (p = 0003 and p = 0.0056, respectively).
Figure 7.
Relative species frequency recorded using the following protocols: long cold incubation (LCI), phenolic enrichment (PE), and direct plating (DP). The species frequency is expressed as percentage (x axis).
3.2.3. Diversity of New Fungal Taxa
Novel strains accounted for 12% of total isolates (22/181) and 15.3% (19/124) if the general data are filtered to exclude duplicated strains. These unidentified strains belong to 16 different taxa, each generally represented by a single strain. Their higher taxonomic rank distribution (Figure S8) evidenced the Eurotiomycetes prevalence (57.9%). The most frequent families were Herpotrichiellaceae (42.1%) and Sympoventuriaceae (31.6%); the latter was represented exclusively by the Scolecobasidium species.
Herpotrichiellaceae sp. CCFEE 6392 possibly belong to a new Minimelanolacus species due its position in the ITS tree (Figure S3), and the 98.19% LSU sequences identity with Minimelanolacus asiaticus MFLUCC 15-0237 and Minimelanolacus curvatus MFLUCC 15-0259. Cyphellophora sp. CCFEE 6028 recorded an ITS best match with Cyphellophora reptans CBS 113.85 (96.43%) and with Cyphellophora europaea CBS 129.96 when LSU was used for comparison (98.76%). Sequencing blasting identified Cladophialophora chaetospira as the closest known species to Cladophialophora sp. CCFEE 6390 and LSU confirmed this position. Otherwise, Teratosphaeriaceae sp. CCFEE 6137 showed very similar ITS identities, 97.53% and 97.97%, with the ex-type strains of Mycocalicium victoriae (Eurotiomycetes) and Constantinomyces macerans (Dothideomycetes) respectively, and 96.25% (coverage 99%) with Patellariopsis dennisii (Leotiomycetes). However, LSU sequence comparison evidenced Catenulostroma elginense CBS 111030 (Dothideomycetes) as the best match (99.34%). No Mycocalicium victoriae LSU sequences are available in GenBank, but neither Eurotiomycetes nor Leotiomycetes strains were recorded in the LSU top 100 identity records. The new Scolecobasidium species fall into three groups, their closest species are S. robustum (Scolecobasidium sp. CCFEE 6152, CCFEE 6153, CCFEE 6154), S. constrictum (Scolecobasidium sp. CCFEE 6388), and S. globale, respectively. Two different taxa belonged to this last group, CCFEE 6391 and CCFEE 6363, characterized by different identity scores (Figure S6).
4. Discussion
The high-throughput sequencing of environmental DNA has become the dominant method to study microbial diversity in a given habitat. The behavior of individual species can, in fact, be frequently explained in a community context by its interaction with other organisms, which are often unculturable under laboratory conditions [44]. Only a small fraction of the world’s microbial diversity can be cultured under standard microbiological tools and media [45]. If, for bacteria, the “1% culturability paradigm” (recently revised by Martiny [46,47]) is generally accepted (according to which around 99% of them are unculturable), for fungi, it is somehow different. Magnuson and Lasure suggested that 70–90% of soil fungi cannot be obtained by culturing methods [48], but this number is far from being fixed due to the parameters used for evaluation, the habitat considered, and the estimated number of the world’s fungal species [49,50,51,52,53,54]). Whatever this fraction actually is, living pure cultured microorganisms are still necessary for several omics technologies and biotechnological processes and represent the foundation of many in silico applications and for barcoded identifications [55,56].
Three different protocols were used to assess the most suitable method and, additionally, to increase the chances of isolation. Direct plating on DRBC agar (a medium used to inhibit bacteria and slow fast-growing mold) gave poor results due to its insufficient inhibitory power in controlling the spreading of yeasts and sporulating fungi on agar plates at the tested temperature. The PE, already used to isolate black fungi from gasoline car tanks [24], did not give the expected results. The suitability of PE in gasoline car tanks was likely facilitated by the confined conditions and the high accumulation of alkylbenzenes, which are predominant in gasoline and characterize this environment. Despite their resistance to abiotic stresses, black fungi are believed to be potentially vulnerable because of their poor competitive abilities. They indeed may easily be hidden/overwhelmed when conditions become more permissive due to the introduction of a more competitive species [57]. Diesel fuels produce less stringent conditions due to the lower content in volatile aromatics (prevalence of polycyclic aromatic hydrocarbon, PAH) and a variable percentage of hydrotreated vegetal oils (HVO) [58,59]. Fuel dispensers, unlike car fuel tanks, are exposed to the open air, a condition favoring the evaporation of the volatile fraction along with a continuous load with high sporulating fast-growing airborne fungi. The significantly higher isolation yields using LCI (p < 0.0001) could be due to a favoring effect on oligotrophic slow-growing organisms able to cope with, and even use, aromatics at low temperatures. Some black fungi such as E. xenobiotica MA2853 and Knufia perforans MA1299—both chaetothyrialean fungi belonging to Herpotrichiellaceae and Trichomeriaceae families, respectively—are still metabolically active at 1 °C with no significant variations in the number of expressed proteins when compared to optimal temperature conditions [60].
The diversity of black fungi at higher taxonomic ranks is dominated by members of the Eurotiomycetes (class), Chaetothyriales (order), and Herpotrichielaceae (family), similar to other hydrocarbon-contaminated sites [32,61,62]. Concerning the species found (32), the majority were recorded in DCT (21), followed by GPD (14) and DPD (13). Half of the taxa found (50%, 16/32) could not be assigned to known species, resulting occasionally in low identity scores even when a more conserved target is used. These findings highlight how our knowledge of black fungal diversity is still incomplete and largely fragmentary. Interestingly, all 16 unknown taxa were isolated using the LCI protocol, underlining the suitability of this new method in covering the current biodiversity knowledge gap. Unlike standard protocols that are generally rich in carbon sources, LCI maintains oligotrophic conditions; the water availability allows metabolism, and the xenobiotics already present in the sample could work both as a limiting factor and as a carbon source. Moreover, low temperatures slow down the fast-growing cosmopolitan fungi, and the long-period incubation allows black fungi to thrive, thus broadening the isolation chances (Figure S2). The PE might indeed be more selective towards the subset of black fungal strains that most effectively assimilate phenolic compounds, similar to the enrichment of solid state-like fungal cultures on toluene [63].
The species distribution within the three investigated sites evidenced the predominance of well-known species, but also the presence of a number of relatively rare and sometimes unknown species. Seven taxa were common to all sites and included the cosmopolitan genera Cladosporium, represented by the herbarum and cladosporioides groups, and Aureobasidium. Isolates belonging to Cl. herbarum and Cl. cladosporioides species complexes are among the most frequently isolated fungi from the air, being common saprotrophs occurring in an extremely wide range of natural and man-made substrates [64]. The ubiquitous distribution and their growth rate can explain the significant prevalence (p < 0.05) of these cosmopolitan fungi when DP (a low-selective method) was used.
As for Aureobasidium, all isolated strains shared high sequence identities (up to 100%) with Aureobasidium melanogenum, a species previously isolated from oil finished wood and also reported as an occasional pathogen [65,66]. Interestingly, a comparative genomic study on this species has pointed out the possible existence of genes associated with the degradation of plastic and aromatic compounds [67]. This aromatic-degrading potential could explain its significantly higher isolation rate when PE was used as compared to LCI (p = 0.0001). Besides their phylogenetic affiliation to the Dothydeomycetes, both Cladosporium and Aureobasidium species are characterized by an ability to grow under osmophilic conditions [64] and by a relatively hydrophobic cell wall that promotes the adhesion to hydrophobic surfaces [68,69]. These traits may favor the fungal colonization of oily fuel surfaces having a low affinity for water. Cyphellophora reptans had previously been isolated from food, human nails and skin scrapings, bark, soil, water [70], and now from fuel-contaminated sites. Three known Exophiala species completed the core of fungi found in all the studied sites. Exophiala xenobiotica, which is already known for its affinity towards hydrocarbon-rich environments, was the most frequent species representing 40% of total isolates (74/183) recorded in 22 out 25 DCT (88%). The isolation frequencies recorded in the studied sites evidenced that LCI (p = 0.0003) and PE (0.0056) can significantly increase the chance of finding E. xenobiotica when compared to DP. Additionally, E. heteromorpha isolates deserve attention since they were obtained mainly through the PE protocol (5/7, p < 0.05), suggesting a possible biodegradative assimilation potential for aromatic compounds. Assimilation of alkylbenzenes has not been demonstrated for E. heteromorpha but, just like E. xenobiotica, this fungus has been repeatedly isolated from environments rich in hydrocarbons and aromatic compounds [8,37]. The isolation of E. crusticola, a soil-associated fungus, was not surprising since it was previously isolated from a creosoted railway tie in cold climates [29,71,72].
Notably, 13 out of the 32 species recorded belong to the polyphyletic genus Exophiala. This group is represented by six known species (E. bonariae, E. crusticola, E. heteromorpha, E. oligosperma, E. phaeomuriformis, and E. xenobiotica), five unknown species (CCFEE 6135, 6334, 6348, 6370, and 6387), and two allied taxa (Rhinocladiella similis CCFEE 6361, and Herpotrichieallaceae sp. CCFEE 6392) (Table 1). Since 46% of the Exophiala-related taxa are unknown, it is reasonable to believe that fuel-contaminated sites could be considered a favorable niche for their finding.
Similarly, Scolecobasidium, a genus previously isolated from wet cells and laundry machines [24,73], resulted quite recurrently and is here represented by four different putative new species. There are no previous records on the isolation of Scolecobasidium spp. from fuel contaminated sites, and they were mainly isolated using LCI, also suggesting in this case that low temperatures are a factor favoring their occurrence.
The presence of Coniosporium uncinatum (CCFEE 6149) and Knufia epidermidis (CCFEE 6138, 6198, and 6366) in our study was quite rare, but confirmed previous observations on gasoline car tanks [23]. Knufia tsunedae (CCFEE 6411), described as a soil-associated fungus [74], is recorded here for the first time ever in Italy.
In light of the distinct chemical nature of fuels, diesel with predominantly long-chain alkanes and gasoline rich in BTEX, it would be expected that the type of fuel exerts selection pressure on the species characterizing the different populations. Different factors may have concurred regarding the number of isolates and species results, such as the isolation method, the number of samples taken, the geographic area, and the composition of fuels, which can also vary significantly with the brand considered and also with local regulations. Moreover, the environmental influence on the composition of airborne particles cannot be disregarded, as suggested by the finding of plant-associated species such as Aulographina pinorum [75] and rock-associated species such as Extremus antarcticus, the latter of which was found for the first time in temperate climates [76,77]. Consequently, more samples coming from different countries would be beneficial for gaining a broader knowledge on these contaminated sites in terms of the ecological traits of species, but also from a phylogenetic and taxonomical standpoint. Several authors have demonstrated that Chaetothyriales taxonomy is likely incomplete and deserves a detailed revision [78], but one of the major problems in reconstructing this puzzle is that a huge number of its pieces are still missing. In this context, the hydrocarbon-contaminated niches studied here represent a reservoir for unknown species, which might be brought to light using increasingly specific isolation methods.
Further studies are also necessary to assess the possible dual behavior reported in chaetothyrialean fungi. In this study, we found some species for which a hydrocarbonoclastic potential had already been reported, such as E. xenobiotica and E. oligosperma, and others whose abilities deserve to be assessed, such as for E. heteromorpha, an epiphytic species frequently isolated after phenolic enrichment. Moreover, E. bonariae (CCFEE 6041) deserves attention because its type of strain, which was isolated from a marble sculpture, was able to cope with toluene and to grow even at 35 °C (the highest temperature tested) [24,79,80]. Several members of the genus Exophiala are potential agents of human and animal mycoses and for that are considered to be in the Bio Safety Level- 2 (BSL-2) group. In this study, together with E. xenobiotica, we found other species associated with clinical cases, including E. oligosperma a sporadic agent of phaeohyphomycosis [81,82], E. phaeomuriformis mentioned in relation to keratitis [83], Rhinocladiella similis recently found in a nosocomial infection [84], and Knufia epidermidis associated with mild skin infections [85].
Safety issues should also be considered in light of the frequency with which some species have been found. Above all, the case of E. xenobiotica, reported as the second most frequent Exophiala species in U.S. and Brazilian clinical samples [86,87], is generally associated with opportunistic cutaneous phaeohyphomycoses of a mostly traumatic nature judging from the frequent occurrence of eye, wound, and (sub)cutaneous lesions, mainly in patients with immunodepression or major debilitating diseases [37]. In this study, E. xenobiotica was recorded in 73% (22/30) of the sampled cars, and so it is not a surprise if it has been reported recently (maybe by chance) as responsible for cutaneous infections in a car mechanics [88]. The profuse isolation of Exophiala dermatitidis (a species associated with deep mycoses in immunocompromised and healthy individuals) [89,90] in a car garage [32] highlights the potential risks for automotive engine and fuel manipulation. Further studies focused on the biodiversity of fungi that colonize devices subjected to both car fuel exposure and human manipulation are essential for a better understanding of pathogenicity and biohazard assessment, and also for their potential use as bioremediation agents.
5. Conclusions
The long cold incubation proved to be a powerful tool with which to explore the black fungal diversity in fuel-contaminated sites. Aside from the frequent isolation of E. xenobiotica, the finding of several novel taxa and species known for their pathogenic and biodegradative potential further increased interest in hydrocarbon-rich sites. Investigations on fuel-contaminated niches are indeed crucial for defining taxonomic boundaries within Chaethothyriales/Herpotrichiellaceae and assessing the evolutionary route leading to pathogenicity. This research is also essential in evaluating the associated biological risks and selecting species that exhibit a biotechnological potential associated with a low biohazard.
Acknowledgments
The authors wish to thank Emma Aronne for her support during sampling.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7100817/s1, Table S1. List of the species used to infer the phylogenetic position of the strains in study by ML ITS trees, reported respectively in Figure S3–S6. T indicates ex-type strains. Figure S1. Example of DRBC isolation plates after long cold incubation. Figure S2. Black fungi: general isolation results. (a) Percentage of positive samples (green) by site: DCT: diesel car tank, DPD: diesel pump dispenser, GPD: gasoline pump dispenser; (b) number of isolates by site; for DCT, the different surfaces sampled were reported as IN: internal (red), EXT: external (blue), and: lid (LID, gray). Figure S3 Herpotrichiellaceae ML ITS-based tree generated using the Tamura–Nei model; robustness was estimated using the bootstrap method with 1000 pseudoreplicates; values above 70% are shown. Cyphellophora clade was used as outgroup. T indicates type strains; DCT diesel car tanks; DPD diesel pump dispensers; and GPD gasoline pump dispensers. Figure S4. Trichomeriaceae ML ITS-based tree generated using the Tamura–Nei model; robustness was estimated using the bootstrap method with 1000 pseudoreplicates; values above 80% are shown. Cladophialophora clade was used as outgroup. T indicates type strains; DCT diesel car tanks; DPD diesel pump dispensers; and GPD gasoline pump dispensers. Figure S5. Mycosphaerellaceae ML ITS-based tree generated using the Tamura–Nei model; robustness was estimated using the bootstrap method with 1000 pseudoreplicates; values above 60% are shown. Cladosporium was used as outgroup. T indicates type strains; DCT diesel car tanks; DPD diesel pump dispensers; and GPD gasoline pump dispensers. Figure S6. Sympoventuriaceae ML ITS-based tree generated using the Tamura–Nei model; robustness was estimated using the bootstrap method with 1000 pseudoreplicates. Verruconis was used as outgroup. T indicates type strains; DCT diesel car tanks, and GPD gasoline pump dispensers. Figure S7. Hierarchical clustering analysis of the black fungal populations found in diesel car tanks (DCT), diesel pump dispensers (DPD), and gasoline pump dispensers (GPD) using UPGMA algorithm and Bray–Curtis similarity index with 100,000 bootstrap replicates. Cophen. corr. 0.9271. Figure S8. High taxonomic rank of 19, not duplicated, unknown strains. Different shades of the same color in the chart indicate different families within each order and class. The frequency is expressed as percentage (x axis).
Author Contributions
Conceptualization, D.I. and L.Z.; methodology, D.I. and A.S.; investigation, D.I., A.S. and G.O.; resources, G.O. and L.Z.; data curation, D.I. and F.X.P.-B.; writing—original draft preparation, D.I.; writing—review and editing, D.I., F.X.P.-B., L.Z., G.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grube M., Muggia L., Gostinčar C. Niches and adaptations of polyextremotolerant black fungi. In: Seckbach J., Oren A., Stan-Lotter H., editors. Polyextremophiles. Springer; Dordrecht, The Netherlands: 2013. pp. 551–566. [Google Scholar]
- 2.Selbmann L., Isola D., Egidi E., Zucconi L., Gueidan C., de Hoog G.S., Onofri S. Mountain tips as reservoirs for new rock-fungal entities: Saxomyces gen. nov. and four new species from the Alps. Fungal Divers. 2014;65:167–182. doi: 10.1007/s13225-013-0234-9. [DOI] [Google Scholar]
- 3.Sterflinger K. Black yeasts and meristematic fungi: Ecology, diversity and identification. In: Seckbach J., editor. The Yeast Handbook. Springer; Berlin, Germany: 2005. pp. 501–514. Biodiversity and Ecophysiology of Yeasts. [Google Scholar]
- 4.Gandra S., Ram M.S., Levitz S.M. The “Black Fungus” in India: The emerging syndemic of COVID-19–Associated Mucormycosis. Ann. Intern. Med. 2021;174:1301–1302. doi: 10.7326/M21-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selbmann L., de Hoog G.S., Zucconi L., Isola D., Onofri S. Black yeasts in cold habitats. In: Buzzini P., Margesin R., editors. Cold-adapted Yeasts. Springer; Berlin/Heidelberg, Germany: 2014. pp. 173–189. [DOI] [Google Scholar]
- 6.Prenafeta-Boldú F.X., Armjio-Medina C., Isola D. Black fungi in the built environment–the good, the bad, and the ugly. In: Pacheco-Torgal F., Ivanov V., Falkinham J.O., editors. Viruses, Bacteria, and Fungi in the Built Environment. Designing Healthy Indoor Environments. Elsevier- Woodhead Publishing Science &Technology books; Duxford, UK: 2021. in press. [Google Scholar]
- 7.de Hoog G., Vicente V., Najafzadeh M.J., Harrak M., Badali H., Seyedmousavi S. Waterborne Exophiala species causing disease in cold-blooded animals. Persoonia—Mol. Phylogeny Evol. Fungi. 2011;27:46–72. doi: 10.3767/003158511X614258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prenafeta-Boldú F.X., Summerbell R., de Hoog G.S. Fungi growing on aromatic hydrocarbons: Biotechnology’s unexpected encounter with biohazard? FEMS Microbiol. Rev. 2006;30:109–130. doi: 10.1111/j.1574-6976.2005.00007.x. [DOI] [PubMed] [Google Scholar]
- 9.Sav H., Ozakkas F., Altınbas R., Kiraz N., Tümgör A., Gümral R., Döğen A., Ilkit M., de Hoog G.S. Virulence markers of opportunistic black yeast in Exophiala. Mycoses. 2016;59:343–350. doi: 10.1111/myc.12478. [DOI] [PubMed] [Google Scholar]
- 10.Prenafeta-Boldú F.X., de Hoog G.S., Summerbell R.C. Fungal communities in hydrocarbon degradation. In: McGenity T.J., editor. Microbial Communities Utilizing Hydrocarbons and Lipids: Members, Metagenomics and Ecophysiology. Springer; Cham, Switzerland: 2018. pp. 1–36. [DOI] [Google Scholar]
- 11.Babič M.N., Zupančič J., Gunde-Cimerman N., de Hoog S., Zalar P. Ecology of the human opportunistic black yeast Exophiala dermatitidis indicates preference for human-made habitats. Mycopathol. 2018;183:201–212. doi: 10.1007/s11046-017-0134-8. [DOI] [PubMed] [Google Scholar]
- 12.Babič M.N., Zalar P., Ženko B., Džeroski S., Gunde-Cimerman N. Yeasts and yeast-like fungi in tap water and groundwater, and their transmission to household appliances. Fungal Ecol. 2016;20:30–39. doi: 10.1016/j.funeco.2015.10.001. [DOI] [Google Scholar]
- 13.Döğen A., Kaplan E., Öksüz Z., Serin M.S., Ilkit M., de Hoog G.S. Dishwashers are a major source of human opportunistic yeast-like fungi in indoor environments in Mersin, Turkey. Med. Mycol. 2013;51:493–498. doi: 10.3109/13693786.2012.738313. [DOI] [PubMed] [Google Scholar]
- 14.Raghupathi P.K., Zupančič J., Brejnrod A.D., Jacquiod S., Houf K., Burmølle M., Gunde-Cimerman N., Sørensen S.J. Microbial diversity and putative opportunistic pathogens in dishwasher biofilm communities. Appl. Environ. Microbiol. 2018;84:84. doi: 10.1128/AEM.02755-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zalar P., Novak M., de Hoog G.S., Gunde-Cimerman N. Dishwashers—A man-made ecological niche accommodating human opportunistic fungal pathogens. Fungal Biol. 2011;115:997–1007. doi: 10.1016/j.funbio.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Zupančič J., Babič M.N., Zalar P., Gunde-Cimerman N. The black yeast Exophiala dermatitidis and other selected opportunistic human fungal pathogens spread from dishwashers to kitchens. PLoS ONE. 2016;11:e0148166. doi: 10.1371/journal.pone.0148166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamada N., Abe N. Physiological characteristics of 13 common fungal species in bathrooms. Mycoscience. 2009;50:421–429. doi: 10.1007/S10267-009-0500-6. [DOI] [Google Scholar]
- 18.Hamada N., Abe N. Growth characteristics of four fungal species in bathrooms. Biocontrol Sci. 2010;15:111–115. doi: 10.4265/bio.15.111. [DOI] [PubMed] [Google Scholar]
- 19.Lian X., de Hoog G.S. Indoor wet cells harbour melanized agents of cutaneous infection. Med. Mycol. 2010;48:622–628. doi: 10.3109/13693780903405774. [DOI] [PubMed] [Google Scholar]
- 20.Samerpitak K., Gloyna K., Gerrits van den Ende A.H.G., de Hoog G.S. A novel species of the oligotrophic genus Ochroconis colonizing indoor wet cells. Mycoscience. 2017;58:290–296. doi: 10.1016/j.myc.2017.04.002. [DOI] [Google Scholar]
- 21.Wang X., Cai W., Gerrits van den Ende A.H.G., Zhang J., Xie T., Xi L., Li X., Sun J., de Hoog G.S. Indoor wet cells as a habitat for melanized fungi, opportunistic pathogens on humans and other vertebrates. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-26071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamada N. Growth on various detergent components of fungi found in washing machines. Seikatsu Eisei. 2005;49:161–167. doi: 10.11468/seikatsueisei.49.161. [DOI] [Google Scholar]
- 23.Hamada N. Characteristics of fungi growing inside washing machines. Seikatsu Eisei. 2005;49:108–113. doi: 10.11468/seikatsueisei.49.108. [DOI] [Google Scholar]
- 24.Isola D., Selbmann L., de Hoog G.S., Fenice M., Onofri S., Prenafeta-Boldú F.X., Zucconi L. Isolation and screening of black fungi as degraders of volatile aromatic hydrocarbons. Mycopathologia. 2013;175:369–379. doi: 10.1007/s11046-013-9635-2. [DOI] [PubMed] [Google Scholar]
- 25.Nucci M., Akiti T., Barreiros G., Silveira F., Revankar S.G., Wickes B., Sutton D.A., Patterson T.F. Nosocomial outbreak of Exophiala jeanselmei fungemia associated with contamination of hospital water. Clin. Infect. Dis. 2002;34:1475–1480. doi: 10.1086/340344. [DOI] [PubMed] [Google Scholar]
- 26.Nishimura K., Miyaji M., Taguchi H., Tanaka R. Fungi in bathwater and sludge of bathroom drainpipes. Mycopathologia. 1987;97:17–23. doi: 10.1007/BF00437326. [DOI] [PubMed] [Google Scholar]
- 27.Matos T., de Hoog G.S., de Boer A.G., de Crom I., Haase G.M. High prevalence of the neurotrope Exophiala dermatitidis and related oligotrophic black yeasts in sauna facilities. Mycoses. 2002;45:373–377. doi: 10.1046/j.1439-0507.2002.00779.x. [DOI] [PubMed] [Google Scholar]
- 28.Döğen A., Ilkit M., de Hoog G.S. Black yeast habitat choices and species spectrum on high altitude creosote-treated railway ties. Fungal Biol. 2013;117:692–696. doi: 10.1016/j.funbio.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Döğen A., Kaplan E., Ilkit M., de Hoog G.S. Massive contamination of Exophiala dermatitidis and E. phaeomuriformis in railway stations in subtropical Turkey. Mycopathologia. 2012;175:381–386. doi: 10.1007/s11046-012-9594-z. [DOI] [PubMed] [Google Scholar]
- 30.Satow M., Attili-Angelis D., de Hoog G., Vicente V. Selective factors involved in oil flotation isolation of black yeasts from the environment. Stud. Mycol. 2008;61:157–163. doi: 10.3114/sim.2008.61.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicente V., Attili-Angelis D., Pie M., Queiroz-Telles F., Cruz L., Najafzadeh M.J., de Hoog G., Zhao J., Pizzirani-Kleiner A. Environmental isolation of black yeast-like fungi involved in human infection. Stud. Mycol. 2008;61:137–144. doi: 10.3114/sim.2008.61.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron N., Pagnocca F., Otsuka A., Prenafeta-Boldú F., Vicente V., de Angelis D.A. Black Fungi and Hydrocarbons: An Environmental Survey for Alkylbenzene Assimilation. Microorganisms. 2021;9:1008. doi: 10.3390/microorganisms9051008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sudhadham M., Prakitsin S., Sivichai S., Chaiyarat R., Dorrestein G., Menken S., de Hoog G.S. The neurotropic black yeast Exophiala dermatitidis has a possible origin in the tropical rain forest. Stud. Mycol. 2008;61:145–155. doi: 10.3114/sim.2008.61.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prenafeta-Boldú F.X., Kuhn A., Luykx D.M.A.M., Heidrun A., van Groenestijn J.W., de Bont J.A.M. Isolation and characterisation of fungi growing on volatile aromatic hydrocarbons as their sole carbon and energy source. Mycol. Res. 2001;105:477–484. doi: 10.1017/S0953756201003719. [DOI] [Google Scholar]
- 35.Quan Y., Gerrits van den Ende A.H.G., Shi D., Prenafeta-Boldú F.X., Liu Z., Al-Hatmi A.M.S., Ahmed S.A., Verweij P., Kang Y., de Hoog G.S. A Comparison of isolation methods for black fungi degrading aromatic toxins. Mycopathologia. 2019;184:653–660. doi: 10.1007/s11046-019-00382-3. [DOI] [PubMed] [Google Scholar]
- 36.Zhao J., Zeng J., de Hoog G.S., Attili-Angelis D., Prenafeta-Boldú F.X. Isolation and identification of black yeasts by enrichment on atmospheres of monoaromatic hydrocarbons. Microb. Ecol. 2010;60:149–156. doi: 10.1007/s00248-010-9651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Hoog G.S., Zeng J.S., Harrak M.J., Sutton D.A. Exophiala xenobiotica sp. nov., an opportunistic black yeast inhabiting environments rich in hydrocarbons. Antonie van Leeuwenhoek. 2006;90:257–268. doi: 10.1007/s10482-006-9080-z. [DOI] [PubMed] [Google Scholar]
- 38.Isola D., Selbmann L., Egidi E., Onofri S., Zucconi L. Black fungi from contaminated sites: One year later; Proceedings of the fifth meeting of the ISHAM working groups on black yeasts and chromoblastomycosis; Guanzhou, China. 29 November–1 December 2013. [Google Scholar]
- 39.Isola D., Zucconi L., Cecchini A., Caneva G. Dark-pigmented biodeteriogenic fungi in Etruscan hypogeal tombs: New data on their culture-dependent diversity, favouring conditions, and resistance to biocidal treatments. Fungal Biol. 2021;125:609–620. doi: 10.1016/j.funbio.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 42.Agresti A., Coull B.A. Approximate is better than "exact" for interval estimation of binomial proportions. Am. Stat. 1998;52:119. doi: 10.2307/2685469. [DOI] [Google Scholar]
- 43.Hammer Ø., Harper D.A., Ryan P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001;4:9. [Google Scholar]
- 44.Cheeptham N. Advances and challenges in studying cave microbial diversity. In: Cheeptham N., editor. Cave Microbiomes: A Novel Resource for Drug Discovery. Volume 1. Springer; New York, NY, USA: 2013. pp. 1–34. Springer Briefs in Microbiology. [Google Scholar]
- 45.Molina-Menor E., Gimeno-Valero H., Pascual J., Peretó J., Porcar M. High culturable bacterial diversity from a European desert: The Tabernas desert. Front. Microbiol. 2021;11:583120. doi: 10.3389/fmicb.2020.583120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martiny A.C. High proportions of bacteria are culturable across major biomes. ISME J. 2019;13:2125–2128. doi: 10.1038/s41396-019-0410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martiny A.C. The ‘1% culturability paradigm’ needs to be carefully defined. ISME J. 2020;14:10–11. doi: 10.1038/s41396-019-0507-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magnuson J.K., Lasure L.L. Fungal diversity in soils as assessed by direct culture and molecular techniques; Proceedings of the 102nd General Meeting of the American Society for Microbiology; Salt Lake City, UT, USA. 19–23 May 2002; pp. 19–23. [Google Scholar]
- 49.Gams W. Biodiversity of soil-inhabiting fungi. Biodivers. Conserv. 2006;16:69–72. doi: 10.1007/s10531-006-9121-y. [DOI] [Google Scholar]
- 50.Hawksworth D.L. The fungal dimension of biodiversity: Magnitude, significance, and conservation. Mycol. Res. 1991;95:641–655. doi: 10.1016/S0953-7562(09)80810-1. [DOI] [Google Scholar]
- 51.Hawksworth D.L. The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycol. Res. 2001;105:1422–1432. doi: 10.1017/S0953756201004725. [DOI] [Google Scholar]
- 52.Blackwell M. The Fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 53.Hawksworth D.L., Lücking R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.FUNK-0052-2016. [DOI] [PubMed] [Google Scholar]
- 54.Wu B., Hussain M., Zhang W., Stadler M., Liu X., Xiang M. Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology. 2019;10:127–140. doi: 10.1080/21501203.2019.1614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirchman D.L. Processes in Microbial Ecology. Volume 1. Oxford University Press (OUP); Oxford, UK: 2018. Genomes and meta-omics for microbes; pp. 73–91. [Google Scholar]
- 56.Hyde K.D., Xu J., Rapior S., Jeewon R., Lumyong S., Niego A.G.T., Abeywickrama P.D., Aluthmuhandiram J.V.S., Brahamanage R.S., Brooks S., et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019;97:1–136. doi: 10.1007/s13225-019-00430-9. [DOI] [Google Scholar]
- 57.Selbmann L., Isola D., Fenice M., Zucconi L., Sterflinger K., Onofri S. Potential extinction of Antarctic endemic fungal species as a consequence of global warming. Sci. Total. Environ. 2012;438:127–134. doi: 10.1016/j.scitotenv.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 58.Dobrzyńska E., Szewczynska M., Pośniak M., Szczotka A., Puchałka B., Woodburn J. Exhaust emissions from diesel engines fueled by different blends with the addition of nanomodifiers and hydrotreated vegetable oil HVO. Environ. Pollut. 2019;259:113772. doi: 10.1016/j.envpol.2019.113772. [DOI] [PubMed] [Google Scholar]
- 59.Directive 2009/30/EC of the European Parliament and of the Council of 23 April 2009 Amending Directive 98/70/EC as Regards the Specification of Petrol, Diesel and Gas-Oil and Introducing a Mechanism to Monitor and Reduce Greenhouse Gas Emissions and Amending Council Directive 1999/32/EC as Regards the Specification of Fuel Used by Inland Waterway Vessels and Repealing Directive 93/12/EEC (Text with EEA relevance)—Document 32009L0030. [(accessed on 16 July 2021)]. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32009L0030.
- 60.Tesei D., Marzban G., Zakharova K., Isola D., Selbmann L., Sterflinger K. Alteration of protein patterns in black rock inhabiting fungi as a response to different temperatures. Fungal Biol. 2012;116:932–940. doi: 10.1016/j.funbio.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwarz A., Adetutu E.M., Juhasz A.L., Aburto-Medina A., Ball A.S., Shahsavari E. Response of the fungal community to chronic petrogenic contamination in surface and subsurface soils. Geoderma. 2018;338:206–215. doi: 10.1016/j.geoderma.2018.12.004. [DOI] [Google Scholar]
- 62.Teixeira M.M., Moreno L.F., Stielow B., Muszewska A., Hainaut M., Gonzaga L., Abouelleil A., Patané J.S.L., Priest M., Souza R., et al. Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota) Stud. Mycol. 2017;86:1–28. doi: 10.1016/j.simyco.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nascimento M.M., Vicente V.A., Bittencourt J.V., Gelinski J.M.L., Prenafeta-Boldú F.X., Romero-Güiza M., Fornari G., Gomes R.R., Santos G.D., Gerrits van den Ende A.H.G., et al. Diversity of opportunistic black fungi on babassu coconut shells, a rich source of esters and hydrocarbons. Fungal Biol. 2017;121:488–500. doi: 10.1016/j.funbio.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 64.Domsch K.H., Gams W., Anderson T.H. Compendium of Soil Fungi. 2nd ed. Academic Press; London, UK: 1980. [Google Scholar]
- 65.Chen W.-T., Tu M.-E., Sun P.-L. Superficial phaeohyphomycosis caused by Aureobasidium melanogenum mimicking tinea nigra in an immunocompetent patient and review of published reports. Mycopathologia. 2016;181:555–560. doi: 10.1007/s11046-016-9989-3. [DOI] [PubMed] [Google Scholar]
- 66.Van Nieuwenhuijzen E.J., Houbraken J.A.M.P., Meijer M., Adan O.C.G., Samson R.A. Aureobasidium melanogenum: A native of dark biofinishes on oil treated wood. Antonie van Leeuwenhoek. 2016;109:661–683. doi: 10.1007/s10482-016-0668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gostinčar C., Ohm R.A., Kogej T., Sonjak S., Turk M., Zajc J., Zalar P., Grube M., Sun H., Han J., et al. Genome sequencing of four Aureobasidium pullulans varieties: Biotechnological potential, stress tolerance, and description of new species. BMC Genom. 2014;15:549. doi: 10.1186/1471-2164-15-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chau H., Si B.C., Goh Y.K., Vujanovic V. A novel method for identifying hydrophobicity on fungal surfaces. Mycol. Res. 2009;113:1046–1052. doi: 10.1016/j.mycres.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 69.Webb J., Van der Mei H.C., Nixon M., Eastwood I.M., Greenhalgh M., Read S.J., Robson G.D., Handley P.S. Plasticizers increase adhesion of the deteriogenic fungus Aureobasidium pullulans to polyvinyl chloride. Appl. Environ. Microbiol. 1999;65:3575–3581. doi: 10.1128/AEM.65.8.3575-3581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Réblová M., Untereiner W.A., Réblová K. Novel evolutionary lineages revealed in the Chaetothyriales (Fungi) based on multigene phylogenetic analyses and comparison of ITS secondary structure. PLoS ONE. 2013;8:e63547. doi: 10.1371/journal.pone.0063547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gümral R., Tümgör A., Saraçli M.A., Yildiran S.T., Ilkit M., de Hoog G.S., Saraçlı M.A., Yıldıran Ş.T. Black yeast diversity on creosoted railway sleepers changes with ambient climatic conditions. Microb. Ecol. 2014;68:699–707. doi: 10.1007/s00248-014-0459-5. [DOI] [PubMed] [Google Scholar]
- 72.Bates S.T., Reddy G.S.N., Garcia-Pichel F. Exophiala crusticola anam. nov. (affinity Herpotrichiellaceae), a novel black yeast from biological soil crusts in the Western United States. Int. J. Syst. Evol. Microbiol. 2006;56:2697–2702. doi: 10.1099/ijs.0.64332-0. [DOI] [PubMed] [Google Scholar]
- 73.Samerpitak K., Duarte A.P.M., Attili-Angelis D., Pagnocca F.C., Heinrichs G., Rijs A.J.M.M., Alfjorden A., Gerrits van den Ende A.H.G., Menken S.B.J., de Hoog G.S. A new species of the oligotrophic genus Ochroconis (Sympoventuriaceae) Mycol. Prog. 2015;14:1–10. doi: 10.1007/s11557-015-1023-5. [DOI] [Google Scholar]
- 74.Madrid H., Hernández-Restrepo M., Gené J., Cano J., Guarro J., Silva V. New and interesting chaetothyrialean fungi from Spain. Mycol. Prog. 2016;15:1179–1201. doi: 10.1007/s11557-016-1239-z. [DOI] [Google Scholar]
- 75.Cheewangkoon R., Groenewald J.Z., Hyde K.D., To-Anun C., Crous P.W. Chocolate spot disease of Eucalyptus. Mycol. Prog. 2010;11:61–69. doi: 10.1007/s11557-010-0728-8. [DOI] [Google Scholar]
- 76.Selbmann L., Zucconi L., Isola D., Onofri S. Rock black fungi: Excellence in the extremes, from the Antarctic to space. Curr. Genet. 2014;61:335–345. doi: 10.1007/s00294-014-0457-7. [DOI] [PubMed] [Google Scholar]
- 77.Schiaparelli S., Selbmann L., Onofri S., Zucconi L., Isola D., Rottigni M., Ghiglione C., Piazza P., Alvaro M.C. Distributional records of Antarctic fungi based on strains preserved in the Culture Collection of Fungi from Extreme Environments (CCFEE) Mycological Section associated with the Italian National Antarctic Museum (MNA) MycoKeys. 2015;10:57–71. doi: 10.3897/mycokeys.10.5343. [DOI] [Google Scholar]
- 78.Quan Y., Muggia L., Moreno L.F., Wang M., Al-Hatmi A.M.S., Menezes N.D.S., Shi D., Deng S., Ahmed S., Hyde K.D., et al. A re-evaluation of the Chaetothyriales using criteria of comparative biology. Fungal Divers. 2020;103:47–85. doi: 10.1007/s13225-020-00452-8. [DOI] [Google Scholar]
- 79.Isola D., Zucconi L., Onofri S., Caneva G., de Hoog G.S., Selbmann L. Extremotolerant rock inhabiting black fungi from Italian monumental sites. Fungal Divers. 2016;76:75–96. doi: 10.1007/s13225-015-0342-9. [DOI] [Google Scholar]
- 80.Isola D., Selbmann L., Meloni P., Maracci E., Onofri S., Zucconi L. Detrimental rock black fungi and biocides: A study on the monumental cemetery of Cagliari. In: Rogerio-Candelera M.A., Lazzari M., Cano E., editors. Science and Technology for the Conservation of Cultural Heritage. CRC Press; London, UK: 2013. pp. 83–86. [Google Scholar]
- 81.González-López M.A., Salesa R., González-Vela M.C., Fernández-Llaca H., Val-Bernal J.F., Cano-Lira J.F. Subcutaneous phaeohyphomycosis caused by Exophiala oligosperma in a renal transplant recipient. Br. J. Dermatol. 2007;156:762–764. doi: 10.1111/j.1365-2133.2006.07732.x. [DOI] [PubMed] [Google Scholar]
- 82.Venkateshwar S., Ambroise M.M., Asir G.J., Mudhigeti N., Ramdas A., Authy K., Shivaprakash M.R., Kanungo R. A rare case report of subcutaneous phaeohyphomycotic cyst caused by Exophiala oligosperma in an immunocompetent host with literature review. Mycopathologia. 2014;178:117–121. doi: 10.1007/s11046-014-9762-4. [DOI] [PubMed] [Google Scholar]
- 83.Vicente A., Domellöf F.P., Byström B. Exophiala phaeomuriformis keratitis in a subarctic climate region: A case report. Acta Ophthalmol. 2018;96:425–428. doi: 10.1111/aos.13624. [DOI] [PubMed] [Google Scholar]
- 84.Abdolrasouli A., Gibani M.M., de Groot T., Borman A.M., Hoffman P., Azadian B.S., Mughal N., Moore L.S., Johnson E.M., Meis J.F. A pseudo-outbreak of Rhinocladiella similis in a bronchoscopy unit of a tertiary care teaching hospital in London, United Kingdom. Mycoses. 2021;64:394–404. doi: 10.1111/myc.13227. [DOI] [PubMed] [Google Scholar]
- 85.Chowdhary A., Perfect J., de Hoog G.S. Black molds and melanized yeasts pathogenic to humans. Cold Spring Harb. Perspect. Med. 2014;5:a019570. doi: 10.1101/cshperspect.a019570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zeng J.S., Sutton D.A., Fothergill A.W., Rinaldi M.G., Harrak M.J., de Hoog G.S. Spectrum of clinically relevant Exophiala species in the United States. J. Clin. Microbiol. 2007;45:3713–3720. doi: 10.1128/JCM.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Silva W.C., Gonçalves S.S., Santos D.W.C.L., Padovan A.C.B., Bizerra F.C., Melo A.S.A. Species diversity, antifungal susceptibility and phenotypic and genotypic characterisation of Exophiala spp. infecting patients in different medical centres in Brazil. Mycoses. 2017;60:328–337. doi: 10.1111/myc.12597. [DOI] [PubMed] [Google Scholar]
- 88.Espanhol C.M., Recuero J.K., Pagani D.M., Ribeiro A.C., Vettorato G., Duquia R.P., Luzzatto L., Scroferneker M.L. Cutaneous phaeohyphomycosis caused by Exophiala xenobiotica: A case report. Med. Mycol. Case Rep. 2019;27:39–41. doi: 10.1016/j.mmcr.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moreno L.F., Vicente V., de Hoog G.S. Black yeasts in the omics era: Achievements and challenges. Med. Mycol. 2018;56:S32–S41. doi: 10.1093/mmy/myx129. [DOI] [PubMed] [Google Scholar]
- 90.Sood S., Vaid V., Sharma M., Bhartiya H. Cerebral phaeohyphomycosis by Exophiala dermatitidis. Indian J. Med. Microbiol. 2014;32:188–190. doi: 10.4103/0255-0857.129830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.