Abstract
The relationship between the phenotypic and genotypic characteristics of 105 penicillin-intermediate or -resistant Streptococcus pneumoniae isolates saved during 1994 to 1997 at the Prince of Wales Hospital and Pamela Youde Nethersole Eastern Hospital, Hong Kong, was studied. The pbp genes for penicillin-binding proteins 1a, 2b, and 2x for each isolate were amplified by PCR, and the products were digested with restriction enzymes HinfI and AluI. A combination of the pulsed-field gel electrophoresis (PFGE) profiles, pbp fingerprints, and phenotypic characteristics of capsular types and antibiograms enabled these isolates to be divided into four major groups. Seventy-four percent (78 of 105) of the strains, belonging to serotypes 23F, 19F, and 14, showed indistinguishable pbp fingerprint patterns (group A1, 1-1-1, 1-1-1), with PFGE patterns belonging to group A and its subtypes, suggesting that these strains were closely related. Eighty-three percent (65 of 78) of these isolates were also resistant to tetracycline, erythromycin, chloramphenicol, and trimethoprim. The type 23F isolates were indistinguishable from representative strains of the Spanish 23F clone by these molecular methods, indicating that these strains may be variants of the Spanish 23F clone. Serotype 6B accounted for 19% (20 of 105) of the isolates with reduced penicillin susceptibility and was made up of variants belonging to four different pbp fingerprint groups with the PFGE pattern group B, the predominant group being indistinguishable from that of the Spanish 6B clone. Other PFGE and fingerprint groups were mainly obtained from penicillin-susceptible strains of various serotypes. The results suggest that the rapid emergence of drug-resistant S. pneumoniae in Hong Kong has been due to the rapid dissemination of several successful clones.
Streptococcus pneumoniae is a major cause of respiratory infection, septicemia, and meningitis worldwide and is associated with high morbidity and mortality, especially at the extremes of age. Non-penicillin-susceptible strains (penicillin MICs of ≥0.1 μg/ml), first described in the late 1960s in Australia (15), have increased in prevalence in the last decade in many parts of the world and currently account for more than 10% of pneumococcal isolates in regions of North and South America, Europe, Africa, and Asia (1). The percentage rose to 40% in Spain (24) and 48% in France (13) in the 1990s. In the United States, nationwide surveys showed that 27.8% of S. pneumoniae isolates had reduced susceptibility to penicillin and 16.0% were resistant to penicillin (10). In the Western Pacific region, recent studies have shown rapidly rising rates of resistance in Korea (34, 35), Singapore (21), Taiwan (6), Hong Kong (18, 25), and mainland China (37). Most of the isolates in Korea and Japan belonged to serogroup 19 or 23 (34, 35, 38). In Hong Kong, at the Prince of Wales Hospital (PWH), a 1,400-bed hospital in the New Territories of Hong Kong, the rapid emergence of penicillin-resistant pneumococci has been described, with the resistance rate to penicillin in sputum pneumococcal isolates rising from 6.6% in early 1993 to 55% in mid-1995 (25). Most of these isolates were also resistant to co-trimoxazole, tetracycline, chloramphenicol, and erythromycin.
Two main mechanisms have been postulated for the worldwide dissemination of resistance genes in pneumococci, clonal and horizontal spread. There is good evidence to show the rapid intercontinental spread of some successful clones, in particular the type 23F and 19F clones originating in Spain (26). Another distinct multidrug-resistant Spanish-Icelandic serotype 6B clone has spread extensively from Spain to Iceland and from Spain to other parts of Europe, including France, Finland, the former West Germany, and Hungary (28, 33). Altered forms of penicillin-binding proteins (PBPs), in particular, PBPs 1a, 2b, and 2x, have been implicated in the development of penicillin and cephalosporin resistance (7). The nucleotide sequences of the genes coding for PBP 2b and PBP 2x in penicillin-resistant isolates differ extensively from those present in susceptible strains and may have arisen via interspecies recombinational events (11, 23) with the creation of novel resistant isolates with altered (mosaic) pbp genes which spread horizontally. Pneumococcal isolates apparently belonging to the Spanish type 23F clone have been found to express the capsular types 19F and 14 (8), suggesting that the genes specifying the capsular type can be replaced by genetic transformation. Recent work has provided evidence for the exchange of capsular biosynthetic genes by recombination (9).
In Hong Kong, the majority of the penicillin-resistant pneumococcal isolates had penicillin MICs of 1 or 2 μg/ml, were multidrug resistant, and had either of the capsular types 23F and 19F (25). This suggests a fairly high level of similarity among the isolates and may suggest the presence of one or several main clones within the population. In this study, we aimed to further define the degree of relatedness of multidrug-resistant S. pneumoniae in Hong Kong, in order to understand better the rapid emergence of drug-resistant pneumococci in this territory.
(This work was presented at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, September 1998, San Diego, Calif. [16].)
MATERIALS AND METHODS
Selection of isolates for DNA fingerprinting.
A total of 105 strains of S. pneumoniae intermediate or resistant to penicillin isolated at the PWH, New Territories, and Pamela Youde Nethersole Eastern Hospital (PYNEH) during the period January 1994 to December 1997 were studied. The majority of the PWH and PYNEH strains were from sputum (>95%), and the remainder were from cerebrospinal fluid and blood. S. pneumoniae isolates were screened for penicillin resistance by the 1-μg oxacillin disk method on Mueller-Hinton agar supplemented with 5% horse blood. Isolates with an oxacillin disk zone of inhibition of ≤19 mm were defined as being of reduced susceptibility and were saved. The PWH strains used were isolated from hospitalized patients during the first 48 h after admission and were therefore regarded as community-acquired strains. Only isolates with penicillin MICs of ≥0.1 μg/ml were analyzed. The two hospitals are located in the northern and southern parts of Hong Kong, and the isolates were thus deemed to be representative of strains from patients with pneumococcal infections requiring hospital admissions in Hong Kong. Representatives of well-defined international clones (Spanish serotype 23F [26H; gift from T. J. Coffey] and Spanish serotypes 23F and 6B [Sp267 and Sp681, respectively; gifts from K. P. Klugman]) and ATCC 49619 were included for comparison. Penicillin-susceptible clinical S. pneumoniae isolates of known serotypes including 23F, 19F, and 14 and the penicillin-susceptible ATCC 6305 strain were also included. Capsular serotyping of the isolates was by the chessboard agglutination or the quellung reaction method with Pneumotest antisera (Statens Seruminstitut, Copenhagen, Denmark).
Antimicrobial resistance profiles.
The MICs of penicillin, cefotaxime, trimethoprim, chloramphenicol, tetracycline, and erythromycin were determined by the National Committee for Clinical Laboratory Standards agar dilution method (27). An inoculum of approximately 104 CFU per spot was inoculated onto Mueller-Hinton agar supplemented with 5% defibrinated horse blood and incubated at 35°C for 18 h. S. pneumoniae ATCC 6305 and ATCC 49619 and Staphylococcus aureus NCTC 6571 were included as controls.
Restriction fragment length polymorphism of pbp genes.
PCR amplification of pbp 1a, pbp 2b, and pbp 2x genes was performed with primers as described previously (26). The target sequence was amplified in a 25-μl reaction mixture containing 2 μl of sample DNA, a 5.0 mM mixture of the four deoxynucleoside triphosphates, 50 pM (each) oligonucleotide primers, and 1 U of Expand high-fidelity DNA polymerase (Boehringer, Mannheim, Germany) in the reaction buffer (10 mM KCl, 1.5 mM MgCl2, 10 mM KCl, 0.1 mM dithiothreitol, 0.01 mM EDTA, and 2 mM Tris-HCl, pH 7.5) provided. The reaction mixture was denatured at 94°C for 5 min, followed by 35 cycles of amplification and final extension at 72°C for 10 min on a programmable OmniGene DNA thermal cycler (Hybaid, Middlesex, United Kingdom). Each cycle consisted of 30 s at 94°C, 30 s at the 50°C annealing temperature, and 90 s at 72°C for the PCR. Fingerprinting of pbp genes was performed with AluI and HinfI enzymes, respectively, in a 10-μl reaction mixture containing reaction buffer (10 mM magnesium acetate, 50 mM potassium acetate, 10 mM Tris-acetate, pH 7.5) and incubated at 37°C overnight as recommended by the manufacturer. The mixture was electrophoresed on 3% high-resolution agarose (Metaphor; FMC BioProducts, Rockland, Maine) at 60 V for 3 h.
PFGE.
Isolates of S. pneumoniae were cultured in Todd-Hewitt broth at 37°C in 5% CO2 for 16 h. The bacterial cells were pelleted, washed once, and resuspended in SE buffer (75 mM NaCl, 25 mM EDTA, pH 7.5). The turbidity was adjusted to a 1.0 McFarland standard. One milliliter of this suspension was repelleted, resuspended in 50 μl of SE buffer, mixed with an equal volume of 2% SeaPlaque GTG agarose (FMC BioProducts), and dispensed into plug molds. Plugs were prepared for pulsed-field gel electrophoresis (PFGE) as previously described (19). The chromosomal DNA was digested with restriction enzymes, SmaI and ApaI, respectively. The restriction fragments were resolved on a 1% SeaKem Gold agarose (FMC BioProducts) gel run on a PFGE apparatus, Chef Mapper (Bio-Rad, Richmond, Calif.), at 6 V/cm for 22 h, with switching times ramped from 1 to 30 s. An including angle of 120°C was used. The conditions used for the electrophoresis were modified from those of the work of Hall et al. (14). A lambda DNA-PFGE molecular size standard (Pharmacia Biotech, Piscataway, N.J.) was run on the two outside lanes of the gel and after every five samples. Control strains, Enterococcus faecium ATCC 51558 and S. pneumoniae of the Spanish clone 23F, were included in each gel to act as procedure controls. The gel was stained with ethidium bromide (1 μg/ml) and destained, and the fragments were visualized under UV transillumination and scanned into a computer with the ImageMaster VDS gel documentation system (Pharmacia Biotech, Uppsala, Sweden). Two controls were included in each gel to confirm the reproducibility of the procedures, and if these profiles were not identical, the remaining profiles were repeated.
Computer analysis of PFGE profiles.
The PFGE fingerprints were analyzed by computer comparisons with the GelCompar (version 4) software (Applied Maths, Kortrijk, Belgium), and dendrograms were calculated by the Dice method and by clustering by the unweighted pair group method with averages. Estimates of fragment sizes were made, and the banding patterns of PFGE profiles were compared visually. For closely related patterns, a type, e.g., A, was designated and further subclassified into subtypes A1, A2, A3, etc. Unique PFGE profiles were designated types B, C, and D, etc., if four or more band differences were observed.
RESULTS
Antimicrobial resistance profiles.
The MICs at which 50 and 90% of the isolates were inhibited and MIC range for the tested antimicrobial agents for the strains with reduced susceptibility to penicillin are shown in Table 1. Sixty-two percent (66 of 105) had intermediate susceptibility to penicillin with MICs between 0.12 and 1.0 μg/ml, while the remaining 38% (39 of 105) had MICs of 2.0 μg/ml and were resistant to penicillin. Ten percent (11 of 105) of these isolates were susceptible to cefotaxime with MICs of ≤0.5 μg/ml. Eighty-four percent (88 of 105) had MICs of 1.0 μg/ml and had intermediate susceptibility to cefotaxime, and 5% (6 of 105) were resistant and had MICs of 2.0 μg/ml (Table 2). In the penicillin-resistant group, all the isolates either were intermediately susceptible or resistant to cefotaxime and 87% (34 of 39) of these strains were also resistant to erythromycin, trimethoprim, chloramphenicol, and tetracycline, whereas 62% (41 of 66) of the penicillin-intermediate group were similarly multidrug resistant (P < 0.01, chi-square test).
TABLE 1.
MICs for 105 S. pneumoniae isolates with reduced susceptibility to penicillin (penicillin MIC ≥ 0.12 μg/ml)
| Drug | MIC50a (μg/ml) | MIC90a (μg/ml) | MIC range (μg/ml) |
|---|---|---|---|
| Penicillin | 1.0 | 2.0 | 0.25–2.0 |
| Cefotaxime | 1.0 | 1.0 | 0.12–2.0 |
| Erythromycin | 8.0 | ≥64 | 0.12–>64 |
| Trimethoprim | ≥64 | ≥64 | 2.0–>64 |
| Chloramphenicol | 16 | 16 | 2.0–16 |
| Tetracycline | 16 | 32 | 0.25–>64 |
MIC50 and MIC90, MICs at which 50 and 90%, respectively, are inhibited.
TABLE 2.
Relationship of penicillin susceptibility category to cefotaxime susceptibility and multidrug resistance in 105 S. pneumoniae isolates with penicillin MICs of ≥ 0.12 μg/ml
| Resistance typea | No. of isolates (%)
|
||
|---|---|---|---|
| Penicillin intermediateb | Penicillin resistantc | Totald | |
| Cefotaxime I (MIC, 1.0 μg/ml) | 53 (80) | 35 (90) | 88 (84) |
| Cefotaxime R (MIC, ≥2.0 μg/ml) | 2 (3) | 4 (10) | 6 (6) |
| MDR | 48 (73) | 34 (87) | 82 (78) |
| MDR plus cefotaxime I or R | 41 (62) | 34 (87) | 75 (71) |
I, intermediate; R, resistant; MDR, multidrug resistant (MICs of tetracycline, erythromycin, chloramphenicol, and trimethoprim of ≥4.0, ≥0.5, ≥8.0, and ≥1.0 μg/ml, respectively).
MIC, 0.1 to 1.0 μg/ml; n = 66.
MIC, ≥2.0 μg/ml; n = 39.
n = 105.
Capsular types.
One hundred five strains of S. pneumoniae with intermediate susceptibility or resistance to penicillin were serotyped. Only five serotypes, namely, 23F, 19F, 6B, 14, and 9V, were identified. The most frequent serotypes were 23F (51 of 105) and 19F (30 of 105), constituting 49 and 29% of all the strains, respectively. The remainder of the strains belonged to serotypes 6B (18%, 19 of 105), 14 (2.9%, 4 of 105), and 9V (1%, 1 of 105).
pbp fingerprints and PFGE profiles.
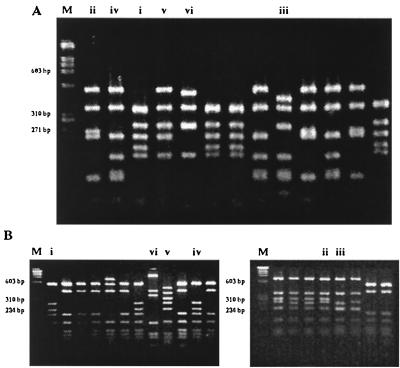
Four to six distinct patterns were obtained for each of the pbp 1a, 2b, and 2x genes when digested with HinfI or AluI. The combination of the six patterns obtained for each strain of S. pneumoniae allowed classification into 13 fingerprint subgroups, A1, A2, B1 to B4, C, D, E1 to E4, and F. The penicillin- and multidrug-resistant pneumococci gave fingerprints which were clearly different from those of the penicillin-susceptible isolates. Seventy-five percent (79 of 105) of the strains were assigned to fingerprint group A1 (1-1-1, 1-1-1), which included strains with capsular serotypes 23F, 19F, 14, and 9V and strains from the Spanish serotype 23F clone (Table 3). Serotype 6B accounted for 18% (19 of 105) of the isolates and was made up of variants belonging to four different fingerprint groups, B1 to B4. Fingerprint groups E1 to E4 and F were obtained from penicillin-susceptible strains of various serotypes. Figure 1 shows the restriction patterns obtained when the pbp 2b and 2x genes were amplified by PCR and the products were digested with restriction endonuclease HinfI.
TABLE 3.
pbp fingerprints, PFGE profiles, and phenotypic characteristics of penicillin-intermediate or -resistant S. pneumoniae isolates
|
pbp gene fingerprinta
|
PFGE profile
|
Phenotypic characteristic
|
No. of isolates (total no. of 105) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group |
HinfI
|
AluI
|
SmaI | ApaI | Serotype | MIC (μg/ml)b
|
Resistance profilec | ||||||
| 1a | 2b | 2x | 1a | 2b | 2x | P | C | ||||||
| A1 | 1 | 1 | 1 | 1 | 1 | 1 | sA (sA1 to -5) | pA (pA1 to -9) | 23F | 1.0–2.0 | 0.5–2.0 | P, C, T, E, Ch, Tm | 35 |
| P, C, T, Ch, Tm | 7 (2)d | ||||||||||||
| P, C, E, Ch, Tm | 3 | ||||||||||||
| P, C, T, E, Tm | 2 | ||||||||||||
| P, T, E, Ch, Tm | 1 | ||||||||||||
| 19F | P, C, T, E, Ch, Tm | 22 | |||||||||||
| P, T, E, Ch, Tm | 3 | ||||||||||||
| P, C, E, Ch, Tm | 1 | ||||||||||||
| 14 | P, C, T, E, Ch, Tm | 4 | |||||||||||
| sF | pF | 9V | P, C, T, Tm | 1 | |||||||||
| A2 | 1 | 2 | 1 | 1 | 2 | 1 | sE | pE, pG | 19F | 0.25–0.5 | 0.25–0.5 | P, T, E, Tm | 2 |
| P, T, E, Ch, Tm | 1 | ||||||||||||
| B1, B2, B3 | 1 | 1 | 4 | 5 | 2 | 3 | sB (sB1 to -3) | pB (pB1 to -5) | 6B | 0.25–2.0 | 0.5–1.0 | P, C, T, E, Ch, Tm | 14 (1)e |
| B2, B3 | 1 | 1 | 4 | 6 | 2 | 3 | P, T, E, Ch, Tm | 2 | |||||
| B1, B2 | 1 | 4 | 4 | 5 | 6 | 3 | P, C, T, E, Tm | 2 | |||||
| B4 | 1 | 6 | 4 | 5 | 2 | 3 | P, T, E, Tm | 1 | |||||
| C | 4 | 3 | 6 | 4 | 4 | 4 | sC | pC | 19F | 0.25 | 0.06 | P, Tm | 1f |
| D | 1 | 5 | 1 | 6 | 5 | 3 | sD | pD | 23F | 1.0–2.0 | 1.0 | P, C, T, E, Tm | 3 |
| sG | pD | 19F | P, C, T, E, Tm | 1 | |||||||||
In the columns marked “HinfI” and “AluI,” rows 3 to 6 indicate pbp patterns of groups B1 to B4, respectively.
P, penicillin; C, cefotaxime.
P, penicillin (MIC, ≥0.1 μg/ml); C, cefotaxime (MIC, ≥0.5 μg/ml); T, tetracycline (MIC, ≥4.0 μg/ml); E, erythromycin (MIC, ≥0.5 μg/ml); Ch, chloramphenicol (MIC, ≥8.0 μg/ml); Tm, trimethoprim (MIC, ≥1.0 μg/ml).
Two Spanish 23F isolates, 26H and Sp267.
One Spanish 6B isolate, Sp681.
ATCC 49619.
FIG. 1.
Fingerprints of the pbp genes. HinfI restriction patterns of the pbp 2b (A) and 2x (B) genes after PCR. Lanes i, ii, iii, etc., indicate the different fingerprint patterns for each gene. Lanes M are molecular size standards.
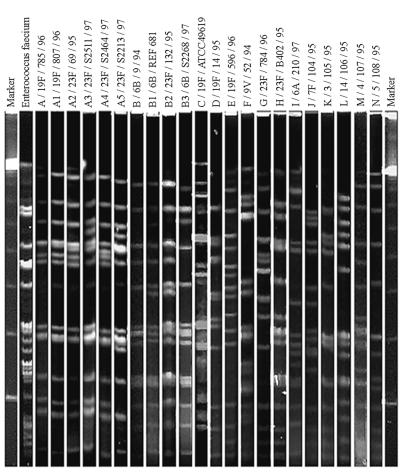
Seven major PFGE profiles, A to G, and their subtypes were identified among the penicillin-intermediate or -resistant pneumococcal strains when either SmaI or ApaI was used (Table 3). The most frequent PFGE pattern was reported as type sA (representing the pattern with SmaI as the enzyme for digestion) or pA (with ApaI for digestion). Closely related patterns were designated subtypes sA1, sA2, sA3, etc. Strains which were considered unique if PFGE patterns differed by more than three bands were designated types B, C, D, etc. The two restriction enzymes enabled all the strains to be sorted into their corresponding groups, except for patterns pD and sE, where two different patterns were obtained when the other enzyme was used. All the isolates with type A PFGE profiles belonged to serotype 23F, 19F, or 14. The penicillin-susceptible group gave unique PFGE patterns (types H to N) different from those of the penicillin-intermediate and -resistant group. A typical SmaI digestion PFGE profile is shown in Fig. 2.
FIG. 2.
SmaI digestion PFGE profiles for S. pneumoniae isolates. Gel strips A to G represent patterns from penicillin-intermediate or -resistant S. pneumoniae isolates, and strips H to N represent patterns from penicillin-sensitive strains. Markers are molecular size standards.
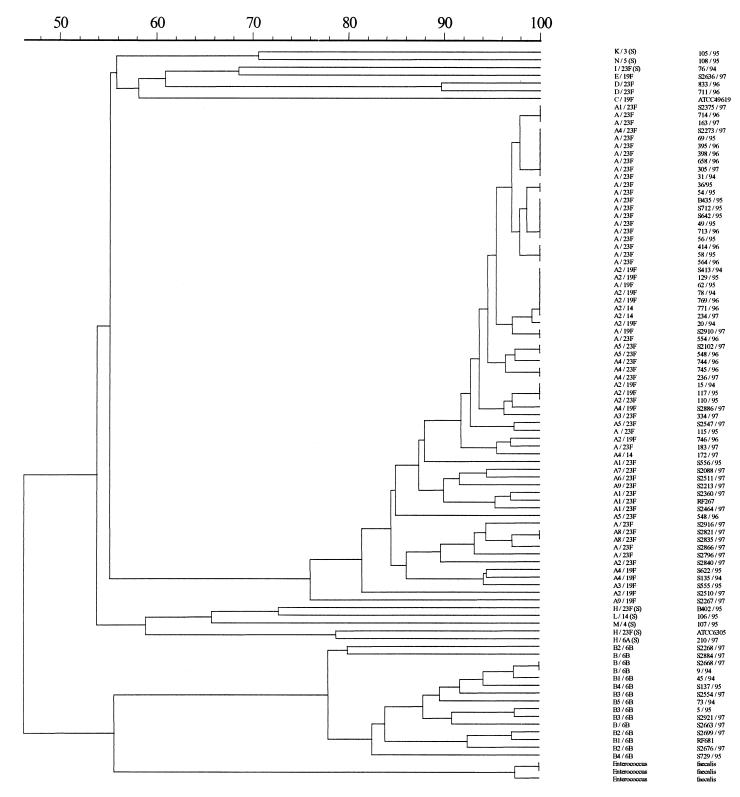
Figure 3 shows a dendrogram of the cluster analysis of the ApaI digestion PFGE profiles. The isolates segregated into two major clusters. The major cluster of isolates fell within a cutoff value of 80% similarity and consisted of strains with PFGE type A profiles, suggesting that these strains were closely related genetically. The other small cluster of isolates belonged to the PFGE type B group.
FIG. 3.
Dendrogram of ApaI digestion electrophoretic profiles of S. pneumoniae isolates made by the Dice method and clustering by unweighted pair group method with averages.
Overall, 74% (78 of 105) of the strains belonged to pbp group A1, which included serotypes 23F, 19F, and 14 and which had PFGE type A profiles, suggesting that these strains were closely related. These isolates were also indistinguishable from isolates of the Spanish 23F clone, except that the majority differed by having resistance to erythromycin. The predominant subgroups were seen in strains from both PWH and PYNEH.
One strain with serotype 9V carried an altered pbp fingerprint pattern (group A1) identical to that of the predominant type but differed in PFGE pattern. This suggests that the altered pbp genes may have arisen by horizontal transfer of the pbp genes from the predominant resistant clone.
Seven isolates also belonging to serotypes 23F and 19F gave three different pbp fingerprint groups, namely, A2 (1-2-1, 1-2-1), C (4-3-6, 4-4-4), and D (1-5-1, 6-5-3), and had unique PFGE profiles. Fingerprint groups A2 and C included only isolates with reduced susceptibility to penicillin with MIC ranges of 0.25 to 0.5 μg/ml and with sensitivity to cefotaxime with a MIC of 0.06 to 0.5 μg/ml. Only one isolate, ATCC 49619, belonged to fingerprint group C, indicating that the alterations in the pbp genes in this strain probably originated quite differently from those of the other penicillin-intermediate or -resistant isolates. pbp fingerprint group A2 had pbp 1a and 2x patterns identical to those of group A1 and differed only in pbp 2b restriction patterns. Fingerprint group D had different restriction patterns from pbp 1a, 2b, and 2x genes when AluI was used. The most likely explanation for this is that recombinations with de novo mutations of the pbp genes occurred and the number of isolates involved has remained small.
DISCUSSION
The isolation of penicillin-resistant pneumococci has significantly increased in many places worldwide during the 1990s. Some of the countries which have seen the highest rates of penicillin resistance have been in the Far East, in particular, South Korea (34), Taiwan (6), and Japan (36). These countries have seen some of the most rapidly rising rates of penicillin-resistant pneumococci; most of these were multidrug resistant and often belonged to a few successful clones, in particular, those of serotypes 23F and 19 (6, 34–36, 38). Our data from Hong Kong provides more on the rapid emergence of penicillin-resistant pneumococci in this region.
The study detected a major group of highly related isolates (74%) which was indistinguishable from the Spanish serotype 23F antibiotic-resistant clone (26H and Sp267), suggesting that these isolates may represent a variant of the Spanish 23F clone. Identical clones of multidrug-resistant serotype 23F S. pneumoniae have been identified in South Africa, the United States, Western Europe, Hungary, and Asia (12, 21, 26, 28, 34, 35). However, our strains expressed three different serotypes, 23F, 19F, and 14. This could be explained in two ways. Firstly, these could represent different clones of pneumococcal isolates that were genetically closely related. Using multilocus sequence typing, Shi et al. (31) in Taiwan in 1998 found that their penicillin-resistant isolates belonged to three prevalent clones, a Taiwanese 19F clone, a Taiwanese 23F clone, and a second serotype 23F clone which was indistinguishable from the Spanish multidrug-resistant serotype 23F clone. They postulated that the Taiwanese clones may have originated in East Asia. As was also the case in Hong Kong, they noted the rapid spread of clones of penicillin-resistant S. pneumoniae during 1994 and 1995.
The second possible explanation for these highly similar isolates expressing three different serotypes is that horizontal transfer of capsular biosynthetic genes may have occurred between isolates which had the genetic background of the Spanish 23F clone. Examples of clonally related organisms manifesting different capsular serotypes have previously been reported (20, 32). Recombinational exchanges have been shown to occur at the capsular polysaccharide biosynthetic gene and have led to serotype changes from serotype 14 to serotype 23F (3) and from serotype 19 to serotype 23F in penicillin-resistant S. pneumoniae (9). We also found our predominant serotype 6B profile to be indistinguishable from the Spanish 6B clone (Sp681) which has been identified also in Iceland, France, Finland, Germany, and Hungary (12), providing further support for the hypothesis of the spread of international clones in Hong Kong. Further assessment of the population structure of our isolates, e.g., by multilocus sequence typing, will be required to distinguish these possibilities for Hong Kong isolates.
Certain common factors may have contributed to this rapid spread of clones in Hong Kong and in South Korea, Japan, and Taiwan. Using population genetic methods and epidemiological observations, Austin et al. (2) showed that consistent antibiotic consumption above a critical level can trigger the emergence of resistance and that the rapidity of emergence and final level of resistance are proportional to the level of consumption. These countries have seen significant economic development, and antibiotics are affordable and readily available. High levels of antibiotic consumption have been noted in the Hong Kong community (22) and are reflected by high levels of resistance to tetracycline in S. pneumoniae in Hong Kong prior to the emergence of penicillin resistance (16, 17). In contrast, areas such as Beijing, China (37), Malaysia (29), and Bangladesh (30) have reported lower levels of penicillin resistance and a more heterogeneous pattern of penicillin-resistant S. pneumoniae. In the United States, institutions such as day care centers and nursing homes have been implicated as the source of these resistant pneumococci (4, 5). In Hong Kong, and likewise in Taiwan and Japan, the cities are densely populated. Under such circumstances, these multidrug-resistant S. pneumoniae strains may become highly successful and disseminate widely.
Knowledge of the genetic basis of resistance is essential for development of effective control strategies for the prevention of drug-resistant S. pneumoniae. Our data adds to the literature documenting the rapid dissemination of successful clones of drug-resistant S. pneumoniae in regions of East Asia.
ACKNOWLEDGMENTS
We thank T. J. Coffey and K. P. Klugman for the provision of isolates of the Spanish 23F and 6B clones and T. Pitt, CPHL, Colindale, London, United Kingdom, for help with PFGE protocols.
The study was supported by Hong Kong RGC grant no. CUHK4215/97M.
REFERENCES
- 1.Appelbaum P C. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin Infect Dis. 1992;15:77–83. doi: 10.1093/clinids/15.1.77. [DOI] [PubMed] [Google Scholar]
- 2.Austin D J, Kristinsson K G, Anderson R M. The relationship between the volume of antimicrobial consumption in human communities and the frequency of resistance. Proc Natl Acad Sci USA. 1999;96:1152–1156. doi: 10.1073/pnas.96.3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;17:1890–1896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Outbreaks of pneumococcal pneumonia among unvaccinated residents in chronic care facilities—Massachusetts, October 1995; Oklahoma, February 1996; and Maryland, May–June 1996. Morbid Mortal Weekly Rep. 1997;46:60–62. [PubMed] [Google Scholar]
- 5.Cherian T, Steinhoff M C, Harrison L H, Rohn D, McDougal L K, Dick J. A cluster of invasive pneumococcal disease in young children in child care. JAMA. 1994;271:695–697. [PubMed] [Google Scholar]
- 6.Chiou C C C, Liu Y C, Huang T S, Hwang W K, Wang J H, Lin H H, Yen M Y, Hsieh K S. Extremely high prevalence of nasopharyngeal carriage of penicillin-resistant Streptococcus pneumoniae among children in Kaohsiung, Taiwan. J Clin Microbiol. 1998;36:1933–1937. doi: 10.1128/jcm.36.7.1933-1937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffey T J, Dowson C G, Daniels M, Spratt B G. Genetics and molecular biology of β-lactam-resistant pneumococci. Microb Drug Resist. 1995;1:29–34. doi: 10.1089/mdr.1995.1.29. [DOI] [PubMed] [Google Scholar]
- 8.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 9.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 10.Doern G V, Pfaller M A, Kugler K, Freeman J, Jones R N. Prevalence of antimicrobial resistance among respiratory tract isolates of Streptococcus pneumoniae in North America: 1997 results from the SENTRY antimicrobial surveillance program. Clin Infect Dis. 1998;27:764–770. doi: 10.1086/514953. [DOI] [PubMed] [Google Scholar]
- 11.Dowson C G, Hutchison A, Spratt G. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 12.Feldman C, Klugman K. Pneumococcal infections. Curr Opin Infect Dis. 1997;10:109–115. [Google Scholar]
- 13.Geslin P, Buu-Hoi A, Fremaux A, Acar J F. Antimicrobial resistance in Streptococcus pneumoniae: an epidemiological survey in France, 1970–1990. Clin Infect Dis. 1992;15:95–98. doi: 10.1093/clinids/15.1.95. [DOI] [PubMed] [Google Scholar]
- 14.Hall L M C, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansman D, Bullen M M. A resistant pneumococcus. Lancet. 1967;ii:264–265. doi: 10.1016/s0140-6736(75)91547-0. [DOI] [PubMed] [Google Scholar]
- 16.Ip M, Lyon D J, Yung R W H, Chan C, Cheng A F. Abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Molecular epidemiology of penicillin- and multidrug-resistant Streptococcus pneumoniae in Hong Kong, abstr. C20; p. 74. [Google Scholar]
- 17.Kam K M, Luey K Y. Vaccine coverage of Streptococcus pneumoniae in Hong Kong with attention to the multiple-antibiotic-resistant strains. Vaccine. 1996;14:1573–1580. doi: 10.1016/s0264-410x(96)00156-9. [DOI] [PubMed] [Google Scholar]
- 18.Kam K M, Luey K Y, Fung S M, Yiu P P, Harden T J, Cheung M M. Emergence of multiple-antibiotic-resistant Streptococcus pneumoniae in Hong Kong. Antimicrob Agents Chemother. 1995;39:2667–2670. doi: 10.1128/aac.39.12.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufmann M E, Pitt T L. Pulsed-field gel electrophoresis of bacterial DNA. In: Chart H, editor. Methods in practical laboratory bacteriology. Boca Raton, Fla: CRC Press; 1994. pp. 83–92. [Google Scholar]
- 20.Kell C M, Jordens J Z, Daniels M, Coffey T J, Bates J, Paul J, Gilks C, Spratt B G. Molecular epidemiology of penicillin-resistant pneumococci isolated in Nairobi, Kenya. Infect Immun. 1993;61:4382–4391. doi: 10.1128/iai.61.10.4382-4391.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koh T H, Sng L H, Ngan C. Molecular typing of multiresistant Streptococcus pneumoniae serogroup 19 in Singapore. Pathology. 1998;30:395–398. doi: 10.1080/00313029800169696. [DOI] [PubMed] [Google Scholar]
- 22.Kumana C R, Li K Y, Chau P Y. Worldwide variation in use of chloramphenicol. Lancet. 1987;ii:449–450. doi: 10.1016/s0140-6736(87)90979-2. [DOI] [PubMed] [Google Scholar]
- 23.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP 2X genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 24.Linares J, Pallares R, Alonso T, Perez J L, Ayats J, Gudiol F, Viladrich P F, Martin R. Trends in antimicrobial resistance of clinical isolates of Streptococcus pneumoniae in Bellvitge Hospital, Barcelona, Spain (1979–1990) Clin Infect Dis. 1992;15:99–105. doi: 10.1093/clinids/15.1.99. [DOI] [PubMed] [Google Scholar]
- 25.Lyon D J, Scheel O, Fung K S, Cheng A F B, Henrichsen J. Rapid emergence of penicillin-resistant pneumococci in Hong Kong. Scand J Infect Dis. 1996;28:375–376. doi: 10.3109/00365549609037922. [DOI] [PubMed] [Google Scholar]
- 26.Munoz R, Coffey T J, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, 8th informational supplement, 5th ed. M100-S8. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 28.Reichmann R, Varon E, Guther E, Reinert R R, Luttiken R, Marton A, Geslin P, Wagner J, Hakenbeck R. Penicillin-resistant Streptococcus pneumoniae in Germany: genetic relationship to clones from other European countries. J Med Microbiol. 1995;43:377–385. doi: 10.1099/00222615-43-5-377. [DOI] [PubMed] [Google Scholar]
- 29.Rohani M Y, Raudzah A, Ng A J, Ng P P, Zaidatul A A R, Asmah I, Murtaza M, Parasakthy N, Mohd Yasmin M Y, Cheong Y M. Epidemiology of Streptococcus pneumoniae infection in Malaysia. Epidemiol Infect. 1999;122:77–82. doi: 10.1017/s0950268898001605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saha S K, Rikitomi N, Ruhulamin M, Masaki H, Hanif M, Islam M, Watanabe K, Ahmed K, Matsumoto K, Sack R B, Nagatake T. Antimicrobial resistance and serotype distribution of Streptococcus pneumoniae strains causing childhood infections in Bangladesh, 1993 to 1997. J Clin Microbiol. 1999;37:798–800. doi: 10.1128/jcm.37.3.798-800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Z Y, Enright M C, Wilkinson P, Griffiths D, Spratt B G. Identification of three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals by multilocus sequence typing. J Clin Microbiol. 1998;36:3514–3519. doi: 10.1128/jcm.36.12.3514-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sibold C, Wang J, Henrichsen J, Hakenbeck R. Genetic relationship of penicillin-susceptible and -resistant Streptococcus pneumoniae strains isolated on different continents. Infect Immun. 1992;60:4119–4126. doi: 10.1128/iai.60.10.4119-4126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980’s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 34.Song J H, Yang J W, Peck K R, Kim S, Lee N Y, Jacobs M R, Appelbaum P C, Pai C H. Spread of multidrug-resistant Streptococcus pneumoniae in South Korea. Clin Infect Dis. 1997;25:747–749. doi: 10.1086/516945. [DOI] [PubMed] [Google Scholar]
- 35.Tarasi A, Chong Y, Lee K, Tomasz A. Spread of the serotype 23F multidrug-resistant Streptococcus pneumoniae clone to South Korea. Microb Drug Resist. 1997;3:105–109. doi: 10.1089/mdr.1997.3.105. [DOI] [PubMed] [Google Scholar]
- 36.Ubukata K, Asahi Y, Okuzumi K, Konno M. The Working Group for Penicillin-Resistant S. pneumoniae. Incidence of penicillin-resistant Streptococcus pneumoniae in Japan, 1993–1995. J Infect Chemother. 1996;1:166–176. doi: 10.1007/BF02350645. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Huebner R, Chen M, Klugman K. Antibiotic susceptibility patterns of Streptococcus pneumoniae in China and comparison of MICs by agar dilution and E-test methods. Antimicrob Agents Chemother. 1998;42:2633–2636. doi: 10.1128/aac.42.10.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida R, Hirakata Y, Kaku M, Takemura H, Tanaka H, Tomono K, Koga H, Kohno S, Kamihira S. Genetic relationship of penicillin resistant Streptococcus pneumoniae serotype 19B strains in Japan. Epidemiol Infect. 1997;118:105–110. doi: 10.1017/s0950268896007273. [DOI] [PMC free article] [PubMed] [Google Scholar]