Abstract
AML1/RUNX1 belongs to the runt domain transcription factors that are important regulators of hematopoiesis and osteogenesis. Expression of AML1 is regulated at the level of transcription by two promoters, distal (D) and proximal (P), that give rise to mRNAs bearing two distinct 5′ untranslated regions (5′UTRs) (D-UTR and P-UTR). Here we show that these 5′UTRs act as translation regulators in vivo. AML1 mRNAs bearing the uncommonly long (1,631-bp) P-UTR are poorly translated, whereas those with the shorter (452-bp) D-UTR are readily translated. The low translational efficiency of the P-UTR is attributed to its length and the cis-acting elements along it. Transfections and in vitro assays with bicistronic constructs demonstrate that the D-UTR mediates cap-dependent translation whereas the P-UTR mediates cap-independent translation and contains a functional internal ribosome entry site (IRES). The IRES-containing bicistronic constructs are more active in hematopoietic cell lines that normally express the P-UTR-containing mRNAs. Furthermore, we show that the IRES-dependent translation increases during megakaryocytic differentiation but not during erythroid differentiation, of K562 cells. These results strongly suggest that the function of the P-UTR IRES-dependent translation in vivo is to tightly regulate the translation of AML1 mRNAs. The data show that AML1 expression is regulated through usage of alternative promoters coupled with IRES-mediated translation control. This IRES-mediated translation regulation adds an important new dimension to the fine-tuned control of AML1 expression.
The AML/RUNX genes (previously termed CBFA or PEBP2α [61]) belong to a gene family of heterodimeric transcription factors. Family members encode a protein with a highly conserved region of 128 amino acids designated the runt domain (RD), because of its homology to a region in the Drosophila Runt protein (21). The RD mediates both AML heterodimerization with the CBFβ protein and binding of AML/RUNX to its consensus DNA sequence, PyGPyGGT (17, 21, 36, 61). In humans and mice, three highly conserved AML genes have been identified: AML1/RUNX1 on chromosome 21q22.1, AML2/RUNX3 on chromosome 1p36, and AML3/RUNX2 on chromosome 6p21 (16, 31). Homozygous disruption of AML1/RUNX1 and AML3/RUNX2 in mice indicated that AML1/RUNX1 plays a crucial role in hematopoiesis (44, 45, 69) while AML3/RUNX2 is essential for osteogenesis (51). Importantly, the AML1/RUNX1 and CBFβ genes are the most frequent targets for leukemia-associated translocations (32), further highlighting the pivotal role these genes play in hematopoiesis. AML1/RUNX1 is abbreviated AML1 in this paper.
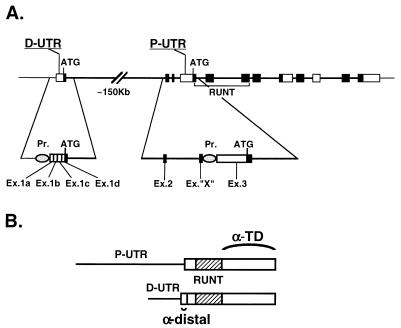
Transcription of AML1 is initiated at two promoter regions, designated the proximal (P) promoter and the distal (D) promoter (12). Genomic analysis revealed that the distance between them is ∼150 kb (Fig. 1) (unpublished data). Transcription yields mRNAs that differ in their 5′ untranslated region (5′UTR), the transcripts carry either the D-UTR or the P-UTR (Fig. 1) (12). The P-UTR is remarkably long (1,631 bp) but is nevertheless carried on a single exon. It contains 15 AUG codons upstream of the authentic initiator AUG (uAUG), several of which are followed by short open reading frames (ORFs) (29). Such uAUGs were shown to inhibit translation initiation (23). Two GC-rich islands, which could form stable stem-loop structures, are present at the 5′ and 3′ parts of this 5′UTR (29). These features suggested that translation of P-UTR-bearing mRNAs via the ribosome-scanning mechanism would be inefficient (24). The D-UTR, while much shorter than the P-UTR, consists of four exons that are alternatively spliced (Fig. 1). When all four exons are included, the length of the D-UTR adds up to 452 bp. It contains only two uAUGs and lacks GC-rich elements. The striking differences in size and structure between the two UTRs and the presence in the P-UTR of structural elements characterizing an internal ribosome entry site (IRES) (reviewed in reference 18) prompted us to investigate the possibility that AML1 expression is regulated at the level of mRNA translation.
FIG. 1.
Schemes depicting the structure of the AML1 gene and two of the mRNA species. (A) Genomic organization of the gene. Exons are presented as boxes; coding regions are dark, and the 3′UTR and 5′UTR are light. Introns are drawn as a thick line. The proximal (P) and distal (D) promoters (Pr.) are indicated, as well as the P-UTR and D-UTR. (B) The two AML1 mRNA families made of the P- and D-promoters, indicating the coding domains recognized by the α-TD and α-distal antibodies.
Since the initial identification of IRES regions that allow cap-independent translation in picornavirus mRNAs (19, 47), IRES elements have been found in several cellular mRNAs including those encoding the human immunoglobulin heavy-chain binding protein (BiP) (33), the Drosophila Antennapedia and Ultrabithorax proteins (70), fibroblast growth factor 2 (FGF2) (67), platelet-derived growth factor 2 (PDGF-2/c-sis) (4), insulin-like growth factor II (IGF-II) (65), translation initiation factor eIF4G (11), human c-myc (42, 63), cardiac voltage-gated potassium channel Kv1.4 (43), nervous system-specific DNA-binding protein-MYT2 (22), vascular endothelial growth factor (VEGF) (1, 15, 39, 62), and, more recently, the X-linked inhibitor of apoptosis XIAP (14). It is believed that cap-dependent translation of cellular mRNAs is regulated through modulation of eIF4E activity (58) and that IRES-containing mRNAs are translated at times when cap-dependent translation is inhibited (6, 25, 66).
Expression of AML1 is strictly regulated during development and in adult life, but surprisingly little is known about the molecular mechanisms regulating the expression of the gene. In mouse embryos it is expressed in a number of tissues (44, 57), while in adults it is expressed mainly in the hemapoietic system (40, 55). Here we demonstrate that AML1 expression is regulated through usage of alternative promoters coupled to translation control by either cap- or IRES-dependent mechanisms.
MATERIALS AND METHODS
Plasmid constructions.
Plasmid M-CAT (pOS14) (5) contains the Escherichia coli chloramphenicol acetyltransferase (CAT) gene, placed between the bacteriophage T7 promoter and transcription terminator. The unique NcoI site of pOS14 is located downstream of the T7 promoter and harbors the translational initiation codon of the CAT coding sequence. Plasmids T7-MAP and T7-MAD were constructed by inserting an NcoI fragment of 1,631 or 452 bp, containing the proximal (P) or distal (D) AML1 5′UTRs, respectively, into the NcoI site of pOS14. This resulted in placement of the 5′UTR segments between T7 promoter and the CAT gene in both orientations. The 5′UTR fragments were generated by PCR, using plasmids containing the relevant 5′UTR (29) as the template and the sense oligonucleotide primer 5′-GGCGCCCCATGGCCGGACCCAGCC-3′ (T7-MAP) or 5′-AAACAACCATGGAACCACAAGTTGGGTAGC-3′ (T7-MAD) and the antisense oligonucleotide primer 3′-GAACAACGGTACCCATAGGGGCATCTACGG-5′ (T7-MAP) or 3′-GTAAAAGTCCTCCTTCGGTACCGAAGTC-5′ (T7-MAD) to yield a 1.63- or 0.45-kb product with 5′- and 3′-terminal NcoI sites.
To construct BAP, BhAP and BAP-Rev, a 2-kb EcoRI-ApaI fragment containing the P-UTR and 370 bp of the AML1 coding region was inserted into EcoRI-ApaI-digested pBS-SK to generated pBS-AML1/Apa. This plasmid was cut with SmaI, yielding three fragments, of 260, 1,560, and 3,100 bp. The 260-bp fragment, which contains the 3′ end of the P-UTR and the 5′ end of the AML1 coding region, was inserted into SmaI-EheI-digested pGL2-promoter plasmid. The resulting construct was further digested with SmaI, and the 1,560-bp fragment containing the rest of the P-UTR was ligated, to generate the pGL-P-UTR/AML1-LUC plasmid. A 131-bp polylinker containing the SpeI, BamHI, SmaI, and PstI restriction sites was inserted between the EcoRI site at the 5′ end of the P-UTR and the SmaI site a few nucleotide upstream. A fragment containing 752 bp of the 5′ end of the luciferase (LUC) region with the NdeI site at the initiation AUG codon was generated by PCR. A fragment of the P-UTR linked to the LUC ORF was used as a template with the sense oligonucleotide primer 5′-CCGGTACTGTTGGTCATATGGAAGACGCC-3′ and the antisense oligonucleotide primer 3′-ACACCTAAAGCTCAGCAGAATT-5′. The PCR product was digested with NdeI-XcmI and inserted into NdeI-XcmI-cut and the modified pGL-P-UTR/AML1-LUC plasmid, resulting in the pGUL-Del plasmid, which is missing 602 bp from the 3′ end of the P-UTR. This 602-bp fragment with an NdeI site at the 3′ end was generated by PCR using a plasmid containing AML1 cDNA as a template (29) and with the sense oligonucleotide primer 5′-GCTCAGCACTGCTCCAACTACTG-3′ and the antisense oligonucleotide primer 3′-GCCGAACAAGTATACGCATAGGGGCATCTA-5′. The PCR product was digested with NdeI and inserted into NdeI-linearized pGUL-Del in both orientations, resulting in pGUL and pGUL-Rev, respectively. Two 850-bp fragments containing CAT and CAT preceded by a 30-bp hairpin structure were excised from the pBI-FGF and pHP-FGF plasmids, respectively (gift of Anne-Catherine Prats [67]), using BamHI. These fragments were inserted into BamHI-linearized pcDNA3 (Invitrogen), resulting in plasmids pcDNA-CAT and pcDNA-HpCAT. Plasmids pGUL and pGUL-Rev were digested with BamHI, and the P-UTR-LUC-containing fragment was blunted by treatment with Klenow enzyme (Biolabs) and inserted into EcoRV-linearized pcDNA-CAT and pcDNA-HpCAT to generate plasmids BAP, BhAP, and BAP-Rev.
Plasmid T7-BiEMC, a gift of Peter Sarnow (University of Colorado), contains a bicistronic unit composed of CAT and LUC ORFs separated by the encephalomyocarditis virus (EMCV) 5′UTR and driven by the T7 promoter. It was digested with EcoRI, and a 1.7-kb fragment containing the EMCV 5′UTR between the 5′ end of the LUC coding region and the 3′ end of the CAT ORF was generated. This fragment was inserted into the EcoRI-linearized BAP plasmid to generate the BiEMC plasmid.
Plasmids pCL, pCPL, and M-EMCV were constructed as previously described (4). pCL contains the CAT ORF linked through a short linker to the LUC coding region. pCPL contains the 5′UTR of PDGF2/c-sis placed between the CAT and LUC regions. M-EMCV contains a T7 promoter-driven cassette of the EMCV 5′UTR linked to CAT.
In vitro transcription-translation assays.
Plasmids were linearized downstream of the T7 transcription terminator, and capped RNAs were generated by using T7 RNA polymerase (Promega) in a transcription reaction mixture containing an excess of m7G(5′)ppp(5′)G (54). The integrity of RNA products was confirmed by agarose gel electrophoresis (1.5% agarose) and quantified by GeneQuant II (Pharmacia Biotech). In vitro translation was performed with nuclease-treated rabbit reticulocyte lysate (RRL) (Promega) as specified by the manufacturer, with or without [35S]methionine (>1,000 Ci/mmol) (Amersham). Aliquots of 35S-labeled translation products were analyzed by polyacrylamide gel electrophoresis (PAGE) (12.5% polyacrylamide).
In vitro translation in the presence of 2A protease employed a plasmid containing the 2A coding region downstream of the EMCV IRES (gift of Chaim Kahana, Weizmann Institute of Science). Viral 2A protease was produced in nuclease-treated TNT RRL (Promega) as specified by the manufacturer. The reaction mixture was incubated for 1 h at 37°C, after which aliquots were removed and added to translation assays. A 12-μl reaction mixture containing 8.4 μl of nuclease-treated RRL, 3 μl of 5× buffer (10 mM HEPES [pH 7.5], 1 mM MgCl2, 0.1 mM EDTA, 7 mM β-mercaptoethanol, 100 mM KCl), 0.3 μl of 1 mM amino acid mixture (minus Met), and 0.3 μl of 1 mM amino acid mixture (minus Leu) was preincubated for 30 min at 37°C with 0, 1, or 2 μl of in vitro-translated 2A plus 2, 1, or 0 μl of in vitro-translated β-galactosidase, respectively. Then equimolar amounts of in vitro-transcribed RNA were added to a final concentration of 0.9 μM. The reaction mixtures were further incubated for 1 h at 30°C prior to analysis. CAT activity was measured by a phase extraction assay. The amount of 2A protease was determined by TNT RRL translation in the presence of [35S]methionine. Labeled protein products were subjected to PAGE (12.5% polyacrylamide), radioactivity was measured with a Phosphorimager and quantitated by comparison to the specific activity of [35S]Met, taking into consideration the number of methionine residues in the 2A protease. Reaction mixtures with 0, 1, or 2 μl of in vitro-translated 2A protease contained final concentrations of 0, 17.5, and 35 pM 2A protein, respectively.
Cell cultures.
Cell lines 293, SV80, and HeLa were maintained in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum (Bet-Haemek). Leukocytes, 1 day old, were obtained from the blood bank. Lymphocytes were isolated as described previously (29). Thymocytes were collected from mouse thymus. Human umbilical vein endothelial cells (HUVEC) were obtained from Naomi Lanir (Department of Hematology, Rambam Medical Center, Haifa, Israel) and were cultured as described previously (38). Daudi, K562, Jurkat, SUP-T1, Bjab, and SKW6.4 cells, murine thymocytes, and human lymphocytes were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (Bet-Haemek for Jurkat cells and Sigma for the others) at 37°C under 5% CO2. Erythroid differentiation of K562 was scored by benzidine staining. Benzidine dihydrochloride (2 mg/ml in 0.5% acetic acid) was mixed with 30% hydrogen peroxidase (5 μl/ml), added directly to an equal volume (10 μl) of cell suspension in an Eppendorf tube, and incubated for 5 min at room temperature. Cells were scored as benzidine positive (blue) or benzidine negative (yellow) using a Leitz-Wetzlar microscope at a magnification of ×100.
Transfection into cell cultures.
Plasmid DNAs were prepared using the Qiagen plasmid purification kit. Exponentially growing SV80, HeLa, or 293 cells were transfected with plasmid DNA (15 μg for SV80, 25 μg for HeLa, and 2.3 μg for 293) using the standard method of calcium phosphate-mediated transfection followed by incubation for 48 h at 37°C prior to analysis.
Electroporation.
Electroporation was employed for the hematopoietic cell lines. Cells (2 × 107) in log phase were transfected using 25 μg of plasmid DNA as specified by the manufacturer (BTX), using a capacitor discharge of 250 V, 1,700 μF, and 13 Ω (R1) for K562 cells or 250 V, 1,700 μF, and 72 Ω (R4) for Jurkat cells. SUP-T1, Bjab, and SKW6.4 cells were electroporated as above (but without addition of salmon sperm and HEBS buffer), with a capacitor discharge of 110 V, 2,750 μF and 129 Ω (R5) for SUP-T1 and Bjab cells and 250 V 1,700 μF, and 13 Ω (R1) for SKW6.4 cells. Following electroporation, the cells were immediately transferred to 30 ml of growth medium and incubated for 48 h prior to analysis.
Cell extracts.
At 48 h after treatment, cells were collected, washed with phosphate-buffered saline (Ca and Mg free), and centrifuged. Cell pellets were resuspended in 0.1 ml of 0.1 M Tris-HCl (pH 8.0) and lysed by four freeze-thaw cycles (30 s in liquid N2, 1 to 2 min at 30°C in a water bath). Suspensions were centrifuged at room temperature, and supernatants were assayed for LUC and CAT activities. LUC signals were monitored with a TD-200 luminometer (Turner). CAT activity was determined by a phase extraction assay and counted in the 14C channel of a 1500-TRI-CARB liquid scintillation analyzer (Packard) as described previously (4).
Protein and RNA analysis.
Cells were harvested and washed once in phosphate-buffered saline, and proteins or RNAs were extracted as described previously (3). Protein extracts were subjected to Western blot analysis as described previously (3). Poly(A)+ RNA was purified from 150 μl of total RNA using oligo(dT) magnetic beads (Dynal, Oslo, Norway) and then was subjected to Northern blot analysis as previously described (29).
RESULTS
Expression of AML1 is modulated in vivo by P-UTR and D-UTR.
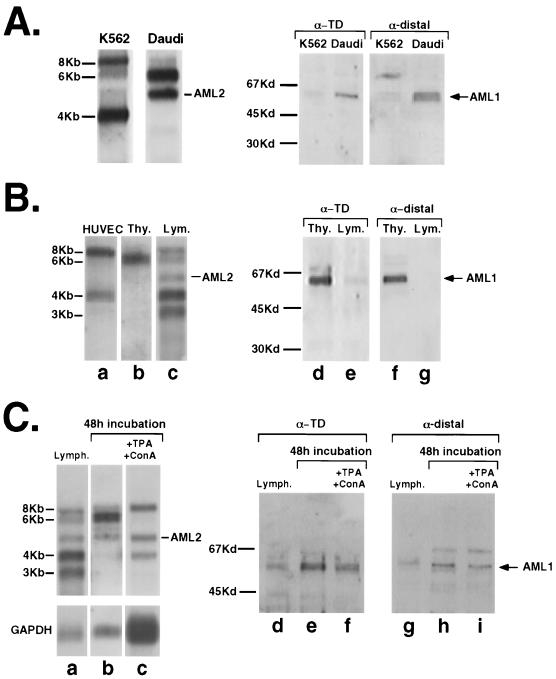
AML1 expresses several size classes of mRNAs (Fig. 2A, left). Transcription of the 6-kb species is regulated by the D promoter, and hence this species carries the shorter (0.45-kb) D-UTR, while the 8- and 4-kb mRNAs are transcribed via the P-promoter and carry the long (1.6-kb) P-UTR (Fig. 1A) (12). In addition to differential promoter usage, the various AML1 mRNAs are generated through alternative splicing and by the use of spaced polyadenylation signals. The difference in transcript length results mainly from a different combination of alternatively spliced 5′UTRs and 3′UTRs (29). The initiator AUG codon in mRNAs bearing the D-UTR resides in exon 1d, while in mRNAs harboring the P-UTR it is in exon 3 (Fig. 1A). Hence, proteins translated from the 6-kb mRNAs containing the D-UTR possess an additional 32 amino acids, encoded by exons 1d and 2, that are not included in the 8- and 4-kb mRNAs generated from the P-promoter (Fig. 1). To distinguish between these two classes of protein products, we used antibodies raised against the 17-amino-acid peptide encoded by exon 1d (designated α-distal) (Fig. 1B). The other type of antibodies (α-TD) are directed against the transactivation domain (TD) of AML1 (3) and were used to detect all the isoforms that contain the TD.
FIG. 2.
Expression pattern of AML1 mRNAs and the corresponding protein products. (A) Profiles of AML1 mRNAs in K562 and Daudi cell lines are shown in the left panel. Northern blots of poly(A)+ mRNA from K562 and Daudi cells were probed with runt domain (SmaI fragment encompassing nucleotides 1745 to 2135 of AML1a cDNA [29]). Of note, the AML2 gene, a member of the runt domain gene family, which is carried on chromosome 1p36, is highly expressed in Daudi cells but not at all in K562 cells (31). Profiles of AML1 proteins in K562 and Daudi cells are shown in the right panel. Proteins extracted from the cell lines were subjected to immunoblotting with the α-TD or α-distal antibodies described in the legend to Fig. 1B. (B) Profiles of AML1 mRNAs and proteins in lymphocytes, thymocytes, and HUVEC. Poly(A)+ mRNA and proteins were extracted and analyzed as in panel A. (C) Expression of AML1 mRNAs and proteins in lymphocytes freshly isolated from blood samples and after a 48-h incubation with or without TPA (1.5 ng/ml) and ConA (3.6 mg/ml). Following hybridization, the blot was stripped and rehybridized with the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe. Films were scanned using a UMAX PowerLOOK II scanner, and band densities were determined by the Image Gauge V3.0 program. With nontreated lymphocytes (lanes d and g) as a reference (i.e., 1.0), lanes e and f gave densities of 1.98 and 1.52, respectively, and lanes h and i gave densities of 1.46 and 1.12, respectively. The probe used in panels B and C was a 2.5-kd SalI fragment encompassing nucleotides 1 to 2500 of AML1a (29). Due to the relative stability of AML1 mRNAs (29), the reduction seen following TPA and ConA treatment in panel C, lanes h and i, is very small. The slower-migrating protein bands in lanes h and i reflect a nonspecific reaction of the α-distal antibodies.
Northern analysis of RNA from the human erythroleukemia cell line K562 and the human B-lymphoblast cell line Daudi is presented in Fig. 2A, left. K562 cells express predominantly the 8- and 4-kb mRNA species, transcribed by the P-promoter, while in Daudi cells the major mRNA was the D-promoter-directed 6-kb mRNA (Fig. 2A, left). Parallel analysis of the protein products showed a very small amount of AML1 proteins in K562 and a relatively large amount of the protein of the expected size (53 kDa) in Daudi cells (Fig. 2A, right). Importantly, the AML1 proteins in Daudi cells reacted with both the α-TD and α-distal antibodies, indicating that they were translated from mRNAs containing the D-UTR. These data are consistent with the possibility that the P-UTR-containing mRNAs (the 8- and 4-kb species) in K562 were poorly translated compared to the D-UTR-containing mRNA (the 6-kb species) in Daudi cells. Differential levels of AML1 D- and P-proteins were also seen in thymocytes and blood lymphocytes (Fig. 2B). Thymocytes expressed exclusively the D-UTR-bearing mRNAs (lane b), which gave rise to AML1 proteins containing the distal peptide (lanes d and f). Lymphocytes and human umbilical vein endothelial cells (HUVEC), on the other hand, expressed mainly P-UTR-containing mRNAs (the 8-, 4-, and 3-kb species) (lanes a and c), which gave rise to a low level of the P-protein (lanes 3 and g). Expression of AML1 was also affected by various exogenous stimuli. Incubation of fresh human lymphocytes for 48 h under culture conditions caused a promoter switch linked to a marked increase in the 6-kb mRNA species (Fig. 2C, lanes a and b) and a consequent increase in the production of the 53-kd AML1 protein (compare lanes d and e and compare lanes g and h). Mitogenic stimulation with tetradecanoyl phorbol acetate and concanavalin A (TPA/ConA) abolished transcription of the 6-kb mRNA (compare lanes b and c) and reduced the production of the 53-kd protein (lanes e and f and lanes h and i). Taken together, the data show that AML1 expression is regulated in vivo, in different cell types and in response to physiological stimuli, at both the transcriptional and posttranscriptional levels. This raised the possibility that the two 5′-UTRs are involved in translation regulation of AML1 expression.
Transcripts bearing the P-UTR are poorly translated in vitro, while mRNAs bearing the D-UTR are readily translatable.
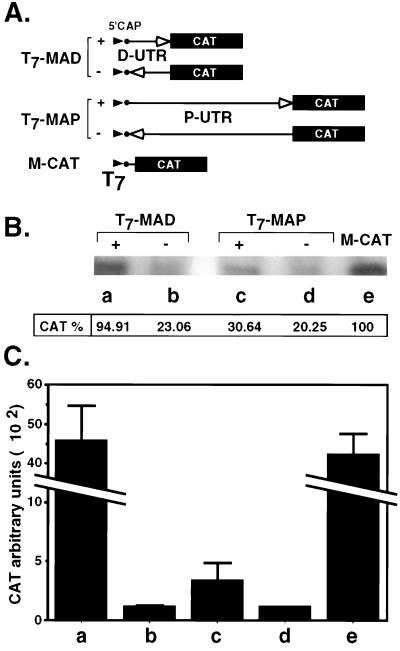
To investigate the involvement of the P- and D-UTRs in translation control of AML1, the translation efficiency of the two UTRs was first assayed in vitro. The P-UTR and D-UTR were cloned into the CAT expression vector between the T7 promoter and CAT to generate monocistronic AML1 P-UTR (T7-MAP) and AML1 D-UTR (T7-MAD), respectively (Fig. 3A). In vitro translation reactions were performed with RRL. Two types of assays were done, in vitro translation in the presence of [35S]methionine (Fig. 3B) and translation followed by a CAT activity assay (Fig. 3C). The results demonstrated that D-UTR mediates translation of the downstream cistron with an efficiency similar to that of the control construct containing the authentic CAT 5′UTR (Fig. 3B and C, lanes a and e); however, its translation-promoting activity was orientation dependent (lane b). The P-UTR, on the other hand, caused marked inhibition of translation of the downstream CAT relative to control (lanes c and e). These results indicate that in vitro, the template activity of P-UTR-CAT is markedly lower than that of the D-UTR-CAT construct and support the thesis that the P-UTR has the potential to negatively regulate the translation of AML1 mRNA.
FIG. 3.
In vitro translation assays of T7-UTR-CAT constructs. (A) Schematic representation of the monocistronic constructs. Plasmids T7-UTR-CAT contain the D-UTR (T7-MAD) in the correct and reverse orientation, the P-UTR (T7-MAP) in both orientations, and the control M-CAT with the authentic CAT 5′UTR. Arrowheads represent the T7 promoter, boxes represent the CAT coding region, and arrows represent 5′UTRs. (B) In vitro translation assays. mRNAs (0.5 μM) were generated as described in Materials and Methods, using the plasmids shown in panel A, and translated in RRL in the presence of [35S]methionine. Labeled proteins were analyzed by PAGE, followed by autoradiography. Dried gels were exposed to a phosphorimaging plate, and radioactivity was recorded with a phosphorimager (FUJIX BAS 2500) using the Image Gauge V3.0 program. (C) Activity assay of in vitro translation products. CAT activity was measured in nonradioactive reactions by the phase extraction assay as described in Materials and Methods. The values correspond to arbitrary units given by the scintillation counter. Bars represent the mean and standard error for three independent experiments.
Picornavirus 2A protease abrogates D-UTR- and not P-UTR-mediated translation.
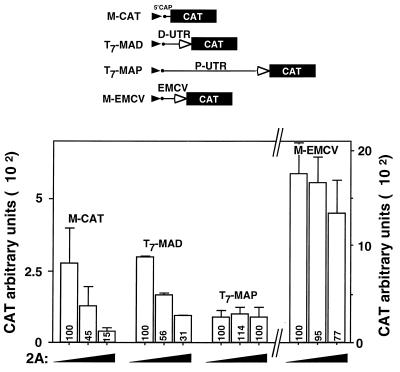
We next questioned whether the P- and D-UTRs are capable of mediating cap-dependent translation. Monocistronic capped mRNAs of T7-MAP and T7-MAD (Fig. 4), as well as of M-CAT and M-EMCV, which contained the highly active IRES of EMCV (7, 19), were made. The four mRNAs were translated in vitro in the presence of increasing amounts of 2A protease (Fig. 4). The picornavirus 2A protease cleaves the initiation factor eIF-4G, causing inhibition of cap-dependent translation of cellular mRNA while promoting the IRES-dependent translation of viral mRNA (59). Translation of the T7-MAD transcript, containing the D-UTR, was inhibited by the 2A protease in a dose-dependent manner. Similar inhibition was observed with control M-CAT mRNA, which contained the authentic CAT 5′UTR. On the other hand, translation of the T7-MAP mRNA, which contained the P-UTR, was not affected by the addition of 2A protease, and translation mediated by the IRES-containing EMCV 5′UTR was only slightly reduced. The latter may reflect the involvement of eIF-4G in EMCV 5′UTR-mediated translation initiation (48). Taken together, the data strongly suggest that the P-UTR directs translation via a cap-independent mechanism and raised the possibility that it contains an IRES.
FIG. 4.
Differential effect of picornavirus 2A protease on translation mediated by different 5′UTRs. A plasmid containing the 2A coding region downstream of the EMCV IRES was translated in vitro in a separate tube. RRL translation assay mixtures were preincubated for 30 min at 37°C with increasing amounts of the in vitro-made 2A protease. At this time point, mRNAs generated as in the experiment in Fig. 3, using the plasmids M-CAT, T7-MAP, T7-MAD, and M-EMCV, were added and reaction mixtures were incubated for an additional 1 h (see Materials and Methods). CAT activity was measured as indicated in the legend to Fig. 3. Values represent arbitrary units given by the scintillation counter. Numbers inside rectangles are relative CAT activities out of 100% attained in assays with no 2A protease. Bars represent the mean and standard error for three independent experiments.
The P-UTR contains IRES characteristic motifs and directs IRES-dependent translation in a bicistronic construct.
All in all, there are no sequence similarities among IRES regions of viral and cellular origins (27). However, they have common structural motifs, a few of which could be detected in AML1 P-UTR. These included an oligopyrimidine tract located at the 3′ part of the 5′UTR and a short sequence, UUUCC, which is complementary to the 3′ end of the 18S rRNA. The latter is considered a functional analog of the known Shine-Delgarno sequence (26, 49). A Y-shaped stem-loop structure upstream of the AUG codon is an additional motif common to many viral and cellular IRESes (1, 4, 26, 27). A typical hairpin structure was found in these IRESes between the Y-shaped structure and the initiator AUG (27). Using a computerized RNA-folding program (28), the overall secondary structure of the P-UTR was deduced. It folds into a highly stable array of stem-loop structures which will most probably inhibit translation via the ribosome-scanning mechanism (results not shown). The program predicted that AML1 P-UTR folds into an array of hairpins and Y-shaped structures which are spaced along the UTR. Of note, a region of ∼100 nucleotides immediately upstream of the initiator AUG, which is highly conserved in the mouse homologue of AML1, the CBFA2/RUNX1 mRNA, can form the characteristic Y-shaped stem-loop structures mentioned above (data not shown).
Bicistronic constructs have been previously used to identify both viral and cellular IRESs (reviewed in reference 18). To investigate the possibility that the P-UTR possesses an IRES activity, it was cloned into the bicistronic vector BAP (bicistronic AML1 P-UTR), in which transcription is mediated by the T7 promoter or the highly active promoter-enhancer region of cytomegalovirus and the first cistron CAT is followed by the P-UTR linked to LUC (Fig. 5A). Transcription generates capped transcripts that direct cap-dependent translation of CAT, whereas LUC is produced only when the intercistronic P-UTR confers an IRES-dependent translation. BAP directed an efficient translation of LUC in the RRL system (Fig. 5B), comparable to other cellular IRESs that confer translation in RRL (22, 42, 43, 67). In comparison, the P-UTR IRES was as active as the EMCV IRES (Fig. 5B) and much more active than the PDGF/c-sis IRES, which is known to perform poorly in RRL (4, 56). To further characterize the IRES activity of P-UTR and to rule out the possibility that the BAP-directed translation of LUC was due to a reinitiation past the CAT cistron, two new bicistronic plasmids were constructed (Fig. 6A): BhAP, in which a hairpin structure (ΔG = −40 kcal/mol) was inserted into BAP upstream of the CAT cistron, and BAP-Rev, in which a fragment bearing the most 3′ 600 bp of P-UTR was inverted. BAP and BhAP were used to program RRL, and the resulting products were analyzed by enzymatic assays of CAT and LUC (Fig. 6B). Constructs were also transfected into K562 cells, a hematopoietic cell line expressing AML1 (29) (Fig. 6C). Comparison of BAP and BhAP showed that CAT production was strongly inhibited by the hairpin structure whereas P-UTR-directed LUC production was hardly affected (Fig. 6B to D). These opposite effects of the hairpin on the activities of the CAT and LUC cistrons in BhAP indicated that the two cistrons were independently translated and excluded the possibility of reinitiation past the CAT region. Consistent with this, modification of the P-UTR in BAP-Rev significantly reduced LUC activity in transfected K562 cells (Fig. 6E).
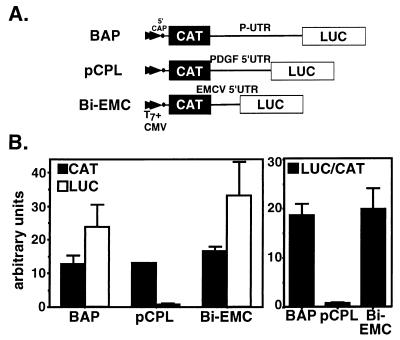
FIG. 5.
Translation of bicistronic CAT and LUC vectors in RRL. (A) Schematic representation of the bicistronic constructs. BAP contained the proximal 5′UTR, pCPL contained the PDGF/c-sis 5′UTR, and BiEMC contained the EMCV 5′UTR. (B) Equimolar capped bicistronic mRNAs were translated in RRL, and CAT and LUC activities were measured as described in Materials and Methods. Data are presented as arbitrary CAT and LUC values. Bars represent the mean and standard error for two independent experiments.
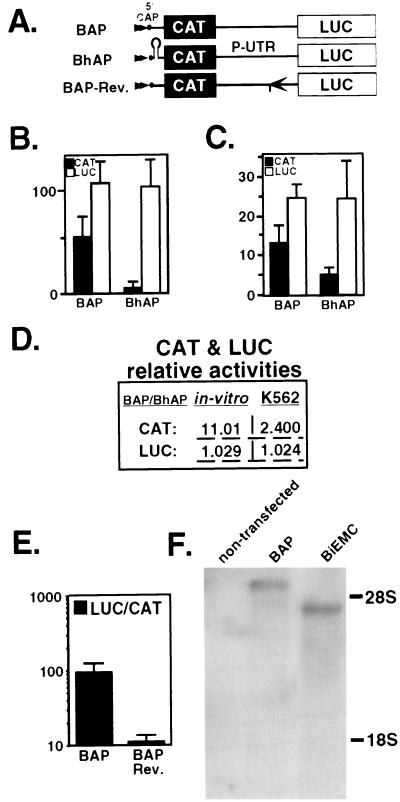
FIG. 6.
Translation of the bicistronic constructs in vitro and in transfected cells is affected by the 5′ hairpin structure. (A) Schematic representation of the bicistronic constructs. Plasmids BAP and BAP-Rev contain the P-UTR and P-UTR/Rev, respectively, in the intercistronic space (in P-UTR/Rev, the 600 bp at the 3′ end is reversed). A hairpin structure with stability of ΔG = −40 kcal mol−1 was inserted upstream of CAT in BhAP. (B) BAP and BhAP were translated in RRL. CAT and LUC activities were measured in nonradioactive reactions as described in Materials and Methods. Data represent the mean and standard error for four independent experiments. (C) K562 cells were transfected with plasmids BAP and BhAP. At 48 h after electroporation, CAT and LUC activities were determined. The bars represent the mean and standard error for three independent experiments done in triplicate. (D) CAT and LUC activities of BAP relative to those of BhAP. Values of CAT or LUC generated by BAP, in vitro (B) or in K562 cells (C), were divided by the values generated by BhAP in the same system. (E) Plasmids BAP and BAP-Rev were transfected into K562 cells, and their activities were analyzed as above. Bars represent the mean LUC/CAT ratios and standard error for three independent experiments done in triplicate. (F) Analysis of mRNAs extracted from transfected K562 cells. Total RNA was purified from cells transfected with BAP and BiEMC (see Fig. 5A). Poly(A)+ mRNAs were subjected to Northern blot analysis using a 32P-labeled cDNA probe corresponding to the 590 bp at the 5′ terminus of the LUC coding sequence. Size standard RNAs are indicated (28S, 4.7 kb; 18S, 1.8 kb). Following hybridization, the blots were exposed for 48 h at −80°C.
The observed LUC activity in K562 cells could have originated from truncated monocistronic templates that were produced through cleavage of the original transcript or by the use of a cryptic promoter within the UTR. To examine the intactness of the RNA, mRNA from transfected K562 cells was extracted and analyzed by Northern blotting with a 590-bp probe from the 5′ region of the LUC coding region. Prominent mRNA bands of the expected sizes (5.1 and 4.0 kb, respectively) were detected in poly(A)+ mRNAs extracted from cells transfected with BAP and BiEMC (Fig. 6F). As a control, poly(A)+ mRNA from nontransfected cells was analyzed and showed no specific hybridization to the LUC probe. Taken together, the results for LUC production by bicistronic BAP, translation inhibition by the hairpin, reduced LUC production by BAP-Rev, and the mRNAs analysis demonstrate that CAT and LUC production occurred through translation of the bicistronic templates and indicate that the P-UTR contains an IRES activity.
The P-UTR IRES displays differential activities in various cell lines.
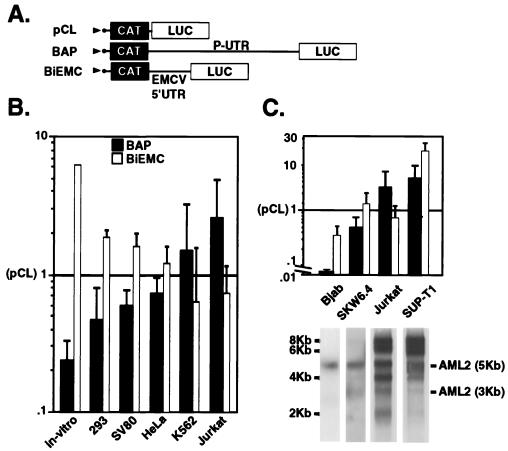
As indicated above, the presence of two 5′UTRs in AML1 mRNAs, one of which has a highly structured IRES, raised the possibility that like the viral IRES (8), the P-UTR is involved in the cell-specific expression documented in Fig. 2. To further address this issue, BAP-directed LUC activity was assayed in several cell lines. The repertoire included two hematopoietic cell lines that express high levels of the AML1 P-UTR-bearing mRNAs, i.e., K562 (early myeloblast/erythroblast) and Jurkat (T cells) and three nonhematopoietic cell lines, 293 (embryonic kidney cells), HeLa (epithelial), and SV80 (fibroblasts), which express very low levels, if any, of AML1. Each cell line was independently transfected with each of the three plasmids pCL, BAP, and BiEMC (Fig. 7A). In each case, the CAT activity served as a direct measure of the overall transfection-transcription efficiency while the LUC/CAT ratio monitored IRES activity. In addition, the performance of the P-UTR IRES in the transfected cells was compared with that of EMCV IRES, which was previously shown to be active in a variety of cell lines (8). The results showed that the P-UTR IRES displayed cell specific activity (Fig. 7B). In the nonhematopoietic cells (293, HeLa, and SV80) and in vitro, the BAP-directed LUC activity was significantly lower (low LUC/CAT ratio) than that of the control pCL plasmid or BiEMC (Fig. 7B). On the other hand, in Jurkat and K562 cells, the BAP IRES not only was more active than that of pCL but also was even more active than that of BiEMC (Fig. 7B). We conclude that the P-UTR IRES functions best in cells that normally express AML1 mRNAs bearing the P-UTR. This conclusion is supported by the observation that the AML1 IRES activity was also low in hematopoietic B cells, such as Bjab and SKW6.4, that do not endogenously express the P-UTR mRNAs, compared to the T-cell lines Jurkat and SUP-T1 (Fig. 7C).
FIG. 7.
IRES activity of AML1 P-UTR in several cell lines. (A) Schematic representation of the bicistronic constructs used in these experiments. pCL contained a short linker between the two reporter genes. BAP contained the P-UTR, and BiEMC contained the EMCV IRES. (B and C) Plasmids were either translated in vitro (in-vitro) or transfected into the various cell lines. CAT and LUC activities were determined. The data shown are the LUC/CAT ratios obtained in each cell line relative to the LUC/CAT ratio obtained in the control pCL-transfected cells. Bars represent the mean and standard deviation for at least three independent experiments done in triplicate. Poly(A)+ mRNAs extracted from the B- and T-cell lines were subjected to Northern blot analysis using a 32P-labeled cDNA probe corresponding to the SmaI fragment of the runt domain.
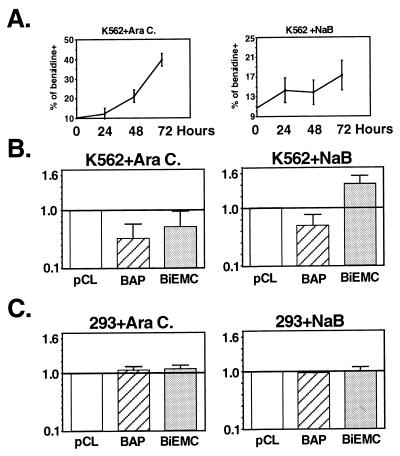
AML1 IRES activity is modulated during differentiation of K562 cells. (i) The activity of P-UTR is enhanced upon megakaryocytic differentiation.
As shown above, K562 cells express relatively high levels of AML1 mRNAs, the majority of which are the P-UTR-bearing species (Fig. 2A). Despite the high mRNA level, the level of AML1 proteins was low (Fig. 2A). This was attributed to the relatively low translatability of the P-UTR-containing mRNAs (Fig. 2 and 3). K562 cells display both megakaryocytic and erythrocytic antigens and can be induced to differentiate into either lineage (52). We questioned whether during these differentiation processes alterations occur in the IRES-dependent translation of AML1 mRNAs. To address this issue, cells were treated with TPA, which induces K562 differentiation toward the megakaryocytic lineage (64); the cells became large and contained multiple separated nuclei that occasionally were lobulated and irregularly shaped. The IRES activity of P-UTR was assayed in TPA-treated and untreated cells. Each of the four plasmids shown in Fig. 8A was transfected into cells, and the LUC/CAT ratio was used as a measure of IRES activity. Results for each individual experiment were normalized to values obtained with pCL. Megakaryocytic differentiation of K562 cells was associated with a 2.5-fold increase in the IRES activity of P-UTR (Fig. 8B). This value is comparable to that reported for the differentiation-linked IRES activity of PDGF/c-sis mRNA (4). The activity of the positive control IRES (BiEMC) was raised fivefold. Significantly, none of the tested plasmids showed increased activity in TPA-treated 293 cells (Fig. 8B).
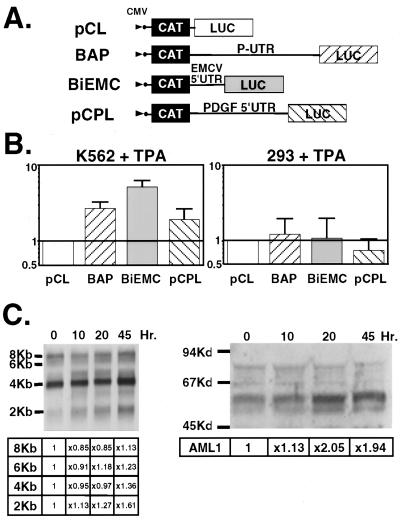
FIG. 8.
IRES activity upon megakaryocytic differentiation of K562 cells. (A) Schematic representation of the bicistronic constructs containing the AML1 P-UTR, EMCV, and PDGF2/c-sis 5′UTRs in the intercistronic space. (B) IRES activity. K562 and 293 cells were transfected with each of the indicated plasmids. After 12 h, TPA (5 nM) was added to half of the cultures and the other half served as a control. Based on the morphological and adherence characteristics of megakaryocytes, we estimated that after 48 h, ∼50% of the cells underwent megakaryocytic differentiation. CAT and LUC activities were determined 48 h after TPA addition. The data represent the LUC/CAT ratio in TPA-treated cells relative to the control cells. Bars indicate the mean and standard error for at least three independent experiments done in triplicate. (C) Induction of endogenous AML1 mRNAs and protein products by TPA. Megakaryocytic differentiation was induced by treatment of K562 cells with 5 nM TPA, and the cells were harvested at 10, 20, and 45 h. RNA was isolated from cells at each time point, and equal amounts of poly(A)+ RNA were analyzed by methylene blue staining (results not shown) and by Northern blotting. The protein concentration was determined by the Bradford assay. Equal amounts of cell lysates were loaded on a polyacrylamide gel, and quantification was confirmed by Ponceau staining (results not shown). Immunoblotting was carried out with antibodies raised against the AML1 transactivation domain (α-TD in Fig. 1B). Films were scanned using a UMAX PowerLOOK II scanner, and band densities were determined by the Image Gauge V3.0 program.
To correlate the above results with the endogenous situation, changes in the profile of AML1 mRNAs and proteins during TPA-induced differentiation were examined. As noted above, the prominent AML1 mRNAs in K562 are the P-UTR-bearing 8- and 4-kb species. The size difference of these mRNAs is mainly due to their different 3′UTRs (29). Upon TPA treatment, an increase in the amount of these mRNA species was noticed after 45 h (Fig. 8C). Western blot analysis of proteins extracted from TPA-treated cell cultures showed that the level of the 53-kDa AML1 protein started to increase earlier, at 10 to 20 h after TPA addition, and continued through the 45-h time point (Fig. 8C). Considering that this AML1 protein product did not react with the α-distal antibodies, this suggests that it was translated from a P-UTR-containing mRNA. Taken together, the results of the transfection assays and the in vivo studies indicate that megakaryocytic differentiation of K562 involves an increase in IRES-dependent translation of AML1 mRNA.
(ii) Erythroid differentiation of K562 abrogates the IRES activity of P-UTR.
We next assessed the changes in IRES activity during differentiation of K562 into erythrocytes (53). In this study, we used sodium n-butyrate (NaB) and arabinosylcytosine (Ara-C), two reagents that stimulate K562 differentiation toward the erythroid lineage. When applied to cells, they both induced hemaglobinization in a time-dependent manner (Fig. 9A). IRES activity in differentiated K562 cells was monitored and normalized to the pCL standard, as mentioned above (Fig. 8). Induction of erythroid differentiation resulted in reduced activity of BAP (Fig. 9B). Ara-C, the more potent differentiation inducer (Fig. 9A), caused 50% reduction in the IRES activity of P-UTR, as well as that of BiEMC, while NaB, the less effective reagent, reduced BAP activity by 25% and increased BiEMC activity (Fig. 9B). No consistent, quantifiable changes in IRES activity were noted in 293 cells treated with either Ara-C or NaB (Fig. 9C). The data show that in contrast to megakaryocytic differentiation of K562, erythroid differentiation was associated with a diminished IRES-dependent translation of AML1 mRNAs. All in all, the above results indicate that translational regulation through an IRES-dependent activity of the P-UTR plays a role in AML1 expression during differentiation of K562 cells and suggest the involvement of AML1 5′UTRs in the regulated expression of the gene during hematopoiesis.
FIG. 9.
IRES activity upon erythroid differentiation of K562 cells. K562 and 293 cells were transfected with three of the plasmids in Fig. 8A. At 12 h, Ara-C (0.36 mM) or NaB (0.4 mM) was added to half of the cultures and the other half served as control. (B and C) At 48 h later, CAT and LUC activities were determined. The data represent the LUC/CAT ratio in the reagent-treated cells relative to the control cells. Bars indicate the mean and standard error for at least three independent experiments done in triplicates. (A) Monitoring of erythroid differentiation. Aliquots of transfected, inducer treated cells were removed every 24 h and scored for benzidine staining.
DISCUSSION
The runt domain transcription factors AML/RUNX are key regulators of gene expression in diverse biological processes such as hematopoiesis and osteogenesis (17, 36, 45, 51, 61). The three runt domain proteins AML1/RUNX1, AML2/RUNX3, and AML3/RUNX2 bind to the same DNA sequence (37); therefore, their pleiotropic functions must result from a regulated, temporally specific and tissue-specific expression of the genes (10, 29, 51, 57). However, surprisingly little is known about the molecular mechanisms regulating their temporally specific and cell type-specific expression. We have previously reported that transcription of AML1 is initiated at two promoter regions generating several mRNA species that differ at their 5′ leader exons (12, 29). In this study, we demonstrate that alternative promoter usage coupled to translation control are key features regulating expression of AML1.
Translational initiation is usually the rate-limiting step in protein synthesis (35). Association of mRNA with the initiation complex is modulated by the phosphorylation state of participating components including several initiation factors, certain ribosomal proteins, and the specific inhibitors 4E-BP1 and 4E-BP2 (9, 20, 41, 58). Translation initiation is also influenced by cis-acting elements such as the 5′ cap structure, the sequences flanking the initiator AUG, the secondary structures within the 5′UTR, and the uAUG codons that are followed by uORFs (35). In AML1, the usage of alternative promoters gives rise to mRNAs with two grossly distinct 5′UTRs. The shorter (452-bp) D-UTR directs efficient translation of the downstream ORF, whereas the longer (1,631-bp) P-UTR is much less active and supports tightly regulated translation. The low translational efficiency of P-UTR was attributed to its length and the cis-acting elements along it. The latter consist of several long GC-rich regions, which form highly stable secondary structures, and 15 cryptic uAUG codons, some of which are followed by short uORFs. These two elements were previously implicated in down regulation of translation (reference 35 and references therein).
The P-UTR mediates a cap-independent translation. It is not affected by the 2A protease and directs internal initiation and LUC translation from a bicistronic mRNA in the presence of an upstream hairpin structure. Activity of the P-UTR is also orientation dependent; translation of the internal LUC cistron is abolished by inversion of the proximal half of this 5′UTR. Based on the above data, we concluded that the P-UTR-bearing AML1 mRNAs belong to the growing group of cellular mRNAs with established 5′UTR-containing IRES. The D-UTR, on the other hand, directs cap-dependent initiation. It is highly active both in vivo and in the RRL, and its activity is inhibited by the 2A protease. When placed within the intercistronic region of the bicistronic construct, the D-UTR allowed some translation of the downstream LUC cistron; however, insertion of a hairpin structure in front of the first cistron diminished the translation of both CAT and LUC (data not shown). These data indicate that the D-UTR lacks IRES activity and emphasize the importance of using the hairpin-containing plasmids as controls.
Comparison of IRES regions of picornaviruses and of several cellular mRNAs (Bip, FGF-2, and PDGF-2) revealed the existence of conserved Y-shaped elements (27, 56). Similar Y-shaped structures were identified in the P-UTR of AML1 and its mouse homologue in this study. Analysis of picornavirus IRES activity with respect to sequence and structure conservation, translation efficiency in RRL, and effect on activity by the 2A/L proteases classified these IRESes into three different groups (8). The AML1 initiator AUG codon is located immediately at the 3′ end of the postulated Y-shaped structure. Furthermore, the P-UTR IRES was relatively active in RRL and was hardly affected by the 2A protease. It meets the criteria of type II IRES found in EMCV and foot-and-mouth disease virus (8). The IRESes of picornavirus and hepatitis C virus were also compared for their ability to support internal initiation in six different cell lines. IRES efficiency varied considerably from one cell type to another, presumably reflecting the presence or absence of IRES-specific factors (8). In this study we observed that in certain hematopoietic cell lines (K562 and Jurkat) the AML1 IRES was more active than that of EMCV while in nonhematopoietic cells (293, SV80, and HeLa) and in B cells (Bjab and SKW6.4) it was less active than the EMCV IRES. The lymphoblastoid T-cell line SUP-T1 supported a high IRES activity of both EMCV and P-UTR and may therefore contain IRES-specific factors needed for both of them. Assuming that IRES-specific proteins exist, one can speculate that their activity will vary in different cell types and in response to specific stimuli during the cell cycle and differentiation. As was previously shown for PDGF/c-sis (4), the AML1 IRES displays differentiation-dependent activity which conforms with the in vivo expression pattern of the gene. The molecular mechanisms underlying this differentiation-dependent IRES activity are not known. They may involve de novo synthesis of megakaryocytic IRES-promoting proteins or posttranslational modifications of preexisting IRES regulatory proteins, which upon differentiation enhance the activity of the P-UTR IRES.
For picornaviruses, the cap-independent, IRES-dependent translation grants an obvious advantage. The virus-encoded 2A protease destroys the cap-ribosome bridging factor eIF-4G, shuts off the cap-dependent translation of cell mRNA, and directs the cell machinery to translate the uncapped IRES-containing viral mRNA (59). The role of cap-independent translation in cell metabolism is less clear. Nevertheless, picornavirus IRES elements function in noninfected cells in the absence of any virus-encoded proteins (46), and it was reported (13, 34) that cellular proteins were identified to specifically interact with IRES elements. These observations support the notion that IRES-mediated translation is part of the cell control mechanism as well. IRES-bearing mRNAs could be translated under conditions when the common cap-dependent translation is diminished, for example when the eIF-4F holoenzyme complex is inactivated by heat shock (25), growth arrest (50), serum starvation and γ-irradiation (14), or conditions of hypoxia (66). In addition, cap-dependent translation is extremely inefficient during mitosis, due to the presence of underphosphorylated and therefore nonfunctional eIF-4E (6). Thus, IRES-dependent translation provides an important regulatory element in the cascade of cellular gene expression. The highly structured 5′UTR may also play a role in mRNA localization and stability and serve to discriminate among various mRNA isoforms (see the discussion of AML1 isoforms below). In such cases, translation via the IRES can circumvent the translation inhibition imposed by the structured 5′UTRs while preserving their unique functional properties. A long structured 5′UTR with cryptic uAUGs is a common feature in the mRNAs of many growth-regulatory genes (those encoding transcription factors, growth factors, cytokines, and receptors) (23). It is believed that the presence of IRESs in these UTRs provides a means for fine-tuning of their translation in time and space and in response to relevant stimuli (4, 68). The findings reported here strongly suggest that the major function of the P-UTR IRES-dependent translation is to tightly regulate the translation of AML1 mRNAs. We show that expression of AML1 is controlled at both the transcriptional and translational levels by alternative promoters and by cap-dependent or -independent translation mechanisms. This provides the necessary flexibility for production of the relevant type of AML1 protein isoforms in the appropriate amounts, at the proper time, and in the correct cell type. As documented here, the D-promoter, which mediates production of mRNAs bearing the D-UTR, is active during defined time windows in specific cell types, such as thymocytes and Daudi cells, that produce large amounts of AML1 proteins. The P-promoter, on the other hand, is more active in certain hematopoietic cell lines (early myeloid) and in HUVEC. Moreover, the P-UTR IRES-dependent translation is increased during megakaryocytic differentiation of K562 cells during a time window when only the P-promoter is active, further highlighting the coupling between transcription regulation and translation control. The observed modulation of AML1 expression during differentiation of K562 may be related to the more recent finding that haploinsufficiency of AML1 impairs megakaryopoiesis (60).
IRES-dependent translation may provoke the use of alternative initiation sites and lead to translation from the same mRNA molecule of different proteins with distinct functions (2, 42, 67, 68). A related issue emerged from our previous observation that AML1 D-UTR- and P-UTR-containing mRNAs encode proteins that differ at their N termini (12, 29), further highlighting the significance of the P-UTR IRES. In addition, the large repertoire of AML1 transcripts include mRNA species encoding proteins that differ at their C termini as well. It is possible that the various N and C termini interact with other proteins of the transcription machinery to regulate the expression of target genes that function at a certain differentiation stage or at a defined time point. IRES-dependent translation can provide an effective solution to the demand for such a fine modulation. We recently demonstrated that the isoform AML1-b with the C terminus VWRPY interacts with the corepressors TLE and Gro (30), implying a putative function for AML1 in transcriptional repression. Notably, the almost identical isoform AML1-a, lacking VWRPY, failed to interact with TLE and Gro or to repress AML1-mediated transactivation (30). We also found that one of the short isoforms, AML1-d, but not the full-length isoform, AML1-b, is capable of suppressing the in vivo growth and differentiation of tumors derived from embryonic stem cells (3).
In summary, in this work we show that expression of AML1 is controlled in vivo at both the transcriptional and translational levels through the usage of alternative promoters coupled to translation control. Translation is mediated by both cap- and IRES-dependent mechanisms. The AML1 IRES displays differentiation-dependent activity which conforms with the expression pattern of the gene. We argue that this cascade provide the necessary flexibility for production of the relevant AML1 protein isoforms in the appropriate amounts, at the proper time, and in the correct cell type.
ACKNOWLEDGMENTS
We thank Chaim Kahana for plasmids, advice, and discussions.
This work was supported by grants from the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ) and Israel's Ministry of Science (MOS); the Commission of the European Community's Biomedicine and Health research program (BIOMED II grant PL963039); Bernard Sabrier, Geneva, Switzerland; and the Shapell Family Biomedical Research Foundation at the Weizmann Institute.
REFERENCES
- 1.Akiri G, Nahari D, Finkelstein Y, Le S Y, Elroy-Stein O, Levi B-Z. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- 2.Arnaud E, Touriol C, Boutonnet C, Gensac M C, Vagner S, Prats H, Prats A C. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG codon and behaves as a survival factor. Mol Cell Biol. 1999;19:505–514. doi: 10.1128/mcb.19.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben Aziz-Aloya R, Levanon D, Karn H, Kidron D, Goldenberg D, Lotem J, Polak-Chaklon S, Groner Y. Expression of AML1-d, a short human AML1 isoform, in embryonic stem cells suppresses in vivo tumor growth and differentiation. Cell Death Differ. 1998;5:765–773. doi: 10.1038/sj.cdd.4400415. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein J, Sella O, Le S-Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein J, Shefler I, Elroy-Stein O. The translational repression mediated by the platelet-derived growth factor 2/c-sis mRNA is relieved during megakaryocytic differentiation. J Biol Chem. 1995;270:10559–10565. doi: 10.1074/jbc.270.18.10559. [DOI] [PubMed] [Google Scholar]
- 6.Bonneau A M, Sonenberg N. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J Biol Chem. 1987;262:11134–11139. [PubMed] [Google Scholar]
- 7.Borman A M, Bailly J L, Girard M, Kean K M. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borman A M, Mercier P L, Girard M, Kean K M. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown E J, Schreiber S L. A signaling pathway to translational control. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 10.Corsetti M T, Calabi F. Lineage- and stage-specific expression of Runt-box polypeptides in primitive and definitive hematopoiesis. Blood. 1997;89:2359–2368. [PubMed] [Google Scholar]
- 11.Gan W, Rhoads R E. Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- 12.Ghozi M C, Bernstein Y, Negreanu V, Levanon D, Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci USA. 1996;93:1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellen C U, Witherell G W, Schmid M, Shin S H, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk R G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 15.Huez I, Creancier L, Audigier S, Gensac M C, Prats A C, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito Y. Structural alterations in the transcription factor PEBP2/CBF linked to four different types of leukemia. J Cancer Res Clin Oncol. 1996;122:266–274. doi: 10.1007/BF01261402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito Y, Bae S-C. The Runt domain transcription factor, PEBP2/CBF, and its involvement in human leukemia. In: Yaniv M, Ghysdael J, editors. Oncogenes as transcriptional regulators. Vol. 2. Basel, Switzerland: Birkhauser Verlag; 1997. pp. 107–132. [Google Scholar]
- 18.Jackson R J. A comparitive view of initiation site selection mechanisms. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translation control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 71–112. [Google Scholar]
- 19.Jang S K, Krausslich H G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jefferies H B J, Thomas G. Ribosomal protein S6 phosphorylation and signal transduction. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translation control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 389–410. [Google Scholar]
- 21.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. The runt domain identifies a new family of heteromeric DNA-binding transcriptional regulatory proteins. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 22.Kim J G, Armstrong R C, Berndt J A, Kim N W, Hudson L D. A secreted DNA-binding protein that is translated through an internal ribosome entry site (IRES) and distributed in a discrete pattern in the central nervous system. Mol Cell Neurosci. 1998;12:119–140. doi: 10.1006/mcne.1998.0701. [DOI] [PubMed] [Google Scholar]
- 23.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamphear B J, Panniers R. Heat shock impairs the interaction of cap-binding protein complex with 5′ mRNA cap. J Biol Chem. 1991;266:2789–2794. [PubMed] [Google Scholar]
- 26.Le S-Y, Chen J-H, Sonenberg N, Maizel J V., Jr Conserved tertiary structure elements in the 5′ untranslated region of human enteroviruses and rhinoviruses. Virology. 1992;191:858–866. doi: 10.1016/0042-6822(92)90261-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le S-Y, Maizel J V., Jr A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le S-Y, Maizel J V., Jr A method for assessing the statistical significance of RNA folding. J Theor Biol. 1989;138:495–510. doi: 10.1016/s0022-5193(89)80047-5. [DOI] [PubMed] [Google Scholar]
- 29.Levanon D, Bernstein Y, Negreanu V, Ghozi M C, Bar-Am I, Aloya R, Goldenberg D, Lotem J, Groner Y. A large variety of alternatively spliced and differentially expressed mRNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol. 1996;15:175–185. doi: 10.1089/dna.1996.15.175. [DOI] [PubMed] [Google Scholar]
- 30.Levanon D, Goldstein R E, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA. 1998;2995:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 32.Look A T. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 33.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 34.Meerovitch K, Svitkin U V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 31–70. [Google Scholar]
- 36.Meyers S, Hiebert S W. Indirect and direct disruption of transcriptional regulation in cancer: E2F and AML-1. Crit Rev Eukaryotic Gene Expression. 1995;5:365–383. doi: 10.1615/critreveukargeneexpr.v5.i3-4.70. [DOI] [PubMed] [Google Scholar]
- 37.Meyers S, Lenny N, Sun W-H, Hiebert S W. AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- 38.Miller A, Lanir N, Shapiro S, Revel M, Honigman S, Kinarty A, Lahat N. Immunoregulatory effects of interferon-β and interacting cytokines on human vascular endothelial cells. Implications for sclerosis and other autoimmune diseases. J Neuroimmunol. 1996;64:151–161. doi: 10.1016/0165-5728(95)00164-6. [DOI] [PubMed] [Google Scholar]
- 39.Miller D L, Dibbens J A, Damert A, Risau W, Vadas M A, Goodall G J. The vascular endothelial growth factor mRNA contains an internal ribosome entry site. FEBS Lett. 1998;434:417–420. doi: 10.1016/s0014-5793(98)01025-4. [DOI] [PubMed] [Google Scholar]
- 40.Miyoshi H, Ohira M, Shimizu K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morley S J, Pain V M. Translational regulation during activation of porcine peripheral blood lymphocytes: association and phosphorylation of the alpha and gamma subunits of the initiation factor complex eIF-4F. Biochem J. 1995;312:627–635. doi: 10.1042/bj3120627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 43.Negulescu D, Leong L E-C, Chandy K G, Semler B L, Gutman G A. Translation initiation of a cardiac voltage-gated potassium channel by internal ribosome entry. J Biol Chem. 1998;273:20109–20113. doi: 10.1074/jbc.273.32.20109. [DOI] [PubMed] [Google Scholar]
- 44.North T, Gu T-L, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck N A. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 45.Okuda T, Van Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocation in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 46.Pelletier J, Sonemberg N. Internal binding of eukaryotic ribosomes on poliovirus RNA: translation in HeLa cell extracts. J Virol. 1989;63:441–444. doi: 10.1128/jvi.63.1.441-444.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 48.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pilipenko E V, Gmyl A P, Maslova S V, Svitkin Y V, Sinyakov A N, Agol V I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- 50.Rhoads R E. Regulation of eukaryotic protein synthesis by initiation factors. J Biol Chem. 1993;268:3017–3020. [PubMed] [Google Scholar]
- 51.Rodan G A, Harada S I. The missing bone. Cell. 1997;89:677–680. doi: 10.1016/s0092-8674(00)80249-4. [DOI] [PubMed] [Google Scholar]
- 52.Rowley P T, Farley B A, LaBella S, Giuliano R, Leary J F. Single K562 human leukemia cells express and are inducible for both erythroid and megakaryocytic antigens. Int J Cell Cloning. 1992;10:232–240. doi: 10.1002/stem.5530100406. [DOI] [PubMed] [Google Scholar]
- 53.Rowley P T, Ohlsson-Wilhelm B M, Farley B A, LaBella S. Inducers of erythroid differentiation in K562 human leukemia cells. Exp Hematol. 1981;9:32–37. [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 55.Satake M, Nomura S, Yamaguchi-Iwai Y, Takahama Y, Hashimoto Y, Niki M, Kitamura Y, Ito Y. Expression of the runt domain-encoding PEBP2α genes in T cells during thymic development. Mol Cell Biol. 1995;15:1662–1670. doi: 10.1128/mcb.15.3.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sella O, Gerlitz G, Le S-Y, Elroy-Stein O. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol Cell Biol. 1999;19:5429–5440. doi: 10.1128/mcb.19.8.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simeone A, Daga A, Calabi F. Expression of runt in the mouse embryo. Dev Dyn. 1995;203:61–70. doi: 10.1002/aja.1002030107. [DOI] [PubMed] [Google Scholar]
- 58.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translation control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. pp. 245–270. [Google Scholar]
- 59.Sonenberg N. Poliovirus translation. Curr Top Microbiol Immunol. 1990;161:23–47. doi: 10.1007/978-3-642-75602-3_2. [DOI] [PubMed] [Google Scholar]
- 60.Song W-J, Sullivan M G, Legare R D, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende I C, Haworth C, Hock R, Loh M, Felix C, Roy D-C, Busque L, Kurnit D, Willamn C, Gerwitz A M, Speck N A, Bushweller J H, Li F P, Gardiner K, Poncz M, Maris J M, Gilliland D G. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develope acute meylogenous leukemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 61.Speck N A, Stacy T. A new transcription factor family associated with human leukemias. Crit Rev Eukaryotic Gene Expression. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 62.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implication for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stoneley M, Paulin F E M, Le Quesne J P C, Chappell S A, Willis A E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 64.Tabillio A, Pelicci P G, Vinci G, Mannoni P, Civin C I, Vainchenker U, Lipinski T M, Rochant H, Breton-Gorius J. Myeloid and megakaryocytic properties of K562 cell lines. Cancer Res. 1983;43:4569–4574. [PubMed] [Google Scholar]
- 65.Teerink H, Voorma H O, Thomas A A. The human insulin-like growth factor II leader 1 contains an internal ribosomal entry site. Biochim Biophys Acta. 1995;1264:403–408. doi: 10.1016/0167-4781(95)00185-9. [DOI] [PubMed] [Google Scholar]
- 66.Tinton S A, Buc-Calderon P M. Hypoxia increases the association of 4E-binding protein1 with the initiation factor 4E in isolated rat hepatocytes. FEBS Lett. 1999;446:55–59. doi: 10.1016/s0014-5793(99)00185-4. [DOI] [PubMed] [Google Scholar]
- 67.Vagner S, Gensac M C, Maret A, Bayard F, Amalric F, Prats H, Prats A C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vagner S, Touriol C, Galy B, Audigier S, Gensac M C, Amalric F, Bayard F, Prats H, Prats A C. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Disruption of the Cbfα2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye X, Fong P, Iizuka N, Choate D, Cavener D R. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol Cell Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]