Abstract
Background and Objectives: Calcium (Ca2+) signaling is critical for the normal functioning of various cellular activities. However, abnormal changes in cellular Ca2+ can contribute to pathological conditions, including various types of cancer. The maintenance of intracellular Ca2+ levels is achieved through tightly regulated processes that help maintain Ca2+ homeostasis. Several types of regulatory proteins are involved in controlling intracellular Ca2+ levels, including the sarco/endoplasmic reticulum (SR/ER) Ca2+ ATPase pump (SERCA), which maintains Ca2+ levels released from the SR/ER. In total, three ATPase SR/ER Ca2+-transporting (ATP2A) 1-3 genes exist, which encode for several isoforms whose expression profiles are tissue-specific. Recently, it has become clear that abnormal SERCA expression and activity are associated with various types of cancer, including breast cancer. Breast carcinomas represent 40% of all cancer types that affect women, with a wide variety of pathological and clinical conditions. Materials and methods: Using cBioPortal breast cancer patient data, Kaplan–Meier plots demonstrated that high ATP2A1 and ATP2A3 expression was associated with reduced patient survival. Results: The present study found significantly different SERCA specific-type expressions in a series of breast cancer cell lines. Moreover, bioinformatics analysis indicated that ATP2A1 and ATP2A3 expression was highly altered in patients with breast cancer. Conclusion: Overall, the present data suggest that SERCA gene-specific expressioncan possibly be considered as a crucial target for the control of breast cancer development and progression.
Keywords: SERCA pump, SERCA genes, breast cancer, SERCA alterations
1. Introduction
Intracellular calcium ion (Ca2+) signaling [1] pathways are involved in key cellular processes, including excitation–contraction coupling, stimulus–secretion coupling, gene expression, control of the cell cycle, cell motility, autophagy, and apoptosis [2,3,4]. The cytosolic Ca2+ concentration is tightly maintained (~10−7 mol/L), with small amounts being released from several organelles (~10−5 mol/L) or through influx from the extracellular reservoir (~10−3 mol/L), that can generate marked signals to activate downstream signaling cascades [5]. However, prolonged intracellular Ca2+ elevation can be harmful and induces cell death [6]. Alteration/deregulation of Ca2+ signaling is a feature of numerous types of diseases, including cancer, heart failure, and neurodegenerative diseases [1]. The sarco/endoplasmic reticulum (SR/ER) Ca2+ ATPase pump (SERCA) belongs to the P-type pump family, which includes a large number of evolutionarily related Ca2+ pumps. There are three SERCA ATPase SR/ER Ca2+-transporting (ATP2A)1-3 eukaryotic genes that exist, which encode for 14 isoforms with tissue-specific expression profiles [7,8,9,10]. All of the isoforms have 75% homology and are cell-type-dependent [11]. SERCA1 has two transcripts, 1a and 1b, which are present in adult and fetal skeletal muscle, respectively [12]. SERCA 2 consists of two transcripts, 2a, which is mostly found in cardiac and adult skeletal muscle, and 2b, which is abundant in both adult and fetal cardiac and skeletal muscles as well as in non-muscle cells. SERCA3 can be transcribed to three isoforms, SERCA3a–3c, all expressed in non-muscle cells [13]. All SERCA isoforms have been reported to be key players in cancer progression, proliferation, and survival [13,14]. Several studies have reported alterations in SERCA gene-specific expression in various types of cancer, suggesting that levels of SERCA may be potential therapeutic targets and biomarkers in cancer [1].
Cancer development and progression towards metastasis are known to be associated with alterations in post-translational modifications, gene copy numbers, epigenetics, and metabolic changes [15,16,17,18]. These changes are frequently observed as a result of alterations of the Ca2+ flux across the plasma membrane or across intracellular organelles [1].
Alterations in the structure of the ATP2A2 gene have been reported in colon and lung cancer. Specifically, 13 different novel alterations of the ATP2A2 gene have been reported in 27 of 416 alleles in patients, including intronic deletions, single-nucleotide alterations, missense mutations, and intronic insertions. Conversely, reduced ATP2A2 expression levels due to alterations in gene-promoter activity have been found in patient tissue biopsies and may be an early event in tumorigenesis [7]. Additionally, ATP2A2 expression levels are significantly correlated with tumor grade and survival rates, where high ATP2A2 expression has been detected in patients with glioblastoma [9]. In a separate study, ATP2A3 gene downregulation has been reported in gastric and colon cancer types. In contrast, ATP2A3 overexpression reduces cell viability by promoting apoptosis in breast cancer cell lines [19].
A known SERCA inhibitor originally extracted from the plant Thapsia garganica is thapsigargin, which is known to induce cell death in human hepatoma cells [19,20,21,22]. Thapsigargin has also been reported to reduce cell proliferation and induce apoptosis through a caspase-dependent pathway [23]. Moreover, a modified version of thapsigargin, mipsagarin, is undergoing clinical trials and is considered to be a strong candidate for the treatment of hepatocellular carcinoma [24]. Another study in breast cancer indicated the involvement of SERCA3 in early stage lobular dysplasia, with low expression levels at more advanced stages of lobular tumorigenesis [25]. In invasive breast carcinoma, SERCA3 expression levels are significantly decreased compared with that in normal patients, indicating that SERCA3 expression is inversely associated with tumor differentiation and the degree of aggressiveness/malignancy of ductal carcinoma [18,24,25,26,27,28].
There is a continued need to identify novel targeted therapies for the treatment of breast cancer, in combination with low toxicity chemotherapy, especially for chemoresistant triple-negative breast cancer (TNBC). This paper investigates the role and significance of different calcium-dependent SERCA ATPase proteins in breast cancer development.
2. Materials and Methods
2.1. Cell Culture and Reagents
MCF10A (Michigan Cancer Foundation-10 human breast epithelial cells), MCF7 (Michigan Cancer Foundation-7), MDA231 (M.D. Anderson Metastasis Breast Cancer, Houston, TX, USA”), T47D (mammary gland; derived from metastatic site: pleural effusion) cells were provided by Barbara Ann Karmanos Cancer Institute (Detroit, MI, USA) and cultured in Dulbecco’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco, Invitrogen, Waltham, MA, USA). Cells were grown in a humidified atmosphere at 37 °C containing 5% CO2 and were routinely tested for mycoplasma contamination.
2.2. Western Blotting
Total protein was isolated from cultured MCF10A, MCF7, MDA-MB-231, and T47D cells with lysis buffer RIPA plus protease inhibitor cocktail. The homogenates were centrifuged at 17.000× g for 15 min at 4 °C. The supernatants were collected, and protein concentration was measured with Bradford protein assay (Bio-Rad protein assay, Hercules, CA, USA). A total of 30 μg of protein samples was loaded each time into SDS-PAGE gels and transferred to nitrocellulose membranes (Whatman, Maidstone, UK) using the semi-dry transfer system (Bio-Rad). The membranes were blocked with 5% dry milk dissolved in Tris-buffered saline (1×) containing 0.1% Tween-20 for 1 h at room temperature (RT). The membranes were incubated with primary antibodies at 4 °C overnight, followed by secondary antibodies for 1.30 h at RT. The primary antibodies in the Western blot were mouse anti-Serca 1 (Abcam, Cambridge, UK, ab-2819), mouse anti-Serca 2 (Abcam, ab-2861), rabbit anti-Serca 3 (alomone labs, ACP-014), and mouse anti-beta actin (Sigma, Kawasaki, Kanagawa, A5441). The secondary antibodies were rabbit anti-mouse IgG (Sigma, A9044) (1:20.000 dilution) and goat anti-rabbit IgG (Sigma, A6154) (1:10.000 dilution). The protein bands were detected using the Western Luminescent Detection Kit. Western blot images were analyzed using ChemiDoc XRS system (BioRad) with actin used as the protein control, as previously described.
2.3. Cancer OMICS Data Analysis
UALCAN is a web-based resource platform using pancancer gene expression analysis from TCGA OMICS data. Tumor Subgroup Gene Expression and Survival Analyses were downloaded from UALCAN [29]. The expression of ATP2A1, ATP2A2, and ATP2A3 was analyzed in order to explore their expressions in breast cancer.
2.4. Data Analysis from Patients with Breast Cancer from cBioPortal
For TCGA pancancer atlas, expression data n = 1084 samples (publicly available cases) of breast cancer downloaded from the cBioPortal website https://www.cbioportal.org, accessed on 7 May 2021 in the form of z-score-transformed data were used.
2.5. Statistical Analysis
Statistical analysis was performed using ANOVA followed by a Dunnett’s post hoc test. p < 0.05 considered statistically significant.
3. Results
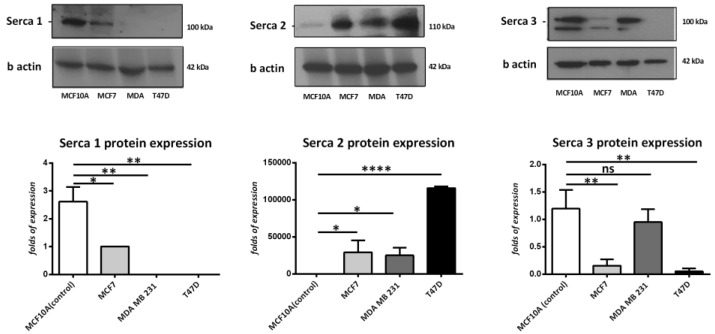
To determine the expression levels of SERCA-A1, A2, and A3 in breast cancer cells, the present study examined the protein expression profiles in MCF-7, MDA 231, and T47D cells and compared them to a non-transformed mammary breast cell line, MCF-10A. As Figure 1 demonstrates, SERCA-A1 expression was significantly higher in the MCF10A cells vs. the MDA 231 and T47D cell lines. SERCA-A2 was also significantly higher in the MCF-7, T47D, and MDA231 cells vs. the MCF10A cells. In contrast, SERCA-A3 expression levels were reduced in the MCF-7 and T47D cells. These findings suggested a differential expression profile of SERCA-A1-3 in various subtypes of breast cancer cell lines.
Figure 1.
Western blot analysis of SERCA isoforms in breast cancer cell lines (MDA-231, MCF-7, MCF10A, and T47D). The analysis was performed in three independent experiments (n = 3), except in the case of Serca1, where n = 2. * p < 0.05, ** p < 0.01, **** p < 0.0001.
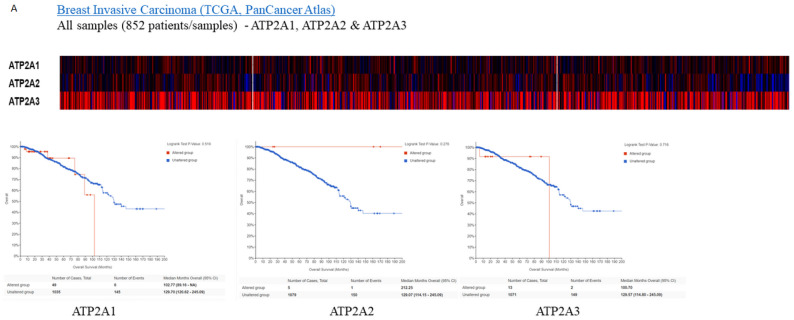
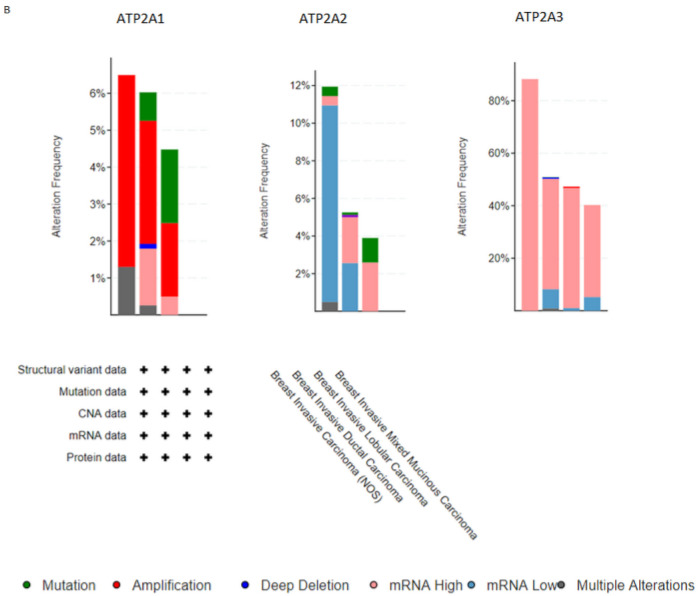
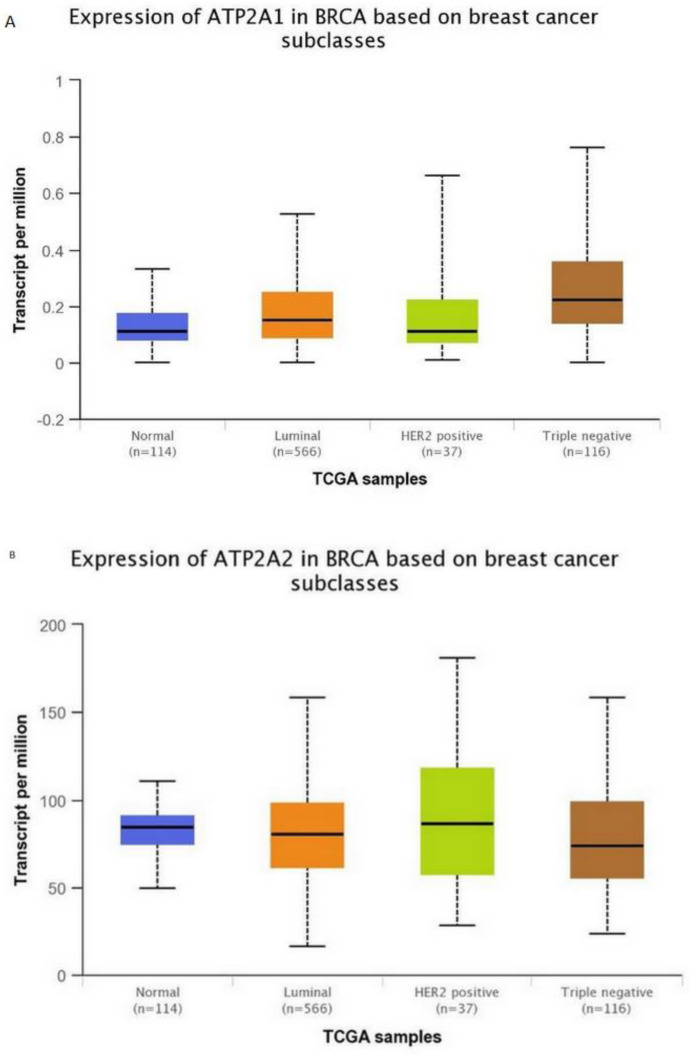
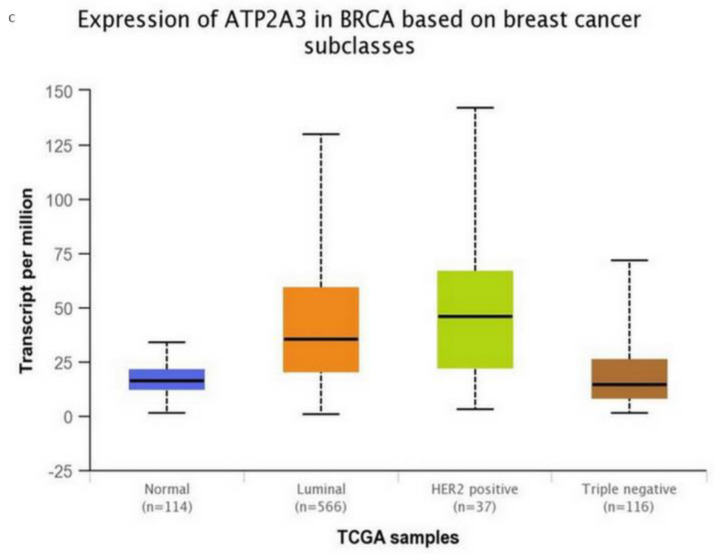
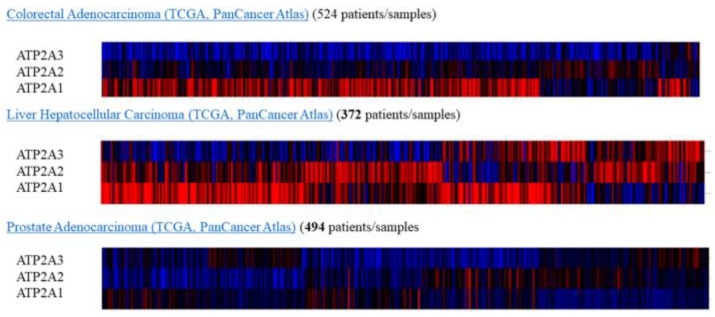
Subsequently, for the present study, we performed bioinformatics analysis using the cBioPortal database of clinical samples. As indicated in Figure 2A,B SERCA-A1-3 expression levels were elevated and altered in patients with breast cancer, where further analysis (Figure 3A–C) showed that SERCA-A1, A2, and A3 had a differential expression profile in breast cancer subclasses, including luminal, HER2-positive, and TNBC. As such, ATP2A1 was found to be expressed at slightly higher levels in luminal, HER2-positive, and TNBC types vs. normal samples. ATP2A2 expression levels were not altered in any of the subclasses of breast cancer vs. normal samples. Interestingly, ATP2A3 expression levels exhibited a trend of increased levels in the luminal and HER2-positive samples vs. the normal controls and marginally increased in TNBC vs. normal samples (Figure 3A–C). Moreover, SERCA-A1, A2, and A3 differential expressions were observed in individual patients with colorectal adenocarcinoma, liver hepatocellular carcinoma, and prostate adenocarcinoma (Figure 4).
Figure 2.
Expressions heatmap of SERCA isoforms in breast cancer patients. Kaplan–Maier plots. (A) ATP2A1 KM survival plots with expressions and alterations. p = 0.149. (B) ATP2A2 KM survival plots with expressions and alterations. p = 0.455.
Figure 3.
SERCA isoforms expressions. (A) ATP2A1 expression in breast cancer subclasses. (B) ATP2A2 expression in breast cancer subclasses. (C) ATP2A3 expression in breast cancer subclasses. Normal n = 114, luminal n = 566, HER-positive n = 37, and triple-negative n = 116. Data collected from: http://ualcan.path.uab.edu/.
Figure 4.
Expressions heatmap of SERCA isoforms in colorectal adenocarcinoma, liver hepatocellular carcinoma, and prostate adenocarcinoma. All data (TCGA PanCancer Atlas) collected from https://www.cbioportal.org/.
Finally, a strong association between the specific expression profiles of the various SERCA genes and the survival rates of patients with different breast cancer subclasses was observed, as shown by Kaplan–Meier plots (Figure 2A). These data indicated that the reduced survival rates were associated with differential expression of the SERCA pump subunits. The high expression level of ATP2A3 was associated with a significant reduction in the monthly survival rates. Similarly, the ATP1A1 lever was elevated and affected breast cancer patients’ survival, whereas ATP2A2 seemed to be neutral in respect to survival.
Potentially, the present results could be considered as indicative of a crucial role of SERCA in breast cancer and thus a possible target for a new generation of treatments.
4. Discussion
The present study investigated the association between altered SERCA-specific gene expression levels in various breast cancer subtypes.
Analyzing TCGA and RNA sequencing data of patients with breast cancer showed that SERCA-A1 and SERCA-A3 levels were increased in tumor samples vs. normal samples. Additionally, this SERCA-specific gene expression was correlated with the malignancy and prognosis of the breast cancer cell lines. These results supported the potential role of SERCA in the development of breast cancer. However, in order to reveal the soundness of these data, those should be validated in vitro on cell lines originated from each subtype and also in large cohorts of patients.
Several previous studies support the findings of the present study, including the link between ATP2B2 and ATP2A3 in cancer, as well as the hypothesis that ATP2B3 might serve as a possible cancer biomarker [7,18,29,30].
Intracellular Ca2+ signaling plays a key role in mediating pathways such as cell proliferation, differentiation, and survival. Ca2+-dependent release of Ca2+ from the ER that is mediated by secondary messengers, such as inositol-1,4,5-tris-phosphate, depends entirely on SERCA activity [31,32,33,34]. Therefore, SERCA-dependent Ca2+ transport and SERCA activity constitute a major negative feedback mechanism in the mobilization of Ca2+ within the cell. As such, SERCA-regulated Ca2+-concentration-dependent pathways are a key component to cellular Ca2+-related signal transduction pathways, that if altered, can lead to a cascade of events that can affect various cellular processes, including cellular proliferation.
A number of experimental studies have shown that changes in Ca2+ levels play an important role in breast cancer prevention and breast cancer cell proliferation [35]. Moreover, SERCA activity and Ca2+ uptake are critical for the development of the differentiated breast acinar phenotype, and defects in Ca2+ accumulation in the ER are associated with altered SERCA expression. These alterations may be involved in the early steps of breast cancer tumorigenesis [36]. Furthermore, the finding that altered SERCA3 expression is associated with several histological and molecular markers of ductal carcinogenesis indicates that altered Ca2+ levels are associated with remodeling during tumorigenesis in the breast epithelium [37].
TNBCs are the most malignant form of breast cancer, and numerous pharmacological trials on patients with TNBC have failed to yield beneficial results. Moreover, the molecular changes underlying the switch between the different breast cancer subtypes towards TNBC and their metastasis need to be further characterized. An improved understanding could possibly lead to novel combinatorial therapies that could reduce the high and sometimes fatal toxicity of chemotherapeutic drugs.
Altered SERCA isoform expression and SERCA-specific gene mutations have been documented in various types of cancer, including lung, colon, and leukemia [7,38,39,40,41,42]. Progress in SERCA-specific isoform activities and their role in Ca2+ signaling will enhance the understanding of how dysregulation in SERCA activity plays a role in tumorigenesis. The present findings reinforce this knowledge in relation to the involvement of altered SERCA-specific gene expression in breast cancer and highlight the need to identify specific and selective pharmacological SERCA isoform inhibitors. SERCA isoforms are differentially expressed in cancer vs. normal samples. ATP2A1 expression negatively affected patient survival rates, especially in luminal, HER2-positive cancer and TNBC vs. normal controls, whereas ATP2A2 levels remained unaltered. ATP2A3 expression was significantly increased in luminal and HER2-positive vs. normal samples. Overall, high expression levels were associated with a reduction in the survival rate. However, further clinical studies on a larger sample of cancer patients are required to evaluate the present results. The type of human sample used as well as the sample size are considered as the limitations of the present study.
5. Conclusions
Concluding, our findings suggested that SERCA pump and altered Ca2+ signaling, as well as dysregulation of Ca2+ homeostasis, had an effective relation, contributing to breast cancer development and progression. A potential future challenge will involve the development of novel therapeutic approaches targeting specific SERCA-related malfunctions with the aim of limiting cancer development and progression.
Author Contributions
Conceptualization: A.Y., P.K.P. and I.P.; Data collection and interpretation: P.C., A.Y., M.-I.C. and A.M.; Manuscript drafting: A.Y., A.M., P.C. and I.P.; Revision of the manuscript for important intellectual content: all authors. Approval of the final version: all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in https://www.cbioportal.org/ and http://ualcan.path.uab.edu/.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Monteith G.R., Davis F.M., Roberts-Thomson S.J. Calcium Channels and Pumps in Cancer: Changes and Consequences. J. Biol. Chem. 2012;287:31666–31673. doi: 10.1074/jbc.R112.343061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui C., Merritt R., Fu L., Pan Z. Targeting calcium signaling in cancer therapy. Acta Pharm. Sin. B. 2017;7:3–17. doi: 10.1016/j.apsb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berridge M.J., Lipp P., Bootman M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 4.Clapham D.E. Calcium Signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 5.Parkash J., Asotra K. Calcium wave signaling in cancer cells. Life Sci. 2010;87:587–595. doi: 10.1016/j.lfs.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McConkey D.J., Orrenius S. The role of calcium in the regulation of apoptosis. Biochem. Biophys. Res. Commun. 1997;239:357–366. doi: 10.1006/bbrc.1997.7409. [DOI] [PubMed] [Google Scholar]
- 7.Korošec B., Glavač D., Rott T., Ravnik-Glavač M. Alterations in the ATP2A2 gene in correlation with colon and lung cancer. Cancer Genet. Cytogenet. 2006;171:105–111. doi: 10.1016/j.cancergencyto.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Boettner B. A Notch further with SERCA. Nat. Med. 2013;19:546. doi: 10.1038/nm.3208. [DOI] [Google Scholar]
- 9.Li W.-Q., Zhong N.-Z., He J., Li Y.-M., Hou L.-J., Liu H.-M., Xia C.-Y., Wang L.-Z., Lu Y.-C. High ATP2A2 expression correlates with better prognosis of diffuse astrocytic tumor patients. Oncol. Rep. 2017;37:2865–2874. doi: 10.3892/or.2017.5528. [DOI] [PubMed] [Google Scholar]
- 10.Pan Y., Zvaritch E., Tupling A.R., Rice W., de Leon S., Rudnicki M., McKerlie C., Banwell B.L., MacLennan D.H. Targeted disruption of the ATP2A1 gene encoding the sarco(endo)plasmic reticulum Ca2+ ATPase isoform 1 (SERCA1) impairs diaphragm function and is lethal in neonatal mice. J. Biol. Chem. 2003;278:13367–13375. doi: 10.1074/jbc.M213228200. [DOI] [PubMed] [Google Scholar]
- 11.Wootton L.L., Michelangeli F. The Effects of the Phenylalanine 256 to Valine Mutation on the Sensitivity of Sarcoplasmic/Endoplasmic Reticulum Ca 2+ ATPase (SERCA) Ca 2+ Pump Isoforms 1, 2, and 3 to Thapsigargin and Other Inhibitors. J. Biol. Chem. 2006;281:6970–6976. doi: 10.1074/jbc.M510978200. [DOI] [PubMed] [Google Scholar]
- 12.Brandl C.J., deLeon S., Martin D.R., MacLennan D.H. Adult forms of the Ca2+ATPase of sarcoplasmic reticulum. Expression in developing skeletal muscle. J. Biol. Chem. 1987;262:3768–3774. doi: 10.1016/S0021-9258(18)61421-8. [DOI] [PubMed] [Google Scholar]
- 13.Martin V., Bredoux R., Corvazier E., Van Gorp R., Kovacs T., Gelebart P., Enouf J. Three novel sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) 3 isoforms. Expression, regulation, and function of the membranes of the SERCA3 family. J. Biol. Chem. 2002;277:24442–24452. doi: 10.1074/jbc.M202011200. [DOI] [PubMed] [Google Scholar]
- 14.Dang D., Rao R. Calcium-ATPases: Gene Disorders and Dysregulation in Cancer. Biochim. Biophys. Acta. 2016;1863:1344–1350. doi: 10.1016/j.bbamcr.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miranda-Gonçalves V., Lameirinhas A., Henrique R., Jerónimo C. Metabolism and Epigenetic Interplay in Cancer: Regulation and Putative Therapeutic Targets. Front. Genet. 2018;9:427. doi: 10.3389/fgene.2018.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baylin S.B., Jones P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveira D.M., Santamaria G., Laudanna C., Migliozzi S., Zoppoli P., Quist M., Grasso C., Mignogna C., Elia L., Faniello M.C., et al. Identification of copy number alterations in colon cancer from analysis of amplicon-based next generation sequencing data. Oncotarget. 2018;9:20409–20425. doi: 10.18632/oncotarget.24912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammoudi N., Ahmed K.B., Garcia-Prieto C., Huang P. Metabolic alterations in cancer cells and therapeutic implications. Chin. J. Cancer. 2011;30:508–525. doi: 10.5732/cjc.011.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izquierdo-Torres E., Rodríguez G., Meneses-Morales I., Zarain-Herzberg A. ATP2A3gene as an important player for resveratrol anticancer activity in breast cancer cells. Mol. Carcinog. 2017;56:1703–1711. doi: 10.1002/mc.22625. [DOI] [PubMed] [Google Scholar]
- 20.Michelangeli F., East J.M. A diversity of SERCA Ca2+ pump inhibitors. Biochem. Soc. Trans. 2011;39:789–797. doi: 10.1042/BST0390789. [DOI] [PubMed] [Google Scholar]
- 21.Doan N.T.Q., Paulsen E.S., Sehgal P., Møller J.V., Nissen P., Denmeade S.R., Isaacs J.T., Dionne C.A., Christensen S.B. Targeting thapsigargin towards tumors. Steroids. 2015;97:2–7. doi: 10.1016/j.steroids.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denmeade S.R., Jakobsen C.M., Janssen S., Khan S.R., Garrett E.S., Lilja H., Christensen S.B., Isaacs J.T. Prostate-specific antigen-activated thapsigargin prodrug as targeted therapy for prostate cancer. J. Natl. Cancer Inst. 2003;95:990–1000. doi: 10.1093/jnci/95.13.990. [DOI] [PubMed] [Google Scholar]
- 23.Huang F., Wang P., Wang X. Thapsigargin induces apoptosis of prostate cancer through cofilin-1 and paxillin. Oncol. Lett. 2018;16:1975–1980. doi: 10.3892/ol.2018.8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahalingam D., Wilding G., Denmeade S., Sarantopoulas J., Cosgrove D., Cetnar J., Azad N., Bruce J., Kurman M., Allgood V.E., et al. Mipsagargin, a novel thapsigargin-based PSMA-activated prodrug: Results of a first-in-man phase I clinical trial in patients with refractory, advanced or metastatic solid tumours. Br. J. Cancer. 2016;114:986–994. doi: 10.1038/bjc.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papp B., Brouland J.-P. Altered Endoplasmic Reticulum Calcium Pump Expression during Breast Tumorigenesis. Breast Cancer. 2011;5:163–174. doi: 10.4137/BCBCR.S7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernández-Oliveras A., Izquierdo-Torres E., Meneses-Morales I., Rodríguez G., Zarain-Herzberg Á., Santiago-García J. Histone deacetylase inhibitors promote ATP2A3 gene expression in hepatocellular carcinoma cells: p300 as a transcriptional regulator. Int. J. Biochem. Cell Biol. 2019;113:8–16. doi: 10.1016/j.biocel.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Flores-Peredo L., Rodríguez G., Zarain-Herzberg A. Induction of cell differentiation activates transcription of the Sarco/Endoplasmic Reticulum calcium-ATPase 3 gene (ATP2A3) in gastric and colon cancer cells. Mol. Carcinog. 2017;56:735–750. doi: 10.1002/mc.22529. [DOI] [PubMed] [Google Scholar]
- 28.Varga K., Hollósi A., Pászty K., Hegedűs L., Szakács G., Tímár J., Papp B., Enyedi Á., Padányi R. Expression of calcium pumps is differentially regulated by histone deacetylase inhibitors and estrogen receptor alpha in breast cancer cells. BMC Cancer. 2018;18:1029. doi: 10.1186/s12885-018-4945-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Rodriguez I.P., Chakravarthi B.V.S.K., Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pimenta P.H.C., Silva C.L.M., Noël F. Ivermectin is a nonselective inhibitor of mammalian P-type ATPases. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2010;381:147–152. doi: 10.1007/s00210-009-0483-z. [DOI] [PubMed] [Google Scholar]
- 31.Stroik D.R., Yuen S.L., Janicek K.A., Schaaf T.M., Li J., Ceholski D.K., Hajjar R.J., Cornea R.L., Thomas D.D. Targeting protein-protein interactions for therapeutic discovery via FRET-based high-throughput screening in living cells. Sci. Rep. 2018;8:12560. doi: 10.1038/s41598-018-29685-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bilmen J.G., Wootton L.L., Michelangeli F. The inhibition of the sarcoplasmic/endoplasmic reticulum Ca2+-ATPase by macrocyclic lactones and cyclosporin A. Biochem. J. 2002;366:255–263. doi: 10.1042/bj20020431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papp B., Launay S., Gélébart P., Arbabian A., Enyedi A., Brouland J.-P., Carosella E.D., Adle-Biassette H. Endoplasmic Reticulum Calcium Pumps and Tumor Cell Differentiation. Int. J. Mol. Sci. 2020;21:3351. doi: 10.3390/ijms21093351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinton P., Giorgi C., Siviero R., Zecchini E., Rizzuto R. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene. 2008;27:6407–6418. doi: 10.1038/onc.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speers C., Brown P. Breast Cancer Prevention Using Calcium and Vitamin D: A Bright Future? J. Natl. Cancer Inst. 2008;100:1562–1564. doi: 10.1093/jnci/djn390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cross B.M., Breitwieser G., Reinhardt T.A., Rao R. Cellular calcium dynamics in lactation and breast cancer: From physiology to pathology. Am. J. Physiol. Physiol. 2014;306:C515–C526. doi: 10.1152/ajpcell.00330.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart T., Yapa K.T., Monteith G.R. Altered calcium signaling in cancer cells. Biochim. Biophys. Acta (BBA)—Biomembr. 2015;1848:2502–2511. doi: 10.1016/j.bbamem.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Meneses-Morales I., Izquierdo-Torres E., Flores-Peredo L., Rodríguez G., Hernández-Oliveras A., Zarain-Herzberg Á. Epigenetic regulation of the human ATP2A3 gene promoter in gastric and colon cancer cell lines. Mol. Carcinog. 2019;58:887–897. doi: 10.1002/mc.22978. [DOI] [PubMed] [Google Scholar]
- 39.Pérez-Riesgo E., Gutiérrez L.G., Ubierna D., Acedo A., Moyer M.P., Núñez L., Villalobos C. Transcriptomic Analysis of Calcium Remodeling in Colorectal Cancer. Int. J. Mol. Sci. 2017;18:922. doi: 10.3390/ijms18050922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roti G., Carlton A., Ross K.N., Markstein M., Pajcini K., Su A.H., Perrimon N., Pear W.S., Kung A., Blacklow S.C., et al. Complementary Genomic Screens Identify SERCA as a Therapeutic Target in NOTCH1 Mutated Cancer. Cancer Cell. 2013;23:390–405. doi: 10.1016/j.ccr.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aït Ghezali L., Arbabian A., Roudot H., Brouland J.P., Baran-Marszak F., Salvaris E., Boyd A., Drexler H.G., Enyedi A., Letestu R., et al. Induction of endoplasmic reticulum calcium pump expression during early leukemic B cell differentiation. J. Exp. Clin. Cancer Res. 2017;36:87. doi: 10.1186/s13046-017-0556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fierro L., Parekh A.B. Substantial depletion of the intracellular Ca2+ stores is required for macroscopic activation of the Ca2+ release-activated Ca2+ current in rat basophilic leukaemia cells. J. Physiol. 2000;522:247–257. doi: 10.1111/j.1469-7793.2000.t01-1-00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are openly available in https://www.cbioportal.org/ and http://ualcan.path.uab.edu/.