Abstract
Airborne transmission of SARS-CoV-2 has been object of debate in the scientific community since the beginning of COVID-19 pandemic. This mechanism of transmission could arise from virus-laden aerosol released by infected individuals and it is influenced by several factors. Among these, the concentration and size distribution of virus-laden particles play an important role. The knowledge regarding aerosol transmission increases as new evidence is collected in different studies, even if it is not yet available a standard protocol regarding air sampling and analysis, which can create difficulties in the interpretation and application of results. This work reports a systematic review of current knowledge gained by 73 published papers on experimental determination of SARS-CoV-2 RNA in air comparing different environments: outdoors, indoor hospitals and healthcare settings, and public community indoors. Selected papers furnished 77 datasets: outdoor studies (9/77, 11.7%) and indoor studies (68/77. 88.3%). The indoor datasets in hospitals were the vast majority (58/68, 85.3%), and the remaining (10/68, 14.7%) were classified as community indoors. The fraction of studies having positive samples, as well as positivity rates (i.e. ratios between positive and total samples) are significantly larger in hospitals compared to the other typologies of sites. Contamination of surfaces was more frequent (in indoor datasets) compared to contamination of air samples; however, the average positivity rate was lower compared to that of air. Concentrations of SARS-CoV-2 RNA in air were highly variables and, on average, lower in outdoors compared to indoors. Among indoors, concentrations in community indoors appear to be lower than those in hospitals and healthcare settings.
Keywords: SARS-CoV-2 in air, COVID-19, Airborne transmission, Outdoor, Indoor
Graphical abstract
1. Introduction
Since the beginning of the global pandemic in 2020, several studies raised a scientific debate on the possible role of airborne transmission of the disease in the spread of COVID-19 (Contini and Costabile, 2020; Domingo et al., 2020; Ishmatov, 2021; Klompas et al., 2020; Morawska and Cao, 2020; Prather et al., 2020; Ram et al., 2021). This mechanism of transmission could arise from virus-laden coarse and fine aerosols emitted by infected individuals during cough, sneezes, respiration, speaking, singing, and shouting. These could remain suspended in air, following different dynamics in indoor and outdoor environments, and being potentially inhaled by other susceptible individuals (Allen and Marr, 2020; Asadi et al., 2020; Belosi et al., 2021; Borouiba, 2020; Tang et al., 2021).
Several parameters are important to determine risks of airborne transmission: concentration and size distribution of virus-laden particles; fraction of infectious (viable) virus in air; minimum dose necessary to transmit infection to a susceptible individual. The first two parameters are depending on meteorological conditions (with differences between indoor and in outdoor), on the dynamics of air currents and on physical-chemical properties of droplets (Niazi et al., 2021a, Niazi et al., 2021b; Ratnesar-Shumate et al., 2020); the third parameter is influenced by specific vulnerabilities of susceptible individuals and vary strongly (Buonanno et al., 2020). Knowledge regarding airborne transmission of SARS-CoV-2 is continuously evolving as new evidence accumulates. This knowledge would directly impact on policy decisions regarding adequate mitigation measures to be implemented for efficient reduction of COVID-19 spread (Morawska and Cao, 2020; Morawska and Milton, 2020).
Mini-reviews about air detection methods for coronavirus in general and specific for SARS-CoV-2 have been published discussing problems and controversies, showing that more studies are necessary to find the method with best performance for SARS-CoV-2 detection in air and that it would be useful to develop a standard procedure for air sampling and analysis (Borges et al., 2021; Pena et al., 2021; Rahmani et al., 2020; Ratnesar-Shumate et al., 2021; Robotto et al., 2021; Yun et al., 2020). Other reviews (Heneghan et al., 2021; Tang et al., 2020) focused on the role of airborne transmission concluding that SARS-CoV-2 genetic material (RNA) is observed intermittently in air in different indoors environments and that the lack of detailed information on recoverable virus cultures prevents solid conclusions on the weight of airborne transmission. Finally, some review papers are focused on results, available during the first wave of pandemic during 2020, relative to detection of SARS-CoV-2 genetic material in air in hospital settings and working places finding that about 6% of air and surface samples in hospital were positive, although the data is very limited for non-healthcare settings and that only a few of the studies report quantification of concentrations in air (Anand et al., 2021; Birgand et al., 2020; Cherrie et al., 2021).
This work is aimed at presenting a systematic review of current knowledge, from the beginning of pandemic and until 31/08/2021, regarding identification/quantification of SARS-CoV-2 RNA in airborne samples comparing different sites: outdoor sites, indoors in hospitals and healthcare settings, and community indoor locations. The analysis will also investigate positivity rates and concentrations comparing the different environments and the current methodologies used for samples collection and analysis. Conclusions about risks of airborne transmission and efficiency of mitigation measures are included, together with a discussion of the aspects that need further studies.
2. Methodology
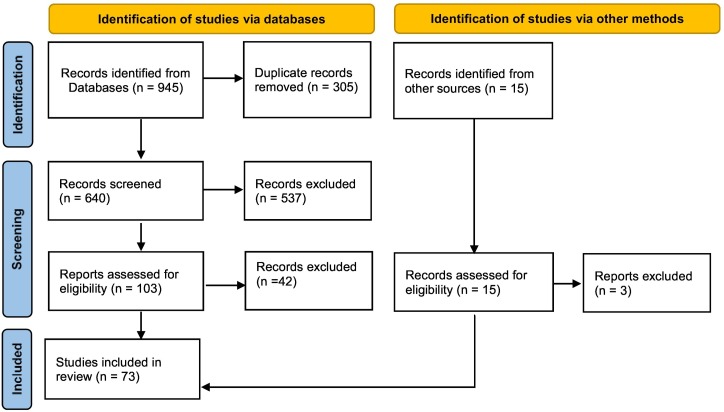
The records used in this review include paper published since the start of COVID-19 pandemic until 31/08/2021. Published papers were selected from the Web of Science, Scopus, and Google Scholar databases using two search strings: “covid-19 and airborne” and “SARS-CoV-2 and airborne transmission”. This gave to 945 entries plus an additional 15 entries that were obtained from different sources, mainly specific searches and reading of the authors. Fig. 1 reports the flowchart summarising the identification and selection of the records using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Moher et al., 2009; Page et al., 2021). After the exclusion of 305 duplicate records, the entries were screened to select records complying with the goal of this review, i.e. studies containing analysis of the presence/concentration of SARS-CoV-2 genetic material through measurements in both indoor and outdoor environments. Successively, the papers were assessed to select cases based on active air stationary sampling (i.e. excluding passive sampling and samples collected with personal samplers), irrespective of the approach/equipment used, and in which the identification/quantification of SARS-CoV-2 RNA traces was done by polymerase chain reaction (PCR). In this step only original studies reporting measurement results were maintained, i.e. review papers and commentaries were excluded. This left a final number of 73 papers included in this review and six of them were preprints.
Fig. 1.
Flowchart of the identification, screen, and assessment of the records included in this review according to PRISMA statement (Page et al., 2021).
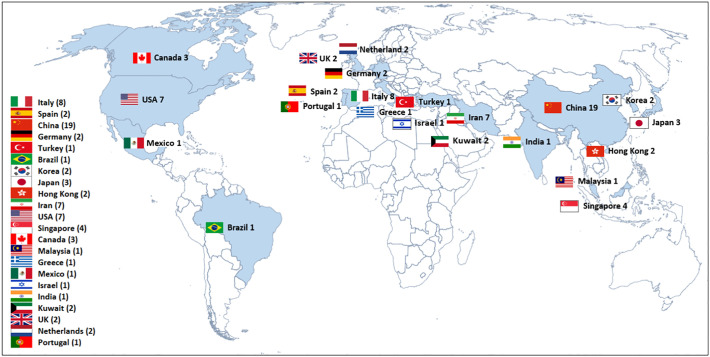
Fig. 2 shows the geographical distribution of the 73 studies that were performed in 22 different Countries. The majority of the studies (26.0%) were done in China, in Europe (24.6%), and in North America (13.7%). Limited or no information is available from South America, Africa, India, and Oceania.
Fig. 2.
Geographical distribution of the 73 studies included in this review.
The selected papers furnished 77 datasets because three papers (Hu et al., 2020; Liu et al., 2020; Passos et al., 2021) included outdoor and indoor datasets, and one paper (Habibi et al., 2021) included datasets collected in different typologies of indoor environments. The datasets were separated into two categories: outdoor studies (9/77, 11.7%) and indoor studies (68/77, 88.3%). The indoor datasets were additionally separated into two groups: datasets dealing with measurements in hospitals, health care structures, and quarantine areas, that were the vast majority (58/68, 85.3%), and studies dealing with measurements taken at indoor community sites (10/68, 14.7%) such as commercial centres and markets, schools and universities, pharmacies, hair salon, stations, and vehicles of public transport. In the latter category it has been included a study on a mink farm (de Rooij et al., 2021a), in a meat processing plant (de Rooij et al., 2021b), in the Diamond Princess cruise ship (Yamagishi et al., 2020), and in two houses of a residential building in China (at Guangzhou) by Xie et al. (2020).
The different studies selected were analysed to extract the main information and results regarding: the methodology used for air sampling; the methodology used for RNA detection; the number of positive samples found in air and the eventual other results found on environmental samples (surface swabs) collected; the concentrations found, when reported, in air samples.
3. Results
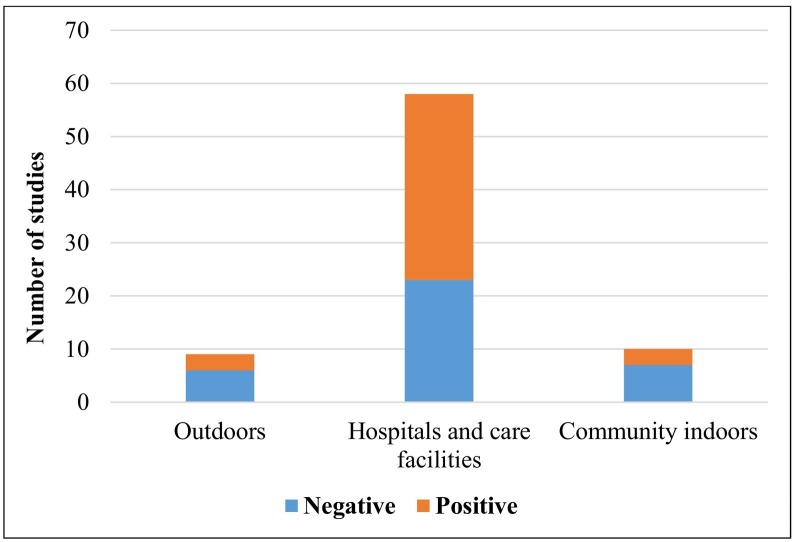
The first analysis was made to estimate the number of datasets that reports negative (i.e. SARS-CoV-2 not detected in any of the air samples) and positive (at least one sample found positive to SARS-CoV-2). Results show (Fig. 3 ) that the majority of the datasets refers to hospitals and care settings where the datasets reporting positive samples were 35 out of 58 (60.3%). Significantly lower percentages were observed for datasets in outdoors (3/9, 33.3%) and in indoor community environments (3/10, 30%) even if statistics is more limited for these last two categories.
Fig. 3.
Number of datasets reviewed for the different typologies of sites. Negative indicates dataset with all air samples that gave negative results for the presence of SARS-CoV-2 and positive indicates the datasets in which at least one of the air samples tested positive.
3.1. Results in outdoor environments
The results of the papers dealing with measurements of airborne SARS-CoV-2 RNA traces in outdoor sites are reported in Table 1 . Table 1 reports a summary of the methodology used and the results found in the different papers that include outdoor measurement in public areas. Results are available for different areas in Europe (Italy, Germany, and Spain), China, Turkey, and Brazil. All studies are based on samples collected in different periods of the first wave of pandemic. In three cases, the study includes both outdoor and indoor measurements (Hu et al., 2020; Liu et al., 2020; Passos et al., 2021). Five studies out of the eight available (62.5%) reported negative results with SARS-CoV-2 concentrations lower than the detection limit (LOD). Of the three studies reporting positive samples, one (Setti et al., 2020) does not report concentrations, the other two (Liu et al., 2020; Kayalar et al., 2021) quantifies concentrations of viral copies in air. Liu et al. (2020) in Wuhan (China) collected samples mainly in indoor environments (different locations hospitals) but eight of them were collected outdoor in public spaces. Authors concluded that these samples presented undetectable or very low concentrations (<3 copies m−3) with the exclusion of the crowded sites in which traces of SARS-CoV-2 were observed up to 11 copies m−3. These concentrations appear to be lower than those found in indoor in the same study (i.e. 1–42 copies m−3 with a positivity rate of 67%). Kayalar et al. (2021) report 3% of samples (2 out of 68) positive in urban or urban background outdoor sites and the rate of positivity increases to 15% at hospital gardens, outdoor sites near SARS-CoV-2 sources, even if concentrations are not significantly different. The average positivity rate among the three datasets, i.e. the ratio between positive and total samples collected summed in the three datasets, was 17.6%. Concentrations were estimated only in two of the three datasets in which positive outdoor samples were identified. The concentration range considering all the data of the two datasets were 0.1–23 copies m−3 and the average value of all quantified samples was 7.9 copies m−3 (median 7.2 copies m−3).
Table 1.
Summary of the methodology used and of the results found in different datasets focused on measuring airborne concentrations of SARS-CoV-2 genetic material (RNA) in different outdoor sites in public areas.
| Reference | Sites | Sampling | Method | Results | Notes |
|---|---|---|---|---|---|
|
Chirizzi et al. (2021) Italy |
Two urban background sites: Veneto (Venice, North Italy) and Apulia (Lecce, South Italy) simultaneously studied, period 13/05/2020–27/05/2020. | At each site, 6 PM10 samples on quartz fibre filters (48 h at 38.3 L min−1) and 24 multi-stage impactor samples (6d at 30 L min−1, size range from D < 0.056 μm up to D > 18 μm. Volumes 110 m3 or 250 m3. | RT-PCR targeting E and RdRp genes. dd-PCR targeting N1 and N2 genes. |
100% of samples negative with both methods RT-PCR and dd-PCR. | LOD - PM10 0.8 copies m−3. LOD - impactor 0.4 copies m−3. Recovery 49%. |
|
Dunker et al. (2021) Germany |
University of Leipzig Medical centre. Samples collected between 11/03/2020 and 28/05/2020. | 7 weekly air samples and one 14 days sample. Sampling at 15 L min−1 with a cyclone trap directly into 1.5 mL micro centrifuge tube. | RT-PCR targeting E gene or N and RdRp genes. | 100% of samples negative | LOD and recovery not reported. Fresh pollen samples were also collected finding no presence of SARS-CoV-2. |
|
Hu et al. (2020) China |
Public areas in Wuhan. | 20 samples collected with a centrifugal sampler WA-400 at 400 L min−1 in PBS. Volumes 12 m3. | qRT-PCR targeting ORF1ab gene. | 100% of samples negative. | Recovery not reported. |
|
Kayalar et al. (2021) Turkey |
Samples from 13 locations in 10 towns, period 13/05/2020–14/06/2020. Hospital garden (HG) sites; urban (U) and urban background (UB) sites. |
80 TSP, 19 PM10, 23 PM2.5–10, 33 PM2.5 samples with different samplers and filters (PTFE, quartz and glass fibre, polycarbonate. Volumes 7.2–360 m3. 48 size segregated (6 sizes) samples on glass fibre filters, volume 1422 m3. |
RT-PCR targeting N1 and RdRp genes. 3D-dPCR targeting N1 gene. |
HG sites 13/87 samples (14.9%) positive. U and UB sites 2/68 samples (3%) positive. U (Istanbul) 5/48 size segregated samples (10%) positive. |
Near hospitals 5–23 copies m−3. Urban and urban background 7–21 copies m−3. Urban size segregated <0.2 copies m−3. Recovery not reported. |
|
Linillos-Pradillo et al. (2021) Spain |
Madrid (district 09) university area in the period 04/05–22/05/2020. | 6 PM10, 6 PM2.5, and 6 PM1 simultaneous samples on quartz fibre filters at 30 m3 h−1 for 17.5–24 h. Volumes 525–720 m3. | RT-PCR targeting N1 and N2 genes and control of human RNase P (RP) gene. | 100% of samples negative | LOD not reported. Recovery not reported. |
|
Liu et al. (2020) China (Wuhan) |
Different sites near hospital, community check point, department stores and supermarket and residential buildings. | TSP sampled on gelatine substrate at 5 L min-1. Volumes 1.5–5 m3. | dd-PCR targeting Orf1ab and N genes. | 3/8 (37%) of samples positive collected near hospital and near the door of a busy department store. | Outdoor concentration non-detectable or very low (<3 copies m−3) but at crowded sites that arrived at 11 copies m−3. Recovery not reported. |
|
Passos et al. (2021) Brazil |
Metropolitan area of Belo Horizonte. Period 25/05/2020–06/08/2020. | 2 PM2.5 and 7 PM10 samples in total at: car parking of a COVID-19 hospital, sidewalk near hospital, busy bus station. Quartz fibre filter sampled at 1130 L min−1. Volumes 7–4500 m3. | RT-PCR targeting N1 and N2 genes. | 100% of samples negative | LOD not reported. Recovery ~100% |
|
Pivato et al. (2021) Italy |
10 sites (urban-rural background, traffic, industrial). NE Italy (Padua province) period 24/02/2020–09/03/2020. | 25 PM10 and 19 PM2.5 samples were collected in total over the 10 sites, on quartz fibre filters (24 h at 38.3 L min−1). Volume 55.2 m3. | RT-PCR targeting N and Orf1b-14nsp genes. | 100% of samples negative | LOD 1.2 copies m−3. Recovery not reported. |
|
Setti et al. (2020) Italy |
Industrial area of Bergamo over a continuous 3-week period 21/02/2020–13/03/2020. |
34 PM10 samples on quartz fibre filters (24 h at 38.3 L min−1). Volume 55.2 m3. | RT-PCR targeting E, RdRp, and N genes. | 58.8% of samples (20/34) positive in 1 gene; 11.8% (4/34) for 2 genes; none for 3 genes. | Concentrations not reported. LOD not reported. Recovery not reported. |
de Rooij et al. (2021a) investigated airborne SARS-CoV-2 concentrations indoor in mink farms in Netherlands collecting samples of total suspended particles as well as inhalable dust mainly in indoor. Some samples were also collected outdoors finding either negative or low concentrations cases at 1.5 m from the open entrance of the farm and negative samples at 20 m distance. This suggests that in outdoor conditions, virus-laden particles are quickly transported and dispersed by wind lowering concentrations when distance from the source increases. Hu et al. (2020) also collected outdoor samples at 10 m from the doors of in-patients and out-patients buildings of a hospital in Wuhan (China) finding 3 positive samples out of 20 (15%) while they did not detect SARS-CoV-2 in residential and open public areas. Stern et al. (2021b) collected outdoor air samples at the gates of COVID-19 hospitals in Kuwait finding 5 positive samples out of 33 (15%) with concentrations in the range 3–17 copies m−3. Habibi et al. (2021) studied indoor locations in Kuwait but also collected two outdoor samples in residential areas for control and both resulted negative.
The results discussed give the indication that, in outdoor conditions excluding crowded areas or zones located near the sources (close contact), virus-laden particles are quickly transported and dispersed by winds resulting in low or undetectable concentrations. In these conditions, airborne transmission of COVID-19 disease is very unlikely or negligible. However, the analysis reported reinforce the importance of avoiding crowds and large gathering of people, as well as to maintain social distancing from the sources to minimise the risks of airborne transmission in outdoor. This aspect is also supported by the simulations in outdoor for Milano and Bergamo areas, the epicentre of the COVID-19 outbreaks in Italy in winter 2020, reported in Belosi et al. (2021). The work of Belosi et al. (2021) focuses on these areas in Lombardia region (northern Italy) located in the “Po Valley” that is an atmospheric pollution hot-spot due to the local meteorological and micrometeorological conditions that favour shallow boundary layer, stable atmospheric conditions, and limited ventilation, especially during winter. Simulation of Belosi et al. (2021) showed low average concentrations (<0.6 copies m−3) even in the grim hypothesis of a 10% of population currently infected and in the worst-case scenario for pollution dispersal.
It must be mentioned that different sampling approaches have been used, as well as different extraction procedures and the LODs are not clearly reported in all studies so that it could be difficult to interpret what a negative result really means. In addition, the recovery, i.e. the efficiency with which SARS-CoV-2 RNA is extracted and identified by PCR in collected samples, is not often reported. Moreover, it is analysed with specific tests only in a few studies and this also could influence results found and comparability of different studies. It was not observed a direct correlation between positivity rate and sampling volumes or sampling supports used. Positive samples were obtained from samples on different typologies of filters (Kayalar et al., 2021; Setti et al., 2020) as well as on gelatine substrates (Liu et al., 2020). On the other hand, datasets with all negative samples were obtained from studies with sampling on filters (Chirizzi et al., 2021; Linillos-Pradillo et al., 2021; Passos et al., 2021; Pivato et al., 2021), using cyclone for sampling in centrifugal tubes (Dunker et al., 2021), as well as using high volume centrifugal sampling on phosphate buffered saline (PBS) (Hu et al., 2020).
3.2. Results of indoor measurements in hospitals, quarantine areas, and COVID-19 health care facilities
The summary of results obtained from the 58 datasets dealing with measurements of airborne SARS-CoV-2 RNA traces in hospitals and care facilities are reported in Table 2, Table 3 . Table 2 deals with the 41 datasets that have negative (i.e. all air sampled tested negative for presence of SARS-CoV-2) or positive (i.e. at least one air sample tested positive at PCR analysis) without quantification of concentrations. Table 3 deals with the 17 datasets (29.3% of the total) that have quantification of concentrations in air samples in hospitals and health care facilities.
Table 2.
Summary of the methodology used and of the results found in different datasets focused on detecting SARS-CoV-2 genetic material (RNA), without quantification of concentrations in indoor sites in hospitals and healthcare settings in which were present COVID-19 patients.
| Reference | Sites | Sampling | Method | Results for air samples | Notes |
|---|---|---|---|---|---|
|
Ahn et al. (2020) Korea |
Negative pressure isolation rooms (12 air exchange h−1) with confirmed COVID-19 patients. | Samples collected with SKC BioSampler at 1 m from patients at 12.5 L min−1. Volumes 0.25 m3. | RT-PCR targeting E and RdRp genes. | All of the 3 air samples tested negative. | LOD and recovery not reported. 15/76 surface samples positive. |
|
Cai et al. (2020) China |
General wards in Wuhuan hospital | Sampling with a Bobcat sampler on electrect filters at 200 L min−1. Volume 12 m3. | RT-PCR targeting ORF1ab gene. | All of the 15 air samples tested negative. | LOD and recovery not reported. 2/128 surface samples positive. |
|
Cheng et al. (2020a) Hong Kong |
Hospital, room of Covid-19 patient. | TSP collected with a SAS sampler at 180 L min−1 at 10 cm from the face of patient with and without facemask. Volume 1 m3. | RT-PCR targeting RdRp gene. | All of the 8 air samples tested negative. | LOD and recovery not reported. 1/13 surface samples positive. |
|
Cheng et al. (2020b) Hong Kong |
Airborne infection isolation rooms (AIIRs) with 12 air exchanges per h. | TSP using MD8 portable sampler at 50 L min−1 on gelatine. Volume 1 m3. | RT-PCR targeting RdRp gene. | All of the 6 air samples tested negative. | LOD and recovery not reported. 19/377 surface samples positive. |
|
Declementi et al. (2020) Italy |
Non-intensive care units in northern Italy. | Sampling with a SKC pump on PTFE filters at 15 L min.1. Volume 5.1 m3. | RT-PCR, not clear the targets. | All of the 8 air samples tested negative, one outdoor control sample negative. | LOD and recovery not reported. 0/24 surface samples positive. |
|
Döhla et al. (2020) Germany |
21 quarantine households in the Bonn area. | Air samples with Coriolis cyclone sampler at 300 L min−1 in 15 mL of 0.9% NaCl, with no close contact (< 2 m) of patients. Volume 3 m3. | RT-PCR targeting E and RdRp genes. | All of the 15 air samples tested negative. | LOD and recovery not reported. 4/119 surface samples positive. No infectious virus under cell cultures. |
|
Dumont-Leblond et al. (2021) Canada |
31 rooms in 7 long-term care settings in major cities of Quebec. | Sampling with IOM Multidust sampler on gelatine filters, at about 2 m from the residents, at 3 L min−1. Volume 0.72 m3. | RT-PCR targeting ORF1b gene. | All of the 31 air samples tested negative. | LOD and recovery not reported. 20/62 surface samples positive. Cultures of virus from surface samples were negative. |
|
Faridi et al. (2020) Iran |
Hospital ward in Theran with confirmed COVID-19 patients. | Sampling with impinger in 20 mL solution between 2 and 5 m from patients. Volume 0.09 m3. | RT-PCR targeting E and RdRp genes. | All of the 10 air samples tested negative. | LOD and recovery not reported. |
|
Kim et al. (2020) Korea |
Four health care facilities with COVID-19 patients. | TSP using MD8 portable sampler at 50 L min−1 on gelatine. Volume 1 m3. | rRT-PCR targeting E and RdRp genes. | All of the 52 air samples tested negative. | LOD and recovery not reported. 89/320 surface samples positive. |
|
Lane et al. (2020a) USA |
Infection isolation room of a ventilated COVID-19 patient in Atlanta. | Air sampling with 2 NIOSH 251 2 BCE-stage samplers for separation in three size fractions at 3.5 L min−1. Volume 1.26 m3. | RT-PCR targeting N and human RNase P genes. | All of the 28 air samples tested negative. | LOD and recovery not reported. |
|
Lane et al. (2020b) USA |
Hospital, different sites outside patients' rooms. | 8 NIOSH 251 2 BCE-stage samplers for separation in three size fractions at 3.5 L min−1. Volume 1.26 m3. | rRT-PCR targeting N1, N2, N3 genes or N2, E, RNAase P. | All of 528 air samples tested negative. | LOD 8 copies m−3. Recovery not reported. |
|
Li et al. (2020) China |
Different wards in a hospital for Covid-19 patients. | Impinger sampler (BIO-Capturer-6) in a sampling buffer at 80 L min-1. Volume 2.4 m3. Distances 1–5 m from patients. | RT-PCR not reported the targets. | All of the 135 air samples tested negative. | LOD and recovery not reported. 2/90 surface swab positive (facemasks of patients). |
|
Masoumbeigi et al. (2020) Iran |
Different wards of a Covid-hospital in Tehran. | Samples taken with all-glass impinger (AGI) at 5–40 L min−1. Volumes 0.1–1 m3. | rRT-PCR, not clear the targets. | All of the 31 samples tested negative. | LOD and recovery not reported. |
|
Nakamura et al. (2020) Japan |
Health care facility, different indoor locations. | TSP using MD8 portable sampler at 50 L min−1 on gelatine. Volume 1 m3. | RT-PCR targeting N gene. | All of the 11 air samples tested negative. | LOD and recovery not reported. 4/141 surface samples positive. |
|
Ong et al. (2020) Singapore |
AIIR in a Covid-19 dedicated center in Singapore. | SKC pump on PTFE filters at 5 L min−1 and a MD8 sampler on gelatine at 100 L min−1. Volumes 1.2–1.5 m3. | RT-PCR targeting E and RNA polymerase genes. | All of 5 air samples tested negative. | LOD and recovery not reported. 17/27 surface samples positive. |
|
Song et al. (2020) China |
Infection isolation rooms (AIIRs) in a Covid-hospital in Shanghai. | Samples in clean, semi-contaminated and contaminated areas with a Derenda (PNS-16 T) sampler at 1 m3 h−1. Volume 1.5 m3. | RT-PCR targeting RdRp gene. | All of the 42 samples tested negative. | LOD and recovery not reported. 25/1502 surface samples positive. |
|
Vosoughi et al. (2021) Iran |
8 hospital locations in Ardabil. | Impinger at 28 L min−1 at 2–5 m from patients' bed. Volumes 1.68–5.04 m3. | RT-PCR targeting ORF1ab, N genes. | All of the 33 air samples tested negative. | LOD and recovery not reported. |
|
Wei et al. (2020a) China |
Hospital rooms of Covid-19 confirmed patients in Chengdou. | Samples taken with microbiological sampler (FSC-1 V) on filter membranes at 100 L min−1. Volume 1.5 m3. | RT-PCR targeting ORF1ab and N genes. | All of the 34 samples tested negative. | LOD and recovery not reported. 3/88 surface samples positive. 0/55 samples from PPE positive. |
|
Wei et al. (2020b) China |
Six negative pressure non-ICU rooms in isolation ward in Chengdou. | Samples taken with microbiological sampler (FSC-1 V) on filter membranes at 100 L min−1. Volume 1.5 m3. | RT-PCR targeting ORF1ab and N genes. | All of the 6 samples tested negative. | LOD and recovery not reported. 44/112 surface samples positive. |
|
Wong et al. (2020) Singapore |
18 sites in four rooms of quarantine non-healthcare settings. | Coriolis cyclonic air sampler at 300 L min−1 in viral transport media (VTM). Volume 9 m3. | RT-PCR targeting RdRp gene. | All of the 6 air samples tested negative. | LOD and recovery not reported. 2/428 surface samples positive. |
|
Zhang et al. (2020) China |
Fangcang shelter hospital in Wuhan. | Air sampling with NingBo IGene Tec at 6 m3 h−1 on gelatine filters in clean, buffer, and contaminated areas. Volume 1 m3. | RT-PCR targeting ORF1ab gene. | All of the 24 air samples tested negative. | LOD 100 copies mL−1 (liquid phase). Recovery not reported. 0/24 surface samples positive. |
|
Zhou et al. (2020b) China |
Different sites in four hospitals in Wuhuan. | Two impingers (WA-15 and WA-400) at 15 and 400 L min−1. Volumes 0.6 m3 and 16 m3. | RT-PCR targeting ORF1ab and N genes. | All of the 44 air samples tested negative. | LOD 100 copies μL−1 (liquid phase). Recovery not reported. 4/318 surface samples positive. |
|
Baboli et al. (2021) Iran |
Different areas of a Covid-19 hospital in Ahvaz. | Glass impinger, SKC pump with PFTE filters, QuickTake30 kit at 4 L min−1, 0.12 m3 volume, 1–3 m from patient beds. | RT-PCR targeting RdRp and N genes. | 5/51 air samples positive. | LOD and recovery not reported. |
|
Barbieri et al. (2021) Italy |
Geriatric ward in a Hospital in Trieste. | PM10 samples on quartz fibre filters at 10 L min−1. Volume 14.4 m3. | RT-qPCR targeting RdRp gene. | 1/5 air sample positive for all replicates. | LOD and recovery not reported. |
| Ben-Shmuel et al. (2020), Israel | Two hospitals and one quarantine facility. | MD8 sampler at 50 L min−1 on gelatine filters. Volume 1 m3. | RT-qPCR targeting E gene. | 3/8 air samples tested positive. | LOD and recovery not reported. 42/89 surface samples positive. Virus cultures negative. |
|
Binder et al. (2020) USA |
Single-occupant rooms at Duke University hospital. | 8 NIOSH BC251 samplers, 1–3.2 m from patients, at 3.5 L min−1. Volume 0.84 m3. | RT-PCR targeting N gene. | 3/143 samples positive at 1.4–2.2 m from patients. | LOD and recovery not reported. 6/70 surface samples positive. Virus cultures negative. |
|
Ding et al. (2021) China |
4 three-bed isolation rooms of Covid-19 hospital in Nanjing and other indoor areas. | 4 bioaerosol samplers QuickTake-30, a MD8 sampler, an impinge WA-15, an ASE100 sampling at different flow-rates. | 1/46 samples were positive. | LOD and recovery not reported. 7/107 surface samples positive. |
|
|
Dubey et al. (2021) India |
Different wards of a COVID-19 dedicated hospital in Delhi. | Sampling at 1.5, 16.7, 27 L min-1 on PVDF filters at 1–3 m distance from patients. Volumes 0.09, 1, 1.62 m3. | RT-PCR targeting RdRp and E genes. | 54/126 air samples positive. | LOD and recovery not reported. 14/18 surface samples positive. |
|
Ge et al. (2020) China |
6 sites in three hospitals in Changsha, Changzhou, and Shaoyang. | NIOSH biosampler (BC251) at 3.5 L min-1. Volume 0.1 m3. | qRT-PCR targeting NP gene. | 4/33 samples were positive. | LOD and recovery not reported. 17/112 surface samples positive. |
|
Hemati et al., 2021 Iran |
Different wards of an hospital in Shahrekord. | Sampling with impinger (SKC) at 2 L min−1. Volume 0.48 m3. | RT-PCR targeting RdRp and N genes | 6/45 samples were positive. | LOD and recovery not reported. |
| Kenarkoohi et al. (2020), Iran | ICU and wards of Covid-19 hospital in Ilam province. | Liquid impinger (SKC) at 12 L min-1. Volume 2.16 m3. | RT-PCR targeting ORF1ab, N genes. | 2/14 air samples were positive. | LOD 200 copies mL−1 (liquid phase). Recovery not reported. |
|
Kotwa et al. (2021) Canada |
Six acute care hospitals in Toronto. | 3 samplers at 3.5 L min−1 on PFTE, Polycarbonate, gelatine filters. A NIOSH bioaerosol sampler. Volume 0.42 m3. | RT-qPCR targeting UTR and E genes. | 3/146 air samples positive. | LOD and recovery not reported. 125/474 surface samples positive Virus cultures negative for air but positive on 6 of 36 surfaces. |
|
Jin et al. (2020) China |
Isolation room and PPE dressing room of a hospital in Guiyang. | WA400 impinger at 400 L min−1. Volume 6 m3. | qRT-PCR targeting ORF1ab, NP genes | 1/2 sample was positive. | LOD and recovery not reported. 0/5 surface samples positive. |
|
Lei et al., 2020 China |
An ICU and an isolation ward of hospital in Guangzhou. | A two-stage cyclonic NIOSH sampler and an aerosol particle liquid concentrator (W-15, DingBlue) operating at 3.5 L min−1. Volume 0.84 m3. | RT-PCR targeting ORF1 and N genes. | All samples collected at ICU were negative. 2 air samples positive in isolation ward. Not clear the total number of air samples collected. |
LOD and recovery not reported. 0/218 ICU samples (air and surfaces) positive. 9/182 air and surfaces at isolation ward positive. |
|
López et al. (2021) Mexico |
Two hospitals in Hermosillo (Sonora). | Sampling on Millipore filters at 9.6 L min−1. Volume 1.73 m3. | RT-PCR targeting E and RdRp genes. | 3/10 samples were positive. | LOD and recovery not reported. |
|
Ma et al. (2021) China |
Hospitals and quarantine hotels in Beijing. | Two impingers: WA-15 (15 L min-1) and WA-400 (400 L min-1). Volumes 0.6–16 m3. | RT-PCR targeting ORF1ab, N genes. | 1/26 sample were positive. | LOD and recovery not reported. 13/242 surface samples positive. |
|
Morioka et al. (2020) Japan |
Tertiary care hospital in Tokyo. | Air samples collected on gelatine filters with a MD8 sampler at 50 L min−1. Volume 1 m3. | RT-PCR targeting N gene. | 0/4 air sample were positive. | LOD and recovery not reported. 1/42 surface samples positive. |
| Mouchtouri et al. (2020), Greece | Three isolation wards and a long-term care facility. | Samples collected with a MD8 sampler on gelatine filters at 50 L min−1. Volume 0.5 m3. | RT-PCR not clear the targeted genes. | 1/12 sample positive at 2.5 m from patient without mask. | LOD and recovery not reported. 3/35 surface samples positive |
|
Nor et al. (2021) Malaysia |
4 hospital wards hosting Covid-19 patients in Kuala Lampur. | PM2.5 samples collected with a Minovol sampler on glass microfiber filters at 5 L min−1. Volume 14.4 m3. | RT-qPCR targeting N1 and N2 genes. | Positive samples were observed in 2/4 wards. | LOD and recovery not reported. |
|
Razzini et al. (2020) Italy |
Covid-19 isolation ward of a hospital in Milan. | Samples collected with a MD8 sampler on gelatin filters at 50 L min−1. Volume 2 m3. | RT-PCR not clear the targeted genes. | 2/5 samples were positive. | LOD and recovery not reported. 9/37 surface samples positive. |
|
Tan et al. (2020) China |
Covid-19 isolation wards and ICUs in Wuhuan. | Air samples collected at 5 L min-1. Volume 0.3 m3 at <1 m from patients plus samples in clean areas. | RT-PCR targeting ORF1ab gene. | 1/12 sample positive taken during intubation. 0/17 in clean area were positive. |
LOD and recovery not reported. 11/341 surface samples positive. |
Table 3.
Summary of the methodology used and of the results found in different datasets focused on detecting SARS-CoV-2 genetic material (RNA), with quantification of concentrations in indoor sites in hospitals and healthcare setting in which were present COVID-19 patients.
| Reference | Sites | Sampling | Method | Results for air samples | Notes |
|---|---|---|---|---|---|
|
Chia et al. (2020) Singapore |
3 AIIRs in the ICU and 27 AIIRS in the wards of a hospital. | 6 NIOSH BC251 bioaerosol samplers at 3.5 L min−1. Volume 5.04 m3. | RT-PCR targeting E, ORF1ab genes. | 4/10 air samples positive, D > 1 μm. Conc. 916–2000 copies m−3. | Recovery not reported. 56/245 surface samples positive. |
| Dumont-Leblond et al. (2020), Canada | Acute care hospital rooms in Quebec. | Two plastic IOM (SKC) samplers with gelatine or polycarbonate filters at 10 L min−1. SASS 3100 dry sampler at 300 L min−1. Volumes 2.4–10.8 m3. | RT-qPCR targeting ORF1b gene. | 11/100 air samples positive. Conc. 10–514 copies m−3. |
Recovery not reported. |
|
Feng et al. (2021) China |
Hospital in Zhejiang. | NIOSH sampler at 3.5 L min−1 at 0.2 m from the bed of patients (head position). Volume 0.105 m3. | RT-qPCR but not clear the gene target. | 1/12 air sample positive. Conc. 1857 copies m−3. |
Recovery not reported. 4/202 surface samples positive. |
|
Guo et al. (2020) China |
Hospital: intensive care unit (ICU) and a general COVID-19 Ward in Wuhuan. |
Samples collected with SASS 2300 wetted cyclone sampler at 300 L min−1 on viral transport medium. Volume 9 m3. | qRT-PCR targeting ORF1ab, N genes. | 4/81 positive in ICU. 0/38 positive in general ward. Conc. 520–3800 copies m−3. |
Recovery not reported. 23/161 surface positive in ICU. 2/134 surface positive in general ward. |
|
Habibi et al. (2021) Kuwait |
Three major hospitals in Kuwait dealing with Covid-19 patients. | Sampling at 30 L min-1 in wash bottles with TRIzol (APB Bioscience). Volume 3.6 m3. | RT-qPCR targeting ORF1ab, N genes. | 5/13 air samples positive. Conc. 12–99 copies m−3. |
Recovery not reported. |
|
Hu et al. (2020) China |
Various sites in different health facilities of Wuhan. | Centrifugal sampler WA-400 at 400 L min−1 in PBS. Volumes 12 m3. | qRT-PCR targeting ORF1ab gene. | 9/81 air samples positive. Conc. 1110–11,200 copies m−3. |
Recovery not reported. Virus cultures were negative. |
|
Lednicky et al. (2020a) USA |
Student health care centre in Florida for Covid-19 patients. | VIVAS sampler on PBS at 6.5 L min−1. Volume 0.39 m3. | RT-PCR targeting N gene. | 1/2 air sample positive. Conc. 870 copies m−3. |
LOD 37.5 copies μL−1. Recovery not reported. Virus cultures were negative. |
|
Lednicky et al. (2020b) USA |
A two patient room in a hospital in Florida with 6 air exch. h−1. | VIVAS sampler and a BioSpot-VIVAS BSS300P on PBS at 8 L min−1. Volume 1.44 m3. | RT-PCR targeting N gene. | 4/4 air samples positive. Conc. 1600–94,000 copies m−3. |
LOD 37.5 copies μL−1 (liquid phase). Recovery not reported. One sample showed positive virus culture. |
|
Liu et al. (2020) China |
Different sites in three hospitals of Wuhuan. | TSP sampled on gelatine substrate at 5 L min−1. 3 size-segregated samples. Volumes 1.5–5 m3. | dd-PCR targeting Orf1ab and N genes. | 16/25 samples positive. Conc. 1–42 copies m−3. |
Recovery not reported. |
|
Moore et al. (2021) UK |
8 hospitals, different locations (11 AIIRS), 11 neutral pressure side rooms, six ICU/HDU, open cohorts and 12 non-ICU sites. | Coriolis sampler at 300 L min−1 in PBS and a MD8 sampler in gelatine filters at 50 L min−1. Volumes 0.5–3 m3. <1 m from patients. |
qRT-PCR targeting N gene. | 4/55 samples positive with Coriolis sampler. 0/34 samples positive with MD8 Conc. 10–460 copies m−3. |
Recovery not reported. 30/336 surface samples positive. Virus cultures were negative. |
|
Ong et al. (2021) Singapore |
AIIRs and a community isolation facility (CIF). | Samples using a BioSpot-VIVAS BSS300-P at 8 L min-1 not clear sampling time. | qRT-PCR targeting E, ORF1ab genes. | 6/12 positive samples in AIIRs conc. 179–2738 copies m−3. 1/9 positive sample in CIF, conc. 978 copies m−3. |
Recovery not reported. |
|
Passos et al. (2021) Brazil |
2 hospitals in the area of Belo Horizonte. | Different low and high volume samplers on cellulose, quartz, and PTFE filters. Volumes 0.12–250 m3. | RT-PCR targeting N1 and N2 genes. | 3/33 samples positive. Conc. 0.14–0.33 copies m−3. |
Recovery ~100% 0/5 surface samples positive. |
|
Santarpia et al. (2020) USA |
Rooms and hallways of quarantine and isolation care areas in Nebraska. | MD8 sampler on gelatine filters at 50 L min−1. Volume 0.75 m3. | RT-PCR targeting E gene. | 12/19 samples in rooms positive. Conc. 2420–8340 copies m-3. 14/24 hallway samples positive. Conc. 2080–8690 copies m−3. |
LOD 5 copies μL−1 (liquid phase). Recovery not reported. 60/74 surface samples positive. Cultivation of virus was tried but not confirmed. |
|
Stern et al. (2021a) USA |
Different sites at a hospital in Boston. | Cascade impactor, 3 stages (<2.5 μm, 2.5–10 μm, >10 μm) using polyurethane foam and glass fibre filters (for PM2.5) at 5 L min−1, volume 14.4 m3. | RT-qPCR targeting N gene. | 8/90 air samples positive. Conc. 5–51 copies m-3. |
Recovery not reported. Positive samples span all size fraction. |
|
Stern et al. (2021b) Kuwait |
A Covid-19 hospital and a temporary quarantine facility (TQF). | Cascade impactor, 3 stages (<2.5 μm, 2.5–10 μm, >10 μm) using polyurethane foam and glass fibre filters (for PM2.5) at 5 L min−1, volume 14.4 m3. | RT-qPCR targeting N gene. | 8/98 air samples positive in hospital. Conc. 8–25 copies m−3. 0/39 air samples positive at TQF. |
Recovery not reported. Positive samples span all size fraction. |
|
Zhou et al. (2020a) UK |
7 clinical areas (Covid-19), a public area of hospital in London. | Coriolis sampler in 5 mL DMEM, volume 1 m3. | RT-PCR targeting E gene. | 2/31 samples positive. Conc. 404–7048 copies m−3. |
Recovery not reported. 23/218 surface samples positive. |
|
Zhou et al. (2021a) China |
4 hospitals with natural ventilation in Wuhan. | Air-nCOV-Watch samplers (impingers) at 15 L min-1 and 400 L min-1 on virus sampling liquid. Volumes 0.6–16 m3. | RT-PCR targeting ORF1ab, N genes and dd-PCR. | 3/44 air samples positive. Conc. 9–219 copies m−3. |
Recovery not reported. 10/318 surface samples positive. |
It is interesting to observe that several studies in this category of sites also provided measurements of surface collected (swabs) samples. The datasets having at least one positive air sample are 60.3% of the total, instead, for surface samples the datasets reporting positive samples are 89.5% (i.e. 34 out of 38). This means that, in this category of indoor environments, SARS-CoV-2 is found more frequently on surface rather than in air. There are 15 studies (Table 2, Table 3) in which air samples tested negative, but traces of SARS-CoV-2 were found on different surfaces; on the contrary only one paper (Jin et al., 2020) reports one positive sample in air and all negative samples on surfaces. Considering that surface stability of SARS-CoV-2 on different kind of surfaces could be of several hours, arriving up to a few days (van Doremalen et al., 2020; Marquès and Domingo, 2021), indirect transmission of the virus through contaminated surfaces (i.e. fomites), in these indoor environments, cannot be ruled out. This could happen by touching contaminated surfaces and objects followed by touching the mouth, nose, or eyes. However, it is extremely difficult, in real cases, to ascertain the mechanism of transmission and the exact role of transmission via fomites in the spread of COVID-19. Epidemiological investigations, or structured analysis (like randomized controlled trials), are not feasible to investigate the role of fomites transmission because fomite-mediated contagions are likely a rare event and it is difficult to decouple from other, more likely, transmission routes (Pitol and Julian, 2021). Different studies indicate that the risks of SARSCoV-2 infection from contact with a fomite are estimated to be low (Marquès and Domingo, 2021; Pitol and Julian, 2021; Zhou et al., 2021b). In addition, these risks could be furtherly reduced by washing hands and with regular disinfection practices of indoor surfaces (Gonçalves et al., 2021; Marquès and Domingo, 2021).
The difference between air and surface samples results could originate from several factors. One factor is that surfaces could be contaminated also by direct contact from infected individuals and not only from respiratory aerosol. A second factor is that virus-laden particles that deposit on surfaces are large respiration droplets that could have a different viral load compared to the fine and ultrafine particles remaining in suspension. Different studies suggest that pathogens are typically enriched in specific size ranges, according to the particle formation mechanisms and sites, in relation to the site of infection. Thus, expiration particles may carry higher viral load if the site of infection is the same or very close to the site of particle formation (Pöhlker et al., 2021).
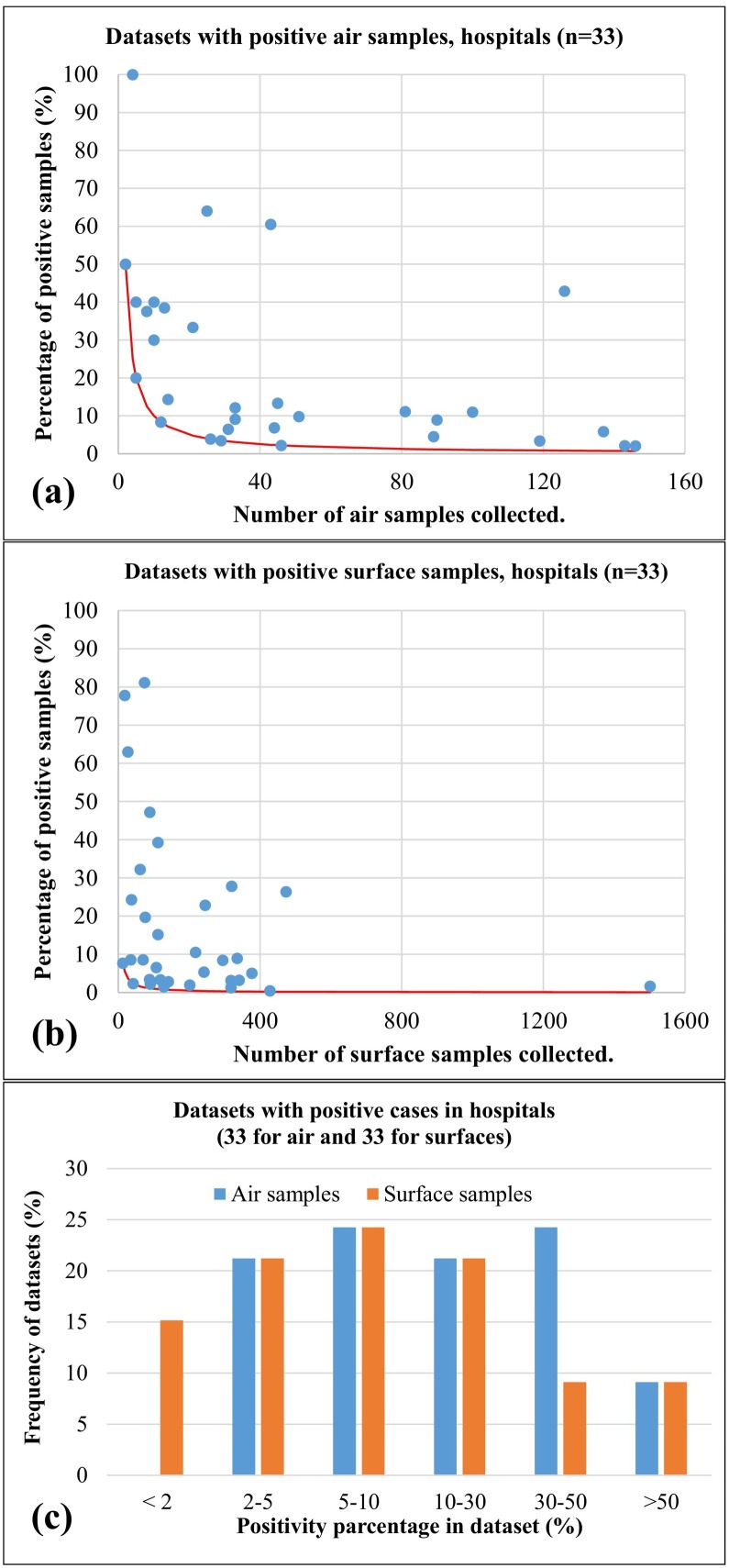
The different datasets showing positive air samples and/or positive surface samples, were analysed to investigate the positivity rate as function of the number of samples available in the dataset (Fig. 4 ). Results show a quite large variability for both kind of environmental samples, especially for cases with small number of samples, reflecting variabilities in source strength (i.e. number of infected people and their viral load), in geometries and ventilations of the different environments, and in the mitigation policies applied. The average positivity rate, considering all available datasets, was 22.8% (median 11.1%) for air samples and it was 12.4% (median 8.5%) for surface samples. This indicate that, even if contamination of surface samples is more frequent compared to air samples, the average positivity rate is lower in surface compared to air. It must be said that there are relevant differences in the total number of air and surface samples because, in the different datasets, the number of air samples is much lower than that of surface samples. This could create a bias in the evaluation of positivity rate because, for example, when only two samples are present the minimum positivity rate is 50% (i.e. one of the two positive). The red curves in Fig. 4a and b report the minimum number as function of the total number of samples in the dataset. If all air samples of Fig. 4a are considered together as if they was a single dataset, the positivity rate would be 13.3% against the value of 10% for the surface samples taken together. This suggest that positivity rates in air and surface samples could be more similar in datasets having comparable number of samples.
Fig. 4.
Positivity rates as function of the total number of samples collected for the different datasets having positive samples collected in hospitals, care facilities, and quarantine areas. (a) refers to air samples; (b) refers to surface (swab) samples; (c) comparison of frequency distributions of positivity rates for the datasets of air and surface samples collected in hospital and care facilities. Red continuous lines represent the minimum positivity rate (i.e. one positive sample) as a function of the total number of samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4c shows the comparison of the frequency distributions of positivity rate for air and surface samples collected in hospital and care facilities. This shows that the larger positivity rate in air samples is due to two aspects: the absence of datasets with low positivity rate (i.e. < 2%) in air samples that are instead present in surface samples; a significantly larger number of air samples datasets having positivity rate in the range 30%–50% compared to surface samples.
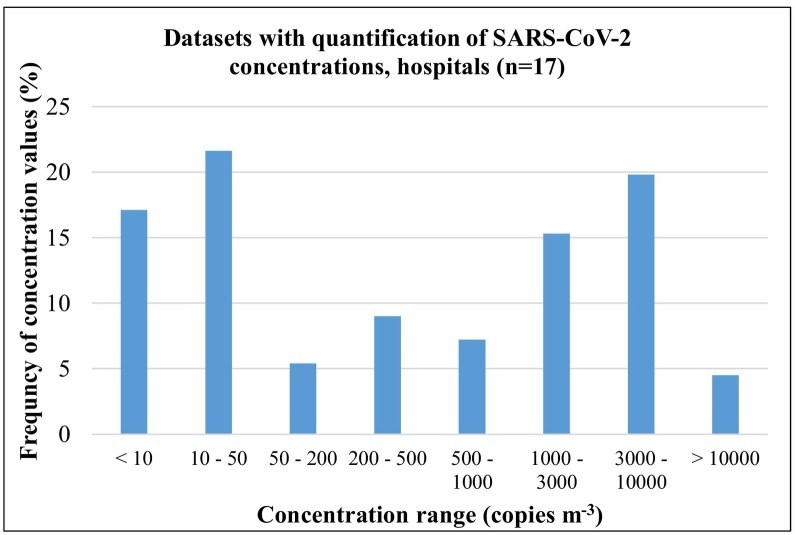
The concentrations of SARS-CoV-2 RNA traces in air samples in these environments observed in the 17 datasets having quantification, span a quite large range: 0.1–94,000 copies m−3 with an average value of 3606 copies m−3. The median value of the samples collected in all available dataset was 540 copies m−3 (17.5–2890 copies m−3 was the inter-quartile range between 25th and 75th percentiles). The frequency distribution of measured concentrations is reported in Fig. 5 . The histogram shows essentially three peaks: a large one at low concentrations <50 copies m−3, a second peak (less frequent) centred in the range 200–500 copies m−3, and a third peak at larger concentrations in the range 1000–10,000 copies m−3. These values indicate a large variability, as expected, mainly of the variability in the intensity of the sources (i.e. number of infected people indoor and their viral load), of the different sizes and ventilation conditions of the indoors environment studied. Dubey et al. (2021) collected, in a systematic way, air samples in hospital settings at two distances from patients: 1 m and 3 m. They observed that at 1 m from patient the positivity rate was 94.4% but it decreased at 22.2% at 3 m distance. This suggests that distance of sampling from infected individuals could also be an issue in comparison of different studies as well as in the risk of airborne transmission of contagion. As a consequence, the physical distancing among people is a measure likely efficient to be maintained for reducing risks of airborne transmission.
Fig. 5.
Frequency distribution of SARS-CoV-2 concentrations measured in air samples in the different datasets referring to hospital and care facilities sites.
A direct correlation, among all the datasets studied, between sampling method (volumes and typologies of substrates used) and positivity rate was not observed similarly to the results discussed for outdoor datasets. However, Dubey et al. (2021) compared systematically air samples collected at different flow rates (for the same duration) giving three different sampling volumes: 0.09 m3, 1 m3, and 1.6 m3. Samples were collected at distances between 1 m and 3 m from patients in a hospital setting. Results show that positivity rate increases when sampling volume increase, going from 28.6% (samples of 0.09 m3) to 45.2% (samples of 1 m3) and reaching 54.8% for samples of 1.6 m3. This suggests that LOD and recovery could be an issue for estimates of positivity rate, especially if low sampling volumes are used. As a consequence, it would be necessary to include estimates of these parameters in the future studies reinforcing the necessity to develop a standard methodology for collection of air sampling devoted to detection/quantification of SARS-CoV-2 RNA. Likely other dedicated studies are necessary to define a protocol, however, the results analysed in this work seems to suggest that a standard operating procedure should include: an evaluation of recovery in the effective sampling conditions; an optimisation (also considering the recovery value) of the sampling substrate, sampling flow, and sampling volume in order to achieve a LOD as low as 1–2 copies m−3; low-temperature conservation of collected samples before the analysis and some results indicate that −25 °C or lower could be suitable (Conte et al., 2021).
It is important to mention that the detection of RNA traces of the SARS-CoV-2 virus does not imply infectivity of these airborne virus-laden particles. Some of the studies, summarized in Table 2, Table 3, tried to perform cultures of the SARS-CoV-2 in collected positive samples, both in air and on surfaces, to have a better insights of the eventual vitality. Infectious virus particles were not observed in environmental (air and/or surface) samples collected and analysed in most of the available studies (Binder et al., 2020; Döhla et al., 2020; Dumont-Leblond et al., 2020, Dumont-Leblond et al., 2021; Hu et al., 2020; Moore et al., 2021). Santarpia et al. (2020) attempted virus culture but, given the low concentrations found in collected samples, cultivation of virus was not successful. Kotwa et al. (2021) investigated viability of positive samples (Ct < 34 on PCR analysis) and found that none of the positive air samples yielded viable virus, however, viable virus was observed in 6 out of 36 surface samples cultured. Virus cultures done in samples collected at a COVID-19 University health care facility in Florida were negative (Lednicky et al., 2020a) but resulted positive the cultures done in samples collected at a hospital in Florida with viable viral concentrations between 6000 and 74,000 TCID50 m−3 (Median Tissue Culture Infectious Dose) (Lednicky et al., 2020b). Ben-Shmuel et al. (2020) used 97 positive air and surface samples to test viability of SARS-CoV-2 and none were found to contain infectious titres of the virus. These results suggest that cultural viable virus is present in a limited fraction of positive air samples, however, there is need of additional studies before to reach a robust conclusion on this aspect.
3.3. Results of measurements in community indoor sites
The summary of results obtained from the 10 datasets dealing with measurements of airborne SARS-CoV-2 RNA traces in community indoor environments are reported in Tables 4 together with details of sampling methods used and results found. A fraction of 30% of available datasets includes at least one positive air sample; this fraction increases up to 67% when surface samples are considered. This means that contamination from SARS-CoV-2 RNA is found more frequently over surfaces rather than on air samples similarly to what has been observed for hospitals and healthcare settings. The positivity rate for air samples in community indoors ranges from 11.1% (de Rooij et al., 2021a) to 64.3% (Hadei et al., 2021). The positivity rate for surface samples ranges between 3% (de Rooij et al., 2021b) up to 42.2% (Moreno et al., 2021). Therefore, even if contamination is found more frequently on surfaces, the positivity rates are lower on surfaces compared to air samples, exactly the same results obtained also for hospitals and healthcare settings. In addition, the positivity rates found for air and surfaces in indoor community environments are similar to those observed in hospitals and healthcare settings.
Table 4.
Summary of the methodology used and of the results found in different datasets focused on detecting SARS-CoV-2 genetic material (RNA), in different community indoor sites.
| Reference | Sites | Sampling | Method | Results for air samples | Notes |
|---|---|---|---|---|---|
|
Conte et al. (2021) Italy |
2 supermarket, 1 train station, 1 canteen, 1 commercial centre, 1 pharmacy, 1 hair salon in different cities in north, central, and south Italy. | PM10 sampling at 38.3 L min−1 on quartz fibre filters, volumes 22.9–29.7 m3. TSP sampling at 13.4 L min−1 and at 61.7 L min−1 on quartz filters, volumes 6.2–9.3 m3. |
RT-PCR targeting RdRp, N, E genes. dd-PCR targeting RdRp and RNAse P genes. |
All of 56 PM10 samples tested negative. All of 13 TSP samples tested negative. |
LOD 1.3 copies m−3 for PM10 and 4 copies m−3 for TSP. Recovery 54%. |
|
de Rooij et al. (2021a) Netherlands |
Measurements taken inside different mink farms | Sampling of inhalable dust (at 3.5 L min−1) on Teflon filters. Volume 1.26 m3. | RT-qPCR targeting E gene. | 3/27 air samples positive. Conc. 2400–4900 copies m−3. |
LOD 10–28 copies m−3. Recovery not reported. |
|
de Rooij et al. (2021b) Netherlands |
Measurements in a meat processing plant experiencing COVID-19 clusters. | Inhalable dust sampled on Teflon filters at 3.5 L min−1. Volume 1.266 m3. | RT-qPCR targeting RdRp and E genes. | All of 14 air samples tested negative. | LOD 3.2 copies/reaction. Recovery not reported. 6/203 surface samples positive. |
|
Di Carlo et al. (2020) Italy |
A bus operating in Chieti (central Italy) in May 2020. | Sampler (AMS Analitica) on gelatine filters at 24 L min−1. Volume 18.72 m3. | RT-PCR targeting ORF1ab, N, S genes. | All of 14 air samples tested negative. | LOD and recovery not reported. 0/90 surface samples positive. |
|
Habibi et al. (2021) Kuwait |
Samples in different indoors at the Kuwait Institute of Research. | Sampling at 30 L min-1 in wash bottles with TRIzol (APB Bioscience). Volume 3.6 m3. | RT-qPCR targeting ORF1ab, N genes. | All of 5 air samples tested negative. | Recovery not reported. |
|
Hadei et al. (2021) Iran |
Public sites in Theran: 3 banks, shopping centres, post office and a governmental building, airport, subway station and train, bus. | One TSP AV100 sampler at 40 L min-1 on PTFE filters, volumes 1.27–3.5 m3. A SKC pump at 3.5 L min−1 for bus and train sampling, volumes 0.2–0.24 m3. | RT-PCR targeting ORF1ab and N genes. | 18/28 air samples positive. | Recovery 20%. |
|
Moreno et al. (2021) Spain |
Subway trains and buses in Barcelona. | PM2.5 sampler at 10 L min−1 on Teflon filters. Volumes 5.2–6.3 m3. | RT-qPCR targeting RNA polymerase (IP2, IP4) and N genes. | 2/6 subway air samples positive. Conc. 18.8–23.4 copies m−3. 1/6 bus air samples positive. Conc. 1.44 copies m−3. |
6/15 surface samples in subway positive. 13/30 surface samples in buses positive. |
|
Viegas et al. (2021) Portugal |
Ten higher education facility in Lisbon area. | Coriolis μ sampler at 300 L min−1 on 5 mL vial with buffer NVL. Volume 0.6 m3. | RT-PCR not reported the targets. | All of 48 air samples tested negative. | LOD and recovery not reported. 0/106 surface sample positive. |
|
Xie et al. (2020) China |
2 houses with confirmed Covid-19 patients in a residential building in Guangzhou. | Not specified details of the sampling approach used. | RT-PCR not clear the target genes. | All of 3 air samples tested negative. | LOD and recovery not reported. 1/31 surface sample positive (door handle). |
|
Yamagishi et al. (2020) Japan |
7 cabins of the Diamond Princess cruise ship. | 2 MD8 samplers at 50 L min-1 on gelatine filters. Volume 1 m3. | rRT-PCR not clear the target genes. | All of 14 air samples tested negative. | LOD and recovery not reported. 58/587 surface samples positive. |
Concentrations were quantified in 2 out of the 3 datasets having positive air samples and showed important differences due to the same causes already mentioned for hospitals and healthcare setting. An average concentration of 14.5 copies m−3 was found by Moreno et al. (2021) and an average of 3700 copies m−3 was reported by de Rooij et al. (2021a). The average of all available samples was about 1857 copies m−3. This suggests that the expected average concentrations in indoor community environments are larger than those typically found in outdoor and lower than those found in hospitals and healthcare settings even if the number of datasets in indoor community environments is quite limited and further studies are needed to confirm this conclusion.
The analysis done in public transport in Barcelona (Moreno et al., 2021) showed that positivity rate and concentrations in air samples are larger for subways compared to surface buses; instead, positivity rates for surface samples are comparable. The results found in a bus in central Italy (Di Carlo et al., 2020) during the first wave of pandemic showed all negative air and surface samples interpreted as a possible consequence of strict measures against COVID-19 spread applied. Hadei et al. (2021) performed logistic regressions between positivity rates and other parameters such as the number of people present during sampling, the percentage of use of facemasks, air temperature, and volumes of indoor sampling sites finding no statistically significant correlations.
The results of positivity rates and concentrations found in the datasets collected in community indoor environments, even if they are limited in numbers, suggest that there could be an intermediate risk of airborne transmissions between outdoor and hospital and health care. In additions, most of the study suggests that volumes and ventilations of indoor environments and the use of facemasks are factors strongly influencing the presence of and concentrations of SARS-CoV-2 RNA in these environments and, consequently, influencing the risks of airborne transmission in general public.
4. Conclusions
This work reviewed 73 papers dealing with identification/quantification of SARS-CoV-2 genetic material in air by means of active sampling and PCR detection, published since the start of pandemic and until 31/08/2021. Selected papers furnished 77 datasets in different environments. Only 9 datasets (11.8% of the total) were obtained in outdoor sites and all of them were collected during the first wave of pandemic in 2020. The remaining datasets were collected in hospitals and healthcare settings (75.1% of the total) and in community indoor environments (13.1% of the total).
The fraction of datasets having at least one air sample positive to SARS-CoV-2 was 33.3% in outdoor, comparable to the 30% observed in community indoors and significantly lower than the value of 60.3% observed in hospitals and healthcare settings. The average positivity rate of air samples (i.e. the ratio between positive and total samples) was lower in outdoor (17.6%) compared to indoor (23.7%) sites. Among the indoor sites, the positivity rates in hospitals and healthcare settings varied in an interval comparable with that of indoor community sites.
Several of indoor studies also included datasets of surface samples (swabs collected on different indoor surfaces). The fraction of datasets having at least one positive surface sample were larger than the fractions observed for air samples in both typologies of sites (i.e. hospitals and community indoors). Despite the fact that contaminated surfaces were found more frequently than contamination in air samples, the positivity rate was lower on indoor surfaces (12.4% in hospitals and 9.7% in community indoors) compared to that of indoor air (23.7%), even if these numbers are calculated using a significantly different number of samples. This suggests that indirect SARS-CoV-2 transmission (i.e. via fomites) could not be ruled out even if its role in the spread of contagion is not actually clear and quantified. However, measures such as increased frequency of disinfection of indoor surfaces and washing hands could be useful to reduce these risks and should be maintained during the next phases of pandemic.
The concentrations observed in outdoor are relatively low, compared to those observed in indoor sites, and positive samples are mainly observed near sources (i.e. crowds or hospital settings). This suggest that risks of airborne transmission in outdoor sites are relatively limited providing that physical distance is maintained avoiding crowds and vicinity to potentially large sources. Concentration in hospital and healthcare setting shows a large variability with average values in the different studies covering five orders of magnitude. This suggests a larger level of risk compared to outdoor and a strong influence of source (i.e. number of infected individuals) but also of volumes of rooms, distances from patients, and ventilation rates. A limited number of studies report concentrations for indoor community environments, that are likely the sites that needs to be further investigated in the next future, however, the concentration in these sites seems to be, on average, larger than in outdoors and lower than in hospital and healthcare settings. The studies that tried to culture SARS-CoV-2 from positive samples (air and surface samples) are still relatively limited; it seems to suggest that in the majority of cases airborne and surface virions are likely not viable, however, it would be advisable to have additional studies on this aspect.
There is not yet a standard methodology of sampling, conservation, and analysis of collected samples; however, datasets with positive samples were obtained from samples on different typologies of filters, on gelatine substrates as well as datasets with all negative samples. A direct correlation, among all the datasets studied, between sampling method (volumes and typologies of substrates used) and positivity rate was not thereby observed neither in outdoor nor in indoor sites. However, Dubey et al. (2021) show that positivity rate increases when sampling volume increase. This suggests that LOD and recovery could be an issue for estimates of positivity rate and it would be necessary to include estimates of these parameters in the future studies. In addition, this work reinforces the usefulness of developing a standard methodology for collection of air samples devoted to detection/quantification of SARS-CoV-2 RNA.
The results of positivity rates and concentrations found in community indoor environments, even if the number of datasets is limited, suggest that there could be intermediate risks of airborne transmissions between outdoor and hospital and healthcare settings. The risk appears strongly related to volumes and ventilations of indoor environments and the use of facemasks, both factors influencing the presence/concentrations of SARS-CoV-2 RNA. Therefore, also these mitigation measures should be maintained in the next phase of pandemic for indoor environments.
Despite the uncertainty on the viral load in positive subjects (which depends not only from subject to subject, but also from the lung region where the infection is taking place and its time course) and the difficulties due to the different sampling methods used, this review allowed a statistical evaluation of the presence of the SARS-CoV-2 in air, of the positivity rates, and of detected concentrations, mediated on the different contexts: outdoor, indoor (hospitals and similar) and indoor (living environments). These quantifications (including ranges of variability) represent an important input for epidemiological models (both outdoor and indoor) to estimate the risk of virus transmission, providing the scientific community with summarized data useful for risk assessment and not just for behavioral indications.
CRediT authorship contribution statement
D. Contini, A. Gambaro, F. Belosi, G. La Salandra conceptualized the study design; E. Barbaro, E. Gregoris, M. Feltracco, M. Conte, A. Dinoi, S. Trabucco collected published material and collaborated for statistical analysis; D. Chirizzi, G. Ciccarese, G. and G. La Bella collaborated to investigate protocols used in the different datasets. All authors collaborated to interpretation of results, wrote, read, commented, and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study did not receive specific funding and was performed within the cooperation project AIR-CoV (Evaluation of the concentration and size distribution of SARS-CoV-2 in air in outdoor and indoor environments).
Editor: Jianmin Chen
References
- Ahn J.Y., An S., Sohn Y., Cho Y., Hyun J.H., Baek Y.J., Kim M.H., Jeong S.J., Kim J.H., Ku N.S., Yeom J.S., Smith D.M., Lee H., Yong D., Lee Y.J., Kim J.W., Kim H.R., Hwang J., Choi J.Y. Environmental contamination in the isolation rooms of COVID-19 patients with severe pneumonia requiring mechanical ventilation or high-flow oxygen therapy. J. Hosp. Infect. 2020;106(3):570–576. doi: 10.1016/j.jhin.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.G., Marr L.C. Recognizing and controlling airborne transmission of SARSCoV-2 in indoor environments. Indoor Air. 2020;30:557–558. doi: 10.1111/ina.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand U., Adelodun B., Pivato A., Suresh S., Indari O., Jakhmola S., Chandra Jha H., Jha P.K., Tripathi V., Di Maria F. A review of the presence of SARS-CoV-2 RNA in wastewater and airborne particulates and its use for virus spreading surveillance. Environ. Res. 2021;196 doi: 10.1016/j.envres.2021.110929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadi S., Bouvier N., Wexler A.S., Ristenpart W.D. The coronavirus pandemic and aerosols: does COVID-19 transmit via expiratory particles? Aerosol Sci. Technol. 2020;54(6):635–638. doi: 10.1080/02786826.2020.1749229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baboli Z., Neisi N., Babaei A.A., Ahmadi M., Sorooshian A., Birgani Y.T., Goudarzi G. On the airborne transmission of SARS-CoV-2 and relationship with indoor conditions at a hospital. Atmos. Environ. 2021;261 doi: 10.1016/j.atmosenv.2021.118563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri P., Zupin L., Licen S., Torboli V., Semeraro S., Cozzutto S., Palmisani J., Di Gilio A., de Gennaro G., Fontana F., Omiciuolo C., Pallavicini A., Ruscio M., Crovella S. Molecular detection of SARS-CoV-2 from indoor air samples in environmental monitoring needs adequate temporal coverage and infectivity assessment. Environ. Res. 2021;198 doi: 10.1016/j.envres.2021.111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belosi F., Conte M., Gianelle V., Santachiara G., Contini D. On the concentration of SARS-CoV-2 in outdoor air and the interaction with pre-existing atmospheric particles. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shmuel A., Brosh-Nissimov T., Glinert I., Bar-David E., Sittner A., Poni R., Cohen R., Achdout H., Tamir H., Yahalom-Ronen Y., Politi B., Melamed S., Vitner E., Cherry L., Israeli O., Beth-Din A., Paran N., Israely T., Yitzhaki S., Levy H., Weiss S. Detection and infectivity potential of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) environmental contamination in isolation units and quarantine facilities. Clin. Microbiol. Infect. 2020;26(12):1658–1662. doi: 10.1016/j.cmi.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder R.A., Alarja N.A., Robie E.R., Kochek K.E., Xiu L., Rocha-Melogno L., Goli S.V., Farrell A.S., Coleman K.K., Turner A.L., Lautredou C.C., Lednicky J.A., Lee M.J., Polage C.R., Simmons R.A., Deshusses M.A., Anderson B.D., Gray G.C., Abdelgadir A. Environmental and aerosolized severe acute respiratory syndrome coronavirus 2 among hospitalized coronavirus disease 2019 patients. J. Inf. Dis. 2020;222:1798–1806. doi: 10.1093/infdis/jiaa575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgand G., Peiffer-Smadja N., Fournier S., Kerneis S., Lescure F., Lucet J.C. Assessment of air contamination by SARS-CoV-2 in hospital settings. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges J.T., Nakada L.Y.K., Maniero M.G., Guimarães J.R. SARS-CoV-2: a systematic review of indoor air sampling for virus detection. Environ. Sci. Pollut. Res. Int. 2021;28:40460–40473. doi: 10.1007/s11356-021-13001-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borouiba L. Turbulent gas clouds and respiratory pathogen emissions, potential implications for reducing transmission of COVID-19. JAMA. 2020;323(18):1837–1838. doi: 10.1001/jama.2020.4756. [DOI] [PubMed] [Google Scholar]
- Buonanno G., Stabile L., Morawska L. Estimation of airborne viral emission: quanta emission rate of SARS-CoV-2 for infection risk assessment. Environ. Int. 2020;141 doi: 10.1016/j.envint.2020.105794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Wu X., Zhang Y., Xia J., Li M., Feng Y., Yu X., Duan J., Weng X., Chen Y., Cheng Z., Zhan Q. 2020. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Contamination in Air and Environment in Temporary COVID-19 ICU Wards. PREPRINT (Version 1) available at Research Square. [DOI] [Google Scholar]
- Cheng V.C.C., Wong S.C., Chen J.H.K., Yip C.C.Y., Chuang V.W.M., Tsang O.T.Y., Sridhar S., Chan J.F.W., Ho P.L., Yuen K.Y. Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect. Control Hosp. Epidemiol. 2020;41(5):493–498. doi: 10.1017/ice.2020.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V., Wong S.C., Chan V.W., So S.Y., Chen J.H., Yip C.C., Chan K.H., Chu H., Chung T.W., Sridhar S., To K.K., Chan J.F., Hung I.F., Ho P.L., Yuen K.Y. Air and environmental sampling for SARS-CoV-2 around hospitalized patients with coronavirus disease 2019 (COVID-19) Infection Control Hosp. Epidemiol. 2020;41(11):1258–1265. doi: 10.1017/ice.2020.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrie J.W., Cherrie M.P.C., Davis A., Holmes D., Semple S., Steinle S., MacDonald E., Moore G., Loh M. Contamination of air and surfaces in workplaces with SARS-CoV-2 virus: a systematic review. Ann. Work Expo Health. 2021 doi: 10.1093/annweh/wxab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P.Y., Coleman K.K., Tan Y.K., Ong S.W.X., Gum M., Lau S.K., Lim X.F., Lim A.S., Sutjipto S., Lee P.H., Son T.T., Young B.E., Milton D.K., Gray G.C., Schuster S., Barkham T., De P.P., Vasoo S., Chan M., Ang B.S.P., Tan B.H., Leo Y.S., Ng O.T., Wong M.S.Y., Marimuthu K. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020;11:2800. doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirizzi D., Conte M., Feltracco M., Dinoi A., Gregoris E., Barbaro E., La Bella G., Ciccarese G., La Salandra G., Gambaro A., Contini D. SARS-CoV-2 concentrations and virus-laden aerosol size distributions in outdoor air in north and south of Italy. Environ. Int. 2021;146 doi: 10.1016/j.envint.2020.106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte M., Feltracco M., Chirizzi D., Trabucco S., Dinoi A., Gregoris E., Barbaro E., La Bella G., Ciccarese G., Belosi F., La Salandra G., Gambaro A., Contini D. Airborne concentrations of SARS-CoV-2 in indoor community environments in Italy. Environ. Sci. Pollut. Res. 2021 doi: 10.1007/s11356-021-16737-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contini D., Costabile F. Does air pollution influence COVID-19 outbreaks? Atmosphere. 2020;11:377. doi: 10.3390/atmos11040377. [DOI] [Google Scholar]
- de Rooij M.M.T., Hakze-Van der Honing R.W., Hulst M.M., Harders F., Engelsma M., van de Hoef W., Meliefste K., Nieuwenweg S., Munnink B.B.O., van Schothorst I., Sikkema R.S., van der Spek A.N., Spierenburg M., Spithoven J., Bouwstra R., Molenaar R.J., Koopmans M., Stegeman A., van der Poel W.H.M., Smit L.A.M. medRxiv; 2021. Occupational and Environmental Exposure to SARS-CoV-2 in and Around Infected Mink Farms. 2021.01.06.20248760. [DOI] [PubMed] [Google Scholar]
- de Rooij M.M.T., Sikkema R.S., Bouwknegt M., de Geus Y., Stanoeva K.R., Nieuwenweg S., van Dam A.S.G., Raben C., Dohmen W., Heederik D., Reusken C., Meijer A., Koopmans M.P.G., Franz F., Smit L.A.M. medRxiv; 2021. Potential Environmental Transmission Routes of SARS-CoV-2 Inside a Large Meat Processing Plant Experiencing COVID-19 Clusters. 2021.06.20.21259212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declementi M., Godono A., Mansour I., Milanesio N., Garzaro G., Clari M., Fedele L., Passini V., Bongiorno C., Pira E. Assessment of air and surfaces contamination in a COVID-19 non-intensive care unit. Med. Lav. 2020;111(5):372–378. doi: 10.23749/mdl.v111i5.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo P., Chiacchiaretta P., Sinjari B., Aruffo E., Stuppia L., De Laurenzi V., Di Tomo P., Pelusi L., Potenza F., Veronese A., Vecchiet J., Falasca K., Ucciferri C. Air and surface measurements of SARS-CoV-2 inside a bus during normal operation. PLoS ONE. 2020;15(11) doi: 10.1371/journal.pone.0235943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Qian H., Xu B., Huang Y., Miao T., Yen H.L., Xiao S., Cui L., Wu X., Shao W., Song Y., Sha L., Zhou L., Xu Y., Zhu B., Li Y. Toilets dominate environmental detection of severe acute respiratory syndrome coronavirus 2 in a hospital. Sci. Total Environ. 2021;753 doi: 10.1016/j.scitotenv.2020.141710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhla M., Wilbring G., Schulte B., Kümmerer B.M., Diegmann C., Sib E., Richter E., Haag A., Engelhart S., Hubinger A.M., Exner M., Streeck H., Schmithause R.M. medRxiv preprint; 2020. SARS-CoV-2 in Environmental Samples of Quarantined Households. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo J.L., Marquès M., Rovira J. Influence of airborne transmission of SARS-CoV-2 in COVID-19 pandemic.A review. 2020;188 doi: 10.1016/j.envres.2020.109861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey A., Kotnala G., Mandal T.K., Sonkar S.C., Singh V.K., Guru S.A., Bansal A., Irungbam M., Husain F., Goswami B., Kotnala R.K., Saxena S., Sharma S.K., Saxena K.N., Sharma C., Kumar S., Aswal D.K., Manchanda V., Koner B.C. Evidence of the presence of SARS-CoV-2 virus in atmospheric air and surfaces of a dedicated COVID hospital. J. Med. Virol. 2021;2021(93):5339–5349. doi: 10.1002/jmv.27029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont-Leblond N., Veillette M., Mubareka S., Yip L., Longtin Y., Jouvet P., Paquet Bolduc B., Godbout S., Kobinger G., McGeer A., Mikszewski A., Duchaine C. Low incidence of airborne SARS-CoV-2 in acute care hospital rooms with optimized ventilation. Emerg. Microbes Infect. 2020;9(1):2597–2605. doi: 10.1080/22221751.2020.1850184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont-Leblond N., Veillette M., Bhérer L., Boissoneault K., Mubareka S., Yip L., Dubuis M.E., Longtin Y., Jouvet P., McGeer A., Duchaine C. Positive no-touch surfaces and undetectable SARS-CoV-2 aerosols in long-term care facilities: an attempt to understand the contributing factors and the importance of timing in air sampling campaigns. Am. J. Infect. Control. 2021 doi: 10.1016/j.ajic.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker S., Hornick T., Szczepankiewicz G., Maier M., Bastl M., Bumberger J., Treudler R., Liebert U.G., Simon J.C. No SARS-CoV-2 detected in air samples (pollen and particulate matter) in Leipzig during the first spread. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridi S., Niazi S., Sadeghi K., Naddafi K., Yavarian J., Shamsipour M., Jandaghi N.Z.S., Sadeghniiat K., Nabizadeh R., Yunesian M., Momeniha F., Mokamel A., Hassanvand M.S., MokhtariAzad T. A field indoor air measurement of SARS-CoV-2 in the patient rooms of the largest hospital in Iran. Sci. Total Environ. 2020;725 doi: 10.1016/j.scitotenv.2020.138401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Xu K., Gu S., Zheng S., Zou Q., Xu Y., Yu L., Lou F., Yu F., Jin T., Li Y., Sheng J., Yen H.L., Zhong Z., Wei J., Chen Y. Multi-route transmission potential of SARS-CoV-2 in healthcare facilities. J. Hazard. Mater. 2021;402 doi: 10.1016/j.jhazmat.2020.123771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y., Pu Y., Liao C.H., Huang W.F., Zeng Q., Zhou H., Yi B., Wang A.M., Dou Q.Y., Zhou P.C., Chen H.L., Liu H.X., Xu D.M., Chen X., Huang X. Evaluation of the exposure risk of SARS-CoV-2 in different hospital environment. Sustain. Cities Soc. 2020;61 doi: 10.1016/j.scs.2020.102413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J., da Silva P.G., Reis L., Nascimento M.S.J., Koritnik T., Paragi M., Mesquita J.R. Surface contamination with SARS-CoV-2: a systematic review. Sci. Total Environ. 2021;798 doi: 10.1016/j.scitotenv.2021.149231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z.D., Wang Z.Y., Zhang S.F., Li X., Li L., Li C., Cui Y., Fu R.B., Dong Y.Z., Chi X.Y., Zhang M.Y., Liu K., Cao C., Liu B., Zhang K., Gao Y.W., Lu B., Chen W. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg. Infect. Dis. 2020;26(7):1583–1591. doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi N., Uddin S., Al-Salameen F., Al-Amad S., Kumar V., Al-Otaibi M., Abdul Razzack N., Shajan A., Shirshikar A. SARS-CoV-2, other respiratory viruses and bacteria in aerosols: report from Kuwait's hospitals. Indoor Air. 2021 doi: 10.1111/ina.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadei M., Mohebbi S.R., Hopke P.K., Shahsavani A., Bazazpour S., Alipour M.R., Jafari A.J., Bandpey A.M., Zali A., Yarahmadi M., Farhadi M., Rahmatinia M., Hasanzadeh V., Nazari S.S.H., Asadzadeh-Aghdaei H., Tanhaei M., Zali M.R., Kermani M., Vaziri M.H., Chobineh H. Presence of SARS-CoV-2 in the air of public places and transportation. Atmos. Pollut. Res. 2021;12:302–306. doi: 10.1016/j.apr.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemati S., Mobini G.R., Heidari M., Rahmani F., Soleymani Babadi A., Farhadkhani M., Nourmoradi H., Raeisi A., Ahmadi A., Khodabakhshi A., Sadeghi M., Bagheri M., Validi M., Taghipour S., Mohammadi-Moghadam F. Simultaneous monitoring of SARS-CoV-2, bacteria, and fungi in indoor air of hospital: a study on Hajar Hospital in Shahrekord,Iran. 2021;10:1–11. doi: 10.1007/s11356-021-13628-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan C.J., Spencer E.A., Brassey J., Plüddemann A., Onakpoya I.J., Evans D., Conly J.M., Jefferson T. SARS-CoV-2 and the role of airborne transmission: a systematic review. F1000Research. 2021;10:232. doi: 10.12688/f1000research.51590.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Lei C., Chen Z., Liu W., Hu X., Pei R., Su Z., Deng F., Huang Y., Sun X., Cao J., Guan W. Distribution of airborne SARS-CoV-2 and possible aerosol transmission in Wuhan hospitals,China. 2020;7:1865–1867. doi: 10.1093/nsr/nwaa250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishmatov A. “SARS-CoV-2 is transmitted by particulate air pollution”: misinterpretations of statistical data, skewed citation practices, and misuse of specific terminology spreading the misconception. Environ. Res. 2021;204 doi: 10.1016/j.envres.2021.112116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T., Li J., Yang J., Li J., Hong F., Long H., Deng Q., Qin Y., Jiang J., Zhou X., Song Q., Pan C., Luo P. SARS-CoV-2 presented in the air of an intensive care unit (ICU) Sustain. Cities Soc. 2020;65 doi: 10.1016/j.scs.2020.102446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayalar O., Arı A., Babuççu G., Konyalılar N., Doğan O., Can F., Şahin U.A., Gaga E.O., Kuzu S.L., Arı P.E., Odabaşı M., Taşdemir Y., Cindoruk S.S., Esen F., Sakın E., Çalışkan B., Tecer L.H., Fıçıcı M., Altın A., Onat B., Ayvaz C., Uzun B., Saral A., Döğeroğlu T., Malkoç S., Üzmez O.O., Kunt F., Aydın S., Kara M., Yaman B., Doğan G., Olgun B., Dokumacı E.N., Güllü G., Uzunpınar E.S., Bayram H. Existence of SARS-CoV-2 RNA on ambient particulate matter samples: a nationwide study in Turkey. Sci. Total Environ. 2021;789 doi: 10.1016/j.scitotenv.2021.147976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenarkoohi A., Noorimotlagh Z., Falahi S., Amarloei A., Mirzaee S.A., Pakzad I., Bastani E. Hospital indoor air quality monitoring for the detection of SARS-CoV-2 (COVID-19) virus. Sci. Total Environ. 2020;748 doi: 10.1016/j.scitotenv.2020.141324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U.J., Lee S.Y., Lee J.Y., Lee A., Kim S.E., Choi O.J., Lee J.S., Kee S.J., Jang H.C. Air and environmental contamination caused by COVID-19 patients: a multi-center study. J. Korean Med. Sci. 2020;35 doi: 10.3346/jkms.2020.35.e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klompas M., Baker M.A., Rhee C. Airborne transmission of SARS-CoV-2.Theoretical considerations and available evidence. 2020;324(5):441–442. doi: 10.1001/jama.2020.12458. [DOI] [PubMed] [Google Scholar]
- Kotwa J.D., Jamal A.J., Mbareche H., Yip L., Aftanas P., Barati S., Bell N.G., Bryce E., Coomes E., Crowl G., Duchaine C., Faheem A., Farooqi L., Hiebert R., Katz K., Khan S., Kozak R., Li A.X., Mistry H.P., Mozafarihashjin M., Nasir J.A., Nirmalarajah K., Panousis E.M., Paterson A., Plenderleith S., Powis J., Prost K., Schryer R., Taylor M., Veillette M., Wong T., Zhong X.Z., McArthur A.G., McGeer A.J., Mubareka S. medRxiv; 2021. Surface and Air Contamination With SARS-CoV-2 From Hospitalized COVID-19 Patients in Toronto, Canada. 2021.05.17.21257122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M.A., Brownsword E.A., Morgan J.S., Babiker A., Vanairsdale S.A., Lyon G.M., Metha A.K., Ingersoll J.M., Lindsley W.G., Kraft C.S. Bioaerosol sampling of a ventilated patient with COVID-19. Am.J.Infect.Control. 2020;48:1540–1542. doi: 10.1016/j.ajic.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M.A., Brownsword E.A., Babiker A., Ingersoll J.M., Waggoner J., Ayers M., Klopman M., Uyeki T.M., Lindsley W.G., Kraft C.S. Bioaerosol sampling for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a referral center with critically ill coronavirus disease 2019 (COVID-19) patients March–May 2020. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky J.A., Shankar S.N., Elbadry M.A., Gibson J.C., Alam M.M., Stephenson C.J., Eiguren-Fernandez A., Morris J.G., Mavian C.N., Salemi M., Clugston J.R., Wu C.Y. Collection of SARS-CoV-2 virus from the air of a clinic within a university student health care center and analyses of the viral genomic sequence. Aerosol Air Qual. Res. 2020;20:1167–1171. doi: 10.4209/aaqr.2020.02.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky J.A., Lauzardo M., Fan Z.H., Jutla A., Tilly T.B., Gangwar M., Usmani M., Shankar S.N., Mohamed K., Eiguren-Fernandez A., Stephenson C.J., Alam Md.M., Elbadry M.A., Loeb J.C., Subramaniam K., Waltzek T.B., Cherabuddi K., Morris J.G., Jr., Wu C.Y. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int. J. Infect. Dis. 2020;100:476–482. doi: 10.1016/j.ijid.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H., Ye F., Liu X., Huang Z., Ling S., Jiang Z., Cheng J., Huang X., Wu Q., Wu S., Xie Y., Xiao C., Ye D., Yang Z., Li Y., Leung N.H.L., Cowling B.J., He J., Wong S.S., Zanin M. SARS-CoV-2 environmental contamination associated with persistently infected COVID-19 patients. Influenza Other Respir. Viruses. 2020;14:688–699. doi: 10.1111/irv.12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.H., Fan Y.Z., Jiang L., Wang H.B. Aerosol and environmental surface monitoring for SARS-CoV-2 RNA in a designated hospital for severe COVID-19 patients. Epidemiol. Infect. 2020;148 doi: 10.1017/S0950268820001570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linillos-Pradillo B., Rancan L., Ramiro E.D., Vara E., Artíñano B., Arias J. 2021. Determination of SARS-CoV-2 RNA in Different Particulate Matter Size Fractions of Outdoor Air Samples in Madrid During the Lockdown. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., Liu X., Xu K., Ho K.F., Kan H., Fu Q., Lan K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- López J.H., Romo Á.S., Molina D.C., Hernández G.Á., Cureño Á.B.G., Acosta M.A., Gaxiola C., Félix M., Galván T.G. Detection of Sars-cov-2 in the air of two hospitals in Hermosillo, Sonora, México, utilizing a low-cost environmental monitoring system. Int. J. Infect. Dis. 2021;102:478–482. doi: 10.1016/j.ijid.2020.10.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Qi X., Chen H., Li X., Zhang Z., Wang H. Coronavirus disease 2019 patients in earlier stages exhaled millions of severe acute respiratory syndrome coronavirus 2 per hour. Clin. Infect. Diseases. 2021;72:e652–e654. doi: 10.1093/cid/ciaa1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquès M., Domingo J.L. Contamination of inert surfaces by SARS-CoV-2: persistence, stability and infectivity.A review. 2021;193 doi: 10.1016/j.envres.2020.110559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoumbeigi H., Ghanizadeh G., Arfaei Y.R., Heydari S., Goodarzi H., Sari D.R., Tat M. Investigation of hospital indoor air quality for the presence of SARS-cov-2. J. Environ. Health Sci. Eng. 2020;18(2):1–5. doi: 10.1007/s40201-020-00543-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D.G., Altman D., Antes G., Atkins D., Barbour V., Barrowman N., Berlin J.A., Clark J., Clarke M., Cook D., D’Amico R., Deeks J.J., Devereaux P.J., Dickersin K., Egger M., Ernst E., Gøtzsche P.C., Grimshaw J., Guyatt G., Higgins J., Ioannidis J.P.A., Kleijnen J., Lang T., Magrini N., McNamee D., Moja L., Mulrow C., Napoli M., Oxman A., Pham B., Rennie D., Sampson M., Schulz K.F., Shekelle P.G., Tovey D., Tugwell P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G., Rickard H., Stevenson D., Aranega-Bou P., Pitman J., Crook A., Davies K., Spencer A., Burton C., Easterbrook L., Love H.E., Summers S., Welch S.R., Wand N., Thompson K.A., Pottage T., Richards K.S., Dunning J., Bennett A. Detection of SARS-CoV-2 within the healthcare environment: a multi-Centre study conducted during the first wave of the COVID-19 outbreak in England. J. Hosp. Infect. 2021;108:189–196. doi: 10.1016/j.jhin.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska L., Milton D.K. It is time to address airborne transmission of coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2020;71(9):2311–2313. doi: 10.1093/cid/ciaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno T., Pintó R.M., Bosch A., Moreno N., Alastuey A., Minguillón M.C., Anfruns-Estrada E., Guix S., Fuentes C., Buonanno G., Stabile L., Morawska L., Querol X. Tracing surface and airborne SARS-CoV-2 RNA inside public buses and subway trains. Environ. Int. 2021;147 doi: 10.1016/j.envint.2020.106326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka S., Nakamura K., Iida S., Kutsuna S., Kinoshita N., Suzuki T., Suzuki T., Yamamoto K., Hayakawa K., Saito S., Ohmagari N. Possibility of transmission of severe acute respiratory syndrome coronavirus 2 in a tertiary care hospital setting: a case study. Infect. Prev. Pract. 2020;2 doi: 10.1016/j.infpip.2020.100079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchtouri V.A., Koureas M., Kyritsi M., Vontas A., Kourentis L., Sapounas S., Rigakos G., Petinaki E., Tsiodras S., Hadjichristodoulou C. Environmental contamination of SARS-CoV-2 on surfaces, air- conditioner and ventilation system. Int. J. Dent. Hyg. 2020;230 doi: 10.1016/j.ijheh.2020.113599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Morioka S., Kutsuna S., Iida S., Suzuki T., Kinoshita N., Sezuki T., Suzuki T., Sugiki Y., Okuhama A., Kanda K., Wakimoto Y., Ujiie M., Yamamoto K., Ishikane M., Moriyama Y., Ota M., Nakamoto T., Ide S., Nomoto H., Akiyama Y., Miyazato Y., Hayakawa K., Saito S., Ohmagari N. Environmental surface and air contamination in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) patient rooms by disease severity. Infect. Prev. Pract. 2020;2 doi: 10.1016/j.infpip.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi S., Short K.R., Groth R., Cravigan L., Spann K., Ristovski Z., Johnson G.R. Humidity-dependent survival of an airborne influenza a virus: practical implications for controlling airborne viruses. Environ. Sci. Technol. Lett. 2021;8:412–418. [Google Scholar]
- Niazi S., Groth R., Spann K., Johnson G.R. The role of respiratory droplet physicochemistry in limiting and promoting the airborne transmission of human coronaviruses: a critical review. Environ. Pollut. 2021;276 doi: 10.1016/j.envpol.2020.115767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nor N.S.M., Yip C.W., Ibrahim N., Jaafar M.H., Rashid Z.Z., Mustafa N., Hamid H.H.A., K. Chandru K., Latif M.T., Saw P.E., Lin C.Y., Alhasa K.M., Hashim J.H., Nadzir M.S.M. Particulate matter (PM2.5) as a potential SARS-CoV-2 carrier. Sci. Rep. 2021;11(1):2508. doi: 10.1038/s41598-021-81935-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.W.X., Tan Y.K., Chia P.K., Lee T.H., Ng O.T., Wong M.S.Y., Marimuthu K. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323(16):1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.W.X., Tan Y.K., Coleman K.K., Tan B.H., Leo Y.S., Wang D.L., Ng C.G., Ng O.T., Wong M.S.Y., Marimuthu K. Lack of viable severe acute respiratory coronavirus (SARS-CoV-2) among PCR-positive air samples from hospital rooms and community isolation facilities. Infect. Control Hosp. Epidemiol. 2021 doi: 10.1017/ice.2021.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., Chou R., Glanville J., Grimshaw J.M., Hróbjartsson A.Q., Lalu M.M., Li T., Loder E.W., Mayo-Wilson E., McDonald S., McGuinness L.A., Stewart L.A., Thomas J., Tricco A.C., Welch V.A., Whiting P., Moher D. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372 doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos R.G., Silveira M.B., Abrahão J.S. Exploratory assessment of the occurrence of SARS-CoV-2 in aerosols in hospital facilities and public spaces of a metropolitan center in Brazil. Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena P., Morais J., Quintal Gomes A., Viegas C. Sampling methods and assays applied in SARS-CoV-2 exposure assessment. Sci. Total Environ. 2021;775 [Google Scholar]
- Pitol A.K., Julian T.R. Community transmission of SARS-CoV-2 by surfaces: risks and risk reduction strategies. Environ. Sci. Technol. Lett. 2021;8:263–269. doi: 10.1021/acs.estlett.0c00966. [DOI] [PubMed] [Google Scholar]
- Pivato A., Amoruso I., Formenton G., Di Maria F., Bonato T., Vanin S., Marion A., Baldovin T. Evaluating the presence of SARS-CoV-2 RNA in the particulate matters during the peak of COVID-19 in Padua, northern Italy. Sci. Total Environ. 2021;784 doi: 10.1016/j.scitotenv.2021.147129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöhlker M.L., Kruger O.O., Förster J.D., Berkemeier T., Elbert W., Fröhlich-Nowoisky J., Pöschl U., Pöhlkery C. 2021. Respiratory Aerosols and Droplets in the Transmission of Infectious Diseases. arXiv:2103.01188v4 [physics.med-ph] [Google Scholar]
- Prather K.A., Wang C.C., Schooley R.T. Reducing transmission of SARS-COV-2. Science. 2020 doi: 10.1126/science.abc6197. [DOI] [PubMed] [Google Scholar]
- Rahmani A.R., Leili M., Azarian G., Poormohammadi A. Sampling and detection of corona viruses in air: a mini review. Sci. Total Environ. 2020;740 doi: 10.1016/j.scitotenv.2020.140207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram K., Thakur R.C., Singh D.K., Kawamura K., Shimouchi A., Sekine Y., Nishimura H., Singh S.K., Pavuluri C.M., Singh R.S., Tripathi S.N. Why airborne transmission hasn't been conclusive in case of COVID-19? An atmospheric science perspective. Sci. Total Environ. 2021;15 doi: 10.1016/j.scitotenv.2021.145525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnesar-Shumate S., Williams G., Green B., Krause M., Holland B., Wood S., Bohannon J., Boydston J., Freeburger D., Hooper I., Beck K., Yeager L., Altamura L.A., Biryukov J., Yolitz J., Schuit M., Wahl V., Hevey M., Dabisch P. Simulated sunlight rapidly inactivates SARS-CoV-2 on surfaces. J. Infect. Dis. 2020;222:214–222. doi: 10.1093/infdis/jiaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnesar-Shumate S., Bohannon K., Williams G., Holland B., Krause M., Green B., Freeburger D., Dabisch P. Comparison of the performance of aerosol sampling devices for measuring infectious SARS-CoV-2 aerosols. Aerosol Sci. Technol. 2021;55:975–986. doi: 10.1080/02786826.2021.1910137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzini K., Castrica M., Menchetti L., Maggi L., Negroni L., Orfeo N.V., Pizzoccheri A., Stocco M., Muttini S., Balzaretti C.M. SARS-CoV-2 RNA detection in the air and on surfaces in the COVID-19 ward of a hospital in Milan,Italy. 2020;742 doi: 10.1016/j.scitotenv.2020.140540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robotto A., Quaglino P., Lembo D., Morello M., Brizio E., Bardi L., Civra A. SARS-CoV-2 and indoor/outdoor air samples: a methodological approach to have consistent and comparable results. Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia J.L., Rivera D.N., Herrera V.L., Morwitzer M.J., Creager H.M., Santarpia G.W., Crown K.K., Brett-Major D.M., Schnaubelt E.R., Broadhurst M.J., Lawler J.V., Lowe J.J., Reid St.P. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci. Rep. 2020;29(10(1)) doi: 10.1038/s41598-020-69286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setti L., Passarini F., De Gennaro G., Baribieri P., Perrone M.G., Borelli M., Palmisani J., Di Gilio A., Torboli V., Fontana F., Clemente L., Pallavicini A., Ruscio M., Piscitelli P., Miani A. SARS-Cov-2 RNA found on particulate matter of Bergamo in Northern Italy: first evidence. Environ. Res. 2020;188 doi: 10.1016/j.envres.2020.109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z.G., Chen Y.M., Wu F., Xu L., Wang B.F., Shi L., Chen X., Dai F.H., She J.L., Chen J.M., Holmes E.C., Zhu T.Y., Zhang Y.Z. Identifying the risk of SARS-CoV-2 infection and environmental monitoring in airborne infectious isolation rooms (AIIRs) Virol. Sin. 2020;35(6):785–792. doi: 10.1007/s12250-020-00301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern R.A., Koutrakis P., Martins G., Lemos B., Dowd E.S., Sunderland E.M., Garshick E. Characterization of hospital airborne SARS-CoV-2. Respir. Res. 2021;22:73. doi: 10.1186/s12931-021-01637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern R.A., Al-Hemoud A., Alahmad B., Koutrakis P. Levels and particle size distribution of airborne SARS-CoV-2 at a healthcare facility in Kuwait. Sci. Total Environ. 2021;782 [Google Scholar]
- Tan L., Ma B., Lai X., Han L., Cao P., Zhang J., Fu J., Zhou Q., Wei S., Wang Z., Peng W., Yang L., Zhang X. Air and surface contamination by SARS-CoV-2 virus in a tertiary hospital I Wuhan,China. 2020;99:3–7. doi: 10.1016/j.ijid.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Mao Y., Jones R.M., Tan Q., Ji J.S., Li N., Shen J., Lv Y., Pan L., Ding P., Wang X., Wang Y., MacIntyre C.R., Shi X. Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ. Int. 2020;144 doi: 10.1016/j.envint.2020.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.W., Bahnfleth W.P., Bluyssen P.M., Buonanno G., Jimenez J.L., Kurnitski J., Li Y., Miller S., Sekhar C., Morawska L., Marr L.C., Melikov A.K., Nazaroff W.W., Nielsen P.V., Tellier R., Wargocki P.I., Dancer S.J. Dismantling myths on the airborne transmission of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) J. Hosp. Infect. 2021;110:89–96. doi: 10.1016/j.jhin.2020.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd- Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas C., Pimenta R., Dias M., Gomes B., Brito M., Caetano L.A., Carolino E., Gomes A. Microbiological contamination assessment in higher education institutes. Atmosphere. 2021;12(8):1079. [Google Scholar]
- Vosoughi M., Karami C., Dargahi A., Jeddi F., Jalali K.M., Hadisi A., Haghighi S.B., Dogahe H.P., Noorimotlagh Z., Mirzaee A.S. Investigation of SARS-CoV-2 in hospital indoor air of COVID-19 patients' ward with impinger method. Environ. Sci. Pollut. Res. 2021 doi: 10.1007/s11356-021-14260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Huang W., Lu X., Wang Y., Cheng L., Deng R., Long H., Zong Z. Contamination of SARS-CoV-2 in patient surroundings and on personal protective equipment in a non-ICU isolation ward for COVID-19 patients with prolonged PCR positive status. Antimicrob. Resist. Infect. Control. 2020;9(1):167. doi: 10.1186/s13756-020-00839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Lin J., Duan X., Huang W., Lu X., Zhou J., Zong Z. Asymptomatic COVID-19 patients can contaminate their surroundings: an environment sampling study. mSphere. 2020;5(3) doi: 10.1128/mSphere.00442-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.C.C., Hapuarachchi H.C., Arivalan S., Tien W.P., Koo C., Mailepessov D., et al. Environmental contamination of SARS-CoV-2 in a non-healthcare setting. Int. J. Environ. Res. Public Health. 2020;18(1):117. doi: 10.3390/ijerph18010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Zhao H., Li K., Zhang Z., Lu X., Peng H., Wang D., Chen J., Zhang X., Wu D., Gu Y., Yuan J., Zhang L., Lu J. The evidence of indirect transmission of SARS-CoV-2 reported in Guangzhou,China. 2020;20:1202. doi: 10.1186/s12889-020-09296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi T., Ohnishi M., Matsunaga N., Kakimoto K., Kamiya H., Okamoto K., Suzuki M., Gu Y., Sakaguchi M., Tajima T., Takaya S., Ohmagari N., Takeda M., Matsuyama S., Shirato K., Nao N., Hasegawa H., Kageyama T., Takayama I., Saito S., Wada K., Fujita R., Saito H., Okinaka K., Griffith M., Parry A.E., Barnetson B., Leonard J., Wakita T., Taskforce for the COVID-19 Cruise Ship Outbreak Environmental sampling for severe acute respiratory syndrome coronavirus 2 during a COVID-19 outbreak on the diamond princess cruise ship. J. Infect. Dis. 2020;222(7):1098–1102. doi: 10.1093/infdis/jiaa437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H., Yang J., Seo J.H., Sohn J.R. Methodology for sampling and detection of airborne coronavirus including SARS-CoV-2. Indoor Built Environ. 2020 doi: 10.1177/1420326X20980160. [DOI] [Google Scholar]
- Zhang M., Wang L., Yu S., Sun G., Lei H., Wu W. Status of occupational protection in the COVID-19 fangcang shelter Hospital in Wuhan,China. 2020;9(1):1835–1842. doi: 10.1080/22221751.2020.1803145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Otter J.A., Price J.R., Cimpeanu C., Garcia D.M., Kinross J., Boshier P.R., Mason S., Bolt F., Holmes A.H., Barclay W.S. 2020. Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Yao M., Zhang X., Hu B., Li X., Chen H., Zhang L., Liu Y., Du M., Sun B., Jiang Y., Zhou K., Hong J., Yu N., Ding Z., Xu Y., Hu M., Morawska L., Grinshpun S.A., Biswas P., Flagan C.R., Zhu B., Liu W., Yuanhang Zhang Y. 2020. Detection of SARS-CoV-2 in Exhaled Breath From COVID-19 Patients Ready for Hospital Discharge. [DOI] [Google Scholar]
- Zhou L., Yao M., Zhang X., Hu B., Li X., Chen H., Zhang L., Liu Y., Du M., Sun B., Jiang Y., Zhou K., Hong J., Yu N., Ding Z., Xu Y., Hu M., Morawska L., Grinshpun S.A., Biswas P., Flagan R.C., Zhu B., Liu W., Zhang Y. Breath-, air- and surface-borne SARS-CoV-2 in hospitals. J. Aerosol Sci. 2021;152 doi: 10.1016/j.jaerosci.2020.105693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Ayeh S.K., Chidambaram V., Karakousis P.C. Modes of transmission of SARS-CoV-2 and evidence for preventive behavioral interventions. Infect. Dis. 2021;21:496. doi: 10.1186/s12879-021-06222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]