Abstract
Although primary transcripts are polycistronic in the mitochondria of Trypanosoma brucei, steady-state levels of mature, monocistronic RNAs change throughout the parasitic life cycle. This indicates that steady-state RNA abundance is controlled by posttranscriptional mechanisms involving differential RNA stability. In this study, in organello pulse-chase labeling experiments were used to analyze the stability of different T. brucei mitochondrial RNA populations. In this system, total RNA and rRNA are stable for many hours. In contrast, mRNAs can be degraded by two biochemically distinct turnover pathways. The first pathway results in the rapid degradation of mRNA (half-life [t1/2] of 11 to 18 min) and is dependent upon the presence of an mRNA poly(A) tail. Remarkably, this pathway also requires the addition of UTP and therefore is termed UTP dependent. The second pathway results in slow turnover of mitochondrial mRNA (t1/2 of ∼3 h) and is not dependent upon the presence of an mRNA poly(A) tail or the addition of exogenous UTP. In summary, these results demonstrate the presence of a novel, UTP-dependent degradation pathway for T. brucei mitochondrial mRNAs and reveal an unprecedented role for both UTP and mRNA polyadenylation in T. brucei mitochondrial gene expression.
The regulation of RNA stability plays a pivotal role in controlling gene expression in many organisms. Although RNA stability in bacteria (12), chloroplasts (23), and the eukaryotic cytoplasm (43, 47) has been extensively studied, little is known about turnover mechanisms of mitochondrial mRNAs. A common theme in all systems examined to date is the regulation of mRNA stability through polyadenylation (32). In the eukaryotic cytoplasm, polyadenylation increases the stability of mRNAs in a mechanism dependent upon the poly(A)-binding protein I (25, 48). Conversely, in bacteria and chloroplasts, polyadenylation destabilizes mRNAs and their endonucleolytic cleavage products (13, 23, 49). Although mitochondrial mRNAs are polyadenylated in many systems (6), little is known about the role of mitochondrial polyadenylation and its possible effect on RNA turnover. The only two studies, to our knowledge, that directly address this subject are the recent reports by Gagliardi and Leaver (18) and Lupold et al. (31), which provide evidence that polyadenylation destabilizes mitochondrial mRNAs in plants.
Both mitochondrial and nuclear gene expression in the protozoan parasite Trypanosoma brucei are primarily regulated at the posttranscriptional level (42, 55). Reverse transcriptase PCR and Northern blot experiments have demonstrated that primary transcripts in the mitochondria are polycistronic and contain at least two different open reading frames (17, 28, 35, 44, 53). The generation of mature mitochondrial RNAs requires several posttranscriptional processing events, and the order of these events is variable. Polycistronic RNAs require cleavage to release individual, monocistronic RNAs with correct 5′ and 3′ ends. mRNAs are also modified by the addition of either a short (20-nucleotide) or long (120- to 200-nucleotide) 3′ poly(A) tail (7, 8, 17, 26, 44, 45). The function of the two different poly(A) tail lengths is unknown; however, previous results from our laboratory indicate that increased poly(A) tail length is not used to mark specific RNAs for translation, as it is in some systems (36). Finally, some T. brucei mitochondrial mRNAs are modified by the posttranscriptional addition and deletion of uridylate residues by the process of RNA editing (52). RNA editing creates start codons, stop codons, and open reading frames for otherwise untranslatable RNAs.
Although immature T. brucei mitochondrial transcripts are polycistronic, mature, monocistronic RNAs are present at different levels throughout the development of the parasite (8, 14, 17, 26, 34, 44, 51, 53). Regulated RNA abundance has been observed for all types of mitochondrial RNAs, including rRNAs, mRNAs that do not require editing (never-edited RNAs), and mRNAs that require editing (edited RNAs). For example, the steady-state level of the never-edited cytochrome oxidase subunit I mRNA is considerably higher in procyclic-form than in bloodstream-form parasites, whereas the never-edited NADH dehydrogenase subunit 4 RNA is more abundant in bloodstream-form parasites (8). Likewise, the edited C-rich region 4 mRNA is found solely in bloodstream-form parasites (14), while edited apocytochrome b and cytochrome oxidase subunit II mRNAs are found almost exclusively in procyclic-form parasites (8). Since mitochondrial transcription generates polycistronic transcripts, little opportunity exists for transcriptional control of individual genes. Therefore, the differential accumulation of rRNAs and never-edited mRNAs can be explained only by differential RNA stability. It is not known whether differential accumulation of edited RNAs is due to differential RNA stability and/or regulation of the RNA editing process itself. Although differential RNA stability has never been directly demonstrated for any T. brucei mitochondrial RNA, RNA stability has been indirectly implicated in regulating mitochondrial rRNA levels. This is based on the observation that the transcription rates of both the 9S and 12S mitochondrial rRNAs are similar in both life cycle stages (35), while 30-fold-higher levels of steady-state mitochondrial 9S and 12S rRNAs are present in procyclic-form parasites than in bloodstream-form parasites (34). To date, however, little is known about the role of RNA stability in regulating steady-state mRNA levels in T. brucei mitochondria. Furthermore, nothing is known about the mechanisms used to degrade RNAs in trypanosome mitochondria.
To begin to investigate the role of mRNA stability and its possible regulation in T. brucei mitochondrial gene expression, in organello pulse-chase labeling experiments were employed. Herein, two different RNA turnover pathways for T. brucei mitochondrial mRNAs that differ in degradation kinetics, nucleotide requirements, and substrate specificity are described. Remarkably, one of the two pathways is dependent upon the addition of UTP and an mRNA poly(A) tail. The second pathway is independent of both of these parameters. These experiments reveal a novel role for UTP in T. brucei mitochondrial gene expression. Also, these data suggest that destabilization of mitochondrial mRNA by polyadenylation is a general phenomenon.
MATERIALS AND METHODS
Cell culture and mitochondria isolation.
Procyclic-form T. brucei brucei clone IsTaR1 from stock EATRO 164 (54) was cultivated in SDM-79 medium supplemented with 10% inactivated fetal bovine serum (11). Mitochondrial vesicles were isolated on linear, 20 to 35% Percoll gradients and were stored at −80°C as previously described (22). Mitochondria were always used within 1 month of isolation.
Metabolic labeling of mitochondrial vesicles.
Metabolic labeling of mitochondrial vesicles was performed essentially as described by Harris et al. (22). Mitochondrial vesicles were collected by centrifugation for 15 min at 14,000 rpm in a Biofuge pico (Heraeus Instruments) at 4°C and were resuspended at a concentration of 1 mg of protein/ml in transcription buffer (5 mM HEPES [pH 7.6], 3 mM potassium phosphate [pH 7.7], 125 mM sucrose, 6 mM KCl, 10 mM MgCl2, 1 mM EDTA, 2 mM 2-mercaptoethanol) containing 0.1 mM ATP. The vesicles were incubated at 27°C for 30 min to reduce endogenous nucleotide pools. Vesicles were labeled at 27°C with 100 μCi of [α-32P]UTP/ml or 100 μCi of [α-32P]CTP/ml (400 Ci/mmol; Amersham) for various times. All labeling reaction mixtures contained 0.1 mM concentrations of the remaining 3 unlabeled nucleotides to allow transcription. Typical reaction volumes were 250 or 500 μl per experimental point.
For chase experiments, labeled vesicles were collected by centrifugation for 4 min at 14,000 rpm at room temperature, resuspended in the same initial reaction volume of transcription buffer containing the unlabeled nucleotide(s) stated in the text, and incubated at 27°C for the indicated times. Typically, 2 mM unlabeled nucleotide was used for chase experiments. To stop pulse or pulse-chase reactions, vesicles were collected by centrifugation for 4 min at 14,000 rpm at room temperature, resuspended in half of the original reaction volume of stop buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 10 μg of proteinase K/ml), and incubated 15 min at 37°C. Samples were extracted with phenol-chloroform. The labeled RNA present at this stage was designated total RNA. All experiments were performed with at least two different preparations of mitochondria for a total of two to four experiments. Since results were extremely consistent between experiments, all figures shown present data from single, representative experiments except for the experimental data in Fig. 6B and Table 1, which are the averages of data from three independent experiments.
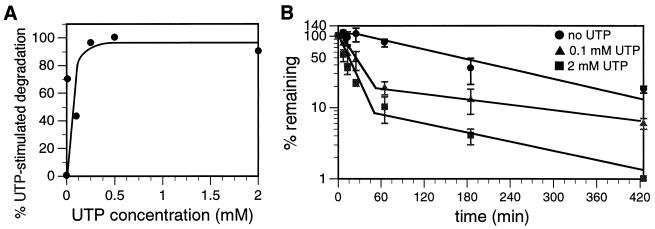
FIG. 6.
Concentration dependence of UTP-dependent degradation of mitochondrial polyadenylated RNA. (A) Mitochondrial vesicles were labeled with [α-32P]CTP for 8.5 min and chased for 13 min in transcription buffer containing 2 mM unlabeled CTP plus increasing amounts of UTP. Incorporation of [α-32P]CTP into poly(A)+ RNA was determined, and the amount of UTP-dependent poly(A)+ RNA degradation was calculated. The small amount of poly(A)+ RNA degradation observed in the absence of UTP was subtracted from the amount observed in the presence of UTP to calculate UTP-dependent poly(A)+ RNA turnover. The maximal amount of poly(A)+ RNA degradation observed was designated 100% UTP-stimulated degradation. (B) Mitochondrial vesicles were pulsed with [α-32P]CTP for 8.5 min and chased for 425 min in transcription buffer containing 2 mM unlabeled CTP and either 0, 0.1, or 2 mM UTP. At the times indicated, incorporation of [α-32P]CTP into poly(A)+ RNA was measured. Error bars represent 1 standard deviation.
TABLE 1.
Effect of UTP concentration on poly(A)+ RNA degradation rate, t1/2, and percent degradeda
| [UTP] (mM) | k (min−1) | t1/2 (min) | % Degraded after 7 h |
|---|---|---|---|
| 0 | 4.6 × 10−3 | 176 | 82 |
| 0.1 | 2.7 × 10−2 | 26 | 94 |
| 2 | 7.0 × 10−2 | 11 | 99 |
The degradation rates (k) and t1/2 values for the 0.1 and 2 mM UTP samples are for the initial, rapid degradation event from 0 to 65 min. The k value for the 0 mM UTP sample is for the single degradation event from 25 to 425 min. The t1/2 value for the 0 mM UTP sample includes the 25-min lag period before degradation begins.
RNA analysis.
Poly(A)+ RNA was isolated from total RNA using magnetic oligo(dT)25 beads (Dynal) according to the manufacturer's instructions. [α-32P]UTP- and [α-32P]CTP-labeled total and poly(A)+ RNA was spotted onto DE81 paper (Whatman), washed three times for 2 min at room temperature with 0.5 M Na2HPO4, and rinsed with water and then 95% ethanol. The paper was dried, and incorporation of the isotope into RNA was measured by scintillation counting. For gel electrophoresis analysis, equal amounts (measured in disintegrations per minute) from each RNA sample were separated by gel electrophoresis on 6% polyacrylamide–7 M urea gels and detected by autoradiography. Incorporation of [α-32P]UTP into 9S and 12S rRNA was measured using a Bio-Rad GS-700 imaging densitometer with Molecular Analyst version 1.5 software. Kinetic parameters of RNA degradation were calculated from semilogarithmic plots (50). Since poly(A)+ RNAs are captured by the binding of the 3′ poly(A) tail to oligo(dT) beads, degradation events that are initiated at the 3′ end of the RNA may appear to be more rapid than degradation events that are initiated at the 5′ end of the RNA.
Dot blot analysis of RPS12 RNA.
Three different plasmids were used for the dot blot analysis: pBluescribe (Stratagene), pRPS12-U (a plasmid containing the entire 221-bp unedited RPS12 gene inserted in the EcoRI/BamHI site of pBluescribe), and pRPS12-E (a plasmid containing the entire 325-bp fully edited RPS12 gene inserted in the EcoRI/BamHI site of pBluescribe). Plasmid DNA (0.5 pmol) was linearized with EcoRI, denatured, and transferred to Nytran (Schleicher and Schuell) by using a Bio-Dot apparatus (Bio-Rad) (10). DNA was fixed to the nylon membrane using UV light generated from a UV Stratalinker 2400 (Stratagene). Hybridization experiments were performed essentially as described by Poyton et al. (41). Membranes were prehybridized in 0.15 ml of hybridization solution (65% formamide, 0.2% SDS, 0.4 M NaCl, 20 mM piperazine-N, N′-bis(2-ethanesulfonic acid) [PIPES] [pH 6.4]/cm2, 0.1 mg of torula yeast RNA/ml, and 0.1 mg of denatured salmon sperm DNA/ml) for 30 min at 50°C. Pulse-labeled RNA probes were hybridized to the membranes in 0.15 ml of hybridization solution/cm2 for 36 to 48 h at 50°C. RNA probes were heated at 65°C for 3 min in 65% formamide prior to hybridization. After hybridization, filters were washed 10 times at 65°C with 0.5 to 1 ml of wash buffer (10 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 1 mM EDTA, 0.5% SDS)/cm2. Subsequently, filters were washed twice at 65°C with 0.5 ml of wash buffer/cm2 without SDS. In some cases, membranes were treated for 10 min with 10 μg of RNase A/ml in 0.3 M NaCl at 37°C to digest unhybridized RNA and finally were rinsed twice with 0.3 M NaCl at room temperature. RNase A treatment of membranes had no effect on the outcome of the in organello pulse-chase dot blot hybridization experiments. Membranes were exposed to film at −80°C with an intensifying screen. Autoradiograms were quantified by densitometry as described above.
In vitro transcription.
[α-32P]UTP-labeled RPS12 RNAs containing 3′ 20-nucleotide poly(A) tails were synthesized with T7 RNA polymerase using the Megascript in vitro transcription system (Ambion). Templates for transcriptions were generated by PCR using the primers CR6-5′T7 (5′TGTAATACGACTCACTATAGGGCTAATACACTTTTGATAACAAACTAAAGTAAA3′) and CR6-A20 (5′TTTTTTTTTTTTTTTTTTTTAAAAACATATCTTAT3′). The CR6-5′T7 primer is complementary to the 5′ never-edited region of RPS12 RNA and contains a T7 promoter (underlined above). The CR6-A20 primer is complementary to the 3′ never-edited region of RPS12 and contains a sequence encoding a 20-nucleotide poly(A) tail. The plasmids pRPS12-U, pRPS12-PE, and pRPS12-E served as templates for these PCRs. pRPS12-PE, clone 2p-21 from a previous study in our laboratory (36), contains a partially edited RPS12 DNA sequence. The 3′ half of this clone contains edited RPS12 sequence, whereas the 5′ half of this clone contains unedited RPS12 sequence. These PCR products were used as templates for transcription reactions, and labeled RPS12 RNAs were recovered by ethanol precipitation. The integrity of the labeled RPS12 RNAs was monitored before use by electrophoresis on 4% acrylamide–7 M urea gels and autoradiography.
RESULTS
Metabolic labeling of mitochondrial RNA.
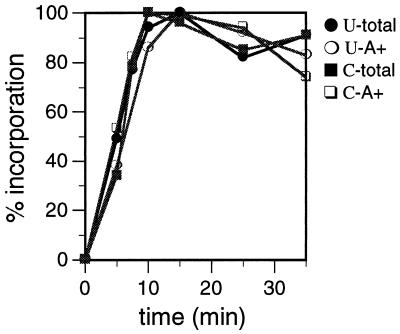
The kinetics of [α-32P]UTP and [α-32P]CTP incorporation into mitochondrial RNA were characterized. Both [α-32P]UTP and [α-32P]CTP were incorporated rapidly into total and poly(A)+ RNA, with incorporation peaking at 10 min (Fig. 1). A typical 1-ml labeling reaction resulted in the incorporation of 200 to 700 fmol of UTP into total RNA and the incorporation of 8 to 12 fmol of UTP into poly(A)+ RNA. The incorporation of CTP into mitochondrial RNA was approximately four- to fivefold less efficient than the incorporation of UTP, as a 1-ml CTP-labeling reaction resulted in the incorporation of 80 to 110 fmol of CTP into total RNA and the incorporation of 1 to 2 fmol of CTP into poly(A)+ RNA. The greater incorporation of UTP into mitochondrial poly(A)+ RNA may result from the high level of thymidines found in T. brucei mitochondrial genes (26), posttranscriptional uridine addition (3, 22, 40; K. T. Militello and L. K. Read, unpublished data), and/or higher levels of UTP transport into the mitochondrial vesicles. The UTP- and CTP-labeled poly(A)+ material was sensitive to NaOH treatment (20), demonstrating that this labeled material is RNA (data not shown). Using an identical metabolic labeling protocol, Harris et al. also demonstrated that the total labeled material generated in the in organello system is RNA, since the labeled material was sensitive to RNase A digestion (22). Based on the time necessary for labeled nucleotides to be incorporated into RNA using this system, mitochondria were pulsed with radiolabeled nucleotides for either 8.5 or 25 min for pulse-chase RNA stability analysis. The length of the pulse period (either 8.5 or 25 min) did not affect the turnover rates of any of the individual RNA populations (data not shown).
FIG. 1.
Kinetics of [α-32P]UTP and [α-32P]CTP incorporation into mitochondrial RNA. Mitochondrial vesicles were labeled with [α-32P]UTP (U) or [α-32P]CTP (C) for the indicated times. Total RNA (total) and poly(A)+ RNA (A+) were isolated, and incorporation of radioisotope was measured by scintillation counting. The maximal amount of labeled nucleotide incorporation observed for each sample was designated 100% incorporation.
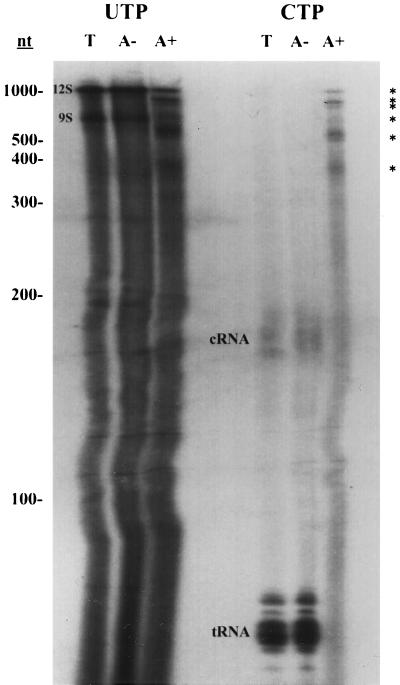
To determine if similar RNA populations were labeled by different nucleotides, UTP- and CTP-labeled RNAs were analyzed by gel electrophoresis (Fig. 2). Both the UTP- and CTP-labeled total RNA populations contained RNAs of heterogeneous sizes. UTP also labeled the 9S and 12S rRNAs strongly and labeled the guide RNAs weakly, presumably due to posttranscriptional labeling of rRNA and guide RNA 3′ poly(U) tails (3, 22, 29; Militello and Read, unpublished data). CTP efficiently labeled tRNAs and the tRNA precursors of 170 to 180 nucleotides (C-RNAs) due to the posttranscriptional addition of the 3′ CCA sequence (21). The patterns of total RNA labeling were similar to those previously described by Harris et al. (22). The UTP-labeled poly(A)− fraction appeared to be identical in composition to the UTP-labeled total RNA population. Likewise, no difference was observed between CTP-labeled poly(A)− and total RNA populations.
FIG. 2.
Gel electrophoresis analysis of mitochondrial RNAs synthesized in organello. Mitochondrial vesicles were labeled with [α-32P]UTP or [α-32P]CTP for 25 min. Total RNA (T), poly(A)− RNA (A−), and poly(A)+ RNA (A+) were isolated and analyzed on a denaturing 6% acrylamide gel. Designations for labeled RNAs: 9S, 9S rRNA; 12S, 12S rRNA; ∗, major polyadenylated RNAs. tRNAs and cRNAs (21) are also labeled. Guide RNAs were labeled weakly by [α-32P]UTP and are not indicated. RNA size standards are indicated on the left (nt, nucleotides).
The CTP- and UTP-labeled poly(A)+ populations were also analyzed by gel electrophoresis (Fig. 2). Both poly(A)+ RNA populations were characterized by heterogeneously sized RNAs and six distinct RNAs of approximately 380, 510, 600, 750, 800, and 1,100 nucleotides (Fig. 2). The size distribution of the major poly(A)+ RNAs detected by pulse-labeling is very similar to that of the major bands observed previously by Northern blot analysis of T. brucei mitochondrial RNA using labeled T. brucei maxicircle (mitochondrial) DNA fragments as probes (24). This strongly suggests that the major in organello-labeled poly(A)+ RNAs are the major native steady-state RNAs. The poly(A)+ RNA fractions were not appreciably contaminated by poly(A)− RNA, since no tRNAs or C-RNAs were found in the CTP-labeled poly(A)+ fraction. Although one of the major polyadenylated RNAs appears to be the same size as 12S rRNA, it is highly unlikely that this band represents rRNA contamination. If this RNA were 12S rRNA, we would not expect to observe it in the CTP-labeled poly(A)+ fraction, since rRNAs are not labeled efficiently by CTP. From these data, we conclude that UTP and CTP label similar, if not identical, poly(A)+ RNA populations and that these RNAs are of mitochondrial origin (see also Fig. 8 and 9).
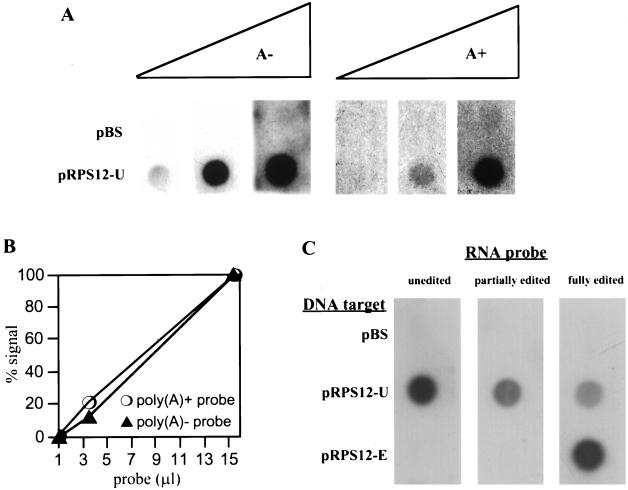
FIG. 8.
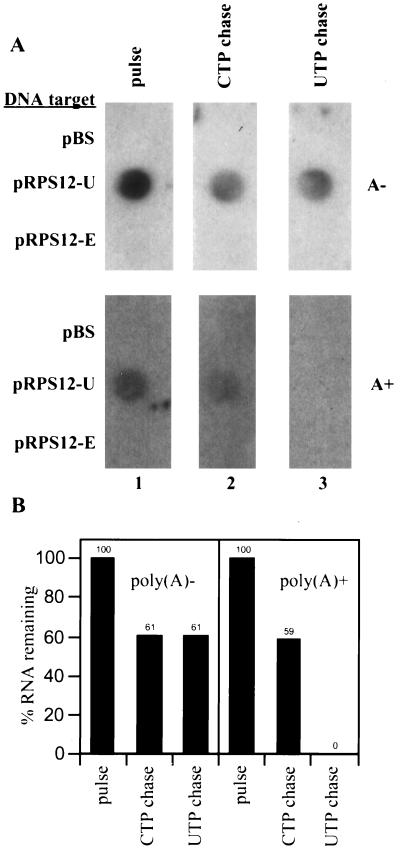
Linearity and sequence specificity of unedited RPS12 RNA dot blot hybridization assay. (A) Mitochondria were labeled with [α-32P]CTP for 8.5 min, and RNAs were isolated. Labeled RNAs were separated into poly(A)− and poly(A)+ fractions by oligo(dT) chromatography, and increasing amounts of both fractions were used to probe filters containing a control pBluescribe plasmid (pBS) or a plasmid containing unedited RPS12 DNA (pRPS12-U). Filters were processed as described in Materials and Methods, and hybridization signals were detected by autoradiography. (B) Hybridization signals in panel A were quantified by densitometry. (C) Radiolabeled, in vitro-synthesized unedited, partially edited (halfway toward the 5′ end), and fully edited RPS12 RNAs with 20-nucleotide poly(A) tails were used to probe filters containing a control pBluescribe plasmid (pBS), a plasmid containing unedited RPS12 DNA (pRPS12-U), and a plasmid containing fully edited RPS12 DNA (pRPS12-E). Signals were detected by autoradiography.
FIG. 9.
UTP-independent and UTP-dependent degradation of RPS12 RNA. (A) Mitochondrial vesicles were labeled with [α-32P]CTP for 8.5 min (column 1) and chased for 65 min in buffer containing 2 mM CTP (column 2) or buffer containing 2 mM CTP plus 2 mM UTP (column 3). Labeled mitochondrial RNA was isolated and separated into poly(A)− and poly(A)+ fractions by oligo(dT) chromatography. Labeled RNAs were used to probe filters containing a control pBS plasmid, a plasmid containing unedited RPS12 DNA (pRPS12-U), and a plasmid containing fully edited RPS12 DNA (pRPS12-E). Labeled poly(A)− RNA (A−) was used to probe the top filters, and labeled poly(A)+ RNA (A+) was used to probe the bottom filters. Hybridization signals were detected by autoradiography. (B) The percentage of unedited RPS12 RNA remaining under different chase conditions was determined by densitometry and displayed in the bar graph. The amount of RNA in each pulse sample was designated 100% RNA remaining.
Pulse-chase analysis of mitochondrial RNAs.
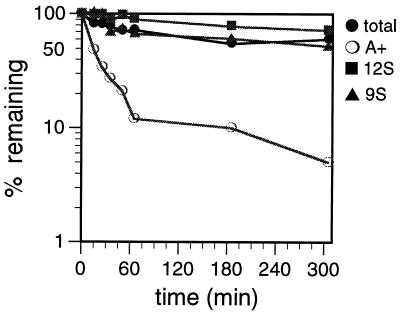
The relative stabilities of total RNA, poly(A)+ RNA, 9S rRNA, and 12S rRNA were determined by pulse-chase experiments (47). Mitochondrial vesicles were labeled with [α-32P]UTP for 25 min and then chased in buffer containing 2 mM UTP for up to 305 min (Fig. 3). The 9S rRNA, 12S rRNA, and total RNA populations degraded slowly over the chase period, never reaching more than 35% degradation, even after a 5-h chase period. In contrast to the rRNAs and total RNAs, the vast majority (90%) of the poly(A)+ RNAs were degraded rapidly, with a half-life (t1/2) of 18 min. The remaining poly(A)+ RNA degraded slowly over the remaining chase period. The greater stability of rRNAs in comparison to mRNAs is consistent with observations from other systems, including human and yeast mitochondria (2, 27, 38).
FIG. 3.
Degradation kinetics of mitochondrial RNAs labeled with [α-32P]UTP. Mitochondrial vesicles were labeled with [α-32P]UTP for 25 min and chased for 305 min in transcription buffer containing 2 mM unlabeled UTP. At the times indicated, incorporation of [α-32P]UTP into total RNA (total), poly(A)+ RNA (A+), 9S rRNA (9S), and 12S rRNA (12S) was measured.
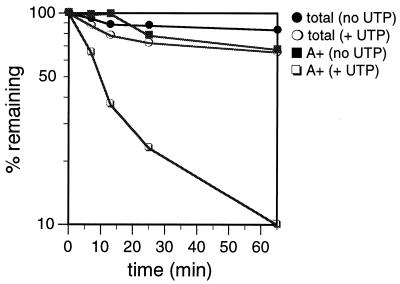
To ensure that the UTP-labeled poly(A)+ RNAs are a representative population of poly(A)+ RNAs, the degradation kinetics of poly(A)+ RNAs labeled with CTP and chased in the presence of unlabeled CTP were determined (Fig. 4). Surprisingly, under these conditions, the CTP-labeled poly(A)+ RNA did not show the rapid decay kinetics of the UTP-labeled poly(A)+ RNA [compare the poly(A)+ (no UTP) curve in Fig. 4 with the poly(A)+ curve in Fig. 3]. Since gel electrophoresis demonstrated that [α-32P]UTP and [α-32P]CTP labeled similar poly(A)+ RNA populations (Fig. 2), we predicted that differences in the UTP and CTP chase conditions influenced the degradation rates of poly(A)+ RNAs. Because a large amount of UTP (2 mM) is added to the system as the chase nucleotide in the UTP labeling experiments, we tested whether UTP was required for rapid degradation of poly(A)+ RNAs. Thus, mitochondria were labeled with [α-32P]CTP and chased with unlabeled CTP plus 2 mM unlabeled UTP for 65 min (Fig. 4). Remarkably, in the presence of 2 mM UTP, the majority of the CTP-labeled poly(A)+ RNA fraction (90%) degraded rapidly, with a t1/2 of 11 min (Fig. 4), suggesting the presence of a UTP-dependent degradation pathway. The ability of UTP to enhance RNA degradation in T. brucei mitochondria is specific for the poly(A)+ RNAs, since exogenous UTP did not substantially affect the rate of total CTP-labeled-RNA degradation (Fig. 4). The small amount of UTP-stimulated degradation of the total CTP-labeled RNA may reflect degradation of the poly(A)+ RNA in this fraction, which constitutes 2% of the total CTP-labeled RNA (data not shown). Neither the 9S rRNA nor the 12S rRNA was rapidly degraded in the presence of UTP, further demonstrating the specificity of the UTP-stimulated turnover pathway for polyadenylated RNAs (Fig. 3).
FIG. 4.
Degradation kinetics of mitochondrial RNAs labeled with [α-32P]CTP. Mitochondrial vesicles were labeled with [α-32P]CTP for 8.5 min and chased for 65 min in transcription buffer containing 2 mM unlabeled CTP (no UTP) or in transcription buffer containing 2 mM unlabeled CTP plus 2 mM unlabeled UTP (+ UTP). At the times indicated, incorporation of [α-32P]CTP into total RNA (total) and poly(A)+ RNA (A+) was measured.
Characterization of UTP-dependent degradation of poly(A)+ RNAs.
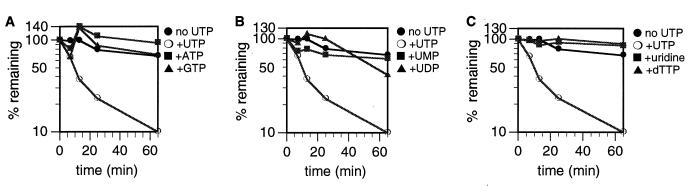
To determine if the effect of UTP on degradation of poly(A)+ RNAs was specific to this nucleotide, mitochondria were labeled with CTP and chased in the presence of unlabeled CTP plus various unlabeled nucleotides at a concentration of 2 mM (Fig. 5). Neither ATP nor GTP could substitute for UTP to enhance degradation of poly(A)+ RNAs (Fig. 5A). Uridine, UMP, and dTTP were also ineffective in stimulating poly(A)+ RNA degradation (Fig. 5B and C). UDP consistently exhibited a minor enhancement of degradation at the 65-min time point (Fig. 5B). While it is possible that UDP itself can enhance mRNA degradation, it is more likely that the effect seen is due either to the slow conversion of UDP to UTP in the mitochondrial vesicles or to UTP contamination in the UDP preparation. These results demonstrate that the effect of UTP on the degradation of poly(A)+ RNA is specific to UTP.
FIG. 5.
Effects of different nucleotides on degradation of mitochondrial polyadenylated RNA. Mitochondrial vesicles were labeled with [α-32P]CTP for 8.5 min and chased for 65 min in transcription buffer containing 2 mM unlabeled CTP plus the indicated nucleotide at a concentration of 2 mM. At the times indicated, incorporation of [α-32P]CTP into poly(A)+ RNA was determined. (A) ATP and GTP; (B) UMP and UDP; (C) uridine and dTTP. As a reference, the same degradation curves of poly(A)+ RNAs chased in the absence and presence of 2 mM UTP from Fig. 4 are shown on each graph.
The concentration of exogenous UTP required to enhance degradation of poly(A)+ RNA was determined. Mitochondria were labeled with CTP and chased in the presence of 2 mM unlabeled CTP and increasing amounts of unlabeled UTP for 13 min (Fig. 6A). Half-maximal stimulation of UTP-dependent poly(A)+ RNA degradation was effected by approximately 0.1 mM UTP.
Mechanistically, UTP may either increase the rate of poly(A)+ RNA degradation or increase the total amount of poly(A)+ RNA that is degraded. To address this question, CTP-labeled mitochondria were chased for 7 h in buffer containing unlabeled CTP and either 0 mM, 0.1 mM (limiting), or 2 mM (maximal) UTP. The initial rates of degradation and the percentages of poly(A)+ RNAs degraded were determined (Fig. 6B; Table 1). The rates of poly(A)+ RNA degradation varied substantially with UTP concentration. Poly(A)+ RNAs chased in the presence of 2 mM UTP degraded at a rate of 7.0 × 10−2/min, whereas poly(A)+ RNAs chased in the presence of 0.1 mM UTP degraded at a rate of 2.7 × 10−2/min. Degradation of poly(A)+ RNAs chased in the absence of added UTP was more complex, as there was a lag period of approximately 25 min before degradation began. After the lag period, poly(A)+ RNA degraded at a rate of 4.6 × 10−3/min, 15-fold slower than in the presence of 2 mM UTP. Factoring in the 25-min lag period, the half-life of poly(A)+ RNA (176 min) was 16-fold longer than in the presence of 2 mM UTP (11 min). While initial degradation rates increased with UTP concentration, after approximately 60 min, the rate of turnover of the remaining RNA was similar under all conditions. In contrast to the effect of UTP on the poly(A)+ RNA degradation rate, the presence of UTP had little effect on the amount of poly(A)+ RNA degraded over a long chase period. After 7 h, 94% of the poly(A)+ RNA chased in the presence of 0.1 mM UTP and 99% of the poly(A)+ RNA chased in the presence of 2 mM UTP had been degraded. Even in the absence of UTP in the chase buffer, 82% of the poly(A)+ RNA had been degraded after 7 h. In summary, these data clearly indicate that UTP increases the rate of poly(A)+ RNA degradation but has little effect on the percentage of poly(A)+ RNA ultimately degraded. These data strongly suggest that mRNA can be degraded by either of two biochemically distinct turnover pathways, and these pathways differ in both kinetics and UTP dependence.
There is evidence that transcription and RNA turnover are coupled in human mitochondria (2). In this model, the presence of active transcription is absolutely required for RNA turnover. Therefore, we investigated whether the same phenomenon occurs in T. brucei mitochondria and whether this could account for the observed UTP stimulation of poly(A)+ RNA turnover. This possibility seemed plausible, since previous studies in our laboratory have demonstrated that in the in organello transcription system there is no transcriptional activity in the absence of added UTP, and the addition of UTP alone can support moderate levels of transcription (Millitello and Read, unpublished data). Therefore, the addition of UTP to chase reactions could restore mitochondrial transcription, which might be required for poly(A)+ RNA turnover. This could account for the UTP dependence of rapid poly(A)+ RNA turnover. If the transcriptional coupling hypothesis were valid, inhibition of transcription should reduce or abolish UTP-dependent poly(A)+ RNA turnover.
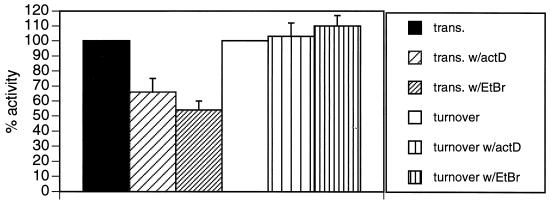
To test this hypothesis, mitochondria were labeled with [α-32P]CTP and chased for 65 min in buffer containing 2 mM unlabeled CTP and 0.1 mM unlabeled UTP. The transcription inhibitors actinomycin D and ethidium bromide were added to the chase reactions, and UTP-dependent poly(A)+ RNA turnover was quantified (Fig. 7). As expected, both actinomycin D and ethidium bromide inhibited the synthesis of CTP-labeled poly(A)+ RNA to some extent in pulse-labeling experiments. Although drug inhibition of poly(A)+ RNA transcription was not 100% efficient, the efficacy of these drugs with in organello poly(A)+ RNA synthesis was similar to the efficacy observed with total mitochondrial transcription (Millitello and Read, unpublished data). Despite their 35 to 45% inhibition of transcription, neither actinomycin D nor ethidium bromide inhibited UTP-dependent turnover of poly(A)+ RNA. Although we cannot rule out that moderate transcriptional activity supports maximal UTP-dependent RNA degradation, these results strongly suggest that the UTP requirement for this turnover pathway is not due simply to transcriptional coupling.
FIG. 7.
Effects of transcriptional inhibitors on RNA synthesis and UTP-dependent turnover of mitochondrial polyadenylated RNAs. To measure the effects of transcriptional inhibitors on in organello poly(A)+ RNA synthesis (trans.), mitochondrial vesicles were labeled with [α-32P]CTP for 10 min in the absence or presence of actinomycin D (50 μg/ml) or ethidium bromide (20 μg/ml). The inhibitors were added 10 min prior to the addition of nucleoside triphosphates. Incorporation of [α-32P]CTP into poly(A)+ RNA was determined. To measure the effect of transcriptional inhibitors on UTP-dependent poly(A)+ RNA turnover (turnover), mitochondrial vesicles were labeled with [α-32P]CTP for 10 min and chased for 65 min in transcription buffer containing 2 mM CTP and 0.1 mM UTP. Actinomycin D (50 μg/ml) or ethidium bromide (20 μg/ml) was added directly to the chase reaction mixtures. Incorporation of [α-32P]CTP into poly(A)+ RNA was determined. To calculate UTP-dependent turnover, the amount of poly(A)+ RNA degradation observed in the absence of UTP was subtracted from the amount observed in the presence of UTP. The amount of transcription or UTP-dependent RNA turnover observed in the absence of drug treatment was defined as 100% activity. Error bars represent 1 standard deviation.
Analysis of mitochondrial RPS12 RNA degradation.
To demonstrate that the UTP-dependent RNA turnover pathway exists for a specific, bona fide mitochondrial RNA, dot blot hybridization experiments were employed. Total RNA was labeled in organello with CTP, and this RNA was used to probe membranes containing specific T. brucei mitochondrial sequences. The most robust hybridization was seen with the unedited ribosomal protein S12 (RPS12) gene (data not shown), and this RNA was chosen for further study. Hybridization to a DNA target representing the fully edited RPS12 RNA sequence was never observed (see Fig. 9; also data not shown). In order to demonstrate that the hybridization experiments were quantitative for unedited RPS12 RNA, increasing amounts of in organello-generated CTP-labeled poly(A)− RNA and poly(A)+ RNA were hybridized to the unedited RPS12 DNA target, and the signals were quantified by densitometry (Fig. 8A and B). Both CTP-labeled poly(A)− and poly(A)+ RNA probes were used, since we have previously observed that a significant fraction of steady-state unedited RPS12 RNA is incapable of binding oligo(dT) columns (36). Furthermore, unedited, nonpolyadenylated cytochrome oxidase subunit III RNAs have been observed by RT-PCR in T. brucei mitochondria (15). Therefore, the generation of poly(A)− and poly(A)+ unedited RPS12 RNA from this in organello system is expected and is probably not an artifact of in organello labeling or oligo(dT) chromatography. Indeed, hybridization of both the poly(A)− and poly(A)+ RNA probes to unedited RPS12 DNA was observed (Fig. 8A). The amount of hybridization to the unedited RPS12 gene was linear with respect to the amount of radiolabeled RNA probe used (Fig. 8B). Therefore, this hybridization technique can be used to accurately quantify the amount of unedited RPS12 RNA present in the in organello system. The amount of radiolabeled RNA probe used for subsequent hybridization experiments was always in the linear range.
Control hybridization experiments were performed to ensure that unedited, partially edited, and fully edited RPS12 RNAs would be detected by our dot blot hybridization system if they were generated in the in organello system. Radiolabeled unedited, partially edited (edited halfway toward the 5′ end), and fully edited RPS12 RNAs with 20-nucleotide 3′ poly(A) tails were synthesized in vitro. Each of the three RNAs was hybridized to membranes containing a control plasmid (pBluescribe), a plasmid containing unedited RPS12 DNA (pRPS12-U), or a plasmid containing fully edited RPS12 DNA (pRPS12-E) (Fig. 8C). None of the RNAs hybridized to the control DNA target. Unedited RPS12 RNA hybridized to unedited RPS12 DNA but not to fully edited RPS12 DNA. Partially edited RPS12 RNA, edited through its 3′ half, also hybridized to the unedited RPS12 DNA target but not the edited DNA target. Fully edited RPS12 RNA hybridized to both the unedited and the fully edited RPS12 DNA targets, with an obvious preference for the fully edited RPS12 DNA target. These experiments demonstrate that unedited, partially edited, and fully edited RPS12 RNAs can be detected by our dot blot hybridization system using a combination of unedited and fully edited RPS12 DNA targets.
Once we had verified the linearity and sequence specificity of the dot blot hybridization assay for RPS12 RNA, we examined the degradation parameters of in organello-synthesized unedited RPS12 RNA. To determine if unedited RPS12 RNAs were rapidly degraded in the presence of UTP, mitochondria were labeled with CTP and chased in buffer containing only unlabeled 2 mM CTP or buffer containing unlabeled 2 mM CTP plus 2 mM UTP for 65 min. Poly(A)− and poly(A)+ RNA fractions were isolated and used to probe membranes containing the unedited and edited RPS12 genes (denatured, double-stranded phagemid DNA) (Fig. 9). In the absence of UTP, a small amount of both poly(A)− (39%) and poly(A)+ (41%) RPS12 RNA was degraded (Fig. 9). The amount of degradation observed in the absence of UTP was similar for the poly(A)− and poly(A)+ unedited RPS12 RNAs throughout several experiments. This indicates that both poly(A)− and poly(A)+ RNAs are equivalent substrates for the UTP-independent turnover pathway. When RNAs were chased in the presence of UTP, 100% of the poly(A)+ unedited RPS12 RNA was degraded (Fig. 9). In contrast, the majority of the poly(A)− unedited RPS12 RNA (61%) was not specifically degraded in the presence of UTP, as the same amount of degradation was observed in the absence of UTP (CTP-only chase). Similar results were obtained when the poly(A)− and poly(A)+ RNA probes generated in this experiment were hybridized to filters containing single-stranded M13 DNA engineered to detect sense-strand unedited RPS12 RNA (data not shown).
Importantly, in the presence of UTP, the loss of the poly(A)+ unedited RPS12 RNA signal is not accompanied by an increase in the amount of the poly(A)− unedited RPS12 signal. This demonstrates that the loss of the poly(A)+ unedited RPS12 signal under UTP chase conditions is not due simply to a loss of hybridization to the oligo(dT) beads, which would cause an increase in the poly(A)− unedited RPS12 signal. Therefore, UTP-dependent degradation of RNA is not caused exclusively by mRNA deadenylation or by uridine insertion into the poly(A) tract (53). Moreover, the degradation of unedited RPS12 RNA under all conditions tested in this system cannot be attributed to the loss of hybridization efficiency due to RNA editing. If partially edited RPS12 RNAs were generated in this system, they would still hybridize to the unedited RPS12 DNA target, as demonstrated in control experiments (Fig. 8C). If fully edited RPS12 RNAs were generated in this system, they would hybridize to the fully edited RPS12 DNA target (Fig. 8C), although hybridization to the fully edited RPS12 DNA target was never observed (Fig. 9A). Therefore, these experiments demonstrate that UTP stimulates the degradation of RNAs that bind to oligo(dT) beads, strongly suggesting that the poly(A) tail is necessary for this pathway. In summary, our data provide strong evidence for two biochemically distinct mRNA turnover pathways in T. brucei mitochondria.
DISCUSSION
In this study, the degradation of several T. brucei mitochondrial RNA populations was analyzed, with specific emphasis on the mRNA population. Two distinct mRNA turnover pathways were identified that differ in degradation kinetics, nucleotide requirements, and substrate specificity. The UTP-independent pathway results in slow degradation of mRNA and does not require UTP or an mRNA poly(A) tail. This pathway may also account for the slow turnover of rRNA observed in this system. The UTP-dependent pathway requires the addition of UTP and results in the rapid degradation of only poly(A)+ mRNA. In many systems, including the yeast cytoplasm (5), bacteria (16), chloroplasts (23), and Drosophila (4), multiple nucleases and RNA turnover pathways are employed to ensure the proper and complete degradation of RNAs. We now describe a similar scenario in T. brucei mitochondria, where at least two biochemically distinct turnover pathways are able to degrade mRNA. The function of these pathways in T. brucei RNA metabolism is currently under investigation.
A novel feature of the rapid RNA turnover pathway is its strict dependence upon the addition of UTP. To our knowledge, this is the only RNA turnover pathway known that specifically requires UTP. The only other reported case of an unusual nucleotide dependence for RNA turnover is for the yeast ω intron of the mitochondrial 21S rRNA (38). Rapid degradation of the ω intron is dependent upon the addition of any one of the eight standard ribo- or deoxyribonucleotide triphosphates. The nucleotide dependence of ω intron degradation is due to the involvement of a yeast nucleoside triphosphate-dependent 3′ exoribonuclease that requires nucleotide hydrolysis for function (33, 37). The UTP-dependent poly(A)+ RNA degradation pathway in our system is mechanistically different from degradation of the yeast mitochondrial ω intron, since UTP is the only nucleotide that supports rapid RNA degradation (Fig. 5), and this effect is restricted to poly(A)+ RNA (Fig. 3 and 4).
There are three general mechanisms by which UTP could stimulate the degradation of poly(A)+ RNAs. First, UTP could directly activate an enzyme involved in RNA degradation. For example, UTP may act as a cofactor or an energy source for an enzyme, as observed for the yeast mitochondrial nucleoside triphosphate-dependent 3′ exoribonuclease (33, 37). An enzyme in the RNA degradation pathway could also be activated by uridylylation (covalent addition of UMP), using UTP as a donor. Uridylylation has been shown to modulate the binding properties of PII proteins, which are involved in regulating glutamine synthesis in Escherichia coli and several other bacteria (46). Second, UTP may stimulate degradation of poly(A)+ RNAs indirectly by modification of the RNA, essentially marking it as a target for rapid degradation. In bacteria and chloroplasts, RNAs that require rapid degradation are marked by polyadenylation, which promotes their rapid turnover (13, 23, 49). In T. brucei mitochondria, RNAs may be polyuridylylated, perhaps by the existing terminal uridylyltransferase (3, 40), as a signal for rapid degradation. Indeed, a small number of polyuridylylated mRNAs have been described for T. brucei mitochondria (15), but the significance of this modification is unclear. Other potential UTP-specific modifications that may decrease the stability of poly(A)+ RNAs in T. brucei mitochondria include the generation of edited RNA sequences, insertion of uridines into the mRNA poly(A) tail (53), or the formation of a uridine cyclic phosphate at the 3′ end of the RNAs, as seen for U6 small nuclear RNA (30). Third, UTP may be required for a process that is coupled to RNA degradation, and therefore RNA turnover would show UTP dependence. However, results presented here strongly argue against coupling between transcription and UTP-dependent RNA degradation (Fig. 7). It is possible that UTP-dependent RNA degradation is coupled to RNA editing. The use of different nonhydrolyzable UTP analogs to study UTP-dependent RNA turnover should differentiate between these different mechanisms. These studies are currently in progress in our laboratory.
It is interesting that the UTP-dependent RNA turnover pathway is specific for RNAs that bind to oligo(dT) beads (Fig. 3, 4, and 9). This is strong evidence that RNAs must be polyadenylated in order to be substrates for the UTP-specific turnover pathway. We cannot rule out that RNAs with a poly(A) tail contain another feature required for UTP-dependent degradation, such as a 5′ cap structure. The establishment of a faithful T. brucei in vitro mitochondrial RNA turnover system will be necessary to confirm the hypothesis that the poly(A) tail is required for UTP-dependent RNA turnover. However, since polyadenylation plays a role in regulating RNA stability in all systems analyzed to date, it is conceptually satisfying to imagine that polyadenylation is also used to regulate the stability of mitochondrial RNAs. In trypanosome mitochondria, the poly(A) tail is apparently required for rapid degradation by the UTP-dependent degradation pathway. This model is particularly appealing in light of recent experiments reported by Gagliardi and Leaver (18) and Lupold et al. (31) that provide strong evidence that polyadenylation has a destabilizing effect on mitochondrial mRNAs in plants. Thus, the destabilizing effect of polyadenylation on mitochondrial RNAs may be a general phenomenon, similar to RNA degradation mechanisms described for bacteria (13, 49) and chloroplasts (23). This is in sharp contrast to the stabilizing effect of polyadenylation on eukaryotic cytosolic RNAs (25, 48). The similar relationship between polyadenylation and RNA stability in bacteria, chloroplasts, and mitochondria may reflect the bacterial origin of both organelles (19).
Unusual processes involving UTP occur at all levels of gene expression in the mitochondria of T. brucei. There is presumably a large UTP requirement for transcription, since many mitochondrial mRNA transcripts consist of more than 45% uridine residues (26). RNA processing involves UTP both in the RNA editing process (52) and for the posttranscriptional addition of 3′ oligo(U) tails to the guide RNAs that direct editing (9, 39). In addition, uridylate residues are commonly found in the poly(A) tails of mitochondrial mRNAs (53). Finally, UTP may be important in translation, since the mature 9S and 12S rRNAs possess nonencoded 3′ poly(U) tails of defined lengths (1). We now report that UTP is required for the rapid turnover of mitochondrial poly(A)+ RNAs. It is striking that T. brucei has evolved multiple mitochondrial gene expression events in which UTP plays a prominent role. It is unclear if the UTP requirement for many aspects of T. brucei mitochondrial RNA metabolism is simply a coincidence. It is tempting to hypothesize that UTP availability is a way to regulate multiple mitochondrial processes through a common requirement.
ACKNOWLEDGMENTS
We thank V. James Hernandez for helpful discussions throughout the course of this work. In addition, we thank Tom Melendy, V. J. Hernandez, and members of the Read lab for critical reading and discussion of the manuscript.
This work was supported in part by NIH grant GM53502 to L.K.R., who is also a recipient of the Burroughs Wellcome Fund New Investigator Award in Molecular Parasitology.
REFERENCES
- 1.Adler B K, Harris M E, Bertrand K I, Hajduk S L. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol Cell Biol. 1991;11:5878–5884. doi: 10.1128/mcb.11.12.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attardi G, Cantatore P, Chomyn A, Crews S, Gelfand R, Merkel C, Montoya J, Ojala D. A comprehensive view of mitochondrial gene expression in human cells. In: Slonimski P, Borst P, Attardi G, editors. Mitochondrial genes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 51–71. [Google Scholar]
- 3.Bakalara N, Simpson A M, Simpson L. The Leishmania kinetoplast-mitochondrion contains terminal uridylyltransferase and RNA ligase activities. J Biol Chem. 1989;264:18679–18686. [PubMed] [Google Scholar]
- 4.Bashirullah A, Halsell S R, Cooperstock R L, Kloc M, Karaiskakis A, Fisher W W, Fu W, Hamilton J K, Etkin L D, Lipshitz H D. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 1999;18:2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 6.Beutow D E, Wood W M. Mitochondrial translation system. In: Roodyn D B, editor. Subcellular biochemistry. Vol. 5. New York, N.Y: Plenum Press; 1978. pp. 29–38. [Google Scholar]
- 7.Bhat G J, Myler P J, Stuart K. The two ATPase 6 mRNAs of Leishmania tarentolae differ at their 3′ ends. Mol Biochem Parasitol. 1991;48:139–149. doi: 10.1016/0166-6851(91)90110-r. [DOI] [PubMed] [Google Scholar]
- 8.Bhat G J, Souza A E, Feagin J E, Stuart K. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol Biochem Parasitol. 1992;52:231–240. doi: 10.1016/0166-6851(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 9.Blum B, Simpson L. Guide RNAs in kinetoplast mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- 10.Brown T. Dot and slot blotting of DNA. In: Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1999. pp. 2.9.16–2.9.20. [Google Scholar]
- 11.Brun R, Schönenberger M. Cultivation and in vitro cloning of procyclic forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 12.Carpousis A J, Vanzo N F, Raynal L C. mRNA degradation: a tail of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 13.Cohen S N. Surprises at the 3′ end of prokaryotic RNA. Cell. 1995;80:829–832. doi: 10.1016/0092-8674(95)90284-8. [DOI] [PubMed] [Google Scholar]
- 14.Corell R A, Myler P J, Stuart K. Trypanosoma brucei mitochondrial CR4 gene encodes an extensively edited mRNA with completely edited sequence only in bloodstream forms. Mol Biochem Parasitol. 1994;64:65–74. doi: 10.1016/0166-6851(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 15.Decker C J, Sollner-Webb B. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell. 1990;61:1001–1011. doi: 10.1016/0092-8674(90)90065-m. [DOI] [PubMed] [Google Scholar]
- 16.Ehretsmann C P, Carpousis A J, Krisch H M. mRNA degradation in procaryotes. FASEB J. 1992;6:3186–3192. doi: 10.1096/fasebj.6.13.1397840. [DOI] [PubMed] [Google Scholar]
- 17.Feagin J E, Jasmer D P, Stuart K. Apocytochrome b and other mitochondrial DNA sequences are differentially expressed during the life cycle of Trypanosoma brucei. Nucleic Acids Res. 1985;13:4577–4596. doi: 10.1093/nar/13.12.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gagliardi D, Leaver C J. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 1999;18:3757–3766. doi: 10.1093/emboj/18.13.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray M W. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 20.Gupta J G, Li Q, Thomson A B, Hunt A G. Characterization of a novel plant poly(A) polymerase. Plant Sci. 1995;110:215–226. [Google Scholar]
- 21.Hancock K, LeBlanc A J, Donze D, Hajduk S L. Identification of nuclear encoded precursor tRNAs within the mitochondrion of Trypanosoma brucei. J Biol Chem. 1992;267:23963–23971. [PubMed] [Google Scholar]
- 22.Harris M E, Moore D R, Hajduk S L. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J Biol Chem. 1990;265:11368–11376. [PubMed] [Google Scholar]
- 23.Hayes R, Kudla J, Gruissem W. Degrading the chloroplast mRNA: the role of polyadenylation. Trends Biochem Sci. 1999;24:199–202. doi: 10.1016/s0968-0004(99)01388-2. [DOI] [PubMed] [Google Scholar]
- 24.Hoeijmakers J H J, Snijders A, Janssen J W G, Borst P. Transcription of kinetoplast DNA in Trypanosoma brucei bloodstream and culture forms. Plasmid. 1981;5:329–350. doi: 10.1016/0147-619x(81)90009-3. [DOI] [PubMed] [Google Scholar]
- 25.Jackson R J, Standart N. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 26.Jasmer D P, Feagin J E, Stuart K. Diverse patterns of expression of the cytochrome c oxidase subunit I gene and unassigned reading frames 4 and 5 during the life cycle of Trypanosoma brucei. Mol Cell Biol. 1985;5:3041–3047. doi: 10.1128/mcb.5.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King T C, Sirdeskmukh R, Schlessinger D. Nucleolytic processing of ribonucleic acid transcripts in procaryotes. Microbiol Rev. 1986;50:428–451. doi: 10.1128/mr.50.4.428-451.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koslowsky D J, Yahampath G. Mitochondrial mRNA 3′ cleavage/polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol Biochem Parasitol. 1997;90:81–94. doi: 10.1016/s0166-6851(97)00133-3. [DOI] [PubMed] [Google Scholar]
- 29.Leegwater P, Speijer D, Benne R. Identification by UV cross-linking of oligo(U)-binding proteins in mitochondria of the insect trypanosomatid Crithidia fasciculata. Eur J Biochem. 1995;227:780–786. doi: 10.1111/j.1432-1033.1995.tb20201.x. [DOI] [PubMed] [Google Scholar]
- 30.Lund E, Dahlberg J E. Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science. 1992;255:327–330. doi: 10.1126/science.1549778. [DOI] [PubMed] [Google Scholar]
- 31.Lupold D S, Caoile A G F S, Stern D B. Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell. 1999;11:1565–1577. doi: 10.1105/tpc.11.8.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manley J L. Messenger RNA polyadenylation: a universal modification. Proc Natl Acad Sci USA. 1995;92:1800–1801. doi: 10.1073/pnas.92.6.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margossian S P, Li H, Zassenhaus H P, Butow R A. The DExH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell. 1996;84:199–209. doi: 10.1016/s0092-8674(00)80975-7. [DOI] [PubMed] [Google Scholar]
- 34.Michelotti E F, Hajduk S L. Developmental regulation of trypanosome mitochondrial gene expression. J Biol Chem. 1987;262:927–932. [PubMed] [Google Scholar]
- 35.Michelotti E F, Harris M E, Adler B, Torri A F, Hajduk S L. Trypanosoma brucei mitochondrial ribosomal RNA synthesis, processing and developmentally regulated expression. Mol Biochem Parasitol. 1992;54:31–42. doi: 10.1016/0166-6851(92)90092-x. [DOI] [PubMed] [Google Scholar]
- 36.Militello K T, Read L K. Coordination of kRNA editing and polyadenylation in Trypanosoma brucei mitochondria: complete editing is not required for long poly(A) tail addition. Nucleic Acids Res. 1999;27:1377–1385. doi: 10.1093/nar/27.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min J, Heuertz R M, Zassenhaus H P. Isolation and characterization of an NTP-dependent 3′-exoribonuclease from mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1993;268:7350–7357. [PubMed] [Google Scholar]
- 38.Min J, Zassenhaus H P. A nucleoside triphosphate-regulated, 3′ exonucleolytic mechanism is involved in turnover of yeast mitochondrial RNAs. J Bacteriol. 1993;175:6245–6253. doi: 10.1128/jb.175.19.6245-6253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollard V W, Hajduk S L. Trypanosoma equiperdum minicircles encode three distinct primary transcripts which exhibit guide RNA characteristics. Mol Cell Biol. 1991;11:1668–1675. doi: 10.1128/mcb.11.3.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard V W, Harris M E, Hajduk S L. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 1992;11:4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poyton R O, Bellus G, McKee E E, Sevarine K A, Goehring B. In organello mitochondrial protein and RNA synthesis systems from Saccharomyces cerevisiae. Methods Enzymol. 1996;264B:36–42. doi: 10.1016/s0076-6879(96)64006-3. [DOI] [PubMed] [Google Scholar]
- 42.Priest J W, Hajduk S L. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J Bioenerg Biomembr. 1994;26:179–191. doi: 10.1007/BF00763067. [DOI] [PubMed] [Google Scholar]
- 43.Rajagopalan L E, Malter J S. Regulation of eukaryotic messenger RNA turnover. Prog Nucleic Acid Res. 1997;56:257–286. doi: 10.1016/s0079-6603(08)61007-7. [DOI] [PubMed] [Google Scholar]
- 44.Read L K, Myler P J, Stuart K. Extensive editing of both processed and preprocessed maxicircle CR6 transcripts in Trypanosoma brucei mitochondria. J Biol Chem. 1992;267:1123–1128. [PubMed] [Google Scholar]
- 45.Read L K, Stankey K A, Fish W R, Muthiani A M, Stuart K. Developmental regulation of RNA editing and polyadenylation in four life cycle stages of Trypanosoma congolense. Mol Biochem Parasitol. 1994;68:297–306. doi: 10.1016/0166-6851(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 46.Rhee S G, Chock P B, Stadtman E R. Regulation of Escherichia coli glutamine synthetase. Adv Enzymol. 1989;62:37–92. doi: 10.1002/9780470123089.ch2. [DOI] [PubMed] [Google Scholar]
- 47.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sachs A, Wahle E. Poly(A) tail metabolism and function in eucaryotes. J Biol Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 49.Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu Rev Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- 50.Segel I H. Biochemical calculations. 2nd ed. New York, N.Y: John Wiley & Sons; 1976. pp. 378–379. [Google Scholar]
- 51.Souza A E, Shu H-H, Read L K, Myler P J, Stuart K D. Extensive editing of CR2 maxicircle transcripts of Trypanosoma brucei predicts a protein with homology to a subunit of NADH dehydrogenase. Mol Cell Biol. 1993;13:6832–6840. doi: 10.1128/mcb.13.11.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuart K, Allen T E, Heidmann S, Seiwert S. RNA editing in kinetoplastid protozoa. Microbiol Mol Biol Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart K, Feagin J E. Mitochondrial DNA of kinetoplastids. Int Rev Cytol. 1992;141:65–88. doi: 10.1016/s0074-7696(08)62063-x. [DOI] [PubMed] [Google Scholar]
- 54.Stuart K, Gobright E, Jenni L, Milhausen M, Thomashow L, Agabian N. The IsTaR 1 serodeme of Trypanosoma brucei: development of a new serodeme. J Parasitol. 1984;70:747–754. [PubMed] [Google Scholar]
- 55.Vanhamme L, Pays E. Control of gene expression in trypanosomes. Microbiol Rev. 1995;59:223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]