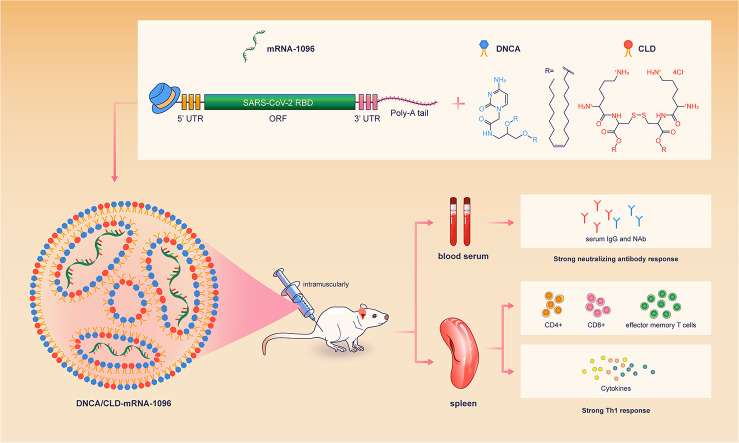
Abstract
The messenger RNA (mRNA)-based therapy, especially mRNA vaccines, has shown its superiorities in versatile design, rapid development and scale production, since the outbreak of coronavirus disease 2019 (COVID-19). Although the Pfizer-BioNTech and Moderna COVID-19 mRNA vaccines had been approved for application, unexpected adverse events were reported to be most likely associated with the mRNA delivery systems. Thus, the development of mRNA delivery system with good efficacy and safety remains a challenge. Here, for the first time, we report that the neutral cytidinyl lipid, 2-(4-amino-2-oxopyrimidin-1-yl)-N-(2,3-dioleoyl-oxypropyl) acetamide (DNCA), and the cationic lipid, dioleoyl-3,3′-disulfanediylbis-[2-(2,6-diaminohexanamido)] propanoate (CLD), could encapsulate and deliver the COVID-19 mRNA-1096 into the cytoplasm to induce robust adaptive immune response. In the formulation, the molar ratio of DNCA/CLD to a single nucleotide of COVID-19 mRNA-1096 was about 0.9: 0.5: 1 (the N/P ratio was about 7: 1). The DNCA/CLD-mRNA-1096 lipoplexes were rationally prepared by the combination of the lipids DNCA/CLD with the aqueous mRNA solution under mild sonication to stimulate multiple interactions, including H-bonding, π-stacking and electrostatic force between the lipids and the mRNA. After intramuscular applications of the DNCA/CLD-mRNA-1096 lipoplexes, robust neutralizing antibodies and long-lived Th1-biased SARS-CoV-2-specific cell immunity were detected in the immunized mice, thus suggesting the DNCA/CLD a promising mRNA delivery system. Moreover, our study might also inspire better ideas for developing mRNA delivery systems.
Keywords: mRNA delivery, Lipids, Lipoplexes, COVID-19
Graphical abstract
1. Introduction
The sudden emergence of the coronavirus disease 2019 (COVID-19) global pandemic offered a great opportunity for mRNA vaccines to outstand from other types of vaccines [1]. Since the first COVID-19 case was reported in the late December 2019, it only took as short as about three months for the first COVID-19 mRNA vaccine candidate, the Moderna COVID-19 mRNA-1273, to start clinical trials, which was also the first-born COVID-19 vaccine candidate throughout the world [2,3]. Surprisingly, by the end of 2020, the Pfizer-BioNTech COVID-19 mRNA vaccine candidate BNT162b2 became the first globally approved COVID-19 mRNA vaccine for emergency use in UK, thereafter FDA US issued an Emergency Use Authorization (EUA) for the Moderna COVID-19 mRNA-1273 vaccine [[2], [3], [4], [5]]. With the advantages of rapid development and potent immunogenicity, mRNA vaccines have gained great victory in the race for COVID-19 vaccines. Though mRNA vaccines seem to be with good safety profiles in theory, adverse reactions were still reported after vaccine receipts [2,4,6,7]. Pfizer-BioNTech and Moderna COVID-19 vaccines based on lipid nanoparticles (LNP) are associated with side effects, which have been reported often linked to inflammation [8,9], such as pain, swelling, fever, and fatigue, and also related to allergic reactions [6,7,10], thus obstacles to developing the mRNA-based therapy still exist. Therefore, developing mRNA delivery systems with desired efficacy and safety has been commonly agreed to encourage more therapeutic mRNA into the rescue [11].
Many classes of materials have been developed as mRNA carriers, including, virus stimulating particles (VSP), lipids, polymers, cell-penetrating peptides, etc., in which lipids are most widely used in clinical research for mRNA therapy for their simplicity in fabrication and lower immunogenicity, compared with VSP [12]. However, given the thread running through many of these options has to be the theme of safety, the bio-derived materials with their innate biocompatibility are most preferred. Nucleic acids, based on nucleoside-nucleoside interactions, could be used as biocompatible carrier as well [13]. Nucleoside phospholipids comprising a covalent combination of molecules with lipid moieties and nucleosides have been developed as novel therapeutic agents [14,15]. Other than the classical amphiphilic lipids, they are constructed with a highly specialized polar head group (adenosine, thymidine, cytidine, guanosine, uracil, or their analogs), facilitating additional hydrogen bonding with complementary nucleobases via Watson-Crick base paring and π-stacking, and spontaneously, these lipids self-assemble in water with low toxicity and immunogenicity [[16], [17], [18], [19], [20]]. A series of similar amphiphilic nucleoside-based lipids have been devised as carriers for nucleic acid drugs [20,21]. The nucleotide moiety promoted the transfection efficacy, where the cationic nucleoside-based lipid, [Tosylate salt of 1′-(2′,3′-dioleyl-5′-trimethylammonium-α, β-D-ribofuranosyl)-3-nitropyrrole] (TRN), significantly enhanced siRNA permeation to silence the expression upon mouse fibroblas (NIH 3T3) at N/P ratio of 10 [22]. In our previous work, a novel type of zwitterionic nucleolipids was designed, which was found to be able to interact with oligonucleotides and self-assemble in water [23]. However, these aminonucleoside phospholipids failed in polyA transfection, probably due to the electrostatic repulsion of phosphate anions between polyA and the phospholipids relatively hindering the interaction of gene and materials [23]. To transfect mRNA, we synthesized a neutral nucleobase lipid, 2-(4-amino-2-oxopyrimidin-1-yl)-N-(2,3-dioleoyl-oxypropyl) acetamide (DNCA), which excluded the phosphate moiety, but included cytosine, glycerinum and oley alcohol. With the aid of mild sonication to promote the assembling, the lipids DNCA and CLD were able to boost the expression of mRNA encoding the enhanced green fluorescent protein (eGFP) in various cell lines, including the somatic cells and antigen presenting cells.
Thus, mRNA-1096 was designed to encode the receptor-binding domain (RBD) of SARS-CoV-2 spike protein for evaluating the application of DNCA/CLD as mRNA vaccine carrier. RBD engages human angiotensin-converting enzyme 2 (hACE2) as the receptor for initial viral attachment, thus it has been widely selected as the antigen target for COVID-19 vaccine development [[24], [25], [26], [27], [28], [29], [30], [31]]. mRNA-1096 encoding the RBD as the vaccine candidate molecule might condense the immune response on interference of receptor binding and theoretically reduce the risk of inducing antibodies that readily mediated antibody-dependent enhancement of infection (ADE), with bright aspects to inducing robust neutralizing antibodies and cellular immunity [26,27,[32], [33], [34]].
2. Materials and methods
2.1. Ethics statement
All animal experiment procedures were reviewed and performed under appropriate licenses proved by the Animal Experiment Committee of Laboratory Animal Center, Academy of Military Medical Sciences (AMMS), China (Assurance Number: IACUC-DWZX-2020-624), and were consistent with both local and national animal experimentation ethics.
2.2. Cells and animals
HEK293T (ATCC, CRL-3216) and RD (ATCC, CCL-136) cells were maintained in Dulbecco's Modified Eagle Medium (DMEM; Thermo Fisher Scientific) with 10% Fetal Bovine Serum (FBS; Thermo Fisher Scientific) and 1% antibiotics (penicillin 100 U/ml-streptomycin 100 μg/ml; Thermo Fisher Scientific). Immortalized DC2.4 (a murine bone marrow derived dendritic cell line) cells, kindly provided by StemiRNA, were propagated in RPMI 1640 complete medium with 10% FBS. Female specific-pathogen-free (SPF) BALB/c mice (6–8 weeks old) were commercially purchased from Beijing Vital River Animal Technology Co., Ltd. (licensed by Charles River), and were housed and bred in the temperature-; humidity- and light cycle-controlled SPF mouse facilities (20 ± 2 °C; 50 ± 10%; light, 7:00–19:00; dark, 19:00–7:00) in AMMS.
2.3. Preparation of mRNA
The unmodified COVID-19 RBD mRNA-1096 was synthesized by T7-polymerase-based in vitro transcription using T7-FlashScribe™ Transcription kit (CELLSCRIPT) and capped using ScriptCap™ Cap 1 Capping System kit with ScriptCap Capping Enzyme and 2'-O-Methyltransferase (CELLSCRIPT) to produce Cap 1 structure. The mRNA was purified by ammonium acetate precipitation. Briefly, the synthesized mRNA was precipitated by 5 M ammonium acetate (Sigma-Aldrich) by mixing well and incubating in ice for 15 min. Then, the mRNA was pelleted by centrifugation at >10,000 ×g for 15 min at 4 °C, after which the supernatant was removed, and the mRNA pellet was gently rinsed with 70% ethanol. After the 70% ethanol was removed without disturbing the mRNA pellet, the air-dried mRNA pellet was resuspended in RNase-Free water for further analysis and application. The quality and concentration of the synthesized mRNA-1096 were authenticated using Agilent 2100 Bioanalyzer and RNA Nano 6000 Assay Kit (Agilent). The mRNA products encoding the eGFP and the firefly luciferase (FLuc) were purchased from TriLink Bio Technologies.
2.4. Synthesis and characterization of DNCA/CLD-mRNA-1096
The lipids DNCA and CLD were synthesized according to previous reports [35,36], then dissolved in ethanol at 25 mM as stock solutions. The mRNA-1096 was stocked in RNase-free deionized H2O at 1 μg/μl. Then, the lipids (DNCA/CLD, molar ratio, 9:5) combined with mRNA-1096 at a N/P ratio of 7:1 were dispersed in RNase-free deionized H2O, and mildly sonicated for 15 min at room temperature (40 kHz, 40% maximum power) in a KQ-800DE ultrasonic cleaner (Kun Shan Ultrasonic Instruments Co., Ltd) at room temperature for 15 min before characterization and application. To test the enzymatic stability, DNCA/CLD-mRNA-1096 was challenged with RNase A (10 μg/ml, Beyotime) at 37 °C for 30 min, 60 min, 90 min and 120 min, then the samples were analyzed by 1% agarose gel retardation assay. The morphology of DNCA/CLD-mRNA-1096 was observed with a Hitachi H7650 transmission electron microscope (TEM) after negatively stained with a sodium phosphotungstate solution (pH 7.2), and images were obtained with a CCD camera system and analyzed with Image J software. The particle size and ζ potential of DNCA/CLD-mRNA-1096 were measured by a Litesizer 500 (Anton Paar) instrument. Data were analyzed using an Anton Paar Kalliope software package. Then, mRNA-1096 was fluorescently labeled with MFP488 using the Label IT Nucleic Acid Labeling Reagent kit (Mirus). The encapsulation efficiency (EE) of the nanocomplexes was detected from the supernatant for the unpacked MFP488-labeled mRNA-1096 by an I-control Infinite 200 PRO microplate reader (TECAN) with excitation at 501 nm and emission at 523 nm after centrifugation.
2.5. Cytotoxicity of DNCA/CLD-mRNA-1096 in vitro
To test the cytotoxicity, the DNCA/CLD-mRNA-1096 (mRNA-1096 = 1 μg) was incubated with different cell lines (HEK293T, DC 2.4 and RD cells) at 80–90% confluency in 96-well plates at 37 °C for 24 h in a 5% CO2 environment, then tested with a cell counting kit- 8 (CCK- 8; Dojindo). The ultraviolet (UV) absorbance was measured at 450 nm by an I-control Infinite 200 PRO microplate reader (TECAN). The cell viability was calculated by the equation, cell viability (%) = (As - Ab)/ (A0 - Ab) × 100%, where the As, A0, and Ab were defined as the absorbance of the experimental sample treated cells, untreated cells and blank controls.
2.6. Cellular uptake of DNCA/CLD-mRNA-1096 in vitro
To detect cellular uptake, mRNA-1096 was fluorescently labeled with MFP488. DC 2.4 and RD cells were seeded at a cell density of 1/3 million in 12-well plates and treated with the MFP488-labeled DNCA/CLD-mRNA-1096 (MFP488-labeled mRNA-1096 = 1 μg) on 80–90% confluency. After incubated for different time periods at 37 °C in a 5% CO2 environment, cells were collected, fixed in 4% paraformaldehyde, and washed with PBS (pH = 7.4) prior to analysis on a BD FACS Aria II flow cytometer.
To determine the route of cellular internalization of DNCA/CLD-mRNA-1096, DC 2.4 and RD cells (at 80–90% confluency) were preincubated with chemical inhibitors, Genistein (50 μM; Beyotime) [37,38], Chlorpromazine (15 μg/ml; Solarbio) [38], Dynasore (50 μM; Glpbio) [39], Nocodazole (1.25 μg/ml; Beyotime) [40], Amiloride (100 μM; Solarbio) [41], for 30 min before transfected with the MFP488-labeled DNCA/CLD-mRNA-1096 (MFP488-labeled mRNA-1096 = 1 μg). After incubation for 3 h, cells were harvested, fixed in 4% paraformaldehyde, and washed with PBS before the flow cytometric analysis.
2.7. Visualization of the release of mRNA-1096 in vitro
DC 2.4 and RD cells (at 50–60% confluency) were transfected with CellLight® Late Endosomes-RFP, BacMam 2.0 (Thermo Fisher Scientific) and incubated overnight at 37 °C in a 5% CO2 environment. Then, cells were treated with the MFP488-labeled DNCA/CLD-mRNA-1096 (MFP488-labeled mRNA-1096 = 1 μg) and incubated for 3 h at 37 °C in a 5% CO2 environment. MFP488-labeled Lipofectamine 2000-mRNA-1096 (MFP488-labeled mRNA-1096 = 1 μg) was set as reference. After removal of the supernatants, cells were washed with PBS, sequentially stained with Lyso-Tracker deep Red (50 nM; Thermo Fisher Scientific) and Hoechst 33342 (1:100; Beyotime) at 37 °C in a 5% CO2 environment for 45 min and 5 min, respectively. After final washes with PBS, cells were observed by Operetta CLS™ High Content System (PerkinElmer), and images were obtained with Harmony 4.9 software and analyzed with Image J software.
2.8. Protein expression of DNCA/CLD-mRNA in vitro
To test the in vitro delivery capability of DNCA/CLD, mRNA-eGFP (1 μg) and mRNA-1096 (1 μg) were encapsulated with DNCA/CLD before applied to different cell lines (HEK293T, DC 2.4 and RD cells). Protein expression was detected at 24 h post transfection. Enhanced green fluorescent protein expression was visualized under an Olympus IX71 fluorescent microscope, and images were obtained by DP controller software. SARS-CoV-2 RBD protein expression was analyzed by Western blotting.
2.9. Validation of DNCA/CLD for mRNA delivery in vivo
To test the capability of DNCA/CLD-mRNA delivery and biodistribution in vivo, mRNA-FLuc (10 μg) was encapsulated with DNCA/CLD. Female BALB/c (6–8 weeks old) mice were inoculated with DNCA/CLD-mRNA-FLuc via intramuscular (i.m., n = 3), subcutaneous (s.c., n = 3), intradermal (i.d., n = 3) or intravenous (i.v., n = 3) route, and another batch was intramuscularly inoculated with the same dose of mRNA-FLuc encapsulated in the commercially available in vivo-jetRNA® mRNA delivery reagent (Polyplus transfection) or Lipofectamine 2000 (Thermo Fisher Scientific). At different time post inoculation, mice were intraperitoneally injected with luciferase substrate (PerkinElmer). After reaction for 3 min, bioluminescence signals were collected by an IVIS Spectrum instrument (PerkinElmer) for 60 s. For ex vivo imaging, at 6 h post inoculation, mice were dissected before bioluminescence signals were collected by IVIS Spectrum instrument (PerkinElmer) for 60 s from the isolated tissues, including heart, liver, spleen, lung, kidney and muscle. The bioluminescence signals in regions of interest (ROIs) were quantified using Living Image 3.5.
2.10. Vaccination of DNCA/CLD-mRNA-1096 in mice
Female BALB/c mice were intramuscularly vaccinated with mRNA-1096 (10 μg) encapsulated with DNCA/CLD (n = 5) or PBS as negative control (n = 5) every 14 days post initial immunization using a three-dose vaccination strategy. Sera were collected on the 10th day post each vaccination, specifically on days 10, 24, 38 post initial immunization, for the detection of SARS-CoV-2 RBD-specific IgG binding antibody and neutralizing antibody against pseudotyped SARS-CoV-2 infection as detailed below. Splenocytes were collected at 50 days post initial immunization for evaluation of cellular immune response by enzyme linked immunospot (ELISpot) and flow cytometry as demonstrated below.
2.11. Evaluation of serum antibody and cellular response
Mice serum samples were heated at 56 °C for 30 min before detection. The SARS-CoV-2 RBD specific IgG antibody titer was detected by enzyme linked immunosorbent assay (ELISA), and neutralizing antibody titer was determined by a SARS-CoV-2-spike protein pseudotyped neutralization assay. Mice spleen tissues were isolated and the splenocytes were collected for cellular immune response analysis by ELISpot.
(a) ELISA analysis for SARS-CoV-2 RBD-specific IgG antibody
Serial 2-fold gradient dilutions of the inactivated serum, starting at 1:100, were added to 2% BSA blocked 96-well plates coated with recombinant SARS-CoV-2 RBD antigen (100 ng per well, SinoBiological). After incubated at 37 °C for 120 min, plates were washed and reacted with horseradish peroxidase (HRP) conjugated goat anti-mouse IgG (1:250; Abclonal) for 60 min at 37 °C. Plates were then washed and incubated with the substrate, tetramethyl benzidine (TMB; TIANGEN), for 20 min at room temperature in dark, followed by reaction termination by HCl (2 M; Solarbio). The absorbance (450/630 nm) was analyzed using an I-control Infinite 200 PRO microplate reader (TECAN). The endpoint titers were defined as the vaccinated serum dilution at which absorbance was read no less than 2.1-fold the average negative sera (1:100).
(b) Detection of neutralizing antibody against pseudotyped SARS-CoV-2 infection
The neutralizing antibody was detected by the reduction of luciferase gene expression, as described previously [31,42]. In brief, Sserial 3-fold diluted serum, starting at 1:30, together with the virus control and cell control were incubated with 650 TCID50 of pseudovirus for 1 h at 37 °C in 96-well plates, then freshly trypsinized Huh-7 cells were added to each well (20,000 cells per well). Following incubation for 24 h in a 5% CO2 environment at 37 °C, luciferase substrate (Perkinelmer, 6066769) was added to each well and incubated for 2 min at room temperature, before 150 μl of the lysate was transferred to white solid 96-well plates for the detection of luminescence using an I-control Infinite 200 PRO microplate reader (TECAN). The determination to the neutralizing activity of serum antibody was expressed as 50% pseudovirus neutralizing antibody titer (NT50), which was defined as the serum dilution where the relative light units (RLUs) were reduced by half of the virus-only control wells and analyzed by non-linear regression, i.e., log (inhibitor) vs. normalized response (Variable slope), using GraphPad Prism 6.0 software.
(c) Cellular response analyzed by ELSpot assay
The DNCA/CLD-mRNA-1096-induced cellular immune response was evaluated using IFN-γ, TNF-α, IL-2, IL-4, or IL-6 precoated ELISpot kits (MabTech), according to the manufacturer’ s protocol. Briefly, plates were washed with PBS and blocked using RPMI 1640 (Thermo Fisher Scientific) with 10% FBS for 30 min at room temperature. Immunized mouse splenocytes (300,000 cells per well) were stimulated with peptide pool for SARS-CoV-2 RBD protein (5 μg/ml of each peptide; Miltenyi Biotec), PMA and Ionomycin (Dakewe) as positive control and RPMI 1640 media as negative control for 24 h, in a 5% CO2 environment at 37 °C. Following washes with PBS, plates were incubated with biotinylated anti-mouse IFN-γ, TNF-α, IL-2, IL-4, or IL-6 antibody for 2 h at room temperature. Spots were exposed on chromogenic reaction following the addition of TMB substrate solution and rinsed with deionized H2O. The air-dried spots were scanned and counted using an automated VSR07 ELISpot reader (AID). The numbers of spot-forming cells (SFU) per million cells were obtained and calculated by vSpot 7.0 software.
(d) Effector memory T cell proliferation detection by flow cytometry
Effector memory T cell proliferation in immunized mice were analyzed on a FACS Aria II flow cytometer (BD Biosciences). Briefly, a total of 300,000 mouse splenocytes was stimulated with SARS-CoV-2 RBD peptide pool (5 μg/ml of each peptide; Miltenyi Biotec) for 6 h in a 5% CO2 environment at 37 °C. Brefeldin A (5 μg/ml; Biolegend) was supplied into splenocytes and cells were continuously incubated for another 4 h. Following washes with PBS, Fc receptors of cells were blocked using CD16/CD32 antibodies (Mouse BD Fc Block; BD Biosciences) for 15 min at 4 °C. Next, dead cells were stained with Fixable Viability Dye eFluor™ 780 (Thermo Fisher Scientific) for 30 min at 4 °C in dark. After washes with cell staining buffer (BD Biosciences), splenocytes were stained with a cocktail of fluorescently conjugated antibodies to CD3 (PE; Biolegend), CD4/CD8 (APC; Biolegend), CD44 (FITC; Biolegend) and CD62L (PE/Cyanine7) for another 30 min at 4 °C in dark. Data were obtained by flow cytometry and analyzed by Flow J software, followed by final washes with cell staining buffer. The effector memory CD4+ and CD8+ T-cell response were represented as Tem, CD44+/CD62L−.
2.12. Safety evaluation of DNCA/CLD-mRNA-1096 in vivo
To assess the safety, the body weights of mice receiving vaccinations were recorded, and another batch of female BALB/c mice of 6–8 weeks old was involved in the tests. Sera were collected from the DNCA/CLD-mRNA-1096-vaccinated (mRNA-1096 = 10 μg) mice (n = 5) at 24 h post vaccination for liver and kidney function analyses using Chemray 240 and Chemray 800 (Rayto) automated Biochemical Analyzer, and the rest for immune activated cytokine analyses using IL-6 ELISA kit (Thermo Fisher Scientific) and TNF-α ELISA kit (Thermo Fisher Scientific). For histopathology analysis, organ tissues, including heart, liver, spleen, lung and kidney, were collected at 48 h post inoculation, and fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) before analysis by a NIKON Eclipse CI microscope.
2.13. Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.0 software package. All data were presented as the mean ± SEM. Difference between groups were analyzed as detailed in figure legends. Statistical significances were demonstrated in figures.
3. Results
3.1. The design of mRNA-1096 as the COVID-19 mRNA candidate molecule
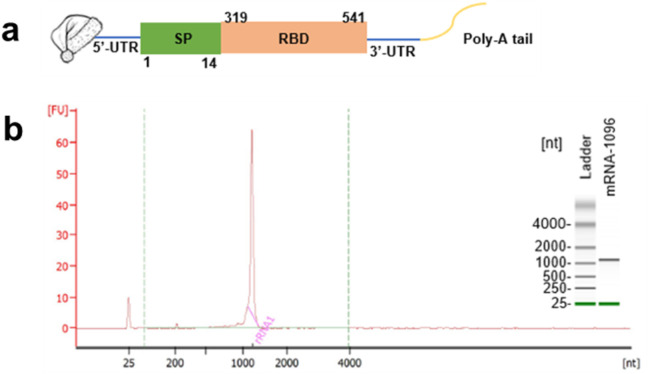
The COVID-19 mRNA molecule, mRNA-1096, began with a 5’-Cap 1, and incorporated 5′-and 3′- untranslated regions [43], then ended up with a 120 nt-Poly-A tail. As shown in Fig. 1a, the coding sequence region starting with AUG encoded the signal sequence (residues 1–14) and RBD (residues 319–541) of the spike glycoprotein genes of the SARS-CoV-2 Wuhan-Hu-1 isolate (GenBank accession number, NC_045512). The incorporation of the 5′- and 3′- untranslated regions as well as a poly-A tail improved the stability of mRNA-1096 and boosted antigen protein expression [[44], [45], [46]]. Briefly, the linearized templates for in vitro transcription were obtained from the chemically synthesized sequence recombined in PcDNA 3.1 (+) vector system containing a T7 promoter site, thus allowing for in vitro transcription in the sense orientation. Multiple portions of the purified mRNA-1096 were authenticated, as shown in Fig. 1b.
Fig. 1.
The design and Agilent 2100 bioanalysis of mRNA-1096. (a) mRNA-1096 was constructed by the optimized codons encoding the signal peptide (SP, 1–14 residues) and the receptor binding domain (RBD, 319–541 residues) of SARS-CoV-2 spike protein, flanked with the untranslated regions at both 5′- and 3′- terminus, and jointed with a Cap 1 at the 5′- terminal end and a Poly-A tail at the 3′- terminal end. (b) Multiple portions of in vitro synthesized and purified mRNA-1096 with desired size were authenticated and quantified by comparison with the heat-denatured Agilent RNA 6000 ladder using the Agilent 2100 bioanalyzer system.
3.2. The characteristics and bioactivities of DNCA/CLD-mRNA-1096 lipoplexes
The lipids, DNCA and CLD, could interact with mRNA to form lipoplexes. The optimization process of DNCA/CLD mRNA delivery system was shown in the supplementary data. As shown in Fig. S1 (a-b), the optimal formulation of the system, DNCA/CLD-mRNA-1096 (Formulation 3: N:P, 7:1) was able to retard mRNA completely in the gel slot and encapsulate mRNA with efficiency excessing 80%, demonstrating better applicability than the other formulations in the test. In addition to formulation, the optimal process of preparation was also considered. The particle size and polydispersity of DNCA/CLD-mRNA complexes (Formulation 3) prepared by sonication for 15 min at room temperature (Process 2: 40 kHz, 40% maximum power) were smaller and more even than the other processes in the test, as shown in Fig. S1 (c-d). After transfection in HEK293T cells, as shown in Fig. S1 (e-f), more robust expression of fluorescent protein was observed 24 h later from the DNCA/CLD-mRNA-eGFP complexed by Process 2. Similar with in vitro transfection, DNCA/CLD-mRNA-FLuc prepared by Process 2 resulted in more luciferase expression in mice, as shown in Fig. S1 (g-h). These results contributed to the optimal formulation and process of preparation of DNCA/CLD for the mRNA delivery in our test. However, it may not be the best, but it can be applied.
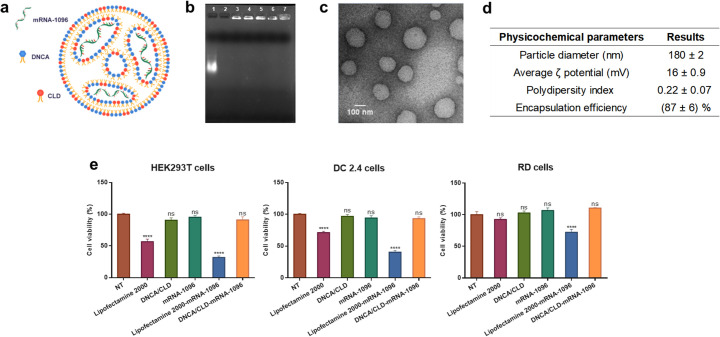
In the optimal formulation, the N/P molar ratio of lipids with mRNA was about 7: 1, and the molar ratio of DNCA/CLD to a single nucleotide of COVID-19 mRNA-1096 was about 0.9: 0.5: 1, as predicted in Fig. 2a. DNCA/CLD-mRNA-1096 lipoplexes were prepared by sonication for 15 min at room temperature (40 kHz, 40% maximum power) in a KQ-800DE ultrasonic cleaner (Kun Shan Ultrasonic Instruments Co., Ltd) at room temperature for 15 min before use. The lipoplexes were proved stable to RNase A challenge within the tested time periods, as shown in Fig. 2b. The morphologic TEM image in Fig. 2c suggested that mRNA-1096 was able to form spherical shaped and homogenously distributed nanocomplexes on encapsulation with DNCA/CLD. As summarized in Fig. 2d, the particle size of DNCA/CLD-mRNA-1096 (mRNA-1096 = 10 μg) was approximately 180 nm with a polydispersity index (PDI) of 0.22. The particle surface was slightly positively-charged, which might help to prevent aggregation. The optimal preparation also achieved mRNA encapsulation efficiency (EE) higher than 80%. All these results suggested DNCA/CLD was able to encapsulate mRNA-1096 to form stable lipoplexes. To evaluate the cytotoxicity of DNCA/CLD-mRNA-1096, cell viabilities of various types of cells were detected, after cells were treated for 24 h. As shown in Fig. 2e, the viabilities of the DNCA/CLD-mRNA-1096-, the DNCA/CLD- and the mRNA-1096-treated cells showed no significant loss compared with the non-treated cells, but reductions were detected from the Lipofectamine 2000- and the Lipofectamine 2000-mRNA-1096-treated cells. Therefore, DNCA/CLD-mRNA-1096 showed little cytotoxicity to the tested cell lines.
Fig. 2.
The characteristics and bioactivities of DNCA/CLD-mRNA-1096 lipoplexes. (a) Predicted illustration of DNCA/CLD-mRNA-1096. (b) RNase A resistance of DNCA/CLD-mRNA-1096 was tested by gel retardation assay. Samples were loaded as the following order: 1-naked mRNA-1096, 2-naked mRNA-1096 challenged with RNase A for 30 min, 3-DNCA/CLD-mRNA-1096, 4–7- DNCA/CLD-mRNA-1096 challenged with RNase A for 30 min, 60 min, 90 min, and 120 min, respectively. (c) A representative transmission electronic microscopic (TEM) image presented the morphology of DNCA/CLD-mRNA-1096. Scale bar = 100 nm, 8000 ×. (d) The physicochemical parameters of DNCA/CLD-mRNA-1096. (e) The viabilities of HEK293T, DC 2.4 and RD cells after transfection. Cells were analyzed at 24 h post treatment of the commercially available Lipofectamine 2000, the DNCA/CLD lipids, the naked mRNA-1096 (mRNA-1096 = 1 μg), the DNCA/CLD-mRNA-1096 (mRNA-1096 = 1 μg) and the Lipofectamine 2000-mRNA-1096 (mRNA-1096 = 1 μg). Data represented means ± standard error of means (SEM) of triplicate independent experiments with triplicate repeats in each. Statistical significance was calculated by a one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparison tests with non-treated (NT) group, (**** p < 0.0001; no significance (ns): p > 0.05).
3.3. The cellular internalization and protein expression of DNCA/CLD-mRNA-1096 lipoplexes
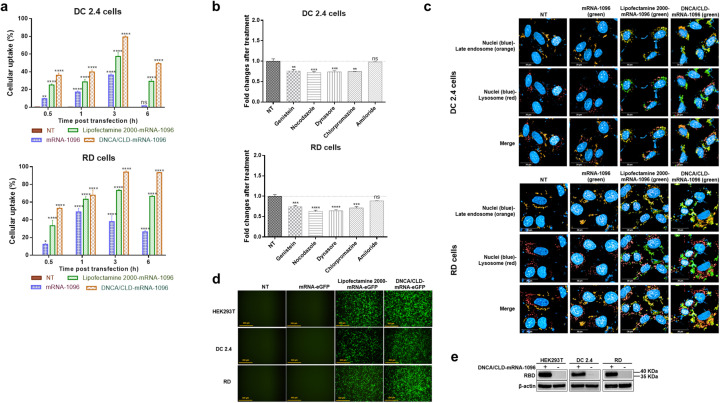
To understand the intracellular mechanisms of DNCA/CLD allowing mRNA translation, the cellular uptake of DNCA/CLD-mRNA by DC 2.4 and RD cells was monitored. DC 2.4 and RD cells were transfected with the MFP488 fluorescently labeled DNCA/CLD-mRNA-1096. As shown in Fig. 3a, at the early stage after transfection, the cellular uptake by DC 2.4 and RD cells both showed an increased tendency with time in all mRNA-transfected groups. At 3 h post transfection, the cellular uptake of the DNCA/CLD-mRNA-1096 group was about 1.4 folds higher in DC 2.4 cells and 1.3 folds higher in RD cells than the Lipofectamine 2000-mRNA-1096 group, and about 2 folds higher in DC 2.4 cells and 2.5 folds higher in RD cells than the naked mRNA-1096 group. These results might imply multiple portions of mRNA-1096 might already reach the cytoplasm where the expression initiated.
Fig. 3.
The cell entry and protein expression of DNCA/CLD-mRNA-1096. (a) The cellular uptake of DNCA/CLD-mRNA-1096 by DC 2.4 and RD cells at indicated time post transfection in comparison with the naked mRNA-1096 and Lipofectamine 2000-mRNA-1096. Statistical significance was calculated by a two-way ANOVA analysis followed by Dunnett's multiple comparison tests with the NT group, (**** p < 0.0001, ** p < 0.01; ns: p > 0.05). Error bars were shown as the SEM of triplicate experiments. (b) Analysis of the cell internalization mechanisms of DNCA/CLD-mRNA-1096 by DC 2.4 and RD cells in the presence of endocytosis pathway inhibitors at 3 h post transfection. The value of the non-transfected (NT) group was set as one. Statistical significance was calculated by one-way ANOVA analysis followed by Dunnett's multiple comparison tests with the NT group, (**** p < 0.0001, *** p < 0.001, ** p < 0.01; ns: p > 0.05). Error bars were shown as the SEM of triplicate experiments. (c) Representative images of cytosolic mRNA-1096 in DC 2.4 and RD cells at 3 h post transfection. Scale bar = 20 μm, 63 ×. (d) Representative fluorescent microscopic images of the eGFP protein expression at 24 h post transfection. Scale bar = 500 μm, 10 ×. (e) Western blotting analysis of the SARS-CoV-2 RBD protein expression from cell lysates at 24 h post transfection.
Then, the mechanisms of internalization of DNCA/CLD-mRNA-1096 were studied by treating DC2.4 and RD cells with various endocytosis inhibitors [47]. The targets and mechanisms of action of each inhibitor were described in supplementary Table 1. Similar results were obtained from both cell types, as shown in Fig. 3b. Except for the lack of inhibition by amiloride, different levels of inhibition were observed in cells treated with other inhibitors, which suggested the macropinocytosis was not essential for the uptake. On the contrary, the clathrin- and caveolae-dependent endocytosis were probably the main paths of the cellular uptake for the complexes. However, these pathways would induce the formation of the early endosomes with vesicular structures trapping the complexes coming from the cell surface [48]. Then the late endosomes receive the internalized complexes from the early endosomes and they are thought to mediate a final set of sorting events before interaction with lysosomes [49]. Lysosomes as the last parts of the endocytic pathway contain the hydrolytic enzymes digesting the contents of the late endosomes [49]. Therefore, DNCA/CLD-mRNA complexes must escape the endosomes before lysosome fusion or escape from lysosomes before the lysosome mediated digestion of the complexes, to allow mRNA reach the cytoplasm to be translated.
To confirm whether mRNA reached cytoplasm, DC 2.4 and RD cells were genetically modified to express the fluorescent RFP fused to Rab 7, so that the late endosomes-RFP could be observed in live cells [48]. Then the MFP488 fluorescently labeled DNCA-CLD-mRNA was added into the cells, and incubated. After removal of the cell supernatants, cells were stained with Lysotracker deep red for visualization of lysosomes, then stained with Hoechst to exhibit nuclei. At 3 h post transfection, cells were observed by Operetta CLS™ High Content System. As shown in Fig. 3c, the naked MFP488 labeled mRNA-1096 was not shown in the test, but both the DNCA/CLD- and Lipofectamine 2000-encapsulated MFP488 labeled mRNA-1096 appeared in the cytoplasm and co-existed with the late endosomes and lysosomes, which might suggest the escape from endosomes before lysosome fusion, or the escape from lysosomes before the degradation of the transfection complexes.
As mRNA was observed in the cytoplasm, the in vitro protein expression was detected on different cells after transfected with the mRNA encoding eGFP and SARS-CoV-2 RBD and cultured for 24 h. The green fluorescent signals of eGFP were generally intense on the tested cells, as shown in Fig. 3d. Receptor binding domain of SARS-CoV-2 spike protein was also expressed on these cell types, detected by the Western blotting analysis, as shown in Fig. 3e. Based on these results, the protein expression capability of DNCA/CLD-mRNA in various cells was strongly confirmed. To some extents, the efficacy of DNCA/CLD-mRNA-triggered protein expression was even more robust than the Lipofectamine 2000-mRNA reference group.
3.4. The in vivo delivery and biodistribution of DNCA/CLD-mRNA-Fluc in mice
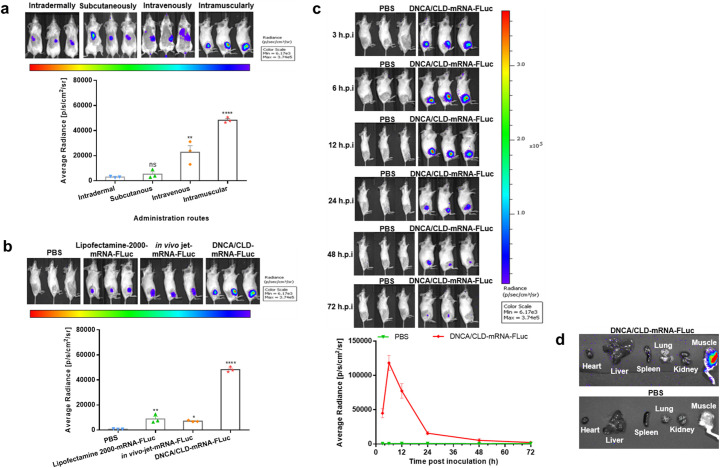
The capability of DNCA/CLD-mRNA delivery in mice was evaluated. To visualize the biodistribution, mRNA-FLuc (10 μg) was prepared with DNCA/CLD and subjected to bioluminescent imaging analysis via different inoculation routes. After intramuscular injection, bioluminescent signals of firefly luciferase expression were seen at the injection site of BALB/c mice as early as 3 h post injection, while weaker signals were detected in mice receiving subcutaneous, intradermal, or intravenous injection, as shown in Fig. 4a. According to Fig. 4b, the bioluminescent signals released by the DNCA/CLD-mRNA-FLuc inoculated mice were more intense than those released by the in vivo-jet-mRNA-FLuc inoculated or the Lipofectamine 2000-mRNA-FLuc inoculated ones. Then, the mice receiving intramuscular inoculation were real-time monitored, as shown in Fig. 4c. The photon flux began to decline since peak at 6 h post injection, then faded hardly detectable at 72 h post injection. Ex vivo imaging analysis of the DNCA/CLD-mRNA-FLuc, as shown in Fig. 4d, further proved that the mouse muscle at the injection site represented the most abundant protein expression tissue, while almost no bioluminescence was detected in other organ tissues via intramuscular inoculation. Meanwhile, ex vivo imaging showed the intravenous administration resulted in DNCA/CLD-mRNA-FLuc trafficking to spleen and lung, as shown in Fig. S2, suggesting DNCA/CLD might be a versatile delivery system for different applications of mRNA. All the data demonstrated that DNCA/CLD could improve the efficacy of mRNA delivery in vivo.
Fig. 4.
The in vivo delivery of the firefly reporter DNCA/CLD-mRNA in mice. Female BALB/c mice of 6–8 weeks old were inoculated with DNCA/CLD-mRNA-FLuc (mRNA-FLuc = 10 μg). (a) Representative bioluminescent images were obtained and analyzed at 6 h post inoculation of DNCA/CLD-mRNA-FLuc via different administration routes. (b) Representative bioluminescent images were obtained at 6 h post inoculation, and the photon flux intensities released from the commercially available in vivo jet-mRNA-FLuc-, Lipofectamine 2000-mRNA-FLuc-, and the DNCA/CLD-mRNA-FLuc-inoculated mice were analyzed after reaction with luciferase substrate. (c) Real-time monitor of the firefly luciferase expression in the DNCA/CLD-mRNA-FLuc-intramuscularly inoculated mice at indicated hours post inoculation (h.p.i). (d) Representative tissue biodistribution of DNCA/CLD-mRNA-FLuc in mice after intramuscular inoculation at 6 h, and mice treated with PBS were set as negative control. The intensity of photon flux was expressed by average radiance. Statistical significance was analyzed by a one-way ANOVA analysis followed by Dunnett's multiple comparisons (**** p < 0.0001, ** p < 0.01; ns: p > 0.05). Error bars were shown as the SEM of the quantified photon flux intensity released by the three tested mice.
3.5. The in vivo efficacy and immunogenicity of DNCA/CLD-mRNA-1096 in mice
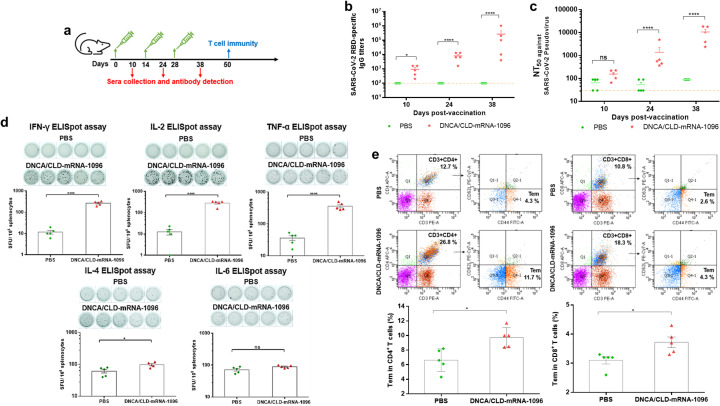
The efficacy and immunogenicity of DNCA/CLD-mRNA-1096 was evaluated with immunocompetent female BALB/c mice of 6–8 weeks old. DNCA/CLD-mRNA-1096 (mRNA-1096 = 10 μg) was prepared. BALB/c mice were intramuscularly vaccinated with DNCA/CLD-mRNA-1096, according to the immunization process illustrated in Fig. 5a. Following injection, no obvious local inflammation response was observed at the site of injection, and the body weight resumed at 2 days post injection. After boost immunization for twice with the same dose, the serum SARS-CoV-2 RBD-specific IgG antibody level in the DNCA/CLD-mRNA-1096-vaccinated mice remarkably increased. At 38 days post vaccination, as shown in Fig. 5 b and c, the SARS-CoV-2 RBD-specific IgG antibody titer and neutralizing antibody titer against pseudovirus (NT50) approached ~1:262,800 and ~1:10,591, respectively. Given the robust antibody level detected in mouse sera, it could be speculated that DNCA/CLD-mRNA-1096 had elicited potent humoral immune response in mice. As mRNA vaccines are able to elicit both humoral and cellular immune response, whether SARS-CoV-2 RBD-specific T cell immune response was evoked by boosting two doses of immunization of DNCA/CLD-mRNA-1096 in mice through intramuscular administration was further studied. ELISpot assays, as shown in Fig. 5d, showed that on stimulation with peptide pools covering SARS-CoV-2 RBD, the secretion of IFN-γ (Th1), IL-2 (Th1), TNF-α (Th1) in splenocytes of the DNCA/CLD-mRNA-1096-vaccinated mice was significantly more abundant in comparison with those treated with PBS, whereas no remarkable increase in IL-4 (Th2) and IL-6 (Th2) secretion was detected between the DNCA/CLD-mRNA-1096- and the PBS-treated mice. To further explore whether the DNCA/CLD-mRNA-1096-induced T cellular immune response produced memory effect, flow cytometry was performed to analyze the SARS-CoV-2 RBD-specific CD4+ and CD8+ effector memory T cells (Tem). As shown in Fig. 5e, flow cytometric results showed a noteworthy increase in CD4+ Tem and CD8+ Tem in the splenocytes of the DNCA/CLD-mRNA-1096-vaccinated mice. To analyze from these results, DNCA/CLD-mRNA-1096 had elicited long-lived Th1-biased SARS-CoV-2 RBD-specific cellular immune response in mice.
Fig. 5.
The in vivo efficacy and immunogenicity of DNCA/CLD-mRNA-1096. Female BALB/c mice of 6–8 weeks old were vaccinated with DNCA/CLD-mRNA-1096 (mRNA-1096 = 10 μg) or PBS via intramuscular route of administration. (a) The schematic diagram depicted the strategy of immunization, and the plans of sera collection and immunogenicity detection. (b) The SARS-CoV-2 RBD-specific IgG titer in the sera of the vaccinated mice was tested by ELISA. (c) The serum neutralization antibody titer (NT50) was tested against SARS-CoV-2-spike protein pseudovirus. The dashed line showed the limit of the test. (d) ELISpot detection of IFN-γ, IL-2, TNF-α, IL-4, or IL-6 release from the SARS-CoV-2 RBD peptide stimulated splenocytes. (e) Flow cytometric detection of the SARS-CoV-2 RBD-specific CD4+ and CD8+ effector memory T cells (CD44+CD62L−) in splenocytes. Data was shown as means ± SEM. Statistical significance (b-c) was analyzed by a two-way ANOVA analysis followed by Bonferroni's multiple comparisons between the PBS and DNCA/CLD-mRNA-1096 group, (**** p < 0.0001, * p < 0.05; ns: p > 0.05). Statistical significance (d-e) was analyzed by the two-tailed unpaired Student's t-tests, for the comparison of the differences between the PBS and DNCA/CLD-mRNA-1096 group, (**** p < 0.0001, *** P < 0.001, * p < 0.05; ns: p > 0.05).
3.6. The safety of DNCA/CLD-mRNA-1096 in mice
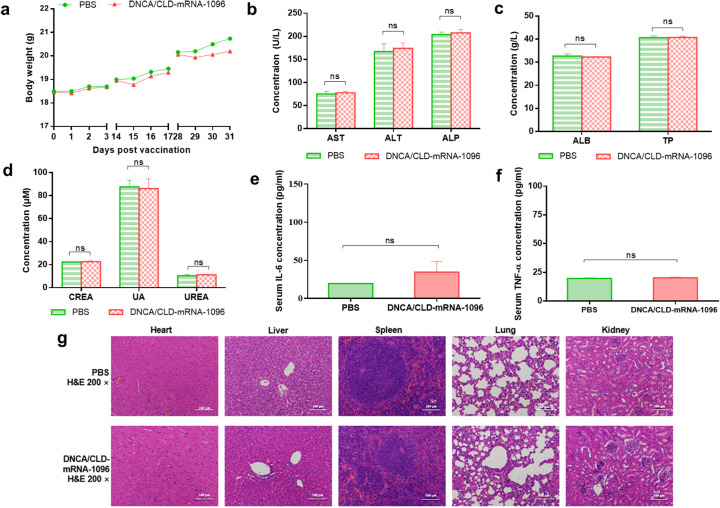
To evaluate the in vivo safety of DNCA/CLD-mRNA-1096, many aspects were concerned, including mice body weight records, biochemical indexes, immune activated factors and tissue histopathological changes. The body weight records on DNCA/CLD-mRNA-1096-vaccinated mice recovered at 2 days post immunization, since then, no more loss of body weight was recorded, as shown in Fig. 6a. In addition, all mice survived until sacrificed for T cellular response analysis at 50 days post initial immunization. The mouse liver and kidney functions were evaluated by blood biochemical indexes, and all data were shown within the normal extent, as exhibited in Fig. 6 (b-d). The levels of proinflammatory cytokines, including IL-6 and TNF-α, were measured, as a result shown in Fig. 6 (e-f), and no significant immune stimulation was observed in DNCA/CLD-mRNA-1096-vaccinated mice. Since no difference among the H&E-stained histopathological tissue sections, as illustrated in Fig. 6g, the safety of DNCA/CLD-mRNA-1096 in mice was further confirmed. These data proved that DNCA/CLD could be used for safe and effective mRNA delivery in vivo.
Fig. 6.
The safety evaluation of DNCA/CLD-mRNA-1096 in mice. Female BALB/c mice of 6–8 weeks old were inoculated with DNCA/CLD-mRNA-1096 (mRNA-1096 = 10 μg) or PBS via intramuscular route of administration. (a) The body weight records of mice on the first three days after each vaccination. (b-d) The liver and kidney function were determined by blood biochemical indexes (n = 5). ALT, AST, ALP, TP and ALB represented the liver function, while CREA-J, UA and UREA represented kidney function. (e-f) The immune activation effects in mice were represented by analysis of the serum proinflammatory cytokines IL-6 and TNF-α (n = 5). Data were shown as means ± SEM. Statistical significance was calculated with the unpaired Student's t-test (two-tailed) for comparing the difference between the PBS and the DNCA/CLD-mRNA-1096 group (ns: p > 0.05). (g) Representative histopathology (H&E) of different tissues, heart, liver, spleen, lung and kidney, in the PBS or the DNCA/CLD-mRNA-1096 group. H&E-stained sections shown in the data were one representative result of the three tested mice at 48 h post inoculation. Scale bar = 100 μm, 200 × .
4. Discussions
The therapeutic mRNA has recently emerged as a promising novel drug class with the potential to be applied to various therapeutic modalities, including protein replacement and vaccination against cancer and infectious diseases. Numerous approaches have been pursued to develop potent mRNA vaccines, many of them have proved to be successful with convincing potentials as alternatives or complementarities to conventional vaccine strategies [50]. With advance in major biotechnological innovation and research investment for developing mRNA as an immunotherapeutic tool, mRNA therapies, especially mRNA-based vaccines, are currently developing fast. and some are being applied to build the herd immunity against the ongoing COVID-19 pandemic [51,52]. Despite substantial advances that have been made in the rational design and chemical modification of mRNA have solved the key challenges regarding to the adjustment of its inherent instability and immune activation, another major challenge for the application of mRNA is to achieve efficient and safe in vivo delivery [53]. Inspired by the previous experience from gene and RNA interference (RNAi) therapy, non-viral vectors were explored as mRNA delivery [16,[53], [54], [55], [56]], especially those based on biocompatible nanotechnologies, due to their elasticity on synthesis and modification. Thus, we synthesized the neutral cytidinyl-lipid, DNCA, and a cationic lipid, CLD, aiming at mRNA delivery. Moreover, mRNA-1096, a COVID-19 mRNA candidate molecule, was designed and synthesized by in vitro transcription, for the evaluation of the application of DNCA/CLD.
According to the optimization process, the protection of mRNA from degradation before transfection is the most important, thus effective encapsulation may be one of the key factors of effective mRNA transfection. Then, the physicochemical characteristics of the DNCA/CLD-mRNA complexes, such as particle diameters and polydispersity, are also key factors. When prepared not by the optimal process, the DNCA/CLD-mRNA complexes might be poorly generated, for maybe either insufficient or over combination, thus producing large size and uneven distribution, and finally led to weaker protein expression after transfection.
By the rational preparation, DNCA/CLD was made more customized for mRNA encapsulation. The neutral lipid DNCA with a highly specialized polar head, cytidine, provided additional hydrogen bonding with the complementary nucleobases via Watson-Crick base pairing and π-stacking. The lipid CLD with the cationic amino head electrostatically interacted with the phosphodiester backbone of the mRNA. DNCA with the assistance of CLD (DNCA/CLD, molar ratio, 9:5) effectively encapsulated and compacted the single-stranded mRNA-1096, and meanwhile the lipids spontaneously self-assembled in water with low toxicity and immunogenicity, and so the spherical-shaped and slightly cationic DNCA/CLD-mRNA-1096 nanocomplexes with particle size at approximately 180 nm (mRNA-1096 = 10 μg) were formed and homogenously distributed under mild sonication.
According to the analysis from in vitro application, DNCA/CLD demonstrated better efficiency at mRNA cellular uptake than the commercial liposome Lipofectamine 2000. Then, mRNA-1096 reached the cytoplasm to initiate robust protein expression in various cell lines, including the somatic cells and the antigen-presenting cells. In addition, DNCA/CLD was also found more feasible to deliver mRNA in vivo than the commercial in vivo-jetRNA® mRNA delivery reagent and Lipofectamine 2000. The stronger luciferase expression in the intramuscularly inoculated mice might due to the simplicity of the application, and the large number of cells that take up the DNCA/CLD-mRNA nanocomplexes to efficiently uptake and translate the mRNA. Also, the distinctive features of size and composition of DNCA/CLD-mRNA allowed the retention and the expression at the site of injection, thus avoiding organ-specific side effects, such as the liver accumulating effect observed with LNP mRNA formulations [57,58], and this biodistribution pattern may trigger the activation of liver-resident cells that subsequently lead to tissue damage and inflammation, and finally contribute to high reactogenicity [58]. Moreover, the skeletal muscle represents an excellent site of vaccination, for an abundant existence of dendritic cells, macrophages and other immune cells also provide good antigen presentation, thus DNCA/CLD might deliver mRNA-1096 encoding the RBD of SARS-CoV-2 spike protein in vivo to evoke immunity.
Thereafter, the DNCA/CLD-mRNA-1096 lipoplexes were intramuscularly applied to mice. After boost immunization for twice, the DNCA/CLD-mRNA-1096-induced SARS-CoV-2 RBD-specific IgG antibody (~1:262,800) and neutralizing antibody (~1:10,591) against pseudotyped SARS-CoV-2 infection were detected in sera. Long-lived Th1-biased cellular response was confirmed by the proliferation of CD4+ and CD8+ Tem cells in addition to the significantly increased secretion of Th1-biased cytokines, including IFN-γ, TNF-α and IL-2, in mouse SARS-CoV-2 RBD-stimulated splenocytes. It could be speculated that DNCA/CLD-mRNA-1096 lipoplexes were taken by various types of cells and expressed as RBD antigens, recognized by innate receptors, thus evoking the innate immune system including antigen-presenting cells. The activated antigen-presenting cells then presented peptides derived from the endogenously expressed RBD or the phagocytosed RBD antigens on MHC-I or MHC-II, respectively, which induced an efficient priming of the adaptive immune system and led to the expansion of antigen-specific T and B cells, and elicited a balanced humoral and cellular immune response. Meanwhile, the DNCA/CLD-mRNA-1096-vaccinated mice kept in good health, and generally, no significant adverse events were observed, thus suggesting DNCA/CLD a novel mRNA delivery with good efficacy and safety.
However, further optimization on the formulation and preparation, such as the inclusion of other biocompatible excipients and the application of more feasible lipid-mRNA blending process, may further improve the efficacy of DNCA/CLD-mRNA-1096 as a COVID-19 vaccine candidate and reduce immunization doses to a great extent. Overall, our study not only proposed DNCA/CLD as a promising mRNA delivery system, but also brought a new idea for developing efficacious and safe mRNA delivery systems.
Author contributions
S.Q.W., J.Y. and Z.J.Y. conceived the project. L.L., J.Y., Z.J.Y. and S.Q.W. designed the experiment. L.L. and J.R.L. performed the main experiments. Y.S., X.W., X.Y.Z., Y.F.P., Y.M.C. and H.Y.H. provided experimental support. L.L. analyzed the data and wrote the manuscript. J.Y., Z.J.Y. and S.Q.W supervised the research and revised the manuscript.
CRediT authorship contribution statement
Lei Li: Investigation, Methodology, Formal analysis, Data curation, Writing-original draft, Funding acquisition. Jinrong Long: Investigation, Methodology, Resources, Formal analysis, Data curation. Ye Sang: Investigation, Methodology, Resources. Xin Wang: Investigation, Methodology, Resources. Xinyang Zhou: Methodology, Resources. Yufei Pan: Methodology, Resources. Yiming Cao: Investigation, Methodology. Huiyuan Huang: Investigation, Methodology. Zhenjun Yang: Conceptualization, Methodology, Resources, Formal analysis, Supervision, Funding acquisition. Jing Yang: Conceptualization, Methodology, Formal analysis, Data curation, Writing - review & editing, Visualization, Supervision, Funding acquisition. Shengqi Wang: Conceptualization, Methodology, Formal analysis, Data curation, Writing - review & editing, Visualization, Supervision, Funding acquisition.
Declaration of Competing Interest
No potential conflict of competing interest was reported by the authors.
Acknowledgements
The study was supported by the National Natural Science Foundation of China (Grant No. 81830101) and the National Science and Technology major Project (Grant No. 2017ZX09303013-003). L.L. was supported by the China Postdoctoral Science Foundation (Grant No: 2020T130136ZX).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jconrel.2021.10.023.
Appendix A. Supplementary data
Supplementary material 1
References
- 1.Boni M.F., Lemey P., Jiang X., Lam T.T.-Y., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5(11):1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 2.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 — Preliminary Report. N. Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., Nagata B.M., Andersen H., Martinez D.R., Noe A.T., Douek N., Donaldson M.M., Nji N.N., Alvarado G.S., Edwards D.K., Flebbe D.R., Lamb E., Doria-Rose N.A., Lin B.C., Louder M.K., O’Dell S., Schmidt S.D., Phung E., Chang L.A., Yap C., Todd J.-P.M., Pessaint L., Van Ry A., Browne S., Greenhouse J., Putman-Taylor T., Strasbaugh A., Campbell T.-A., Cook A., Dodson A., Steingrebe K., Shi W., Zhang Y., Abiona O.M., Wang L., Pegu A., Yang E.S., Leung K., Zhou T., Teng I.T., Widge A., Gordon I., Novik L., Gillespie R.A., Loomis R.J., Moliva J.I., Stewart-Jones G., Himansu S., Kong W.-P., Nason M.C., Morabito K.M., Ruckwardt T.J., Ledgerwood J.E., Gaudinski M.R., Kwong P.D., Mascola J.R., Carfi A., Lewis M.G., Baric R.S., McDermott A., Moore I.N., Sullivan N.J., Roederer M., Seder R.A., Graham B.S. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., Cooper D., Weaver S.C., Muik A., Sahin U., Jansen K.U., Xie X., Dormitzer P.R., Shi P.Y. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021;384(15):1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner P.J., Ansotegui I.J., Campbell D.E., Cardona V., Ebisawa M., El-Gamal Y., Fineman S., Geller M., Gonzalez-Estrada A., Greenberger P.A., Leung A.S.Y., Levin M.E., Muraro A., Sánchez Borges M., Senna G., Tanno L.K., Yu-Hor Thong B., Worm M. COVID-19 vaccine-associated anaphylaxis: a statement of the world allergy organization anaphylaxis committee. World Allergy Organiz. J. 2021;14(2):100517. doi: 10.1016/j.waojou.2021.100517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerji A., Wickner P.G., Saff R., Stone C.A., Jr., Robinson L.B., Long A.A., Wolfson A.R., Williams P., Khan D.A., Phillips E., Blumenthal K.G. mRNA vaccines to prevent COVID-19 disease and reported allergic reactions: current evidence and suggested approach. J Allergy Clin Immunol Pract. 2021;9(4):1423–1437. doi: 10.1016/j.jaip.2020.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wadman M. Public needs to prep for vaccine side effects. Science. 2020;370(6520):1022. doi: 10.1126/science.370.6520.1022. [DOI] [PubMed] [Google Scholar]
- 9.Ndeupen S., Qin Z., Jacobsen S., Estanbouli H., Bouteau A., Igyártó B.Z. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. bioRxiv. July 23 2021 doi: 10.1016/j.isci.2021.103479. 2021.03.04.430128, [Accessed on September 20 2021] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selvaraj G., Kaliamurthi S., Peslherbe G.H., Wei D.-Q. Are the allergic reactions of COVID-19 vaccines caused by mRNA constructs or nanocarriers? Immunol. Insights Interdisc. Sci. Comput. Life Sci. 2021;13(2):344–347. doi: 10.1007/s12539-021-00438-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13(10):759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 12.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Therapy: J. Am. Soc. Gene Therapy. 2019;27(4):710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanderson K. Bioengineering: what to make with DNA origami. Nature. 2010;464(7286):158–159. doi: 10.1038/464158a. [DOI] [PubMed] [Google Scholar]
- 14.Simeone L., Mangiapia G., Irace C., Di Pascale A., Colonna A., Ortona O., De Napoli L., Montesarchio D., Paduano L. Nucleolipid nanovectors as molecular carriers for potential applications in drug delivery. Mol. BioSyst. 2011;7(11):3075–3086. doi: 10.1039/c1mb05143a. [DOI] [PubMed] [Google Scholar]
- 15.Cuomo F., Ceglie A., Lopez F. Specific interactions between nucleolipid doped liposomes and DNA allow a more efficient polynucleotide condensation. J. Colloid Interface Sci. 2012;365(1):184–190. doi: 10.1016/j.jcis.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Gissot A., Camplo M., Grinstaff M.W., Barthélémy P. Nucleoside, nucleotide and oligonucleotide based amphiphiles: a successful marriage of nucleic acids with lipids. Org. Biomol. Chem. 2008;6(8):1324–1333. doi: 10.1039/b719280k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaManna C.M., Lusic H., Camplo M., McIntosh T.J., Barthélémy P., Grinstaff M.W. Charge-reversal lipids, peptide-based lipids, and nucleoside-based lipids for gene delivery. Acc. Chem. Res. 2012;45(7):1026–1038. doi: 10.1021/ar200228y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohli A.G., Kierstead P.H., Venditto V.J., Walsh C.L., Szoka F.C. Designer lipids for drug delivery: From heads to tails. J. Control. Release. 2014;190:274–287. doi: 10.1016/j.jconrel.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulet X., Kaasgaard T., Conn C.E., Waddington L.J., Kennedy D.F., Weerawardena A., Drummond C.J. Nanostructured nonionic thymidine nucleolipid self-assembly materials. Langmuir. 2010;26(23):18415–18423. doi: 10.1021/la103370q. [DOI] [PubMed] [Google Scholar]
- 20.Latxague L., Patwa A., Amigues E., Barthélémy P. Glycosyl-nucleolipids as new bioinspired amphiphiles. Molecules. 2013;18(10):12241–12263. doi: 10.3390/molecules181012241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X., Wang S., Zhu Y., Pan Y., Zhang L., Yang Z. Overcoming the delivery barrier of oligonucleotide drugs and enhancing nucleoside drug efficiency: the use of nucleolipids. Med. Res. Rev. 2020;40(4):1178–1199. doi: 10.1002/med.21652. [DOI] [PubMed] [Google Scholar]
- 22.Ceballos C., Prata C.A.H., Giorgio S., Garzino F., Payet D., Barthélémy P., Grinstaff M.W., Camplo M. Cationic nucleoside lipids based on a 3-nitropyrrole universal base for siRNA delivery. Bioconjug. Chem. 2009;20(2):193–196. doi: 10.1021/bc800432n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan D., Sun J., Jin H., Li Y., Li L., Wu Y., Zhang L., Yang Z. Supramolecular assemblies of novel aminonucleoside phospholipids and their bonding to nucleic acids. Chem. Commun. (Camb.) 2015;51(3):469–472. doi: 10.1039/c4cc07538b. [DOI] [PubMed] [Google Scholar]
- 24.Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., Qu Y., Li F., Lv Q., Wang W., Xue J., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao L., Liu P., Zhao L., Ye F., Wang H., Zhou W., Zhu N., Zhen W., Yu H., Zhang X., Guo L., Chen L., Wang C., Wang Y., Wang X., Xiao Y., Sun Q., Liu H., Zhu F., Ma C., Yan L., Yang M., Han J., Xu W., Tan W., Peng X., Jin Q., Wu G., Qin C. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 25.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.-S., Wang Q., Gao G.F., Yuan Z., Yan J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 26.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.-d., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X.S. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182(1) doi: 10.1016/j.cell.2020.05.025. 73–84.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Martinez D.R., Williamson L.E., Chen E.C., Jones T., Day S., Myers L., Hassan A.O., Kafai N.M., Winkler E.S., Fox J.M., Shrihari S., Mueller B.K., Meiler J., Chandrashekar A., Mercado N.B., Steinhardt J.J., Ren K., Loo Y.-M., Kallewaard N.L., McCune B.T., Keeler S.P., Holtzman M.J., Barouch D.H., Gralinski L.E., Baric R.S., Thackray L.B., Diamond M.S., Carnahan R.H., Crowe J.E. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584(7821):443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J., Lu G., Tan S., Xia J., Xiong H., Yu X., Qi Q., Yu X., Li L., Yu H., Xia N., Zhang T., Xu Y., Lin J. A COVID-19 mRNA vaccine encoding SARS-CoV-2 virus-like particles induces a strong antiviral-like immune response in mice. Cell Res. 2020;30(10):936–939. doi: 10.1038/s41422-020-00392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai W., Zhang X., Drelich A., Shi J., Hsu J.C., Luchsinger L., Hillyer C.D., Tseng C.-T.K., Jiang S., Du L. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020;30(10):932–935. doi: 10.1038/s41422-020-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., Hong W., Yang Y., Zhao Y., Ye F., Lin S., Deng W., Chen H., Lei H., Zhang Z., Luo M., Gao H., Zheng Y., Gong Y., Jiang X., Xu Y., Lv Q., Li D., Wang M., Li F., Wang S., Wang G., Yu P., Qu Y., Yang L., Deng H., Tong A., Li J., Wang Z., Yang J., Shen G., Zhao Z., Li Y., Luo J., Liu H., Yu W., Yang M., Xu J., Wang J., Li H., Wang H., Kuang D., Lin P., Hu Z., Guo W., Cheng W., He Y., Song X., Chen C., Xue Z., Yao S., Chen L., Ma X., Chen S., Gou M., Huang W., Wang Y., Fan C., Tian Z., Shi M., Wang F.-S., Dai L., Wu M., Li G., Wang G., Peng Y., Qian Z., Huang C., Lau J.Y.-N., Yang Z., Wei Y., Cen X., Peng X., Qin C., Zhang K., Lu G., Wei X. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 31.Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G., Huang W.-J., Gao P., Zhou C., Zhang R.-R., Guo Y., Sun S.-H., Fan H., Zu S.-L., Chen Q., He Q., Cao T.-S., Huang X.-Y., Qiu H.-Y., Nie J.-H., Jiang Y., Yan H.-Y., Ye Q., Zhong X., Xue X.-L., Zha Z.-Y., Zhou D., Yang X., Wang Y.-C., Ying B., Qin C.-F. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182(5) doi: 10.1016/j.cell.2020.07.024. 1271–1283.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5(10):1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- 33.Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M., Yang M., Li Y., Cheng H., Yuan Y., Zhang W., Ke C., Wong G., Qi J., Qin C., Yan J., Gao G.F. A universal design of Betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182(3) doi: 10.1016/j.cell.2020.06.035. 722–733.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Q., Ji K., Tian S., Wang F., Huang B., Tong Z., Tan S., Hao J., Wang Q., Tan W., Gao G.F., Yan J. Publisher correction: a single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat. Commun. 2021;12(1):2355. doi: 10.1038/s41467-021-22820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma Y., Zhu Y., Wang C., Pan D., Liu S., Yang M., Xiao Z., Yang X., Zhao W., Zhou X., Li Y., Pan Y., Sun J., Wang S., Guan Z., Zhang L., Yang Z. Annealing novel nucleobase-lipids with oligonucleotides or plasmid DNA based on H-bonding or π-π interaction: assemblies and transfections. Biomaterials. 2018;178:147–157. doi: 10.1016/j.biomaterials.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Zheng Y., Guo Y., Li Y., Wu Y., Zhang L., Yang Z. A novel gemini-like cationic lipid for the efficient delivery of siRNA. New J. Chem. 2014;38(10):4952–4962. [Google Scholar]
- 37.Wang L.H., Rothberg K.G., Anderson R.G. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J. Cell Biol. 1993;123(5):1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khalil I.A., Kogure K., Akita H., Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol. Rev. 2006;58(1):32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 39.Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell. 2006;10(6):839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Idilli A.I., Morandini P., Onelli E., Rodighiero S., Caccianiga M., Moscatelli A. Microtubule depolymerization affects endocytosis and exocytosis in the tip and influences endosome movement in tobacco pollen tubes. Mol. Plant. 2013;6(4):1109–1130. doi: 10.1093/mp/sst099. [DOI] [PubMed] [Google Scholar]
- 41.Lim J.P., Gleeson P.A. Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol. Cell Biol. 2011;89(8):836–843. doi: 10.1038/icb.2011.20. [DOI] [PubMed] [Google Scholar]
- 42.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Fan C., Huang W., Xu M., Wang Y. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microb. Infect. 2020;9(1):680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168(6) doi: 10.1016/j.cell.2017.02.017. 1114–1125.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strong T.V., Hampton T.A., Louro I., Bilbao G., Conry R.M., Curiel D.T. Incorporation of β-globin untranslated regions into a Sindbis virus vector for augmentation of heterologous mRNA expression. Gene Ther. 1997;4(6):624–627. doi: 10.1038/sj.gt.3300423. [DOI] [PubMed] [Google Scholar]
- 45.Jemielity J., Kowalska J., Rydzik A.M., Darzynkiewicz E. ChemInform abstract: synthetic mRNA cap analogues with a modified triphosphate bridge — synthesis, applications and prospects. ChemInform. 2010;41(34) [Google Scholar]
- 46.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.-J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9(11):1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu E., Saltzman W.M., Piotrowski-Daspit A.S. Escaping the endosome: assessing cellular trafficking mechanisms of non-viral vehicles. J. Control. Release. 2021;335:465–480. doi: 10.1016/j.jconrel.2021.05.038. [DOI] [PubMed] [Google Scholar]
- 48.Midoux P., Pichon C. Lipid-based mRNA vaccine delivery systems. Expert Rev. Vacc. 2015;14(2):221–234. doi: 10.1586/14760584.2015.986104. [DOI] [PubMed] [Google Scholar]
- 49.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151(3):220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Xiong Q., Lee G.Y., Ding J., Li W., Shi J. Biomedical applications of mRNA nanomedicine. Nano Res. 2018;11(10):5281–5309. doi: 10.1007/s12274-018-2146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng Z., O'Keeffe-Ahern J., Lyu J., Pierucci L., Zhou D., Wang W. A new developing class of gene delivery: messenger RNA-based therapeutics. Biomater. Sci. 2017;5(12):2381–2392. doi: 10.1039/c7bm00712d. [DOI] [PubMed] [Google Scholar]
- 52.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nature Rev. Mater. 2017;2(10):17056. [Google Scholar]
- 54.Cho S.-W., Goldberg M., Son S.M., Xu Q., Yang F., Mei Y., Bogatyrev S., Langer R., Anderson D.G. Lipid-like nanoparticles for small interfering RNA delivery to endothelial cells. Adv. Funct. Mater. 2009;19(19):3112–3118. doi: 10.1002/adfm.200900519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhi D., Bai Y., Yang J., Cui S., Zhao Y., Chen H., Zhang S. A review on cationic lipids with different linkers for gene delivery. Adv. Colloid Interf. Sci. 2018;253:117–140. doi: 10.1016/j.cis.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 56.Tan L., Sun X. Recent advances in mRNA vaccine delivery. Nano Res. 2018;11(10):5338–5354. [Google Scholar]
- 57.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang R., Deng Y., Huang B., Huang L., Lin A., Li Y., Wang W., Liu J., Lu S., Zhan Z., Wang Y., A R., Wang W., Niu P., Zhao L., Li S., Ma X., Zhang L., Zhang Y., Yao W., Liang X., Zhao J., Liu Z., Peng X., Li H., Tan W. A core-shell structured COVID-19 mRNA vaccine with favorable biodistribution pattern and promising immunity. Signal Transduct. Target. Therapy. 2021;6(1):213. doi: 10.1038/s41392-021-00634-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1