Abstract
The COVID-19 pandemic has demanded a range of biotechnological products for detection of SARS-CoV-2 variants and evaluation of human seroconversion after infection or vaccination. In this work, we describe an easy pipeline for expression of SARS-CoV-2 nucleocapsid (N) protein in insect cells followed by its purification via affinity chromatography. The N gene was cloned into the genome of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) via transposition and the resulting recombinant baculovirus was used for infection of lepidopteran Sf9 cells adapted to high-density suspension. Using Tris−HCl pH 8.0 buffer as mobile phase and eluting bound proteins with 175 mM imidazole as part of a three-step gradient, an average of 1 mg N protein could be purified from each 50 mg of total protein from clarified supernatant. Such protein amount allows the manufacturing of serological tests and the development of basic studies on cellular responses to SARS-CoV-2.
Keywords: Affinity chromatography, Baculovirus, COVID-19, Nucleocapsid protein, One-step purification, SARS-CoV-2
Gene expression systems based on baculoviruses generate high amounts of recombinant proteins in insect cells (Chambers et al., 2018). These systems also allow the introgression of large DNA fragments and have undergone various improvements along the years that have made molecular cloning easier (Possee et al., 2019). Baculoviruses contain a circular double-stranded DNA genome and two distinct infection forms: occlusion-derived virus (ODV) and budded virus (BV) (Harrison et al., 2018). While the ODV is responsible for primary infection and virions are embedded in a protein matrix of polyhedrin, forming an occlusion body (OB), the BVs are released from host cells and are responsible for spreading viruses in insect bodies systemically.
For in vitro expression of heterologous proteins, DNA from recombinant baculovirus produced in Escherichia coli (E.coli) by site-specific transposition are transfected in insect cells and the resulting budded virions are then used for infection of new culture batches and titration (Galleno and Sick, 1999). The polyhedrin (polh) promoter, which contains transcription elements for high expression rates, often regulates the expression of recombinant genes in baculovirus-based systems (Galleno and Sick, 1999; Possee and Howard, 1987). Commonly, the gene of interest substitutes the polh gene, which is non-essential for viral replication and results in the production of virus without OB. In summary, the cloning of recombinant baculoviruses and their transfection/infection in insect cell culture batches have been simplified and are straightforward procedures for heterologous protein expression. The main bottleneck, as observed for other expression systems, is to obtain pure and stable proteins.
The current coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has demanded several biotechnological products and services for detection, treatment, and prevention of virus infection and disease. Besides the molecular tools for virus detection such as the reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR), serological techniques have been highly employed to evaluate the seroconversion of infected and immunized patients (Gong et al., 2021; Machado et al., 2020). Therefore, the production of SARS-CoV-2 antigens has become indispensable for manufacturing serological tests [e.g., enzyme-linked immunosorbent assay (ELISA)] or use in basic studies, aiming a better understanding of SARS-CoV-2 infection and host-triggered immune responses. In this work, we provide a straightforward protocol for one-step purification of SARS-CoV-2 nucleocapsid (N) protein expressed in insect cells using a recombinant baculovirus.
The SARS-CoV-2 N gene was amplified by RT-PCR using purified SARS-CoV-2 RNA from a nasopharyngeal swab sample kindly supplied by the Central Laboratory of Public Health in the Federal District, Brazil. The primers N-COV-F (5′- GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GTC TGA TAA TGG ACC CCA AAA –3′) and N-COV-His-R (5′- GGG GAC CAC TTT GTA CAA GAA AGC TGG GTC CTA GTG GTG ATG GTG ATG ATG GGC CTG AGT TGA GTC AGC AC -3′) were used for cDNA synthesis and PCR. Both primers contain attachment (att) sites for Gateway cloning and the latter an additional coding sequence for 6xHis-tag before the stop codon. cDNA first strand was synthesized with SuperScript IV [Thermo Fisher Scientific (TFS), Waltham, Massachusetts, U.S.A.] and the PCR product was amplified with Phusion Taq DNA polymerase (TFS). The PCR product was recombined into the entry vector pDONR207 with BP Clonase Enzyme Mix (TFS), and the resulting pDONR207-SARS-CoV-2-N-gene was recombined into the destination vector pDEST8 with LR Clonase Enzyme Mix (TFS). In the end, pDEST8-SARS-CoV-2-N-gene was transformed in E. coli DH10Bac cells (TFS) by electroporation and the N gene, under control of the polh promoter, was inserted in the genome of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) via transposition (Bac-to-Bac®, Baculovirus Expression Systems, TFS), generating the recombinant bacmid vAc-N-6xHis. All procedures followed the manufacturers’ recommendations.
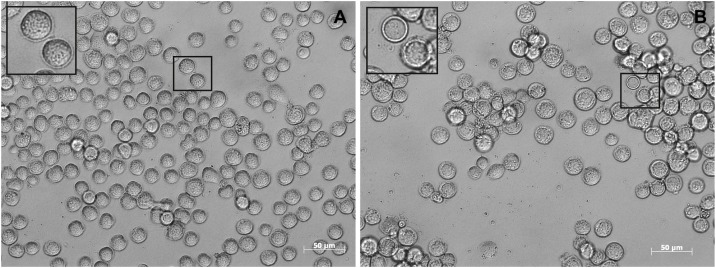
For recovery of infectious baculoviruses, one microgram of vAc-N-6xHis DNA was transfected into lepidopteran (Spodoptera frugiperda) Sf9 cells (106) grown in a six-well plate using FuGENE® HD Transfection Reagent (Promega, Madison, Wisconsin, U.S.A.) and following the manufacturer’s protocol. Sf9 cells were maintained in TC-100 medium (Vitrocell, Campinas, São Paulo, Brazil) supplemented with 10 % fetal bovine serum, 25 μg/mL amphotericin B, and 50 mg/L gentamycin sulfate. Cells were passaged twice a week and kept at 27 °C. Seven days post-transfection, the supernatant was collected and the recombinant virus was titrated as described elsewhere (O’Reilly et al., 1992). Then, Sf9 cells (1.5 × 107) grown in 75 cm2 flasks were infected at a multiplicity of infection (M.O.I.) of 1 to increase the virus titer as described in O’Reilly et al. (O’Reilly et al., 1992). Infected Sf9 cells presented typical cytopathic effects such as increasing nucleus size, loss of attachment, and irregular shapes (Fig. 1 ).
Fig. 1.
Sf9 cells infected with the recombinant baculovirus vAc-N-6xHis. (A) Mock and (B) infected cells. The inset in B zooms in cells presenting hypertrophied nucleus with amorphous mass accumulation.
For bulk protein production, anchored Sf9 cells were adapted to grow in suspension. Cells were cultured in 50 mL supplemented TC-100 medium (in 100-ml glass flasks) under constant shaking of 100 rpm in minitron incubator shaker (INFORS HT). They were passaged every 3 days, maintaining a density between 0.5 and 4 × 106 cells/mL. Before infection, cells were cultured in 100 mL supplemented TC-100 medium (in 250-ml glass flasks) at density varying between 1.5–2 × 106 cells/mL and then infected at M.O.I of 10. After 72 h post-infection (h.p.i.), cells were harvested by centrifugation at 2455 x g.
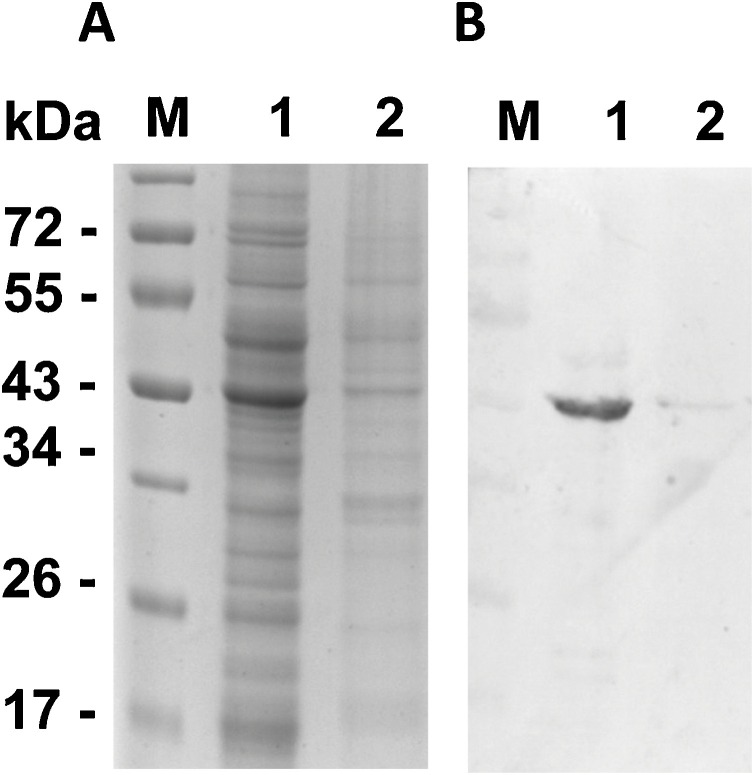
Pellet from each flask was resuspended with one ml lysis buffer [2.5 mL of 20 mM Tris−HCl buffer, pH 8, with 500 mM NaCl and 1 mM phenylmethylsulfonyl fluoride (PMSF) protease inhibitor] and sonicated twice for 30 s at 25 % amplitude and then centrifuged at 10,000 x g for 30 min at 4 °C. Both soluble and insoluble fractions were submitted to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by western blot. Mouse monoclonal anti-6xHis and alkaline-phosphatase (ALP)-conjugated goat anti-mouse, both from Abcam (Cambridge, U.K), were used as primary and secondary antibodies for immunoblotting, respectively. In the end, N proteins accumulated substantially more in soluble fractions (Fig. 2 ). Therefore, insoluble fractions were not considered for chromatography to avoid further protein refolding steps.
Fig. 2.
Detection of heterologous SARS-CoV-2 N protein from disrupted Sf9 cells. (A) SDS-PAGE of soluble (1) and insoluble (2) fractions from disrupted Sf9 cells expressing SARS-CoV-2 N protein. (B) These fractions were also submitted to western blot using mouse monoclonal anti-6xHis as primary antibody. M stands for the molecular marker PageRuler Prestained Protein Ladder (TFS).
For SDS-PAGE, a volume of 15 μL per sample in 4X protein loading buffer (0.25 M Tris-Cl, pH 6.8, 4% SDS, 20 % glycerol, 10 % 2-mercaptoethanol, and 0.02 % bromophenol blue) was heated (100 °C) for 5 min and run through a 12 % polyacrylamide gel. Proteins were visualized by Coomassie brilliant Blue R-250 staining. For Western blotting, proteins were transferred onto Immobilon-P transfer membrane (MilliporeSigma, Burlington, Massachusetts, U.S.A.) using a Trans-Blot Semi-Dry Transfer Cell (Bio-Rad, Hercules, California, U.S.A.). Membranes were then blocked in 1X PBS Buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) containing 3% bovine serum albumin for 16 h at 4 °C. Then, they were washed three times with 1X PBS Tween (0.05 %). Primary and secondary antibodies were incubated for one hour and washed three times with 1X PBS Tween (0.05 %) between incubations. Chromogenic substrate NBT-BCIP (Promega) was used for the detection of N protein according to the manufacturer’s protocol.
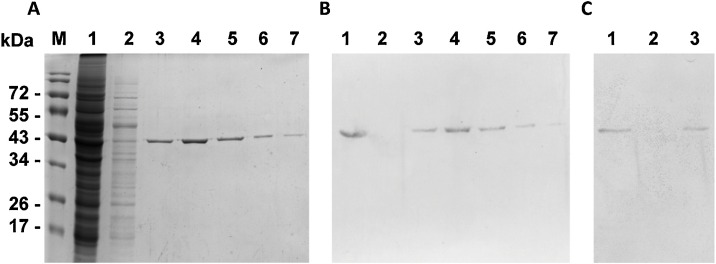
All purification runs were performed on AKTA lab-scale purification system, model AKTA Pure (GE Healthcare, Chicago, Illinois, U.S.A.). Data were collected and analyzed using UNICORN 6.3 software. A 5 mL HisTrap HP column (GE Healthcare) was first equilibrated with 10 mL of buffer A (500 mM of NaCl, 20 mM TrisHCl buffer, pH 8.0) at 4 mL/min flow-rate. About 6 mL of clarified supernatant was loaded to the system superloop with a syringe and flowed through the column at 1 mL/min. Unbound proteins were washed out of the column with 25 mL buffer A at 2 mL/min flow-rate. To elute bound proteins, 65 mL buffer B (buffer A with 500 mM imidazole) flowed through the column as a three-step gradient of 100 mM, 175 mM, and 500 mM imidazole concentrations at 4 mL/min. Fractions of 2 mL were collected and submitted to SDS-PAGE and Western blot. Pure recombinant SARS-CoV-2 N protein was obtained in fractions of bound proteins that were eluted with 175 mM imidazole (Fig. 3 A,B). These fractions were then pooled in SnakeSkin Dialysis Tubing 10,000 MWCO (TFS), and desalted against 1X PBS for 12 h. Protein concentration was measure with Quant-iT™ Protein assay kit (Invitrogen) according to the manufacturer’s protocol. An average of 1 mg N protein was purified from 50 mg of total protein in clarified supernatant. The recombinant protein was also subjected to immunoblotting with rabbit monoclonal anti-SARS-CoV-2 N protein (TFS) and ALP-conjugated goat anti-rabbit (TFS) as primary and secondary antibodies, respectively (Fig. 3C).
Fig. 3.
Visualization of protein fractions for and after HisTrap chromatography. (A) Protein fractions loaded for SDS-PAGE. M protein marker, 1 soluble fraction (input), 2 unbound protein fraction (wash step), 3-7 bound protein fractions (eluted with 175 mM imidazole). (B) western blot of the same fractions using mouse monoclonal anti-6xHis as primary antibody. (C) western blot of fractions 1, 2 and 3 using rabbit monoclonal anti-SARS-CoV-2 N protein as primary antibody. Secondary antibodies were conjugated with alkaline phosphatase.
Several strategies were explored without success in the first attempts for SARS-CoV-2 N protein purification by affinity chromatography. Following some published technical manuals, Na3PO4 pH 7.4 buffer, for example, was used as mobile phase (buffer A), and imidazole elution was programmed to occur as a linear gradient. These methods failed to produce pure fractions containing the N protein. The mobile phase was then changed to Tris−HCl pH 8.0 buffer and imidazole elution was set up to occur as a three-step gradient as described above. Changing the mobile phase increased the binding of proteins in the column and the imidazole step-gradient resulted in fractions containing pure N protein in a higher yield (Fig. 3).
Ease of scale-up by adapting cell growth to high-density suspension and less demanding culture conditions make insect cells more advantageous than mammalian cells for expression of certain animal or animal-infecting virus proteins. Therefore, baculovirus expression systems offer more advantages than other systems for combining robust protein expression with post-translational modifications that are more comparable to mammals (Tsai et al., 2020). Baculoviruses have been used for manufacturing viral vaccines based on subunits or virus-like particles (mimicking virions) against human and veterinary pathogenic viruses (Felberbaum, 2015). During the COVID-19 pandemic, baculoviruses have been used for expression of SARS-CoV-2 spike (S) protein versions as subunits for the development of supplies, and virus-like particles and baculoviral vector-based DNA as vaccine candidates (Cho et al., 2021; Fujita et al., 2020; Li et al., 2020b; Mi et al., 2021). None of these studies included the N gene, and public protocols for heterologous expression and purification of SARS-CoV-2 N protein seem to be restricted to E. coli (Djukic et al., 2021; Garg et al., 2020; Li et al., 2021).
Although S protein mediates SARS-CoV-2 entry in host cells and is targeted by most known neutralizing antibodies, N protein plays important roles during virus infection and host response (Dai and Gao, 2021). In charge of genome packaging, N is the most abundant protein in coronaviruses and has been shown as a major target for humoral response during SARS-CoV-2 infection, and many T cells epitopes have been mapped to it (Oliveira et al., 2020; Sariol and Perlman, 2020; Wu et al., 2021). As a result, this protein has been widely used for quantifying IgM and IgG from COVID-19 patients (Burbelo et al., 2020; Wu et al., 2021). N protein has also key functions during SARS-CoV-2 infectivity, such as suppressing RNA interference and inhibiting interferon signaling pathways in mammalian cells (Chen et al., 2020; Li et al., 2020a; Mu et al., 2020b, 2020a). In summary, expression and purification of N protein are relevant for studies on SARS-CoV-2 infectivity and pathogenicity in addition to the development of diagnostic supplies. Here, we present a protocol that can be easily scaled up and reproduced in laboratories that use baculovirus-based systems for expression of recombinant proteins.
Authors’ contributions
Conceptualization: B.R.C., L.A.S., A.S.O., and B.M.R.; methodology: B.R.C., L.A.S, and A.S.O.; investigation: B.R.C., L.A.S., and A.S.O.; resources: B.M.R; Writing – original draft preparation: B.R.C., L.A.S., and A.S.O.; Writing – review and editing: B.M.R.; funding: B.M.R.
Funding information
We acknowledge Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Grant number 88887.504570/2020-00), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Grant number 305756/2017-6), and Fundação de Apoio à Pesquisa do Distrito Federal (FAP-DF, Grant numbers 193.001532/2016 and 19300000525/2020-41) for financial support.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
References
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Jr., Cohen J.I. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers A.C., Aksular M., Graves L.P., Irons S.L., Possee R.D., King L.A. Overview of the baculovirus expression system. Curr. Protoc. Protein Sci. 2018;91:5.4.1–5.4.6. doi: 10.1002/CPPS.47. [DOI] [PubMed] [Google Scholar]
- Chen K., Xiao F., Hu D., Ge W., Tian M., Wang W., Pan P., Wu K., Wu J. SARS-CoV-2 nucleocapsid protein interacts with RIG-I and represses RIG-Mediated IFN-β production. Viruses. 2020;13 doi: 10.3390/v13010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Jang Y., Park K.-H., Choi H., Nowakowska A., Lee H.-J., Kim M., Kang M.-H., Kim J.-H., Shin H.Y., Oh Y.-K., Kim Y.B. Human endogenous retrovirus-enveloped baculoviral DNA vaccines against MERS-CoV and SARS-CoV2. Npj Vaccines. 2021;6:37. doi: 10.1038/s41541-021-00303-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djukic T., Mladenovic M., Stanic-Vucinic D., Radosavljevic J., Smiljanic K., Sabljic L., Devic M., Cujic D., Vasovic T., Simovic A., Radomirovic M., Cirkovic Velickovic T. Expression, purification and immunological characterization of recombinant nucleocapsid protein fragment from SARS-CoV-2. Virology. 2021;557:15–22. doi: 10.1016/j.virol.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felberbaum R.S. The baculovirus expression vector system: a commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnol. J. 2015;10:702–714. doi: 10.1002/biot.201400438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita R., Hino M., Ebihara T., Nagasato T., Masuda A., Lee J.M., Fujii T., Mon H., Kakino K., Nagai R., Tanaka M., Tonooka Y., Moriyama T., Kusakabe T. Efficient production of recombinant SARS-CoV-2 spike protein using the baculovirus-silkworm system. Biochem. Biophys. Res. Commun. 2020;529:257–262. doi: 10.1016/j.bbrc.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galleno M., Sick A.J. Gene Expression Systems. Elsevier; 1999. Baculovirus expression vector system; pp. 331–363. [Google Scholar]
- Garg A., Liu L., Ho D.D., Joshua-Tor L. Heterologous expression and purification of SARS-CoV2 nucleocapsid protein. Bioprotocol. 2020;10:e5005. doi: 10.21769/BioProtoc.5005. [DOI] [Google Scholar]
- Gong F., Wei H.-X., Li Q., Liu L., Li B. Evaluation and comparison of serological methods for COVID-19 diagnosis. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.682405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R.L., Herniou E.A., Jehle J.A., Theilmann D.A., Burand J.P., Becnel J.J., Krell P.J., Oers M.M.Van, Mowery J.D., Bauchan G.R., Report I. 2018. ICTV ICTV Virus Taxonomy Profile: Baculoviridae; pp. 1185–1186. [DOI] [PubMed] [Google Scholar]
- Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., Ge X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Zheng Q., Yu H., Wu D., Xue W., Xiong H., Huang X., Nie M., Yue M., Rong R., Zhang S., Zhang Y., Wu Y., Wang S., Zha Z., Chen T., Deng T., Wang Y., Zhang T., Chen Y., Yuan Q., Zhao Q., Zhang J., Gu Y., Li S., Xia N. SARS-CoV-2 spike produced in insect cells elicits high neutralization titres in non-human primates. Emerg. Microbes Infect. 2020;9:2076–2090. doi: 10.1080/22221751.2020.1821583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Li W., Fang X., Song X., Teng S., Ren Z., Hu D., Zhou S., Wu G., Li K. Expression and purification of recombinant SARS-CoV-2 nucleocapsid protein in inclusion bodies and its application in serological detection. Protein Expr. Purif. 2021;186 doi: 10.1016/j.pep.2021.105908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado B.A.S., Hodel K.V.S., Barbosa-Júnior V.G., Soares M.B.P., Badaró R. The main molecular and serological methods for diagnosing COVID-19: an overview based on the literature. Viruses. 2020;13 doi: 10.3390/v13010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi Y., Xie T., Zhu B., Tan J., Li X., Luo Y., Li F., Niu H., Han J., Lv W., Wang J. Production of SARS-CoV-2 virus-like particles in insect cells. Vaccines. 2021 doi: 10.3390/vaccines9060554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Fang Y., Yang Q., Shu T., Wang A., Huang M., Jin L., Deng F., Qiu Y., Zhou X. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6:65. doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Xu J., Zhang L., Shu T., Wu D., Huang M., Ren Y., Li X., Geng Q., Xu Y., Qiu Y., Zhou X. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci. China Life Sci. 2020 doi: 10.1007/s11427-020-1692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly D.R., Miller L.K., Luckow V.A. A Laboratory Manual WH Freeman and Company; New York: 1992. Baculovirus Expression Vector. [Google Scholar]
- Oliveira S.C., de Magalhães M.T.Q., Homan E.J. Immunoinformatic analysis of SARS-CoV-2 nucleocapsid protein and identification of COVID-19 vaccine targets. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.587615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possee R.D., Howard S.C. Analysis of the polyhedrin gene promoter of the Autographa californica nuclear polyhedrosis virus. Nucleic Acids Res. 1987;15:10233–10248. doi: 10.1093/nar/15.24.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possee R.D., Chambers A.C., Graves L.P., Aksular M., King L.A. Recent developments in the use of baculovirus expression vectors. Curr. Issues Mol. Biol. 2019 doi: 10.21775/cimb.034.215. [DOI] [PubMed] [Google Scholar]
- Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-H., Wei S.-C., Lo H.-R., Chao Y.-C. Baculovirus as versatile vectors for protein display and biotechnological applications. Curr. Issues Mol. Biol. 2020;34:231–256. doi: 10.21775/cimb.034.231. [DOI] [PubMed] [Google Scholar]
- Wu J., Liang B., Chen C., Wang Hua, Fang Y., Shen S., Yang Xiaoli, Wang B., Chen L., Chen Q., Wu Y., Liu J., Yang Xuecheng, Li W., Zhu B., Zhou W., Wang Huan, Li S., Lu S., Liu D., Li H., Krawczyk A. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat. Commun. 2021:1–9. doi: 10.1038/s41467-021-22034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]