Abstract
Western Australia (WA) has been able to prevent methicillin-resistant Staphylococcus aureus (MRSA) strains from outside of the state from becoming established in its hospitals. Recently, a single-strain outbreak of MRSA occurred in a WA metropolitan teaching hospital following admission of an infected patient from a remote community. The strain responsible for the outbreak was unrelated to any imported strains and spread rapidly in the hospital. Screening of two remote communities in the region from which the index case came revealed that 42% of the people in one community and 24% in the other carried MRSA. Isolates were typed by resistance pattern, plasmid analysis, contour-clamped homogeneous electric field electrophoresis, bacteriophage pattern, and coagulase gene restriction fragment length polymorphism. It was found that of the people carrying MRSA, 39% in the former community and 17% in the latter community were carrying an MRSA strain which was indistinguishable from the strain that caused the hospital outbreak.
There have been an increasing number of reports of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) (4). Boyce has recently reviewed the evidence for community-acquired MRSA and pointed out the difficulties in demonstrating that the organism was not acquired as the result of a visit to a health care facility (4). He also pointed out that many community MRSA strains are not multiply resistant and that some of these may be borderline methicillin-resistant strains, which do not contain the mecA gene (4).
Community MRSA strains were first reported in Australia in the state of Western Australia (WA) in 1993 (32). These strains were isolated from hospital patients who resided in isolated communities hundreds of kilometers from any small town and thousands of kilometers from any urban area. The strains differed from other MRSA strains previously isolated in WA (29), based on their resistance to antimicrobial agents, plasmid content, and contour-clamped homogeneous electric field (CHEF) electrophoresis patterns (32) and have been referred to as WA MRSA (33).
WA has been able to monitor MRSA in the state because of its geographical isolation. The state has a landmass of approximately one-third of Australia but a population of only 1.8 million. A total of 1.35 million of these people live in Perth, the capital city. Perth is on the shores of the Indian Ocean and is separated from the main population centers to the east by a distance of 2,000 km, which includes 1,100 km of desert.
All MRSA strains isolated in WA are sent to a reference center. Initially, MRSA strains were isolated only from people who had been in hospitals outside WA and the strains were prevented from spreading in WA hospitals (17, 18). Occasionally an imported MRSA strain was not detected on a person resulting in a few single-strain outbreaks in hospitals, which were terminated only by a rigorous screening and containment policy (17, 18, 29). WA MRSA strains, however, had not caused outbreaks when patients carrying them were admitted to the hospital. The situation has now changed, and a community MRSA has caused a single-strain outbreak in a Perth hospital.
Screening of two remote communities in the vicinity where the index patient of the outbreak lived has revealed a high prevalence of MRSA strains, many of which were indistinguishable from the outbreak strain.
MATERIALS AND METHODS
Screening.
All consenting members of the communities were screened. The nostrils, throat, finger webs, and any skin lesions were swabbed with moistened cotton wool swabs which were placed in Amies transport medium (Interpath Services, Pty. Ltd., West Heidelberg, Australia) and transported in a cool, insulated container to the laboratory in Perth. All swabs were processed within 48 h of collection.
Laboratory processing.
Swabs were plated onto mannitol salt agar (MSA) (Oxoid, Basingstoke, England) and methicillin aztreonam mannitol salt agar (MAMSA) (21). MSA and MAMSA plates were incubated at 35°C for 48 and 20 h, respectively. Mannitol-fermenting colonies were cultured overnight in brain heart infusion broth (Gibco Diagnostics, Gaithersburg, Md.), presumptively identified as S. aureus by the tube coagulase test, and confirmed by PCR amplification of the nuc gene (5). One colony from each positive site was characterized.
Susceptibility testing.
Susceptibility testing was performed on Mueller-Hinton agar (BBL, Becton Dickinson, Cockeysville, Md.) (7) by disk diffusion according to National Committee for Clinical Laboratory Standards guidelines (15) by using antibiotic disks (Oxoid). Colonies were screened for susceptibility to oxacillin (1 μg), and resistance was confirmed by PCR for mecA (5). Samples with methicillin-sensitive S. aureus (MSSA) and/or MRSA colonies were recorded, and colonies were selected for further studies. MRSA colonies were tested for susceptibility to gentamicin, kanamycin, neomycin, streptomycin, erythromycin, lincomycin, chloramphenicol, minocycline, tetracycline, trimethoprim, sulfamethoxazole, fusidic acid, rifampin, novobiocin, vancomycin, mupirocin, and ciprofloxacin. Resistogram testing for susceptibility to cadmium acetate (10−2 M), mercuric chloride (0.4 μM), and phenyl mercuric acetate (5 mM), all from Ajax Chemicals, Sydney, Australia, ethidium bromide (15 mM) and sodium arsenate (0.2 μM) from Sigma Chemical Company, St. Louis, Mo., and propamidine isethionate (2% [wt/vol]) from May and Baker Limited, Dagenham, England, was performed as previously described (29, 30). Beta-lactamase production was detected by using Nitrocefin disks (BBL, Becton Dickinson) according to the manufacturer’s instructions. MICs were determined by using the E-Test (AB Biodisk, Solna, Sweden).
Isolation and analysis of plasmid DNA.
MRSA plasmids were isolated by using the cetyltrimethylammonium bromide method as previously described (27) and separated by horizontal-gel electrophoresis in 0.6% (wt/vol) molecular biology-grade agarose (Promega Corporation, Madison, Wis.). Plasmid size was calculated by using the MacVector 4.0 program with the plasmids of WBG4483 used as the size standard (27). When possible, individual plasmids were digested with the restriction endonucleases EcoRI and HindIII (Promega) according to the manufacturer’s directions. The fragments were separated in 1% agarose and sized by Molmatch (UV Products, Cambridge, United Kingdom). Phage λ DNA (Pharmacia, Uppsala, Sweden) digested with HindIII was used as the size standard.
CHEF electrophoresis of chromosomal DNA.
The method for preparation of agarose blocks for CHEF electrophoresis was adapted from methods previously described (8, 35). Cells from 3 ml of overnight Trypticase soy broth (Gibco Diagnostics) cultures were harvested by centrifugation (1,089 × g for 10 min) and washed twice by resuspension in 5 ml of 50 mM EDTA, pH 8.0. The cell pellets were then resuspended in EC buffer (6 mM Tris, 1 M NaCl, 100 mM EDTA, 0.5% Brij 58, 0.2% sodium deoxycholate, and 0.5% sodium lauroyl sarcosine [pH 7.5]) to a final concentration of 1.8 × 109 cells/ml. Fifty microliters of the cell suspension was then mixed with 50 μl of lysostaphin (400 μg/ml) (Sigma Chemical Company) and 100 μl of molten 1% agarose (chromosomal grade; Bio-Rad Laboratories, Hercules, Calif.) held at 50°C and then loaded into agarose block formers (Bio-Rad Laboratories). The blocks were allowed to set at room temperature for 10 min and then placed in 500 μl of EC buffer and incubated at 37°C for 3 to 4 h. The EC buffer was then replaced with 300 μl of EST buffer (5 mM Tris, 0.5 M EDTA, and 1% sodium lauroyl sarcosine [pH 7.5]) and 30 μl of proteinase K (20 mg/ml) (Boehringer, Mannheim, Germany), and the mixtures were incubated overnight at 50°C. The DNA blocks were then washed in 1 ml of 50 mM EDTA (pH 8.0) for 2 h, with gentle shaking, and the EDTA was replaced every 30 min. The prepared blocks were stored in 50 mM EDTA (pH 8.0) at 4°C until required. The agarose blocks were then washed in 1.5 ml of sterile water (18.2 MΩ-cm resistivity at 25°C), followed by equilibration in buffer J (Promega) on ice for 30 min and then digested with 40 U of SmaI (Promega) at 22°C for 4 h. Electrophoresis was performed in 1% agarose slabs (pulsed field certified; Bio-Rad Laboratories) by using the CHEF-DRII system (Bio-Rad Laboratories) in 0.5× Tris-borate-EDTA (45 mM Tris, 45 mM boric acid, 1 mM EDTA [pH 8.0]) at 12°C, for 20 h at 200 V. The initial pulse time was 1 s, and the final pulse time was 40 s. Chromosomal banding patterns were scanned with a Fluor-S MultiImager and analyzed by Multi-Analyst/PC (Bio-Rad Laboratories). S. aureus NCTC8325 was used as the size standard (16). CHEF patterns were grouped according to the criteria of Tenover et al. (26).
Phage typing.
Phage typing of the index strain and all CHEF pattern 10 community isolates was performed as previously described (2) by using the Basic International set of phages and the Australian Supplementary set of phages (1, 34).
Coagulase gene typing.
Coagulase gene restriction fragment length polymorphism typing of the index strain and all CHEF pattern 10 community isolates was performed as previously described (9).
Mixed-culture transfer.
Mixed-culture transfer of plasmids was performed on representative isolates as previously described (28). Recipients were S. aureus WBG2110 (strain RN450 mutated to chromosomal streptomycin and novobiocin resistance and lysogenized with phage J) and WBG1876 (28). WBG2110 transcipients were selected on media containing streptomycin (25 μg/ml) and novobiocin (5 μg/ml) (Sigma Chemical Company) with either cadmium acetate (135 μg/ml) or erythromycin (5 μg/ml) (Sigma Chemical Company). WBG1876 transcipients were selected on media containing fusidic acid (5 μg/ml) and rifampin (25 μg/ml) (Sigma Chemical Company) with either sodium arsenate (31 μg/ml) or cadmium acetate (135 μg/ml).
Plasmid curing.
Plasmid curing was performed at 43.5°C as previously described (30). Isolated colonies were screened by replication onto brain heart infusion agar containing either cadmium acetate (135 μg/ml) or erythromycin (5 μg/ml). Colonies showing loss of resistance were further characterized by antibiogram and resistogram testing, plasmid isolation, and CHEF analysis.
RESULTS
The inhabitants of two remote communities, 10 km (community X) and 200 km (community Y) from a regional hospital 600 km from Perth, were screened. The people in both communities had high rates of carriage of both MSSA and MRSA (Table 1). Forty-two percent of the people in community X and 24% in community Y carried MRSA (Table 1). There was considerable variation in the number of sites from which MRSA strains were isolated (Table 2). Although only one isolate was characterized from each positive site, six persons were found to be colonized with slightly different strains isolated from different sites (Table 3). The characteristics and prevalence of the isolates in the communities are presented in Table 4.
TABLE 1.
Prevalence of MRSA and MSSA
| Parameter | No. (%) for indicated community
|
|
|---|---|---|
| X | Y | |
| No. of persons sampleda | 43 | 74 |
| MRSA and/or MSSA | 28 (68) | 48 (65) |
| MSSA | 17 (40) | 45 (61) |
| MRSA | 18 (42) | 18 (24) |
| MRSA and MSSA | 7 (16) | 15 (20) |
Total populations of X and Y were 80 and 200, respectively, at the time of screening.
TABLE 2.
Sites of MRSA carriage
| Site of carriage | No. of MRSA-positive persons (% of persons MRSA positive) in indicated community
|
|
|---|---|---|
| X | Y | |
| Throat only | 3 (17) | 2 (11) |
| Nose only | 3 (17) | 3 (17) |
| Skin only | 4 (22) | 9 (50) |
| Nose and throat | 2 (11) | 2 (11) |
| Nose and skin | 3 (17) | 2 (11) |
| Nose, throat, and skin | 3 (17) | 0 (0) |
TABLE 3.
Persons carrying more than one type of MRSAa
| Person | CHEF pattern | % Similarity of CHEF typesb | Resistancec | Plasmid size(s) (kb) |
|---|---|---|---|---|
| X1 | 10 | 94 | EI, F, Cd | 19.6, 2.0 |
| 10d | EI, Cd | 19.6, 2.0 | ||
| X2 | 9 | 97 | Cd, Asa | 24.2 |
| 9a | Cd, Asa | 24.2 | ||
| Y1 | 12 | 96 | EI | 41.3, 21.6, 2.0 |
| 12c | EI, Cd | 37.8, 21.6, 2.0 | ||
| Y2 | 10 | 52 | EI, F, Cd | 19.6, 2.0 |
| 12a | M only | 33.0, 21.6, 2.3 | ||
| Y3 | 10 | 56 | EI, F, Cd | 19.6, 2.0 |
| 9 | Cd, Asa | 24.2 | ||
| Y4 | 9 | 100 | Cd, Asa | 32.0, 24.2 |
| 9 | Cd, Asa | 24.2 |
Resistant to all beta-lactams.
Calculated as Dice coefficients by Multi-Analyst/PC (Bio-Rad).
Abbreviations: Asa, sodium arsenate; Cd, cadmium acetate; E, erythromycin; F, fusidic acid; M, methicillin; superscript I, inducible.
TABLE 4.
Characteristics of community MRSAa isolates
| No. of persons | Community | CHEF pattern | Resistanceb | Plasmid size(s) (kb) |
|---|---|---|---|---|
| 7 | X | 10 | EI, F, Cd | 19.6, 2.0 |
| 1 | X | 10d | EI, Cd | 19.6, 2.0 |
| 2 | X | 12b | EI, Cd | 34.0, 2.0 |
| 2 | X | 8 | EI, Cd | 21.8, 2.0 |
| 2 | X | 9 | Cd, Asa | 24.2 |
| 3 | X | 9a | Cd, Asa | 24.2 |
| 1 | X | 9a | Cd, Asa, Pi | 24.2 |
| 2 | X | 9b | EI, C, Cd | 19.6, 4.2, 2.0 |
| 3 | Y | 10 | EI, F, Cd | 19.6, 2.0 |
| 2 | Y | 10a | EI, Cd | 19.6, 2.0 |
| 3 | Y | 10a | Cd | 19.6 |
| 1 | Y | 10e | EI, Cd | 37.8, 21.6, 2.0 |
| 1 | Y | 12 | EI | 41.3, 21.6, 2.0 |
| 1 | Y | 12a | 33.0, 2.3 | |
| 1 | Y | 12a | Mp | 33.0 |
| 1 | Y | 12a | Mp | 33.0, 21.6, 2.3 |
| 1 | Y | 12c | E, Cd | 37.8, 21.6, 2.0 |
| 1 | Y | 9 | Cd, Asa | 32.0, 24.2, 2.0 |
| 4 | Y | 9 | Cd, Asa | 24.2 |
| 2 | Y | 9a | Cd, Asa | 24.2 |
Resistant to all beta-lactams.
Abbreviations: Asa, sodium arsenate; Cd, cadmium acetate; C, chloramphenicol; E, erythromycin; F, fusidic acid; Pi, propamidine isethionate; Mp, mupirocin; superscript I, inducible.
Resistance to antimicrobial agents.
All the MRSA strains were mecA positive, produced beta-lactamase, and were resistant to the antimicrobial agents shown in Table 4. CHEF pattern 9 and 9a isolates were all low-expression class MRSA strains (7).
CHEF electrophoresis.
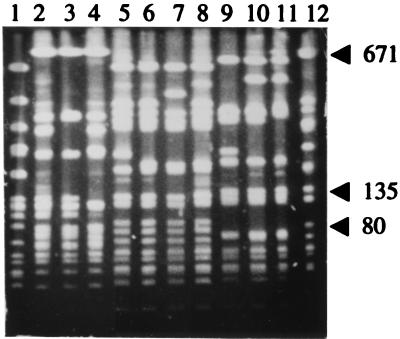
The isolates were divided into four CHEF types, 8, 9, 10, and 12, which were regarded as epidemiologically unrelated (26). These were then divided into subtypes which were regarded as epidemiologically related (26). CHEF pattern 9 contained two subtypes, 9a and 9b, CHEF pattern 10 had three subtypes, 10a, 10d, and 10e, and CHEF pattern 12 had three subtypes, 12a, 12b, and 12c. Representatives of the different CHEF patterns are shown in Fig. 1, and their distribution in the communities is shown in Table 4. CHEF pattern 10 was the most prevalent. A dendrogram of the CHEF patterns (Fig. 2) reveals two clusters of strains. Patterns 10, 10a, 10d, 10e, 12, 12a, 12b, and 12c clustered together with 80% similarity to each other, and CHEF patterns 9, 9a, and 9b clustered with 82% similarity to each other. However, these two clusters were only 49% similar to each other. CHEF pattern 8 was not closely related to either cluster.
FIG. 1.
CHEF patterns of community MRSA isolates. Lanes: 1, CHEF pattern 8; 2, CHEF pattern 9; 3, CHEF pattern 9a; 4, CHEF pattern 9b; 5, CHEF pattern 10; 6, CHEF pattern 10a; 7, CHEF pattern 10d; 8, CHEF pattern 10e; 9, CHEF pattern 12; 10, CHEF pattern 12a; 11, CHEF pattern 12c; 12, NCTC8325.
FIG. 2.
Dendrogram of WA MRSA strains isolated from communities X and Y.
Plasmid content of isolates.
All of the isolates contained plasmids, whose profiles are presented in Table 4. The plasmid profiles were not completely consistent with the CHEF patterns. CHEF patterns 10, 10a, and 10d carried plasmids of 19.6 and 2.0 kb, except for three CHEF pattern 10a strains that carried only the 19.6-kb plasmid. The one CHEF pattern 10e isolate carried the 2.0-kb plasmid and two different plasmids, of 37.8 and 21.6 kb. Mixed-culture transfer and curing experiments revealed that the 19.6-kb plasmid encoded beta-lactamase production and resistance to cadmium, and the 2.0-kb plasmid encoded inducible resistance to erythromycin. The plasmid profiles of CHEF patterns 12, 12a, 12b, and 12c isolates were varied, although uncharacterized plasmids of 33.0 and 21.6 kb predominated (Table 4). CHEF patterns 9 and 9a carried a 24.2-kb plasmid which encoded resistance to cadmium and arsenate. One CHEF pattern 9 isolate carried additional 32- and 2.0-kb plasmids. CHEF pattern 9b isolates carried plasmids of 19.6, 4.4, and 2.0 kb.
Comparison with the hospital outbreak strain.
The strain (EMRSA-WA95/1) which caused the hospital outbreak (20) was found to be indistinguishable from the most prevalent community isolate, CHEF pattern 10 (Table 5).
TABLE 5.
Comparison of the most prevalent community isolate with the outbreak straina
| Parameter | Community isolate | EMRSA-WA95/1 |
|---|---|---|
| CHEF pattern | 10 | 10 |
| Plasmid size in kb (phenotype) | 19.6 (Cdr, Bla), 2.0 (Er) | 19.6 (Cdr, Bla), 2.0 (Er) |
| Resistance | M, EI, F, Cd | M, EI, F, Cd |
| Phage type (100 × RTD) | 52A/79/6/42E/47/53/(54)/83A//56B/56C/67R | 52A/79/6/42E/47/53/(54)/83A//56B/56C/67R |
| Coagulase gene | 20 | 20 |
Abbreviations: Bla, beta-lactamase; Cd, cadmium acetate; E, erythromycin; F, fusidic acid; M, methicillin; RTD, routine test dilution; RFLP, restriction fragment length polymorphism; superscript I and r, inducible and resistant, respectively. Parentheses indicate a weak reaction.
In communities X and Y, CHEF pattern 10 was isolated from 39 and 17%, respectively, of people carrying an MRSA strain. When closely related CHEF patterns are included, 44% of the people carrying an MRSA strain in both communities were colonized by a CHEF pattern 10 or a closely related strain.
DISCUSSION
Although MRSA strains are endemic in the hospitals of the eastern seaboard of Australia (31), WA has been able to prevent imported strains of MRSA from becoming established in its hospitals because of its geographical isolation and because of a statewide policy to screen all patients and staff who have been in a hospital or a nursing home outside the state in the previous 12 months (17, 18). The screening policy has not prevented imported MRSA strains from causing an occasional single-strain outbreak in WA hospitals, but on every occasion the MRSA strains have been eliminated from the hospital (17–19).
However, over the last decade, MRSA strains have been isolated from an increasing number of patients from remote communities who have not been hospitalized outside the State. These MRSA strains were first isolated in the Kimberley, a remote region 2,000 to 2,500 km north of Perth. They were not multiply resistant (24, 32) like the imported MRSA strains from eastern Australia, Asia, and Europe (10, 18, 23, 29, 32), and genetic studies have shown that they are distinct from them (32, 33). Consequently, they have been referred to as WA MRSA strains (23, 33). A panel of eight antibiotics (gentamicin, erythromycin, tetracyline, trimethoprim, rifampin, fusidic acid, ciprofloxacin, and mupirocin) has been used to provisionally distinguish WA MRSA strains from the multiply resistant MRSA strains. Strains resistant to no more than two of the antibiotics are regarded as nonmultiresistant, and strains resistant to three or more antibiotics are regarded as multiresistant (19, 24).
WA MRSA strains cause infections but initially did not cause outbreaks in hospitals. However, the first large single-strain hospital outbreak caused by a WA MRSA strain occurred in a Perth teaching hospital in 1995 to 1996 (20). The index case was a patient who had been transferred from a regional hospital 600 km east of Perth (20). As there was no evidence that MRSA was endemic in the regional hospital, communities X and Y in the region where the index case originated were screened for MRSA strains, and the isolates were typed and compared with the hospital outbreak strain.
The carriage rates of S. aureus in communities X and Y were 68 and 65%, respectively. These rates are high based on published carriage rates (11). The carriage rates for MRSA in communities X and Y were 42 and 24%, respectively. The high rates of staphylococcal carriage may be related to a high prevalence of skin lesions in remote communities (25). However, there was no obvious explanation for the high carriage rates of MRSA. Antibiotic audits of these communities are planned to determine whether antibiotic consumption is related to the high prevalence of MRSA.
The majority of people were colonized at only one site; however, the site was not the same in each person. Consequently, if only one site was swabbed, MRSA would have escaped detection in many of the people who came for screening. This result emphasizes the need to screen more than one site to reliably detect MRSA carriage.
In addition to the high carriage rates of MRSA, a large number of the MRSA strains were indistinguishable from the hospital outbreak strain—39% in community X and 17% in community Y. Including closely related CHEF patterns, 44% of the MRSA strains isolated from each community were related to the hospital outbreak strain. The other MRSA strains isolated could be related to the outbreak strain, since plasmids can be lost or gained and restriction patterns can change due to transpositions, mutations, recombination, and changes in lysogeny (3, 26, 35). Likewise, although these isolates differed from other WA MRSA strains regarding CHEF pattern and plasmid content (32, 33), they could be related but have undergone changes over time.
The results also demonstrated that different MRSA strains can be isolated from different sites on the same person. In some cases, the variation involved only a single CHEF fragment or a different plasmid or resistance determinant. In other cases, the differences were quite marked. Two people carried isolates which varied in resistance profile, plasmid content, and CHEF pattern. One individual had two types of MRSA which had only 52% similarity in CHEF patterns, while another carried two isolates which had 56% similarity. Whether these people acquired different MRSA strains or whether the isolates represent variations which have occurred in a strain in vivo cannot be determined. Also, it must be remembered that only one MRSA strain from each site was examined, and that if more than one isolate from each site had been examined, further variations in the isolates would likely have been detected. This situation illustrates the problems that can occur in the study of epidemiology if only one isolate from each site is typed.
We have demonstrated that a community strain of MRSA was responsible for a single-strain hospital outbreak. A single-strain outbreak in a hospital is defined as an outbreak when three or more patients are infected or colonized by isolates of MRSA which are indistinguishable by at least two typing techniques and the strain has been transmitted to at least two patients within the hospital. Epidemic strains of MRSA (EMRSA) are strains which have caused single-strain outbreaks in two or more hospitals (13). In 1995, a single-strain outbreak of MRSA occurred in a Perth metropolitan hospital (20), and the strain was subsequently found to be indistinguishable, by five typing techniques, from the predominant community strain described in this article. In 1996, a strain which was indistinguishable from this strain also caused a small outbreak in a country hospital in WA (20). Consequently, this WA MRSA strain has been called an epidemic strain and has been designated EMRSA-WA95/1 (20).
Community MRSA strains with resistance profiles similar to those described for WA MRSA strains have been reported from the Northern Territory (NT) of Australia, which borders WA (12). It has been suggested that NT strains may be the same as WA strains (12), but there is no molecular evidence to support this possibility. Even more recently, community MRSA strains have been reported from the more populated eastern seaboard of Australia. However, there is no evidence that these strains are related to WA MRSA, and it has been suggested that they are related to the Western Samoan strains of New Zealand (6, 14, 22).
There have been a number of reports of community MRSA from other parts of the world (4). However, it has not always been clear whether these strains have arisen in the community or are hospital strains which have spread in the community (4). Our results have demonstrated that MRSA indistinguishable from a strain that caused an outbreak in a metropolitan hospital is prevalent in remote communities. As MRSA strains are not endemic in WA hospitals, and the WA MRSA strains are quite different from any MRSA strains studied so far in Australia, the evidence suggests that they have a community origin. Further screening of other WA communities and comparing WA MRSA with the community MRSA strains which have recently been reported in other parts of Australia will help elucidate the epidemiology and origin of WA MRSA.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Health and Medical Research Council of Australia.
We acknowledge F. Quadros, senior medical officer of the regional public health unit, for his assistance, Peta Williams, Sandra Rodgers, Delia Riley, and Eleanor Sullivan for assistance in the field, Ian Kay for performing the multiplex mecA/nuc PCR and coagulase gene typing, and Geoff Coombs for phage typing the isolates. Senior members of the communities are thanked for their assistance. The Health Department of Western Australia provided facilities and logistic support.
REFERENCES
- 1.Beard-Pegler M A, Vickery A M. Lysogenicity of methicillin-resistant strains of Staphylococcus aureus. J Med Microbiol. 1985;20:147–155. doi: 10.1099/00222615-20-2-147. [DOI] [PubMed] [Google Scholar]
- 2.Blair J E, Williams R E O. Phage typing of staphylococci. Bull W H O. 1961;24:771–784. [PMC free article] [PubMed] [Google Scholar]
- 3.Borecka P, Rosypal S, Patucek R, Doskar J. Localization of prophages of serological group B and F on restriction fragments defined in the restriction map of Staphylococcus aureus NCTC8325. FEMS Microbiol Lett. 1996;143:203–210. doi: 10.1111/j.1574-6968.1996.tb08481.x. [DOI] [PubMed] [Google Scholar]
- 4.Boyce J M. Are the epidemiology and microbiology of methicillin-resistant Staphylococcus aureus changing? JAMA. 1998;279:623–624. doi: 10.1001/jama.279.8.623. [DOI] [PubMed] [Google Scholar]
- 5.Brakstad O D, Maeland J E, Tveten Y. Multiplex polymerase chain reaction for detection of genes for Staphylococcus aureus thermonuclease and methicillin resistance and correlation with oxacillin resistance. APMIS. 1993;101:681–688. doi: 10.1111/j.1699-0463.1993.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 6.Collignon P, Gosbell I, Vickery A, Nimmo G, Stylianopoulos T, Gottleib T. Community-acquired methicillin-resistant Staphylococcus aureus in Australia. Lancet. 1998;352:145–146. doi: 10.1016/s0140-6736(98)85051-4. [DOI] [PubMed] [Google Scholar]
- 7.Coombs G W, Pearman J W, Khinsoe C H, Boehm J D. The problems in detecting low-expression-class methicillin resistance in Staphylococcus aureus with batches of Oxoid Mueller-Hinton agar. J Antimicrob Chemother. 1996;38:551–553. doi: 10.1093/jac/38.3.551. [DOI] [PubMed] [Google Scholar]
- 8.Goering R V, Winters M A. Rapid method for epidemiological evaluation of gram-positive cocci by field inversion electrophoresis. J Clin Microbiol. 1992;30:577–580. doi: 10.1128/jcm.30.3.577-580.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goh S-H, Byrne S K, Zhang J L, Chow A W. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992;30:1642–1645. doi: 10.1128/jcm.30.7.1642-1645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grubb W B. Molecular epidemiology of methicillin-resistant Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 595–606. [Google Scholar]
- 11.Kauffman C A, Bradley S F. Epidemiology of community-acquired infection. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 287–301. [Google Scholar]
- 12.Maguire G P, Arthur A D, Boustead P J, Dwyer B, Currie B J. Clinical experience and outcomes of community-acquired and nosocomial methicillin-resistant Staphylococcus aureus in a northern Australian hospital. J Hosp Infect. 1998;38:273–281. doi: 10.1016/s0195-6701(98)90076-7. [DOI] [PubMed] [Google Scholar]
- 13.Marples R R, Reith S. Epidemic methicillin-resistant Staphylococcus aureus. CDR Weekly. 1996;6:197. [PubMed] [Google Scholar]
- 14.Mitchell J M, MacCullough D, Morris A J. MRSA in the community. N Z Med J. 1996;109:411. [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. 6th ed. 1997. Approved standard. M2-A6. National Committee for Clinical Laboratory Standards, Villanova, Pa. [Google Scholar]
- 16.Pattee P A, Lee H C, Bannantine J P. Genetic and physical mapping of the chromosome of Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 41–58. [Google Scholar]
- 17.Pearman J W, Christiansen K J, Annear D I, Goodwin C S, Metcalf C, Donovan F P, Macey K L, Bassett L D, Powell I M, Green J M, Harper W E, McKelvie M S. Control of methicillin-resistant Staphylococcus aureus (MRSA) in an Australian metropolitan teaching hospital complex. Med J Aust. 1985;142:103–108. [PubMed] [Google Scholar]
- 18.Pearman J W, Grubb W B. Preventing the importation and establishment of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in Western Australia. APUA Newsl. 1993;11(3):1–3. , 8. [Google Scholar]
- 19.Pearman J W, Grubb W B. Emerging strains of multiresistant methicillin-resistant Staphylococcus aureus threaten success of screening policy. APUA Newsl. 1993;11(4):1–3. , 8. [Google Scholar]
- 20.Pearman, J. W., P. L. Perry, G. W. Coombs, R. C. Lee, F. G. O’Brien, and W. B. Grubb. Outbreak of a community strain of methicillin-resistant Staphylococcus aureus in a Western Australian metropolitan hospital. Submitted for publication.
- 21.Perry P L, Coombs G W, Boehm J D, Pearman J W. A rapid (20 h) solid screening medium for detecting methicillin-resistant Staphylococcus aureus. J Hosp Infect. 1998;40:67–72. doi: 10.1016/s0195-6701(98)90027-5. [DOI] [PubMed] [Google Scholar]
- 22.Riley D, MacCullough D, Morris A J. Methicillin-resistant S. aureus in the suburbs. N Z Med J. 1998;111:59. [PubMed] [Google Scholar]
- 23.Riley T V, Pearman J W, Rouse I L. Changing epidemiology of methicillin-resistant Staphylococcus aureus in Western Australia. Med J Aust. 1995;163:412–414. doi: 10.5694/j.1326-5377.1995.tb124656.x. [DOI] [PubMed] [Google Scholar]
- 24.Riley T V, Rouse I L. Methicillin-resistant Staphylococcus aureus in Western Australia, 1983–1992. J Hosp Infect. 1995;29:172–188. doi: 10.1016/0195-6701(95)90327-5. [DOI] [PubMed] [Google Scholar]
- 25.Skull S A, Krause V, Coombs G, Pearman J W, Roberts L A. Investigation of a cluster of Staphylococcus aureus invasive infection in the Top End of the Northern Territory. Aust N Z J Med. 1999;29:66–72. doi: 10.1111/j.1445-5994.1999.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Townsend D E, Ashdown N, Bolton S, Grubb W B. The use of cetyltrimethylammonium bromide for the isolation from Staphylococcus aureus of relaxable and non-relaxable plasmid DNA for in vitro manipulation. Lett Appl Microbiol. 1985;1:207–212. [Google Scholar]
- 28.Townsend D E, Ashdown N, Greed L C, Grubb W B. Analysis of plasmids mediating gentamicin resistance in methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1984;13:347–352. doi: 10.1093/jac/13.4.347. [DOI] [PubMed] [Google Scholar]
- 29.Townsend D E, Ashdown N, Pearman J W, Annear D I, Grubb W B. Genetics and epidemiology of methicillin-resistant Staphylococcus aureus in a Western Australian hospital. Med J Aust. 1985;142:108–111. doi: 10.5694/j.1326-5377.1985.tb133045.x. [DOI] [PubMed] [Google Scholar]
- 30.Townsend D E, Grubb W B, Ashdown N. Gentamicin resistance in methicillin-resistant Staphylococcus aureus. Pathology. 1983;15:169–174. doi: 10.3109/00313028309084707. [DOI] [PubMed] [Google Scholar]
- 31.Turnidge J D, Nimmo G R, Francis G. Evolution of resistance in Staphylococcus aureus in Australian teaching hospitals. Med J Aust. 1996;164:68–71. [PubMed] [Google Scholar]
- 32.Udo E E, Pearman J W, Grubb W B. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- 33.Udo E E, Pearman J W, Grubb W B. Emergence of high-level mupirocin resistance in methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1994;26:157–165. doi: 10.1016/0195-6701(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 34.Vickery A M, Beard-Pegler M A. Strain differentiation in methicillin-resistant Staphylococcus aureus. Pathology. 1983;15:235–240. doi: 10.3109/00313028309083499. [DOI] [PubMed] [Google Scholar]
- 35.Wei M Q, Wang F, Grubb W B. Use of contour-clamped homogeneous electric field (CHEF) electrophoresis to type methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1992;36:172–176. doi: 10.1099/00222615-36-3-172. [DOI] [PubMed] [Google Scholar]