Abstract
The global pandemic of coronavirus disease 2019 (COVID-19) is continuing to have a significant effect on the well-being of the global population, thus increasing the demand for rapid testing, diagnosis, and treatment. As COVID-19 can cause severe pneumonia, early diagnosis is essential for correct treatment, as well as to reduce the stress on the healthcare system. Along with COVID-19, other etiologies of pneumonia and Tuberculosis (TB) constitute additional challenges to the medical system. Pneumonia (viral as well as bacterial) kills about 2 million infants every year and is consistently estimated as one of the most important factor of childhood mortality (according to the World Health Organization). Chest X-ray (CXR) and computed tomography (CT) scans are the primary imaging modalities for diagnosing respiratory diseases. Although CT scans are the gold standard, they are more expensive, time consuming, and are associated with a small but significant dose of radiation. Hence, CXR have become more widespread as a first line investigation. In this regard, the objective of this work is to develop a new deep transfer learning pipeline, named DenResCov-19, to diagnose patients with COVID-19, pneumonia, TB or healthy based on CXR images. The pipeline consists of the existing DenseNet-121 and the ResNet-50 networks. Since the DenseNet and ResNet have orthogonal performances in some instances, in the proposed model we have created an extra layer with convolutional neural network (CNN) blocks to join these two models together to establish superior performance as compared to the two individual networks. This strategy can be applied universally in cases where two competing networks are observed. We have tested the performance of our proposed network on two-class (pneumonia and healthy), three-class (COVID-19 positive, healthy, and pneumonia), as well as four-class (COVID-19 positive, healthy, TB, and pneumonia) classification problems. We have validated that our proposed network has been able to successfully classify these lung-diseases on our four datasets and this is one of our novel findings. In particular, the AUC-ROC are 99.60, 96.51, 93.70, 96.40% and the F1 values are 98.21, 87.29, 76.09, 83.17% on our Dataset X-Ray 1, 2, 3, and 4 (DXR1, DXR2, DXR3, DXR4), respectively.
Keywords: COVID-19, Pneumonia, Chest X-rays, Deep transfer learning network, Automatic classification, Tuberculosis
Graphical Abstract
1. Introduction
Coronavirus 2019 (COVID-19), a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus, has affected the health of populations globally (Gorbalenya et al., 2020). In order to control the COVID-19 pandemic, there is an urgent need for rapid and accurate diagnostic testing in healthcare (Cheng et al., 2020; Tang et al., 2020). Since SARS-CoV-2 can cause COVID-19 pneumonia and severe lung damage, differentiating viral from bacterial pneumonia and other respiratory infections such as Tuberculosis (TB) using chest imaging technology is essential for managing infection control decisions and diagnosis and for planning treatment regimes (Qin et al., 2019).
Many infectious respiratory diseases present in a similar manner, with symptoms such as difficulty in breathing, persistent cough, and fever. Pneumonia, an infection affecting the airspaces in the lung, is caused by various etiological agents such as bacteria, viruses, and fungi. There is a wide range of symptoms associated with the infection, which include shortness of breath, fever, phlegm production, and cough. The progression of the disease is marked by the air space opacification, which can be detected using imaging diagnostics (Suzuki et al., 2019; Reittner et al., 2003). Despite the availability of antimicrobials, pneumonias contribute to the most common cause of mortality, especially in childhood (World Health Organization, 2011). In addition, TB induces a persistent cough and breathlessness with symptoms that overlap those of pneumonia and COVID-19. The mortality rates have also risen due to drug-resistant pulmonary TB, caused by Mycobacterium tuberculosis (World Health Organization, 2020). There is therefore a clear need for a robust artificial intelligence (AI) system that can detect and classify the various respiratory diseases that have overlapping presentations to the clinic, so that the right course of treatment regime can be prescribed.
The standard imaging modalities for lung disease diagnosis include magnetic resonance imaging (MRI), chest X-ray (CXR), and computed tomography (CT) scan. Although MRI and CT scan are the gold standard for assessing lung diseases, they are more expensive, involve radiation exposure, and are not readily available globally (Soltan et al., 2020). In comparison, CXR is less expensive, readily available, and is one of the most common diagnostic imaging techniques for cardiothoracic and pulmonary disorders.
CXR patterns of lung disease present differentiation challenges and often result in high inter-reader variability across radiologists (Williams et al., 2013). With potential future waves of the pandemic, radiologists’ workloads will increase and there is an urgent need for new automated image analysis tools that can enhance the radiologists’ qualitative assessment. These tools will classify or segment sections of the CXR in order to support the diagnostic workflow. Decision support systems are designed to aid the clinical decision-making process and have established themselves as emerging research trend in healthcare (Stacey et al., 2017). Over recent months of the pandemic, automated detection of pneumonia or other lung diseases, specifically their early detection and classification, have gained significant attention from both clinical and the AI researchers.
The development of AI-based medical systems, as well as their translation to medical practice, is playing an increasingly prominent role in the treatment and therapy of patients (Greenspan et al., 2020). Along with the automated methods that rely on the blood test results or biomarkers for diagnosis (Banerjee et al., 2020; Song et al., 2020a; Lalmuanawma et al., 2020), an increasing number of deep learning-based methods, specifically the convolution neural network (CNN)-based models (Pereira et al., 2020; Das et al., 2020; Sarker et al., 2020; Li et al., 2020a; Ozturk et al., 2020a), are being implemented and used in order to develop accurate, robust, and fast detection techniques to fight against COVID-19 and other respiratory diseases.
In this regard, the aim of the current study is to test the feasibility of early automated detection and distinction between COVID-19, pneumonia, TB, and healthy patients based on CXR scans. We have developed a deep transfer learning pipeline, named DenResCov-19, to diagnose if a patient is healthy or has a lung disease. The proposed network optimally combines the DenseNet-121 and ResNet-50 networks. This combination unifies the simplicity of ResNet structure and the complexity of DenseNet blocks and delivers a well-balanced result of accuracy and increased specificity and sensitivity. Pretrained networks on the ImageNet cohort are used as transfer learning techniques. We have tested the adaptability of our proposed network for two-class (pneumonia and healthy), three-class (COVID-19 positive, pneumonia, and healthy), as well as four-class (COVID-19 positive, pneumonia, TB, and healthy) classification problems. To the best of our knowledge, this is the first work to examine the feasibility of early automatic detection and distinction between COVID-19 positive, pneumonia, TB, and healthy patients based only on CXR scans using a deep-learning (DL) network. The proposed DenResCov-19 network has been able to perform optimally in different multi-class problems and has achieved robust and improved performance over the state-of-the-art methods for the classification of lung-diseases in all our datasets. The main contributions of this paper are as follows:
-
1.
The development of a new deep-learning network, named DenResCov-19, for robust and accurate classification.
-
2.
Evaluating the accuracy and robustness of DenResCov-19 over heterogeneous CXR image datasets with binary and multi-class labels (COVID-19, pneumonia, TB, and healthy).
-
3.
Evaluating the robustness of DenResCov-19 network over a Monte Carlo cross validation scheme for multi-class classification.
-
4.
The comparison of DenResCov-19 with established networks of ResNet-50, DenseNet-121, VGG-16, and Inception-V3.
-
5.
Developing a pre-screening fast-track decision network to detect COVID-19 and other lung pathologies.
The rest of the paper is organized as follows: Section 2 gives a brief overview of the related works. Section 3 presents the development of the proposed methodology, while Section 4 summarizes its implementation and the description of clinical datasets. Numerical results of the application of our method on four different datasets are presented in Section 5. The final conclusions are presented in Section 6.
2. Related works
In this section, we present a brief overview of pneumonia and COVID-19 diagnosis studies based on CXR/CT scans and the impact of artificial intelligence in clinical management of COVID-19.
2.1. Review of pneumonia detection in CXR images
There exists a significant body of literature on the application of deep learning networks on CXR images for detecting pneumonia in patients (Bustos et al., 2020; Jaiswal et al., 2019; Varshni et al., 2019; Varela-Santos and Melin, 2021). Here we give a summary of the most important approaches.
Jaiswal et al. (2019) used Mask-Region-based CNN (He et al., 2017) model to automatically identify potential pneumonia cases from CXR images. Bharati et al. (2020) proposed a hybrid deep learning framework by combining VGG (Ozturk et al., 2020b), data augmentation, and spatial transformer network (STN) with CNN. They trained their model in NIH CXR dataset (Kermany et al., 2018) with 73% accuracy. Even though their approach did not achieve a high accuracy, their network required training time of only 431 s on their full dataset. Bustos et al. (2020) presented a comprehensive study on a significantly large dataset of 160,000 CXR images, including 19 different classes of lung diseases. They compared four models, namely CNN, recurrent neural network (RNN) composed of bi-directional long short-term memory (LSTM) cells (Hochreiter and Schmidhuber, 1997), CNN with per-label attention mechanism (CNN-ATT) (Mullenbach et al., 2018), and RNN composed of bi-directional LSTM cells with per-label attention mechanism (RNN-ATT). Among the four models, the RNN-ATT model achieved the best results with 86.4% accuracy with only 41 epochs training. Varela-Santos and Melin (2021) implemented an automated system for future detection of COVID-19 and pneumonia diseases in CXR and CT lung images. They efficiently utilized the image texture feature descriptors from CXR images in feed-forward and convolutional neural networks for detecting COVID-19, pneumonia, and healthy individuals.
2.2. Review of COVID-19 detection in CXR and CT images
Prior to COVID-19, deep learning (DL) models have been used extensively for the classification of pneumonia and other lung diseases. Following their successes, a range of DL approaches have been developed for diagnosing and differentiating COVID-19 lung infections (He et al., 2021; Gilanie et al., 2021). Most of these new approaches are based on CXR and CT modalities, which are the most widely used imaging modalities for diagnosing pneumonia and COVID-19 (Das et al., 2020; Sarker et al., 2020; Li et al., 2020a). Here we review the performance of some of these studies.
Ozturk et al. (2020a) proposed the DarkCovidNet model to assist clinicians and radiologists to diagnose COVID-19. Their network, inspired by DarkNet, achieved accuracy of 98.08% and 87.02% respectively, for binary (COVID-19 vs healthy) and multi-class classification (COVID-19, pneumonia or healthy). DarkCovidNet is based on the DarkNet, which is good for fast performance (e.g., with self-driving cars); but in our case, time is not really a critical issue. Moreover, the network was only tested on a limited number of cases. Larger datasets will be able to test its robustness.
Pereira et al. (2020) designed the network model RYDLS-20, that achieved F1 value of 89% for COVID-19 diagnosis. Their dataset was highly imbalanced, with 1000 healthy cases and 90 patients affected by COVID-19. More importantly, their classification performance was presented without any cross-validation step.
Yoo et al. (2020) proposed a combination of three decision-tree classifiers for pre-screening fast-track decision making in order to detect COVID-19. Their pipeline was a combination of three binary decision trees, each trained by a deep learning model with CNN. The accuracies of the binary decision trees ranged between 80% and 98%. However, their network did not test any pathologically confirmed data. In addition, they did not incorporate any data augmentation technique during training in order to reduce the overfitting effects. A large dataset of 5000 CXR scans was used by (Minaee et al., 2020) for classification of healthy and COVID-19 cases. They used four different models, including ResNet18 (He et al., 2016), ResNet50 (He et al., 2016), SqueezeNet (Iandola et al., 2016), and DenseNet-161 (Huang et al., 2017), and achieved on average sensitivity of 98% and specificity of 92%.
Another set of studies have presented satisfactory outcomes in the classification of COVID-19 and healthy cases from CT images (Chen et al., 2020; Harmon et al., 2020; Li et al., 2020b; Song et al., 2020b; Wang et al., 2020). Li et al. (2020b) studied CT images deployed at sixteen different hospitals. They used a U-net to first segment the lung regions and then applied a ResNet-50 to classify the patient as COVID-19 affected or not. Their pipeline achieved a good accuracy, because there was no noise in peripheral organ regions due to the segmentation of lungs. One of the limitations of their study is that their dataset had higher number of positive cases that made the prediction biased (723/1136). However, the major achievements of this study are the incorporation of inter-hospital variations in datasets and the use of six independent experts to arrive at the ground truth.
In another study, Li et al. (2020a) used statistical methods, which included ‘total severity score’ to classify healthy and unhealthy patients based on CXR images. The authors applied the Wilcoxon-rank test to predict the level of severity of the patients. They computed their ground truth using inter-scan and inter-observer variability and also provided thorough details on how the severity level was computed. However, their severity dataset was not large enough and also, they did not incorporate any data splitting based on advanced age, underlying diseases, and pleural effusions.
Song et al. (2020b) implemented a deep learning-based CT diagnosis system, named Deep-Pneumonia, to identify patients with COVID-19. They manually segmented the lung region and then classified COVID-19 or healthy cases using a DL network. This network, named DRE-Net, is a combination of ResNet-50, feature pyramid network (FPN), and an attention module. The main advantages of this study are the multi-vendor datasets from three different hospitals, the very high sensitivity (95%) and specificity (96%) values, and the fast diagnosis time per patient (30 s). However, its drawbacks include: the need of semi-automatic lung segmentation, the classification of datasets based only on CT images without any splitting depending on advanced age, underlying diseases, or pleural effusions, and the absence of any reference in inter-observer variability of the ground truth.
Chen et al. (2020) trained their deep network using 46,096 anonymous images from 106 admitted patients, including 51 patients with laboratory confirmed COVID-19 pneumonia and 55 control patients with other diseases, in Renmin Hospital of Wuhan University. They used a U-net++ network to segment the lungs and classified whether the region had a scar area. The two-tailed paired Student’s t-test with 0.05 significance level was used for time comparison between radiologist and the model. The key advantages of this study include: the large and well-balanced training dataset, the high classification accuracy (over 95%), and the use of three expert radiologists accounting for the inter-observer variability to extract the ground truth. The main limitations of this study were that the dataset was collected from only one hospital and the classification was based only on CT images without any data-splitting based on advanced age, underlying diseases, or pleural effusions. Also, their lung segmentation step, being in a new cohort, can potentially decrease the total classification accuracy.
2.3. Review of artificial intelligence in clinical management of COVID-19 pandemic
During the global pandemic of COVID-19, a lot of different problems were introduced from the scientific community. Greenspan et al. (2020) highlighted the importance of artificial intelligence (AI) (Chassagnon et al., 2021) in the early disease detection, the severity risk management of COVID-19 disease in the hospitals, and the importance of implemented patient-specific predictive models based on imaging and additional clinical features.
Regarding the early stage of COVID-19 detection, Gao et al. (2021) implemented a segmentation-classification network to classify COVID-19 or healthy patient. They used a cross-institute protocol validation of internal and external validation datasets. Di et al. (2021) implemented hypergraph learning to classify COVID-19 or community acquired pneumonia (CAP) from CT imaging. They used a multi-center dataset of 3330 CT images with COVID-19 and CAP cases. A combination of weakly supervised active learning, 2D U-Net and a 3D residual network by Wu et al. (2021) delivered impressive results of the lung region segmentation and COVID-19 detection.
The severity risk level of COVID-19 pathologies was studied by Zhu et al. (2021) and Xue et al. (2021). Zhu et al. (2021) proposed a joint classification and regression method to determine the severity level and the conversion time for patients. Xue et al. (2021) introduced a severity detection of COVID-19 based on lung ultrasound and clinical features.
Even though the above studies including the works by Yang et al. (2021) and Goncharov et al. (2021) delivered impressive robust and accurate results of binary COVID-19 classification, the lack of use of networks to predict multi-class classification of different lung pathologies (including COVID-19) is still a main challenge.
2.4. Limitations of the existing studies
As discussed, some studies have attempted to solve the problem of automated diagnosis of pneumonia and COVID-19, based on existing deep learning networks (Minaee et al., 2020; Ozturk et al., 2020a; Pereira et al., 2020) on CT (Chen et al., 2020; Harmon et al., 2020; Li et al., 2020b; Song et al., 2020b; Wang et al., 2020) or on CXR (Ozturk et al., 2020a; Yoo et al., 2020; Das et al., 2020; Li et al., 2020a; Sarker et al., 2020) image cohorts. However, the noted algorithms suffer from the following limitations and challenges:
-
1.
The lack of regularization techniques (data augmentation, penalty norms, etc.) used in models to avoid possible overfitting.
-
2.
Lack of balance in the models between the speed and the robustness and accuracy.
-
3.
The lack of generalization techniques, such as cross-validation, for accurate predictions of the models.
-
4.
The need of manual segmentation of the lung region from experts to deliver a robust semi-automatic classification result.
-
5.
The validation of the models for only binary classification (Minaee et al., 2020; Pereira et al., 2020; Yoo et al., 2020; Gao et al., 2021; Chassagnon et al., 2021) or three-class (COVID-19, pneumonia, and healthy) classification tasks (Ozturk et al., 2020a; Das et al., 2020; Li et al., 2020a; Sarker et al., 2020).
-
6.
The validation of the models in one specific cohort (i.e. no cross-vendor or cross-institute validation).
3. Methodology and background
In the current study, we propose to train a deep learning network, named DenResCov-19, to solve a multi-class problem, namely, whether a patient is healthy or has pneumonia, COVID-19, or tuberculosis.
3.1. Background
Our approach is based on two state-of-the-art networks: ResNet (He et al., 2016) and DenseNet (Huang et al., 2017). They have recently been used to solve similar multi-class problems.
ResNet-L is inspired by the structure of VGG nets (Simonyan and Zisserman, 2015). The network comprises of L layers, each of which implements a non-linear transformation. In the majority of ResNet-based networks, the convolutional layers have 3 × 3 filters. Downsampling is performed by convolutional layers with a stride of 2. The last two layers of the network are an average pooling layer, followed by a 1000-way fully-connected (FC) layer. The main rule of this deep network is that the layers have the same number of filters as the number of the output feature map size. In case the feature map size is halved, the number of filters is doubled, thus reducing the time complexity per layer. CNN feed-forward inputs x i are the outputs x i−1 of the previous layer, so the transition layer is given by x i = H i(x i−1). In particular, ResNet adds a skip-connection and the identity function is given by:
| (1) |
DenseNet-L is a convolutional network. The network comprises of L layers, each of which implements a non-linear transformation. These transformations can be different function operations, such as Batch Normalization, rectified linear units (ReLU), Pooling, and Convolution. Huang et al. (2017) introduced a unique connectivity pattern information flow between layers to direct connecting any layer to all subsequent layers. As a result, the ith layer includes the feature-maps of all previous layers. The input of ith layer is given by the equation:
| (2) |
where [x 0, x 1, ⋯ , x i−1] refers to the concatenation of the feature-maps produced in layers 0, …, i−1. All inputs of a composite function H i(⋅) are concatenated into a single tensor. Each composite function is a combination of batch normalization (BN), followed by a rectified linear unit (ReLU) and a 3 × 3 convolution (Conv).
3.2. Network architecture
To evaluate the state-of-the-art networks before we train and test them in the CXR cohorts, we initially test them in the open-source and widely used CT cohort of Zhao et al. (2020). Since there is currently a lack of existing publicly available dataset of CXR images relating to COVID-19 cases, we have tested the behavior of benchmark models in the CT cohort in order to check if the expected behavior of the proposed network can be observed (i.e. achieve high F1 and AUC-ROC values).
Table 1 highlights the results of DenseNet-121, ResNet-50, and VGG-16 networks for classification of pneumonia, COVID-19, and healthy cases in CT images. From the results, it can be observed that, while the ResNet has better precision, recall, and F1 metrics than the DenseNet, DenseNet has better AUC-ROC. ResNet also achieves higher precision, AUC-ROC, and F1 metrics than the VGG, while VGG attains higher recall values. Based on these observations, we hypothesize that a combination of the two models, ResNet and DenseNet, can deliver a well-balanced AUC-ROC and F1 metric results.
Table 1.
Metrics of deep learning networks to classify pneumonia, COVID-19, and healthy cases in CT images.
| CT dataset (Zhao et al. (2020)) | |||
|---|---|---|---|
| Metric (%) | DenseNet-121 | ResNet-50 | VGG-16 |
| Recall | 44.0 | 71.2 | 100.0 |
| Precision | 81.2 | 91.0 | 50.0 |
| AUC-ROC | 86.4 | 64.0 | 51.0 |
| F1 | 58.4 | 81.0 | 71.4 |
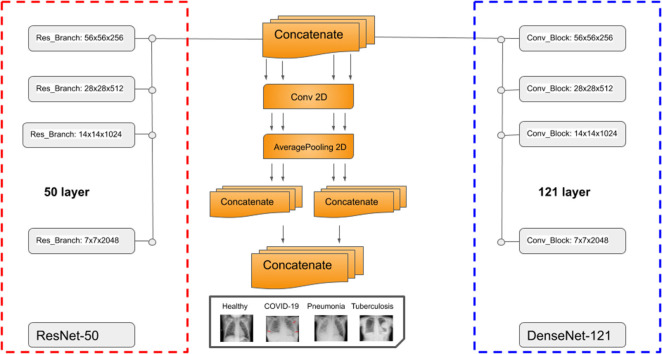
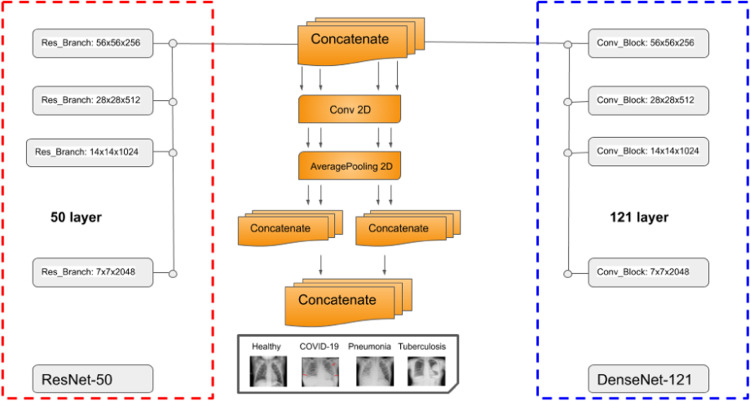
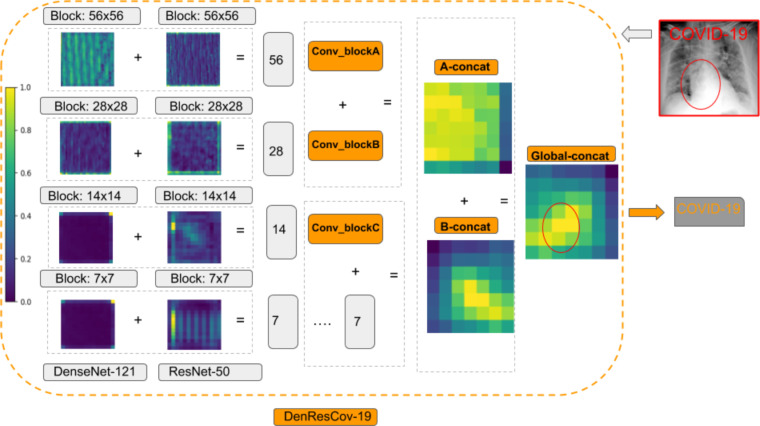
The architecture of our proposed DenResCov-19 network is presented in Fig. 1. DenResCov-19 network is a concatenation of four blocks from ResNet-50 and DenseNet-121 with width, height, and frames of 58 × 58 × 256, 28 × 28 × 512, 14 × 14 × 1024, and 7 × 7 × 2048, respectively. We chose these specific blocks from the networks, as we needed layers with the same width × height × frames, so that the information of both models can be combined. As a result, we used four different layers of 58, 28, 14, and 7 size kernels, as we wanted to concatenate the information of the two networks in different regions of interest. Each of the four outputs feed a block of convolution and average pooling layers. Thus, the initial concatenated information can be translated into the convolution space. After that, we used some levels of concatenation-CNN block techniques to create kernels that will deliver a final layer of soft-max regression, so that the network can conclude in the classification decision.
Fig. 1.
DenResCov-19: a deep transfer learning pipeline to classify if a patient has COVID-19, pneumonia, or tuberculosis, based on CXR.
The convolution layer is defined as:
| (3) |
where is a unit in layer l, ω ab is an M × M filter, and is the nonlinearity of previous convolutional layer given by:
| (4) |
The average pooling layer is defined over a K × K region and outputs a single value, which is the average over that region. The inputs of the lth (l = 1, 2, 3, 4) layer block are provided according to the equation:
| (5) |
where is the composite function of lth ResNet layer and is composite function of lth DenseNet layer. The last step of the pipeline is the combination of two pair concatenation and a global concatenation followed by a 512-way fully-connected softmax layer.
3.3. Evaluation metrics
The most common metrics for evaluating classification performance are the precision, recall, and F1-Score, which follow the standard definitions:
| (6) |
| (7) |
| (8) |
where TP, TN, FP, and FN are the true positive, true negative, false positive, and false negative values, respectively. The F1-score is defined as the harmonic mean of the precision and recall, as follows
| (9) |
Besides these metrics, we have also used the AUC-ROC metric values (Davis and Goadrich, 2006) for evaluation. The AUC (area under the curve)-ROC value can be computed by integrating over the receiver operating characteristic (ROC) curve, plotting the true positive rate against the false positive rate.
4. Implementation
This section describes the implementation details of the proposed DenResCov-19 pipeline.
4.1. Cohort details
In order to train and validate our proposed network, we have used three different publicly available open-source cohorts of CXR images, namely, the Pediatric CXRs dataset to detect pneumonia vs healthy cases (Kermany et al., 2018) (source-1), the IEEE COVID-19 CXRs dataset (Cohen et al., 2020) (source-2), and the Tuberculosis CXRs from Shenzhen Hospital x-ray dataset (Jaeger et al., 2014) (source-3). It is important to mention that there were no multi-label cases, such as pneumonia and COVID-19 findings in the same patient, in any of these datasets.
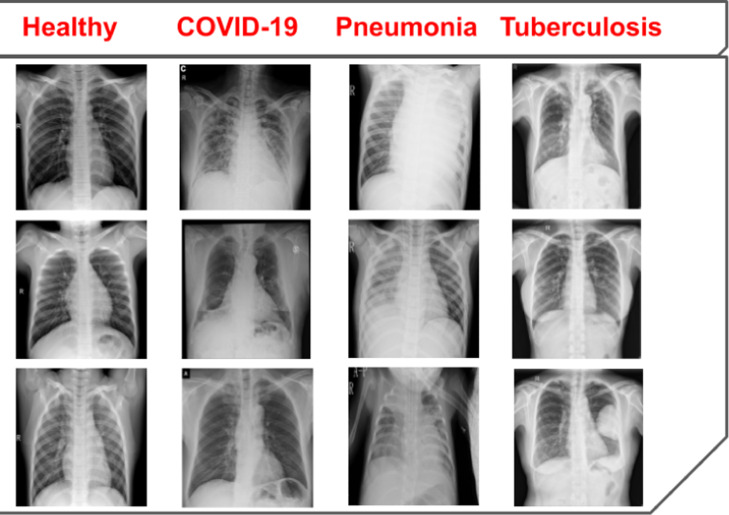
In order to demonstrate the adaptability of our model in multi-class datasets, we created four different datasets, namely DXR1, DXR2, DXR3, and DXR4. The DXR1 dataset was based on source-1 cohort with 3883 pneumonia images and 1350 healthy images. This dataset is a binary classification dataset to detect pneumonia and healthy cases. The source-1 cohort is collected based on pediatric populations. Next, in DXR2, we have trained and tested the models for classification of COVID-19, pneumonia, and healthy patients in the IEEE COVID-19 x-rays dataset (source-2) with 69 COVID-19 images, 79 pneumonia images, and 79 healthy cases. In the third dataset (DXR3) of our study, we have trained and validated our network on source-2 and the tuberculosis (TB) cases of Shenzhen Hospital x-ray dataset (source-3) to detect TB, COVID-19, pneumonia, and healthy cases. As the source-3 had more than 300 CXR images for both TB and healthy classes, the combination of the two sources would end up with an unbalanced dataset. Thus, we randomly selected 79 tuberculosis images from source-3 and 69 COVID-19 images, 79 pneumonia images, and 79 healthy cases from source-2, in order to generate the DXR3. In the DXR4 dataset of our study, we have trained and validated our network on a combination of source-1, source-2, and source-3 to detect TB, COVID-19, pneumonia, and healthy cases. Only in this case, we mixed the pediatric and adult patients populations of the three sources, in order to test the robustness of our proposed model in multi-class dataset with a variation of the patient’s age. To avoid the bias effects, we created the dataset with randomly selected balanced number of images. In the healthy class, we included 110 images from each source to generate a total of 330 healthy cases. In the pneumonia class, we included 79 images from source-2 and 221 images from source-1 to generate 300 pneumonia images. Finally, 310 tuberculosis images source-3 and 69 COVID-19 images from source-2 were included to prepare the final DXR4 dataset. To the best of our knowledge, there was no other COVID-19 CXR open-source dataset available, in order to balance the number of images in the COVID-19 class. Fig. 2 depicts a sample of the CXR scans from the healthy, COVID-19, pneumonia, and tuberculosis patients, as determined by expert radiologists. Summarizing the four different cases:
-
•
DXR1: 3883 pneumonia and 1350 healthy cases (Kermany et al., 2018).
-
•
DXR2: 69 COVID-19 images, 79 pneumonia images, and 79 healthy cases (Cohen et al., 2020).
-
•
DXR3: 69 COVID-19 images, 79 pneumonia images, 79 tuberculosis images, and 79 healthy cases (Cohen et al., 2020; Jaeger et al., 2014).
-
•
DXR4: 69 COVID-19 images, 300 pneumonia images, 310 tuberculosis images, and 330 healthy cases (Kermany et al., 2018; Cohen et al., 2020; Jaeger et al., 2014).
Here, the DXR4 is simply an extended version of DXR3, generated using more images from the datasets of (Kermany et al., 2018; Cohen et al., 2020; Jaeger et al., 2014).
Fig. 2.
A sample of healthy, COVID-19, pneumonia, and tuberculosis cases from the CXR image dataset.
4.2. Cohort’s pre-processing image analysis
Image analysis techniques have been applied on all slices to reduce the effect of noise and increase the signal-to-noise ratio (SNR). We have used noise filters such as binomial deconvolution, Landweber deconvolution (Vonesch and Unser, 2008), and curvature anisotropic diffusion image filters (Perona and Malik, 1990) to reduce noise in the images. We have normalized the images by subtracting the mean value from each image and dividing by its standard deviation. Finally, we have used data augmentation techniques including rotation (rotation around the center of image by a random angle in the range of −15∘ to 15∘), width shift range (width shift of image by up to 20 pixels), height shift range (height shift of image by up to 20 pixels), and ZCA whitening (add noise in each image) (Koivunen and Kostinski, 1999).
4.3. Hyper-parameters initialization
After random shuffling, each dataset has been partitioned into 70% and 30% of the total CXR images using the repeated random subsampling validation technique (also known as the Monte Carlo cross-validation split), before training and testing the models, respectively. We have used the categorical cross-entropy as cost function. The loss function is optimized using the stochastic gradient descent (SGD) method with learning rate of 0.001 and with 30 epochs (the models converged after 20–25 epochs). We have applied transfer learning techniques on the ResNet-50 and DenseNet-121 networks using the ImageNet dataset (Deng et al., 2009) (http://www.image-net.org). It consists of over 14 million images and the task is to classify the images into one of almost 22,000 different categories (cat, sailboat, etc.).
4.4. Software
The code developed in this study is written in the Python programming language using Keras/TensorFlow (Python) libraries. For training and testing of the deep learning networks, we have used an NVIDIA cluster, with 4 GPUs and 64 GB RAM memory. The code implementation is available on a public repository with url: https://github.com/team-globs/COVID-19_CXR.
5. Performance analysis and discussions
This section presents the performance of our proposed DenResCov-19 network, along with a quantitative performance comparison with established DL networks, on four different datasets. The underlying reason behind choosing the DenseNet-121 and ResNet-50 networks in our study is that we wanted to combine the advantages of both networks to develop a new network with well-balanced AUC-ROC and F1 metric values. VGG-16 is a network with relatively faster training time and, in the majority of cases, it has very good AUC-ROC, but comparatively poor F1-value. Hence, we wished to check if the performance of our network is superior enough from VGG, to compensate for the relatively slower training procedure. We preferred to choose ResNet-50 as a well-balanced choice regarding the training time and accuracy of the network, since ResNet is very fast in low layers (such as ResNet-18), but the accuracy improves as the layers of the structure increase (50, 110 etc.). The same approach was followed for the DenseNet too.
In addition, in the study presented in Bressem et al. (2020), the DL structures with superior performance in classification were determined as ResNet, DenseNet, AlexNet, Inception, VGG, and SqueezeNet. Among these networks, the most superior AUC-ROC value in COVID-19 image data collection and CXR cohort were the ResNet-50, DenseNet-161, VGG-19, and AlexNet. Regarding the Area Under the Precision Recall Curve and Sensitivity and Specificity, the best networks were the ResNet-50, DenseNet-161, VGG-16, and Alex-Net. Since ResNet-50 and DenseNet-161 presented satisfactory performance in the majority of the cases, we preferred to consider them as the benchmark networks. However, instead of DenseNet-161, we used the DenseNet-121 due to its significantly less computation time during training.
5.1. Evaluating the classification performance
As explained in Section 4.1, we have created four different CXR image collections to evaluate the performance of the models in binary and multiclass classification. Table 2 summarizes the metrics for the different networks and datasets. Our initial hypothesis that our network DenResCov-19 will have more balanced AUC-ROC and F1 measurements compared to the DenseNet-121 and ResNet-50 networks, has been verified in all four datasets.
Table 2.
Comparative performance metrics of the different deep learning networks performing classification of pneumonia, TB, COVID-19, and healthy cases. Boldface indicates the best metric among the networks.
| DXR1 dataset: pneumonia and healthy | ||||
|---|---|---|---|---|
| Metric | DenResCov-19 | DenseNet-121 | ResNet-50 | Inception-V3 |
| Recall (%) | 98.12 | 97.80 | 97.71 | 93.32 |
| Precision (%) | 98.31 | 94.62 | 95.01 | 90.10 |
| AUC-ROC (%) | 99.60 | 99.10 | 98.95 | 92.80 |
| F1 (%) | 98.21 | 96.27 | 96.34 | 91.68 |
| DXR2 dataset: COVID-19, pneumonia and healthy | ||||
|---|---|---|---|---|
| Metric | DenResCov-19 | DenseNet-121 | ResNet-50 | VGG-16 |
| Recall (%) | 89.38 | 83.54 | 83.53 | 99.83 |
| Precision (%) | 85.28 | 77.45 | 73.35 | 33.38 |
| AUC-ROC (%) | 96.51 | 93.2 | 92.39 | 50.07 |
| F1 (%) | 87.29 | 80.37 | 78.11 | 49.51 |
| DXR3 dataset: COVID-19, pneumonia, tuberculosis and healthy | ||||
|---|---|---|---|---|
| Metric | DenResCov-19 | DenseNet-121 | ResNet-50 | VGG-16 |
| Recall (%) | 59.28 | 57.71 | 56.66 | 66.53 |
| Precision (%) | 79.56 | 74.87 | 74.00 | 26.53 |
| AUC-ROC (%) | 91.77 | 89.49 | 92.12 | 53.11 |
| F1 (%) | 68.09 | 65.17 | 64.17 | 38.00 |
| DXR4 dataset: COVID-19, pneumonia, tuberculosis and healthy | ||||
|---|---|---|---|---|
| Metric | DenResCov-19 | DenseNet-121 | ResNet-50 | VGG-16 |
| Recall (%) | 69.7 | 62.70 | 62.00 | 93.69 |
| Precision (%) | 82.90 | 79.35 | 78.60 | 27.17 |
| AUC-ROC (%) | 95.00 | 91.00 | 93.21 | 54.99 |
| F1 (%) | 75.75 | 70.07 | 69.51 | 42.13 |
In particular, DenResCov-19 has AUC-ROC of 99.60%, 96.51%, and 95.00%, contrary to the 98.95%, 92.12%, and 93.21% of ResNet-50 and 99.10%, 93.20%, and 91.00% of DenseNet-121 for the DXR1, DXR2, and DXR4 datasets, respectively. In addition, DenResCov-19 has F1 values of 98.21%, 87.29%, and 75.75%, contrary to the 96.34%, 78.11%, and 69.51% of ResNet-50 and 96.27%, 80.37%, and 70.07% of DenseNet-121 for the DXR1, DXR2, and DXR4 datasets, respectively. Our network has achieved more than 98% in all metrics in the binary label classification (pneumonia or healthy) of the DXR1 dataset. With the exception of the recall values in DXR2 and DXR4 datasets of VGG-16, our approach outperforms all other networks for all four metrics in all four datasets.
From the results presented in Table 2, it is clear that as the number of label classes increases, the accuracies of evaluation metrics decrease. In our DenResCov-19 network, the recall value of 98.12% in DXR1 has decreased in DXR2, DXR3, and DXR4 datasets with a variation between 59.28% and 89.38%. In a similar way, the precision value has reduced from 98.31% to 79.56–85.28%, AUC-ROC from 99.60% to 91.77–96.51%, and the F1-value from 98.21% to 68.09–87.29%. However, as previously discussed, the results or our network are still better than the state-of-the-art networks. It should also be noted that the metric results in DXR4 dataset are better than the results in DXR3, although the numbers of label classes in two datasets are the same (COVID-19, pneumonia, tuberculosis, and healthy). This happens as the number of training data has increased from almost 80 images to almost 300 images per class (except for the COVID-19 cases, which remains at 69). It is worth mentioning that, since the number of labeled COVID-19 x-ray images is very limited (69 images), it has affected the quantitative results of both precision and recall values in DXR2, DXR3, and DXR4 datasets. Incorporation of additional labeled data in future would significantly improve the performance with respect to these two indices.
5.2. Evaluating the cross validation results
For any classification task, it is very important to minimize the bias effects generated from a fixed validation scheme (70% training, 30% testing). Thus, we have compared the three networks (DenseNet-121, ResNet-50, and DenResCov-19) in DXR4 dataset for classification over four randomly shuffled fixed ratio validation schemes (also known as Monte Carlo cross-validation method). For each cross-validation set, we have calculated the F1 and AUC-ROC metrics and the ‘micro’, ‘macro’, and ‘weighted’ versions of the indices. The ‘micro’ version is calculated by counting the total number of true positives, false negatives, and false positives. The ‘macro’ version computes the metric for each class and finds their unweighted means. The ‘weighted’ version measures the metric for each class and determines their weighted means. Table 3 summarizes the results of four different cross-validations in the DXR4 dataset for the DenseNet-121, ResNet-50, and DenResCov-19 networks. DenResCov-19 achieves the highest score in all average, higher, and lower values of the metrics. DenseNet-121 has higher recall and precision average values (62.7, 79.3% against 62.0, 78.6%) and lower AUC-ROC (91.0 against 93.2%) as compared the ResNet-50 network.
Table 3.
Quantitative evaluation metrics for four cross-validation cases on the DXR4 dataset (Kermany et al., 2018; Cohen et al., 2020; Jaeger et al., 2014), and the resulting average and standard deviation. Superscript max/min indicates the highest/lowest score among the cross validation sets.
| Classification performance of ResNet-50 in DXR4 dataset: COVID-19, pneumonia, tuberculosis and healthy | |||||
|---|---|---|---|---|---|
| Metric (%) | Cross validation #1 | Cross validation #2 | Cross validation #3 | Cross validation #4 | Average |
| Recall | 62.7 | 61.9 | 62.4 | 61.5 | 62.0 ± 0.5 |
| Precision | 81.0 | 78.9 | 77.1 | 77.6 | 78.6 ± 1.7 |
| AUC-ROC | 94.1 | 93.2 | 92.7 | 92.6 | 93.2 ± 0.7 |
| AUC-ROC macro | 89.9 | 89.0 | 91.0 | 89.5 | 89.9 ± 0.9 |
| AUC-ROC micro | 88.1 | 88.8 | 90.4 | 88.8 | 89.0 ± 1.0 |
| AUC-ROC weighted | 87.3 | 88.4 | 89.6 | 88.2 | 88.4 ± 0.9 |
| F1 | 70.7 | 69.9 | 69.0 | 68.6 | 69.5 ± 0.9 |
| F1 macro | 70.7 | 70.0 | 69.1 | 68.6 | 69.6 ± 0.9 |
| F1 micro | 70.5 | 69.8 | 68.8 | 68.4 | 69.4 ± 0.9 |
| F1 weighted | 70.7 | 70.1 | 69.0 | 68.7 | 69.6 ± 0.9 |
| Classification performance of DenseNet-121 in DXR4 dataset: COVID-19, pneumonia, tuberculosis and healthy | |||||
|---|---|---|---|---|---|
| Metric (%) | Cross validation #1 | Cross validation #2 | Cross validation #3 | Cross validation #4 | Average |
| Recall | 63.3 | 62.4 | 62.3 | 62.9 | 62.7 ± 0.5 |
| Precision | 80.1 | 80.8 | 79.3 | 76.8 | 79.3 ± 1.7 |
| AUC-ROC | 93.8 | 91.2 | 89.0 | 89.8 | 91.0 ± 2.1 |
| AUC-ROC macro | 90.1 | 92.5 | 90.2 | 91.5 | 91.1 ± 1.1 |
| AUC-ROC micro | 89.2 | 91.2 | 88.8 | 89.2 | 89.6 ± 1.1 |
| AUC-ROC weighted | 88.7 | 90.6 | 87.6 | 87.2 | 88.5 ± 1.5 |
| F1 | 70.7 | 70.4 | 69.8 | 69.2 | 70.0 ± 0.6 |
| F1 macro | 70.0 | 69.6 | 69.0 | 68.3 | 69.3 ± 0.7 |
| F1 micro | 70.7 | 70.4 | 69.8 | 69.2 | 70.0 ± 0.6 |
| F1 weighted | 70.4 | 69.9 | 69.6 | 69.0 | 69.8 ± 0.6 |
| Classification performance of DenResCov-19 in DXR4 dataset: COVID-19, pneumonia, tuberculosis and healthy | |||||
|---|---|---|---|---|---|
| Metric (%) | Cross validation #1 | Cross validation #2 | Cross validation #3 | Cross validation #4 | Average |
| Recall | 70.0 | 71.0 | 67.0 | 70.7 | 69.7 ± 1.8 |
| Precision | 80.0 | 83.0 | 86.0 | 82.6 | 82.9 ± 2.4 |
| AUC-ROC | 93.9 | 95.0 | 96.0 | 95.2 | 95.0 ± 0.8 |
| AUC-ROC macro | 94.7 | 94.7 | 94.7 | 94.8 | 95.6 ± 0.1 |
| AUC-ROC micro | 93.9 | 94.4 | 98.2 | 93.9 | 95.1 ± 2.1 |
| AUC-ROC weighted | 93.3 | 94.1 | 98.0 | 93.6 | 94.7 ± 1.8 |
| F1 | 75.0 | 76.5 | 75.3 | 76.2 | 75.8 ± 0.7 |
| F1 macro | 76.2 | 77.6 | 76.1 | 77.1 | 76.7 ± 0.7 |
| F1 micro | 74.9 | 76.3 | 75.3 | 76.2 | 75.6 ± 0.7 |
| F1 weighted | 75.0 | 76.5 | 75.3 | 76.2 | 75.7 ± 0.7 |
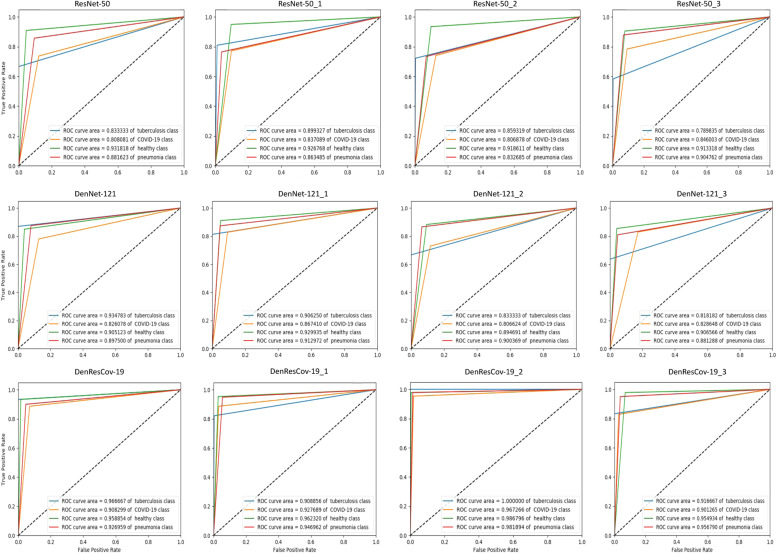
Fig. 3 presents the ROC curves for multi-class classification by ResNet-50, DenseNet-121, and DenResCov-19 networks. The ROC curves are computed in four different cross-validation cases in DXR4 dataset. Based on these figures, it is clear that the true/false positive rate and the ROC curves’ results of the DenResCov-19 network (Fig. 3 third row) are much better in all classes, as compared to the other two networks (Fig. 3 first and second rows). From the results presented in the Fig. 3, we can find the average AUC-ROC values of the four classes for DenseNet-121, ResNet-50, and DenResCov-19 networks. The average AUC-ROC values of TB, COVID-19, healthy, and pneumonia classes for ResNet-50 are 84.8, 82.5, 92.3, 87.1%, while the same for DenseNet-121 are 87.3, 83.1, 90.8, 89.7% and for DenResCov-19 are 94.7, 92.6, 96.4, 95.3%, respectively. Hence, the DenseNet-121 achieves improved true/false positive rate and AUC-ROC values as compared to the ResNet-50 (except for the healthy class). On the other hand, the performance of DenResCov-19 is higher in all average AUC-ROC values of TB, COVID-19, healthy, and pneumonia classes compared to both ResNet-50 and DenseNet-121.
Fig. 3.
The ROC curves for the four cross-validation cases over DXR4 dataset. Top to bottom: ResNet-50, DenseNet-121, and DenResCov-19.
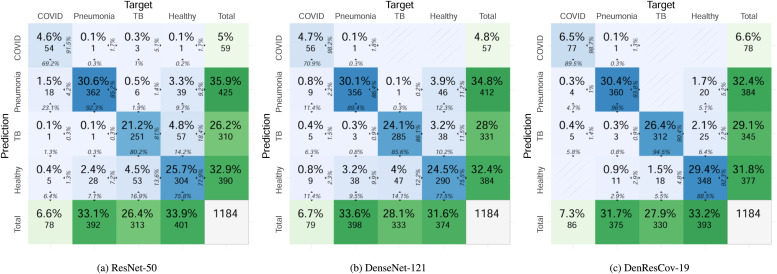
Fig. 4 presents the confusion matrices of multi-class classification by the ResNet-50, DenseNet-121, and DenResCov-19 networks on DXR4 dataset (combined over four cross-validation cases). In the ResNet-50 network, the COVID class has 69.2% true positive and 30.8% false negative predictions among the total number of positive cases, combined over four cross-validation iterations; while in the pneumonia class, the network has 92.3% true positive and 7.7% false negative predictions. In the TB class, the network has 80.2% true positive and 19.8% false negative predictions, and in the healthy class 75.8% true positive and 24.2% false negative predictions. On the other hand, the DenseNet-121 has in the COVID class 70.9% true positive and 29.1% false negative predictions, in the pneumonia class 89.4% true positive and 10.6% false negative predictions, in the TB class 85.6% true positive and 14.4% false negative predictions, and in the healthy class 77.5% true positive and 22.5% false negative predictions. In comparison, the DenResCov-19 network has in the COVID class 89.5% true positive and 10.5% false negative predictions, in the pneumonia class 96.0% true positive and 4.0% false negative predictions, in the TB class 94.5% true positive and 5.5% false negative predictions, and in the healthy class 88.5% true positive and 11.5% false negative predictions. Based on these evidences, we can infer that our proposed network results in higher true positive and lower false negative values as compared to the two established networks. Detailed results for the confusion matrices of individual cross validation cases of the three networks are provided in the supplementary material.
Fig. 4.
Confusion matrices of the three deep learning networks on DXR4 dataset (combined over four cross-validation cases). Each blue-colored cell (i, j) in the matrix denotes the number (and percentage) of cases in target class i that has been classified as class j during prediction. At the right edge of each cell, the percentage of cases in the cell with respect to prediction class j is shown, while the bottom edge presents the percentage with respect to target class i. The last row and last column denote the total number (and percentage) of cases in the target and prediction classes, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
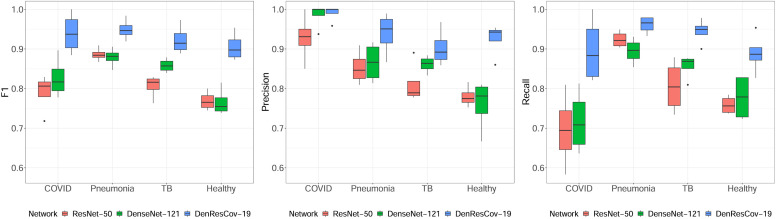
The quantitative performance analysis of the Monte Carlo cross-validation experiment for ResNet-50, DenseNet-121, and DenResCov-19 networks over DXR4 dataset has been presented as box-plots in Fig. 5. Here it is clearly visible that the proposed DenResCov-19 network achieves higher classification performance for all 4 classes, irrespective of the quantitative evaluation indices. For the statistical significance analysis of the classification performance of three networks in terms of the F1-score, precision, and recall values, the linear mixed model analysis has been adopted, where the four different classes, namely COVID-19, pneumonia, tuberculosis, and healthy patients, have been included as random effects in the linear mixed model. Applying the Kenward and Roger’s method for the degrees of freedom of the t-statistic (Kenward and Roger, 1997) and the Tukey’s method for pairwise comparisons (Tukey, 1949), we found that the proposed DenResCov-19 network achieves statistically significantly better classification performance than both DenseNet-121 and ResNet-50 networks in DXR4 dataset in terms of all three quantitative evaluation indices. In terms of F1-score, the DenResCov-19 attains significant p-values of 6.5 ×10-9 and 5.6 × 10-11 as compared to the DenseNet-121 and ResNet-50, respectively, while for the precision index, the p-values are measured as 0.0006 and 2.3 × 10-6 as compared to the same two networks. Similarly with respect to the recall values, the proposed DenResCov-19 has attained significant p-values of 3.5 × 10-6 and 3.1 × 10-7 as compared to the DenseNet-121 and ResNet-50 networks, respectively.
Fig. 5.
Boxplots for the quantitative performance analysis of three deep learning networks on DXR4 dataset for the classification of COVID-19, pneumonia, tuberculosis, and healthy patients.
5.3. Heatmap analysis
The significant results of our network can be clinically validated using a heatmap analysis. We test the classification behavior of the networks (heatmap analysis) in eight randomly selected patients. DenResCov-19 outperformed the ResNet-50 and DenseNet-121 results, as it demonstrates the closest detection pattern with the expert (red ellipses in Fig. 7). This shows our network follows a more human-like approach and it can detect more accurate patterns of the different lung diseases as compared to the two other established networks.
Fig. 7.
Heatmap results of ResNet-50 (left), DenseNet-121 (middle) and DenResCov-19 (right). Red ellipses indicate the (human-generated) detection areas that are correctly identified by the network and black ellipses represent the undetected areas from the corresponding network. The successfully classified images are annotated with green ticks and the wrongly classified images are annotated with red crosses. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6 highlights the main steps of our pipeline. The CXR image initializes the network. The outputs of the four blocks of ResNet-50 and DenseNet-121 are then concatenated. This concatenation creates four new heatmaps 56, 28, 14, and 7 (gray squares). The new heatmaps 56, 28, and 14 pass from a convolution and average max-pool block layer (Conv-blockA, Conv-blockB, and Conv-blockC). The new heatmap 7 continues in the next layer to be combined with Conv-blockC. Following this step, the network combines the outputs of Conv-blockA and Conv-blockB (A-concat) and the outputs of Conv-blockC with the new heatmap 7 (B-concat). The last step is a concatenation of A-concat and B-concat to extract the Global-concat heatmap. Based on that, the model learns to classify the images in the supervised training tasks. Fig. 6 demonstrates the delineation of the COVID-19 detection point, denoted by red ellipse, by our proposed pipeline from a CXR lung image. The delineated region is identical to the area of interest detected by an expert radiologist. Fig. 7 shows the heatmaps of DenseNet-121, ResNet-50, and DenResCov-19 from a total of eight classification cases of the DXR4 dataset. In all cases of Fig. 7, we have highlighted the last heatmap layer of the networks. The red circles in the CXR images are the detection points from our expert radiologist (AS). These points are used to classify the disease in each CXR image. In the top figure of Fig. 7, all images are accurately diagnosed by the three networks. The black and red circles in the heatmap images denote the wrong and accurate detection points, respectively, with respect of the manual annotation. The extraction of the circle is based on a colormap threshold of 0.5. If the average number of the area inside the circle is higher than the threshold, then the detection point assumes correct (red circle); otherwise, it assumes wrong (black circle). In the bottom figure of Fig. 7, the DenseNet-121 accurately diagnoses the last two images (green tick), but wrongly classifies the first two (red cross). On the other hand, the ResNet-50 accurately diagnoses the first two images (green tick), while wrongly classifies the last two (red cross). In comparison, our network diagnoses correctly all images except the first one.
Fig. 6.
Main steps of the DenResCov-19 network to determine the classification decision, represented by heatmaps: The CXR image is the input of network, and the four blocks of ResNet-50 and DenseNet-121 outputs are then concatenated, creating four new heatmaps 56, 28, 14, and 7 (gray squares). The new heatmaps 56, 28, and 14 initialize a convolution and average max-pool block layer (Conv-blockA, Conv-blockB, and Conv-blockC), and the new heatmap 7 is used by the next layer without any further analysis. The network combines the Conv-blockA with the Conv-blockB outputs (A-concat), and the Conv-blockC with the new-heatmap 7 (B-concat). Finally, A-concat and B-concat outputs are concatenated in a Global-concat heatmap. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In the top figure of Fig. 7, the DenseNet-121 cannot detect the left circle annotation (black circle) in the COVID-19 CXR image. Additionally, it cannot clearly detect the right circle annotation either. On the other hand, the ResNet-50 cannot detect the right circle annotation (black circle), while it identifies the left circle. In comparison, DenResCov-19 can detect both circle annotations strongly. In the tuberculosis CXR image, the DenResCov-19 also strongly detects the circle annotation, while the ResNet-50 and DenseNet-121 cannot detect clearly the circle annotation (near the threshold). However, in the pneumonia CXR image, the DenseNet-121 strongly detects the red circle annotation, while the ResNet-50 and DenResCov-19 cannot clearly detect it.
5.4. Discussions
One important limitation of this study is the relatively small cohort size for patients with COVID-19. Due to this, we mixed the pediatric and adult patients populations of the sources-1, 2 and 3 in DXR4 dataset, in order to test the robustness of our proposed model in a dataset with larger cohort size. To avoid the bias effects, we created the dataset with randomly selected balanced number of images. As a result, there are some detected features, for example the pneumonia scars in Fig. 7 (top frame) or the healthy case in Fig. 7 (bottom frame) in DXR4 dataset, which our pipeline cannot strongly detect. The detection of these pathologies in DXR4 is more challenging and a larger dataset with additional demographics is required for further investigation.
Another limitation of this study is the multi-label lung pathology task. In order to further evaluate the generalization and robustness of our pipeline as a whole lung multi-pathologies classifier, we need to provide the multi-class and multi-label classification. Although we have delivered the multi-class challenge in the best possible way based on the available published cohorts, we still face a luck of the generalization from the multi-label aspect. An example of multi-label sample is when a subject has both bacterial pneumonia and COVID-19 diseases. The main reason we could not deliver this aspect is the lack of any publicly available multi-label lung disease datasets.
The main advantage of this study is that, in the majority of lung pathologies, the detection points of radiologist CXR lung images are identified more strongly using the DenResCov-19 network, as compared to the ResNet-50 and DensNet-121. The heatmap results presented in Figs. 6 and 7 justify the accurate classification of our network and validate our initial hypothesis. Moreover, all evaluation metrics of the different classification datasets (from DXR1 to DXR4) demonstrate the robustness and superior performance of the DenResCov-19 network as compared to the benchmark deep learning based approaches.
As we discussed in Section 2, there are some limitations in the majority of the existing studies regarding robust and efficient detection of the COVID-19 and lung diseases. In our current study, we have examined these limitations and tried to solve them. We have trained our model based on regularization techniques, such as data augmentation and penalty L2 norms, to avoid possible overfitting. Furthermore, we have verified the generalization and accurate prediction of our model using Monte Carlo cross-validation technique. The proposed method is fully automated and it does not need any manual segmentation of the lung region from experts to deliver a robust classification result. Finally, we have demonstrated different applications of the model over binary and multi-class classification tasks.
6. Conclusions
In this study, we have implemented a new deep-learning network named DenResCov-19, which can deliver robust classification results in multi-class lung diseases. We have tested the proposed model over three different published datasets with four classes, namely, the COVID-19 positive, pneumonia, TB, and healthy patients. We have also mitigated the class imbalance issue by properly composing the datasets (except for DXR4, where the dataset is imbalanced in COVID-19 positive class due to limited number of available images). Hence, based on our experimental analysis, we can infer a favorable generalization and robust behavior of our proposed model. Our experimental analysis has demonstrated improved classification accuracy of our network, as compared to the state-of-the-art networks such as ResNet-50, DenseNet-121, VGG-16, and Inception-V3. Our initial hypothesis that our network can deliver a well-balanced AUC-ROC and F1 metric results has been verified. In most of the cases, the detection points of our network from heatmaps are in line with the detection points from the expert radiologist. To summarize, we have developed a pre-screening fast-track decision network to detect COVID-19 and other lung pathologies based on CXR images.
In our future study, we will further focus on the generalization of our model with the availability of a significantly larger COVID-19 patients’ cohort. In addition, it will be beneficial to extend the number of classes to include more lung diseases if the corresponding datasets exist. Finally, we wish to evaluate the DenResCov-19 network in different datasets, in order to further evaluate the generalization and robustness of our pipeline in different medical image classification tasks, such as diagnosing multi-label lung diseases and other medical disease classification.
CRediT authorship contribution statement
Michail Mamalakis: Conceptualization, Data curation, Methodology, Software, Visualization, Investigation, Formal analysis, Writing – original draft, Writing – review & editing, Validation. Andrew J. Swift: Conceptualization, Resources, Data curation, Writing – review & editing, Validation. Bart Vorselaars: Conceptualization, Resources, Writing – review & editing, Validation. Surajit Ray: Conceptualization, Writing – review & editing, Validation. Simonne Weeks: Conceptualization, Writing – review & editing. Weiping Ding: Writing – review & editing, Validation. Richard H. Clayton: Conceptualization, Writing – review & editing. Louise S. Mackenzie: Conceptualization, Writing – review & editing. Abhirup Banerjee: Conceptualization, Visualization, Statistical analysis, Writing – original draft, Writing – review & editing, Validation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The work of Andrew J. Swift was supported by the Wellcome Trust UK fellowship grant 205188/Z/16/Z. The work of Surajit Ray was supported by the EPSRC IAA (EP/R511705/1) Finger prick test for early prediction of SARS-CoV-2; a screening method using changes in full blood count parameters. The work of Weiping Ding was supported in part by the National Natural Science Foundation of China under Grant 61976120, in part by the Natural Science Foundation of Jiangsu Province under Grant BK20191445, in part by the Natural Science Key Foundation of Jiangsu Education Department under Grant 21KJA510004, and sponsored by Qing Lan Project of Jiangsu Province. The authors acknowledge the use of facilities of the Research Software Engineering (RSE) Sheffield, UK. The authors express no conflict of interest.
References
- Banerjee A., Ray S., Vorselaars B., Kitson J., Mamalakis M., Weeks S., Baker M., Mackenzie L.S. Use of machine learning and artificial intelligence to predict SARS-CoV-2 infection from full blood counts in a population. Int. Immunopharmacol. 2020;86 doi: 10.1016/j.intimp.2020.106705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharati S., Podder P., Mondal M.R.H. Hybrid deep learning for detecting lung diseases from x-ray images. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressem K.K., Adams L.C., Erxleben C., Hamm B., Niehues S.M., Vahldiek J.L. Comparing different deep learning architectures for classification of chest radiographs. Sci. Rep. 2020;10:13590. doi: 10.1038/s41598-020-70479-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos A., Pertusa A., Salinas J.M., de la Iglesia-Vayá M. Padchest: a large chest x-ray image dataset with multi-label annotated reports. Med. Image Anal. 2020;66 doi: 10.1016/j.media.2020.101797. [DOI] [PubMed] [Google Scholar]
- Chassagnon G., Vakalopoulou M., Battistella E., Christodoulidis S., Hoang-Thi T.N., Dangeard S., Deutsch E., Andre F., Guillo E., Halm N., ElHajj S., Bompard F., Neveu S., Hani C., Saab I., Campredon A., Koulakian H., Bennani S., Freche G., Barat M., Lombard A., Fournier L., Monnier H., Grand T., Gregory J., Nguyen Y., Khalil A., Mahdjoub E., Brillet P.Y., Tran Ba S., Bousson V., Mekki A., Carlier R.Y., Revel M.P., Paragios N. AI-driven quantification, staging and outcome prediction of COVID-19 pneumonia. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Chen Q., Huang S., Yang M., Yang X., Hu S., Wang Y., Hu X., Zheng B., Zhang K., Wu H., Dong Z., Xu Y., Zhu Y., Chen X., Zhang M., Yu L., Cheng F., Yu H. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10:19196. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., Dittrich S., Yansouni C.P. Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2. Ann. Intern.Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J.P., Morrison, P., Dao, L., Roth, K., Duong, T.Q., Ghassemi, M., 2020. COVID-19 image data collection: prospective predictions are the future. arXiv 2006.11988.
- Das D., Santosh K.C., Pal U. Truncated inception net: COVID-19 outbreak screening using chest x-rays. Phys. Eng. Sci. Med. 2020;43:915–925. doi: 10.1007/s13246-020-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J., Goadrich, M., 2006. The relationship between precision-recall and ROC curves. In: Proceedings of the 23rd International Conference on Machine Learning, pp. 233–240.
- Deng, J., Dong, W., Socher, R., Li, L., Kai, L., Fei-Fei, L., 2009. Imagenet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255.
- Di D., Shi F., Yan F., Xia L., Mo Z., Ding Z., Shan F., Song B., Li S., Wei Y., Shao Y., Han M., Gao Y., Sui H., Gao Y., Shen D. Hypergraph learning for identification of COVID-19 with CT imaging. Med. Image Anal. 2021;68 doi: 10.1016/j.media.2020.101910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K., Su J., Jiang Z., Zeng L.L., Feng Z., Shen H., Rong P., Xu X., Qin J., Yang Y., Wang W., Hu D. Dual-branch combination network (DCN): towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilanie G., Bajwa U.I., Waraich M.M., Asghar M., Kousar R., Kashif A., Aslam R.S., Qasim M.M., Rafique H. Coronavirus (COVID-19) detection from chest radiology images using convolutional neural networks. Biomedical Signal Process. Control. 2021;66 doi: 10.1016/j.bspc.2021.102490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncharov M., Pisov M., Shevtsov A., Shirokikh B., Kurmukov A., Blokhin I., Chernina V., Solovev A., Gombolevskiy V., Morozov S., Belyaev M. CT-based COVID-19 triage: deep multitask learning improves joint identification and severity quantification. Med. Image Anal. 2021 doi: 10.1016/j.media.2021.102054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan H., San José Estépar R., Niessen W.J., Siegel E., Nielsen M. Position paper on COVID-19 imaging and AI: From the clinical needs and technological challenges to initial AI solutions at the lab and national level towards a new era for AI in healthcare. Med. Image Anal. 2020;66 doi: 10.1016/j.media.2020.101800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11:4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K., Gkioxari, G., Dollár, P., Girshick, R.B., 2017. Mask R-CNN. arXiv:1703.06870〈http://arxiv.org/abs/1703.06870〉. [DOI] [PubMed]
- He, K., Zhang, X., Ren, S., Sun, J., 2016. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778.
- He, X., Wang, S., Ying, G., Zhang, J., Chu, X., 2021. Efficient multi-objective evolutionary 3D neural architecture search for COVID-19 detection with chest CT scans. arXiv:2101.10667.
- Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q., 2017. Densely connected convolutional networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2261–2269.
- Iandola, F.N., Moskewicz, M.W., Ashraf, K., Han, S., Dally, W.J., Keutzer, K., 2016. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size. CoRR abs/1602.07360. 〈http://arxiv.org/abs/1602.07360〉.
- Jaeger S., Candemir S., Antani S., Wáng Y.X.J., Lu P.X., Thoma G. Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014;4:475–477. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal A.K., Tiwari P., Kumar S., Gupta D., Khanna A., Rodrigues J.J. Identifying pneumonia in chest x-rays: a deep learning approach. Measurement. 2019;145:511–518. [Google Scholar]
- Kenward M.G., Roger J.H. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53:983–997. [PubMed] [Google Scholar]
- Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F., Dong J., Prasadha M.K., Pei J., Ting M.Y., Zhu J., Li C., Hewett S., Dong J., Ziyar I., Shi A., Zhang R., Zheng L., Hou R., Shi W., Fu X., Duan Y., Huu V.A., Wen C., Zhang E.D., Zhang C.L., Li O., Wang X., Singer M.A., Sun X., Xu J., Tafreshi A., Lewis M.A., Xia H., Zhang K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- Koivunen A.C., Kostinski A.B. The feasibility of data whitening to improve performance of weather radar. J. Appl. Meteorol. 1999;38:741–749. [Google Scholar]
- Lalmuanawma S., Hussain J., Chhakchhuak L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: a review. Chaos, Solitons Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Fang Y., Li W., Pan C., et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur. Radiol. 2020;30:4407–4416. doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.D., Arun N.T., Gidwani M., Chang K., et al. Automated assessment and tracking of COVID-19 pulmonary disease severity on chest radiographs using convolutional siamese neural networks. Radiol.: Artif. Intell. 2020;2 doi: 10.1148/ryai.2020200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaee S., Kafieh R., Sonka M., Yazdani S., JamalipourSoufi G. Deep-COVID: predicting COVID-19 from chest x-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenbach, J., Wiegreffe, S., Duke, J., Sun, J., Eisenstein, J., 2018. Explainable prediction of medical codes from clinical text. In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), pp. 1101–1111.
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Deep learning, reusable and problem-based architectures for detection of consolidation on chest x-ray images. Comput. Methods Progr. Biomed. 2020;185 doi: 10.1016/j.cmpb.2019.105162. [DOI] [PubMed] [Google Scholar]
- Pereira R.M., Bertolini D., Teixeira L.O., Silla C.N., Costa Y.M. COVID-19 identification in chest x-ray images on flat and hierarchical classification scenarios. Comput. Methods Progr. Biomed. 2020;194 doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona P., Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 1990;12:629–639. [Google Scholar]
- Qin Z.Z., Sander M.S., Rai B., Titahong C.N., Sudrungrot S., Laah S.N., Adhikari L.M., Carter E.J., Puri L., Codlin A.J., Creswell J. Using artificial intelligence to read chest radiographs for tuberculosis detection: a multi-site evaluation of the diagnostic accuracy of three deep learning systems. Sci. Rep. 2019;9:15000. doi: 10.1038/s41598-019-51503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reittner P., Ward S., Heyneman L., Johkoh T., Müller N.L. Pneumonia: high-resolution CT findings in 114 patients. Eur. Radiol. 2003;13:515–521. doi: 10.1007/s00330-002-1490-3. [DOI] [PubMed] [Google Scholar]
- Sarker, L., Islam, M.M., Hannan, T., Ahmed, Z., 2020. COVID-DenseNet: a deep learning architecture to detect COVID-19 from chest radiology images. Preprints, 2020050151.
- Simonyan, K., Zisserman, A., 2015. Very deep convolutional networks for large-scale image recognition. CoRR abs/1409.1556.
- Soltan, A.A.S., Kouchaki, S., Zhu, T., Kiyasseh, D., Taylor, T., Hussain, Z.B., Peto, T., Brent, A.J., Eyre, D.W., Clifton, D., 2020. Artificial intelligence driven assessment of routinely collected healthcare data is an effective screening test for COVID-19 in patients presenting to hospital. medRxiv.
- Song J.W., Lam S.M., Fan X., Cao W.J., Wang S.Y., Tian H., et al. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 2020;32:188–202.e5. doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., Zheng, S., Li, L., Zhang, X., et al., 2020b. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv. [DOI] [PMC free article] [PubMed]
- Stacey D., Légaré F., Lewis K., Barry M.J., Bennett C.L., Eden K.B., et al. Decision aids for people facing health treatment or screening decisions. Cochrane Database Syst. Rev. 2017;4 doi: 10.1002/14651858.CD001431.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Araki K., Matsubayashi S., Kobayashi K., Morino E., Takasaki J., Iikura M., Izumi S., Takeda Y., Sugiyama H. A case of recurrent hemoptysis caused by pulmonary actinomycosis diagnosed using transbronchial lung biopsy after bronchial artery embolism and a brief review of the literature. Ann. Transla. Med. 2019:7. doi: 10.21037/atm.2019.02.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory diagnosis of COVID-19: Current issues and challenges. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey J.W. Comparing individual means in the analysis of variance. Biometrics. 1949;5:99–114. [PubMed] [Google Scholar]
- Varela-Santos S., Melin P. A new approach for classifying coronavirus COVID-19 based on its manifestation on chest x-rays using texture features and neural networks. Inf. Sci. 2021;545:403–414. doi: 10.1016/j.ins.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshni, D., Thakral, K., Agarwal, L., Nijhawan, R., Mittal, A., 2019. Pneumonia detection using CNN based feature extraction. In: IEEE International Conference on Electrical, Computer and Communication Technologies, pp. 1–7.
- Vonesch C., Unser M. A fast thresholded landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE Trans. Image Process. 2008;17:539–549. doi: 10.1109/TIP.2008.917103. [DOI] [PubMed] [Google Scholar]
- Wang, S., Kang, B., Ma, J., Zeng, X., et al., 2020. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19). medRxiv. [DOI] [PMC free article] [PubMed]
- Williams G.J., Macaskill P., Kerr M., Fitzgerald D.A., Isaacs D., Codarini M., McCaskill M., Prelog K., Craig J.C. Variability and accuracy in interpretation of consolidation on chest radiography for diagnosing pneumonia in children under 5 years of age. Pediatr. Pulmonol. 2013;48:1195–1200. doi: 10.1002/ppul.22806. [DOI] [PubMed] [Google Scholar]
- World Health Organization, 2011. Maternal, newborn, child and adolescent health. 〈https://apps.who.int/iris/handle/10665/44873〉.
- World Health Organization, 2020. Global tuberculosis report. 〈https://www.who.int/publications/i/item/9789240013131〉.
- Wu X., Chen C., Zhong M., Wang J., Shi J. COVID-AL: The diagnosis of COVID-19 with deep active learning. Med. Image Anal. 2021;68 doi: 10.1016/j.media.2020.101913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Cao C., Liu J., Duan Y., Cao H., Wang J., Tao X., Chen Z., Wu M., Zhang J., Sun H., Jin Y., Yang X., Huang R., Xiang F., Song Y., You M., Zhang W., Jiang L., Zhang Z., Kong S., Tian Y., Zhang L., Ni D., Xie M. Modality alignment contrastive learning for severity assessment of COVID-19 from lung ultrasound and clinical information. Med. Image Anal. 2021;69 doi: 10.1016/j.media.2021.101975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Xu Z., Li W., Myronenko A., Roth H.R., Harmon S., Xu S., Turkbey B., Turkbey E., Wang X., Zhu W., Carrafiello G., Patella F., Cariati M., Obinata H., Mori H., Tamura K., An P., Wood B.J., Xu D. Federated semi-supervised learning for COVID region segmentation in chest CT using multi-national data from China, Italy, Japan. Med. Image Anal. 2021;70 doi: 10.1016/j.media.2021.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.H., Geng H., Chiu T.L., Yu S.K., et al. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest x-ray imaging. Front. Med. 2020;7:427. doi: 10.3389/fmed.2020.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Zhang, Y., He, X., Xie, P., 2020. COVID-CT-Dataset: a CT scan dataset about COVID-19. arXiv preprint arXiv:2003.13865.
- Zhu X., Song B., Shi F., Chen Y., Hu R., Gan J., Zhang W., Li M., Wang L., Gao Y., Shan F., Shen D. Joint prediction and time estimation of COVID-19 developing severe symptoms using chest CT scan. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101824. [DOI] [PMC free article] [PubMed] [Google Scholar]