Abstract
Regulation of NF-κB occurs through phosphorylation-dependent ubiquitination of IκBα, which is degraded by the 26S proteasome. Recent studies have shown that ubiquitination of IκBα is carried out by a ubiquitin-ligase enzyme complex called SCFβTrCP. Here we show that Nedd8 modification of the Cul-1 component of SCFβTrCP is important for function of SCFβTrCP in ubiquitination of IκBα. In cells, Nedd8-conjugated Cul-1 was complexed with two substrates of SCFβTrCP, phosphorylated IκBα and β-catenin, indicating that Nedd8–Cul-1 conjugates are part of SCFβTrCP in vivo. Although only a minute fraction of total cellular Cul-1 is modified by Nedd8, the Cul-1 associated with ectopically expressed βTrCP was highly enriched for the Nedd8-conjugated form. Moreover, optimal ubiquitination of IκBα required Nedd8 and the Nedd8-conjugating enzyme, Ubc12. The site of Nedd8 ligation to Cul-1 is essential, as SCFβTrCP containing a K720R mutant of Cul-1 only weakly supported IκBα ubiquitination compared to SCFβTrCP containing WT Cul-1, suggesting that the Nedd8 ligation of Cul-1 affects the ubiquitination activity of SCFβTrCP. These observations provide a functional link between the highly related ubiquitin and Nedd8 pathways of protein modification and show how they operate together to selectively target the signal-dependent degradation of IκBα.
NF-κB is a transcription factor required for inducible expression of a number of proinflammatory mediators including cytokines, chemokines, and leukocyte adhesion molecules (6). In addition, NF-κB regulates the expression of survival genes which prevent cell death in response to tumor necrosis factor alpha (TNF-α) (7, 37, 59, 62). NF-κB is a member of the Rel family of proteins and is typically a heterodimer composed of p50 and p65 subunits. In quiescent cells, NF-κB is retained in the cytosol bound to IκB, a family of inhibitory proteins which mask the nuclear localization and DNA binding sequences on NF-κB (5, 22). Stimulation of these cells with various cytokines, lipopolysaccharide, viruses, antigens, or oxidants triggers signaling events that ultimately lead to the phosphorylation and degradation of IκB, allowing NF-κB to translocate into the nucleus and activate target genes (3, 21, 38, 54).
Phosphorylation of Ser32 and Ser36 has been shown to target IκB for ubiquitination and subsequent proteolysis by the ubiquitin-proteasome pathway (UPP) of protein degradation (2, 8, 45, 49). The UPP is the principal pathway for intracellular protein turnover, including regulatory proteins (9). Protein substrates that enter the UPP are first marked by the covalent ligation of polyubiquitin chains mediated by a cascade of enzymes called E1 (ubiquitin activation enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase) (9). In a reaction requiring ATP, ubiquitin is activated by E1 and charged onto an E2 through a thioester formed between the active-site cysteine residue in the E2 and the C-terminal glycine of ubiquitin. The E3 then directs the transfer of ubiquitin from the E2 onto lysine residues within specific substrate proteins, ultimately resulting in the formation of a ubiquitin-protein conjugate. Polyubiquitinated proteins are then recognized and degraded by the 26S proteasome complex to yield small peptides and monomeric ubiquitin.
Recently, the receptor component of the IκB E3 was identified as a member of the βTrCP (beta-transducin repeat-containing protein) family of proteins called E3RSIκB (39, 53, 63, 65) or HOS (11). βTrCP is a member of a much larger family of F-box domain containing proteins which form SCF complexes. The core components of SCF complexes include Skp-1, which interacts with the F-box domain, and Cul-1, which is linked to the F-box protein via binding to Skp-1 (4, 10, 35, 46, 47, 51). At least two additional SCF components have been described: (i) Rbx1, which is thought to stabilize the interaction between Cul-1 and the E2s, Cdc34, and Ubc5 (25, 26, 43, 50, 52, 56), and (ii) Sgt1, a protein which interacts with Skp-1 (27). SCF complexes were initially described in yeast to function as E3 ligases for a variety of phosphorylated proteins, including the cell cycle regulator, Sic1 (10, 51). In addition to an F-box domain, βTrCP also contains a WD40 repeat domain that specifically recognizes IκBα only when IκBα is phosphorylated on Ser32 and Ser36. Similarly, at least two other proteins are recognized by βTrCP in a phosphorylation-dependent manner, β-catenin (16, 31, 36, 63) and human immunodeficiency virus type 1 Vpu (40). βTrCP in which the F-box is deleted (ΔF-βTrCP) retains its specificity for phosphorylated IκBα but fails to interact with Skp-1 and no longer supports the ubiquitination reaction. Thus, the interaction of the F-box protein with other SCF components is essential for function. The core components of SCFβTrCP alone, however, are not sufficient to support the ubiquitination of phosphorylated IκBα (53, 63, 65). Additional components, supplied by the addition of crude cellular extracts (63) or recombinant proteins (including UbcH5 [43, 53, 65], Cdc34 [56], and Rbx1 [56]), are required for activity, suggesting that essential proteins and/or modifications to existing proteins are needed to support ubiquitination of IκBα by SCFβTrCP. To date, modifications of the cellular components in SCFβTrCP have not been characterized.
In an effort to understand the requirements for SCFβTrCP-mediated ubiquitination of IκBα, we examined SCF core components that associate with βTrCP as well as with IκBα. Remarkably, we observed that endogenous phosphorylated IκBα associated exclusively with a form of Cul-1 that is singly modified by the ubiquitin-like protein Nedd8. Along this line, we found that optimal ubiquitination of IκBα in vitro required the presence of Nedd8 and the Nedd8-conjugating enzyme, Ubc12, as well as two ubiquitin-conjugating enzymes, UbcH5A and Cdc34. Moreover, a Cul-1 point mutant which retains the ability to associate with other SCF components, but lacks the site of the Nedd8 modification, showed a greatly reduced ability to ubiquitinate IκBα in vitro. It is well established that a small percentage of cellular Cul-1 and related cullin proteins form conjugates containing single molecules of Nedd8 in yeast and mammalian cells (30, 44, 60), and the Nedd8 homologue, Rub1, has been genetically linked to SCF components in yeast (30) and plants (14). However, a functional role for Nedd8 in any ubiquitination reaction or cellular process has not been demonstrated. Here we show that the Nedd8 modification of Cul-1 is necessary for the function of SCFβTrCP, linking the ubiquitin and Nedd8 pathways in the regulation of targeted protein degradation.
MATERIALS AND METHODS
Plasmids and antibodies.
βTrCP, Cul-1, Cul-2, Skp-1, and Rbx1 were isolated from the SuperScript human leukocyte cDNA library (GibcoBRL) by PCR using AmpliTaq (Perkin-Elmer) or Pwo (Boehringer Mannheim) DNA polymerase and oligonucleotides purchased from Research Genetics. βTrCP and Cul-1 were subcloned into NotI-BamHI sites, Cul-2 was cloned into EcoRI-BamHI sites, and Skp-1 was cloned into NotI-XbaI sites of pFlag-CMV2 vector. The βTrCP clone corresponds to GenBank accession no. Y14153 (40). The Cul-1 clone encoded the same 24 amino acid insertion as previously reported (42). Cul-1 K720R mutant was generated by PCR using high-fidelity DNA polymerase Pfu (QuickChange site-directed mutagenesis kit; Stratagene). To generate the ΔF-βTrCP, the βTrCP was subjected to site-directed mutagenesis with the primer 5′GATTTCATAACTGCTAAGCTTGCTCGGGGATTGG3′ and its complement, inserting a HindIII site just upstream of the F box. The F-box coding region corresponding to amino acid residues 148 through 190 was removed by restriction digestion with HindIII. Mutagenesis and subsequent removal of the F box were both confirmed by sequence analysis. βTrCP (wild type [WT] and ΔF box) were subcloned into pcDNA (Invitrogen) with a Myc epitope tag. Rbx1 was cloned into the NdeI-Xba sites of pcDNA3 with addition of a hemagglutinin (HA) epitope tag.
GenBank sequences for the E2 cDNAs and Nedd8 were isolated by PCR and subcloned into the following plasmids: UbcH5A, pGEX-4T-2; Ubc12, pT7-7; Cdc34, pT7-7; Nedd8, pT7-7. Active-site cysteines were mutated to serines by site-directed mutagenesis using a QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Antisera to Cul-1 (Rb-042), Cul-2 (Rb-046), and Skp-1 (Rb-040) were purchased from NeoMarker, LabVision Corporation. Anti-Nedd8 was purchased from Alexis Biochemicals. Anti-IκBα (sc-371), anti-SUMO1 (sc-6375), anti-Myc (sc-789), and anti-HA (sc-7392) were purchased from Santa Cruz. Anti-β-catenin (C19220) was purchased from Transduction Laboratories. Mouse anti-FLAG M5 antibody and M2 resin were purchased from Sigma. Horseradish peroxidase-conjugated anti-rabbit and anti-mouse were purchased from Amersham. Anti-Cul-1658–670 was produced in rabbits against the peptide VDEVELKPDTLIKC corresponding to residues 658 to 670 of human Cul-1 and was affinity purified using the peptide coupled to Sulfolink resin (Pierce) according the manufacturer's instructions.
Preparation of proteins.
Plasmids encoding the E2 proteins Ubc12 and Cdc34 were expressed in Escherichia coli BL-21(DE3) (Novagen, Milwaukee, Wis.) in Luria-Bertani medium containing carbenicillin (50 μg/ml; Sigma), and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (Boehringer Mannheim) for 3 h at 37°C. Bacterial cell pellets were resuspended in 50 mM HEPES–0.1% Triton X-100–1 μg of leupeptin per ml–50 μg of lysozyme per ml, lysed by sonication, and clarified by centrifugation at 10,000 × g for 1 h. Ubc12 lysates were subjected to anion-exchange chromatography (Mono Q; Pharmacia), and the flowthrough was separated by size exchange chromatography, resulting in proteins of >95% purity. For Cdc34, protein was purified over Ni-nitrilotriacetic acid agarose (Qiagen) followed by size exclusion chromatography. UbcH5A (pGEX-4T-2) was expressed in E. coli BL-21(DE3)pLysS (Novagen) in Luria-Bertani medium containing carbenicillin (50 μg/ml; Sigma) and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (Boehringer Mannheim) for 3 h at 37°C. Bacterial cell pellets were resuspended in 50 mM HEPES (pH 7.6)–0.5 mM dithiothreitol–1 μg of leupeptin per ml, lysed by sonication, and clarified by centrifugation at 10,000 × g for 1 h. The glutathione S-transferase (GST)-tagged protein was purified over GST-Sepharose 4B (Pharmacia) according to manufacturer's instructions. The fusion protein was subjected to thrombin cleavage using biotinylated thrombin (Novagen), followed by separation over HiTrap heparin-Sepharose (Pierce), using a NaCl gradient.
In vitro ubiquitin and Nedd8 conjugation reactions.
WT or Ser32/36Ala His6-IκBα in pET15b was metabolically labeled with [35S]methionine in B834(DE3) cells (Novagen) (32). Recombinant p652 RHR (residues 1 to 323) (22) was provided by Marc D. Jacobs and was produced in E. coli. The purified radiolabeled His6-IκBα and purified recombinant p652 were combined in equal molar ratios to form trimeric IκBα/p652. The IκBα/p652 was phosphorylated with purified recombinant IKK2 produced in baculovirus (33). For conjugation reactions, the phosphorylated substrate was incubated with fraction I (FI; 20 μg) and fraction II (FII; 40 μg) (18), an ATP regeneration system (8), 60 μM ubiquitin (Sigma), 1 μM microcystin LR (Calbiochem), 0.5 μM ubiquitin aldehyde (19, 41), and 2.5 μM MG273 (15) in a final volume of 20 μl. Unless otherwise indicated, reactions were incubated at 37°C for 90 min, terminated with the addition of sodium dodecyl sulfate (SDS) sample buffer, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on 9% gels. Conjugates were detected by phosphorimager analysis (ImageQuant software, Storm 840).
Nedd8 conjugation reactions of Cul-1 were performed using FLAG epitope-tagged WT and K270R Cul-1 immunoprecipitated from 293 cells as substrates. Substrate proteins were incubated with 1.6 μg of Nedd8, 20 pmol of Ubc12, an ATP regeneration system, and 20 μg of FII (as a source of Nedd8-activating enzyme, APP-BP1/Uba3). Reactions were adjusted to 20 μl with 50 mM Tris-HCl (pH 7.5) and incubated at 30°C for 30 min. Reactions were stopped by the addition of SDS sample buffer, resolved by SDS-PAGE on 9% gels, and transferred to nitrocellulose for Western blotting with anti-FLAG antibody as described.
The heterodimeric Nedd8-activating enzyme, APP-BP1/Uba3, was affinity purified from HeLa S100 by Nedd8 affinity chromatography using activated CH Sepharose 4B (Pharmacia).
Cell culture and transfections.
For studies examining in vivo protein interactions, human embryonic kidney 293 cells were seeded in 100-mm-diameter plates in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Transfection of 293 cells was performed with 4 μg of the indicated DNA and Lipofectamine PLUS as instructed by the manufacturer (Gibco-BRL). Where indicated, 48 h after transfection, cells were treated with 5 μM MG273 (15) for 1 h and then stimulated with 10 ng of recombinant human TNF-α (R&D) per ml for 10 min.
For ubiquitination assays using SCFβTrCP, 293 cells were seeded as above and cotransfected with 4 μg each of the indicated DNAs, using calcium phosphate transfection as instructed by the manufacturer (InVitrogen). Cells were harvested 48 h after transfection as described below.
Immunoprecipitations and immunoblotting.
Cells were rinsed in cold phosphate-buffered saline and lysed in 400 μl of cold lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EGTA, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 2.5 μM MG273, 1 μM microcystin). Lysates cleared by centrifugation at 10,000 × g for 5 min at 4°C were incubated with anti-FLAG M2 resin or 1 μg of primary antibody plus 25 μl Tris-acryl protein A (Pierce) for 4 h with rotation at 4°C. Resins were washed six times with lysis buffer and resuspended in Laemmli SDS sample buffer. Proteins bound to resin were resolved by SDS-PAGE on a 9 or 15% gel and analyzed by Western blotting, using indicated primary antibodies and either horseradish peroxidase-conjugated anti-mouse or anti-rabbit (Amersham) followed by detection by enhanced chemiluminescence (Amersham) according to the manufacturer's instructions.
RESULTS
The Nedd8-modified form of the SCFβTrCP component Cul-1 preferentially associates with phosphorylated IκBα and β-catenin.
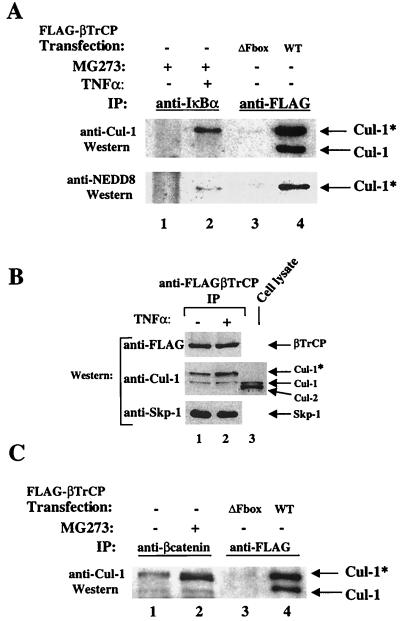
SCFβTrCP was recently identified as a ubiquitin E3 ligase that recognizes phosphorylated IκBα (11, 17, 28, 53, 55, 63, 65). We and others have noticed that multiple cullin protein species cofractionate with IκBα ubiquitin-conjugating activity in cellular extracts (data not shown) (63). Both βTrCP and phosphorylation-specific IκBα ubiquitin ligase activity can be detected in immunoprecipitates of IκBα/NF-κB complexes from cells treated with TNF-α (65). Taking advantage of the high affinity of the phosphorylated substrate for the active enzyme, we investigated the in vivo association of endogenous Cul-1 with endogenous IκBα. The dependence of this interaction on TNF-α induction was examined in 293 cells pretreated with the proteasome inhibitor MG273 (to prevent degradation of phosphorylated IκBα [15]). Lysates from these cells were immunoprecipitated with an antibody to IκBα, and the immunoprecipitates were subjected to Western blotting with anti-Cul-1 antiserum generated against a C-terminal Cul-1 peptide. As expected, IκBα associated with Cul-1 in a TNF-α-dependent manner (Fig. 1A, lanes 1 and 2, upper panel), and only a single anti-Cul-1-reactive species was detected (lane 2). For comparison, anti-FLAG immunoprecipitates from 293 cells transfected with either FLAG-tagged WT or F-box deletion mutant βTrCP were analyzed alongside the IκBα immunoprecipitates (Fig. 1A, lanes 3 and 4, upper panel) by immunoblotting with the same anti-Cul-1 antibody. In contrast to the single anti-Cul-1 reactive band associated with phosphorylated IκBα, two Cul-1 immunoreactive species were associated with the ectopically expressed WT βTrCP (lane 4). The faster-migrating species was consistent with the majority of cellular Cul-1 (Fig. 1B, lane 3), and we refer to the species exhibiting reduced mobility by SDS-PAGE as Cul-1*. The identity of both immunoreactive species was confirmed using a second, independently derived antibody directed against a peptide sequence in Cul-1 that is poorly conserved in other cullin proteins (residues 658 to 670 of Cul-1) and is specific for Cul-1 (data not shown). Cul-1 is the only cullin family member that has been shown to interact with Skp-1 and thus with F-box proteins (42). Concordant with these findings, ΔF-βTrCP, which does not bind to Skp-1 (40), also failed to associate with either Cul-1 or Cul-1* (Fig. 1A, lane 3). Strikingly, only Cul-1* was associated with endogenous IκBα in TNF-α-treated cells, indicating that Cul-1* is the prevalent species of Cul-1 present in SCFβTrCP.
FIG. 1.
Cul-1 associated with IκBα, β-catenin, and SCFβTrCP is modified by Nedd8. (A) Association of endogenous Nedd8 conjugated Cul-1 with IκBα or ectopically expressed βTrCP. Lanes 1 and 2, 293 cells were treated with 5 μM MG273 for 1 h and with TNF-α (10 ng/ml) for 10 min as indicated. Lysates were immunoprecipitated (IP) with anti-IκBα. Lanes 3 and 4, 293 cells were transfected with FLAG-tagged ΔF-βTrCP or WT-βTrCP, and lysates were immunoprecipitated with anti-FLAG resin. In all lanes, immune complexes were resolved by SDS-PAGE on 9% gels and immunoblotted with anti-Cul-1 and anti-Nedd8. (B) Ectopically expressed βTrCP associates with endogenous Cul-1 and Cul-1*. Lanes 1 and 2, 293 cells were transfected with FLAG-tagged WT-βTrCP and treated with TNF-α as indicated, and lysates were immunoprecipitated with anti-FLAG. Immune complexes were resolved by SDS-PAGE and immunoblotted with anti-FLAG, anti-Cul1, or anti-Skp-1. Lane 3, cell lysate (30 μg) from nontransfected 293 cells analyzed as above. (C) Association of endogenous Cul-1 with β-catenin. Lanes 1 and 2, 293 cells were treated with or without 5 μM MG273 for 2 h as indicated. Lysates were immunoprecipitated with anti-β-catenin. Lanes 3 and 4, 293 cells were transfected with FLAG-tagged ΔF-βTrCP or WT-βTrCP, and lysates were immunoprecipitated with anti-FLAG resin. In all lanes, immune complexes were resolved by SDS-PAGE on 9% gels and immunoblotted with anti-Cul-1. Cul-1* = Nedd8-ligated Cul-1.
Several cullin proteins, including Cul-1 (mammalian) and Cdc53 (yeast), are known to be modified by the ubiquitin-like protein Nedd8 (mammalian) or Rub1 (yeast) (26, 30, 34, 44, 60). The mobility of Cul-1* on SDS-PAGE is consistent with Cul-1 that is conjugated to a single molecule of Nedd8. To test whether Cul-1* was in fact a Nedd8-conjugated form of Cul-1, we examined the Cul-1 associated with IκBα and βTrCP with a Nedd8-specific antibody. Cul-1* was immunoreactive with the Nedd8 antibody (Fig. 1A, lower panel), whereas the faster-migrating Cul-1 was not. Thus, Cul-1* is a Nedd8-conjugated form of Cul-1.
We next investigated whether the Nedd8 modification occurred in response to TNF-α by examining endogenous Cul-1 associated with FLAG-βTrCP in control and TNF-α-treated cells. Lysates from 293 cells transfected with FLAG-βTrCP were immunoprecipitated with anti-FLAG resin, and Western blot analyses were performed with antibodies to the SCF components Skp-1 and Cul-1. Endogenous Skp-1, Cul-1, and Cul-1* were associated with βTrCP (Fig. 1B, lanes 1 and 2). The association of Cul-1 and Cul-1* with βTrCP and the relative abundance of the two Cul-1 species was unaffected by TNF-α induction (Fig. 1B, lanes 1 and 2), unlike the association of Cul-1* with phosphorylated IκBα (Fig. 1A, lane 2). We observed that Cul-1* was a minor component in cell extracts relative to the majority of total Cul-1 and was not readily detected in crude cell lysates (Fig. 1B, lane 3). We also noted that the crude cell lysates contained an additional Cul-1-immunoreactive species which was most likely Cul-2 based on its SDS-PAGE mobility and its reactivity with anti-Cul-2 antibodies (data not shown).
We next addressed whether another known substrate of SCFβTrCP, β-catenin, also associated with Nedd8-modified Cul-1 in vivo. To stabilize β-catenin and allow detection of associated proteins, 293 cells were treated with MG273 prior to lysis (data not shown) (2). Lysates from these cells were immunoprecipitated with anti-β-catenin, and the immune complexes were subjected to immunoblotting with anti-Cul-1 (Fig. 1C, lanes 1 and 2). As in Fig. 1A, FLAG-tagged WT and ΔF-box βTrCP immunoprecipitates were included on the same gel for comparison (Fig. 1C, lanes 3 and 4). Similar to results with phosphorylated IκBα, stabilized β-catenin associated strictly with Cul-1*.
Two lines of evidence suggest that Nedd8-Cul-1 is the preferred form of Cul-1 present in cellular SCFβTrCP even though the Nedd8-modified Cul-1 represents only a minor portion of the total cellular Cul-1. First, the relative proportion of Nedd8-Cul-1 (Cul-1*) to unmodified Cul-1 was greatly enriched in association with ectopically expressed βTrCP compared to the total cellular pool of Cul-1. Second, Cul-1* was the sole form of Cul-1 associated with two substrates of SCFβTrCP, IκBα and β-catenin, when examined at physiological levels in cells.
The Nedd8 conjugation pathway is required for IκBα ubiquitination.
The association of Nedd8-conjugated Cul-1 with phosphorylated IκBα in vivo prompted us to ask whether the Nedd8 conjugation pathway is involved in the IκBα ubiquitination reaction. We performed ubiquitination assays using cell extract or recombinant proteins as sources of IκBα-conjugating enzymes. HeLa cells contain a phosphorylation-specific IκBα ubiquitin ligase activity which can be detected in cytosolic extract (see below) (2, 63, 64). When this extract is passed over a Q-Sepharose column, two fractions are obtained: FI, consisting of proteins which flow through the column, and FII, consisting of proteins which are retained on the column and eluted with 300 mM NaCl (18). FII contains the heterodimeric Nedd8-activating enzyme, APP-BP1/Uba3, as well as several SCF components including Skp-1, Rbx1, Cul-1 (data not shown), and Cdc34, the only E2 shown to interact directly with SCF complexes by virtue of its binding to Cul-1 (46). It is well established that in the presence of ubiquitin and ATP, FII alone is insufficient to form ubiquitin conjugates on phosphorylated IκB but requires the addition of FI proteins (see Fig. 2A, lanes 1 to 3) (2, 64). FI contains several ubiquitin-E2s, including members of the Ubc4/5 family (UbcH5A, -B, and -C) (23, 48), the Nedd8-E2 Ubc12 (13, 44), and both ubiquitin and Nedd8. Thus, supplementing FII with the required FI components should faithfully reproduce the conjugation reaction.
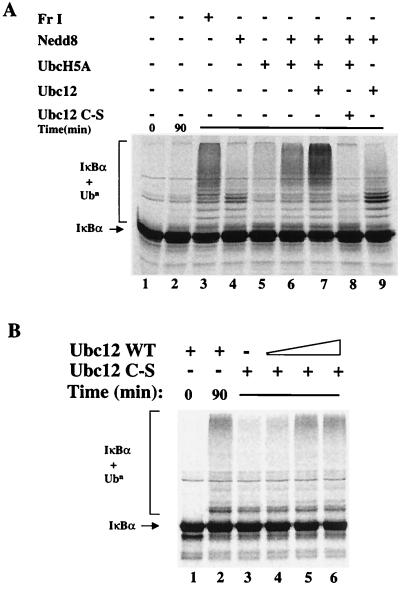
FIG. 2.
Ubiquitination of IκBα requires the Nedd8 pathway. (A) Reconstitution of IκBα ubiquitination activity in vitro. Ubiquitination assays were performed with HeLa FII (40 μg), ubiquitin (60 μM), an ATP-regenerating system, and recombinant 35S-labeled IκBα/p652 phosphorylated on S32 and S36 as described in the text. Into these reactions, HeLa FI (15 μg), UbcH5A (150 nM), Nedd8 (250 nM), and WT Ubc12 (150 nM) or Ubc12C111S (5 μM) was added as indicated. Following incubation at 37°C for 0 or 90 min, reactions were stopped by the addition of SDS sample buffer and resolved on an SDS–9% gel, and the reaction products were detected with a phosphorimager. (B) Ubc12C111S acts as a dominant negative inhibitor of IκBα ubiquitination. Reactions were performed as for panel A, using FII (40 μg), ubiquitin (60 μM), UbcH5A (150 nM), Nedd8 (1 μM), an ATP-regenerating system, and recombinant 35S-labeled IκBα/p652 phosphorylated on S32 and S36 as described in the text. Into these reactions, WT Ubc12 was added at 50 nM (lanes 1, 2 and 4), 500 nM (lane 5), or 1 μM (lane 6). Ubc12C111S (Ubc12 C-5; 5 μM) was added in lanes 3 to 6. Following incubation at 37°C for 0 or 90 min, reactions were stopped by the addition of SDS sample buffer and resolved on an SDS–9% gel, and the reaction products were detected with a phosphorimager.
We performed ubiquitin conjugation reactions with purified recombinant phosphorylated 35S-labeled IκBα/p652 as the substrate and with FII supplemented with FI, or with purified, recombinant FI components, as the source of conjugating enzymes. The combination of FI and FII supported IκBα ubiquitination resulting in the formation of high-molecular-weight conjugates (Fig. 2A, lane 3). When FI was replaced by the addition of either bacterially produced Nedd8 or UbcH5A, only a slight difference in conjugate formation was observed compared to FII alone (Fig. 2A, lanes 2, 4, and 5). The effect of adding Nedd8 together with UbcH5A and FII resulted in a modest enhancement of conjugate formation (lane 6). Strikingly, the presence of Nedd8, UbcH5A, and Ubc12 resulted in the formation of high-molecular-weight conjugates to a greater extent than even observed for FI and FII (compare lanes 3 and 7). Clearly, the presence of both the Nedd8-E2 and ubiquitin E2 activities was required for full conjugation activity in this system since the omission of UbcH5A from reactions containing Nedd8 and Ubc12 resulted in the formation of only low-molecular-weight conjugates (lane 9).
To further define the requirement for the Nedd8-activating pathway in IκBα ubiquitination, recombinant Ubc12 with an active-site Cys-to-Ser mutation was expressed and purified from bacteria. In the presence of Nedd8 and Nedd8-activating enzyme, Ubc12C112S can form stable oxygen esters with Nedd8 but cannot transfer Nedd8 to a target protein (data not shown). Similar ubiquitin-E2 mutants can act as competitive inhibitors in conjugation assays and therefore are useful reagents for discerning the role of specific E2s within crude reaction systems (57). Substitution of WT Ubc12 with the active-site mutant Ubc12C111S in ubiquitination reactions failed to support IκBα conjugate formation and essentially blocked all conjugate formation in the presence of Nedd8 and UbcH5A (Fig. 2A, compare lanes 6 and 8). The Ubc12C112S mutant also inhibited ubiquitination in the presence of WT Ubc12 (Fig. 2B, compare lanes 2 and 4). This effect was reversible when increasing amounts of WT Ubc12 were added back to the reaction (lanes 4 to 6), suggesting that Ubc12C112S exerts its effects by inhibiting Ubc12 through a competitive mechanism. Taken together, these results suggest that the factors in FI responsible for supporting IκBα-conjugating activity by FII include Nedd8 and at least two E2 activities operating in both the Nedd8 (Ubc12) and ubiquitin (UbcH5A) conjugation pathways.
Cul-1 Lys720Arg fails to form conjugates with Nedd8.
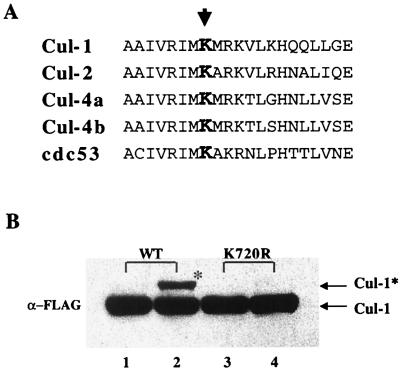
Given that only a minor fraction of Cul-1 is modified by Nedd8 in vivo, the results above suggest that Nedd8 modification may play a significant role in the regulation and function of SCFβTrCP. To examine this possibility further, we prepared a mutant of Cul-1 that does not form conjugates with Nedd8. Recently, a single lysine residue present in the C terminus of Cul-2 was shown to be required for Nedd8–Cul-2 conjugate formation (60). This lysine lies within a region of Cul-2 that is highly conserved in all cullin proteins and is analogous to residue 720 in Cul-1 (Fig. 3A). We thus prepared a FLAG epitope-tagged construct of Cul-1 with the lysine at position 720 changed to arginine, a conservative residue that cannot accept the Nedd8 modification. To establish that K720R Cul-1 was not a substrate for Nedd8 conjugation, FLAG-WT and FLAG-K720R Cul-1 were expressed in 293 cells and immunoprecipitated. The immunoprecipitates were then incubated with recombinant Nedd8, recombinant Nedd8-conjugating enzyme Ubc12, FII (see Materials and Methods) as a source of Nedd8-activating enzyme (APP-BP1/UBA3) (13, 44), and an ATP-regenerating system. The reaction products were resolved by SDS-PAGE and immunoblotted with the anti-FLAG antibody. Nedd8 conjugates of WT Cul-1 were readily detected due to a mobility shift in SDS-PAGE (Fig. 3B, lane 2), which was notably similar to the pattern observed for cellular Cul-1* versus Cul-1 (Fig. 1). Conversely, the K720R mutant Cul-1 was not subject to Nedd8 modification under these conditions, confirming that K720 is a critical residue for the formation of Nedd8–Cul-1.
FIG. 3.
Identification of Nedd8 conjugation site in human Cul-1. (A) Alignment of Cul-1 amino acid sequence with sequences of other cullins. The Nedd8 conjugation site identified in Cul-2 is indicated by an arrowhead. (B) K720R Cul-1 is defective in forming Nedd8 conjugates. FLAG-tagged WT Cul-1 (lanes 1 and 2) or K720R Cul-1 (lanes 3 and 4) was expressed in 293 cells and immunoprecipitated with FLAG resin. The immunoprecipitates were then incubated with recombinant Nedd8 (250 nM), Ubc12 (150 nM), and FII (20 μg) for either 0 (lanes 1 and 3) or 30 (lanes 2 and 4) min at 30°C. The reaction products were resolved by SDS-PAGE (7.5% gel) under reducing conditions and immunoblotted with the anti-FLAG antibody. Cul-1* = Nedd8–Cul-1.
SCFβTrCP containing K720R Cul-1 has reduced IκBα ubiquitination activity.
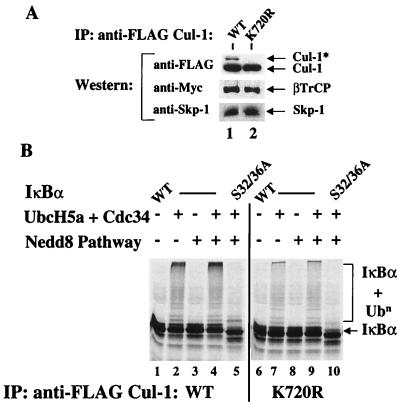
In addition to the results presented here, a growing body evidence from other systems, including yeast (20, 30) and plants (14), suggests that Nedd8–Cul-1 is important for the regulation of SCF function. To address a potential role of Nedd8–Cul-1 in SCFβTrCP assembly, FLAG-WT and FLAG-K720R Cul-1 were tested for the ability to associate with SCF components. WT or K720R Cul-1 was coexpressed in 293 cells with WT βTrCP fused to a Myc epitope tag, and immune complexes were purified from cell lysates with anti-FLAG resin. An aliquot of each complex was resolved by SDS-PAGE and immunoblotted with anti-FLAG, anti-Myc, and anti-Skp-1. Given that mutation of the sequence 755-IVRIMK-760 to polyalanine in the yeast Cul-1 homologue Cdc53 (in which K760 in Cdc53 is analogous to K720 in human Cul-1) did not affect Cdc53 binding with Skp-1, F-box proteins, or Cdc34 (46), we expected that the K720R Cul-1 would also retain its ability to participate in SCF-protein interactions in this system. Indeed, no difference in the interaction of either FLAG-WT or K720R Cul-1 with Skp-1 or Myc-βTrCP was detected (Fig. 4A, lanes 1 and 2). We noted that a portion of FLAG WT Cul-1 was modified by Nedd8 (Cul-1*), while K720R Cul-1 was not (Fig. 4A). We also noted that WT Cul-1 coexpressed with βTrCP showed enhanced Nedd8 ligation compared to WT Cul-1 expressed alone (compare Fig. 3B, lane 1, to Fig. 4A, lane 1).
FIG. 4.
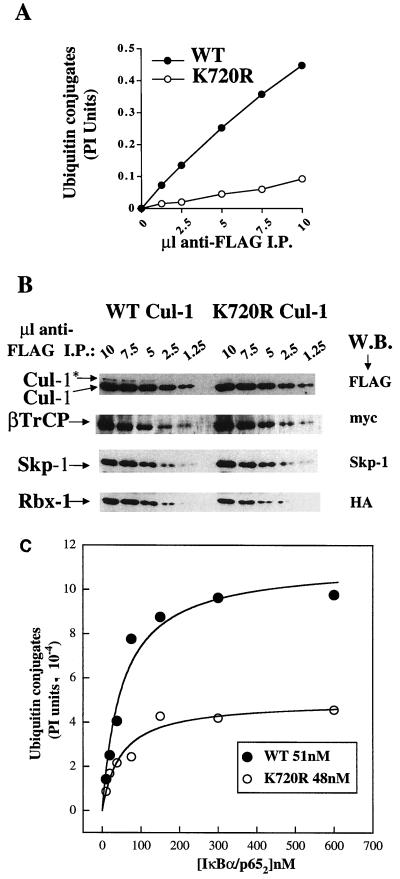
Nedd8 modification of Cul-1 stimulates ubiquitination activity of SCFβTrCP. 293 cells were cotransfected with FLAG-tagged Cul-1 (WT or K720R) and Myc-βTrCP. (A) Anti-FLAG immune complexes from these lysates were immunoblotted with anti-FLAG to detect Cul-1, anti-Myc to detect βTrCP, and anti-Skp-1. Note the presence of Cul-1* in WT Cul-1-transfected cells only. (B) Immune complexes from WT Cul-1 plus WT βTrCP (lanes 1 to 5) or K720R Cul-1 plus WT βTrCP (lanes 6 to 10) were assayed for IκBα ubiquitination activity. Ubiquitination of WT (lanes 1 to 4 and 6 to 9) or S32/36A (lanes 5 and 10) IκBα (150 nM) was assayed in the presence of 100 nM E1, 60 μM ubiquitin, 0.5 μM ubiquitin aldehyde, 2.5 μM MG273, an ATP-regenerating system, and 1 μM microcystin LR. UbcH5a (150 nM), Cdc34 (150 nM), and the Nedd8 pathway (Nedd8 [250 nM], Ubc12 [150 nM], and APP-BP1/Uba3) were added as indicated.
To assess the affect of the Nedd8 pathway on the activity of these SCFβTrCP complexes, ubiquitination reactions were reconstituted using FLAG immunoprecipitates containing either WT or K720R Cul-1 and phosphorylated 35S-labeled IκBα/p652 as the substrate (Fig. 4B). In the absence of added ubiquitin E2, the FLAG immune complexes did not support ubiquitination of IκBα (lanes 1 and 6). Formation of conjugates was stimulated when the ubiquitin E2s Cdc34 and UbcH5, both previously implicated in IκBα ubiquitination (12, 43, 53, 56, 65), were added to SCFβTrCP containing WT Cul-1 (lane 2). In contrast, under the same reaction conditions, the conjugation activity of complexes containing K720R Cul-1 was significantly less compared to WT Cul-1 (compare lanes 2 and 7). When Nedd8 pathway components were added to the FLAG immune complexes in the absence of ubiquitin E2 activity, no conjugates were detected, indicating that addition of the Nedd8 pathway alone was insufficient to promote ubiquitination and that IκBα itself did not form conjugates with Nedd8 (lane 3). Strikingly, Nedd8 pathway components added to FLAG-WT Cul-1 immunoprecipitates in the presence of UbcH5A and Cdc34 resulted in an increase in formation of high-molecular-weight conjugates over that observed with the ubiquitin-E2s alone (compare lanes 2 and 4), and anti-FLAG Western blots of these reactions showed increased ligation of Nedd8 to WT Cul-1 (data not shown). Conversely, addition of the Nedd8 pathway had no effect on the ubiquitination activity of the K720R complexes (compare lanes 7 and 9). Importantly, the effects of the Nedd8 pathway on ubiquitination activity in these reactions retained specificity for phosphorylated IκBα, as no conjugates were detected with S32/36A IκBα (lanes 5 and 10). Together, these results suggest that optimal ubiquitination activity of SCFβTrCP requires Nedd8 and ubiquitin-conjugating enzyme systems, and that K720 of Cul-1 is an important site of Nedd8 conjugation that has profound effects on activity.
We next sought to compare the relative ubiquitin-conjugating activity of SCFβTrCP containing either WT or K720R Cul-1 by titrating similar amounts of the immunoprecipitates into reactions containing 150 nM IκBα/p652, a level well in excess of its Km (see below). These reactions were performed in the presence of the fully reconstituted ubiquitination system and components of the Nedd8 pathway. The activity of both enzyme complexes was linear with time at each enzyme concentration tested (data not shown), and SCFβTrCP containing WT Cul-1 was significantly more active than the K720R Cul-1 mutant (Fig. 5A). Importantly, the levels of Myc-βTrCP, endogenous Skp-1, and HA-Rbx1 associated with WT and K720R Cul-1 were comparable between the enzyme preparations (Fig. 5B); thus, the lower activity of the SCFβTrCP containing K720R Cul-1 is not due to apparent differences in the association of these complex components. These results provide the first biochemical evidence that optimal catalytic activity of an SCF complex ubiquitin ligase requires Cul-1 that is competent for Nedd8 conjugation.
FIG. 5.
K720R Cul-1 affects the ubiquitination activity of SCFβTrCP. 293 cells were cotransfected as in Fig. 4 except that HA-Rbx1 was included. (A) Aliquots of anti-FLAG immune complexes were assayed for IκBα ubiquitination activity for 20 min at 37°C in the presence of 100 nM E1, 60 μM ubiquitin, 0.5 μM ubiquitin aldehyde, 2.5 μM MG273, an ATP-regenerating system, 1 μM microcystin LR, UbcH5a (150 nM), Cdc34 (150 nM), the Nedd8 pathway (Nedd8 [250 nM], Ubc12 [150 nM], 0.5 μl of affinity-purified APP-BP1/Uba3), and 150 nM IκBα/p652. Samples were analyzed by SDS-PAGE on 9% gels and quantified by phosphorimage (PI) analysis. Shown is conjugate formation plotted versus the amount of enzyme added. (B) Indicated amounts of the immune complexes assayed in panel A were separated by SDS-PAGE on 9% gels and immunoblotted with the indicated antisera. (C) Nedd8 conjugation to Cul-1 does not affect the Km for IκBα. Aliquots (5 μl [WT] or 10 μl [K720R]) of the anti-FLAG immune complexes shown in panel B were assayed for IκBα ubiquitination activity as in panel A, using IκBα/p652 ranging in concentration from 9.4 to 600 nM. Shown is conjugate formation plotted versus substrate concentration and fitted to the equation v = Vmax[S]/Km + [S].
Since we observed no obvious difference in the composition of SCFβTrCP formed with either WT or K720R Cul-1, we asked whether the differences in ubiquitination activity could be explained by differences in the affinity of these complexes for phosphorylated IκBα. FLAG immunoprecipitates were prepared from cells transfected with FLAG-Cul-1 (WT or K720R), Myc-βTrCP, and HA-Roc1. Ubiquitination reactions reconstituted as above were conducted using 35S-labeled IκBα/p652 ranging in concentration from 9.4 to 600 nM (using twofold serial dilutions) and resolved by SDS-PAGE. The high-molecular-weight ubiquitin conjugates were quantified by phosphorimage analysis. The Km for IκBα/p652 was determined by fitting conjugate formation versus substrate concentration to the equation v = Vmax[S]/Km + [S] (Fig. 5C). In two independent experiments, the value of Km for IκBα/p652 was essentially the same for both forms of SCFβTrCP and ranged from 30 to 50 nM. Thus, the Nedd8 modification of Cul-1 apparently does not influence substrate binding affinity of SCFβTrCP under our assay conditions.
DISCUSSION
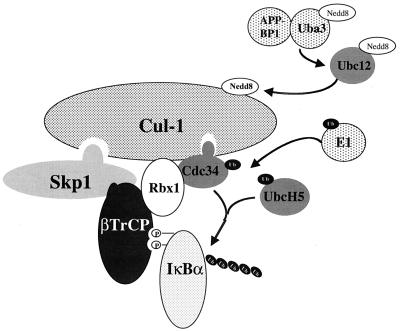
Several recent studies have shown that SCFβTrCP functions as a ubiquitin ligase (E3) responsible for phosphorylation-dependent ubiquitination of IκBα (11, 17, 28, 53, 55, 63, 65). In this report, we present several lines of evidence that Nedd8 modification of Cul-1 and Nedd8-conjugating activity enhance the IκBα ubiquitination activity of SCFβTrCP. First, although only a small portion of the total cellular pool of Cul-1 is modified by Nedd8, Nedd8-Cul-1 was the only form detected in association with the SCFβTrCP substrates phosphorylated IκBα and β-catenin in vivo. Second, a significant portion of endogenous Cul-1 associated with ectopically expressed βTrCP was modified by Nedd8. Third, Ubc12-dependent Nedd8-conjugating activity, in addition to ubiquitin-conjugating activity, was required for robust ubiquitination of IκBα by SCFβTrCP in vitro. Moreover, an active-site mutant of Ubc12 was a potent inhibitor of IκBα ubiquitination. Finally, IκBα ubiquitination activity of SCFβTrCP was stimulated in the presence of Nedd8 pathway components. In contrast, SCFβTrCP assembled with a mutant Cul-1 (K720R) that does not form conjugates with Nedd8 was significantly less active than the WT enzyme and was not stimulated in the presence of the Nedd8 pathway. Taken together, our results suggest that Nedd8 conjugation of Cul-1 has profound effects on SCFβTrCP ubiquitination activity, linking the Nedd8 and ubiquitin pathways in the signal dependent degradation of IκBα (Fig. 6).
FIG. 6.
Ubiquitination of IκBα by SCFβTrCP involves both the ubiquitin and Nedd8 conjugation pathways. SCFβTrCP is composed of Skp-1, Cul-1, Rbx1, and the F-box protein, βTrCP. The ubiquitination activity of SCFβTrCP is potentiated when Cul-1 is modified by Nedd8. Nedd8 modification of Cul-1 occurs through a pathway which includes the heterodimeric Nedd8-activating enzyme, APP-BP1/Uba3, and the Nedd8-conjugating enzyme, Ubc12. The ubiquitin E2, Cdc34, is recruited to SCFβTrCP by interacting with Rbx1 and Cul-1. Both Cdc34 and UbcH5 have been implicated in ubiquitination of IκBα, but their precise relationship is unclear. The ubiquitin-E2s build a polyubiquitin chain on phosphorylated IκBα when IκBα is bound to βTrCP.
Our experiments suggest that Nedd8 conjugation of Cul-1 is important for the function of SCFβTrCP but is not essential for detecting ubiquitination activity in vitro (Fig. 4B and 5). These results are reminiscent of recent studies in which SCFβTrCP activity was reconstituted in the absence of exogenous Nedd8 pathway components (53, 65). While the status of Nedd8–Cul-1 conjugation was not examined in these studies, in light of our findings, at least some portion of the Cul-1 was likely modified by Nedd8. We found that the IκBα ubiquitination activity both in cellular extracts and in isolated SCFβTrCP was significantly enhanced in the presence of the Nedd8 pathway. We also noted that a portion of the WT Cul-1 in immunoprecipitated SCFβTrCP was conjugated to Nedd8 in vivo and that Nedd8–Cul-1 levels increased in reactions containing Nedd8 pathway components, an effect coincident with increasing IκBα ubiquitination activity (Fig. 4).
SCF complexes are implicated in the regulated proteolysis of a growing number of cellular proteins. Recent reports have added the R-box proteins, Rbx-1 (Roc1), Rbx-2, and APC11, as members of SCF and related E3 complexes and have begun to shed light on the mechanism by which these enzymes recognize target substrates and catalyze the formation of Ub conjugates (25, 43, 50, 52, 56) and Nedd8 conjugates (26). Cul-1 and related cullin proteins are known to form conjugates with Nedd8 (60). How Nedd8 conjugation of the cullin proteins impinges on the association and/or activity of these factors with the core components of the SCF and related complexes is not known. Our analysis of SCFβTrCP using coimmunoprecipitation experiments showed that the associations of β-TrCP, Skp-1, and Roc1 with either FLAG-tagged WT or K720R Cul-1 were indistinguishable (Fig. 5B). Moreover, the ability to form Nedd8–Cul-1 conjugates had no obvious affect on the affinity of SCFβTrCP for phosphorylated IκBα, as the apparent Km for this substrate was similar for both forms of the enzyme. The striking difference in ubiquitination activity observed for SCFβTrCP containing WT or K720R Cul-1 could reflect a role for Nedd8 modification in altering the conformation of Cul-1 in a manner that does not grossly affect interaction with other SCF components or substrate but does stimulate ubiquitin transfer. While this report was in preparation, two reports describing Nedd8 conjugation of Cul-2 in an SCF-related E3 ligase (see below) called the von Hippel Lindau-elonginB/C (VBC) complex were published (34, 61). Neither report addressed a role for Nedd8–Cul-2 in ubiquitination activity; however, both showed that formation of Nedd8–Cul-2 in vivo was dependent on the integrity of the VBC complex. Moreover, Wada et al. (61) speculated that differences in their ability to immunoprecipitate WT versus a mutant form of Cul-2 that is analogous to K720R Cul-1 could reflect a conformational effect of forming the Nedd8 conjugate.
SCF complexes, along with the VBC and other cullin-containing complexes, are members of a proposed superfamily of ubiquitin ligases (E3s) (for a review, see reference 58). These complexes share a common architecture which includes the presence of cullin-like and R-box proteins at the core. Cul-1 and related cullin proteins are the only components of these complexes known to form conjugates with Nedd8. Since K720 and the surrounding sequence in Cul-1 is conserved in all cullin proteins (Fig. 3A), and since all cullin proteins tested to date can be modified by a single Nedd8 molecule (44, 60), it seems likely that the requirement for Nedd8 modification at this site will also be a common feature of all cullin protein-containing complexes. This raises the intriguing possibility that formation of Nedd8 conjugates represents a novel and universal mechanism for regulating the activity of SCF and other cullin-containing complexes. Thus, regulation of Nedd8 levels, and/or the associated Nedd8-activating enzyme and E2 activities, may play a fundamental role in controlling levels of key protein targets of these E3s under different physiological states. This idea is supported by the fact that Nedd8 is differentially expressed in a variety of tissue types and is down regulated during cellular differentiation (24, 29).
This study demonstrates that the Cul-1 component of SCFβTrCP is decorated with a single Nedd8 molecule when it is part of an active SCFβTrCP complex engaged with its cellular substrates, phosphorylated IκBα and β-catenin (Fig. 6). While the role of the Nedd8 modification in any biological function has been elusive, the involvement of Nedd8 in SCF-mediated ubiquitination links these two highly related pathways of protein modification. Both pathways are required to ultimately result in degradation of a phosphorylated substrate. Inhibition of the Nedd8-activating pathway in cells could reveal additional pathways where this modification exerts a regulatory role.
ACKNOWLEDGMENTS
Margaret A. Read and James E. Brownell contributed equally to this work.
We are grateful to Luan Dang, David Norse, Sha-Mei Liao, Chunhua Wang, Susan Fish, Laura Faron, Teresa McCormack, and Eric Lightcap for assistance with reagent preparation. We thank Keith Kropp for continued support. We thank Julian Adams, Eric Lightcap, and Tom Maniatis for carefully reading the manuscript.
This work was supported in part by grant GM53136 from the National Institutes of Health.
REFERENCES
- 1.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkalay I A, Yaron A, Hatzubai A, Orian A, Ceichanover A, Ben-Neriah Y. Stimulation-dependent IκBα phosphorylation marks the NF-κB inhibitor for degradation via the Ub-proteasome pathway. Proc Natl Acad Sci USA. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 4.Bai C, Sen P, Hofmann K, Ma L, Goebel M, Harper J W, Elledge S J. Skp-1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin A S. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 6.Barnes P J, Karin M. Nuclear factor-kappa B: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 7.Beg, A. A., and D. Baltimore. An essential role for NF-κB in preventing TNFα-induced cell death. Science 274:782–784. [DOI] [PubMed]
- 8.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 9.Ciechanover A. The ubiquitin-proteasome pathway:on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldman R M, Correl C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs S Y, Chen A, Xiong Y, Pan Z-Q, Ronai Z. HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IκB and β-catenin. Oncogene. 1999;18:2039–2046. doi: 10.1038/sj.onc.1202760. [DOI] [PubMed] [Google Scholar]
- 12.Gonen H, Bercovich B, Orian A, Carrano A, Takizawa C, Yamanaka K, Pagano M, Iwai K, Ciechanover A. Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IκBα. J Biol Chem. 1999;274:14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- 13.Gong L, Yeh E T H. Identification of the activating and conjugating enzymes of the Nedd8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 14.Gray W M, del Pozo J C, Walker L, Hobbie L, Risseeuw E, Banks T, Crosbyk W L, Yang M, Me H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grisham M B, Palombella V J, Elliot P, Conner E M, Brand S, Wong H, Pien C, Destree A. Inhibition of NF-κB activation in vitro and in vivo: role of the 26S proteasome. Methods Enzymol. 1999;300:345–363. doi: 10.1016/s0076-6879(99)00140-8. [DOI] [PubMed] [Google Scholar]
- 16.Hart M, Concordet J-P, Lassot I, Albert I, Del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin R, Banarous R, Polakis P. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 17.Hatakeyama S, Kitagawa M, Nakayama K, Shirane M, Matsumoto M, Hattori K, Higashi H, Nakano H, Okumura K, Onoe K, Good R A, Nakayama K. Ubiquitin-dependent degradation of IκBα is mediated by a ubiquitin ligase Skp1/Cul 1/F-box protein FWD1. Proc Natl Acad Sci USA. 1999;96:3859–3863. doi: 10.1073/pnas.96.7.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;256:8206–8214. [PubMed] [Google Scholar]
- 19.Hershko A, Rose I A. Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc Natl Acad Sci USA. 1987;84:1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochstrasser M. There's the Rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 21.Israel A. IκB kinase all zipped up. Nature. 1997;388:519–521. doi: 10.1038/41433. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs M, Harrison S C. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 23.Jensen J P, Bates P W, Yang M, Vierstra R D, Weissman A M. Identification of a family of closely related human ubiquitin conjugating enzymes. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- 24.Kamitani T, Kito K, Nguyen H P, Yeh E T H. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 25.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kailin W G, Elledge S J, Conaway R C, Harper J W, Conaway J W. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 26.Kamura T, Conrad M N, Yan Q, Conaway R C, Conaway J W. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitagawa K, Skowyra D, Elledge S J, Harper J W, Heiter P. SGT1 encodes an essential component of the yeast kinetichore assemble pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- 28.Kroll M, Margottin F, Kohl A, Renard P, Durand H, Concordet J P, Bachelerie F, Arenzana-Seisdedos F, Benarous R. Inducible degradation of IκBα by the proteasome requires interaction with the F-box protein h-βTrCP. J Biol Chem. 1999;274:7941–7945. doi: 10.1074/jbc.274.12.7941. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 30.Lammer D, Mathias N, Laplaza J M, Juang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latres E, Chiaur D S, Pagano M. The human F box protein β-TrCP associates with the Cul1/Skp1 complex and regulates the stability of β-catenin. Oncogene. 1999;18:849–854. doi: 10.1038/sj.onc.1202653. [DOI] [PubMed] [Google Scholar]
- 32.Leahy D J, Hendrickson W A, Aukhil I, Erickson H P. Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science. 1992;258:987–991. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- 33.Lee F S, Peters R T, Dang L C, Maniatis T. MEKK1 activates both IκB kinase-α and IκB kinase-β. Proc Natl Acad Sci USA. 1998;95:9319–9324. doi: 10.1073/pnas.95.16.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liakopoulos D, Büsgen T, Brychzy A, Jentsch S, Pause A. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc Natl Acad Sci USA. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lisztwan J A, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. Association of human Cul-1 and ubiquitin-conjugating enzyme Cdc34 with the F-box protein p45 (Skp-2): evidence for evolutionary conservation in the subunit composition of the Cdc34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Kato Y, Zhang Z, Do V M, Yankner B A, He X. β-TrCP couples β-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci USA. 1999;96:6273–6278. doi: 10.1073/pnas.96.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z G, Hsu H, Goeddel D V, Karin M. Dissection of TNF receptor effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 38.Maniatis T. Catalysis by a multiprotein IκB kinase complex. Science. 1997;278:818–819. doi: 10.1126/science.278.5339.818. [DOI] [PubMed] [Google Scholar]
- 39.Maniatis T. A ubiquitin ligase complex essential for the NF-κB, Wnt/wingless, and hedgehog signaling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 40.Margottin F, Bour S P, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebeland D, Benarous R. A novel human WD protein, h-βTrCP, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 41.Melandri R, Grenier L, Plamondon L, Huskey W P, Stein R L. Kinetic studies on the inhibition of isopeptidase T by ubiquitin aldehyde. Biochemistry. 1996;35:12893–12900. doi: 10.1021/bi9612935. [DOI] [PubMed] [Google Scholar]
- 42.Michel J J, Xiong Y. Human Cul-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:435–449. [PubMed] [Google Scholar]
- 43.Ohta T, Michel J J, Schottelius S J, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 44.Osaka F, Kawasaki W, Aida N, Saiki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palombella V J, Rando O J, Goldberg A L, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 46.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 48.Scheffner M, Huibregtse J M, Howley P M. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:1259–1263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seol J H, Feldman R M R, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Shevchenki A, Deshaies R J. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 52.Skowyra D, Keopp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 53.Spencer E, Jiang J, Chen Z J. Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stancovski I, Baltimore D. NF-κB activation: the IκB kinase revealed? Cell. 1997;91:299–302. doi: 10.1016/s0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki H, Chiba T, Kobayashi M, Takeuchi M, Suzuki T, Ichiyama A, Ikenoue T, Omata M, Furuichi K, Tanaka K. IκBα ubiquitination is catalyzed by an SCF-like complex containing Skp1, cullin-1, and two F-box/WD40-repeat proteins, βTrCP1 and βTrCP2. Biochem Biophys Res Commun. 1999;256:127–132. doi: 10.1006/bbrc.1999.0289. [DOI] [PubMed] [Google Scholar]
- 56.Tan P, Fuchs S Y, Chen A, Wu K, Gomez C, Ronai Z, Pan Z-Q. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 57.Townsley F M, Aristarkhov A, Beck S, Hershko A, Ruderman J V. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/Ubc10 blocks cells in metaphase. Proc Natl Acad Sci USA. 1997;94:2362–2367. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyers M, Willems A R. One ring to rule a superfamily of E3 ubiquitin ligases. Science. 1999;284:601–603. doi: 10.1126/science.284.5414.601. [DOI] [PubMed] [Google Scholar]
- 59.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Suppression of TNFα induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 60.Wada H, Yeh E T H, Kamitani T. Identification of NEDD8-conjugation site in human cullin-2. Biochem Biophys Res Commun. 1999;257:100–105. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- 61.Wada H, Yeh E T H, Kamitani T. The von Hippel-Lindau tumor suppressor gene product promotes, but is not essential for, Nedd8 conjugation to Cullin-2. J Biol Chem. 1999;274:36025–36029. doi: 10.1074/jbc.274.50.36025. [DOI] [PubMed] [Google Scholar]
- 62.Wang C-Y, Mayo M W, Baldwin A S. TNF-and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 63.Winston J T, Strack P, Beer-Romero P, Chu C Y, Elledge S J, Harper J W. The SCFβ-TrCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yaron A, Gonen H, Alkalay I, Hatzubai A, Jung S, Beyth S, Mercurio F, Manning A M, Ciechanover A, Ben-Neriah Y. Inhibition of NF-κB cellular functions via specific targeting of the IκB-ubiquitin ligase. EMBO J. 1997;16:6486–6494. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning T M, Andersen J S, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]