Abstract
Mitogen-activated protein kinases (MAPKs) are activated through cascades or modules consisting of a MAPK, a MAPK kinase (MAPKK), and a MAPKK kinase (MAPKKK). Investigating the molecular basis of activation of the c-Jun N-terminal kinase (JNK) subgroup of MAPK by the MAPKKK MEKK2, we found that strong and specific JNK1 activation by MEKK2 was mediated by the MAPKK JNK kinase 2 (JNKK2) rather than by JNKK1 through formation of a tripartite complex consisting of MEKK2, JNKK2, and JNK1. No scaffold protein was required for the MEKK2-JNKK2-JNK1 tripartite-complex formation. Expression of JNK1, JNKK2, and MEKK2 significantly augmented the coprecipitation of, respectively, MEKK2-JNKK2, MEKK2-JNK1, and JNKK2-JNK1, indicating that the interaction of MEKK2, JNKK2, and JNK1 is synergistic. Finally, the JNK1 was activated more efficiently in the MEKK2-JNKK2-JNK1 complex than was the JNK1 excluded from the complex. Thus, formation of a signaling complex through synergistic interaction of a MAPKKK, a MAPKK, and a MAPK molecule like MEKK2-JNKK2-JNK1 is likely to be responsible for the efficient, specific flow of information via MAPK cascades.
Mitogen-activated protein kinase (MAPK) cascades are central components of the intracellular signaling networks involved in transducing a wide spectrum of extracellular signals to nuclear and cytoplasmic effectors that control cell growth, differentiation, and apoptosis (for reviews, see references 6, 20, 25, and 26). Multiple MAPK cascades that lead to the activation of extracellular signal-related kinases 1 and 2 (ERK1 and ERK2), c-Jun N-terminal kinases 1 through 3 (JNK1 through JNK3), p38α through p38γ, or ERK5 were identified in eukaryotic cells, and each is believed to respond to a distinct set of extracellular stimuli (3, 11, 18, 29, 44, 61). Each of these MAPKs is activated by a MAPK kinase (MAPKK), usually with rather narrow specificity (6, 19, 33, 43, 46), although individual MAPKKs are believed to respond to several or many MAPKK kinases (MAPKKKs). The MAPKKKs are responsible for responding to a variety of upstream activators that connect them to various cell surface receptors. In addition to amplifying weak receptor-generated signals, MAPK cascades are believed to participate in the generation of signaling specificity (5, 16, 20, 26, 33, 46).
A given MAPK cascade can respond to several extracellular stimuli, and a given stimulus can activate several MAPK cascades, but the response and fidelity of MAPK activation are specific (for reviews, see references 8, 25, 26, and 46). Although MAPK cascades may have ample opportunities in vivo for cross talk at different levels, an individual MAPK cascade is generally insulated from other closely related cascades, and each MAPK cascade is believed to preferentially respond to a distinct set of stimuli (4, 13, 16, 42, 55).
The molecular mechanism of MAPK cascade specificity is best studied in yeast. Specific MAPK activation in response to mating pheromones is conferred by STE5, a protein that acts as a molecular scaffold tethering the MAPKKK STE11, MAPKK STE7, and MAPK FUS3 proteins to form a pheromone-responsive module (4, 32, 39). Direct interaction of the components of a MAPK module has also been observed and suggested to play a role in determining MAPK specificity. The MAPKK STE7 in yeast was shown, for example, to interact with its target FUS3 in the absence of STE5 (1), and the yeast MAPKK PBS2 was shown to assemble a module with the MAPK HOG1 and the MAPKKK STE11 in response to osmotic stress (38).
Although a mammalian homologue of STE5 has not yet been identified, two proteins, MP1 and JIP-1, have been suggested to function as a scaffold for MAPK modules that leads to specific activation of ERK and JNK (41, 52). MP1 binds both MEK1 and ERK1 in activating the ERK pathway (41), whereas JIP-1, a protein originally identified as a JNK1-interacting protein, binds to JNK1, JNKK2/MKK7, and MAPKKK MLK3/DLK, thereby facilitating JNK1 activation by MLK3/DLK (52). A different scheme has been suggested for JNK (or p38) activation in response to the MAPKKK MEKK1, in which the MAPKK JNKK1/MKK4 was shown to be involved in specific and sequential interactions with MEKK1 and JNK1 (55). These interactions were bipartite and sequential, so that formation of a MEKK1-JNKK1 complex resulted in activation of JNKK1 followed by disassembly and formation of a specific JNKK1-JNK1 complex and then by activation of JNK1 (55). No ternary MEKK1-JNKK1-JNK1 complex could be detected, probably because the same interaction surface on JNKK1, its N-terminal region, was used to contact either MEKK1 or JNK1 (55). The N-terminal region of MEKK1 was shown in other studies to be directly associated with the downstream kinase JNK1, suggesting that this region may function as a scaffold in certain situations (56).
The JNK subgroup of MAPKs is activated by a particularly large number of stimuli, including physical stresses, cytokines, T-cell costimulation, and growth factors (24, 25, 35, 46). Two specific JNK-activating MAPKKs, JNKK1/MKK4 and JNKK2/MKK7, were identified (12, 15, 22, 30, 31, 37, 40, 49, 54, 59). Although JNKK1/MKK4 and JNKK2/MKK7 are believed to be able to activate JNK, a recent study suggested that JNKK1 and JNKK2 may differentially phosphorylate their substrate JNK at the conserved Thr and Tyr residues, thus synergizing their effect on JNK activation (28). JNKK1/MKK4 and JNKK2/MKK7 are also differentially activated by tumor necrosis factor alpha and cellular stresses, and distinct upstream activators were proposed for their activation in those responses (21, 37). JNKK1/MKK4 was found to be the preferential substrate of the MAPKKK MEKK1 (55), while JNKK2/MKK7 was shown to be activated by several MAPKKKs, including MEKK1, MEKK2, MEKK3, and MLK3/DLK, in JNK activation ((9, 34, 52, 55; J. Yang and B. Su, unpublished data).
A growing number of MAPKKKs are being identified as capable of JNK activation; they include MEKK1, MEKK2, MEKK3, MEKK4, MEKK5, ASK, TAK, and Tpl (2, 14, 16, 17, 23, 27, 47, 48, 51, 58). These kinases have in common a conserved kinase domain with substantial homology to that of the yeast MAPKKK STE11. These MAPKKKs are believed to be major players involved in sorting distinct cell surface signals to the downstream cytoplasmic and nuclear effectors. While MEKK1 clearly prefers JNKK1/MKK4 to JNKK2/MKK7, it is not known whether other MAPKKKs operate preferentially via JNKK1, JNKK2 or an as yet unidentified JNKK in JNK activation.
We investigated the molecular mechanisms that underlie the strong and specific JNK1 activation by human MEKK2, a molecule we cloned from human T cells. We identified a tripartite molecular complex consisting of MEKK2, JNKK2, and JNK1 that was responsible for strong, specific JNK1 activation by MEKK2.
MATERIALS AND METHODS
Cell culture and transient transfection.
COS-1 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum, 100 U of penicillin per ml, and 100 mg of streptomycin per ml. Plasmid DNA was transfected into COS-1 cells with Lipofectamine (Life Technologies, Gaithersburg, Md.).
Plasmids, proteins, and antibodies.
Hemagglutinin (HA)-tagged JNK1, JNKK1, JNKK1(AL), JNKK2, JNKK2(AL), MEK1, MEK2, MKK6, p38, and MEKK1(CT) were cloned into pSRα3 vector as previously described (30, 36, 45, 59). JNKK1(AL) and JNKK2(AL) are the kinase domain mutants in which Ser-257 and Thr-261 (JNKK1) and Ser-271 and Thr-275 (JNKK2) were replaced with Ala and Lys, respectively, using a PCR-directed mutagenesis method. The HA-tagged MEKK1(CT) expression vector has been described by Minden et al. (36). The HA-tagged MEKK2(CT) expression vector was constructed by introducing an NcoI site at codon 343 by the PCR-based method, followed by subcloning into the expression vector pSRα3HA at the NcoI site, in which MEKK2(CT) fused in frame with the HA tag. The pCMV-Flag-JNK1 expression vector has been described previously (11). Glutathione S-transferase (GST)-tagged MEKK2(FL), MEKK2(CT), MEKK1(CT), and JNKK2 mammalian expression vectors were cloned into the pEGB vector (30) by standard molecular biology techniques. Briefly, MEKK2(FL) and JNKK2 were introduced into an NcoI site at the first Met codon, MEKK2(CT) at codon 343, and MEKK1(CT) at codon 1199 by the PCR-based method, followed by subcloning into the NcoI site in mammalian expression vector pEGB and fused in frame with a GST tag. Bacterial recombinant protein expression vectors for GST–c-Jun, GST-JNK1, and His-p38 have been described previously, as have expression and purification of the recombinant proteins (30, 36, 45). Anti-HA antibody 12CA5 was prepared from the 12CA5 hybridoma (53) and further purified using a protein A-Sepharose column. Anti-Flag antibody M2 was purchased from IBI-Kodak. Anti-MEKK1 antibody (C22) was purchased from Santa Cruz (Santa Cruz, Calif.). MEKK2-specific antibody 1128 was prepared by immunizing rabbits with a GST fusion protein fused in frame with a fragment of human MEKK2 peptide (amino acids 343 to 428) (B. Su and J. Cheng, unpublished data). Anti-JNK1 antibody was from Pharmingen.
Immunoprecipitation and in vitro kinase assay.
Cell lysates were prepared 40 h after transfection, as described previously (45, 55), and incubated with appropriate antibodies for 4 h at 4°C in a rotator; protein A-Sepharose beads were added for another 45 min. The beads were washed four times with lysis buffer (20 mM Tris [pH 7.5], 0.5% Nonidet P-40, 250 mM NaCl, 3 mM EDTA, 3 mM EGTA, 100 mM Na3VO4) and twice with kinase reaction buffer (20 mM HEPES [pH 7.6], 1 mM MgCl2, 10% glycerol). The immunoprecipitates were then subjected to kinase assays in 30 μl of kinase buffer with appropriate substrates in the presence of 0.5 μl of [γ-32P]ATP and 20 μM cold ATP. After 20 min at 30°C, the reactions were terminated by addition of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and boiling for 5 min. The proteins were separated by SDS-PAGE, and 32P incorporation was determined with an FX Phosphorimager (Bio-Rad Laboratories, Richarmond, Calif.).
Coupled-kinase assay.
Cell lysates and immunoprecipitates were prepared as described previously (45). The immunoprecipitates were subjected to a coupled-kinase assay as described by Lin et al. (30). Briefly, the immunoprecipitates were incubated in 30 μl of kinase buffer with two substrates, GST-JNK1 and GST–c-Jun, in the presence of 1 μl of [γ-32P]ATP and 30 μM cold ATP. After 40 min at 30°C, the reactions were terminated by adding SDS-PAGE loading buffer and boiling for 5 min. The proteins were separated by SDS-PAGE, and 32P incorporation into GST-JNK1 and GST–c-Jun was determined.
Coprecipitation assay.
COS-1 cells transfected with HA-, Flag-, or GST-tagged expression vectors were lysed 40 h after transfection by using low-salt lysis buffer (50 mM HEPES [pH 7.6], 150 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 10% glycerol). The nuclei and cell debris were removed from the lysates by centrifugation in a microcentrifuge for 20 min at 4°C. Expressed proteins were precipitated from the clarified lysates with glutathione (GSH)-Sepharose beads at 4°C for a 4-h incubation in a rotator. The beads were washed four times with low-salt lysis buffer, and the precipitates were eluted with sample buffer and resolved by SDS-PAGE. After electrophoresis and transfer, the proteins were analyzed by Western blotting with appropriate antibodies. Western blotting was quantitated with a Bio-Rad G360 Image system.
Gel filtration.
Cell lysates containing 200 μg of proteins were prepared from COS-1 cells expressing GST-MEKK2(CT), HA-JNKK2, or HA-JNK1 and loaded onto a fast protein liquid chromatography (FPLC) Superdex-200 column (Pharmacia). The column was washed with buffer containing 20 mM Tris-HCl and 150 mM NaCl. Then 0.2-ml fractions were collected and subjected to Western blotting and coprecipitation assays as described above.
RESULTS
Strong and specific JNK1 activation by MEKK2 is not mediated by JNKK1.
During our investigation of upstream activators of the JNK MAPK cascade expressed in Jurkat T cells, we isolated a cDNA that encodes human MEKK2 (GenBank accession no. AF111105) and demonstrated its involvement in transducing T-cell-activating signals to JNK (B. Su et al., unpublished data). Human MEKK2 is highly homologous to murine MEKK2, and its expression leads to strong activation of JNKs in many cell lines similar to the murine MEKK2 (2, 9; Su et al., unpublished). Like all other MAPKKKs, MEKK2 does not activate JNK1 directly; such activation requires the involvement of an intermediary MAPKK, although the specific MAPKK through which MEKK2 operates is not known.
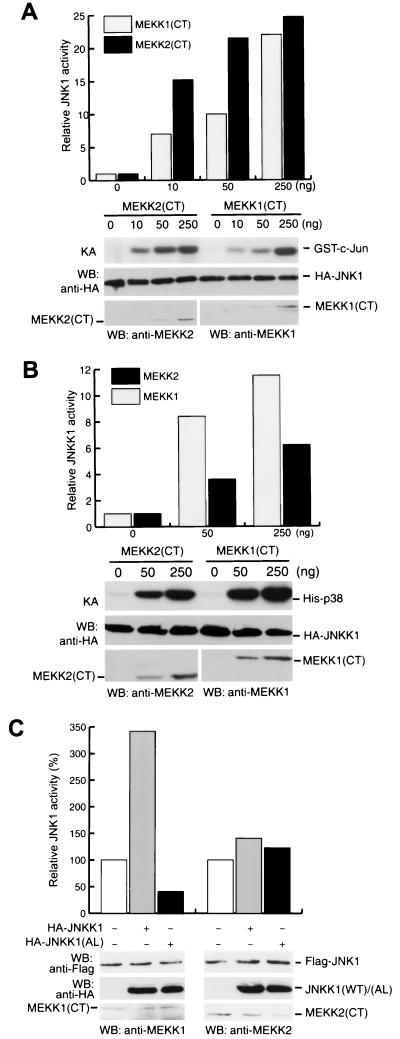
We used cotransfection experiments to compare the abilities of MEKK1 and MEKK2 to activate JNK1 and found that MEKK2 was an even more potent JNK1 activator than was MEKK1, one of the most potent JNK activators reported to date (J. Cheng and B. Su, unpublished results). MEKK1 has a much larger N-terminal domain than MEKK2, and this domain was shown to affect the MEKK1 expression level (55, 57), suggesting that the difference in the abilities of full-length MEKK1 and MEKK2 to activate JNK1 may be due to their different expression levels. Since the expression levels of the kinase domains of MEKK1 (residues 1199 to 1495) and MEKK2 (residues 343 to 618) are compatible and both are potent activators of JNK1 (Fig. 1), we compared the activities of the MEKK1 and MEKK2 kinase domains, which have 49% homology. Cotransfecting JNK1 and JNKK1 expression vectors with different amounts of expression vectors for the kinase domains of MEKK1 [MEKK1(CT)] and MEKK2 [MEKK2(CT)], we determined the enzymatic activities of JNK1 and JNKK1 by an in vitro kinase assay. At low levels of MEKK1(CT) and MEKK2(CT) expression plasmids, we found MEKK2(CT) to be a more effective JNK activator than MEKK1(CT), but the difference was diminished when higher levels of MEKK1(CT) and MEKK2(CT) vectors were used (Fig. 1A). Under the same conditions, however, the patterns of JNKK1 activation by MEKK1(CT) and MEKK2(CT) were reversed: MEKK2(CT) was a less effective activator of JNKK1 than was MEKK1(CT) (Fig. 1B). These results suggested that in activating JNK1, JNKK1 may not be the preferred substrate for MEKK2.
FIG. 1.
MEKK2 is a stronger JNK1 activator but weaker JNKK1 activator than is MEKK1. (A) HA-JNK1 expression vector (0.5 μg/plate) was cotransfected into COS-1 cells with increasing amounts of either MEKK2(CT) or MEKK1(CT) expression vectors. HA-JNK1 activity was measured by an immunocomplex kinase assay (KA) as described in Materials and Methods. Expression of HA-JNK1, MEKK1(CT), and MEKK2(CT) was determined by Western blotting (WB). The relative fold increase in JNK1 activity is shown in the upper panel. (B) HA-JNKK1 expression vector (0.5 μg/plate) was cotransfected into COS-1 cells with increasing amounts of either MEKK2(CT) or MEKK1(CT) expression vectors. HA-JNKK1 activity was measured as described in Materials and Methods. Expression levels of HA-JNKK1, MEKK1(CT), and MEKK2(CT) were determined by Western blotting (WB). The relative JNKK1 activity is shown in the upper panel. (C) Either GST-MEKK1(CT) or GST-MEKK2(CT) (each at 10 ng/plate) and Flag-JNK1 (0.5 μg/plate) expression vectors were cotransfected into COS-1 cells with expression vectors for HA-JNKK1(WT) or HA-JNKK1(AL) as indicated. Flag-JNK1 was immunoprecipitated, and its activity was determined by the immunocomplex kinase assay and quantitated by the Bio-Rad FX Imager system. Relative JNK1 activity is shown after being normalized to its expression level. Expression levels of Flag-JNK1, HA-JNKK1(WT), HA-JNKK1(AL), GST-MEKK1(CT), and GST-MEKK2(CT) were determined by Western blotting (WB).
To further address this possibility, we examined the effect of JNKK1 expression on MEKK1(CT)- and MEKK2(CT)-mediated JNK1 activation. Coexpression of JNKK1 with MEKK1 had been shown to strongly potentiate MEKK1-mediated JNK1 activation (30, 55). We found, similarly, that coexpression of JNKK1 clearly augmented JNK1 activation by MEKK1(CT) (Fig. 1C), although the JNKK1 coexpression had only a marginal effect on JNK1 activation by MEKK2 (CT) (Fig. 1C). Furthermore, overexpression of a kinase-defective JNKK1 mutant, JNKK1(AL), which has mutations of the conserved Ser-257 and Thr-261 to Ala and Leu, respectively, inhibited MEKK1(CT)- but not MEKK2(CT)-mediated JNK1 activation (Fig. 1C). While confirming the previous results that JNK1 activation by MEKK1 is mediated by JNKK1 (30, 55), these data also indicated that activation of JNK1 by MEKK2 is unlikely to be mediated by JNKK1 but may occur through a JNKK1-related MAPKK.
JNKK2 is a major effector MAPKK for MEKK2 in JNK1 activation.
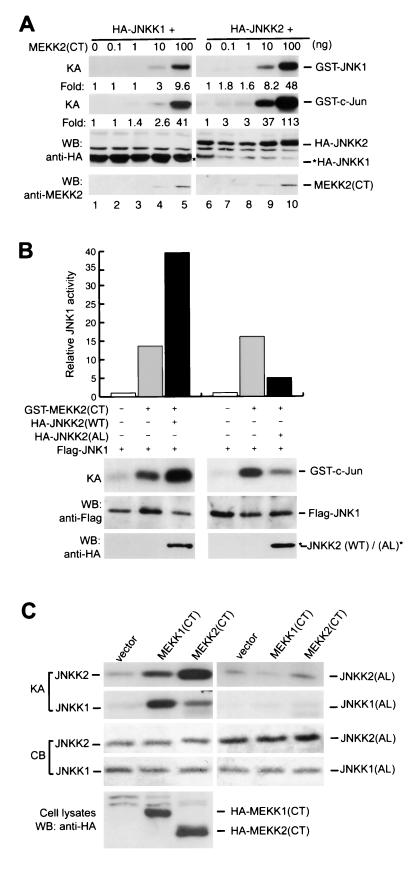
A second JNK-activating MAPKK, JNKK2/MKK7, was recently identified (15, 22, 31, 37, 49, 54, 59). JNKK2 was found not to activate ERK1 and ERK2 and, unlike JNKK1, not to activate p38. To investigate whether JNKK2 serves as the major intermediary MAPKK between MEKK2 and JNK1, we performed a coupled-kinase assay. Cotransfection of JNKK2 with MEKK2(CT) resulted in strong JNKK2 activation even at very low MEKK2(CT) expression levels (Fig. 2A). Although JNKK1 could also be activated by MEKK2(CT) in this assay, we found that almost 10 times more MEKK2(CT) expression vectors were needed (100 ng for JNKK1 activation versus 10 ng for JNKK2 activation) to obtain JNKK1 activation levels similar to those of JNKK2. These results suggested that in activating JNK1, JNKK2 may be a preferred MAPKK for MEKK2. To study this possibility more thoroughly, we examined whether coexpression of wild-type JNKK2 or the JNKK2 kinase-defective mutant JNKK2(AL) could affect MEKK2-mediated JNK1 activation. Coexpression with wild-type JNKK2 augmented JNK1 activation by MEKK2(CT), whereas coexpression with JNKK2(AL) inhibited JNK1 activation by MEKK2(CT) (Fig. 2B). Furthermore, JNKK2 was directly phosphorylated by MEKK2 at Ser-271 and Thr-275 in the SXXXT motif, a highly conserved motif in all members of the MAPKK supergene family. Mutation of these residues abolished JNKK2 phosphorylation by MEKK2 (Fig. 2C). Although JNKK2 was also phosphorylated by MEKK1(CT), this was much less efficient than the phosphorylation mediated by MEKK2(CT). In contrast, JNKK1 was more efficiently phosphorylated by MEKK1(CT) than by MEKK2(CT) on its conserved SXXXT motif (Fig. 2C). Together with the recent finding that JNKK1 but not JNKK2 is the major effector MAPKK for MEKK1 (7, 55), these results suggested that MEKK1 and MEKK2 use different effector MAPKKs in activating JNK1.
FIG. 2.
JNKK2 is a major effector of MEKK2 in JNK1 activation. (A) An increasing amount of GST-MEKK2(CT) expression vectors was cotransfected into COS-1 cells with HA-JNKK2 or HA-JNKK1 expression vectors (0.5 μg/plate each) as indicated. HA-JNKK2 and HA-JNKK1 were immunoprecipitated, and their activities were determined by a coupled-kinase assay. The relative fold increases of JNKK1, JNKK2, and JNK1 activities are indicated. Expression of HA-JNKK2 and HA-JNKK1 was determined by western blotting (WB). (B) GST-MEKK2(CT) (10 ng/plate) and Flag-JNK1 (0.5 μg/plate) expression vectors were cotransfected into COS-1 cells alone or with HA-JNKK2 or HA-JNKK2(AL) expression vectors (0.5 μg/plate) as indicated. Flag-JNK1 activity was measured as described for Fig. 1C, and the expression levels of Flag-JNK1, HA-JNKK2(WT), and HA-JNKK2(AL) were determined by western blotting (WB). Relative JNK1 activity is shown in the top panel. (C) Empty vectors or HA-MEKK1(CT) or HA-MEKK2(CT) expression vectors (0.5 μg/plate) were transfected into COS-1 cells. HA-MEKK1(CT) and HA-MEKK2(CT) were immunoprecipitated, and their activities were measured by an in vitro kinase assay with JNKK1, JNKK1(AL), JNKK2 or JNKK2(AL) as substrates as described in Materials and Methods. The amounts of JNKK1, JNKK1(AL), JNKK2, and JNKK2(AL) used in the in vitro kinase assay were shown by Coomassie blue staining (CB). Expression of HA-MEKK1(CT) and HA-MEKK2(CT) was determined by Western blotting (WB).
JNKK2 interacts with MEKK2 and JNK1.
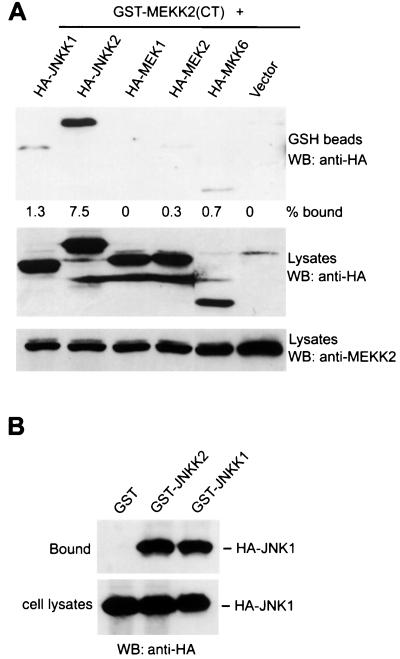
Protein-protein interactions have been suggested to play a critical role in determining the specificity of signal transduction pathways. To investigate whether such a mechanism can explain the selectivity of MEKK2 in JNK1 activation, we performed coprecipitation assays. As shown in Fig. 3A, JNKK2 coprecipitated with MEKK2(CT) about six times more efficiently than JNKK1 did, consistent with the finding that JNKK1 was activated less efficiently than JNKK2 by MEKK2. We also examined the interaction of MEKK2 with MEK1, MEK2, and MKK6 and did not observe any significant complex formation (Fig. 3A). Consistent with a report by Xia et al. (55), MEKK1 preferentially interacted with JNKK1 over other MAPKKs, including JNKK2 (data not shown). In addition to the preformed complex inside cells, JNKK2 and MEKK2 could form a complex in vitro when cell lysates prepared from COS-1 cells transfected separately with JNKK2 and MEKK2 expression vectors were mixed (data not shown). We also found, in a similar coprecipitation assay, that JNKK2, like JNKK1, could interact directly with JNK1 (Fig. 3B), confirming the finding by Holland et al. (22) and Tournier et al. (50). These results suggested that specific JNK1 activation by MEKK2 may be mediated by JNKK2-MEKK2 and JNKK2-JNK1 protein-protein interactions.
FIG. 3.
JNKK2 binds to MEKK2 and JNK1. (A) GST-MEKK2(CT) expression vectors (0.5 μg/plate) were cotransfected into COS-1 cells with 0.5 μg of HA-JNKK1, HA-JNKK2, HA-MEK1, HA-MEK2, or HA-MKK6 expression vector per plate. GST-MEKK2(CT) was precipitated with GSH-Sepharose beads, and its associated proteins were analyzed as described in Materials and Methods. Expression of HA-JNKK1, HA-JNKK1, HA-MEK1, HA-MEK2, HA-MKK6, and GST-MEKK2(CT) was measured by Western blotting (WB) as indicated. The percentages of HA-JNKK1, HA-JNKK2, HA-MEK1, HA-MEK2, and HA-MKK6 bound to GST-MEKK2(CT) were determined with a Bio-Rad G360 Imager system. (B) Equal amounts of cell lysates prepared from COS-1 cells transfected with GST, GST-JNKK1, or GST-JNKK2 vector (0.5 μg/plate) were mixed with lysates prepared from COS-1 cells transfected with the HA-JNK1 expression vector (0.5 μg/plate). The mixtures were incubated for 4 h at 4°C in a rotator, and GST, GST-JNKK1 or GST-JNKK2 was precipitated with GSH-Sepharose beads as described above. The bound and input HA-JNK1 was analyzed by Western blotting (WB).
MEKK2, JNKK2, and JNK1 form a multimolecular complex through synergistic interaction.
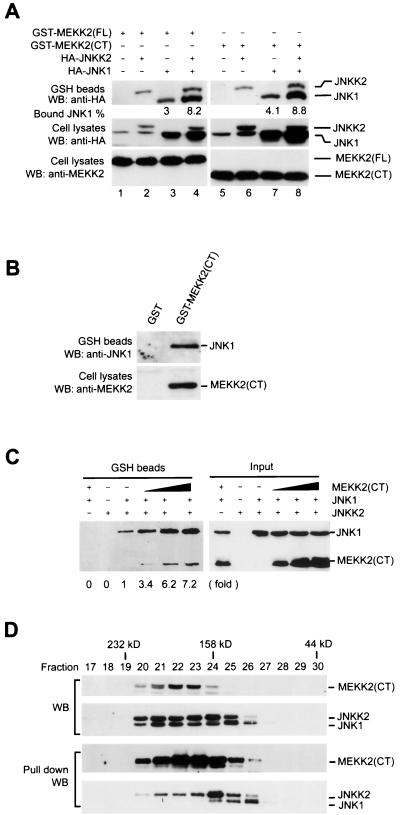
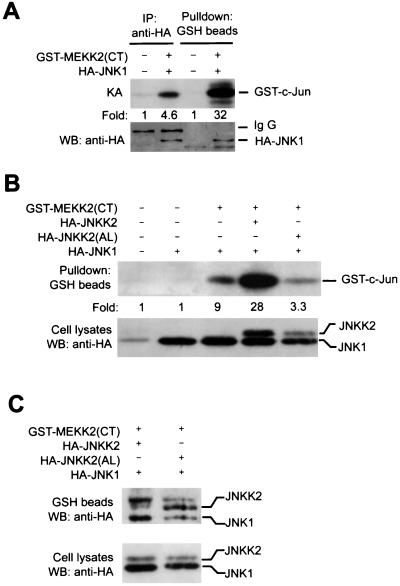
The above results provided strong evidence that JNKK2 is a major effector MAPKK for MEKK2 in JNK activation. It is not clear, however, whether MEKK2-JNKK2 and JNKK2-JNK1 interactions occur sequentially or simultaneously. Recently, sequential protein-protein interactions between MEKK1-JNKK1 and JNKK1-JNK1 were shown to underlie specific JNK activation by MEKK1 (55). MEKK1 was also shown to interact directly with JNK1 (56) and JNKK1 (T. Hunter, personal communication), independent of scaffolding molecules. No tripartite complex of MEKK1, JNKK1, and JNK1 has been demonstrated so far, however. To determine whether MEKK2, JNKK2, and JNK1 interact sequentially or simultaneously, we carried out coprecipitation assays using COS-1 cell lysates expressing variously tagged MEKK2, JNKK2, and JNK1. As shown in Fig. 4A, both full-length MEKK2 and its COOH-terminal kinase domain coprecipitated JNK1 (lanes 3 and 7). Similarly, both full-length MEKK2 and its COOH-terminal kinase domain coprecipitated JNKK2 (Fig. 3A and 4A, lanes 2 and 6). Most importantly, by using a JNK1-specific monoclonal antibody, we found that MEKK2 could coprecipitate not only the transfected HA-JNK1 but also the endogenous JNK1 (Fig. 4B). Since both full-length MEKK2 and the COOH-terminal kinase domain of MEKK2 coprecipitated JNKK2 and JNK1 equally well, the kinase domain of MEKK2 was apparently sufficient for mounting a specific interaction with its downstream components. Consistent with this, we could not detect stable interactions between the truncated N-terminal portion of MEKK2 (without the entire COOH-terminal kinase domain) and either JNKK2 or JNK1 (data not shown).
FIG. 4.
MEKK2, JNKK2, and JNK1 form a tripartite complex through synergistic interaction. (A) COS-1 cells were transfected with various combinations of expression vectors for GST-MEKK2(FL) (1 μg/plate) (lanes 1 to 4), GST-MEKK2(CT) (0.5 μg/plate) (lanes 5 to 8), HA-JNKK2 (0.5 μg/plate) (lanes 2, 4, 6, and 8), and HA-JNK1 (0.5 μg/plate) (lanes 3, 4, 7, and 8) as indicated in the figure. GST-MEKK2(FL) and GST-MEKK2(CT) were precipitated with GSH-Sepharose beads, and their associated proteins were analyzed by Western blotting (WB) as described in the legend to Fig. 3. The percentages of HA-JNK1 bound to the GST-MEKK2(CT)-containing complex were measured. Expression of HA-JNKK2 and HA-JNK1 (middle panel) and of GST-MEKK2(FL) and GST-MEKK2(CT) (bottom panel) was determined by Western blotting. (B) COS-1 cells were transfected with GST-MEKK2(CT). After 40 h, GST-MEKK2(CT) was precipitated with GSH-Sepharose beads and its associated proteins were analyzed by Western blotting (WB) using anti-JNK1 antibody. Expression of GST-MEKK2(CT) was determined by Western blotting. (C) Equal amounts of cell lysates prepared from COS-1 cells transfected with GST-JNKK2 vector were mixed with equal amounts of COS-1 cell lysates expressing HA-JNK1 and increasing amounts of HA-MEKK2(CT). The mixtures were incubated for 4 h at 4°C in a rotator, and GST-JNKK2 was precipitated with GSH-Sepharose beads as described above. JNKK2-bound and input HA-JNK1 and HA-MEKK2(CT) were analyzed by Western blotting. The relative fold increase of JNK1 coprecipitated with JNKK2 is indicated. (D) In gel filtration analysis of the MEKK2-JNKK2-JNK1 complex, cell lysates prepared from COS-1 cells transfected with GST-MEKK2(CT), HA-JNKK2, and HA-JNK1 were loaded on a Superdex-200 column and run on a Pharmacia FPLC system. Then 20 μl of each fraction was analyzed by western blotting (WB) with anti-HA antibody and anti-MEKK2 antibody for HA-JNK1, HA-JNKK2, and MEKK2 respectively (top two panels). The remains of each fractions were subjected to a coprecipitation assay with GSH-Sepharose beads as described in the legend to Fig. 3. The precipitated GST-MEKK2(CT) and GST-MEKK2(CT)-bound HA-JNKK2 and HA-JNK1 were analyzed by western blotting (bottom two panels).
In addition to coprecipitating JNKK2 and JNK1 individually, MEKK2 was found to simultaneously coprecipitate JNKK2 and JNK1 (Fig. 4A, lanes 4 and 8). Most interestingly, MEKK2 (either the full-length or the COOH-terminal kinase domain) coprecipitated both JNKK2 and JNK1 more efficiently than when they were coexpressed with either protein alone (compare lanes 2 and 3 with lane 4, and compare lanes 6 and 7 with lane 8). We consistently observed twofold increases in JNKK2 coprecipitation with MEKK2 upon JNK1 coexpression and two- to threefold increases in JNK1 coprecipitation with MEKK2 upon JNKK2 coexpression. JNK1 was coprecipitated with MEKK2 without JNKK2 cotransfection (lanes 3 and 7), but this coprecipitation may be mediated by the endogenous JNKK2. Thus, these data suggested that coexpression of JNK1 or JNKK2 could significantly affect the interaction between MEKK2 and JNKK2 or between MEKK2 and JNK1, indicating a synergistic interaction among the three kinases MEKK2, JNKK2, and JNK1.
To further investigate whether the interaction of MEKK2, JNKK2, and JNK1 was synergistic, we coprecipitated JNK1 with JNKK2 in the presence of increasing amounts of MEKK2. If a synergistic interaction occurred among MEKK2, JNKK2, and JNK1, we would expect JNK1 coprecipitation with JNKK2 to increase significantly when increasing amounts of MEKK2 were added. In contrast, if the interactions were independent, we would expect JNK1 coprecipitation with JNKK2 to decrease because of the competition of MEKK with JNKK2. As shown in Fig. 4C, while JNKK2 and JNK1 expression levels were constant in all transfections, JNK1 coprecipitation with JNKK2 increased dramatically (up to sevenfold) with the increasing amounts of MEKK2. As expected, MEKK2 also coprecipitated with the JNKK2-JNK1 complex (Fig. 4C). Together with the results described above, these data confirmed that the interaction of MEKK2, JNKK2, and JNK1 is synergistic.
To investigate whether the formation of the MEKK2-JNKK2-JNK1 complex required additional scaffolding or anchoring molecules, we carried out gel filtration experiments to determine the molecular weight of the MEKK2-JNKK2-JNK1 multimolecular complex. Cytosolic extracts were prepared from COS-1 cells transfected with GST-MEKK2(CT), HA-JNKK2, and HA-JNK1 expression vectors and subjected to gel filtration analysis using a Pharmacia FPLC system with a Superdex-200 column. Elution of MEKK2, JNKK2, and JNK1 in each fraction was monitored by Western blotting with the MEKK2-specific antibody 1128 and anti-HA tag antibody 12CA5. The MEKK2-JNKK2-JNK1 complex in each fraction was determined by the coprecipitation assay described for the experiment in Fig. 4A. As shown in Fig. 4D, while MEKK2, JNKK2, and JNK1 were coeluted in fractions with a molecular mass up to 200 kDa, the MEKK2-JNKK2-JNK1 tripartite complex was detected only in fractions corresponding to a molecular mass about 158 kDa, approximately the combined molecular masses of GST-MEKK2(CT), HA-JNKK2, and HA-JNK1. Thus, these data strongly indicated that MEKK2, JNKK2, and JNK1 can form a tripartite complex inside cells independent of scaffolding proteins. The MEKK2, JNKK2, and JNK1 that were eluted at a molecular mass larger than 158 kDa could be from different multimolecular complexes, since no MEKK2-JNKK2-JNK1 tripartite complex was detected in these fractions. Such a high-molecular-mass complex was reported previously by Moriguchi et al. (37); for example, in L5178Y cells, nearly all JNKK2/MKK7 molecules were found by a similar gel filtration assay in fractions peaking at 180 kDa.
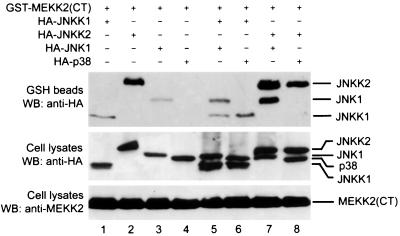
To examine whether formation of this tripartite complex is specific, we used p38, a JNK1-related MAPK, and JNKK1 as controls. Although both JNK1 and p38 were expressed at similar levels (Fig. 5, lanes 3 through 6), only JNK1 (lanes 3, 5, and 7) and not p38 (lanes 4, 6, and 8) was readily coprecipitated by MEKK2. Even when JNKK1 and JNKK2 were coexpressed, p38 could not be coprecipitated by MEKK2. This result suggested that p38 could not form a specific molecular complex with MEKK2 and JNKK2. Importantly, although JNKK1 is able to interact with and activate p38 (30, 55, 60; Cheng and Su, unpublished), its coexpression with p38 did not result in coprecipitation of p38 with MEKK2, even though MEKK2 could coprecipitate JNKK1 (lane 6). Consistent with the results shown in Fig. 3A, MEKK2 also coprecipitated JNKK1 but with a lower affinity than it coprecipitated JNKK2 (lanes 1 and 2). Importantly, this coprecipitation was not affected by the coexpression of JNK1, nor was MEKK2 coprecipitation with JNK1 affected by the expression of JNKK1 (lane 5). These results suggested that coprecipitation of JNK1 and JNKK1 by MEKK2 (lane 5) may be due to differential precipitations of JNK1 and JNKK1 by MEKK2 rather than by a MEKK2-JNKK1-JNK1 tripartite complex.
FIG. 5.
JNK1 but not p38 synergizes with JNKK2 to form a stable multimolecular complex with MEKK2. Equal amounts of COS-1 cell lysates expressing GST-MEKK2(CT) were mixed with lysates of COS-1 cells expressing HA-JNKK1 (lanes 1, 5, and 6), HA-JNKK2 (lanes 2, 7, and 8), HA-JNK1 (lanes 3, 5, and 7), and HA-p38 (lanes 4, 6 and 8) as indicated. The mixtures were incubated in a rotator at 4°C for 4 h. GST-MEKK2(CT) was precipitated by adding GSH-Sepharose beads and analyzed as described in Materials and Methods. The bound (top panel) and input (middle panel) amounts of HA-tagged proteins were determined by Western blotting (WB). The input amount of MEKK2(CT) was determined by Western blotting using an anti-MEKK2 antibody (bottom panel).
Formation of an MEKK2-JNKK2-JNK1 complex leads to efficient JNK activation.
To determine the functional significance of the MEKK2-JNKK2-JNK1 complex, we examined whether JNK1 associated with the complex was activated more strongly and efficiently than were nonassociated forms. Since only a small portion of JNK1 was found in a complex containing MEKK2 (Fig. 4), we used an anti-HA antibody to immunoprecipitate the entire pool of transiently expressed HA-JNK1 from lysates of cells in which HA-JNK1 was coexpressed with MEKK2 and compared its activity to that of JNK1 isolated by coprecipitation with MEKK2 (presumably from the complex). As shown in Fig. 6A, the JNK1 presented in the MEKK2-containing complex was much more active than that of the total-cell lysate, indicating that the JNK1 in the complex was activated more efficiently. Furthermore, we found that cotransfection of a wild-type JNKK2 caused an even greater increase of JNK1 enzymatic activity in the complex (Fig. 6B), probably because more JNK1 was recruited into the complex, as shown in Fig. 4. In contrast, when we cotransfected a JNKK2 mutant, JNKK2(AL), the JNK1 activity decreased (Fig. 6B), suggesting that the mutant could block MEKK2-activating signals in the complex and providing further evidence that JNKK2 is a major intermediate MAPKK for MEKK2 in JNK1 activation. Although wild-type JNKK2 and the mutant JNKK2(AL) were expressed equally in these transfections, a coprecipitation assay showed that more JNKK2(AL) than wild-type JNKK2 was coprecipitated by MEKK2 (Fig. 6C). This could be caused by the inability of JNKK2(AL) to dissociate from MEKK2. On the other hand, wild-type JNKK2 and JNK1 may quickly dissociate from MEKK2 when activated. By competing with wild-type JNKK2 for interaction with MEKK2 and JNK1, the JNKK2(AL) mutant blocks JNK1 activation by MEKK2 (Fig. 2 and 6B). Taken together, these results suggested that the MEKK2-JNKK2-JNK1 complex is an efficient MAPK module that is capable of strong and specific JNK1 activation.
FIG. 6.
Effective JNK1 activation in a multimolecular complex. (A) COS-1 cells were transfected with 0.1 μg of control vector or GST-MEKK2(CT) expression vector per plate, together with 0.5 μg of HA-JNK1 per plate. The cells were lysed 40 h later. Half of the lysates was used to precipitate the MEKK2(CT)-containing complex by adding GSH-Sepharose beads as described above; the other half was used to immunoprecipitate (IP) HA-JNK1 by adding the 12CA5 monoclonal antibody. The amount of HA-JNK1 precipitated by IP and GST-Sepharose beads was determined by Western blotting (WB). Equal amounts of HA-JNK1 from each precipitation were assayed for JNK activity using GST-c-Jun as substrate, as described in the legend to Fig. 1A. The relative fold increases of JNK1 activity are shown. (B) GST-MEKK2CT (10 ng/plate) and HA-JNK1 (0.5 μg/plate) expression vectors were cotransfected into COS-1 cells with expression vectors for HA-JNKK2 (0.5 μg/plate) or HA-JNKK2(AL) (0.5 μg/plate) as indicated. Cells were lysed 40 h later, and GSH-Sepharose beads were added to coprecipitate HA-JNK1 from the lysates as described in the legend to Fig. 4. The coprecipitated HA-JNK1 was assayed for kinase activity as described in the legend to Fig. 1A. Expression levels of HA-JNKK2, HA-JNKK2(AL), and HA-JNK1 were determined by Western blotting (WB). The relative fold increases of JNK1 activity are shown. (C) GST-MEKK2(CT) (0.1 μg/plate) and HA-JNK1 (0.5 μg/plate) expression vectors were cotransfected into COS-1 cells with 0.5 μg of expression vectors for HA-JNKK2 or HA-JNKK2(AL) per plate as shown. The cells were lysed 40 h later, and GSH-Sepharose beads were added to precipitate the MEKK2(CT)-containing complex. Coprecipitated HA-JNKK2, HA-JNKK2(AL), and HA-JNK1 (upper panel) and their expression levels in the cell lysates (lower panel) were determined by Western blotting (WB).
DISCUSSION
To respond to the vast number of extracellular cues, eukaryotic cells have evolved an efficient “antenna” system with specific cell surface receptors to detect distinct stimuli and a complicated intracellular network system with many protein kinase cascades to transduce and integrate the surface signals in order to activate or initiate specific cellular processes that result in cell proliferation, differentiation, or apoptosis. Many of the responses are regulated by members of the MAPK family of signal-transducing protein kinases. The recent identification of multiple MAPKs, MAPKKs, and MAPKKKs that can be expressed within a single cell raises the question of how specific extracellular cues can activate distinct repertoires of MAPK effectors (13, 24, 25, 42).
We investigated how MEKK2, a member of the MEKK/STE11 subgroup of MAPKKKs, leads to specific JNK activation. Overexpression of a modest amount of MEKK2 results in strong JNK1 activation but only marginal activation of ERK1 and ERK2 and of p38 (9; Cheng and Su, unpublished). We found that although both MEKK2 and the closely related MAPKKK MEKK1 activated JNK strongly, MEKK2 was a weaker activator of JNKK1 but a more efficient activator of JNKK2 than was MEKK1. Coexpression of JNKK2 augmented MEKK2-mediated JNK1 activation, whereas coexpression of an inactive JNKK2 mutant inhibited MEKK2-mediated JNK1 activation (Fig. 2B). In contrast, expression of either wild-type or mutant JNKK2 had only a marginal effect on MEKK1-mediated JNK1 activation (data not shown). Previous studies have shown that MEKK1 activates JNK and p38 via JNKK1, which is the preferred substrate for this MAPKKK (55). JNKK1/MKK4, but not other MAPKKs such as MEK1 and MEK2, MKK3, MKK6, and JNKK2/MKK7, is the major effector molecule directly downstream of MEKK1 (30, 55). Thus, MEKK1-JNKK1 specificity has been attributed to a direct physical interaction between the two molecules. We showed, in contrast, that MEKK2 appears to operate via JNKK2 rather than via JNKK1. Since JNKK2 is a more specific JNK kinase than is JNKK1, which is incapable of p38 activation (15, 37, 49, 59), these results also explain why MEKK2 is a less potent activator of p38 than is MEKK1 (9; Cheng and Su, unpublished).
The strong and specific JNK1 and JNKK2 activation by MEKK2 was found to be caused by a synergistic interaction of MEKK2, JNKK2, and JNK1. In both coprecipitation and gel filtration experiments, MEKK2, JNKK2, and JNK1 were found in the same complex. Most notably, the MEKK2-JNKK2-JNK1 interaction seemed synergistic, since the interaction between any two of these three kinases was further strengthened by the presence of the third. We observed two- to threefold augmentation of MEKK2-JNKK2 and MEKK2-JNK1 coprecipitation and up to sevenfold augmentation of JNKK2-JNK1 coprecipitation by expression of JNK1, JNKK2, or MEKK2, respectively (Fig. 4). Although MEKK2 could also interact with JNKK1, expression of JNK1 or JNKK1 could not affect the interaction of MEKK2-JNKK1 or MEKK2-JNK1. Furthermore, p38, a MAPK closely related to JNK1, could not form a multimolecular complex with either MEKK2-JNKK2 or MEKK2-JNKK1, even though p38 could interact with JNKK1, suggesting that the MEKK2-JNKK2-JNK1 complex is specific.
Recently, it was reported that MEKK3, a close homologue of MEKK2 (75% overall homology), also activated JNK1 via JNKK2 (10). Whereas we could not detect a tripartite complex of MEKK3, JNKK2, and JNK1 (data not shown), it remained to be determined whether the kinase domain of MEKK3 could form a tripartite complex with JNKK2 and JNK1, since the homology between the kinase domains of MEKK2 and MEKK3 is much higher than their overall homology (93 and 75%, respectively).
The synergistic interaction of MEKK2-JNKK2-JNK1 is physiologically relevant. Using an in vitro kinase assay, we found that JNK1 in the MEKK2-containing complex was activated more efficiently than JNK1 that was not associated with the complex. Its activation of JNK1 in the MEKK2-containing complex depended on the presence of a functional JNKK2, since such activation could be blocked by expression of a kinase-defective JNKK2 mutant, JNKK2(AL). Inhibition of JNK1 activation in the complex by such a mutant was not caused by the inability of JNK to bind to the complex. Interestingly, significantly more mutant JNKK2(AL) than wild-type JNKK2 was found in the MEKK2-containing complex (Fig. 6C). Such an example was also observed for STE11 and STE7 in yeast (4). This suggested that when JNKK2 and JNK1 in the MEKK2-containing complex are activated, they may quickly disassociate from MEKK2, allowing MEKK2 to phosphorylate and activate more substrates and hence amplify the upstream signals.
Direct and specific protein-protein interaction between kinases in the MAPK module has been demonstrated in both yeast and mammalian cells. Such an interaction is crucial in the formation of multimolecular signaling complexes. In mammals, MEKK1 interacts directly with its preferred MAPKK, JNKK1 (55), and with JNK1 independent of JNKK1 (56). JNKK1 and JNKK2 also interact directly with their substrates, JNK or p38 (22, 50, 55). Although the subject of speculation, a MEKKK, a MAPKK, and a MAPK have not been shown to form a tripartite complex without anchoring or scaffolding molecules in yeast and mammalian cells before. Our present study demonstrated that a mammalian MAPKKK, a MAPKK, and a MAPK could interact synergistically to form a specific signaling complex.
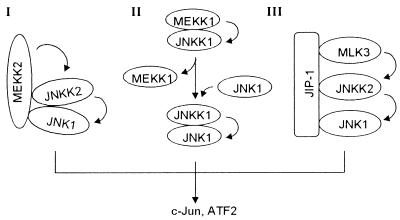
Two mammalian molecules, MP-1 and JIP-1, were shown recently to function as scaffold proteins, like STE5 in yeast, in organizing the ERK-MAPK and JNK-MAPK modules (4, 32, 39, 41, 52). MP1 was shown to significantly increase the complex formation between MEK1 and ERK1, the two kinases in the ERK MAPK module. JIP-1 was shown to bind simultaneously to MLK3/DLK, JNKK2/MKK7, and JNK1, thus organizing a JNK MAPK module. Whether MLK3/DLK, JNKK2/MKK7, and JNK1 in the JIP-1-organized kinase module could synergistically interact with one another was unclear. Studies from several groups including ours have shown that MLK3/DLK and JNKK2/MKK7 could directly interact with one another, as could JNKK2/MKK7 and JNK1 (22, 34, 50; Cheng and Su, unpublished). Such interactions may be crucial for initiating the stable MAPK module formation organized by the scaffolding protein JIP-1 (52). The interactions could also increase the fidelity of individual MAPK pathways. Although our study suggested that coordinated interaction between the kinase molecules in certain MAPK pathways could form distinct signaling modules without an anchoring or scaffolding protein, it did not rule out the possibility that additional molecules could participate in the complex under certain conditions. Their participation could provide additional levels of specificity control for each MAPK module. For instance, participation of such molecules could provide additional surface area for interaction with various upstream activating molecules or could insulate the MAPK module from cross talk with functionally unrelated kinases in the same cell. Thus, both scaffold proteins and direct kinase-kinase interaction within a MAPK module may be required to determine the specificity of the MAPK cascade. Together with the sequential-activation model (55), three distinct JNK1 MAPK modules may be responsible for efficient and specific JNK1 activation in response to distinct upstream stimuli, as illustrated in Fig. 7.
FIG. 7.
Schematic diagram of three postulated mammalian JNK1 MAPK modules.
The functional role of the putative N-terminal regulatory domain of MEKK2 is still not clear. We found that the full-length MEKK2 and COOH-terminal kinase domain of MEKK2 interact with and activate JNKK2 and JNK1 similarly, suggesting that the N-terminal region of MEKK2 may have a more distinct regulatory function than the C-terminal kinase domain. Interestingly, overexpression of the N-terminal region of MEKK2 could block MEKK2 activation of JNK1 (Cheng and Su, unpublished), suggesting that the N-terminal region of MEKK2 may interact with another, as yet unidentified molecule that regulates the activity of the MEKK2-JNKK2-JNK1 signaling complex. By using the yeast two-hybrid system, we recently isolated several candidate genes whose products seem to interact specifically with the N-terminal region of MEKK2. Studies of such interacting molecules should provide further clues to understanding the specificity and regulating mechanisms of MEKK2 and other members of the MEKK/STE11 gene family in general.
ACKNOWLEDGMENTS
We thank T. Hunter for helpful discussion and critical comments that improved the manuscript. We thank R. Davis, G. Johnson, J. Han, T. Deng, and A. Minden for expression plasmids; F. Zhang and Z. Dong for gel filtration analysis; and Sue Adams and Lore Feldman for their excellent assistance in preparation of the manuscript.
B.S. was a Special Fellow of the Leukemia Society of America. This work was partially supported by a grant from the American Association for Cancer Research (RPG-97-090-01) to B.S.
Jinke Cheng and Jianhua Yang contributed equally to this work.
REFERENCES
- 1.Bardwell L, Cook J G, Chang E C, Cairns B R, Thorner J. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blank J L, Gerwins P, Elliott E M, Sather S, Johnson G L. Molecular cloning of mitogen-activated protein/ERK kinase kinases (MEKK) 2 and 3. Regulation of sequential phosphorylation pathways involving mitogen-activated protein kinase and c-Jun kinase. J Biol Chem. 1996;271:5361–5368. doi: 10.1074/jbc.271.10.5361. [DOI] [PubMed] [Google Scholar]
- 3.Boulton T G, Nye S H, Robbins D J, Ip N Y, Radziejewska E, Morgenbesser S D, DePinho R A, Panayotatos N, Cobb M H, Yancopoulos G D. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell. 1991;65:663–675. doi: 10.1016/0092-8674(91)90098-j. [DOI] [PubMed] [Google Scholar]
- 4.Choi K Y, Satterberg B, Lyons D M, Elion E A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 5.Cobb M H. MAP kinase pathways. Prog Biophys Mol Biol. 1999;71:479–500. doi: 10.1016/s0079-6107(98)00056-x. [DOI] [PubMed] [Google Scholar]
- 6.Cobb M H, Goldsmith E J. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 7.Cuenda A, Dorow D S. Differential activation of stress-activated protein kinase kinases SKK4/MKK7 and SKK1/MKK4 by the mixed-lineage kinase-2 and mitogen-activated protein kinase kinase (MKK) kinase-1. Biochem J. 1998;333:11–15. doi: 10.1042/bj3330011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis R J. Signal transduction by the c-Jun N-terminal kinase. Biochem Soc Symp. 1999;64:1–12. doi: 10.1515/9781400865048.1. [DOI] [PubMed] [Google Scholar]
- 9.Deacon K, Blank J L. Characterization of the mitogen-activated protein kinase kinase 4 (MKK4)/c-Jun NH2-terminal kinase 1 and MKK3/p38 pathways regulated by MEK kinases 2 and 3. MEK kinase 3 activates MKK3 but does not cause activation of p38 kinase in vivo. J Biol Chem. 1997;272:14489–14496. doi: 10.1074/jbc.272.22.14489. [DOI] [PubMed] [Google Scholar]
- 10.Deacon K, Blank J L. MEK kinase 3 directly activates MKK6 and MKK7, specific activators of the p38 and c-Jun NH2-terminal kinases. J Biol Chem. 1999;274:16604–16610. doi: 10.1074/jbc.274.23.16604. [DOI] [PubMed] [Google Scholar]
- 11.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 12.Derijard B, Raingeaud J, Barrett T, Wu I H, Han J, Ulevitch R J, Davis R J. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. . (Erratum, 269:17, 1995.) [DOI] [PubMed] [Google Scholar]
- 13.Elion E A. Routing MAP kinase cascades. Science. 1998;281:1625–1626. doi: 10.1126/science.281.5383.1625. [DOI] [PubMed] [Google Scholar]
- 14.Fanger G R, Gerwins P, Widmann C, Jarpe M B, Johnson G L. MEKKs, GCKs, MLKs, PAKs, TAKs, and tpls: upstream regulators of the c-Jun amino-terminal kinases? Curr Opin Genet Dev. 1997;7:67–74. doi: 10.1016/s0959-437x(97)80111-6. [DOI] [PubMed] [Google Scholar]
- 15.Foltz I N, Gerl R E, Wieler J S, Luckach M, Salmon R A, Schrader J W. Human mitogen-activated protein kinase kinase 7 (MKK7) is a highly conserved c-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) activated by environmental stresses and physiological stimuli. J Biol Chem. 1998;273:9344–9351. doi: 10.1074/jbc.273.15.9344. [DOI] [PubMed] [Google Scholar]
- 16.Garrington T P, Johnson G L. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 17.Gerwins P, Blank J L, Johnson G L. Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4, that selectively regulates the c-Jun amino terminal kinase pathway. J Biol Chem. 1997;272:8288–8295. doi: 10.1074/jbc.272.13.8288. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Lee J D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 19.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 20.Hill C S, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 21.Hirai S, Noda K, Moriguchi T, Nishida E, Yamashita A, Deyama T, Fukuyama K, Ohno S. Differential activation of two JNK activators, MKK7 and SEK1, by MKN28-derived nonreceptor serine/threonine kinase/mixed lineage kinase 2. J Biol Chem. 1998;273:7406–7412. doi: 10.1074/jbc.273.13.7406. [DOI] [PubMed] [Google Scholar]
- 22.Holland P M, Suzanne M, Campbell J S, Noselli S, Cooper J A. MKK7 is a stress-activated mitogen-activated protein kinase kinase functionally related to hemipterous. J Biol Chem. 1997;272:24994–24998. doi: 10.1074/jbc.272.40.24994. [DOI] [PubMed] [Google Scholar]
- 23.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 24.Ip Y T, Davis R J. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 25.Karin M. Mitogen-activated protein kinase cascades as regulators of stress responses. Ann N Y Acad Sci. 1998;851:139–146. doi: 10.1111/j.1749-6632.1998.tb08987.x. [DOI] [PubMed] [Google Scholar]
- 26.Karin M, Hunter T. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- 27.Lange-Carter C A, Pleiman C M, Gardner A M, Blumer K J, Johnson G L. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 28.Lawler S, Fleming Y, Goedert M, Cohen P. Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Curr Biol. 1998;8:1387–1390. doi: 10.1016/s0960-9822(98)00019-0. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Jiang Y, Ulevitch R J, Han J. The primary structure of p38 gamma: a new member of p38 group of MAP kinases. Biochem Biophys Res Commun. 1996;228:334–340. doi: 10.1006/bbrc.1996.1662. [DOI] [PubMed] [Google Scholar]
- 30.Lin A, Minden A, Martinetto H, Claret F X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 31.Lu X, Nemoto S, Lin A. Identification of c-Jun NH2-terminal protein kinase (JNK)-activating kinase 2 as an activator of JNK but not p38. J Biol Chem. 1997;272:24751–24754. doi: 10.1074/jbc.272.40.24751. [DOI] [PubMed] [Google Scholar]
- 32.Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 34.Merritt S E, Mata M, Nihalani D, Zhu C, Hu X, Holzman L B. The mixed lineage kinase DLK utilizes MKK7 and not MKK4 as substrate. J Biol Chem. 1999;274:10195–10202. doi: 10.1074/jbc.274.15.10195. [DOI] [PubMed] [Google Scholar]
- 35.Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochim Biophys Acta. 1997;1333:F85–F104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 36.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 37.Moriguchi T, Toyoshima F, Masuyama N, Hanafusa H, Gotoh Y, Nishida E. A novel SAPK/JNK kinase, MKK7, stimulated by TNFalpha and cellular stresses. EMBO J. 1997;16:7045–7053. doi: 10.1093/emboj/16.23.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 39.Printen J A, Sprague G F., Jr Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 41.Schaeffer H J, Catling A D, Eblen S T, Collier L S, Krauss A, Weber M J. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 42.Schlesinger T K, Fanger G R, Yujiri T, Johnson G L. The TAO of MEKK. Front Biosci. 1998;3:D1181–D1186. doi: 10.2741/a354. [DOI] [PubMed] [Google Scholar]
- 43.Seger R, Krebs E G. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 44.Stein B, Yang M X, Young D B, Janknecht R, Hunter T, Murray B W, Barbosa M S. p38-2, a novel mitogen-activated protein kinase with distinct properties. J Biol Chem. 1997;272:19509–19517. doi: 10.1074/jbc.272.31.19509. [DOI] [PubMed] [Google Scholar]
- 45.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 46.Su B, Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr Opin Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 47.Teramoto H, Coso O A, Miyata H, Igishi T, Miki T, Gutkind J S. Signaling from the small GTP-binding proteins Rac1 and Cdc42 to the c-Jun N-terminal kinase/stress-activated protein kinase pathway. A role for mixed lineage kinase 3/protein-tyrosine kinase 1, a novel member of the mixed lineage kinase family. J Biol Chem. 1996;271:27225–27228. doi: 10.1074/jbc.271.44.27225. [DOI] [PubMed] [Google Scholar]
- 48.Tibbles L A, Ing Y L, Kiefer F, Chan J, Iscove N, Woodgett J R, Lassam N J. MLK-3 activates the SAPK/JNK and p38/RK pathways via SEK1 and MKK3/6. EMBO J. 1996;15:7026–7035. [PMC free article] [PubMed] [Google Scholar]
- 49.Tournier C, Whitmarsh A J, Cavanagh J, Barrett T, Davis R J. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc Natl Acad Sci USA. 1997;94:7337–7342. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tournier C, Whitmarsh A J, Cavanagh J, Barrett T, Davis R J. The MKK7 gene encodes a group of c-Jun NH2-terminal kinase kinases. Mol Cell Biol. 1999;19:1569–1581. doi: 10.1128/mcb.19.2.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X S, Diener K, Jannuzzi D, Trollinger D, Tan T H, Lichenstein H, Zukowski M, Yao Z. Molecular cloning and characterization of a novel protein kinase with a catalytic domain homologous to mitogen-activated protein kinase kinase kinase. J Biol Chem. 1996;271:31607–31611. doi: 10.1074/jbc.271.49.31607. [DOI] [PubMed] [Google Scholar]
- 52.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 53.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 54.Wu Z, Wu J, Jacinto E, Karin M. Molecular cloning and characterization of human JNKK2, a novel Jun NH2-terminal kinase-specific kinase. Mol Cell Biol. 1997;17:7407–7416. doi: 10.1128/mcb.17.12.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia Y, Wu Z, Su B, Murray B, Karin M. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 1998;12:3369–3381. doi: 10.1101/gad.12.21.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu S, Cobb M H. MEKK1 binds directly to the c-Jun N-terminal kinases/stress-activated protein kinases. J Biol Chem. 1997;272:32056–32060. doi: 10.1074/jbc.272.51.32056. [DOI] [PubMed] [Google Scholar]
- 57.Xu S, Robbins D J, Christerson L B, English J M, Vanderbilt C A, Cobb M H. Cloning of rat MEK kinase 1 cDNA reveals an endogenous membrane-associated 195-kDa protein with a large regulatory domain. Proc Natl Acad Sci USA. 1996;93:5291–5295. doi: 10.1073/pnas.93.11.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 59.Yang J, New L, Jiang Y, Han J, Su B. Molecular cloning and characterization of a human protein kinase that specifically activates c-Jun N-terminal kinase. Gene. 1998;212:95–102. doi: 10.1016/s0378-1119(98)00158-9. [DOI] [PubMed] [Google Scholar]
- 60.Zanke B W, Rubie E A, Winnett E, Chan J, Randall S, Parsons M, Boudreau K, McInnis M, Yan M, Templeton D J, Woodgett J R. Mammalian mitogen-activated protein kinase pathways are regulated through formation of specific kinase-activator complexes. J Biol Chem. 1996;271:29876–29881. doi: 10.1074/jbc.271.47.29876. [DOI] [PubMed] [Google Scholar]
- 61.Zhou G, Bao Z Q, Dixon J E. Components of a new human protein kinase signal transduction pathway. J Biol Chem. 1995;270:12665–12669. doi: 10.1074/jbc.270.21.12665. [DOI] [PubMed] [Google Scholar]