Abstract
Many Isaria-like species have recently been moved into more appropriate genera. However, more robust molecular phylogenetic analyses are still required for Isaria-like fungi to ensure accurate taxonomic identification. We analyzed these Isaria-like strains using multi-gene phylogenetics. Cryptic diversity was discovered in several Isaria farinosa strains, and two new species, Samsoniella pseudogunnii and S. pupicola, are proposed. Our results reveal that more attention needs to be paid to cryptic intraspecific diversity across different isolates and genotypes of the Isaria-like species, some of which will need to be transferred to Samsoniella. Interestingly, S. hepiali, with a very broad host distribution, has been widely used as a medicinal and edible cordycipitoid fungus.
Keywords: cryptic diversity, intraspecific, Isaria-like, multi-gene analysis
1. Introduction
The genus Isaria was originally establish based on the species Isaria terrestris Fr. [1]. Brown and Smith [2] transferred some species described in Isaria Pers. and Spicaria Harting into Paecilomyces, which possess a conidiogenous structure similar to that of Paecilomyces variotii Bainier. de Hoog [3] redescribed the genus Isaria and chose Isaria felina (DC.) Fr. as the lectotype. Typical characteristics include denticulate conidiogenous cells without elongation that arise in clusters from subtending cells or are solitarily from undifferentiated hyphae; mostly present synnemata; and globose, ellipsoidal, or subcylindrical conidia, mostly with a rounded base [3]. Samson [4] divided the genus Paecilomyces into two sections and all entomogenous species were placed in the section Isarioidea. Hodge et al. [5] reintroduced the genus Isaria with the type species Isaria farinosa (Holmsk.) Fr. and most entomopathogenic mesophilic Paecilomyces species were transferred to Isaria (Hypocreales, Clavicipitaceae) [6,7,8].
Kepler et al. [9] proposed the rejection of Isaria in favor of Cordyceps and transferred Isaria species into Cordyceps. Mongkolsamrit et al. [10] introduced some Isaria-like species and the new genus Samsoniella Mongkols., Noisrip., Thanakitp., Spatafora, and Luangsa-ard. Chen et al. [11,12] reported four Isaria-like species: Akanthomyces araneogenus Z.Q. Liang, W.H. Chen, and Y.F. Han; Samsoniella coleopterorum W.H. Chen, Y.F. Han, and Z.Q. Liang; Samsoniella hymenopterorum W.H. Chen, Y.F. Han, and Z.Q. Liang; and Samsoniella lepidopterorum W.H. Chen, Y.F. Han, and Z.Q. Liang. Currently, many species previously placed in the genus Isaria have been transferred to more appropriate genera. However, robust molecular phylogenetic analyses are still needed for Isaria-like fungi to ensure accurate taxonomic identification with comparable results across different isolates and genotypes [10].
We previously collected many Isaria-like morphs of invertebrate-pathogenic fungi from Guizhou Province, China. Some demonstrated close phylogenetic relationships with Isaria farinosa (Holmsk.) Fr. based on the analysis of associated ITS sequences. In the present study, we applied multi-gene (ITS, LSU, RPB1, RPB2, TEF) phylogenetic analysis to reevaluate the taxonomic position of these strains, as well as the cryptic diversity among the different isolates of I. farinosa, and to describe new taxa to accommodate the cryptic diversity of Isaria-like fungi.
2. Materials and Methods
2.1. Fungal Materials and Identification
The strains used in this study were isolated from infected insect and spider specimens collected in different areas of Guizhou Province, China, including Dali Forest in Rongjiang County, Yaorenshan National Forest Park in Sandu County, Mount Fanjing in Yinjiang County, Tongmuling in Guiyang City, and Doupengshan in Duyun City. Isolation of strains was conducted as described by Chen et al. [13]. Fungal colonies emerging from specimens were isolated and cultured at 25 °C for 14 days under 12 h light/12 h dark conditions following protocols described by Zou et al. [14]. Accordingly, the living isolates were obtained. The specimens and the isolated strains were deposited in the Institute of Fungus Resources, Guizhou University (formally Herbarium of Guizhou Agricultural College; code, GZAC), Guiyang City, Guizhou, China.
Macroscopic and microscopic morphological characteristics of the fungal isolates were examined, especially for the arrangement, shape, and measurement of phialides and conidia, and also the growth rates of cultures incubated at 25 °C for 14 days were determined in Potato Dextrose Agar (PDA) (Potato powder 6%, Agar 20%, Glucose 20%, Beijing Solarbio Technology Co., Ltd., China). Hyphae and conidiogenous structures were mounted in lactophenol cotton blue or 20% lactate solution and observed with an optical microscope (OM, DM4 B, Leica, Germany).
2.2. DNA Extraction, Polymerase Chain Reaction Amplification and Nucleotide Sequencing
DNA extraction was carried out with a fungal genomic DNA extraction kit (DP2033, BioTeke Corporation) in accordance with Liang et al. [15]. The extracted DNA was stored at −20 °C. The amplification of the internal transcribed spacer (ITS) region, the large subunit ribosomal RNA (LSU) gene, the RNA polymerase II largest subunit 1 (RPB1), the RNA polymerase II largest subunit 2 (RPB2), and the translation elongation factor 1 alpha (TEF) by PCR was described by White et al. [16], Rakotonirainy et al. [17], Castlebury et al. [18], and van den Brink et al. [19], respectively. PCR reactions for five loci of all strains were performed in a total volume of 25 μL containing 12.5 μL 2× PowerTaq PCR Master Mix (Tiangen Biotech (Beijing) Co., LTD, China), 1 μL of each primer (10 μM), 1 μL of genomic DNA (20–100 ng), and 9.5 μL of sterile water. Primer sequence information is shown in Table 1. PCR products were purified and sequenced at Sangon Biotech (Shanghai) Co. The resulting sequences were submitted to GenBank (the accession number is shown in Table 2).
Table 1.
Primers information for 5-locus DNA sequences.
| Name | Length | Direction | Sequence 5′-3′ | Optimised PCR Protocols | References | |
|---|---|---|---|---|---|---|
| ITS | ITS5 | 22 | forward | GGAAGTAAAAGTCGTAACAAGG | (95 °C: 30 s, 51 °C: 50 s, 72 °C: 45 s) × 33 cycles | [16] |
| ITS4 | 20 | reverse | TCCTCCGCTTATTGATATGC | |||
| LSU | LROR | 17 | forward | ACCCGCTGAACTTAAGC | (94 °C: 30 s, 51 °C: 1 min, 72 °C: 2 min) × 33 cycles | [17] |
| LR5 | 17 | reverse | TCCTGAGGGAAACTTCG | |||
| RPB1 | CRPB1 | 20 | forward | CAYCCWGGYTTYATCAAGAA | (94 °C: 30 s, 55 °C: 30 s, 72 °C: 1 min) × 33 cycles | [18] |
| RPB1Cr | 23 | reverse | CCNGCDATNTCRTTRTCCATRTA | |||
| RPB2 | RPB2-5F3 | 20 | forward | GACGACCGTGATCACTTTGG | (94 °C: 30 s, 54 °C: 40 s, 72 °C: 1 min 20 s) × 33 cycles | [19] |
| RPB2-7Cr2 | 20 | reverse | CCCATGGCCTGTTTGCCCAT | |||
| TEF | 983F | 23 | forward | GCYCCYGGHCAYCGTGAYTTYAT | (94 °C: 30 s, 58 °C: 1 min 20 s, 72 °C: 1 min) × 33 cycles | [19] |
| 2218R | 23 | reverse | ATGACACCRACRGCRACRGTYTG | |||
Table 2.
List of strains and GenBank accession numbers of sequences used in this study.
| Species | Strain No. | Host/ Substratum | GenBank Accession No. | ||||
|---|---|---|---|---|---|---|---|
| ITS | LSU | RPB1 | RPB2 | TEF | |||
| Akanthomyces aculeatus | HUA 772 | Lepidoptera; Sphingidae | - | KC519370 | - | - | KC519366 |
| A. attenuates | CBS 402.78 | Leaf litter (Acer saccharum) | - | AF339565 | EF468888 | EF468935 | EF468782 |
| A. coccidioperitheciatus | NHJ 6709 | Araneae (Spider) | - | EU369042 | EU369067 | - | EU369025 |
| A. farinosa | CBS 541.81 | - | AY624180 | MF416553 | MF416655 | MF416449 | JQ425686 |
| A. tuberculatus | BCC 16819 | Lepidoptera (Adult moth) | - | GQ249987 | - | - | GQ250037 |
| A. tuberculatus | OSC 111002 | Lepidoptera | - | DQ518767 | DQ522384 | - | DQ522338 |
| Ascopolyporus polychrous | P.C. 546 | Plant | - | DQ118737 | DQ127236 | - | DQ118745 |
| A. villosus | ARSEF 6355 | Plant | - | AY886544 | DQ127241 | - | DQ118750 |
| Beauveria bassiana | ARSEF 1564 | Lepidoptera; Arctiidae | - | - | HQ880833 | HQ880905 | HQ880974 |
| B. brongniartii | ARSEF 617 | Coleoptera; Scarabaeidae | - | AB027381 | HQ880854 | HQ880926 | HQ880991 |
| B. brongniartii | BCC 16585 | Coleoptera (Anomala cuprea) | JN049867 | JF415967 | JN049885 | JF415991 | JF416009 |
| B. caledonica | ARSEF 2567 | Soil | - | AF339520 | HQ880889 | HQ880961 | EF469057 |
| Bionectria ochroleuca | AFTOL-ID187 | - | - | DQ862027 | - | DQ862013 | DQ862029 |
| B. vericulosa | HMAS 183151 | Plant | HM050304 | HM050302 | - | - | - |
| Blackwellomyces cardinalis | OSC 93609 | Lepidoptera; Tineidae (Larva) | - | AY184962 | DQ522370 | - | DQ522325 |
| B. cardinalis | OSC 93610 | Lepidoptera; Tineidae (Larva) | - | AY184963 | EF469088 | - | EF469059 |
| B. pseudomilitaris | NBRC 101409 | Lepidoptera (Larva) | - | JN941393 | JN992482 | - | - |
| B. pseudomilitaris | NBRC 101410 | Lepidoptera (Larva) | - | JN941394 | JN992481 | - | - |
| Calcarisporium arbuscula | CBS 221.73 | - | AY271809 | - | - | - | - |
| C. arbuscula | CBS 900.68 | Hymenomycetes (Agarics sp.) | KT945003 | KX442598 | - | KX442597 | KX442596 |
| C. cordycipiticola | CGMCC 3.17904 | Cordycipitaceae (Cordyceps militaris) | KT945001 | KX442604 | - | KX442607 | KX442605 |
| C. cordycipiticola | CGMCC 3.17905 | Cordycipitaceae (Cordyceps militaris) | KT944999 | KX442599 | - | KX442594 | KX442593 |
| C. xylariicola | HMAS 276836 | Xylariaceae (Xylaria sp.) | KX442603 | KX442601 | - | KX442606 | KX442595 |
| Calonectria ilicicola | CBS 190.50 | Plant | GQ280605 | GQ280727 | - | KM232307 | AY725726 |
| Cephalosporium curtipes | CBS 154.61 | Uredinales (Hemileia vastatrix) | AJ292404 | AF339548 | - | EF468947 | EF468802 |
| Claviceps fusiformis | ATCC 26019 | Poaceae | JN049817 | - | - | - | DQ522320 |
| Clonostachys rosea | GJS 90-227 | Plant | - | AY489716 | - | - | AY489611 |
| Cocoonihabitus sinensis | HMAS 254523 | Saturniidae (Cocoon) | KY924870 | KY924869 | - | - | - |
| C. sinensis | HMAS 254524 | Saturniidae (Cocoon) | MF687395 | MF687396 | - | - | - |
| Cordyceps amoene-rosea | CBS 107.73 | Coleoptera (Pupa) | MH860646 | MH872342 | MF416651 | - | - |
| C. bifusispora | EFCC 5690 | Lepidoptera (Pupa) | - | EF468806 | EF468854 | EF468909 | EF468746 |
| C. cateniannulata | CBS 152.83 | Coleoptera (Adult) | NR_111169 | NG_067333 | - | - | - |
| C. cateniobliqua | CBS 153.83 | Lepidoptera (Adoxophyesprivatana) | NR_111170 | - | - | - | JQ425688 |
| C. cf. farinosa | OSC 111004 | Lepidoptera (Pupa) | - | EF468840 | EF468886 | - | EF468780 |
| C. coleopterorum | CBS 110.73 | Coleoptera (Larva) | AY624177 | JF415988 | JN049903 | JF416006 | JF416028 |
| C. farinosa | CBS 111113 | - | - | MF416554 | MF416656 | MF416450 | MF416499 |
| C. fumosorosea | CBS 107.10 | - | - | MF416556 | MF416659 | MF416453 | MF416502 |
| C. militaris | OSC 93623 | Lepidoptera (Pupa) | - | AY184966 | DQ522377 | AY545732 | DQ522332 |
| Dactylonectria alcacerensis | CBS 129087 | Plant (Vitis vinifera) | JF735333 | KM231629 | - | - | JF735819 |
| Elaphocordyceps ophioglossoides | NBRC106332 | - | JN943322 | JN941409 | - | - | - |
| E. paradoxa | NBRC 106958 | - | JN943324 | JN941411 | - | - | - |
| Engyodontium aranearum | CBS 309.85 | Araneae (Spider) | - | AF339526 | DQ522387 | DQ522439 | DQ522341 |
| Epichloë typhina | ATCC 56429 | Poaceae (Festuca rubra) | JN049832 | U17396 | - | DQ522440 | AF543777 |
| Flammocladiella aceris | CPC 24422 | Plant (Acer platanoides) | KR611883 | KR611901 | - | - | - |
| Flavocillium bifurcatum | YFCC 6101 | Noctuidae (Larva) | - | MN576781 | MN576841 | MN576897 | MN576951 |
| Fusarium circinatum | CBS 405.97 | - | U61677 | - | - | JX171623 | KM231943 |
| F. subluratum | CBS 189.34 | Soil | HQ897830 | KM231680 | - | - | - |
| Gelasinospora tetrasperma | AFTOL-ID 1287 | - | - | DQ470980 | - | DQ470932 | DQ471103 |
| Gibellula longispora | NHJ 12014 | Araneae (Spider) | - | - | EU369055 | - | EU369017 |
| G. pulchra | NHJ 10808 | Araneae (Spider) | - | EU369035 | EU369056 | - | EU369018 |
| G. ratticaudata | ARSEF 1915 | Araneae (Spider) | - | DQ518777 | DQ522408 | - | DQ522360 |
| Haptocillium sinense | CBS 567.95 | Nematode | AJ292417 | AF339545 | - | - | - |
| Harposporium harposporiferum | ARSEF 5472 | - | - | NG_060621 | - | - | - |
| Hevansia arachnophile | NHJ 10469 | Araneae (Spider) | - | EU369031 | EU369047 | - | EU369008 |
| H. cinerea | NHJ 3510 | Araneae (Spider) | - | - | EU369048 | - | EU369009 |
| H. nelumboides | BCC 41864 | Araneae (Spider) | JN201871 | JN201873 | - | - | JN201867 |
| H. novoguineensis | NHJ 11923 | Araneae (Spider) | - | EU369032 | EU369052 | - | EU369013 |
| Hyperdermium pulvinatum | P.C. 602 | Hemiptera (Scale insect) | - | DQ118738 | DQ127237 | - | DQ118746 |
| Hydropisphaera erubescens | ATCC 36093 | - | - | AF193230 | - | AY545731 | DQ518174 |
| H. peziza | GJS 92-101 | Plant (Bark) | - | AY489730 | - | - | AY489625 |
| Hypocrea americana | AFTOL-ID 52 | - | DQ491488 | AY544649 | - | - | DQ471043 |
| H. lutea | ATCC 208838 | On decorticated conifer wood | - | AF543791 | - | DQ522446 | AF543781 |
| H. rufa | DAOM JBT1003 | - | JN942883 | JN938865 | |||
| H. discoidea | BCC 8237 | - | JN049840 | DQ384937 | - | DQ452461 | DQ384977 |
| Hypomyces polyporinus | ATCC 76479 | - | - | AF543793 | - | - | AF543784 |
| Isaria farinosa | CEP 004 | Soil | JN998783 | - | - | - | JN998763 |
| I. farinosa | CEP 005 | Soil | JN998784 | - | - | - | JN998764 |
| I. farinosa | CEP 029 | Trialeurodes vaporariorum | JN998785 | - | - | - | JN998765 |
| I. farinosa | OSC 111005 | Lepidoptera (Pupa) | - | DQ518772 | DQ522394 | - | DQ522348 |
| I. farinosa | OSC 111006 | Lepidoptera (Pupa) | - | EF469080 | EF469094 | - | EF469065 |
| I. farinosa | OSC 111007 | Lepidoptera (Pupa) | - | DQ518773 | DQ522395 | DQ522449 | DQ522349 |
| Lecanicillium antillanum | CBS 350.85 | Hymenomycetes (Agaric sp.) | - | AF339536 | DQ522396 | DQ522450 | DQ522350 |
| L. attenuatum | CBS 402.78 | Leaf litter of Acer saccharum | - | AF339565 | EF468888 | EF468935 | EF468782 |
| L. aranearum | CBS 726.73a | Arachnida (Spider) | - | AF339537 | EF468887 | EF468934 | EF468781 |
| L. fusisporum | CBS 164.70 | Hymenomycetes (Coltricia perennis) | - | AF339549 | EF468889 | - | EF468783 |
| L. psalliotae | CBS 367.86 | Puccinia graminis | - | KM283800 | - | - | KM283823 |
| L. lecanii | CBS101247 | Hemiptera (Coccus viridis) | JN049836 | KM283794 | - | KM283859 | DQ522359 |
| Leptobacillium chinense | LC 1345 | submerged wood | - | JQ410322 | - | - | - |
| L. coffeanum | CDA 734 | Plant (Coffea arabica) | - | MF066032 | - | - | - |
| Liangia sinensis | YFCC 3103 | Fungi (Beauveria yunnanensis) | - | MN576782 | MN576842 | MN576898 | MN576952 |
| L. sinensis | YFCC 3104 | Fungi (Beauveria yunnanensis) | - | MN576783 | MN576843 | MN576899 | MN576953 |
| Metapochonia goniodes | CBS 891.72 | Fungi | AJ292409 | AF339550 | DQ522401 | DQ522458 | DQ522354 |
| Myrotheciomyces corymbiae | CPC 33206 | Plant (Corymbia variegata) | NR_160351 | NG_064542 | - | - | - |
| Myrothecium inundatum | IMI 158855 | Hymenomycetes (Russula nigricans) | - | AY489731 | - | - | AY489626 |
| M. roridum | ATCC 16297 | Soil | - | AY489708 | - | - | AY489603 |
| M. verrucaria | ATCC 9095 | Plant (Gossypium sp.) | - | AY489713 | - | - | AY489608 |
| Nectira cinnabarina | CBS 125165 | Plant (Aesculus sp.) | HM484548 | HM484562 | - | KM232402 | HM484527 |
| N. nigrescens | CBS 125148 | Plant (Dicotyledonous tree) | HM484707 | HM484720 | - | KM232403 | HM484672 |
| Nectriopsis violacea | CBS 424.64 | Fungi (Fuligo sp.) | - | AY489719 | - | - | - |
| Neobarya parasitica | Marson s/n | Fungi (Bertia moriformis) | KP899626 | KP899626 | - | - | - |
| Neonectria candida | CBS 151.29 | Plant (Malus sylvestris) | JF735313 | AY677333 | - | - | JF735791 |
| N. faginata | CBS 217.67 | - | HQ840385 | HQ840382 | - | DQ789797 | JF268746 |
| N. neomacrispora | CBS 118984 | - | HQ840388 | HQ840379 | - | DQ789810 | JF268754 |
| N. ramulariae | CBS 182.36 | - | HM054157 | HM042435 | - | DQ789793 | HM054092 |
| Neurospora crassa | ICMP 6360 | - | AY681193 | AY681158 | - | - | - |
| Niesslia exilis | CBS 560.74 | - | - | AY489720 | - | - | AY489614 |
| Ophiocordyceps heteropoda | EFCC 10125 | Cicadidae (Tibicen bihamatus) | JN049852 | EF468812 | - | EF468914 | EF468752 |
| O. sinensis | EFCC 7287 | Lepidoptera (Ghostmoth) | JN049854 | EF468827 | - | EF468924 | EF468767 |
| O. stylophor | OSC 111000 | Insect (Larvae) | JN049828 | DQ518766 | - | DQ522433 | DQ522337 |
| Peethambara spirostriata | CBS 110115 | Plant (Theobroma cacao) | - | AY489724 | - | EF692516 | AY489619 |
| Purpureocillium lilacinum | CBS 284.36 | Soil | - | AY624227 | EF468898 | EF468941 | EF468792 |
| P. lilacinum | CBS 431.87 | Nematoda (Meloidogyne sp.) | HQ842812 | EF468844 | EF468897 | EF468940 | EF468791 |
| Rosasphaeria moravica | LMM | - | JF440985 | - | - | JF440986 | JF440987 |
| Roumegueriella rufula | GJS 91-64 | - | - | EF469082 | - | EF469116 | EF469070 |
| R. rufula | CBS 346.85 | - | - | DQ518776 | - | DQ522461 | DQ522355 |
| Samsoniella alboaurantium | CBS 240.32 | Lepidoptera (Pupa) | - | JF415979 | JN049895 | JF415999 | JF416019 |
| S. alboaurantium | CBS 262.58 | Soil | - | AB080087 | MF416654 | MF416448 | MF416497 |
| S. alpina | YFCC 5818 | Hepialidae (Hepialus baimaensis) | - | MN576809 | MN576869 | MN576923 | MN576979 |
| S. alpina | YFCC 5831 | Hepialidae (Hepialus baimaensis) | - | MN576810 | MN576870 | MN576924 | MN576980 |
| S. antleroides | YFCC 6016 | Noctuidae (Larvae) | - | MN576803 | MN576863 | MN576917 | MN576973 |
| S. antleroides | YFCC 6113 | Noctuidae (Larvae) | - | MN576804 | MN576864 | MN576918 | MN576974 |
| S. aurantia | TBRC 7271 | Lepidoptera | - | MF140728 | MF140791 | MF140818 | MF140846 |
| S. aurantia | TBRC 7272 | Lepidoptera | - | MF140727 | - | MF140817 | MF140845 |
| S. aurantia | DY10951 | Lepidoptera (Pupa) | MZ827667 | MZ827827 | - | - | MZ855229 |
| S. aurantia | DY10952 | Lepidoptera (Pupa) | MZ827666 | MZ827084 | - | - | MZ855230 |
| S. cardinalis | YFCC 5830 | Limacodidae (Pupa) | - | MN576788 | MN576848 | MN576902 | MN576958 |
| S. cardinalis | YFCC 6144 | Limacodidae (Pupa) | - | MN576786 | MN576846 | MN576900 | MN576956 |
| S. coleopterorum | A19501 | Curculionidae (Snout beetle) | MT626376 | - | MT642600 | MN101585 | MN101586 |
| S. coleopterorum | A19502 | Curculionidae (Snout beetle) | MT626625 | - | MT642603 | MN101587 | MT642602 |
| S. cristata | YFCC 6021 | Saturniidae (Pupa) | - | MN576791 | MN576851 | MN576905 | MN576961 |
| S. cristata | YFCC 6023 | Saturniidae (Pupa) | - | MN576792 | MN576852 | MN576906 | MN576962 |
| S. hepiali | ICMM 82-2 | Fungi (Ophiocordyceps sinensis) | - | MN576794 | MN576854 | MN576908 | MN576964 |
| S. hepiali | ICMM Cs-4 | Fungi (Ophiocordycepssinensis) | - | MN576799 | MN576859 | MN576913 | MN576969 |
| S. hepiali | YFCC 661 | Fungi (Ophiocordycepssinensis) | - | MN576795 | MN576855 | MN576909 | MN576965 |
| S. hepiali | YJ06171 | Formicidae | MZ831866 | MZ831868 | MZ855241 | - | MZ855235 |
| S. hepiali | YJ06172 | Formicidae | MZ831867 | MZ831873 | - | - | MZ855236 |
| S. hymenopterorum | A19521 | Vespidae (Bee) | MN128224 | - | MT642601 | MT642604 | MN101588 |
| S. hymenopterorum | A19522 | Vespidae (Bee) | MN128081 | - | MN101589 | MN101590 | MN101591 |
| S. inthanonensis | TBRC 7915 | Lepidoptera (Pupa) | MF140761 | - | MF140790 | MF140815 | MF140849 |
| S. inthanonensis | TBRC 7916 | Lepidoptera (Pupa) | MF140760 | - | MF140789 | MF140814 | MF140848 |
| S. kunmingensis | YHH 16002 | Lepidoptera (Pupa) | - | MN576802 | MN576862 | MN576916 | MN576972 |
| S. lanmaoa | YFCC 6148 | Lepidoptera (Pupa) | - | MN576789 | MN576849 | MN576903 | MN576959 |
| S. lanmaoa | YFCC 6193 | Lepidoptera (Pupa) | - | MN576790 | MN576850 | MN576904 | MN576960 |
| S. lepidopterorum | DL10071 | Lepidoptera (Pupa) | MN128076 | - | MN101592 | MN101593 | MN101594 |
| S. lepidopterorum | DL10072 | Lepidoptera (Pupa) | MN128084 | - | - | MT642605 | MT642606 |
| S. pseudogunii | GY407201 | Lepidoptera (Larvae) | MZ827470 | MZ827010 | - | MZ855239 | MZ855233 |
| S. pseudogunii | GY407202 | Lepidoptera (Larvae) | MZ831863 | MZ831865 | - | MZ855240 | MZ855234 |
| S. pupicola | DY101681 | Lepidoptera (Pupa) | MZ827085 | MZ827009 | - | MZ855237 | MZ855231 |
| S. pupicola | DY101682 | Lepidoptera (Pupa) | MZ827008 | MZ827635 | - | MZ855238 | MZ855232 |
| S. ramose | YFCC 6020 | Limacodidae (Pupa) | - | MN576805 | MN576865 | MN576919 | MN576975 |
| S. tortricidae | YFCC 6013 | Limacodidae (Pupa) | - | MN576807 | MN576867 | MN576921 | MN576977 |
| S. tortricidae | YFCC 6131 | Limacodidae (Pupa) | - | MN576806 | MN576866 | MN576920 | MN576976 |
| S. yunnanensis | YFCC 1527 | Limacodidae (Pupa) | - | MN576812 | MN576872 | MN576926 | MN576982 |
| S. yunnanensis | YFCC 1824 | Limacodidae (Pupa) | - | MN576813 | MN576873 | MN576927 | MN576983 |
| Sarocladium bacillisporum | CBS 425.67 | Soil | NR_145039 | MH870718 | - | - | - |
| S. dejongiae | CBS 144929 | Soil | NR_161153 | NG_067854 | - | - | - |
| S. implicatum | CBS 959.72 | Soil | HG965023 | MH878470 | - | - | - |
| S. subulatum | CBS 217.35 | Soil | MH855652 | NG_070566 | - | - | - |
| S. terricola | CBS 243.59 | Soil | MH857853 | MH869389 | - | - | - |
| Shimizuomyces paradoxus | EFCC 6279 | Smilacaceae (Smilax sieboldii) | JN049847 | EF469084 | - | EF469117 | EF469071 |
| Simplicillium lamellicola | CBS 116.25 | Hymenomycetes (Agaricus bisporus) | AJ292393 | MH866307 | - | DQ522462 | DQ522356 |
| S. lanosoniveum | CBS 101267 | Uredinales (Hemileia vastatrix) | - | AJ292395 | - | DQ522463 | DQ522357 |
| S. lanosoniveum | CBS 704.86 | Uredinales (Hemileia vastatrix) | AJ292396 | AF339553 | - | DQ522464 | DQ522358 |
| Sordaria fimicola | AFTOL-ID 216 | - | DQ518178 | - | - | - | DQ518175 |
| Sphaerostilbella aureonitens | GJS74-87 | - | FJ442633 | HM466683 | - | FJ442763 | - |
| S. berkeleyana | GJS82-274 | - | - | U00756 | - | - | AF543783 |
| Stachybotrys chlorohalonata | DAOM 235557 | - | JN942888 | JN938870 | - | - | - |
| S. eucylindrospora | ATCC 18851 | - | JN942887 | JN938869 | - | - | - |
| Stephanonectria keithii | GJS92-133 | Plant (Bark) | - | AY489727 | - | - | AY489622 |
| Tilachlidium brachiatum | CBS 363.97 | Hymenomycetes (Agaricus sp.) | KM231838 | KM231719 | - | KM232414 | KM231975 |
| T. brachiatum | CBS 506.67 | Hymenomycetes (Hypholoma fasciculare) | KM231839 | HQ232177 | - | KM232415 | KM231976 |
| Tolypocladium inflatum | SCALT1007-002 | Sclerotium | KC963032 | - | - | - | - |
| Trichoderma aggresivum | CBS 100525 | - | - | JN939837 | - | JQ014130 | - |
| T. viride | GJS89-127 | Plant (Bark) | - | AY489726 | - | - | AY489621 |
| Trichothecium roseum | DUCC 502 | Plant (Solanum lycopersicum) | JN937590 | JX458860 | - | - | - |
| Valetoniellopsis laxa | GJS 96-174 | - | - | AY015635 | - | AY015638 | - |
2.3. Sequence Alignment and Phylogenetic Analyses
Lasergene software (version 6.0, DNASTAR) was applied for the assembling and editing of DNA sequence in this study. The ITS, LSU, RPB1, RPB2, and TEF sequences were downloaded from GenBank, based on Kepler et al. [9], Mongkolsamrit et al. [10,19], Chen et al. [12], Wang et al. [20], and others selected on the basis of BLAST algorithm-based searches in GenBank (Table 2). A single gene data set was aligned and edited by MAFFT v7.037b [21] and MEGA6 [22]. Combined sequences of ITS, LSU, RPB1, RPB2, and TEF were performed by SequenceMatrix v.1.7.8 [23]. The combined datasets (ITS+LSU+RPB2+TEF) and (ITS+LSU+RPB1+RPB2+TEF) were used to determine the family placement of those strains in Hypocreales and the taxonomic position of strains and the cryptic diversity among the different isolates of I. farinosa in Cordycipitaceae
The combined genes were both analyzed using the Bayesian inference (BI) and maximum likelihood (ML) methods. For BI, the model was selected for Bayesian analysis by ModelFinder [24] in the software PhyloSuite [25]. A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v.3.2 [26] for the combined sequence datasets. The Bayesian analysis resulted in 20,001 trees after 10,000,000 generations. The first 4000 trees, representing the burn-in phase of the analyses, were discarded, while the remaining 16,001 trees were used for calculating posterior probabilities in the majority rule consensus tree. After the analysis was finished, each run was examined using the program Tracer v1.5 [27] to determine burn-in, confirming that both runs had converged. ML analyses were constructed with RAxMLGUI [28]. The GTRGAMMA model was used for all partitions, in accordance with recommendations in the RAxML manual against the use of invariant sites.
3. Results
3.1. Phylogenetic Analyses
Gelasinospora tetrasperma Dowding, Neurospora crassa Shear and B.O. Dodge, and Sordaria fimicola (Roberge ex Desm.) Ces. and De Not. were used as the outgroup in analysis 1 (Figure 1) (to determine the family placement of those strains in Hypocreales). Purpureocillium lilacinum (Thom) Luangsa-ard, Houbraken, Hywel-Jones, and Samson was used as the outgroup in analysis 2 (Figure 2) (to determine the taxonomic position of strains and the cryptic diversity among the different isolates of I. farinosa in Cordycipitaceae). The concatenated sequences of analysis 1 and 2 included 77 and 62 taxa, respectively, and consisted of 2396 (ITS, 620; LSU, 712; RPB2, 510; and TEF, 554) and 3309 (ITS, 554; LSU, 677; RPB1, 533; RPB2, 671; and TEF, 874) characters with gaps, respectively.
Figure 1.
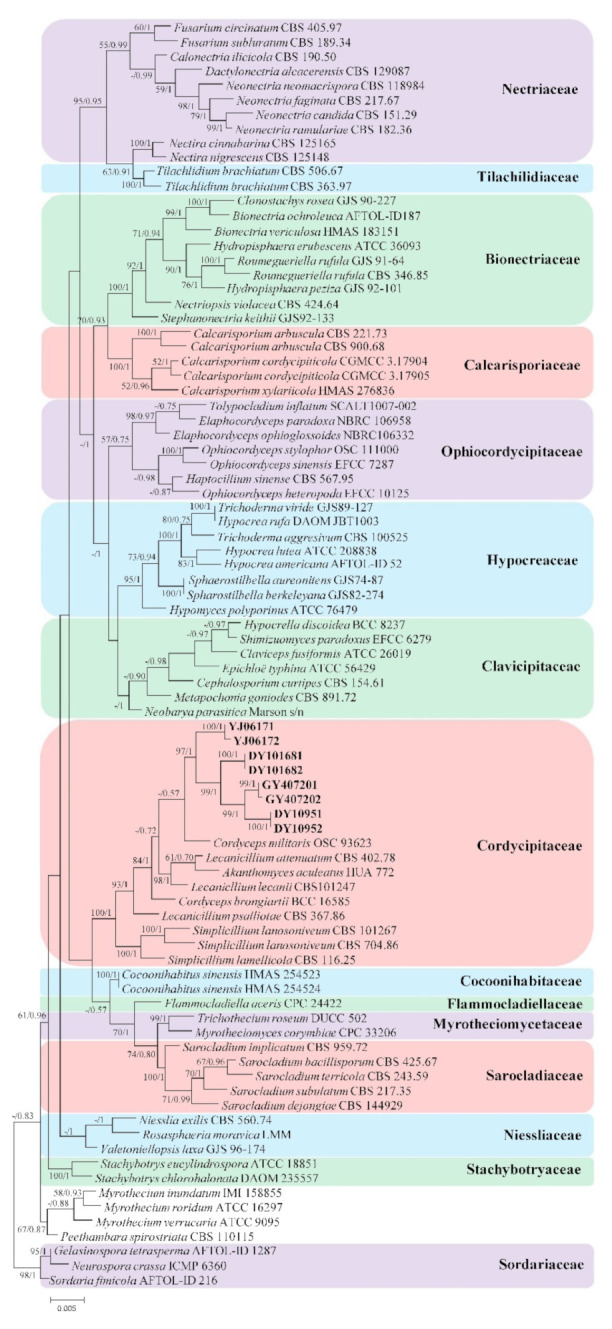
Phylogenetic placement of the new Isaria-like strains in the order of Hypocreales based on multigene dataset (ITS, LSU, RPB2m and TEF). Statistical support values (≥50%/0.5) are shown at the nodes for ML bootstrap support/BI posterior probabilities.
Figure 2.
Phylogenetic placement of the new strains in Cordycipitaceae, based on multigene dataset (ITS, LSU, RPB1, RPB2, and TEF). Statistical support values (≥50%/0.5) are shown at the nodes for ML bootstrap support/BI posterior probabilities.
Analysis 1: The selected models for BI analysis were GTR+F+I+G4 parameters for partition ITS and LSU+RPB2, and GTR+F+G4 parameters for partition TEF. The final value of the highest scoring tree was –37,321.078127, which was obtained from an ML analysis of the dataset (ITS+LSU+RPB2+TEF). The parameters of the general time reversible (GTR) model used to analyze the dataset were estimated using the following frequencies: A = 0.230263, C = 0.272892, G = 0.280445, and T = 0.216401; substitution rates AC = 1.451341, AG = 2.441940, AT = 1.532513, CG = 1.182477, CT = 5.701598, and GT = 1.000000; as well as the gamma distribution shape parameter α = 0.381402. In the phylogenetic tree (Figure 1), both analyses of ML and BI trees were largely congruent, and strongly supported in most branches. DY10951, DY10952, DY101681, DY101682, GY407201, GY407202, YJ06171, and YJ06172 strains had a close relationship with Cordyceps Fr., Akanthomyces Lebert, and Simplicillium W. Gams and Zare, and clustered into Cordycipitaceae.
Analysis 2: The selected models for BI analysis were GTR+F+I+G4 parameters for partition ITS+LSU+RPB2+TEF and GTR+F+G4 parameters for partition RPB1. The final value of the highest scoring tree was –31,206.916701, which was obtained from an ML analysis of the dataset (ITS+LSU+RPB1+RPB2+TEF). The parameters of the general time reversible (GTR) model used to analyze the dataset were estimated using the following frequencies: A = 0.238319, C = 0.279080, G = 0.271674, and T = 0.210926; substitution rates AC = 1.120096, AG = 2.745044, AT = 0.784066, CG = 0.934312, CT = 6.322628, and GT = 1.000000; as well as the gamma distribution shape parameter α = 0.308970. In the phylogenetic tree (Figure 2), both analyses of ML and BI trees were largely congruent, and strongly supported in most branches. The new strains were all clustered within the genus Samsoniella. GY407201 and GY407202 strains clustered with Samsoniella coleopterorum W.H. Chen, Y.F. Han, and Z.Q. Liang in a subclade. DY10951 and DY10952 strains clustered with Samsoniella aurantia Mongkols., Noisrip., Thanakitp., Spatafora, and Luangsa-ard in a subclade. DY101681 and DY101682 strains had a close relationship with Samsoniella alboaurantia (G. Sm.) Mongkols., Noisrip., Thanakitp., Spatafora, and Luangsa-ard; Samsoniella alpina H. Yu, Y.B. Wang, Y. Wang, and Zhu L. Yang; and Samsoniella cardinalis H. Yu, Y.B. Wang, Y. Wang, Q. Fan, and Zhu L. Yang. YJ06171 and YJ06172 strains clustered with Isaria farinosa (Holmsk.) Fr. in a subclade and had close relationship with Samsoniella hepiali (Q.T. Chen and R.Q. Dai ex R.Q. Dai, X.M. Li, A.J. Shao, Shu F. Lin, J.L. Lan, Wei H. Chen, and C.Y. Shen) H. Yu, R.Q. Dai, Y.B. Wang, Y. Wang, and Zhu L. Yang.
3.2. Taxonomy
3.2.1. Samsoniella pseudogunnii W.H. Chen, Y.F. Han, J.D. Liang, and Z.Q. Liang, sp. nov.
MycoBank No.: MB840999
Etymology: referring to similar morphology with Keithomyces neogunnii.
Holotype: CHINA, Guizhou, Guiyang, Tongmuling (N26°23’, E106°40’). On a larva (Lepidoptera), 1 April 2019, Wanhao Chen, GZAC GY40720 (holotype), ex-type living cultures, GY407201, GY407202.
Description: Colonies on PDA, 4.1–4.3 cm diam. after 14 d at 25°C, white, consisting of a basal felt and cottony, floccose hyphal overgrowth, reverse yellowish. Prostrate hyphae smooth, septate, hyaline, 1.0–1.3 μm diam. Erect conidiophores usually arises from aerial hyphae. Phialides are solitary or in whorls of two to nine. Phialides 6.8–11.0 × 2.2–2.4 μm, with a cylindrical basal portion, tapering into a short distinct neck. Conidia in chains, hyaline, fusiform, one-celled, 2.8–3.2 × 1.7–2.1 μm. Chlamydospores and sexual state were not observed. Sizes and shapes of phialides and conidia are similar in culture and on natural substratum.
Known distribution: Tongmuling, Guiyang, Guizhou Province, China.
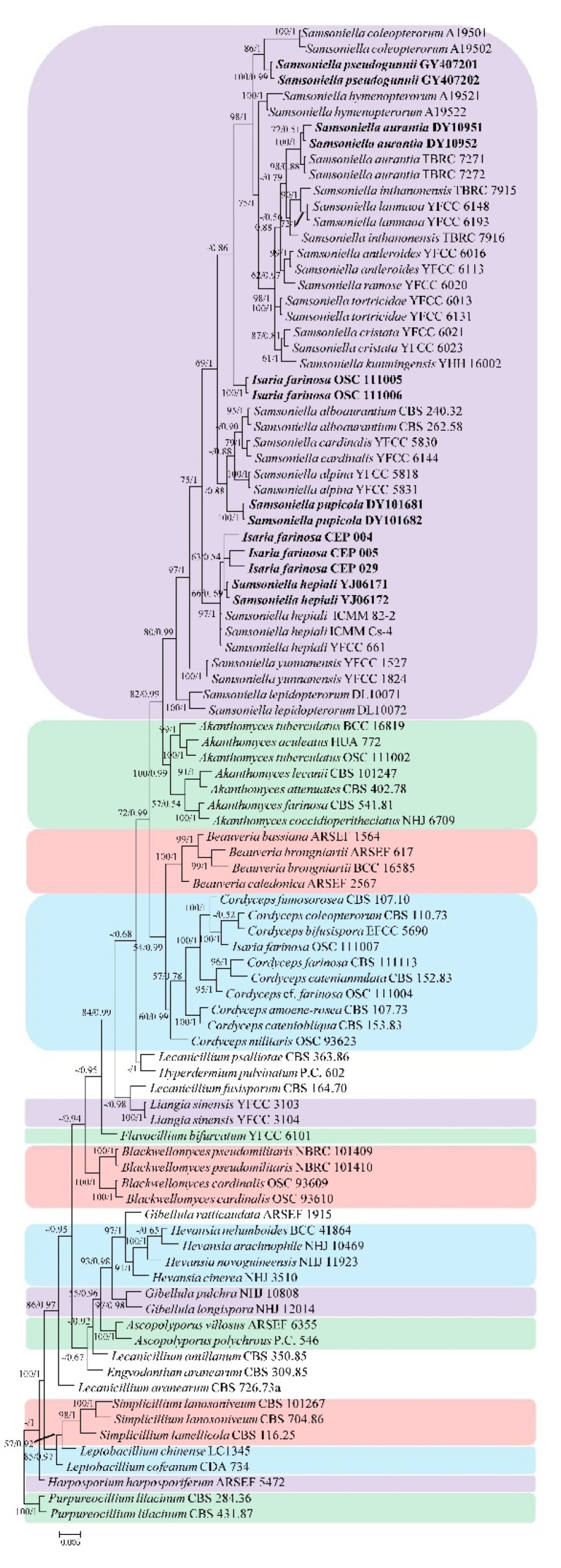
Notes: Samsoniella pseudogunnii was identified as belonging to Samsoniella based on the phylogenetic analyses (Figure 2) and has a close relationship with S. coleopterorum. However, Samsoniella pseudogunnii (Figure 3) has longer phialide, larger conidia, and its larva host belongs to the order Lepidoptera.
Figure 3.
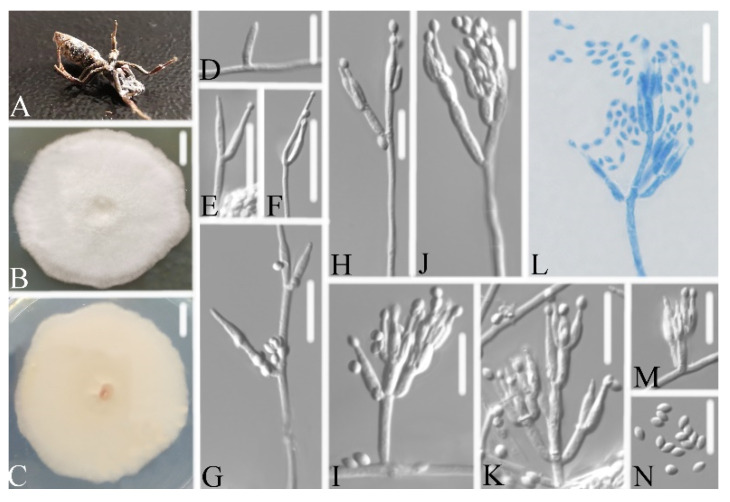
Samsoniella pseudogunnii (A) infected larva (Lepidoptera) (B,C) culture plate, showing the front (B) and the reverse (C) of the colony, cultured on PDA medium (D–I) phialides solitary, conidia adhering ellipsoidal slimy head and conidia (J) conidia. Scale bars: 10 mm (B,C), 10 μm (D–J).
3.2.2. Samsoniella Pupicola W.H. Chen, Y.F. Han, J.D. Liang, and Z.Q. Liang, sp. nov.
MycoBank No.: MB841003
Etymology: referring to its pupa-inhabitor.
Holotype: CHINA, Guizhou, Qiannan Buyi and Miao Autonomous Prefecture, Duyun City (26°21′24.71″ N, 107°22′48.22″ E). On a pupa (Lepidoptera), 1 October 2019, Wanhao Chen, GZAC DY10168 (holotype), ex-type living cultures, DY101681, DY101682.
Description: Colonies on PDA, 2.3–2.4 cm diam. after 14 d at 25°C, white, consisting of a basal felt and cottony, floccose hyphal overgrowth, reverse yellowish. Prostrate hyphae smooth, septate, hyaline, 1.2–2.2 μm diam. Erect conidiophores usually arise from aerial hyphae. Phialides are solitary or in whorls of two to nine. Phialides 7.0–9.2 × 2.5–3.3 μm, with a cylindrical basal portion, tapering into a short distinct neck. Conidia in chains, hyaline, fusiform, one-celled, 2.5–3.3 × 2.2–2.6 μm. Chlamydospores and sexual state were not observed. Sizes and shapes of phialides and conidia are similar in culture and on natural substratum.
Known distribution: Duyun City, Qiannan Buyi and Miao Autonomous Prefecture, Guizhou Province, China.
Additional specimens examined: CHINA, Guizhou, Qiandongnan Miao and Dong Autonomous Prefecture, Rongjiang County (26°01′58.70″ N, 108°24′48.06″ E), on a lepidopteran pupa, 1 October 2018, W.H. Chen, GZAC DL1014.
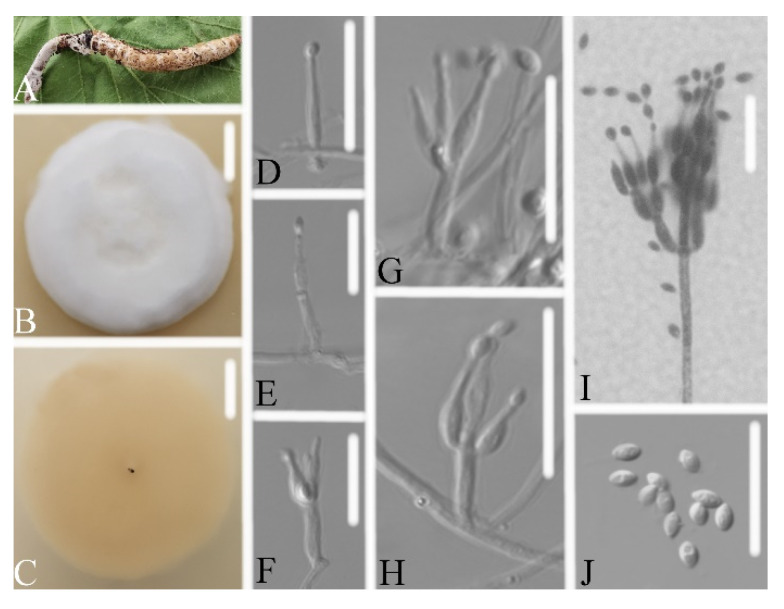
Notes: Samsoniella pupicola was identified as belonging to Samsoniella, based on the phylogenetic analyses (Figure 2) and has a close relationship with S. alboaurantium, S. alpina, and S. cardinalis. However, S. pupicola (Figure 4) is distinguished from S. alboaurantium by having larger fusiform conidia, distinguished from S. alpina by having white colony and fusiform conidia, and distinguished from S. cardinalis by having shorter phialides.
Figure 4.
Samsoniella pupicola (A) infected pupa (Lepidoptera) (B,C) culture plate, showing the front (B) and the reverse (C) of the colony, cultured on PDA medium (D–J) phialides solitary, conidia adhering ellipsoidal slimy head and conidia (K) conidia. Scale bars: 10 mm (B,C), 10 μm (D–K).
3.2.3. Samsoniella aurantia Mongkols., Noisrip., Thanakitp., Spatafora, and Luangsa-ard, Mycologia 110(1): 249
Description: Colonies on PDA, 3.7–4.2 cm diam. after 14 d at 25°C, white, consisting of a basal felt and cottony, floccose hyphal overgrowth, pale green and pale pink in the middle of the colony, reverse yellowish and pale brown in the middle. Prostrate hyphae smooth, septate, hyaline, 1.3–2.6 μm diam. Erect conidiophores usually arise from aerial hyphae. Phialides are solitary or in whorls of two to ten. Phialides 3.6–7.7 × 1.3–1.6 μm, with a cylindrical basal portion, tapering into a short distinct neck. Conidia in chains, hyaline, fusiform, occasionally cylindrical, one-celled, 2.6–3.9 × 1.7–2.2 μm. Chlamydospores and sexual state were not observed. Sizes and shapes of phialides and conidia are similar in culture and on natural substratum.
Specimens examined: CHINA, Guizhou, Qiannan Buyi and Miao Autonomous Prefecture, Duyun City (26°21′24.71″ N, 107°22′48.22″ E). On a pupa (Lepidoptera), 1 October 2019, Wanhao Chen, GZAC DY1095, living cultures, DY10951, DY10952.
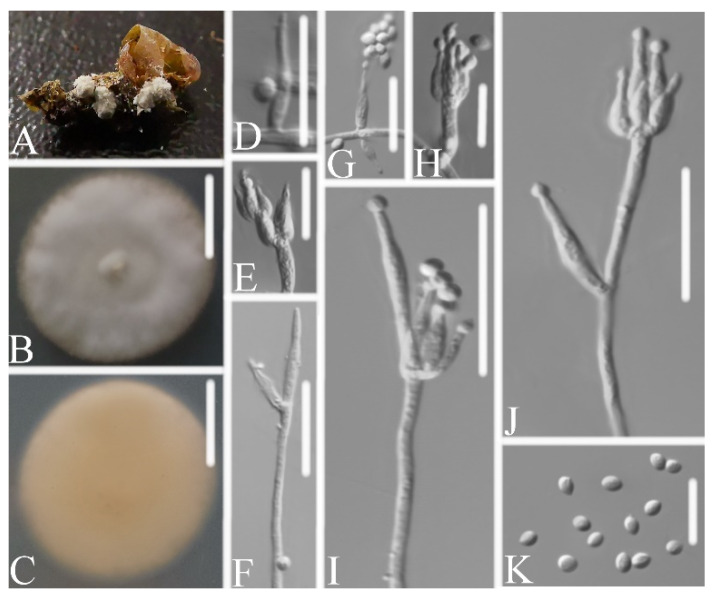
Note: DY10951 and DY10952 strains were identified as belonging to Samsoniella, based on the phylogenetic analyses (Figure 2), and clustered with Samsoniella aurantia in a clade. The characteristics of DY10951 and DY10952 (Figure 5) strains are similar to that of S. aurantia, which had fusiform conidia (2–4 × 1–2 μm) and larger phialide (5–13 × 2–3 μm). Besides, the pairwise dissimilarities of ITS sequences show no difference within 554 bp between DY10951 and S. aurantia. Thus, molecular phylogenetic results and morphologically based conclusions support the idea that DY10951 and DY10952 strains were S. aurantia.
Figure 5.
Samsoniella aurantia (A) infected pupa (Lepidoptera) (B,C) culture plate, showing the front (B) and the reverse (C) of the colony, cultured on PDA medium (D–J) phialides solitary, conidia adhering ellipsoidal slimy head and conidia (K) conidia. Scale bars: 10 mm (B,C), 10 μm (D–K).
3.2.4. Samsoniella hepiali (Q.T. Chen, and R.Q. Dai ex R.Q. Dai, X.M. Li, A.J. Shao, Shu F. Lin, J.L. Lan, Wei H. Chen, and C.Y. Shen) H. Yu, R.Q. Dai, Y.B. Wang, Y. Wang, and Zhu L. Yang, Fungal Diversity 103: 31
Description: Colonies on PDA, 5.8–5.9 cm diam. after 14 d at 25°C, white, consisting of a basal felt and cottony, floccose hyphal overgrowth, reverse yellowish. Prostrate hyphae smooth, septate, hyaline, 1.1–1.8 μm diam. Erect conidiophores usually arise from aerial hyphae. Phialides are solitary or in whorls of two to eight. Phialides 6.0–7.8 × 1.5–1.8 μm, with a cylindrical basal portion, tapering into a short distinct neck. Conidia in chains, hyaline, fusiform, one-celled, 2.1–2.5 × 0.9–1.6 μm. Chlamydospores and sexual state were not observed. Sizes and shapes of phialides and conidia are similar in culture and on natural substratum.
Specimens examined: CHINA, Guizhou, Tongren City, Yinjiang (N 27°55′17.1″, E 108°41′25.2″), on an ant, 1 October 2019, Wanhao Chen, GZAC YJ0617, DY1044, living cultures, YJ06171, YJ06172.
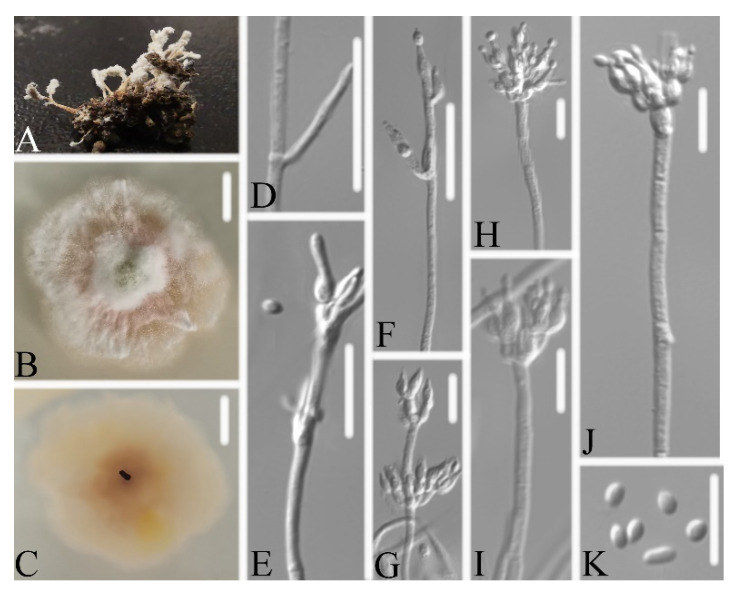
Note: YJ06171 and YJ06172 strains were identified as belonging to Samsoniella, based on the phylogenetic analyses (Figure 2), and clustered with Samsoniella hepiali in a clade. The characteristics of YJ06171 and YJ06172 (Figure 6) strains were very closely linked with S. hepiali, which had fusiform or oval conidia (1.8–3.3 × 1.4–2.2 μm) and larger phialide (3.5–13.6 × 1.3–2.1 μm). Besides, the pairwise dissimilarities of LSU sequences show no difference within 677 bp between YJ06171 and S. hepiali. Thus, molecular phylogenetic results and morphologically based conclusions supported the idea that YJ06171 and YJ06172 strains were S. hepiali.
Figure 6.
Samsoniella hepiali (A) infected ant (Formicidae) (B,C) culture plate, showing the front (B) and the reverse (C) of the colony, cultured on PDA medium (D–J) phialides solitary, conidia adhering ellipsoidal slimy head and conidia (N) conidia. Scale bars: 10 mm (B,C), 10 μm (D–N).
4. Discussion
The taxonomic delimitation of Isaria was originally based on morphological characteristics. However, Isaria shares many morphological characters with other genera in Hypocreales, which has resulted in a turbulent taxonomic history [10,29]. D’Alessandro et al. [30] noted that the morphological characteristics used to classify the genus Isaria frequently do not resolve new isolates into clearly defined species and need additional molecular markers in phylogenetic analyses. In the present study, Isaria-like strains collected from Guizhou Province, China, and previously identified by morphological characteristics, were reanalyzed using multi-gene (ITS, LSU, RPB1, RPB2, TEF) phylogenetic methodology. We proposed two new species of Samsoniella in this study.
The species Isaria farinosa is a well-known entomopathogenic fungi with worldwide distribution and a wide host range [31]. Kepler et al. [9] transferred Isaria farinosa to the genus Cordyceps as C. farinosa (Holmsk.) Kepler, B. Shrestha, and Spatafora based on a phylogenetic analysis of the CBS 111113 strain. We analyzed several strains of Isaria farinosa in the present study. Some properly belonged in the genus Samsoniella. CEP 004, CEP 005, CEP 029, YJ06171, and YJ06172 strains were identified as S. hepiali. Strains DY10951 and DY10952 were identified as S. aurantia. OSC 111005 and OSC 111006 strains were identified as new species but are absent in delineating morphological characteristics. Our results reveal cryptic diversity present in Isaria farinosa (now treated as Cordyceps farinosa) and illustrated that more attention should be paid on cryptic intraspecific diversity across different fungi isolates and genotypes.
The genus Samsoniella was established for the typical species S. inthanonensis Mongkolsamrit, Noisripoom, Thanakitpipattana, Spatafora, and Luangsa-ard, and two other species (S. alboaurantia (G. Sm.) Mongkols., Noisrip., Thanakitp., Spatafora, and Luangsa-ard and S. aurantia Mongkols., Noisrip., Thanakitp., Spatafora, and Luangsa-ard) [10]. Samsoniella species all have Isaria-like morphological characteristics, and cluster in an independent clade with close relationship to the genus Akanthomyces. The species S. alboaurantia was established based on two strains, CBS 240.32 and CBS 262.58, which previously belonged to Isaria farinosa (originally designated Paecilomyces farinosus (Holmsk.) A.H.S. Br. and G. Sm.) [10]. Lin et al. [32] revised the taxonomy of some Isaria-like strains originally identified as Isaria farinosa by morphological characteristics using multi-gene phylogenetic analysis. All the strains were identified as Samsoniella hepiali (Q.T. Chen and R.Q. Dai ex R.Q. Dai, X.M. Li, A.J. Shao, Shu F. Lin, J.L. Lan, Wei H. Chen, and C.Y. Shen) H. Yu, R.Q. Dai, Y.B. Wang, Y. Wang, and Zhu L. Yang. In the present study, YJ06171 and YJ06172 strains were also identified as Samsoniella hepiali. Our results revealed that more isolates and genotypes, originally designated as Isaria, will need to be transferred to Samsoniella.
Samsoniella hepiali (otherwise known as Paecilomyces hepiali) is isolated from a field collection of natural Ophiocordyceps sinensis insect–fungi complex [33], and is widely used as a medicinal and edible cordycipitoid fungus, creating a great economic value [20]. Lin et al. [32] reported six isolates of Samsoniella hepiali from Anhui Province, China, which were isolated from leafhopper, larva, and cicada. CEP 004, CEP 005, CEP 029 strains from Buenos Aires, Argentina, were isolated from whitefly and soil [30]. YJ06171 and YJ06172 strains from Guizhou Province, China were isolated from ant. It is interesting that Samsoniella hepiali and its hosts are widely distributed in China and Argentina. This result will help us to assess the extent and distribution of genetic diversity of Samsoniella hepiali on a large scale, understand its biology and demographic history, and guide biodiversity conservation programs.
Acknowledgments
We thank Steven M. Thompson for editing the English text of a draft of this manuscript.
Author Contributions
Resources, W.C., J.L. and X.R.; data curation, W.C.; writing—original draft preparation, W.C., J.L., X.R.; writing—review and editing, J.L., Y.H.; review and editing, J.Z., Z.L.; funding acquisition, W.C., J.L., X.R., J.Z., Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (31860002, 32060011), High-level Innovative Talents Training Object in Guizhou Province (Qiankehepingtairencai [2020]6005), Science and Technology Foundation of Guizhou Province (Qiankehejichu [2020]1Y060), Program of Innovative Scientific and technological Talent Team of Guizhou Province (2020-5010), Guizhou Science and Technology Support Project (Qiankehezhicheng [2019]2776), The Youth Science and Technology Talent Growth project from Guizhou Provincial Department of Education ([2018]389).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fries E.M. Systema Mycologicum. Volume 1. Ex Officina Berlingiana; Lund & Greifswald, Sweden: 1821. pp. 1–726. [Google Scholar]
- 2.Brown A.H., Smith G. The genus Paecilomyces Bainier and its perfect stage Byssochlamys Westling. Trans. Br. Mycol. Soc. 1957;40:17–89. doi: 10.1016/S0007-1536(57)80066-7. [DOI] [Google Scholar]
- 3.De Hoog G.S. The genera Beauveria, Isaria, Tritirachium and Acrodontium Gen. Nov. Centraalbureau voor Schimmelcultures; Utrecht, The Netherlands: 1972. pp. 1–41. [Google Scholar]
- 4.Samson R.A. Paecilomyces and some allied Hyphomycetes. Volume 6. Centraalbureau voor Schimmelcultures; Utrecht, The Netherlands: 1974. pp. 1–119. [Google Scholar]
- 5.Hodge K.T., Gams W., Samson R.A., Korf R.P., Seifert K.A. Lectotypification and status of Isaria Pers.: Fr. Taxon. 2005;54:485–489. doi: 10.2307/25065379. [DOI] [Google Scholar]
- 6.Luangsa-ard J.J., Hywel-Jones N.L., Samson R.A. The order level polyphyletic nature of Paecilomyces sensu lato as revealed through 18S-generated rRNA phylogeny. Mycologia. 2004;96:773–780. doi: 10.2307/3762111. [DOI] [PubMed] [Google Scholar]
- 7.Luangsa-Ard J.J., Hywel-Jones N.L., Manoch L., Samson R.A. On the relationships of Paecilomyces sect. Isarioidea species. Mycol. Res. 2005;109:581–589. doi: 10.1017/S0953756205002741. [DOI] [PubMed] [Google Scholar]
- 8.Gams W., Hodge K.T., Samson R.A., Korf R.P., Seifert K.A. (1684) Proposal to conserve the name Isaria (anamorphic fungi) with a conserved type. Taxon. 2005;54:537. doi: 10.2307/25065390. [DOI] [Google Scholar]
- 9.Kepler R.M., Luangsa-ard J.J., Hywel-Jones N.L., Quandt A., Sung G.-H., Rehner S.A., Aime M.C., Henkel T.W., Sanjuan T., Zare R., et al. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales) IMA Fungus. 2017;8:335–353. doi: 10.5598/imafungus.2017.08.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mongkolsamrit S., Noisripoom W., Thanakitpipattana D., Wutikhun T., Spatafora J.W., Luangsa-ard J. Disentangling cryptic species with isaria-like morphs in Cordycipitaceae. Mycologia. 2018;110:230–257. doi: 10.1080/00275514.2018.1446651. [DOI] [PubMed] [Google Scholar]
- 11.Chen W.-H., Liu C., Han Y.-F., Liang J.-D., Liang Z.-Q. Akanthomyces araneogenum, a new Isaria-like araneogenous species. Phytotaxa. 2018;379:66–72. doi: 10.11646/phytotaxa.379.1.6. [DOI] [Google Scholar]
- 12.Chen W.H., Han Y.F., Liang J.D., Tian W.Y., Liang Z.Q. Morphological and phylogenetic characterizations reveal three new species of Samsoniella (Cordycipitaceae, Hypocreales) from Guizhou, China. MycoKeys. 2020;74:1–15. doi: 10.3897/mycokeys.74.56655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W.-H., Liu C., Han Y.-F., Liang J.-D., Tian W.-Y., Liang Z.-Q. Three novel insect-associated species of Simplicillium (Cordycipitaceae, Hypocreales) from Southwest China. MycoKeys. 2019;58:83–102. doi: 10.3897/mycokeys.58.37176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou X., Liu A., Liang Z., Han Y., Yang M. Hirsutella liboensis, a new entomopathogenic species affecting Cossidae (Lepidoptera) in China. Mycotaxon. 2010;111:39–44. doi: 10.5248/111.39. [DOI] [Google Scholar]
- 15.Liang J.D., Han Y.F., Zhang J.W., Du W., Liang Z.Q., Li Z.Z. Optimal culture conditions for keratinase production by a novel thermophilic Myceliophthora thermophila strain GZUIFR-H49-1. J. Appl. Microbiol. 2011;110:871–880. doi: 10.1111/j.1365-2672.2011.04949.x. [DOI] [PubMed] [Google Scholar]
- 16.White T., Bruns T., Lee S., Taylor J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; New York, NY, USA: 1990. pp. 315–322. [DOI] [Google Scholar]
- 17.Rakotonirainy M., Cariou M., Brygoo Y., Riba G. Phylogenetic relationships within the genus Metarhizium based on 28S rRNA sequences and isozyme comparison. Mycol. Res. 1994;98:225–230. doi: 10.1016/S0953-7562(09)80190-1. [DOI] [Google Scholar]
- 18.Castlebury L.A., Rossman A.Y., Sung G.-H., Hyten A.S., Spatafora J.W. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 2004;108:864–872. doi: 10.1017/S0953756204000607. [DOI] [PubMed] [Google Scholar]
- 19.Van den Brink J., Samson R.A., Hagen F., Boekhout T., de Vries R.P. Phylogeny of the industrial relevant, thermophilic genera Myceliophthora and Corynascus. Fungal Divers. 2012;52:197–207. doi: 10.1007/s13225-011-0107-z. [DOI] [Google Scholar]
- 20.Wang Y.-B., Wang Y., Fan Q., Duan D.-E., Zhang G.-D., Dai R.-Q., Dai Y.-D., Zeng W.-B., Chen Z.-H., Li D.-D., et al. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 2020;103:1–46. doi: 10.1007/s13225-020-00457-3. [DOI] [Google Scholar]
- 21.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaidya G., Lohman D.J., Meier R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 24.Kalyaanamoorthy S., Minh B.Q., Wong T., Von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D., Gao F., Jakovlić I., Zou H., Zhang J., Li W.X., Wang G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 26.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Hoehna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silvestro D., Michalak I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 29.Sung G.-H., Hywel-Jones N.L., Sung J.-M., Luangsa-ard J.J., Shrestha B., Spatafora J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007;57:5–59. doi: 10.3114/sim.2007.57.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Alessandro C.P., Jones L.R., Humber R.A., López Lastra C.C., Sosa-Gomez D.R. Characterization and phylogeny of Isaria spp. strains (Ascomycota: Hypocreales) using ITS 1-5.8 S-ITS 2 and elongation factor 1-alpha sequences. J. Basic Microbiol. 2014;54:S21–S31. doi: 10.1002/jobm.201300499. [DOI] [PubMed] [Google Scholar]
- 31.Zimmermann G. The entomopathogenic fungi Isaria farinosa (formerly Paecilomyces farinosus) and the Isaria fumosorosea species complex (formerly Paecilomyces fumosoroseus): Biology, ecology and use in biological control. Biocontrol Sci. Technol. 2008;18:865–901. doi: 10.1080/09583150802471812. [DOI] [Google Scholar]
- 32.Lin Y., Liu Y.J., Wang T., Chen M.J. Revision of taxonomic status of several Isaria-like strains. J. Microbiol. China. 2021 doi: 10.13344/j.microbiol.china.210220. (In Chinese) [DOI] [Google Scholar]
- 33.Dai R.-Q., Shen C.-Y., Li X.-M., Lan J.-L., Lin S.-F., Shao A.-J. Response to neotypification of Paecilomyces hepiali (Hypocreales) (Wang & al., 2015) Taxon. 2018;67:784–786. doi: 10.12705/674.7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.