Abstract
We prospectively compared the performance of culture, direct fluorescent-antibody testing (DFA), and an in-house-developed PCR test targeting the repeated insertion sequence IS481 for the detection of Bordetella pertussis in nasopharyngeal swab specimens. We tested 319 consecutive paired specimens on which all three tests were performed. A total of 59 specimens were positive by one or more tests. Of these, 5 were positive by all three tests, 2 were positive by culture and PCR, 16 were positive by PCR and DFA, 28 were positive by PCR only, and 8 were positive by DFA only. Any specimen positive by culture was considered to be a true positive, as were specimens positive by both PCR and DFA. Specimens positive only by PCR or DFA were considered discrepant, and their status was resolved by review of patient histories. Patients with symptoms meeting the Centers for Disease Control and Prevention clinical case definition for pertussis and who had a specimen positive by PCR or DFA were considered to have true B. pertussis infections. Of the 28 patients positive by PCR only, 20 met the clinical case definition for pertussis, while 3 of the 8 patients positive by DFA only met the clinical case definition. After resolution of the status of discrepant specimens, the sensitivity, specificity, positive predictive value, and negative predictive value were 15.2, 100, 100, and 87.5%, respectively, for culture; 93.5, 97.1, 84.3, and 98.9%, respectively, for PCR; and 52.2, 98.2, 82.8, and 92.4%, respectively, for DFA. The actual positive predictive value of PCR was probably greater, as several PCR-positive patients who did not meet the clinical case definition had symptoms consistent with typical or atypical pertussis. PCR is a sensitive and specific method for the detection of B. pertussis.
Pertussis remains an important respiratory disease, afflicting both unvaccinated children and previously vaccinated children and adults in whom immunity has waned. Recent studies have shown that Bordetella pertussis is a common cause of prolonged cough illness in adolescents and adults (2, 6, 25, 30). Many infections in these older individuals lack the typical symptoms of pertussis and are not diagnosed or properly treated.
Accurate laboratory identification of B. pertussis infections remains problematic. Culture, long considered the “gold-standard” diagnostic method, is highly specific, but sensitivity can be affected by a number of factors, including patient age, immunization status, antibiotic treatment and duration of symptoms prior to specimen collection, and specimen transport conditions (28, 29). The turnaround time for culture is typically 7 to 10 days for a negative specimen. Direct fluorescent-antibody testing (DFA) provides a much more rapid result but also has the disadvantage of poor sensitivity, and its specificity varies widely due to the subjective interpretation of test results. Serologic tests are not widely available and provide mainly a retrospective diagnosis. Because of the increased awareness of B. pertussis and the limitations of traditional diagnostic methods, a growing number of laboratories are using PCR to detect B. pertussis DNA in patient specimens (4, 7–11, 13–16, 19, 23, 24, 26, 27, 29). Since no FDA-approved PCR test for B. pertussis is commercially available, laboratories must develop and validate their own PCR tests. As a result, the analytical sensitivity, accuracy, and quality control of PCR-based B. pertussis tests may vary widely among laboratories. Notwithstanding this lack of standardization, PCR has consistently been shown to provide a more sensitive, specific, and rapid means of detecting B. pertussis infections than other methods (4, 7–11, 13–15, 23, 24, 26, 27, 29). While many studies have compared the positivity rates of culture and PCR tests for B. pertussis, relatively few have attempted to resolve the status of specimens that were positive by PCR alone and to determine the true clinical performance of the PCR test, particularly the specificity and positive predictive value (13, 26, 29).
In the present study we compared prospectively the performance of culture, DFA, and PCR for the detection of B. pertussis in nasopharyngeal (NP) swab specimens. Patient histories were reviewed to resolve discrepant test results, after which clinical sensitivity and specificity and predictive values were determined.
MATERIALS AND METHODS
Patients and samples.
From July through early November 1997, specimens were obtained from 319 children and adults throughout the state of Iowa who had clinically suspected pertussis or who were exposed to known or suspected cases of pertussis. The majority of specimens were obtained locally, during a communitywide outbreak of pertussis in Johnson County, Iowa. Two NP specimens (one from each of the nares) were obtained with calcium alginate swabs (Fisher Scientific, Pittsburgh, Pa.). After each swab was rolled into two 1-cm circles on two glass slides, one swab was placed into a vial containing Regan-Lowe transport medium supplemented with 40 μg of cephalexin (Sigma, St. Louis, Mo.) per ml, and the second swab was placed into an empty, sterile 1.5-ml screw-cap microcentrifuge tube. Slides were air dried and heat fixed, then shipped with vials containing swabs at ambient temperature to the State Hygienic Laboratory. The shipping time was usually 1 to 4 days.
Bacterial strains.
Laboratory strains of B. pertussis and Bordetella parapertussis were used to verify the performance of culture, PCR, and DFA. The following strains, obtained from the State Hygienic Laboratory culture collection, were used to evaluate the specificity of the PCR test: Alcaligenes faecalis, Bordetella bronchiseptica, B. parapertussis, Moraxella catarrhalis, Neisseria subflava, Staphylococcus aureus, and Streptococcus pneumoniae. Bacterial strains were grown on routinely used media, under standard conditions.
To evaluate the sensitivity of the PCR test, B. pertussis was suspended in sterile physiological saline at a concentration equivalent to approximately 108 cells per ml, based on McFarland turbidimetric standards. Serial dilutions were made from the suspension, and aliquots were tested as described herein. Additionally, dilutions of B. pertussis kindly provided by George Buck (Alliant Health System, Louisville, Ky.) were tested blindly. To evaluate the specificity of the PCR test, strains grown on solid media were suspended in water (>108 cells per ml, based on turbidity) and centrifuged, and the pellets were extracted and tested as described herein. PCR mixtures contained approx. 105 to 106 cells.
Culture.
Swabs received in Regan-Lowe transport medium were inoculated onto Bordet-Gengou, Regan-Lowe, and Trypticase soy agar plates containing 5% sheep blood. Regan-Lowe plates contained 40 μg of cephalexin per ml. Plates were incubated at 35°C in the presence of CO2 for as long as 10 days and were examined daily for the presence of colonies typical of Bordetella spp. Suspected colonies were confirmed by staining with fluorescein isothiocyanate (FITC)-conjugated B. pertussis or B. parapertussis antiserum (Difco Laboratories, Detroit, Mich.).
DFA.
One-half of each heat-fixed slide was stained with FITC-conjugated B. pertussis antiserum, and the other half was stained with B. parapertussis antiserum. Slides were examined with a 40× objective, and fluorescence was scored from 1+ to 4+; barely visible fluorescence was scored 1+, and brilliant yellow-green fluorescence was scored 4+. Smears with 3+ or 4+ fluorescence were further examined with a 100× oil immersion objective to confirm proper cell morphology.
PCR.
Dry NP swabs were stored in the laboratory at 4°C prior to PCR testing. At the beginning of the evaluation period, PCR was performed twice each week, resulting in specimen storage for as long as 4 days prior to testing. Due to the increased sample load associated with the pertussis outbreak, PCR was performed daily, Monday through Friday, beginning in early October 1997 and continuing through the remainder of the study period. PCR and culture procedures were performed in separate areas of the laboratory.
(i) DNA extraction.
DNA was extracted from NP swabs by a modification of the procedure described by Nelson et al. (21). Four hundred microliters of lysis reagent (5 M guanidine thiocyanate; 50 mM Tris [pH 7.6]; 50 μg of glycogen from mussels [Sigma Chemical Co.] per ml, and 100 mM dithiothreitol) was added to microcentrifuge tubes containing NP swabs. The tubes were vortexed, then incubated at 70°C for 10 min. Following incubation, swabs were removed by using forceps. The forceps were rinsed in 10% bleach, followed by water, after each specimen. After the addition of 500 μl of isopropanol, the tubes were vortexed and centrifuged at approximately 15,000 × g for 15 min. The supernatant was decanted, and 750 μl of 70% ethanol was added to the tube. After 10 min of centrifugation at 15,000 × g, the ethanol was decanted. Air-dried DNA pellets were suspended in 100 μl of sterile distilled H2O. Each extraction run included a positive control consisting of B. pertussis cells and a negative control to which no DNA was added. Extracted DNA from specimens and controls was added to PCR mixtures immediately or stored at −20°C.
(ii) Amplification of B. pertussis DNA.
One microliter of extracted DNA, equivalent to 1/100 of the original NP swab specimen, was added to 0.5-ml thin-walled PCR tubes containing 24 μl of PCR mix and was overlaid with mineral oil. Final PCR mixtures consisted of 10 mM Tris (pH 8.3); 50 mM KCl; 3 mM MgCl2; 125 μM (each) dATP, dCTP, and dGTP; 220 μM dUTP; 30 μM dTTP; 1 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Norwalk, Conn.); and 0.25 μM (each) primers BP1 (5′-biotin-GATTCAATAGGTTGTATGCATGGTT) (10) and BP2 (5′-TTCAGGCACACAAACTTGATGGGCG) (14). Uracil N-glycosylase was not routinely added to PCR mixtures until shortly after this study was completed. PCR tubes were placed in a model 480 thermal cycler (Perkin-Elmer), denatured at 94°C for 8 min, then subjected to 40 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min. Included in each amplification run were positive and negative extraction controls, a positive amplification control (1 μl of a previous positive extraction control), and at least one negative amplification control (1 μl of the water used to prepare the PCR mix). One negative amplification control was usually included for every 10 clinical specimens.
Through October 1997, duplicate PCRs were run on each specimen. One reaction mixture was spiked with 1 μl of the positive amplification control to check for PCR inhibition. A total of 278 specimens in this study were tested for inhibition in this fashion. Following review of the data, testing of spiked duplicate reactions was discontinued in November 1997.
(iii) Detection of PCR products.
Following amplification, 25 μl of denaturation reagent (Roche Diagnostic Systems, Somerville, N.J.) was added to the PCR mixtures. The denatured reaction mixtures were analyzed after a 10-min, room temperature incubation or stored at −20°C for as long as 3 days. Amplification products were detected by using a modification of the plate hybridization procedure from the Amplicor Chlamydia trachomatis test (Roche Diagnostic Systems) as described by Buck (1). A specific capture probe (5′-AATTGCTGGACCATTTCGAGTCGACG), the downstream primer described by Glare et al. (10), was coated on microtiter plate wells by a modification of the procedure described by Cook et al. (5). The capture probe was diluted in 1 M ammonium acetate to a final concentration of 400 ng per ml. One hundred microliters of the probe solution was added to wells of 96-well microtiter plates (catalog no. 2580; Costar Corp., Cambridge, Mass.). After the wells were sealed with tape, the plates were incubated overnight (20 to 24 h) at 37°C. Wells were washed twice with phosphate-buffered saline (PBS) containing 0.1% Tween 20, followed by a single wash with PBS containing 1 mM EDTA. The plates were then air dried completely. They were used immediately or stored at 4°C in sealed bags with dessicant. The liquid reagents in the Amplicor Chlamydia trachomatis Detection Kit (Roche) were used for the detection of the B. pertussis PCR product in microtiter plate wells. Twenty-five microliters of the denatured PCR product was added to wells and detected colorimetrically according to the manufacturer’s instructions. The optical density (OD) of wells was read on a plate reader at 450 nm, and results were scored as positive if the OD was ≥0.5. The total PCR assay time was approximately 6 h. The results of a run started at 8:00 a.m. were usually available by 3:00 p.m. the same day.
Analysis of discrepant results.
Specimens positive by culture were considered to be true positives. If specimens were culture negative, they had to be positive by both PCR and DFA to be considered true positives. Specimens positive by PCR or DFA alone were considered to be potential false positives, and their status was resolved by review of patient medical records. PCR- or DFA-positive specimens from patients with symptoms meeting the Centers for Disease Control and Prevention (CDC) clinical case definition for pertussis were considered to be true positives. The CDC clinical case definition for pertussis is a cough of at least 2 weeks’ duration, accompanied, for cases in areas of endemicity or sporadic cases, by one or more of the following specific symptoms: paroxysms, posttussive emesis, or inspiratory whoop (without any other apparent cause). Patients with outbreak-related cases may present with any cough illness lasting at least 2 weeks (3).
Statistical analyses.
The significance of discordance among the results of culture, DFA, and PCR was measured by McNemar’s test. The sensitivities, specificities, and predictive values of the pertussis tests were determined in two-way frequency tables after the resolution of discrepant results.
RESULTS
Analysis of PCR analytical sensitivity, specificity, and inhibition rate.
Two independently prepared B. pertussis dilution panels tested by the PCR test showed comparable analytical sensitivities. Testing of serial dilutions prepared from a suspension of organisms demonstrated a sensitivity of approximately 0.5 cell per PCR, while testing of dilutions provided by G. Buck showed a sensitivity of 0.3 CFU per PCR (data not shown).
Testing of a limited specificity panel including B. bronchiseptica and B. parapertussis showed no cross-reactivity with the B. pertussis PCR test (data not shown). The primers used in this study were tested against a larger panel of organisms and generated no detectable PCR product (14).
A total of 278 NP swab specimens, including a number of grossly bloody specimens, were tested for PCR inhibition by amplification of a spiked duplicate aliquot. Four specimens (1.4%) showed complete inhibition (OD below cutoff). Of these, two were noninhibitory upon repeat testing, suggesting either a pipetting error during initial testing or freeze-thaw inactivation of inhibitors. It is possible that the inhibition rate was below 1%. Six specimens showed partial inhibition.
Detection of B. pertussis in NP swab specimens.
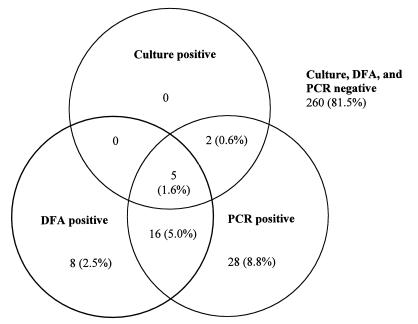
Between 2 July and 7 November 1997, culture, DFA, and PCR were performed on 319 NP swab specimens submitted for B. pertussis testing. During the local pertussis outbreak, two PCR runs were performed every day, Monday through Friday. At the height of the outbreak, 25 to 30 patient specimens (not including spiked controls) were usually tested in each run (50 to 60 patient results per day). All testing was routine, within the normal laboratory work flow. No specimens were frozen and tested retrospectively. An additional 263 specimens submitted during this period were excluded from analysis because they were not tested by all three methods. In many cases only DFA or PCR testing was requested. Effective 8 November 1997, the Hygienic Laboratory discontinued routine performance of DFA for B. pertussis. Of the 319 NP swab specimens tested by all three methods, 59 (18.5%) were positive by at least one test. B. pertussis was detected by culture in 7 (11.9%) of the 59 specimens. B. parapertussis was not isolated from any specimens. Twenty-nine (49.2%) of the 59 specimens were DFA positive, and 51 (86.4%) were PCR positive. The proportion of specimens positive by PCR was significantly greater than those positive by culture (P < 0.001) and by DFA (P < 0.001). Of the 29 DFA-positive specimens, 5 were also positive by PCR and culture, and 16 were positive by PCR, leaving 8 discrepant specimens positive by DFA alone. Of the 51 PCR-positive specimens, 5 were also positive by DFA and culture, 2 were positive by culture, and 16 were positive by DFA, leaving 28 discrepant specimens positive by PCR alone. All culture-positive specimens were also positive by PCR. Agreement among the three tests is shown in Fig. 1. Compared to that of culture, the sensitivities of DFA and PCR were 71.4 and 100%, respectively, and the positive predictive values were 17.2 and 13.7%, respectively (Table 1).
FIG. 1.
Agreement among culture, DFA, and PCR results.
TABLE 1.
Performance characteristics of culture, DFA, and PCR for the detection of B. pertussis in NP swab specimens
| Gold standard | No. (%) of patients positive | Test | Sensitivity (%) | Specificity (%) | PPVb (%) | NPVc (%) |
|---|---|---|---|---|---|---|
| Culture | 7 (2.2) | DFA | 71.4 | 92.3 | 17.2 | 99.3 |
| PCR | 100 | 85.9 | 13.7 | 100 | ||
| Expanded gold standarda | 46 (14.4) | Culture | 15.2 | 100 | 100 | 87.5 |
| DFA | 52.2 | 98.2 | 82.8 | 92.4 | ||
| PCR | 93.5 | 97.1 | 84.3 | 98.9 |
Either (i) culture positive, (ii) PCR and DFA positive, or (iii) PCR or DFA positive, with clinical features indicating pertussis.
PPV, positive predictive value.
NPV, negative predictive value.
Review of patient histories and resolution of discrepant test results.
Patient histories (pertussis surveillance worksheets) were reviewed to resolve discrepant test results. Histories were available for six of the eight patients who tested positive by DFA only. Three patients met the CDC clinical case definition for pertussis and thus were considered to represent true pertussis cases. Two of these three patients were epidemiologically linked to another confirmed case. Of the three DFA-positive patients who did not meet the clinical case definition, one had a nonspecific cough lasting 25 days at the time of the final interview. Linkage of this person to a confirmed case was unknown. A second patient with an unresolved positive DFA result was linked to a confirmed case.
Histories were available for 25 of the 28 patients who tested positive by PCR only. Twenty patients met the pertussis case definition, and 11 of these were epidemiologically linked to confirmed cases. Of the five patients who did not meet the clinical case definition, one, a 7-year-old male with a paroxysmal cough that lasted 10 days, was considered to represent a likely case, and two were considered to represent possible cases: a 39-year-old female with a nonspecific cough that lasted 14 weeks and a 1-month-old, unvaccinated female with a nonspecific cough that lasted 20 days. Both of these possible cases were determined to be sporadic. A fourth patient with an unresolved positive PCR result, a 2-month-old with a paroxysmal cough that lasted 29 days, also tested positive for respiratory syncytial virus (RSV) and therefore was not considered to represent a case.
To determine the sensitivity of the pertussis clinical case definition as a means of defining true infections, the medical histories of patients with nondiscrepant (true) positive test results were reviewed. Histories of 16 patients were available, including 4 who tested positive by all three tests, 10 who tested positive by PCR and DFA, and 2 who tested positive by culture and PCR. All 16 patients met the clinical case definition for pertussis.
The performance characteristics of culture, DFA, and PCR were recalculated after the gold standard definition for pertussis infection was expanded to include patients positive by both PCR and DFA, as well as patients positive by a single test other than culture who met the CDC case definition for pertussis. Relative to the expanded gold standard, the sensitivities of culture, DFA, and PCR were 15.2, 52.2, and 93.5%, respectively. Positive predictive values were 100, 82.8, and 84.3%, respectively (Table 1).
DISCUSSION
In this study, the accuracy of PCR in detecting B. pertussis was compared to those of culture and DFA. PCR has repeatedly been shown to be more sensitive than culture and DFA for the detection of B. pertussis (4, 7–11, 13–15, 23, 24, 26, 27, 29). However, resolution of discrepant test results and assessment of the clinical performance of PCR have been carried out infrequently (13, 26, 29). The PCR test described in this study was shown to have excellent analytical sensitivity (<1 cell per PCR). Two factors that we believe are largely responsible for this level of sensitivity are the nucleic acid extraction method and the use of AmpliTaq Gold DNA polymerase. The use of guanidine thiocyanate to lyse cells, followed by washing and precipitation of nucleic acids with alcohol, results in DNA relatively free of cellular and other debris that reduce amplification efficiency or cause complete inhibition of amplification. Commonly used processing methods for respiratory swab specimens and aspirates consist of boiling or proteinase K digestion followed by heating. These methods do not remove cellular debris from nucleic acids, and the proportion of specimens that are inhibitory to PCR has been reported to range between 6 and 18% (18, 22, 29) for detection of B. pertussis and other respiratory-tract pathogens. The proportion of specimens that were inhibitory with the procedure described in this study was approximately 1%. Grossly bloody specimens were not inhibitory. Because of this low rate of inhibition and the costs associated with testing specimens in duplicate, we elected to discontinue testing for inhibition in specimens near the end of the study period. AmpliTaq Gold DNA polymerase was incorporated into the PCR mixtures because regular AmpliTaq was shown to generate considerable amounts of low-molecular-weight product (primer dimer). Mechanical hot start and use of a wax barrier reduced the amount of primer dimer while increasing the amount of specific product (data not shown). However, these methods were cumbersome and prone to contamination. AmpliTaq Gold also increased the amount of specific product generated, was more sensitive than the wax barrier method, and except for increasing the length of the initial denaturation step by several minutes, was invisible to the user.
Notwithstanding the growing acceptance of PCR as a diagnostic test method during the past decade, concerns remain over false-positive results due to contamination or cross-reactivity. During this study very few positive PCR results (approximately 14%) were confirmed by culture, causing some to doubt the accuracy of a positive PCR result. However, after comparison of the performance of culture, DFA, and PCR against an expanded gold standard defining infection (multiple positive tests or a single positive test other than culture combined with symptoms meeting the CDC clinical case definition), it became apparent that the difference in the PCR and culture positivity rates was due largely to false-negative culture results rather than to false-positive PCR results. Indeed, in this study, culture and PCR were determined to have sensitivities of 15.2 and 93.5%, respectively. The sensitivity of DFA was 52.2%.
The culture sensitivity reported in this study (15.2%) was much lower than that reported by Wadowsky et al. (29) in their comparison of PCR and culture (73.4%). One possible explanation for our lower culture sensitivity was the specimen transport conditions. At our institution, a public health laboratory that serves the entire state, transport of NP swab specimens to the laboratory ranges from several hours for hand-delivered specimens up to 4 days for specimens shipped by mail. Specimens are shipped at room temperature. In comparison, NP swabs in the Wadowsky et al. study were inoculated onto agar plates immediately after collection. B. pertussis is an extremely labile organism, and bedside inoculation of culture media is recommended for optimal recovery of organisms (20). Another possible explanation for the low culture sensitivity was the patient population. In this study, both children and adults were tested, and the majority of patients were older children and adults, while the Wadowsky et al. study was carried out strictly in a pediatric setting. It has been shown that increased age is associated with decreased pertussis culture sensitivity (28). Grimprel et al. (11) reported a culture positivity rate of 54.1% in pediatric index cases versus a positivity rate of only 15.4% in adult contacts of the index cases. The sensitivity of PCR has also been shown to be inversely related to patient age, but to a lesser extent than that of culture (11, 28).
Use of a different gold standard could alter the calculated sensitivities of the tests evaluated in this study. A gold standard based on clinical and epidemiologic criteria rather than laboratory results may enable the identification of additional infections missed by all test methods. Additionally, serologic testing has been used as a reference method against which other laboratory tests have been compared and has been reported to detect recent infections missed by PCR and culture (11, 17, 28). However, the serologic testing for pertussis may lack specificity (12), and criteria that define a recent infection have not been standardized. For these reasons CDC guidelines for laboratory confirmation of pertussis cases do not include serologic testing (3).
The positive predictive values of culture, DFA, and PCR in this study were 100, 82.8, and 84.3%, respectively. The actual positive predictive value of the PCR test may be greater, as the clinical criteria we used to define an infection did not include atypical pertussis. Atypical pertussis is observed in patients of all ages, particularly in previously vaccinated older children and adults, and often presents as a prolonged cough or acute bronchitis syndrome. In this study there were four patients positive by PCR only who presented with cough syndromes that did not meet the pertussis case definition. These included a 39-year-old female with a cough described as severe that lasted 14 weeks, a 1-month-old unvaccinated infant with a cough that lasted 20 days (considered to represent a probable case by local public health officials), and a 7-year-old fully vaccinated male with a paroxysmal cough that lasted 10 days. The cough duration must be at least 14 days to meet the CDC clinical case definition. A fourth patient, a 2-month-old infant, was considered to have a false-positive PCR result because she also tested positive for RSV. In order to meet the CDC clinical case definition for pertussis, no other likely causes for the symptoms may be present. A limitation of this definition is that it does not take into account dual respiratory-tract infections, which are known to occur. The presence of RSV, an RNA virus, in PCR mixtures could not cause a positive result. Alternatively, the positive PCR result, which was just above the cutoff, may have been due to splashing of postamplification reagents during the microtiter plate detection. We now repeat testing of all borderline-positive specimens. Of the 36 patients positive by PCR only or by DFA only, it is possible that others in addition to the patient described may have had RSV or other viral infections at the time of specimen collection. Unfortunately, specimens for viral testing were rarely collected from this diverse patient population. Medical histories were unavailable for three patients who were positive by PCR only, and as a result these were reported as false positives.
Of the five patients with false-positive DFA results, two possibly had atypical pertussis infections: an adult female (age not reported) with a cough lasting 25 days and a 15-year-old female with an unknown cough history who was epidemiologically linked to a laboratory-confirmed case. Medical histories were unavailable for two DFA-positive patients.
The DFA specificity and positive predictive value in this study (98.2 and 82.8%, respectively) were higher than expected. Ewanowich et al. (9) reported that 85% of DFA-positive, culture-negative patients were also PCR negative. Halperin et al. (12) reported a positive predictive value of 56% for DFA. In an earlier study conducted by us, the positive predictive value of DFA was 62% relative to a expanded gold standard consisting of culture, PCR, and DFA (26). Possible explanations for the different positive predictive values observed in our laboratory are a change in the personnel performing the test or in the reagents, since the DFA procedure and interpretive criteria did not change. The subjectivity of DFA is well documented (9, 20). Nonetheless, our data suggest that DFA can, under tightly controlled conditions, provide a specific diagnosis of pertussis. The sensitivity of DFA, while higher than that of culture in our setting, was still well below that of PCR.
In summary, we have demonstrated that as a routine test, PCR offers the specific detection of B. pertussis, even when used at a high volume during an outbreak. This study also showed, in our public health laboratory setting, that pertussis culture is extremely insensitive, while PCR is highly sensitive. PCR has become the primary diagnostic test for pertussis in our institution. Culture is performed routinely on PCR-positive specimens only, in order to obtain isolates for future antibiotic susceptibility testing and molecular strain typing.
ACKNOWLEDGMENTS
We thank Kot Flora and Elizabeth Miller from the Johnson County Department of Public Health for their professional assistance.
REFERENCES
- 1.Buck G E. Detection of Bordetella pertussis by rapid-cycle PCR and colorimetric microwell detection. J Clin Microbiol. 1996;34:1355–1358. doi: 10.1128/jcm.34.6.1355-1358.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cattaneo L A, Reed G W, Haase D H, Wills M J, Edwards K M. The seroepidemiology of Bordetella pertussis infections: a study of persons ages 1–65 years. J Infect Dis. 1996;173:1256–1259. doi: 10.1093/infdis/173.5.1256. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Case definitions for infectious conditions under public health surveillance. Morbid Mortal Weekly Rep. 1997;46(no. RR-10):25. [PubMed] [Google Scholar]
- 4.Cimolai N, Trombley C, O’Neill D. Diagnosis of whooping cough: a new era with rapid molecular diagnostics. Pediatr Emerg Care. 1996;12:91–93. doi: 10.1097/00006565-199604000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Cook A F, Vuocolo E, Brakel C L. Synthesis and hybridization of a series of biotinylated oligonucleotides. Nucleic Acids Res. 1988;16:4077–4095. doi: 10.1093/nar/16.9.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deen J L, Mink C M, Cherry J D, Christenson P D, Pineda E F, Lewis K, Blumberg D A, Ross L A. Household contact study of Bordetella pertussis infections. Clin Infect Dis. 1995;21:1211–1219. doi: 10.1093/clinids/21.5.1211. [DOI] [PubMed] [Google Scholar]
- 7.Doucet-Populaire F, Richardin R, Bellaiche M, Salomon J L, Bertrand D, Pangon B, Berardi-Grassias L, Ghnassia J C. Abstracts of the 98th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1998. Routine application of PCR for whooping cough diagnosis in infants, abstr. C-222; p. 168. [Google Scholar]
- 8.Erlandsson A, Backman A, Tornqvist E, Olsen P. PCR assay or culture for diagnosis of Bordetella pertussis in the routine diagnostic laboratory? J Infect. 1997;35:221–224. doi: 10.1016/s0163-4453(97)92738-9. [DOI] [PubMed] [Google Scholar]
- 9.Ewanowich C A, Chui L W L, Paranchych M G, Peppler M S, Marusyk R G, Albritton W L. Major outbreak of pertussis in northern Alberta, Canada: analysis of discrepant direct fluorescent-antibody and culture results by using polymerase chain reaction methodology. J Clin Microbiol. 1993;31:1715–1725. doi: 10.1128/jcm.31.7.1715-1725.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glare E M, Paton J C, Premier R R, Lawrence A J, Nisbet I T. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J Clin Microbiol. 1990;28:1982–1987. doi: 10.1128/jcm.28.9.1982-1987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimprel E, Begue P, Anjak I, Betsou F, Guiso N. Comparison of polymerase chain reaction, culture, and Western immunoblot serology for diagnosis of Bordetella pertussis infection. J Clin Microbiol. 1993;31:2745–2750. doi: 10.1128/jcm.31.10.2745-2750.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halperin S A, Bortolussi R, Wort A J. Evaluation of culture, immunofluorescence, and serology for the diagnosis of pertussis. J Clin Microbiol. 1989;27:752–757. doi: 10.1128/jcm.27.4.752-757.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartin M S, Sawyer M H, Mueller T, Espina R, Woo C, Billman G F, Bradley J S. Abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Superiority of Bordetella pertussis PCR over culture for diagnosis of pertussis in children, abstr. D-30; p. 135. [Google Scholar]
- 14.He Q, Mertsola J, Soini H, Skurnik M, Ruuskanen O, Viljanen M K. Comparison of polymerase chain reaction with culture and enzyme immunoassay for diagnosis of pertussis. J Clin Microbiol. 1993;31:642–645. doi: 10.1128/jcm.31.3.642-645.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Q, Schmidt-Schlapfer G, Just M, Matter H C, Nikkari S, Viljanen M K, Mertsola J. Impact of polymerase chain reaction on clinical pertussis research: Finnish and Swiss experiences. J Infect Dis. 1996;174:1288–1295. doi: 10.1093/infdis/174.6.1288. [DOI] [PubMed] [Google Scholar]
- 16.Houard S, Hackel C, Herzog A, Bollen A. Specific identification of Bordetella pertussis by the polymerase chain reaction. Res Microbiol. 1989;140:477–487. doi: 10.1016/0923-2508(89)90069-7. [DOI] [PubMed] [Google Scholar]
- 17.Jansen D L, Gray G C, Putnam S D, Lynn F, Meade B D. Evaluation of pertussis in U.S. Marine Corps trainees. Clin Infect Dis. 1997;25:1099–1107. doi: 10.1086/516099. [DOI] [PubMed] [Google Scholar]
- 18.Jaulhac B, Reyrolle M, Sodahlon Y K, Jarraud S, Kubina M, Monteil H, Piemont Y, Etienne J. Comparison of sample preparation methods for detection of Legionella pneumophila in culture-positive bronchoalveolar lavage fluids by PCR. J Clin Microbiol. 1998;36:2120–2122. doi: 10.1128/jcm.36.7.2120-2122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Jansen D L, Finn T M, Halperin S A, Kasina A, O’Connor S P, Aoyama T, Manclark C R, Brennan M J. Identification of Bordetella pertussis infection by shared-primer PCR. J Clin Microbiol. 1994;32:783–789. doi: 10.1128/jcm.32.3.783-789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcone M J. Bordetella. In: Murray P R, Barron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 566–573. [Google Scholar]
- 21.Nelson S, Matlow A, McDowell C, Roscoe M, Karmali M, Penn L, Dyster L. Detection of Bordetella pertussis in clinical specimens by PCR and a microtiter plate-based DNA hybridization assay. J Clin Microbiol. 1997;35:117–120. doi: 10.1128/jcm.35.1.117-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pham D G, Madico G E, Quinn T C, Enzler M J, Smith T F, Gaydos C A. Use of lambda phage DNA as a hybrid internal control in a PCR-enzyme immunoassay to detect Chlamydia pneumoniae. J Clin Microbiol. 1998;36:1919–1922. doi: 10.1128/jcm.36.7.1919-1922.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reizenstein E, Johansson B, Mardin L, Abens J, Mollby R, Hallander H O. Diagnostic evaluation of polymerase chain reaction discriminative for Bordetella pertussis, B. parapertussis, and B. bronchiseptica. Diagn Microbiol Infect Dis. 1993;17:185–191. doi: 10.1016/0732-8893(93)90094-n. [DOI] [PubMed] [Google Scholar]
- 24.Schlapfer G, Cherry J D, Heininger U, Uberall M, Schmitt-Grohe S, Laussucq S, Just M, Stehr K. Polymerase chain reaction identification of Bordetella pertussis infections in vaccinees and family members in a pertussis vaccine efficacy trial in Germany. Pediatr Infect Dis J. 1995;14:209–214. doi: 10.1097/00006454-199503000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Smith S, Tilton R C. Acute Bordetella pertussis infection in an adult. J Clin Microbiol. 1996;34:429–430. doi: 10.1128/jcm.34.2.429-430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson C, Loeffelholz M, Holcomb L, Long K, Gilchrist M. Abstracts of the 97th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. Primary and confirmatory PCR assays for the detection of Bordetella pertussis, abstr. C-24; p. 123. [Google Scholar]
- 27.van der Zee A, Agterberg C, Peeters M, Schellekens J, Mooi F R. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J Clin Microbiol. 1993;31:2134–2140. doi: 10.1128/jcm.31.8.2134-2140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Zee A, Agterberg C, Peeters M, Mooi F, Schellekens J. A clinical validation of Bordetella pertussis and Bordetella parapertussis polymerase chain reaction: comparison with culture and serology using samples from patients with suspected whooping cough from a highly immunized population. J Infect Dis. 1996;174:89–96. doi: 10.1093/infdis/174.1.89. [DOI] [PubMed] [Google Scholar]
- 29.Wadowsky R M, Michaels R H, Libert T, Kingsley L A, Ehrlich G D. Multiplex PCR-based assay for detection of Bordetella pertussis in nasopharyngeal swab specimens. J Clin Microbiol. 1996;34:2645–2649. doi: 10.1128/jcm.34.11.2645-2649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright S W, Edwards K M, Decker M D, Zeldin M H. Pertussis infection in adults with persistent cough. JAMA. 1995;273:1044–1046. [PubMed] [Google Scholar]