Abstract
Wastewater surveillance for SARS-CoV-2 RNA has been a successful indicator of COVID-19 outbreaks in populations prior to clinical testing. However, this has been mostly conducted in high-income countries, which means there is a dearth of performance investigations in low- and middle-income countries with different socio-economic settings. This study evaluated the applicability of SARS-CoV-2 RNA monitoring in wastewater (n = 132) to inform COVID-19 infection in the city of Bangkok, Thailand using CDC N1 and N2 RT-qPCR assays. Wastewater influents (n = 112) and effluents (n = 20) were collected from 19 centralized wastewater treatment plants (WWTPs) comprising four large, four medium, and 11 small WWTPs during seven sampling events from January to April 2021 prior to the third COVID-19 resurgence that was officially declared in April 2021. The CDC N1 assay showed higher detection rates and mostly lower Ct values than the CDC N2. SARS-CoV-2 RNA was first detected at the first event when new reported cases were low. Increased positive detection rates preceded an increase in the number of newly reported cases and increased over time with the reported infection incidence. Wastewater surveillance (both positive rates and viral loads) showed strongest correlation with daily new COVID-19 cases at 22–24 days lag (Spearman's Rho = 0.85–1.00). Large WWTPs (serving 432,000–580,000 of the population) exhibited similar trends of viral loads and new cases to those from all 19 WWTPs, emphasizing that routine monitoring of the four large WWTPs could provide sufficient information for the city-scale dynamics. Higher sampling frequency at fewer sites, i.e., at the four representative WWTPs, is therefore suggested especially during the subsiding period of the outbreak to indicate the prevalence of COVID-19 infection, acting as an early warning of COVID-19 resurgence.
Keywords: Environmental surveillance, Sewage treatment plants, COVID-19 pandemic, Wastewater-based epidemiology, Human sewage
Graphical abstract
1. Introduction
The coronavirus disease 2019 (COVID-19) has continued to impact world populations since its first outbreak in December 2019 (World Health Organization (WHO), 2020a), accounting for 209 million infected populations worldwide and 4.4 million deaths to date (https://covid19.who.int/, access date August 20, 2021). The severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), which is the etiological agent of COVID-19, mainly targets the respiratory system; however, it can also be excreted through the feces and urine, which may lead to wastewater contamination (Cheung et al., 2020; Jones et al., 2020; Parasa et al., 2020). The monitoring of SARS-CoV-2 genetic materials in wastewater was developed at the early stages of the pandemic using reverse-transcription quantitative polymerase chain reaction (RT-qPCR) (Medema et al., 2020). Since then, over 2,428 sampling sites in 55 countries have been under wastewater surveillance (Naughton et al., 2021) (https://ucmerced.maps.arcgis.com/apps/dashboards/c778145ea5bb4daeb58d31afee389082, access date August 22, 2021). Wastewater-based epidemiology (WBE) has previously been used to monitor the circulation of many substances, such as illegal drugs (Choi et al., 2018; Zuccato et al., 2005), pharmaceuticals (Riva et al., 2020; Shao et al., 2021), and microbial agents such as polioviruses (Garg et al., 2018; WHO, 2003) and noroviruses (Kazama et al., 2017) in communities. Notably, SARS-CoV-2 RNA wastewater surveillance has been demonstrated to provide the following advantages that could supplement the clinical surveillance of COVID-19: 1) delivering early warning of outbreaks or resurgences and evaluating public health interventions (Bibby et al., 2021; Gonzalez et al., 2020; Hillary et al., 2021; Róka et al., 2021; Sherchan et al., 2020; F. Wu et al., 2021); 2) decreasing the number of tested samples that cover a similar number of populations by clinical testing, thus reducing costs, time, and the workload of the public health sector (Betancourt et al., 2021; Hart and Halden, 2020); and 3) identifying the circulation of SARS-CoV-2 from asymptomatic patients, who might constitute a high proportion (Betancourt et al., 2021; Schmitz et al., 2021). The WHO supports the use of wastewater surveillance to complement the clinical surveillance of COVID-19 (WHO, 2020b; WHO Regional Office for Europe, 2020, WHO Regional Office for Europe, 2021). Furthermore, a European Union regulation requires its member states to apply WBE in their countries no later than October 2021 (European Commission, 2021). Effective COVID-19 surveillance and early prevention measures are crucial to prevent disease transmission across boundaries, particularly in areas with higher vulnerability.
Inevitably, the unbalanced distribution of SARS-CoV-2 WBE practice has become evident. Majority of the practitioners (72%) are high-income countries (Naughton et al., 2021) despite the fact that low- and middle-income countries would gain higher benefits from SARS-CoV-2 wastewater surveillance, especially with their relatively lower COVID-19 clinical testing and vaccination rates, limited budgets, and lesser economic resilience (Donde et al., 2020; Gwenzi, 2020). However, it is worth mentioning that there are environmental and socio-economic factors that need to be considered prior to introducing SARS-CoV-2 wastewater surveillance into resource-limited regions. For example, the aging and defects of sewage collection infrastructures constitute challenges that could increase uncollected municipal wastewater portions, degree of stormwater dilution, and sewer retention time (F. Ahmed et al., 2021; Okadera et al., 2020; Pillay et al., 2021). The accumulation of SARS-CoV-2 RNA from various drains into larger sewer systems has also been reported, highlighting that the impact of complicated sewer networks could not be neglected (F. Ahmed et al., 2021). Accelerated microbial degradation could occur due to high temperature, especially in tropical climates (Ahmed et al., 2020a; Bivins et al., 2020). Other than the wastewater travel time to the wastewater treatment plants, the potential of WBE as an early indicator of COVID-19 outbreak also depends on asymptomatic infection prevalence, clinical testing accessibility, and result reporting time (Bibby et al., 2021; Hart and Halden, 2020; Olesen et al., 2021). Therefore, to promote the use of WBE as an early warning of COVID-19 worldwide, there is an urgent need to evaluate its use in countries with different settings.
Thailand has experienced three surges of COVID-19 outbreaks. The first outbreak in January 2020 and the second wave in December 2020 caused a total of 28,863 infected cases and 94 deaths before the beginning of the third resurgence in April 2021 (Department of Disease Control, 2021). In this study, wastewater surveillance was conducted from January to April 2021 to investigate the prevalence and abundance of SARS-CoV-2 RNA in the influents (n = 112) and effluents (n = 20) of Bangkok's 19 municipal wastewater treatment plants (WWTPs) with varying sizes: four large (serving 432,000–580,000 of the population); four medium (serving 70,000–250,000 of the population); and 11 small (serving 2,200–36,000 of the population) WWTPs. The relationship of the viral loads circulated in wastewater and the daily new clinical cases was also investigated to determine the applicability of WBE in determining a COVID-19 outbreak in the population. CrAssphage, a human gut bacteriophage, was used for normalization of SARS-CoV-2 loads when comparing data among different WWTPs, as has been recommended in previous studies (CDC, 2020a; Sweetapple et al., 2021; Wilder et al., 2021; J. Wu et al., 2021). Its consistent presence and abundance in Thailand human sewage has been previously validated (Kongprajug et al., 2019). A surrogate virus, bacteriophage φ6, was also used as a whole process control to determine SARS-CoV-2 recovery. As an enveloped virus, lipid layer covering viral particle enhances viral adhesive ability, which results in higher virus removal from aqueous phase due to attachment on suspended solids in wastewater (Ye et al., 2016). Therefore, the use of bacteriophage φ6 could imitate interaction between SARS-CoV-2 and suspended solids during concentration (Flood et al., 2021; Sherchan et al., 2020; Torii et al., 2021). The results of this study could benefit public health authorities in terms of targeted prevention measures, such as movement restriction policies or increased accessibility to clinical surveillance and vaccination in low- or middle-income countries.
2. Materials and methods
2.1. Study sites and sample collection
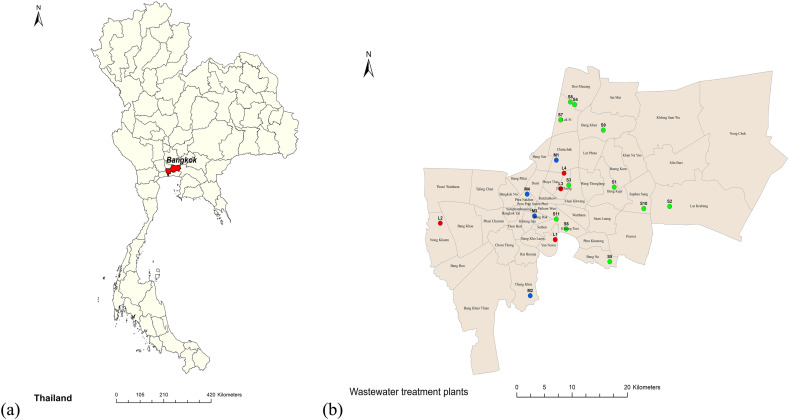
A total of 132 samples were taken comprising 112 influents and 20 effluents from four large (L1 – L4), four medium (M1 – M4), and 11 small (S1 – S11) WWTPs during seven sampling events from January to April 2021 (Table S1 and Fig. 1 ). All sewerage systems in Bangkok are combined sewerage systems receiving sewage and stormwater runoff; however, the precipitation records from January to April 2021 demonstrated very light rains in all sampling events (Table S2). In-sewer travel times of each WWTPs vary by sewershed, with shorter times for small than for medium and large WWTPs. The influent samples were collected after the bar screening and the effluent samples were collected after the secondary treatment processes of each WWTP (Table S3).
Fig. 1.
Sampling locations in (a) Bangkok, Thailand; (b) large (L1 – L4; red), medium (M1 – M4; blue), and small (S1 – S11; green) municipal WWTPs.
The four large WWTPs (L1 – L4) and the two medium WWTPs (M1 and M2) have their own autosamplers, whereas the others do not (Table S1). Composite samples were collected from influents and effluents using an autosampler by combining multiple samples 1 h apart over a 24-h period. Grab and composite samples of 1.5 L were collected between 8:30 and 10:30 AM in a sterile container and transported on ice to the laboratory within 3 h. Therefore, 44 of the 24-h composites and 88 grabbed samples were obtained for this study. The samples were stored at 5 °C for no more than 24 h before further analysis.
2.2. Virus concentration and RNA extraction
Samples of 500–1,000 mL were incubated at 60 °C in a water bath for 1 h prior to concentrating them through electronegative membrane filtration method with magnesium chloride (MgCl2) addition according to the reported highest recovery based on the murine hepatitis virus surrogate (Ahmed et al., 2020b). The magnesium cations help in linking the negatively charged virus particles to the electronegative membrane during filtration (Corpuz et al., 2020; Haramoto et al., 2018; Katayama et al., 2002). In brief, the MgCl2 powder was added to the sample to obtain a final concentration of 25 mM MgCl2. Then, the samples were filtered using a 0.45-μm-pore size mixed cellulose-ester membrane (HAWP04700, Merck Millipore, USA) through vacuum filtration. The membrane was immediately cut into small pieces and put into a 2 mL ZR BashingBead™ Lysis tube (Zymo Research, USA) containing bead sizes of 0.1 and 0.5 mm. Subsequently, 750 μL DNA/RNA Shield™ (Zymo Research, USA) was added into the tube before shaking for 20 min. After shaking, the tube was centrifuged at 13,200 ×g for 30 s. The 140 μL supernatant was collected for viral genome extraction using the QIAamp viral RNA mini- kit (Qiagen, Germany) to retrieve a final volume of 60 μL RNA extract. The extracted RNA was stored at −80 °C until further analysis.
2.3. Process recovery characterization using bacteriophage φ6
A stock solution of bacteriophage φ6 (NRBC 105899) was prepared by mixing 100 μL of bacteriophage φ6 with 1 mL of Pseudomonas syringae (NRBC 14084) in a tube containing nutrient broth liquid (Difco, USA) added with yeast extract, glucose, and magnesium sulfate (MgSO4) according to the manufacturer's procedure. The virus was allowed to grow at 26 °C for 3 days in a cooled shaking incubator. Afterward, bacterial cells were removed from the culture media by centrifugation, followed by filtration through a 0.2-μm-pore size mixed cellulose acetate membrane (Merck, USA). The viral concentration was determined using a single-layer agar method with a mixture molten agar of nutrient broth, yeast extract, glucose, potassium dihydrogen phosphate (KH2PO4), dipotassium hydrogen phosphate (K2HPO4), and pre-grown P. syringae at 26 °C for 24 h. The number of plaques was counted and the concentration of bacteriophage φ6 was reported as plaque-forming unit (PFU) per milliliter. The viral solution was kept at −20 °C until use.
For the recovery experiment, 100–500 μL was inoculated into 100–500 mL of water sample to achieve an initial concentration of around 103–105 most probable number (MPN)/mL. The concentration of bacteriophage φ6 after the virus concentration and RNA extraction was measured through RT-qPCR analysis. The genome copies (GC) were calculated to MPN/mL through end-point dilution assay. The recovery efficiency was calculated by dividing the bacteriophage φ6 in MPN/mL after the concentration with those before the concentration.
2.4. Quantitative PCR assays and standard curves
The US CDC N1 and N2 multiplex RT-qPCR assays targeting two different regions of the SARS-CoV-2 N gene, as well as the bacteriophage φ6 RT-qPCR assay, were performed using the QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, USA), as shown in Table S4. SARS-CoV-2 detection was performed using the multiplex RT-qPCR assay. The 10 μL RT-qPCR reaction mixture for SARS-CoV-2 comprised 2.5 μL of TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific, USA), 0.3 μL each of 100 nM forward and reverse primers, 0.2 μL each of 300 nM hydrolysis probe, 2.5 μL of extracted template, and 3.4 μL of sterile water. The 10 μL RT-qPCR reaction mixture for bacteriophage φ6 comprised 2.5 μL of TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific, USA), 1 μL each of 1000 nM forward and reverse primers, 0.3 μL of 300 nM hydrolysis probe, 2.5 μL of extracted template, and 2.7 μL of sterile water. The crAssphage gene (CPQ_056) was analyzed through a qPCR assay using the QuantStudio3 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA), as shown in Table S4. The 20 μL qPCR mixture was composed of 0.8 μL of each 10 μM forward and reverse primers, 0.4 μL of 10 μM hydrolysis probe, 2 μL of extracted template, 6 μL of 1 μg/μL BSA, and 10 μL of the Luna Universal probe qPCR master mix (New England BioLabs, Inc., USA). The RT-qPCR and qPCR protocols were performed according to the MIQE guideline (Bustin et al., 2009). The RT-qPCR and qPCR results were analyzed using QuantStudio Design & Analysis software (Applied Biosystems, Thermo Fisher Scientific, USA) with automatic baseline and manual adjustments of the threshold value to 0.115, 0.01, and 0.036, for N1, N2, and crAssphage, respectively. Each sample was run in duplicate, and the average Ct was used when the standard deviation of Ct was less than 0.5; otherwise, an additional run was undertaken (Kongprajug et al., 2021). In each instrumental run, the positive and negative controls were included. Inhibition was assessed using the TaqMan™ Exogenous Internal Positive Control Reagents following the manufacturer's instruction (Thermo Fisher Scientific, USA).
The qPCR standard curves were constructed using linearized synthetic plasmid standards for the N1, N2, and the crAssphage markers (Kongprajug et al., 2019) (GeneArt®, Invitrogen, Thermo Fisher Scientific, Waltham, MA USA). The assay limit of quantification (ALOQ) for the N1 marker was expressed as the concentration in GC/reaction that showed positive detection of 8 from all 10 standard replicates (Ct = 35.78, CV = 50%). The assay limit of detection (ALOD) for the N1 marker was determined as the concentration of genomic copies below 1.36 GC/reaction (Ct = 38). The ALOD for the N2 marker was set at Ct of 40. The ALOD and ALOQ for the crAssphage qPCR assay were 20 and 50 GC/reaction, respectively (Kongprajug et al., 2019).
2.5. Viral load and population equivalent calculations and COVID-19 case data
The SARS-CoV-2 concentrations (C SAR) in the wastewater samples were calculated from the RT-qPCR analysis of the N1 marker to the GC/100 mL of the sample by incorporating sample volume and RNA extraction volume. The viral loads in the wastewater samples were calculated by multiplying (C SAR) with the wastewater flow rate (Q) from each treatment plant (data requested from the Department of Drainage and Sewerage, Bangkok Metropolitan Administration [BMA]). CrAssphage was used to normalize the SARS-CoV-2 concentrations in the form of population equivalent (PE) based on Eq. (1) as follows:
| (1) |
where C cr is the crAssphage concentration in a wastewater sample (GC/100 mL) measured by qPCR assay, Q is the influent flow rate to the wastewater treatment plant (m3/d), P w is the wastewater volume produced per person per day or 160 L/person-day according to the recommended value from the Office of Natural Resources and Environmental Policy and Planning, Thailand, and C cr,ave is the averaged concentration of crAssphage in wastewater in Thailand of 2.79 × 106 GC/100 mL (Kongprajug et al., 2019). The normalized SARS-CoV-2 viral loads were calculated by dividing the viral loads with PE. Although the P w and C cr,ave contain uncertainties that were not accounted for in Eq. (1), this fixed-value PE denominator did not induce bias to the normalized viral loads because it was commonly applied to all viral loads. New clinical case numbers in Bangkok were requested from the Division of Communicable Disease Control, Department of Health, BMA, and also retrieved from the publicly available website of the Department of Disease Control, Ministry of Public Health (https://ddc.moph.go.th/viralpneumonia/index.php). The population per district in 2020 was retrieved from the Administrative Strategy Division, BMA.
2.6. Statistical data analysis
Spearman's rank correlation analysis was used to evaluate the temporal relationship between the viral data in wastewater (viral loads and positive detection rates in wastewater) and the new clinical case numbers in the study areas. To evaluate the temporal effect, the viral load and positive detection rate data were paired with 5-day averaged new cases at each lag time from 0 to 40 days lag to calculate Spearman's rank correlation coefficients (Spearman's Rho). Area maps showing viral loads and daily new cases were created by representing data in clusters using the Jenks natural breaks classification method.
3. Results and discussion
3.1. RT-qPCR standard curve characteristics and quality controls
The significance of the reporting standard curve parameters and the associated controls is emphasized for the reliability of SARS-CoV-2 RNA quantitative data in wastewater surveillance (Bivins et al., 2021a; Borchardt et al., 2021). In this study, the RT-qPCR standard curve characteristics for N1, bacteriophage φ6, and crAssphage are shown in Table S5. The slopes ranged from −3.364 to −3.487 and the Y-intercepts from 33.996 to 41.680. The PCR efficiencies were 93.5%–98.3%. The ALOD for N1, bacteriophage φ6, and crAssphage was 1 GC/reaction, 5 MPN/reaction, and 20 GC/reaction, respectively, while the ALOQ was 6 GC/reaction, 36 MPN/reaction, and 50 GC/reaction, respectively.
No inhibition was observed in 56 representative wastewater samples, based on the internal amplification controls. NTCs were negative, indicating the absence of cross-contamination during the RT-qPCR and qPCR assays. The process efficiencies, as identified by bacteriophage φ6, in 76 representative samples (57.6%) indicated averaged recoveries of 1.54%–3.57% when 103 MPN/mL of initial bacteriophage stock was added in the early sampling dates, while showing higher averaged recovery of 36.15% when 105 MPN/mL of initial bacteriophage stock was added in the later date (Table S6). This is similar to the trend previously observed in which the higher initial concentrations expressed higher recoveries (W. Ahmed et al., 2021b). Low recoveries as observed by the spiked surrogate in January, February, and March 2021 (Table S6), therefore, could have increased the likelihood of non-detectable SARS-CoV-2 results. This study followed the concentration method using an adsorption-extraction method with magnesium chloride (MgCl2) pre-addition according to the reported highest recovery of 65.7% ± 23.8% (mean ± S.D.). This is based on the murine hepatitis virus surrogate (Ahmed et al., 2020b) with modification in the membrane elution step by using a 2 mL ZR BashingBead™ Lysis tube (Zymo Research, USA) instead of a beading tube with Precellys Evolution 24 tissue homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) and a different RNA extraction kit. However, another study following a similar concentration method demonstrated very low recovery of the seeded bovine coronavirus surrogate (Jafferali et al., 2021). Nonetheless, the whole process control reflected recoveries from all downstream processes including water concentration, RNA extraction, and RT-qPCR steps, which expressed high variability depending on the microbial target, water matrix, types of laboratory protocols and instruments, and data analysis method (Ahmed et al., 2022; W. Ahmed et al., 2021b; Haramoto et al., 2018; Kongprajug et al., 2020; Rusiñol et al., 2020). It is suggested that the recovery efficiencies be reported separately from the observed concentrations of viruses, rather than incorporating the recoveries into the virus concentrations (Haramoto et al., 2018; Kantor et al., 2021).
3.2. Ct values and detection rates of SARS-CoV-2 RNA in wastewater
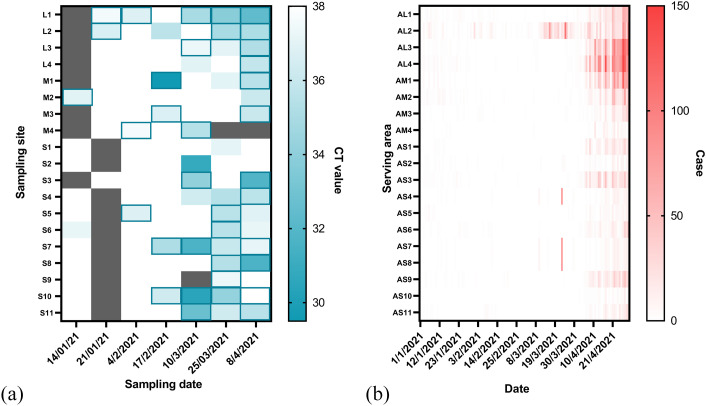
N1 and N2 markers were present at 39.39% and 28.78%, respectively, with 28.03% co-presence of the 132 wastewater samples. N2 was detected less frequently, as previously observed (Medema et al., 2020). The Ct values of the N1 and N2 markers were also moderately correlated in all samples (n = 37, Spearman's Rho = 0.69) and strongly correlated in the wastewater influents (n = 35, Spearman's Rho = 0.74) (Fig. S1). In most samples, N2 showed equal or higher Ct values than N1, which is in agreement with other reports (Chakraborty et al., 2021; Colosi et al., 2021; Wu et al., 2020). In addition, the PCR efficiency of N1 (93.56%) was higher than that of N2 (81.35%). Both assays were used to detect the same N gene; thus, only N1 gene quantification was calculated using the N1 standard curve. Of the 112 wastewater influents, 44.64% were positive for N1 (Ct < 38) (Fig. 2 ).
Fig. 2.
Heat maps showing the (a) RT-qPCR cycle threshold values (Ct) of SARS-CoV-2 N1 gene (Ct < 38) in the wastewater influent samples from the seven sampling events conducted in the four large (L1 – L4), four medium (M1 – M4), and 11 small (S1 – S11) WWTPs with N2 gene positive detection (Ct < 40) marked by framing; (b) number of new COVID-19 cases reported daily in the districts serviced by the four large (AL1 – AL4), four medium (AM1 – AM4), and 11 small (AS1 – AS11) WWTPs. Dark cells indicate no sample collection.
During the first two sampling events, four of 20 influents and two of 20 effluents were detectable but not quantifiable for the N1 gene (i.e., ALOQ < Ct < ALOD). Due to the fact that the COVID-19 outbreak subsided in January 2021 in Thailand and other studies reported lesser frequencies of SARS-CoV-2 RNA presence in wastewater effluents (Gerrity et al., 2021; Hasan et al., 2021; Hemalatha et al., 2021; Sherchan et al., 2020), wastewater effluents were not further collected for subsequent sampling events. Of the four positive influents, either negative detection or higher Ct value was observed in corresponding effluents indicating SARS-cov-2 reduction in activated sludge processes from the four WWTPs (L1, L2, M2, and S6). However, due to small numbers of positive samples and very low concentrations, the removal efficiencies of the treatment processes could not be estimated. The finding in this study was consistent with a previous study observing the reduction of SARS-CoV-2 N gene in the activated sludge process (Serra-Compte et al., 2021).
Higher N1 and N2 positive rates in wastewater influents were also observed in large WWTPs (L1 – L4) at 58.33% compared with 39.13% and 41.54% positive detection in medium (M1 – M4) and small WWTPs (S1 – S11), respectively. There are many confounding factors affecting the detectability of SARS-CoV-2 RNA in wastewater, such as variations in individual viral shedding and duration, sewer system condition and retention time, and environmental factors affecting degradation, particularly water temperature (Ahmed et al., 2020a; Albert et al., 2021; Cheung et al., 2020; Hokajärvi et al., 2021). Moreover, the sampling method has been reported to affect the detectability of SARS-CoV-2 RNA in wastewater (W. Ahmed et al., 2021a; Bivins et al., 2021b; Feng et al., 2021; Rafiee et al., 2021). In this study, six WWTPs (L1 – L4 and M1 – M2) were with their own autosamplers and wastewater samples were composited for 24 h, while other WWTPs delivered grab samples. This study aimed to evaluate the actual situations in sampling sites, so inevitable biases could have confounded the results.
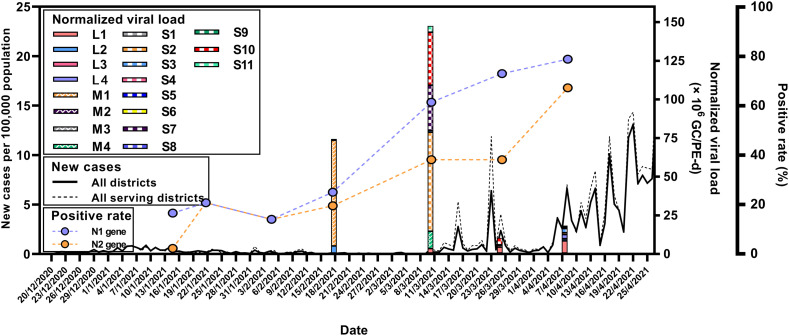
The temporal distributions of the N1 and N2 markers were at higher frequencies compared with those obtained at the first event (January 14, 2021) up to the last event (April 8, 2021), concurring with the higher infection incidences from the districts serviced by each WWTP (Fig. 2). Interestingly, the first positive detection of SARS-CoV-2 RNA was found at the first sampling event from M2 and S6 WWTPs, located on the west and east sides of Bangkok, respectively, while the clinical cases observed in both served areas were lower than 10. Interestingly, the positive detection rates of the wastewater samples at each event exhibited the same upward trend as the total number of new cases in Bangkok, but the increase in the positive detection rates occurred earlier (Fig. 3 ).
Fig. 3.
Normalized SARS-CoV-2 RNA loads (×106 GC/PE-day) in influents of the 19 WWTPs plotted in the bar graph and compared with new clinical confirmed cases per 100,000 population from all districts in Bangkok (solid line) and in the districts served by the 19 WWTPs (dotted line). The positive detection rates of N1 and N2 genes in the wastewater samples are shown by closed circles.
3.3. SARS-CoV-2 RNA viral loads in wastewater and daily new cases
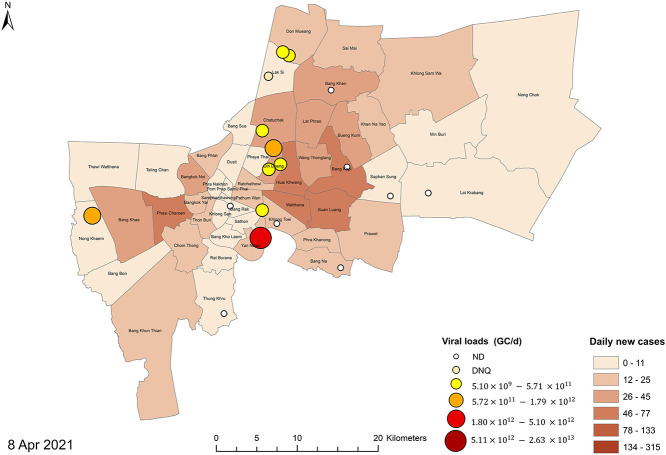
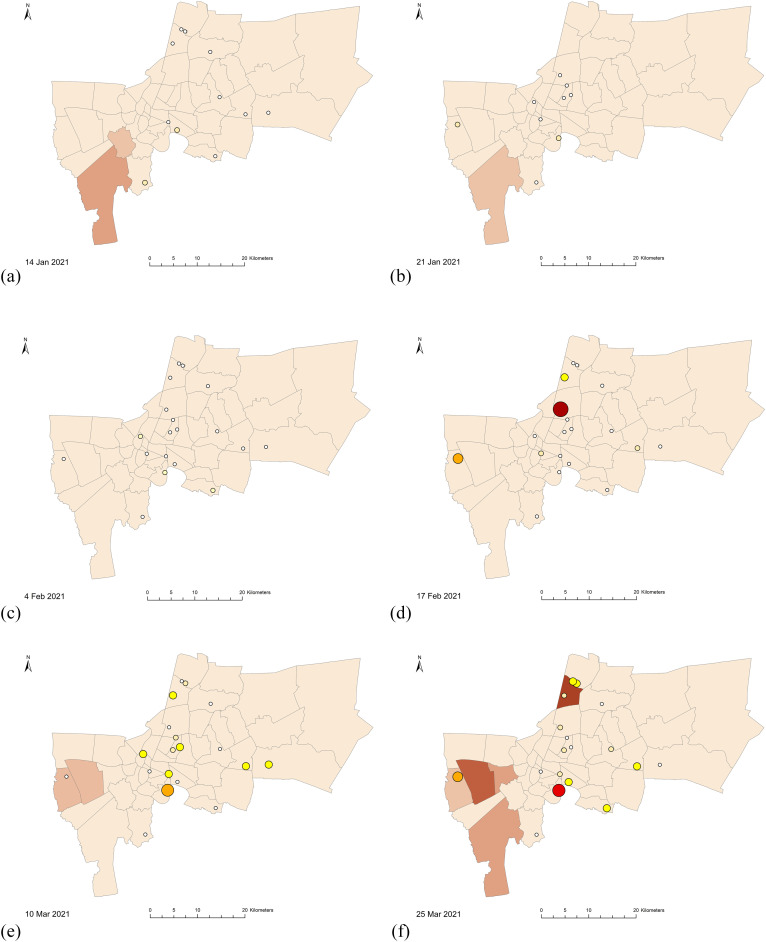
The SARS-CoV2 RNA concentrations in the wastewater influent samples calculated from the N1 marker ranged from 1.77 to 4.35 log10 GC/100 mL (n = 25), comparable with 2–5 log10 GC/100 mL reported levels from other geographical regions (Chakraborty et al., 2021; Pillay et al., 2021; Sherchan et al., 2020; F. Wu et al., 2021). Normalized viral loads of wastewater influent samples were plotted and compared with new cases from all districts in Bangkok and from districts served by the 19 WWTPs (Fig. 3). While SARS-CoV-2 RNA was detected at the first event (January 14, 2021), the viral RNA first became quantifiable at the fourth event (February 17, 2021), with total normalized viral loads from L2, M1, and S7 WWTPs at 7.87 log10 GC/PE-day. However, only one reported case was shown on this day from the serving districts. The spatial distributions of the viral loads from the corresponding WWTPs and the new cases in each district are represented in a map in Fig. 4 . The first peak of new cases from all serving districts occurred on March 16, 2021 at 5.31 new cases/100,000 populations, corresponding to the outbreak cluster of the Bang Khae Market complex, one of the largest fresh food markets on the west side of Bangkok. However, the normalized viral loads from the preceding day (March 10, 2021) were not contributed by the Bang Khae serving area L2. Another new case peak on March 23, 2021 was from the Immigration Bureau's detention centers in Bang Khen (Lak Si District), but the normalized SARS-CoV-2 loads were not detected from the Lak Si District serving area S7 on the subsequent sampling date of March 25, 2021. The rise of infection incidence began on April 4, 2021, which marked the third COVID-19 wave in Thailand. The outbreak was identified from a nightclub cluster, whose wastewater was not included in any of the WWTPs studied. The viral signals on March 25, 2021, 10 days earlier than the start of the outbreak as identified by clinical surveillance, were shown by the L1, S4, S5, S6, S8, and S10 WWTPs. Overall, the SARS-CoV-2 RNA signals did not exactly match the trends with the new cases in the served districts on the same day but appeared prior to the rise of new cases in the served and adjacent districts. The differences in in-sewer travel time and sewage losses among sewersheds could play important roles in virus detectability at each WWTP (Hart and Halden, 2020; Okadera et al., 2020). It should be noted that the clinical cases reported were retrieved from the Health Bureau, Bangkok Metropolitan Administration and indicated the patients' current addresses. Therefore, potential movements or daily commute in different districts could affect the collection of wastewater.
Fig. 4.
Maps of Bangkok showing the viral loads from each of the 19 WWTPs (in circles) and the 5-day averaged new COVID-19 cases on the following sampling dates: (a) January 14, 2021, (b) January 21, 2021, (c) February 4, 2021, (d) February 17, 2021, (e) March 10, 2021, (f) March 25, 2021, and (g) April 8, 2021.
3.4. Correlation between viruses in wastewater (positive rates and viral loads) and daily new cases at different lag times
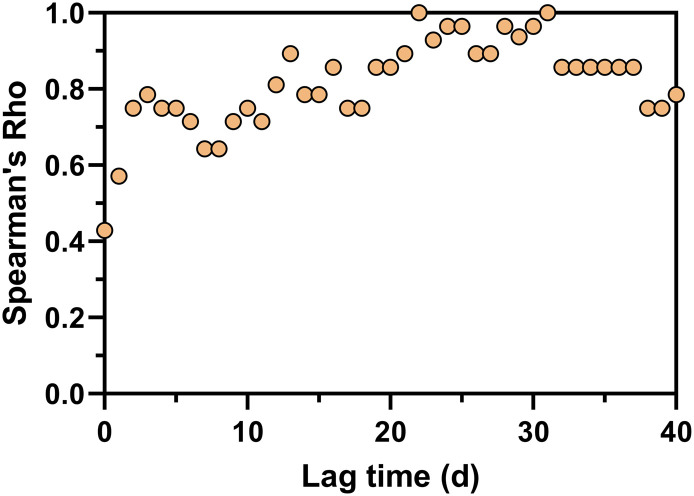
The positive detection rates of SARS-CoV-2 RNA in wastewater were correlated with 5-day averaged new COVID-19 cases (Fig. 5 ). Spearman's rank correlation coefficients showed the highest value at 22 days lag (Spearman's Rho = 1.0). The ability to detect viral loads in WWTPs with different sizes prior to the new case report was further investigated. Spearman's rank correlation analysis was performed between viral loads in wastewater and the new cases at different lag times (Fig. 6 ). When all 19 WWTPs were considered, the strongest correlation was shown at 23–24 days lag time (Spearman's Rho = 0.85) (Fig. 6a). Interestingly, large WWTPs (L1 – L4) serving 432,000–580,000 populations reflected similar trends, showing the strongest correlation at 21–22 days lag time (Spearman's Rho = 0.64) (Fig. 6b). The medium (70,000–250,000 populations) and small (2,200–36,000 populations) WWTPs demonstrated very weak correlations between viral loads and the daily confirmed cases (Fig. 6b). The details of the correlation analysis for each WWTP are presented in Table S7. The weak correlation in medium and small WWTPs may be due to the limited information on new COVID-19 cases, which were only from the facilities in the corresponding sewersheds. In light of the foregoing, the viral loads from large WWTPs could reflect the SARS-CoV-2 circulation and dynamics of the city. Therefore, routinely monitoring large WWTPs could be beneficial as a complement to clinical surveillance at the city scale.
Fig. 5.
Spearman's rank correlation coefficients (Spearman's Rho) between virus positive detection rates and 5-day averaged new cases from all corresponding served districts.
Fig. 6.
Spearman's rank correlation coefficients (Spearman's Rho) between viral loads (log10 GC/d) and 5-day averaged new cases at different lag times from 0 to 40 days for (a) all 19 WWTPs and (b) large, medium, and small WWTPs.
3.5. Implications of SARS-CoV-2 wastewater surveillance
This study demonstrated the successful applicability of SARS-CoV-2 WBE in 19 sewersheds from four large, four medium, and 11 small WWTPs to determine COVID-19 circulation in the population of Bangkok prior to the clinical surveillance. It is recommended that wastewater from large WWTPs be routinely monitored for SARS-CoV-2 RNA because it could reflect the dynamics of the whole city. WWTPs of smaller sizes could also be beneficial in indicating hotspots of outbreaks and recommending appropriate interventions, such as individual clinical testing (Betancourt et al., 2021; Gibas et al., 2021). This study revealed the correlation between viral loads and clinical reported cases at relatively long lag times of 22–24 days, a period that is longer than other reported studies (Bibby et al., 2021; Róka et al., 2021; Zhu et al., 2021). It is possible that asymptomatic infection is responsible for the virus signal in wastewater, as SARS-CoV-2 RNA signals were detected from in-facility sewage systems in Bangkok during reported low COVID-19 prevalence (Wannigama et al., 2021). Furthermore, during the study period of January to April 2021, there was a low prevalence of COVID-19 infection in Thailand. Lesser awareness of COVID-19 symptoms and accessibility to clinical testing led to low clinical surveillance rates. Potentially, in future resurgences, clinical testing accessibility will be improved with the availability of commercial test kits, resulting in lesser lag time.
The frequency of wastewater sampling is another important factor for WBE to indicate the prevalence of COVID-19 in the community. In the United States and in European countries, it is recommended to sample wastewater once or twice a week (CDC, 2020b; European Commission, 2021). However, for resource-constrained regions, balancing the budget with the application of SARS-CoV-2 WBE is crucial, especially during times when the outbreak has subsided. Although this study showed that the surveillance of wastewater influents of the treatment plants in Bangkok every 2–3 weeks could provide adequate information about the prevalence of COVID-19 during the study period, it is suggested that the representative four large WWTPs be monitored at higher frequency, to serve as an early warning of future COVID-19 resurgence.
The significance of WBE at the national level has been increasingly recognized, especially in high-income countries (CDC, 2020b; European Commission, 2021). However, national structures and wastewater surveillance systems in middle- and low-income countries have lesser degrees of availability. All stakeholders should be involved to form a national governance system, with supportive legislation on the integration of public health, wastewater, environmental water, and outbreak management (Takeda et al., 2021). The institutionalization of wastewater surveillance systems will not only benefit the surveillance and control of the COVID-19 pandemic but also future infectious ‘disease x’ outbreak, as defined by the WHO (2018). Wastewater surveillance application will be an important tool to strengthen existing public health interventions to effectively curb COVID-19 transmission in the community (Giné-garriga et al., 2021; Triukose et al., 2021).
4. Conclusions
This study reported the first successful SARS-CoV-2 WBE application in 19 centralized wastewater systems in Bangkok (n = 132) with varying sizes, which serve 49.6% of Bangkok's population (serving 2,200 to 580,000 of the population). The CDC N1 assay yielded higher detection rates and mostly lower Ct values than the CDC N2 assay. High recovery rates (36.15%) of the bacteriophage φ6 surrogate were achieved with the highest seed titer of 5.7 × 105 MPN/mL. Of the 112 wastewater influents, 44.64% were positive for N1. Interestingly, the positive detection rates of SARS-CoV-2 RNA in the wastewater from all of the WWTPs increased along with the clinical cases over time. The strongest correlation between the positive rates and the 5-day averaged new cases from all of the corresponding districts served occurred at 22 days lag time. The viral loads in the wastewater were not associated with the clinical cases from the served or adjacent districts on the present day. However, a significant correlation was observed at 22–24 days lag time when considering combined viral loads and related clinical cases from all of the 19 WWTPs and from the four large WWTPs. Consequently, large WWTPs are recommended for the routine monitoring of SARS-CoV-2 RNA because they reflect the city-scale dynamics of viral loads.
CRediT authorship contribution statement
Jatuwat Sangsanont: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Writing – original draft, Writing – review & editing, Project administration, Funding acquisition. Surapong Rattanakul: Conceptualization, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization, Funding acquisition. Akechai Kongprajug: Formal analysis, Investigation, Visualization. Natcha Chyerochana: Investigation. Montakarn Sresung: Investigation. Nonnarit Sriporatana: Investigation. Nasamon Wanlapakorn: Resources, Writing – review & editing. Yong Poovorawan: Resources, Writing – review & editing, Supervision. Skorn Mongkolsuk: Conceptualization, Resources, Writing – review & editing, Supervision, Funding acquisition. Kwanrawee Sirikanchana: Conceptualization, Formal analysis, Resources, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by the TSRI Fund (CU_FRB640001_01_21_6), Research Strengthening Project of Faculty of Engineering (King Mongkut's University of Technology Thonburi) and the Chulabhorn Research Institute (grant no. 312/3057). Ms. Thitima Srathongneam is thankfully acknowledged for assistance with map preparation. We acknowledge the Department of Drainage and Sewerage, Bangkok Metropolitan Administration (BMA), and the Division of Communicable Disease Control, Department of Health, BMA, for providing data on wastewater flow rates and daily new cases.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.151169.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR- based recovery of murine hepatitis virus, a surrogate for SARS- CoV-2 from untreated wastewater warish. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F., Islam M.A., Kumar M., Hossain M., Bhattacharya P., Islam M.T., Hossen F., Hossain M.S., Islam M.S., Uddin M.M., Islam M.N., Bahadur N.M., Didar-ul-Alam M., Reza H.M., Jakariya M. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation Centre in Bangladesh: variation along the sewer network. Sci. Total Environ. 2021;776 doi: 10.1016/j.scitotenv.2021.145724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Gyawali P., Sherchan S.P., Simpson S.L., Thomas K.V., Verhagen R., Kitajima M., Mueller J.F., Korajkic A. Intraday variability of indicator and pathogenic viruses in 1-h and 24-h composite wastewater samples: implications for wastewater-based epidemiology. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Simpson S.L., Smith W.J.M., Metcalfe S., McMinn B., Symonds E.M., Korajkic A. Comparative analysis of rapid concentration methods for the recovery of SARS-CoV-2 and quantification of human enteric viruses and a sewage-associated marker gene in untreated wastewater. Sci. Total Environ. 2021;799 doi: 10.1016/j.scitotenv.2021.149386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Bibby K., Bivins A., Linda L., Bofill-mas S., Bosch A., Brandão J., Choi P.M., Ciesielski M., Donner E., D’Souza N., Farnleitner A.H., Gerrity D., Gonzalez R., Griffith J.F., Gyawali P., Haas C.N., Hamilton K.A., Hapuarachchi C., Harwood V.J., Haque S.R., Jackson G., Khan S., Khan W., Kitajima M., Korajkic A., Rosa G.La, Layton B.A., Lipp E., McLellan S., McMinn B., Medema G., Metcalfe S., Meijer W., Mueller J., Murphy H., Naughton C.C., Noble R.T., Payyappat S., Petterson S., Pitkänen T., Rajal V.B., Reyneke B., Jr., F.A.R, Rose J.B., Rusiñol M., Sadowsky M., Sala-Comorera L., Setoh Y.X., Sherchan S., Sirikanchana K., Smith W., Steele J., Subburg R., Symonds E.M., Thai P., Thomas K., Tynan J., Toze S., Thompson J., Whitely A.S., Wong J., Sano D., Wuertz S., Xagoraraki I., Zhang Q., Zimmer-Faust A.G., Shanks O. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S., Ruíz A., Pemán J., Salavert M., Domingo-Calap P. Lack of evidence for infectious SARS - CoV - 2 in feces and sewage. Eur. J. Clin. Microbiol. Infect. Dis. 2021;2–4 doi: 10.1007/s10096-021-04304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Bivins A., Wu Z., North D. Making waves: plausible Lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 2021;117438 doi: 10.1016/j.watres.2021.117438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V., Bibby K. Persistence of SARS-CoV - 2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;1–6 doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Kaya D., Bibby K., Simpson S.L., Bustin S.A., Shanks O.C., Ahmed W. Variability in RT-qPCR assay parameters indicates unreliable SARS- CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021;117516 doi: 10.1016/j.watres.2021.117516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Wu Z., Shaffer M., Ahmed W., Bibby K. Within- and between-day variability of SARS-CoV-2 RNA in municipal wastewater during periods of varying COVID-19 prevalence and positivity. ACS ES&T Water. 2021 doi: 10.1021/acsestwater.1c00178. [DOI] [Google Scholar]
- Borchardt M.A., Boehm A.B., Salit M., Spencer S.K., Wigginton K.R., Noble R.T. The environmental microbiology minimum information (EMMI) guidelines: qPCR and dPCR quality and reporting for environmental microbiology. Environ. Sci. Technol. 2021;55:10210–10223. doi: 10.1021/acs.est.1c01767. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J.F., Kubista M., Mueller R.D., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- CDC COVID-19: 2. Wastewater surveillance testing methods [WWW document] 2020. https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/testing-methods.html URL. (accessed 10.16.21)
- CDC . 2020. COVID-19: 1. Developing a Wastewater Surveillance Sampling Strategy. [Google Scholar]
- Chakraborty P., Pasupuleti M., Jai Shankar M.R., Bharat G.K., Krishnasamy S., Dasgupta S.C., Sarkar S.K., Jones K.C. First surveillance of SARS-CoV-2 and organic tracers in community wastewater during post lockdown in Chennai, South India: methods, occurrence and concurrence. Sci. Total Environ. 2021;778 doi: 10.1016/j.scitotenv.2021.146252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., Yip C.C.Y., Leung K.H., Fung A.Y.F., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J.X., To K.K.W., Chan K.H., Yuen K.Y., Leung W.K. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends Anal. Chem. 2018;105:453–469. doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- Colosi L.M., Barry K.E., Kotay S.M., Porter M.D., Poulter M.D., Ratliff C., Simmons W., Steinberg L.I., Wilson D., Morse R., Zmick P., Mathers A.J. Development of wastewater pooled surveillance of SARS-CoV-2 from congregate living settings. Appl. Environ. Microbiol. 2021;1–44 doi: 10.1128/AEM.00433-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpuz M.V.A., Buonerba A., Vigliotta G., Zarra T., Ballesteros F., Campiglia P., Belgiorno V., Korshin G., Naddeo V. Viruses in wastewater: occurrence, abundance and detection methods. Sci. Total Environ. 2020;104743 doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Disease Control Confirmed cases of COVID-19 in Thailand [WWW document] 2021. https://ddc.moph.go.th/viralpneumonia/eng/index.php URL. (accessed 8.22.21)
- Donde O.O., Atoni E., Muia A.W., Yillia P.T. COVID-19 pandemic: water, sanitation and hygiene (WASH) as a critical control measure remains a major challenge in low-income countries. Water Res. 2020;116793 doi: 10.1016/j.watres.2020.116793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Commission Commission recommendation of 17.3.2021 on a common approach to establish a systematic surveillance of SARS-CoV-2 and its variants in wastewaters in the EU. Off. J. C. 2021:1–6. https://ec.europa.eu/environment/pdf/water/recommendation_covid19_monitoring_wastewaters.pdf [Google Scholar]
- Feng S., Roguet A., McClary-Gutirrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. medRxiv; 2021. Evaluation of Sampling Frequency and Normalization of SARS-CoV-2 Wastewater Concentrations for Capturing COVID-19 Burdens in the Community. [Google Scholar]
- Flood M.T., D’Souza N., Rose J.B., Aw T.G. Methods evaluation for rapid concentration and quantification of SARS-CoV-2 in raw wastewater using droplet digital and quantitative RT-PCR. Food Environ. Virol. 2021;2 doi: 10.1007/s12560-021-09488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Pattamadilok S., Bahl S. Successes and challenges of expansion of environmental poliovirus surveillance in the WHO South-East Asia Region. WHO South-East Asia J. Public Health. 2018;7:122–128. doi: 10.4103/2224-3151.239424. [DOI] [PubMed] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Bharati Visva, Barua, Brazell L.R., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Akella S., Tang W., Chen D., Schlueter J., Munir M. Implementing Building Level SARS-CoV-2 Wastewater Surveillance on a University Campus. medRxiv; 2021. pp. 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giné-garriga R., Delepiere A., Ward R., Alvarez-sala J., Alvarez-murillo I., Mariezcurrena V., Göransson H., Saikia P., Avello P., Thakar K., Ibrahim E., Nouvellon A., El O., Hutton G., Jiménez A. COVID-19 water, sanitation, and hygiene response : review of measures and initiatives adopted by governments, regulators, utilities, and other stakeholders in 84 countries. Sci. Total Environ. 2021;795 doi: 10.1016/j.scitotenv.2021.148789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwenzi W. Leaving no stone unturned in light of the COVID-19 faecal-oral hypothesis? A water, sanitation and hygiene (WASH) perspective targeting low-income countries. Sci. Total Environ. 2020;141751 doi: 10.1016/j.scitotenv.2020.141751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Hart Olga E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S.W., Ibrahim Y., Daou M., Kannout H., Jan N., Lopes A., Alsafar H., Yousef A.F. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: surveillance of COVID-19 epidemic in the United Arab Emirates. Sci. Total Environ. 2021;764 doi: 10.1016/j.scitotenv.2020.142929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemalatha M., Kiran U., Kuncha S.K., Kopperi H., Gokulan C.G., Mohan S.V., Mishra R.K. Surveillance of SARS-CoV-2 spread using wastewater-based epidemiology: comprehensive study. Sci. Total Environ. 2021;768 doi: 10.1016/j.scitotenv.2020.144704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary L.S., Farkas K., Maher K.H., Lucaci A., Thorpe J., Distaso M.A., Gaze W.H., Paterson S., Burke T., Connor T.R., Mcdonald J.E., Malham S.K., Jones D.L. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokajärvi A.-M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.-M., Kankaanpää A., Gunnar T., Al-Hello H., Blomqvist S., Miettinen I.T., Savolainen-Kopra C., Pitkänen T. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., Quintela M., Graham D.W., Corbishley A., Mcdonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ. Sci. Technol. 2021 doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Katayama H., Shimasaki A., Ohgaki S. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 2002;68:1033–1039. doi: 10.1128/AEM.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama S., Miura T., Masago Y., Konta Y., Tohma K., Manaka T., Liu X., Nakayama D., Tanno T., Saito M., Oshitani H., Omura T. Environmental surveillance of norovirus genogroups I and II for sensitive detection of epidemic variants. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.03406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongprajug A., Mongkolsuk S., Sirikanchana K. CrAssphage as a potential human sewage marker for microbial source tracking in Southeast Asia. Environ. Sci. Technol. Lett. 2019;6:159–164. doi: 10.1021/acs.estlett.9b00041. [DOI] [Google Scholar]
- Kongprajug A., Chyerochana N., Mongkolsuk S., Sirikanchana K. Effect of quantitative polymerase chain reaction data analysis using sample amplification efficiency on microbial source tracking assay performance and source attribution. Environ. Sci. Technol. 2020;54:8232–8244. doi: 10.1021/acs.est.0c01559. [DOI] [PubMed] [Google Scholar]
- Kongprajug A., Chyerochana N., Rattanakul S., Denpetkul T., Sangkaew W., Somnark P., Patarapongsant Y., Tomyim K., Sresang M., Mongkolsuk S., Sirikanchana K. Integrated analyses of fecal indicator bacteria, microbial source tracking markers, and pathogens for southeast asian beach water quality assessment. Water Res. 2021;203 doi: 10.1016/j.watres.2021.117479. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Naughton C.C., Roman F.A., Alvarado A.G.F., Tariqi A.Q., Deeming M.A., Bibby K., Bivins A., Rose J.B., Medema G., Katsivelis P., Allan V., Sinclair R., Zhang Y., Maureen N. Show us the Data : Global COVID-19 Wastewater Monitoring Efforts , Equity , and Gaps. medRxiv; 2021. pp. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okadera T., Syutsubo K., Yoochatchaval W., Ebie Y., Kubota R. Water volume- and BOD- based flow analysis for domestic wastewater treatment using wastewater inventories of. J. Water Environ. Technol. 2020;18:71–79. doi: 10.2965/jwet.19-064. [DOI] [Google Scholar]
- Olesen S.W., Imakaev M., Duvallet C. Making waves: defining the lead time of wastewater-based epidemiology for COVID-19. Water Res. 2021;117433 doi: 10.1016/j.watres.2021.117433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasa S., Desai M., Chandrasekar V.T., Patel H.K., Kennedy K.F., Roesch T., Spadaccini M., Colombo M., Gabbiadini R., Artifon E.L.A., Repici A., Sharma P. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw. Open. 2020;3(6):1–14. doi: 10.1001/JAMANETWORKOPEN.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay L., Amoah I.D., Deepnarain N., Pillay K., Awolusi O.O., Kumari S., Bux F. Monitoring changes in COVID-19 infection using wastewater-based epidemiology: a south african perspective. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiee M., Isazadeh S., Mohseni-Bandpei A., Mohebbi S.R., Jahangiri-rad M., Eslami A., Dabiri H., Roostaei K., Tanhaei M., Amereh F. Moore swab performs equal to composite and outperforms grab sampling for SARS-CoV-2 monitoring in wastewater. Sci. Total Environ. 2021;790 doi: 10.1016/j.scitotenv.2021.148205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva F., Castiglioni S., Pacciani C., Zuccato E. Testing urban wastewater to assess compliance with prescription data through wastewater-based epidemiology : first case study in Italy. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139741. [DOI] [PubMed] [Google Scholar]
- Róka E., Khayer B., Kis Z., Kovács L.B., Schuler E., Magyar N., Málnási T., Oravecz O., Pályi B., Pándics T., Vargha M. Ahead of the second wave: early warning for COVID-19 by wastewater surveillance in Hungary. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-puchol S., Forés E., Itarte M., Girones R., Bofill-mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Innes G.K., Prasek S.M., Walter Q., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;149794 doi: 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Compte A., Gonzalez S., Arnaldos M., Berlendis S., Courtois S., Loret J.F., Schlosser O., Yanez A.M., Soria-Soria E., Fittipaldi M., Saucedo G., Pinar-Mendez A., Paraira M., Galofre B., Lema J.M., Balboa S., Mauricio-Iglesia M., Bosch A., Pinto R.M., Bertrand I., Gantzer C., Montero C., Litrico X. Elimination of SARS-CoV-2 along wastewater and sludge treatment processes. Water Res. 2021;117435 doi: 10.1016/j.watres.2021.117435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., Ge S., Jones T., Santosh M., Silva L.F.O., Cao Y., Oliveira M.L.S., Zhang M., Bérubé K. The role of airborne particles and environmental considerations in the transmission of SARS-CoV-2. Geosci. Front. 2021;101189 doi: 10.1016/j.gsf.2021.101189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetapple C., Melville-shreeve P., Chen A.S., Grimsley J.M.S., Bunce J.T., Gaze W., Wade M.J. Building knowledge of university campus population dynamics to enhance near-to-source sewage surveillance for SARS-CoV-2 detection. Sci. Total Environ. 2021;150406 doi: 10.1016/j.scitotenv.2021.150406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Kitajima M., Abeynayaka A., Houng N.T.T., Dinh N.Q., Sirikanchana K., Navia M., Sam A.A., Tsudaka M., Setiadi T., Hung D.T., Haramoto E. Environmental Resilience and Transformation in Times of COVID-19: Climate change effects on environmental functionality. Elsevier; 2021. Chapter 11. Governance of wastewater surveillance systems to minimize the impact of COVID-19 and future epidemics: Cases across Asia-Pacific; pp. 115–125. [Google Scholar]
- Torii S., Furumai H., Katayama H. Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total Environ. 2021;756 doi: 10.1016/j.scitotenv.2020.143067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triukose S., Nitinawarat S., Satian P., Somboonsavatdee A., Chotikarn P., Thammasanya T., Wanlapakorn N., Sudhinaraset N., Boonyamalik P., Kakhong B., Poovorawan Y. Effects of public health interventions on the epidemiological spread during the first wave of the COVID-19 outbreak in Thailand. PLoS One. 2021;16:1–18. doi: 10.1371/journal.pone.0246274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannigama D.L., Amarasiri M., Hurst C., Phattharapornjaroen P., Abe S., Hongsing P., Rad S.M.A.H., Pearson L., Saethang T., Luk-In S., Kueakulpattana N., Storer R.J., Ounjai P., Jacquet A., Leelahavanichkul A., Chatsuwan T. Tracking COVID-19 with wastewater to understand asymptomatic transmission. Int. J. Infect. Dis. 2021 doi: 10.1016/j.ijid.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2003. Guidelines for Environmental Surveillance of Poliovirus Circulation Guidelines for Environmental. [Google Scholar]
- WHO . World Heal. Organ; 2018. Prioritizing diseases for research and development in emergency contexts [WWW document]https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts%0Ahttps://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-context%0Ahttps://www.who.int/activities/prioritizing- URL. (accessed 7.31.21) [Google Scholar]
- WHO . 2020. Novel Coronavirus (2019-nCoV) Situation Report-1. 21 January 2020. [Google Scholar]
- WHO Status of environmental surveillance for SARS-CoV-2 virus: Scientific brief 5 August 2020. 2020. https://www.who.int/news-room/commentaries/detail/status-of-environmental-surveillance-for-sars-cov-2-virus Retrieved July 31, 2021, from.
- WHO Regional Office for Europe . 2020. Rapid Expert Consultation on Environmental Surveillance of SARS-CoV-2 in Wastewater. Summary Report. [Google Scholar]
- WHO Regional Office for Europe . 2021. Summary Report: Expert Consultation on Public Health Needs Related to Surveillance of SARS-CoV-2 in Wastewater. [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:1–9. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Rhode S.F., Wuertz S., Thompson J., Alm E.J. Wastewater Surveillance of SARS-CoV-2 across 40 U.S. states from February to June 2020. Water Res. 2021;1–13 doi: 10.1016/j.watres.2021.117400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wang Z., Lin Y., Zhang L., Chen J., Li P., Liu W., Wang Y., Yao C., Yang K. Technical framework for wastewater-based epidemiology of SARS-CoV-2. Sci. Total Environ. 2021;791 doi: 10.1016/j.scitotenv.2021.148271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Oishi W., Maruo C., Saito M., Chen R., Kitajima M., Sano D. Early warning of COVID-19 via wastewater-based epidemiology: potential and bottlenecks. Sci. Total Environ. 2021 doi: 10.1016/j.scitotenv.2021.145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato E., Chiabrando C., Castiglioni S., Calamari D., Bagnati R., Schiarea S., Fanelli R. Cocaine in surface waters: a new evidence-based tool to monitor community drug abuse. Environ. Health Glob. Access Sci. Source. 2005;4:1–7. doi: 10.1186/1476-069X-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material