Abstract
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a global health concern. It is now well established that the spike (S) protein of SARS-CoV-2 interacts with its primary host receptor, the angiotensin converting enzyme 2 (ACE2). Additionally, the interaction of S with the neuropilin (NRP) receptor has been reported to facilitate viral entry. SARS-CoV-2 S protein binds to neuropilin-1 (NRP1) by virtue of a CendR motif which terminates with either an arginine or lysine. Furthermore, a number of different peptide sequences have been reported to bind to the same site in NRP1 including vascular endothelial growth factor A and other viral proteins. To gain a deeper understanding of additional factors besides the C-terminal arginine that may favour high NRP1 binding, several modelled peptides were investigated using triplicate 1 μs molecular dynamics simulations. A C-end histidine failed to exhibit strong NRP1 affinity. Some previously reported factors that increase binding affinity and secure NRP1 receptor activation was observed in the NRP1-peptide complexes studied and such complexes had higher molecular mechanics-generalized Born surface area based free energy of binding. Additionally, the results also highlight the relevance of an exposed arginine at its canonical location as capping it blocked arginine from engaging key residues at the NRP1 receptor site that are indispensable for functional binding; and that the presence of proline reinforces the C-terminal arginine. Given that stable NRP1 binding is crucial for viral uptake, stable interactions should be accounted for in the design of potential drugs and treatment routes to target or disrupt this interface, considering the S1-NRP1 interaction as well as its endogenous VEGF-A ligand that is associated with nociception.
Keywords: Neuropilin, CendR motif, Spike protein, VEGF-A, Molecular dynamics
Neuropilin; CendR motif; Spike protein; VEGF-A; Molecular dynamics.
1. Introduction
The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) leading to coronavirus disease 2019 (COVID19) is under extensive investigation. However, more focus is being directed towards its interaction with its primary host target angiotensin converting enzyme 2 (ACE2) (Moutal et al., 2021) as indicated by the literature. Recently, additional host factors that facilitate SARS-CoV-2 uptake have been uncovered. Among these are neuropilin-1 (NRP1) receptor which mediates viral entry (Daly et al., 2020; Cantuti-Castelvetri et al., 2020); and the serine protease TMPRSS2 which is implicated in S protein priming (Hoffmann et al., 2020).
NRP1 and NRP2, the two neuropilin isoforms, are transmembrane glycoprotein receptors originally discovered in neurons. In humans, neuronal and endothelial cells are rich in these receptors. In recognizing and binding with several membrane proteins, NRPs play diverse biological functions such as angiogenesis, axon pathfinding and vascular permeability. These receptors are also implicated in tumor vascularization and are aimed at in cancer treatment (Guo and Vander Kooi, 2015).
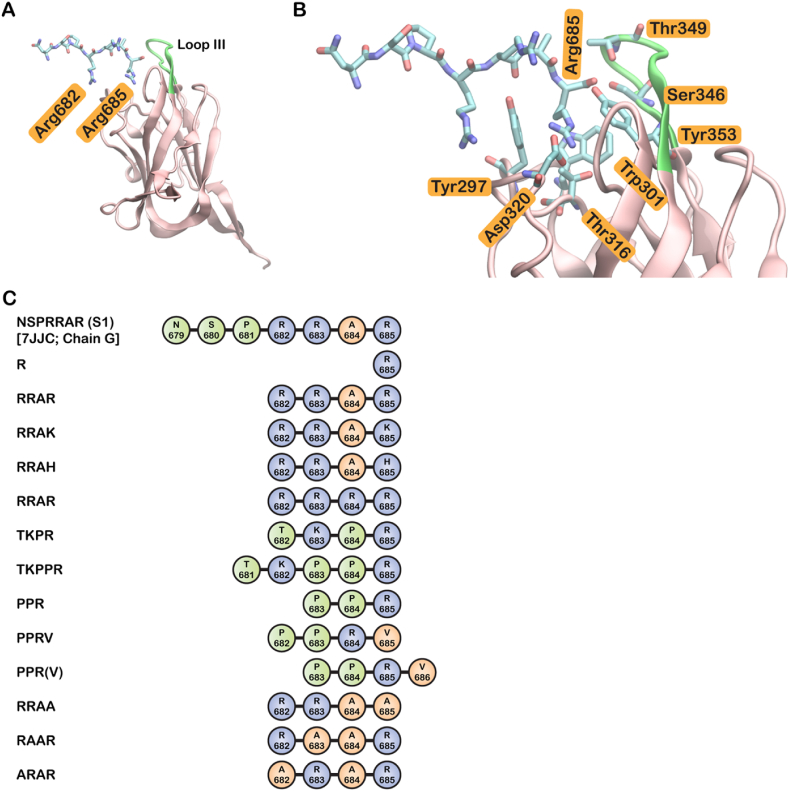
With regard to domain organization, NRP1 consists of an 850 amino acid long N-terminal extracellular domain followed by a short 24 residue long transmembrane domain and a 40 residue long cytoplasmic domain. The ectodomain is sectioned into five individual subdomains, namely a1, a2, b1, b2, and c; where the first four facilitate ligand binding, while the c domain is associated with oligomerization (Lee et al., 2003). The binding region on NRP1 b1 is molded by loops within the protein structure shaping a “receptor” for a C-terminal arginine moiety (Jarvis et al., 2010). This receptor region is termed as the arginine receptor, tuftsin site, or aromatic box (Peng et al., 2019). SARS-CoV-2 spike (S) protein and other endogenous NRP1 ligands achieve binding by virtue of the C-end rule (CendR) which signifies the presence of a polybasic carboxyl-terminal sequence motif [R/K]XX[R/K] that binds to NRP1/2 (Teesalu et al., 2009). The S glycoprotein precursor of SARS-CoV-2 is 10 nm long and comprises of 1273 amino acids (Seyran et al., 2020). Upon cleavage by furin protease, which is key to viral invasion, the S glycoprotein of SARS-CoV-2 presents the CendR motif 682RRAR685 which engages NRP1 (Wrapp et al., 2020). SARS-CoV-2 S1 protein (CendR sequence RRAR) and vascular endothelial growth factor A (VEGF-A; CendR sequence KPRR), an endogenous ligand of NRP1, exhibit like interaction with NRP1 as the residues that bind the terminal arginine in both complexes are the same - Tyr297, Trp301, Thr316, Asp320, Ser346, Thr349 and Tyr353 (Daly et al., 2020) (Figure 1B). In fact, any peptide with a C-end arginine shows affinity of some extent to NRP (Peng et al., 2019).
Figure 1.
Structure of NRP1 in complex with S1 and alignment of the modelled peptides. (A) Structure of NRP1 (pink) in complex with S1 peptide fragment NSPRRAR (blue stick representation) (PDB ID:7JJC). Loop III region is shown in green. The flanking arginine residues of the S1 CendR motif RRAR is labelled in orange (B) Enlargement of A showing the NRP1-S1 binding interface and the residues that interact with both S1 and VEGF-A are labelled in orange. (C) An alignment of the mutated peptides used in this study. Amino acid numbering is based on the S1 peptide (NSPRRAR) bound to NRP1 in the structure 7JJC (chain G). The amino acids are colored based on the polarity of the sidechain; blue - positively charged; green - polar uncharged; orange – nonpolar.
The S glycoprotein precursor of SARS-CoV-2 gets divided into non-covalently associated S1 and S2 fragments upon cleavage by the protease furin which occurs either upon host cell entry or during viral synthesis. Such cleavage exposes the polybasic carboxyl-terminal sequence motif (682RRAR685) on S1, which is crucial for cell uptake and viral invasion through NRP1 receptor binding on the cell surface. S2 triggers membrane fusion as it remains secured on the virus membrane. It then goes through further proteolytic cleavage by TMPRSS2 or other proteases which release its fusion peptide (Jobe and Vijayan, 2021). The interaction of RBD of the SARS-CoV-2 S protein with the primary host receptor ACE2 has been studied recently. The residues Tyr449, Gln493 and Gln498 of SARS-CoV-2 formed three stable hydrogen bonds with Asp38, Glu35 and Lys353 of ACE2 at one extreme, while Lys417 of SARS-CoV-2 S protein engaged in a stable salt bridge interaction with Asp30 of ACE2 at the center; and Tyr473, Ala475 and Phe486 exhibited well-retained hydrophobic contact with Leu79, Met82 and Tyr83 of ACE2 at the other extreme. The above interacting residues noticeably differ from those mediating the S1-NRP1 interaction, but in view of persistent viral interaction with host receptor being key to viral uptake, such lasting interactions along with those featuring the S1-NRP1 interface should be accounted for in the design of prospective therapeutics (Ali and Vijayan, 2020). Since no specific variation has been identified in the region near the C-end motif, the recently emerged SARS-CoV-2 variants are unlikely to alter S-protein-NRP1 interactions. Rather, variations have primarily been identified in the ORF1ab and RBD of S glycoprotein which is implicated in ACE2 interaction (Antony and Vijayan, 2021).
Besides its role in SARS-CoV-2 infection, NRP1 has also been reported to mediate the cellular uptake of two additional viruses, Epstein Barr virus (EBV) through the CendR motif RRRR of its glycoprotein and Human T-Cell Lymphotropic Virus Type 1 (HTLV-1) via its surface unit (SU) residues 85–94 (KKPNR) and SU 304–312 (SRSRR) (Wang et al., 2015).
Though C-terminal arginine of the CendR motif has been highlighted as key to NRP1 binding, other reports hint that it may not hold as much weight as previously thought and that it cannot represent the sole factor behind high-affinity NRP1 interaction. For instance, Baek and colleagues screened a non-CendR peptide library and identified the V12 peptide (HLQESPGKPPRV) fused to Fc (fragment crystallizable) domain that exhibits specific binding to NRP1 b1 domain. Their finding marks the first account of a non-CendR peptide going against the CendR rule. They also found the valine residue at the of Fc-V12 C-terminal end to be critical for NRP1 interaction (Baek et al., 2018).
Surprisingly, the immunostimulatory peptide tuftsin (TKPR) and a stronger affinity inhibitor TKPPR were reported to bind selectively to NRP1 and disrupt VEGF-NRP1 association (von Wronski et al., 2006). The central basic residues in the CendR motif may be implicated in the stability of the NRP1-peptide complex. The fact that TKPPR binds to endothelial cells with around 20-fold higher strength relative to tuftsin (TKPR) reinforces the significance of a separation between the flanking lysine and arginine in NRP1 binding. Additionally, CendR peptides exhibit optimal binding affinity in the event of non-basic amino acids at the two central positions flanked by R/K (Teesalu et al., 2009). RPAR analogs have been evaluated to delve into the significance of the central residues of CendR peptides in NRP1 binding and receptor stimulation (Zanuy et al., 2013). Results revealed that RPAR evidently exhibits strongest NRP1 interaction largely owing to peptide rigidity as most of the analogs that resulted in unsubstantial binding did not bear proline at position 2, and peptide basicity as an additional arginine strengthens receptor binding. However, the binding of RPAR analogs is not synonymous to receptor stimulation. Rather, complexes stabilized via secure binding involving loop III (Ser346, Thr349, Tyr353) (Figure 1) is thought to induce NRP1 activation. Additional receptor regions are possibly of ancillary importance in NRP1 activation as they offer greater flexibility in allowing the binding of non-functional fragments (Zanuy et al., 2013).
To qualify protein-protein interactions, virtual alanine scanning was performed by mutating one position at a time of the RRAR peptide to alanine, the resulting peptides namely RRAA, RAAR and ARAR were simulated with NRP1 (PDB ID:7JJC). The well-documented NRP1/VEGF-A blocker EG00229 (Jarvis et al., 2010) was also studied to compare its NRP1 interaction with that of VEGF-A. Thus, in this study, to explore the stability and binding interactions of CendR peptides, seventeen systems were studied – VEGF-A, 13 peptides, a lone arginine, an arginine analogue and an NRP1 inhibitor (Table 1 and Figure 1C).
Table 1.
Summary of simulation systems used in this study. For systems 4–16 the bound peptide was modelled based on the peptide in system 3. For systems 4–15, the C-terminal amino acid of the bound peptide takes the equivalent position of C-terminal arginine of system number 3, while in system 16, the arginine of the peptide was at the equivalent position of the C-terminal arginine of system 3 and a C-terminal valine residue was added.
| System | Name | PDB ID of NRP1 structure | Bound peptide/amino acid/amino acid analog |
|---|---|---|---|
| 1 | NRP1-VEGF-A | 4DEQ | VEGF-A |
| 2 | NRP1-HRG | 5IJR | Arginine analogue (HRG) |
| 3 | NRP1-S1 | 7JJC | NSPRRAR |
| 4 | NRP1-R | 7JJC | R |
| 5 | NRP1-RRAR | 7JJC | RRAR |
| 6 | NRP1-ARAR | 7JJC | ARAR |
| 7 | NRP1-RRAA | 7JJC | RRAA |
| 8 | NRP1-RAAR | 7JJC | RAAR |
| 9 | NRP1-RRAK | 7JJC | RRAK |
| 10 | NRP1-RRAH | 7JJC | RRAH |
| 11 | NRP1-RRRR | 7JJC | RRRR |
| 12 | NRP1-TKPR | 7JJC | TKPR |
| 13 | NRP1-TKPPR | 7JJC | TKPPR |
| 14 | NRP1-PPR | 7JJC | PPR |
| 15 | NRP1-PPRV | 7JJC | PPRV |
| 16 | NRP1-PPR(V) | 7JJC | PPRV |
| 17 | NRP1-EG00229 | 3I97 | EG00229 (inhibitor) |
Given that NRP1 antibodies featuring a C-terminal histidine are able to fill the same arginine binding pocket as tuftsin (Appleton et al., 2007), it is worth investigating how histidine being a basic residue compares to arginine and lysine at the C-terminal. The peptides TKPR and TKPPR were also modelled with NRP1 to look at the effect of an additional proline residue in supporting the C-terminal arginine (Zanuy et al., 2013). To assess the relevance of the C-terminal arginine and its positioning, two versions of the peptide PPRV were studied – in one, the valine occupied the canonical terminal position of the CendR peptide arginine; and in another case, the valine was positioned beyond the canonical arginine binding location.
Since the binding of the S1 CendR peptide to NRP1 is highly similar to that of its endogenous partner VEGF-A 164, contrasting the two complexes is warranted, particularly when it was reported that S protein blocks VEGF-A/NRP1 signaling, which otherwise signals nociception (Jobe and Vijayan, 2021; Moutal et al., 2021). Such masking of pain signalling could be implicated in the subtle transmission of the virus through asymptomatic individuals, challenging the curb of the pandemic.
It was reported that roughly eight residues preceding the C-terminal arginine in VEGF-A exhibit specific binding with residues within and surrounding the ‘arginine-binding pocket’, ensuring selectivity of the binding interaction (Parker et al., 2012). To explore this aspect in the interaction of S1 with NRP1, a seven, four and one-residue system of the terminal sequence of SARS-CoV-2-NRP1 complex were evaluated in order to contrast the interaction and binding stability of the three systems. Due to the lack of computational studies on the interaction of NRP1 with the CendR motif of the EBV glycoprotein (RRRR), this study also explored the role of the non-C terminal arginine residues in NRP1 binding.
At present, a few high resolution three-dimensional structures of peptide-bound NRP1 b1 domain structures are available in the Protein Data Bank (Daly et al., 2020; Mota et al., 2018; Parker et al., 2012). These X-ray crystallography-based structures offer noteworthy insights into both intermolecular interactions and macromolecular structure and can be extended to test hypotheses on how other peptides could bind. Since molecular recognition and resulting binding are dynamic in nature, molecular dynamics (MD) simulations are typically performed as a complement to conventional structural studies for the purpose of investigating the atom-level dynamics of such molecular processes (De Vivo et al., 2016; Durrant and McCammon, 2011). As an approach, MD simulations reveal insights in terms of structural integrity of macromolecular complexes, the versatility of the involved subunits and residues at the binding interface. By carrying out triplicate 1 μs MD simulations, this study reports the stability, interaction dynamics and energetics of NRP1 in complex with the seventeen peptides presented in Table 1.
2. Materials and methods
Coordinates of the three dimensional X-ray crystal structures of NRP1 complexed with VEGF-A (Protein Data Bank (PDB) ID: 4DEQ), an arginine analogue (PDB ID: 5IJR), the NRP1 inhibitor EG00229 (PDB ID: 3I97), and SARS-CoV-2 CendR peptide (PDB ID: 7JJC) were obtained from RCSB PDB (https://www.rcsb.org). For the structure with PDB ID 7JJC, the NRP1 polypeptide in chain C along with the bound SARS-CoV-2 S protein C-terminal region with the sequence NSPRRAR (chain G) was used. Peptide structures of R, RRAR, RRAK, RRAH, RRRR, TKPR, TKPPR, PPR, RRAA, RAAR, and ARAR based on the backbone of the SARS-CoV-2 CendR peptide (chain G of 7JJC), bound to NRP1 (chain C of 7JJC) were also generated as shown in Figure 1C. Additionally, two structures of the peptide PPRV bound to NRP1 were generated. In one version, the arginine of PPRV corresponded to the terminal arginine of the CendR peptide of SARS-CoV-2 in chain G of 7JJC and a C-terminal valine residue was added. The second variant had the terminal valine of PPRV at the position of the terminal arginine of the CendR peptide. Schrödinger Maestro 2019-4 (Schrödinger, LLC, New York, NY) was used to visualize and prepare the protein structures for simulations. All structures were first pre-processed using the Protein Preparation Wizard (Schrödinger, LLC, New York, NY) of the Schrödinger suite. The protein preparation stage included proper assignment of bond order, adjustment of ionization states, orientation of disorientated groups, creation of disulphide bonds, removal of unwanted water molecules, metal and co-factors, capping of the termini, assignment of partial charges, and addition of missing atoms and side chains. Schrödinger Prime was used to complete missing sidechains of the modelled peptides (Jacobson et al., 2004). In each system, the termini of the NRP1 structure were capped while the termini of the peptide structures were not capped. Hydrogen atoms were incorporated, and standard protonation state at pH 7 was used. Subsequently, hydrogen bonds were optimized and the energy of the structures were minimized. Processed protein-peptide complexes were positioned in orthorhombic boxes of size 80 Å × 80 Å × 80 Å and solvated with single point charge (SPC) water molecules using the Desmond System Builder (Schrödinger, LLC, New York, NY). Simulation systems were neutralized with counterions and a salt concentration of 0.15 M NaCl was maintained and the MD simulations were performed using Desmond (Bowers et al., 2006). The OPLS forcefield was used for all calculations. Prior to initiating the production simulations, Desmond's default eight stage relaxation protocol was used for all systems. 1 μs simulation of each complex was performed in triplicate with a different set of initial velocities. The NRP1-RRAR simulations were further extended to 2 μs. The Nose-Hoover thermostat and the isotropic Martyna-Tobias-Klein barostat were used to maintain the temperature at 300 K and pressure at 1 atm respectively (Martyna et al., 1992; Martyna et al., 1994). Short-range cut-off was set as 9.0 Å and long-range coulombic interactions were evaluated using the smooth particle mesh Ewald method (PME) (Essmann et al., 1995). A time-reversible reference system propagator algorithm (RESPA) integrator was employed with an outer time step 6.0 fs and an inner time step of 2.0 fs (Tuckerman et al., 1992). The molecular mechanics-generalized Born surface area (MM-GBSA) approach was used to calculate the binding free energy of the protein-peptide complexes, using frames extracted every 10 ns from MD simulation trajectories. These free energy calculations were performed using Schrödinger Prime employing the VSGB 2.0 solvation model (Li et al., 2011). Packaged and in-house scripts were used to analyze the simulation data. Visual Molecular Dynamics version 1.9.3 (Humphrey et al., 1996) was used for generating images and R version 3.6.3 (https://www.r-project.org) was used for plotting.
3. Results
3.1. MD simulations of NRP1-peptide complexes
To ensure that findings were not biased by just one structure, 1 μs MD simulations of thirteen NRP1-peptide complexes (Table 1) were carried out in triplicate.
The structural integrity of NRP1 was well-preserved during the entire length of the 1 μs simulations, as well as the extended RRAR 2 μs systems, with a protein Cα root mean square deviation (RMSD) below 2 Å (Supplementary Figures S1 and S5). The composition and compactness of protein secondary structure as indicated by the radius of gyration (Rg) of NRP1 and peptide structures was also well-maintained for the course of the entire simulation in all the systems (Supplementary Figures S2 and S7).
3.2. Comparison of regional fluctuations in the NRP1-peptide complexes
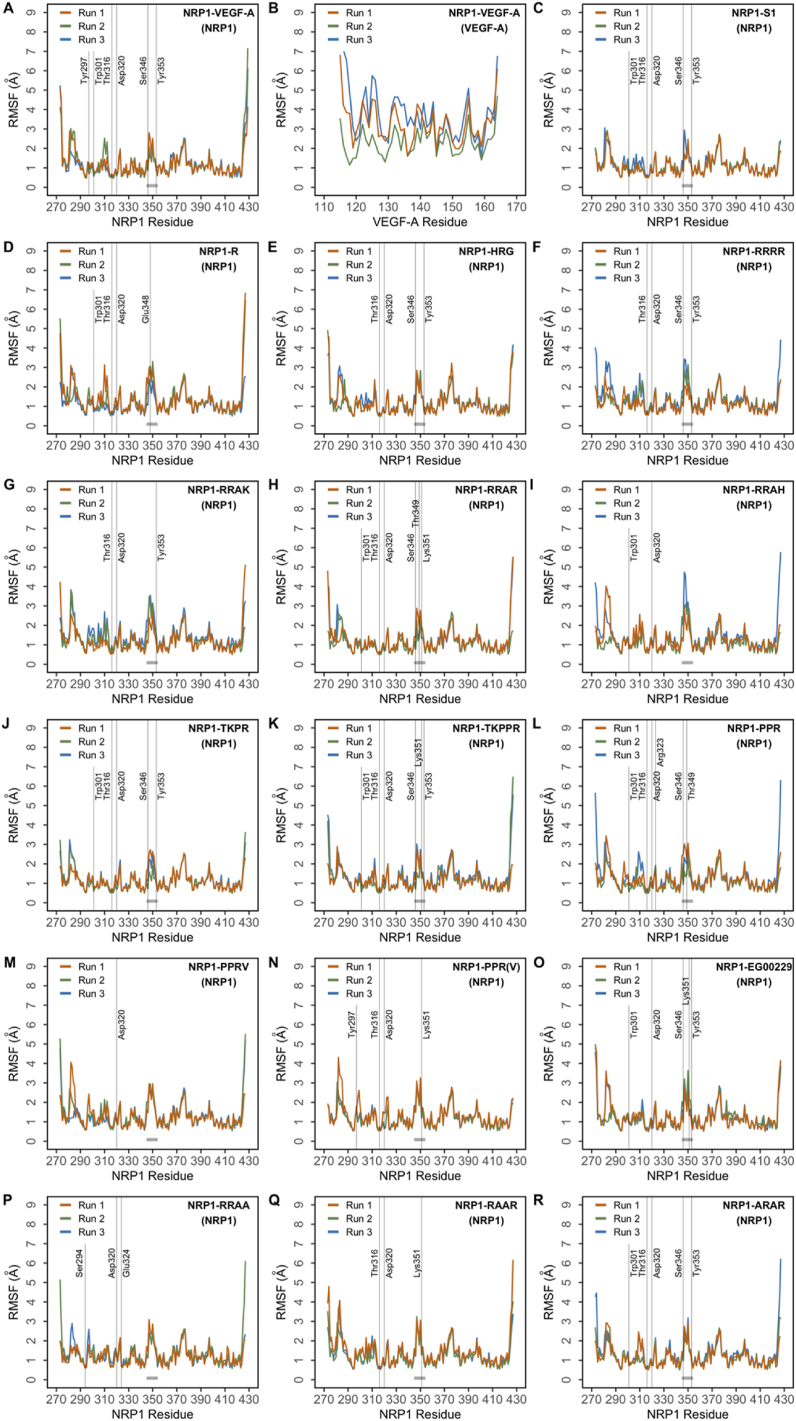
Root mean square fluctuation (RMSF) of backbone Cα atoms were computed and plotted with the purpose of identifying and comparing backbone stability and fluctuations in order to identify highly flexible regions (Figure 2 and Supplementary Figure S6).
Figure 2.
Root mean square fluctuation (RMSF) of protein Cα atoms obtained from three independent runs of the different peptides bound to NRP1. (A) NRP1 protein in the NRP1–VEGF-A complex; (B) VEGF-A ligand in the NRP1–VEGF-A complex; (C) NRP1 protein in the NRP1–S1 complex; (D) NRP1 protein in the NRP1–R complex; (E) NRP1 protein in the NRP1–HRG complex; (F) NRP1 protein in the NRP1–RRRR complex; (G) NRP1 protein in the NRP1–RRAK complex; (H) NRP1 protein in the NRP1–RRAR complex; (I) NRP1 protein in the NRP1–RRAH complex; (J) NRP1 protein in the NRP1–TKPR complex; (K) NRP1 protein in the NRP1–TKPPR complex; (L) NRP1 protein in the NRP1–PPR complex; (M) NRP1 protein in the NRP1–PPRV complex; (N) NRP1 protein in the NRP1–PPR(V) complex; (O) NRP1 protein in the NRP1–EG0029 complex; (P) NRP1 protein in the NRP1– RRAA complex; (Q) NRP1 protein in the NRP1–RAAR complex; (R) NRP1 protein in the NRP1–ARAR complex.
In the NRP1-VEGF-A complex, NRP1 residues exhibit limited fluctuation, with overall RMSF below 3 Å; with regions spanning residues 280–285, 310–312, 375–380 and the loop III region (345–354) showing slightly higher RMSF relative to other regions of the protein (Figure 2A). This pattern is also observed in NRP1-R (Figure 2D) and NRP1-HRG (Figure 2E). Additionally, the NRP1-RRRR (Figure 2F) and NRP1-RRAK (Figure 2G) complexes also show fluctuation in the same regions and exhibit a slightly higher RMSF of approximately under 3.5 Å. Similarly, NRP1-S1 presents RMSF below 3 Å with regions of higher fluctuation being residues 280–285, 375–380 and the loop III region (Figure 2C). A similar pattern was observed for PPR (Figure 2L), RRAR (Figure 2H), TKPR (Figure 2J) and TKPPR (Figure 2K) in complex with NRP1. The NRP1-PPRV complex exhibits minimal fluctuation with RMSF mostly under 3 Å, besides the residues 280–285 in the first run with RMSF of 4Å (Figure 2M). A like trend was observed in the case of NRP1-PPR(V) where the regions of slight fluctuation namely loop III and residues 375–380 were below RMSF of 4.5 Å (Figure 2N); and NRP1-RRAH where the regions of relatively higher fluctuation corresponding to residues 280–285 and loop III was under 5 Å in one simulation (Figure 2I). The complexes NRP1-RRAA, NRP1-RAAR and NRP1-ARAR (Figure 2P,Q,R) exhibit an RMSF of around 3 Å; while the NRP1-EG00229 (Figure 2O) presents a slightly higher value of approximately 3.5 Å. Besides the region covering residues 280–285, the loop III region corresponds to the region of highest fluctuation in all four complexes.
3.3. Duration of interfacial residue contact of the NRP1-peptide complexes
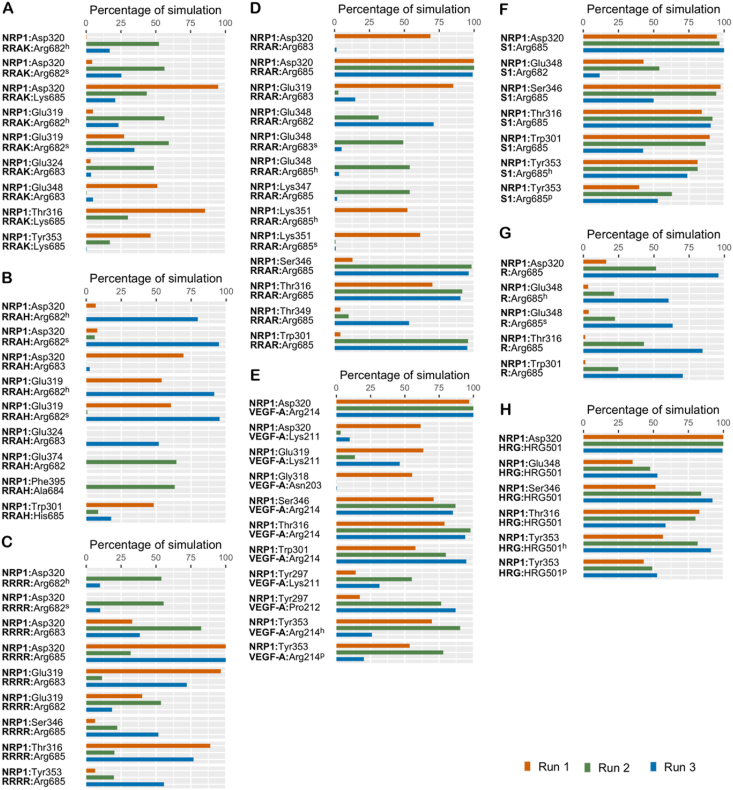
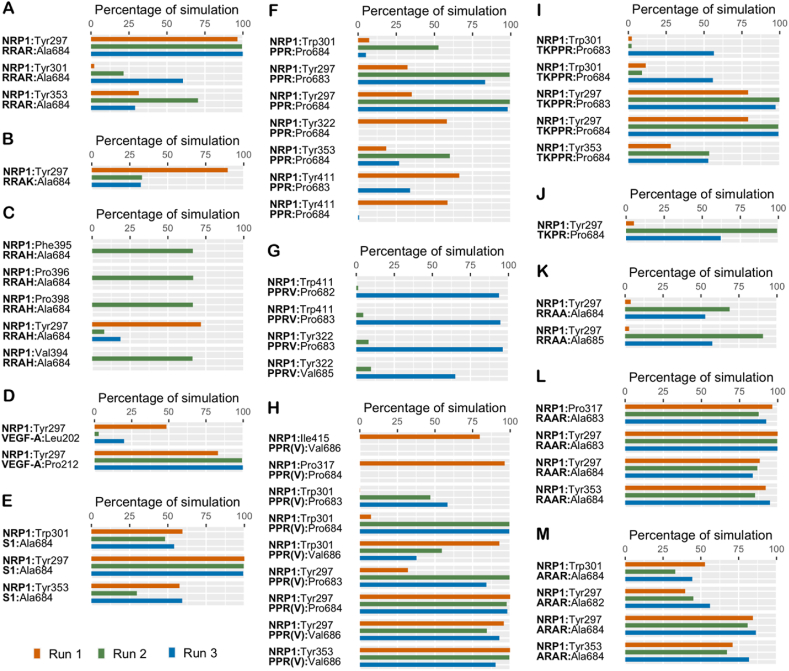
Intermolecular hydrophobic and polar contacts, namely hydrogen bonds, salt bridges, π interactions, were noted to form, break and reform over the course of the simulation with some being more lasting than others. A π–π interaction was observed in the case of NRP1-RRAH for about 63% of the time in one simulation with Trp301; while two π–π contacts were noted in the NRP1-EG00229 complex – one with Trp301 for about 57.17% of the time in all three simulations and the other with Tyr353 for 61.89% of the time in two simulations. Peptides that exhibit persistent interactions for a minimum of 50% of the overall simulation time in at least one run are presented in Figures 3, 4, and 5.
Figure 3.
The percentage of simulation time during which intermolecular polar contacts were retained between NRP1 and the peptides in three independent 1 μs runs of: (A) NRP1–RRAK; (B) NRP1–RRAH; (C) NRP1–RRRR; (D) NRP1–RRAR; (E) NRP1–VEGF-A; (F) NRP1–S1; (G) NRP1–R; (H) NRP1–HRG. When multiple types of polar interactions were observed between two residues, the superscript indicates the interaction type – h: hydrogen bond; s: salt bridge; p: π-cation.
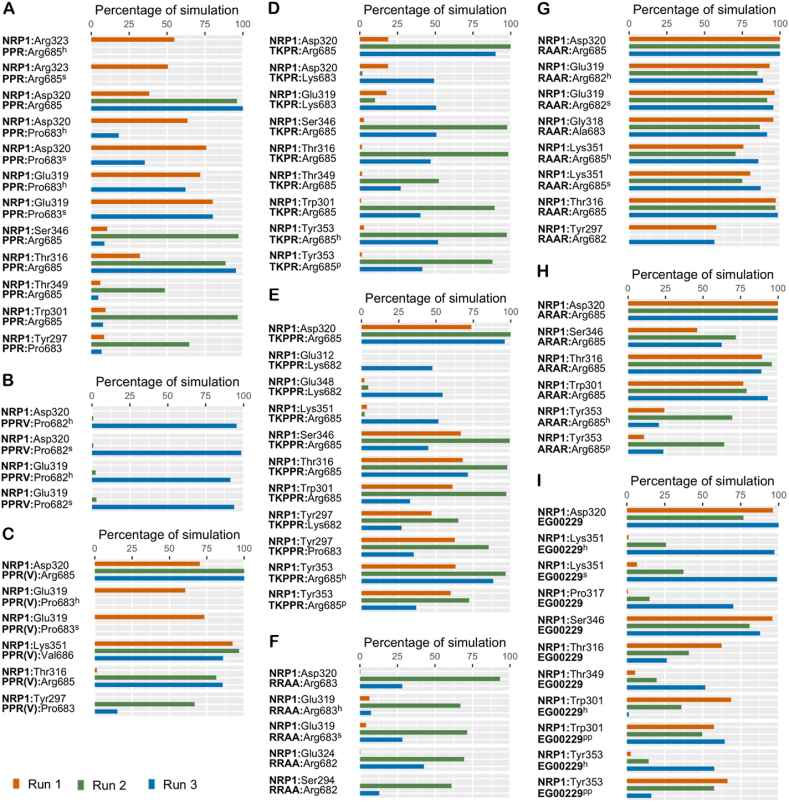
Figure 4.
The percentage of simulation time during which intermolecular polar contacts were retained between NRP1 and the peptides in three independent 1 μs runs of: (A) NRP1–PPR; (B) NRP1–PPRV; (C) NRP1– PPR(V); (D) NRP1–TKPR; (E) NRP1–TKPPR; (F) NRP1–RRAA; (G) NRP1–RAAR; (H) NRP1–ARAR; (I) NRP1–EG00229. When multiple types of polar interactions were observed between two residues, the superscript indicates the interaction type – h: hydrogen bond; s: salt bridge; p: π-cation; pp: π-π.
Figure 5.
The percentage of simulation time during which intermolecular hydrophobic contacts were retained between NRP1 and the peptides in three independent 1 μs runs of: (A) NRP1–RRAR; (B) NRP1–RRAK; (C) NRP1–RRAH; (D) NRP1–VEGF-A; (E) NRP1–S1; (F) NRP1–PPR; (G) NRP1–PPRV; (H) NRP1– PPR(V); (I) NRP1–TKPPR; (J) NRP1–TKPR; (K) NRP1–RRAA; (L) NRP1–RAAR; (M) NRP1–ARAR.
Asp320, one of the receptor site residues of NRP1 was observed to form a salt-bridge in all the complexes (Figures 3 and 4). This interaction was mediated by the N-terminal arginine in both RRAK and RRAH, along with an additional interaction through a hydrogen bond; in RRAA, this salt-bridge was held by the central arginine. The middle arginine of RRAH also forms a salt-bridge interaction with Asp320 (Figure 3A,B). As for RRAR, the Asp320 salt-bridge is well-maintained by the C-terminal arginine and also engages with the middle arginine in one simulation (Figure 3D). Unlike RRAR and RRAK, RRAH does not exhibit significant interaction with any of the other receptor site residues. The C-terminal histidine of RRAH forms no significant polar interaction except with Trp301 via a π–π association in one run (Figure 3B). On the other hand, the terminal arginine of RRAR engages Ser346 and Thr349 at the NRP1 receptor site, while the C-terminal lysine of RRAK engages Tyr353 (Figure 3D,A). As for RRRR, all but the arginine at position 3 form a salt-bridge interaction with Asp320 (Figure 3C). The N-terminal further engages with Asp320 through a hydrogen bond contact while the C-terminal arginine forms additional polar interactions with Ser346 and Tyr353. However, the arginine at position 3 fails to form any marked interaction with NRP1.
The endogenous NRP1 ligand VEGF-A also engages Tyr297 via its proline and lysine residues (Figure 3E) in addition to preserving the Asp320 salt-bridge interaction. The C-terminal arginine binds to Ser346 and to Tyr353 through both a hydrogen bond and cation-π interaction. Similarly, S1 interacts with Ser346 and Tyr353 via the same two interactions but fails to maintain appreciable contact with Tyr297 (Figure 3F).
Solely, arginine (NRP1-R) as a ligand failed to engage any other receptor residue besides Asp320 while the arginine analogue (HRG) not only exhibits well-sustained interaction with Asp320, but also engages Ser346 and interacts with Tyr353 via hydrogen bond and a cation-π interaction (Figure 3G,H).
As a complement to the salt-bridge contact with Asp320, the PPR peptide also engages Tyr297, Ser346, and Thr349 at the NRP1 binding pocket in one simulation through its N-terminal proline in the case of Tyr297 and C-end arginine in the case of Ser346 and Thr349 (Figure 4A). In fact, the Asp320 salt-bridge interaction is observed with both the C-terminal arginine and N-terminal proline. The latter also forms a hydrogen bond with Asp320. In the case of PPRV, only the N-terminal proline interacts with Asp320 (Figure 4B). This is through both a salt-bridge, involving the backbone amino group, and hydrogen bond in one simulation. The same interactions were observed with Glu319 of NRP1. Apart from the Asp320 salt-bridge interaction through its middle arginine, PPR(V) forms an additional interaction with Tyr297 at the NRP1 receptor site through its N-terminal proline in only one simulation (Figure 4C). The central proline of PPR, PPRV and PPR(V) do not form polar interactions, but contribute to hydrophobic interactions instead Figure 5 F,G,H.
Through its terminal arginine, TKPPR engaged NRP1 residues Ser346 via a hydrogen bond and Tyr353 through both a cation-π interaction and a hydrogen bond (Figure 4E). The lysine and proline at position three form an interaction with Tyr297 at the binding pocket. Similarly, the terminal arginine of TKPR engages Tyr353 via a cation-π interaction and a hydrogen bond; and Ser346 through a hydrogen bond (Figure 4D). In lieu of an interaction with Tyr297 as in TKPPR, TKPR interacts with Thr349 of the NRP1 binding pocket through its terminal arginine. However, the interactions formed by TKPPR occur in all three simulations and are overall more lasting while the association of TKPR with residues at the receptor site are observed mostly in two simulations and for quite a short duration, particularly in its interaction with Thr349. Similar to Asp320, Glu319 outside the NRP1 hotspot also forms salt-bridge contact with all the modelled peptides aside from TKPPR, ARAR and EG00229.
The overall count of intermolecular hydrogen bonds within the different complexes was also monitored over the course of the simulations (Supplementary Figures S3 and S8). The hydrogen bond count (mean ± SD) exhibited by NRP1-RRAR (8.2 ± 1.5, 10.2 ± 1.8, 8.8 ± 1.5) and RRRR (7.9 ± 1.4, 6.6 ± 2.4, 9.2 ± 2.0) is comparable and higher than that of RRAK (6.0 ± 2.0, 4.8 ± 1.5, 4.1 ± 1.6) and RRAH (5.1 ± 1.5, 3.2 ± 1.0, 3.9 ± 1.4). RRAK forms significant hydrogen bonds in only one simulation, while RRRR and RRAH form hydrogen bonds in two consecutive simulations and RRAR engages in hydrogen bonds in all three.
In the case of RRAR, the C-end arginine forms a hydrogen bond with the sidechain of Trp301, Thr316, Ser346, Thr349, Lys351 and backbone of Lys347 and Glu348 of NRP1; while the N-terminal arginine forms no significant hydrogen bonds with NRP1. For RRRR however, the sidechain of N-terminal arginine interacts with the backbone of Asp320 while the C-terminal arginine engages the sidechain of Thr316, Ser346, and Tyr353. The C-terminal lysine of RRAK is involved in a sidechain-sidechain hydrogen bond with Thr316 while the N-terminal arginine of both RRAK and RRAH interact with the backbone of Glu319 and Asp320 through their respective sidechains. Distinctly, the central alanine of RRAH is involved in a backbone-backbone hydrogen bond with Phe395.
In terms of overall hydrogen bond formation, VEGF-A (11.8 ± 2.8, 8.8 ± 1.8, 8.7 ± 2.2) and S1 (8.6 ± 1.7, 10.1 ± 2.4, 9.1 ± 2.3) are comparable. Both peptides form hydrogen bond interaction with sidechain of the NRP1 residues Trp301, Thr316, Ser346, and Tyr353; in addition, VEGF-A contacts the sidechain of Tyr297 and Gly318.
Generally, the arginine analog HRG (5.3 ± 1.2, 6.0 ± 1.3, 5.6 ± 1.1) exhibits a higher total hydrogen bond count compared to just a sole arginine (R) (2.1 ± 1.6, 3.4 ± 2.3, 5.7 ± 1.7) which forms hydrogen bond contact with Trp301, Thr316, Glu348 in only one simulation while the HRG sidechain is in quite consistent association with the sidechain of Thr316; and the backbone of Ser346 and Thr349 at the NRP1 hotspot.
The total hydrogen bond count of PPR (4.7 ± 1.6, 6.8 ± 1.3, 5.6 ± 1.3) and PPR(V) (4.3 ± 1.3, 4.6 ± 0.78, 4.9 ± 1.1) are comparable and higher than that of PPRV (2.2 ± 1.2, 2.5 ± 1.2, 3.0 ± 0.86). The sidechain of N-terminal proline of PPRV forms only two significant hydrogen bonds with NRP1, specifically with the backbone of Glu319 and Asp320 while all residues of PPR engage in a hydrogen bond with multiple NRP1 residues, namely Tyr297, Trp301, Thr316, Glu319, Asp320, Arg323, Ser346, Thr349 with significant strength. As for PPR(V), the N-terminal proline and the arginine residue form hydrogen bond contact with the NRP residues Tyr297, Thr316, and Glu319.
TKPPR (7.2 ± 2.4, 8.3 ± 1.0, 7.9 ± 1.5) exhibits a slightly higher number of overall hydrogen bonds relative to TKPR (3.0 ± 1.7, 8.0 ± 1.0, 6.1 ± 2.3). Hence, hydrogen bond formation may not constitute the defining aspect when it comes to the reported twenty-fold higher affinity of TKPPR to NRP1 (Teesalu et al., 2009).
Intermolecular hydrophobic interactions are also important considerations. Hence, this was also investigated, and the ones above 50% in at least one simulation were plotted (Figure 5). RRAR sustained hydrophobic contact with Tyr297 in all simulations and is also associated with Tyr353 though to a much lesser extent (Figure 5A). On the other hand, RRAK forms hydrophobic contact only with Tyr297 while RRAH engages other NRP1 residues namely Val394, Phe395, Pro396 and Pro398 in addition to Tyr297 (Figure 5B,C).
Leu202 and Pro212 of VEGF-A make hydrophobic contact with Tyr297, with the proline residue forming stronger interaction while the alanine of S1 makes strong contact with Tyr297 and engages Trp301 and Tyr353 (Figure 5D,E).
Both proline residues of PPR and PPR(V) form hydrophobic interactions with Tyr297 and the middle proline of PPR interacts with Tyr353 while the terminal valine of PPR(V) engages both Tyr297 and Tyr353 and contributes to the overall higher hydrophobic interaction of PPR(V) (Figure 5F,H). In contrast, PPRV does not feature significant hydrophobic contact with any receptor site residue but with residues Tyr322 and Trp411 in only one simulation (Figure 5G).
Unlike TKPR which only makes hydrophobic contact with Tyr297, TKPPR forms more hydrophobic interactions with NRP1 and exhibits more sustained association with Tyr297 through both its proline residues and also engages Tyr353 (Figure 5I,J).
To evaluate free energy of binding (ΔGbind) of the peptides to NRP1, the MM-GBSA approach was employed using frames from each of the MD simulations. Among the RRXX analogs, RRAR (-72.97 ± 7.26 kcal/mol, -84.60 ± 10.84 kcal/mol, -80.68 ± 7.38 kcal/mol) exhibited the highest estimated ΔGbind followed by RRRR (-79.22 ± 6.48 kcal/mol, -53.71 ± 17.68 kcal/mol, -80.54 ± 12.06 kcal/mol), RRAK (−58.86 ± 11.55 kcal/mol, -48.88 ± 10.65 kcal/mol, -47.68 ± 19.12 kcal/mol) and RRAH (-58.28 ± 12.83 kcal/mol, -43.40 ± 22.11 kcal/mol, -41.66 ± 12.47 kcal/mol). This is expected considering their respective polar and hydrophobic interactions, and the same stands when comparing VEGF-A with S1, HRG with R, TKPPR with TKPR; and to some extent when looking at PPR, PPRV and PPR(V).
PPR and PPR(V) compare in terms of hydrogen bond formation and hydrophobic contacts but the Val of PPR(V) plays into its overall higher hydrophobic interaction and this may partly justify the higher ΔGbind of PPR(V) (-57.49 ± 7.16 kcal/mol, -64.12 ± 5.11 kcal/mol, -68.26 ± 6.97 kcal/mol) relative to PPR (-44.62 ± 10.76 kcal/mol, -61.39 ± 6.14 kcal/mol, -56.56 ± 5.11 kcal/mol). Of the three, PPRV (-36.60 ± 10.45 kcal/mol, -37.90 ± 13.62 kcal/mol, -40.26 ± 4.85 kcal/mol) exhibits the lowest ΔGbind, in accordance with its demonstration of the least hydrogen bond and hydrophobic interaction. The reported ΔGbind scores should be interpreted qualitatively rather than quantitatively, considering the estimations and assumptions in MM-GBSA based binding free energy computations.
Out of all the systems, RAAR forms the most persistent interactions with NRP1 across all three runs. This enhancement in binding affinity could possibly be attributed to the presence of the two non-basic alanine residues separating the flanking basic arginine residues (Teesalu et al., 2009).
However, none of the polar contacts made by RAAR involve the loop III residues, and this could partly justify the overall higher ΔGbind of RRAR (-72.97 ± 7.26 kcal/mol, -84.60 ± 10.84 kcal/mol, -80.68 ± 7.38 kcal/mol) relative to RAAR (-70.05 ± 5.27 kcal/mol, -68.95 ± 4.78 kcal/mol, -71.53 ± 6.08 kcal/mol). The two significant hydrogen bond interactions with Tyr297 involves the N-terminal arginine and not the C-terminal one (Figure 4G). Similarly, Glu319 also associates with the N-terminal arginine in both its hydrogen bond and salt-bridge contact with RAAR. The terminal arginine of RAAR is held by Thr316, Asp320 and Lys351 in persistent interactions. The only contact RAAR makes with the loop III region is mediated by the alanine at position three through hydrophobic interaction with Tyr353 (Figure 5L). Tyr297 at the receptor site interacts with both alanine residues. Unlike the case in RRAR where Tyr301 interacts with Ala684, Pro317 engages Ala683 of RAAR.
Similar to RAAR, ARAR forms stronger hydrophobic interactions relative to RRAR due to the additional alanine which interacts with Tyr297 at the NRP1 binding pocket (Figure 5M). Both RRAR and ARAR engage Ser346, while ARAR engages Tyr353 via both a hydrogen and cation-π interaction instead of Thr349 as the case with RRAR (Figures 3D and 4H). All the marked polar interactions made by ARAR are mediated by the terminal arginine (Figure 4H); the central arginine is not involved in any significant interaction unlike in RRAR where the central arginine makes contact with Glu319, Asp320 and Glu348. The N-terminal arginine of RRAR also engages Glu348 in a salt-bridge contact. Taken together, RRAR (-72.97 ± 7.26 kcal/mol, -84.60 ± 10.84 kcal/mol, - 80.68 ± 7.38 kcal/mol) exhibits a higher ΔGbind relative to ARAR (-72.63 ± 7.79 kcal/mol, -66.50 ± 7.86 kcal/mol, -74.16 ± 6.97 kcal/mol).
RRAA does not engage any of the NRP1 loop III residues. Besides the Asp320 salt-bridge contact via its central arginine, the other contact it makes with the binding pocket is with Tyr297 via both its alanine residues in a hydrophobic interaction (Figure 5K). These interactions are significant in the last two simulations as the peptide briefly dissociates at around 200 ns in the first run (Supplementary Figure 4O) and remains unstable for the rest of the simulation period. Consequently, the NRP1-RRAA complex presents the lowest binding free energy (-45.00 ± 9.49 kcal/mol, -54.63 ± 6.46 kcal/mol, -50.23 ± 10.64 kcal/mol) in comparison to the other two mutated peptides of RRAR, namely RAAR and ARAR.
The EG00229 inhibitor makes polar contact with all loop III residues, the most lasting being a sidechain-sidechain hydrogen bond with Ser346 followed by contact with Tyr353 via a hydrogen bond and π-π interaction (Figure 4I). The hydrogen bond contact with Thr349 is significant in only one simulation. Though EG00229 forms no significant hydrophobic interaction with NRP1, its overall ΔGbind (-78.92 ± 5.70 kcal/mol, -76.74 ± 10.52 kcal/mol, -93.05 ± 7.67 kcal/mol) is comparable to that of VEGF-A (-85.81 ± 18.66 kcal/mol, -67.34 ± 11.27 kcal/mol, -82.07 ± 18.59 kcal/mol) and S1 (-73.14 ± 11.75 kcal/mol, -90.93 ± 16.09 kcal/mol, -86.31 ± 14.15 kcal/mol).
To evaluate whether the ligands stay docked during the simulation, the center of mass (COM) distance between Cα atoms of Asp320 and the Cα atom of the amino acid at the canonical “Arg” position at number 685 (Figure 1C) was plotted (Supplementary Figure S4). The simulations in which the ligands dissociate from the binding pocket and separate from the NRP1 protein are in all three runs of NRP1-R, the first two runs of NRP1-PPRV, run 1 of NRP1-TKPR and NRP1-RRAA, run 2 of NRP1-RRAH, and run 3 of NRP1-RRAK. These simulations correspond to lower binding free energy scores and higher standard deviation as highlighted in Table S1.
4. Discussion
4.1. The relevance of the C-terminal arginine
As illustrated in Table 1 and Figure 1C, PPRV is the only peptide in which the arginine was not positioned at the canonical location of the terminal CendR peptide. This could have an implication in its relatively low performance in terms of polar and hydrophobic interactions and hence overall low ΔGbind score. Infact, the PPRV peptide dissociates from NRP1 in two runs (Supplementary Figure 4L). This supports the relevance of the C-terminal arginine (Haspel et al., 2011) as opposed to the finding that a C-end arginine is dispensable for high-affinity NRP1 binding (Getz et al., 2013; Baek et al., 2018). Furthermore, being capped with a valine residue, the C-end arginine of both PPRV and PPR(V) failed to secure any marked contact with the five receptor site residues, namely Asp320, Tyr297, Ser346, Thr349, and Tyr353 (Parker et al., 2013; Vander Kooi et al., 2007). Comparing the performance of RRAR, RRAK and RRAH presents another support of the significance of a terminal arginine. With its terminal arginine, RRAR interacts with two loop III residues through a hydrogen bond, Ser346 in two runs with average duration of around 97% and Thr349 in one simulation for about 53% of the simulation (Figure 3D). On the other hand, the terminal lysine of RRAK failed to form hydrogen bond with loop III residues but instead forms a cation-π interaction with Tyr353 (Figure 3A); while the terminal histidine of RRAH forms a single considerable interaction in just one simulation, a π-π contact with Tyr301 outside the NRP1 hotspot (Figure 3B). In the case of RRRR, the C-terminal arginine engages the loop III residues Ser346 and Tyr353 (Figure 3C). All but the arginine at the third position formed a salt-bridge with Asp320 at the receptor site, and the N-terminal also forms a hydrogen bond with Asp320. Additionally, the peptides lacking a C-end arginine such as RRAK, RRAH, PPRV and RRAA were found to dissociate from the NRP1 binding pocket in at least one simulation (Supplementary Figure 4F,H, L,O).
4.2. The effect of capping the C-terminal arginine
Upon the introduction of a valine residue to PPR in PPR(V), the hydrogen bonds formed by PPR with loop III residues ceased, but the valine residue sustained polar contact with Lys351 around 92% of the simulation time (Figure 4A,C). Additionally, the valine residue also made hydrophobic contact with Tyr297, Trp301, Tyr353 and Ile415 of NRP1 Figure 5H. Similarly, PPRV exhibited no hydrogen bonds or hydrophobic interactions with any of the NRP1 loop III residues (Figure 4B). However, in PPR(V), where the arginine is located at the canonical arginine location, the arginine still maintained its salt-bridge interaction with Asp320 and further engaged in a sidechain-sidechain hydrogen bond with Thr316 in two simulations for an average period of about 83%; in addition to the other interactions formed by its proline and valine residue (Figure 4C). Capping the C-end arginine with additional C-terminal residues or replacing it with other amino acids was reported to block NRP1 interaction and its mediation of vascular penetration (Parker et al., 2013; Teesalu et al., 2009; Guo et al., 2013). This also highlights the significance of the canonical location of the terminal arginine as this was upheld in PPR(V) unlike in PPRV (Figure 1C). PPRV maintained other interactions such as a salt-bridge and hydrogen bond with Glu319 and Asp320 in one simulation via its two proline residues, but not through its C-terminal arginine (Figure 4B). All the four polar interactions formed by PPRV are mediated by the N-terminal proline, the valine residue mediates only one hydrophobic interaction; whereas the valine residue in PPR(V) mediates four hydrophobic contacts (Figure 5G,H) and seems to be critical in the overall binding of the PPR(V) peptide (Baek et al., 2018).
In terms of binding energy, PPR(V) exhibits an overall higher binding energy (-57.49 ± 7.16 kcal/mol, -64.12 ± 5.11 kcal/mol, -68.26 ± 6.97 kcal/mol) compared to PPR (-44.62 ± 10.76 kcal/mol, -61.39 ± 6.14 kcal/mol, -56.56 ± 5.11 kcal/mol) and PPRV (-36.60 ± 10.45 kcal/mol, -37.90 ± 13.62 kcal/mol, -40.26 ± 4.85 kcal/mol). This may be attributed to the placement of the arginine at position four and the presence of the hydrophobic valine residue beyond this position. Additionally, PPR(V) showed a sustained hydrophobic interaction with one of the loop III residues, namely Tyr353 through its terminal valine residue Figure 5H.
It is quite interesting that the shortest peptide, PPR (-44.62 ± 10.76 kcal/mol, -61.39 ± 6.14 kcal/mol, -56.56 ± 5.11 kcal/mol) exhibits a higher overall binding energy in relation to RRAH (-58.28 ± 12.83 kcal/mol, -43.40 ± 22.11 kcal/mol, -41.66 ± 12.47 kcal/mol), but especially in comparison to RRAK (-58.86 ± 11.55 kcal/mol, -48.88 ± 10.65 kcal/mol, -47.68 ± 19.12 kcal/mol). This further supports the relevance of a C-terminal arginine and its reinforcement by proline residues (Zanuy et al., 2013). Additionally, the distance between the COM of the Cα atom of NRP1:Asp320 and the Cα atom of the arginine of PPR remains quite stable as PPR remains bound to NRP1 throughout the course of the simulation period, unlike the case with one run each of RRAH (run 2) and RRAK (run 3) (Supplementary Figure 4K,H,F). At the receptor site, PPR forms polar interactions with Tyr297, Ser346 and Thr349 in one simulation; and with Asp320 in two runs (Figure 4A). On the other hand, RRAK secures polar contact with only two receptor site residues namely Asp320 and Tyr353 while RRAH only engages Asp320, and for a shorter period (Figure 3A,B). In terms of hydrophobic contacts, both proline residues of PPR engage Tyr297 with high hydrophobicity in two simulations and the middle proline of PPR also presents a significant interaction with Tyr353 in one simulation (Figure 5F); while the alanine of RRAK and RRAH interact with Tyr297 in just one simulation and with lesser strength (Figure 5B,C).
The additional proline in TKPPR adds to its rigidity which greatly enhances NRP1 binding as observed in RPAR (Zanuy et al., 2013). Furthermore, relative to TKPR, the additional proline in TKPPR contributes to higher and more sustained hydrophobic contacts with key residues within the b1 pocket, namely Tyr353 and Tyr297 (Figure 5I,J). Both proline residues of TKPPR maintained hydrophobic contacts with NRP1 in about 83% of the simulation period. Overall, TKPPR exhibits greater binding interaction relative to TKPR (Teesalu et al., 2009). TKPR only makes hydrophobic contact with Tyr297, while TKPPR not only forms more hydrophobic interactions with NRP1 but exhibits more consistent interaction with Tyr297 through both its proline residues and also engages Tyr353. Accordingly, TKPPR (-57.68 ± 14.23 kcal/mol, -63.05 ± 5.14 kcal/mol, -74.50 ± 12.19 kcal/mol) exhibits a higher ΔGbind than TKPR (-23.67 ± 11.80 kcal/mol, -67.33 ± 5.31 kcal/mol, -51.48 ± 19.13 kcal/mol).
The only significant hydrophobic contact exhibited by NRP1-VEGF-A is between Pro212 of VEGF-A and Tyr297 of NRP1 (Figure 5D), this interaction is observed across all three runs. This might partially explain the inhibition of the VEGF-NRP1 interaction by TKPPR and TKPR (von Wronski et al., 2006).
4.3. Functional binding to loop III residues
When RxxR analogs were studied, the strongest interaction holding the C-terminal R of RPAR, was a salt-bridge contact with Asp320 of NRP1 (Zanuy et al., 2013). This interaction was observed in at least two simulations in all the systems (Figures 3 and 4) except in PPRV, where the interaction was between Asp320 of NRP1 and the N-terminal proline of PPRV in just one simulation (Figure 4B). The sidechain of the same proline residue was also in hydrogen bond contact with the Asp320 backbone of NRP1. Though being a requirement for binding and configuration of the peptide-receptor complex, the Asp320 salt-bridge contact between the terminal arginine side chain and carboxyl group of Asp320 at the NRP1 binding pocket does not ensure receptor activation. This may explain why loss of the salt-bridge interaction with Asp320 in the arginine pocket does not eliminate NRP affinity (Peng et al., 2019; Starzec et al., 2014). Similarly, hydrogen bond involving Tyr297 at the receptor site is thought to stabilize the N-terminal portion of a respective ligand with an ancillary significance in securing proper receptor configuration following binding (Haspel et al., 2011).
Functional binding is credited to generation of a hydrogen bond around loop III residues (Ser346, Thr349, Tyr353) within the NRP1 binding pocket via a hydrogen bond interaction between the OH group of these residues with the COOH group of the C-terminal arginine (Zanuy et al., 2013). The complete hydrogen bond triad was observed in the NRP1-TKPR system between 52% to 75% of the time, with Ser346 and Tyr353 involved in two simulations and Thr349 in only one run (Figure 4D) and in NRP1-EG00229 (Figure 4I). Though TKPR has been reported to disrupt the NRP1-VEGF interface (von Wronski, 2006), the results of the present study cannot conclusively support that finding given the inconsistency of binding affinity scores across the three simulations of NRP1-TKPR (-23.67 ± 11.80 kcal/mol, -67.33 ± 5.31 kcal/mol, -51.48 ± 19.13 kcal/mol) and NRP1-VEGF-A (-85.81 ± 18.66 kcal/mol, -67.34 ± 11.27 kcal/mol, -82.07 ± 18.59 kcal/mol). Such inconsistency in the binding energy of TKPR may be due to the dissociation of the peptide in the first simulation (Supplementary Figure 4I). Therefore, based on the findings, the potential interference of tuftsin-NRP1 interaction cannot be conclusively associated with the asymptomatic transmission of COVID-19. Additionally, TKPR is not stable in the first simulation while TKPPR seems to be stable in all three. Thus, the basic residue at the fourth position from the C-terminal end of the CendR motif may have some significance.
TKPPR failed to interact with Thr349 of NRP1 but sustained an interaction with Tyr353 in all three runs around 83% of the time on average, and for almost the same duration with Ser346 in two runs (Figure 4E). Furthermore, the C-terminal arginine of TKPPR showed an additional association with Tyr353 through a cation-π interaction in two continuous runs. Through a salt-bridge contact, the terminal arginine of TKPPR also interacted with Lys351 which facilitates the placement of loop III residues particularly Ser346 and Thr349 (Zanuy et al., 2013). The Lys351 residue was also observed in NRP1-RRAR, forming a interactions with terminal arginine in one simulation (Figure 3D).
The systems that failed to produce hydrogen bond interactions with any of the loop III residues of NRP1 are NRP1-R, NRP1-PPRV, NRP1-PPR(V) and NRP1-RRAA (Figures. 3G, and 4B,C, F). For PPRV, the arginine was not positioned at the canonical arginine location of the CendR motif. While the arginine remained at this position in PPR(V), it was capped by introducing a valine as the terminal residue. Of all the modelled complexes, the NRP1-R complex had the lowest binding energy (-15.68 ± 10.18 kcal/mol, -26.31 ± 17.57 kcal/mol, -43.65 ± 10.86 kcal/mol). The failure of the single arginine in NRP1-R to bind to loop III residues also signifies the relevance of residues preceding the C-terminal arginine in the vicinity of the NRP1 hot spot that contribute to selectivity of the binding association (Parker et al., 2012) as the lone arginine detaches from NRP1 in all three simulations (Supplementary Figure 4C). Out of the five residues shaping the receptor site (Asp320, Tyr297, Ser346, Thr349, and Tyr353) (Parker et al., 2012; Vander Kooi et al., 2007), R only interacted with Asp320 through a salt-bridge in two of the three runs with an average duration of 73%. The other interactions holding the NRP1-R complex were a second salt-bridge interaction between the single arginine and Glu348, along with three hydrogen bond interactions with Trp301, Thr316, and Glu348 of NRP1 (Figure 3G). On its own, R exhibited no interaction with loop III residues that contribute to functional binding necessary to drive receptor activation, and thus ligand internalization.
In comparing the S1-peptide bound structure (PDB ID: 7JJC with peptide NSPRRAR) with its four-residue (RRAR) and one-residue (R) simulation, arginine maintains significant polar interaction with Asp320 in two simulations and with Trp301, Thr316 and Glu348 in only one simulation (Figure 3G); and therefore presents the lowest binding affinity. In terms of polar interaction, the NRP1-S1 interface features a hydrogen bond between the backbone of C-end arginine and the sidechain of Ser346 and Tyr353, with an additional cation-π interaction with Tyr353 (Figure 3F). The other NRP1 residues forming a significant interaction in at least one simulation at the interface are Trp301, Thr316, Asp320, and Glu348. Similarly, the NRP1-RRAR interface presents a hydrogen bond with Ser346 and Thr349 at the receptor site in addition to a higher number of additional NRP1 residues namely Trp301, Thr316, Glu319, Asp320, Lys347, Glu348 and Lys351 (Figure 3D). Both S1 and RRAR form the same hydrophobic interactions, namely with Tyr297, Trp301 and Tyr353 (Figure 5A,E), with S1 exhibiting higher overall hydrophobicity and ΔGbind score.
In comparing the RRAR (1 μs) system to the extended 2 μs system, the major polar and hydrophobic contacts remain preserved after the extension (Supplementary Figures S10 and S11). The slight discrepancies in terms of less significant contacts such as the absence of Asp320-Arg683 and Lys351-Arg685 interaction in the extended version of the NRP1-RRAR simulation and the presence of such contacts in run1 of RRAR (1 μs) (Figure 3D) may be justified by the rearrangement of the peptide in run1 of the extended version roughly past the 1100 ns mark. Beyond this frame, the terminal arginine of RRAR shifts outside the binding pocket and remains protruded for the rest of the simulation period. Instead, the N-terminal region of the peptide remains in contact with Glu319 and Asp320 near the binding pocket. Consequently, the relatively unstable distance between the Cα atom of Asp320 and Arg685 of RRAR in the last-half of the simulation of run1 (Supplementary Figure S9) could be explained by this major rearrangement.
The interaction of EG00229, the well-reported NRP1 inhibitor with all loop III residues and the overall high ΔGbind score of the NRP1-EG00229 complex further adds to the significance of these residues in NRP1 interaction and the relevance of targeting such residues in future clinical interventions.
The first documented non-CendR peptide V12 (HLQESPGKPPRV) exhibited NRP1-guided vascular penetration and enhanced NRP1-guided tumor homing and tissue permeability (Baek et al., 2018). The Fc-V12-NRP1 interactions primarily involve the last five residues of V12 (KPPRV) while the upstream 7 residues markedly confer to the selectivity between the two NRP isoforms. For instance, Fc-V12-33 (RPRPPRQKPPRV), an enhanced V12 (HLQESPGKPPRV) showed nearly 44-fold affinity for NRP1 relative to Fc-V12. Though the present study simulated various forms of the terminal residues of V12 in the form of PPR, PPRV and PPR(V) and reported findings that suggest PPR(V) as the best-binding peptide given its addition of a valine with Arg685 still maintained at its canonical position; it is worth exploring the different V12 variants outlined by Baek and colleagues in a future study as Fc-fused non-CendR peptides hold significance in terms of clinical application considering that the C-terminal arginine and lysine residues of Fc-fused CendR peptides may be cleaved by carboxypeptidases in the circulatory system or in cell culture.
5. Conclusion
In conclusion, the results indicate that a C-terminal histidine exhibits low NRP1 binding affinity relative to the other two basic residues - arginine and lysine - as it fails to effectively retain a salt-bridge. The findings are in line with previous reports on factors that favor NRP1 receptor-peptide complex formation such as the presence of proline as seen in the case of the TKPR and TKPPR and the significance of loop III residues in functional receptor-peptide interaction. Additionally, the findings also highlight the importance of the placement of arginine at the C-terminal as capping it with a valine blocked arginine from forming any functional interaction with loop III residues at the receptor site. Furthermore, as seen in the NRP1-RRRR complex, only the terminal arginine made contact with loop III residues at the receptor site. Though a C-terminal arginine is key to the CendR, the one-residue system of S1, NRP1-R highlights the role of the residues preceding the C-end arginine as the sole arginine made contact with only Asp320 at the NRP1 binding pocket; while the four-residue (NRP1-RRAR) and the seven-residue (NRP1-S1) system engaged with two loop III residues in addition to Asp320. The probability that an exposed arginine at the N-terminal could act as a free C-terminal arginine going by the C-end rule (Kim et al., 2016) requires further investigation in future studies.
Declarations
Author contribution statement
Amie Jobe: Analyzed and interpreted the data; Wrote the paper.
Ranjit Vijayan: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
Ranjit Vijayan was supported by UPAR Grant (31S243 and 12S006) from the United Arab Emirates University.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ali A., Vijayan R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci. Rep. 2020;10:14214. doi: 10.1038/s41598-020-71188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony P., Vijayan R. Role of SARS-CoV-2 and ACE2 variations in COVID-19. Biomed. J. 2021 doi: 10.1016/j.bj.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton B.A. Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding. EMBO J. 2007;26:4902–4912. doi: 10.1038/sj.emboj.7601906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D.-S., Kim J.-H., Kim Y.-J., Kim Y.-S. Immunoglobulin Fc-fused peptide without C-terminal Arg or Lys residue augments neuropilin-1-dependent tumor vascular permeability. Mol. Pharm. 2018;15:394–402. doi: 10.1021/acs.molpharmaceut.7b00761. [DOI] [PubMed] [Google Scholar]
- Bowers K. SC ’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, Florida. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. 43–43. [Google Scholar]
- Cantuti-Castelvetri L. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vivo M., Masetti M., Bottegoni G., Cavalli A. Role of molecular dynamics and related methods in drug discovery. J. Med. Chem. 2016;59:4035–4061. doi: 10.1021/acs.jmedchem.5b01684. [DOI] [PubMed] [Google Scholar]
- Durrant J.D., McCammon J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9:71. doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- Getz J.A., Cheneval O., Craik D.J., Daugherty P.S. Design of a cyclotide antagonist of neuropilin-1 and -2 that potently inhibits endothelial cell migration. ACS Chem. Biol. 2013;8:1147–1154. doi: 10.1021/cb4000585. [DOI] [PubMed] [Google Scholar]
- Guo H.-F. Mechanistic basis for the potent anti-angiogenic activity of semaphorin 3F. Biochemistry. 2013;52:7551–7558. doi: 10.1021/bi401034q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Vander Kooi C. Neuropilin functions as an essential cell surface receptor. J. Biol. Chem. 2015;290:29120–29126. doi: 10.1074/jbc.R115.687327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haspel N. Binding of a C-end rule peptide to the neuropilin-1 receptor: a molecular modeling approach. Biochemistry. 2011;50:1755–1762. doi: 10.1021/bi101662j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD-visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jacobson M.P. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Bioinf. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Jarvis A. Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction. J. Med. Chem. 2010;53:2215–2226. doi: 10.1021/jm901755g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe A., Vijayan R. Neuropilins: C-end rule peptides and their association with nociception and COVID-19. Comput. Struct. Biotechnol. J. 2021;19:1889–1895. doi: 10.1016/j.csbj.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-Y. Necrosis-inducing peptide has the beneficial effect on killing tumor cells through neuropilin (NRP-1) targeting. Oncotarget. 2016;7:32449–32461. doi: 10.18632/oncotarget.8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.C., Kreusch A., McMullan D., Ng K., Spraggon G. Crystal structure of the human neuropilin-1 b1 domain. Structure. 2003;11:99–108. doi: 10.1016/s0969-2126(02)00941-3. [DOI] [PubMed] [Google Scholar]
- Li J. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins Struct. Funct. Bioinf. 2011;79:2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyna G.J., Klein M.L., Tuckerman M. Nosé–Hoover chains: the canonical ensemble via continuous dynamics. J. Chem. Phys. 1992;97:2635–2643. [Google Scholar]
- Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- Mota F. Architecture and hydration of the arginine-binding site of neuropilin-1. FEBS J. 2018;285:1290–1304. doi: 10.1111/febs.14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutal A. SARS-CoV-2 Spike protein co-opts VEGF-A/Neuropilin-1 receptor signaling to induce analgesia. Pain. 2021;162:243–252. doi: 10.1097/j.pain.0000000000002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.W., Linkugel A.D., Vander Kooi C.W. Effect of C-terminal sequence on competitive semaphorin binding to neuropilin-1. J. Mol. Biol. 2013;425:4405–4414. doi: 10.1016/j.jmb.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.W., Xu P., Li X., Vander Kooi C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012;287:11082–11089. doi: 10.1074/jbc.M111.331140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K., Bai Y., Zhu Q., Hu B., Xu Y. Targeting VEGF–neuropilin interactions: a promising antitumor strategy. Drug Discov. Today. 2019;24:656–664. doi: 10.1016/j.drudis.2018.10.004. [DOI] [PubMed] [Google Scholar]
- Seyran M. The structural basis of accelerated host cell entry by SARS-CoV-2. FEBS J. 2020 doi: 10.1111/febs.15651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starzec A., Miteva M.A., Ladam P., Villoutreix B.O., Perret G.Y. Discovery of novel inhibitors of vascular endothelial growth factor-A-Neuropilin-1 interaction by structure-based virtual screening. Bioorg. Med. Chem. 2014;22:4042–4048. doi: 10.1016/j.bmc.2014.05.068. [DOI] [PubMed] [Google Scholar]
- Teesalu T., Sugahara K.N., Kotamraju V.R., Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman M., Berne B.J., Martyna G.J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- Vander Kooi C.W. Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6152–6157. doi: 10.1073/pnas.0700043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wronski M.A. Tuftsin binds neuropilin-1 through a sequence similar to that encoded by exon 8 of vascular endothelial growth factor. J. Biol. Chem. 2006;281:5702–5710. doi: 10.1074/jbc.M511941200. [DOI] [PubMed] [Google Scholar]
- Wrapp D. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-B. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 2015;6:6240. doi: 10.1038/ncomms7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanuy D. Sequence dependence of C-end rule peptides in binding and activation of neuropilin-1 receptor. J. Struct. Biol. 2013;182:78–86. doi: 10.1016/j.jsb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.