Abstract
It has recently been shown that in Xenopus, DNA demethylation at promoter regions may involve protein-DNA interactions, based on the specificity of the demethylated sites. Utilizing a stable episomal system in human cells, we recently mapped the sites and dissected the steps of demethylation at oriP sites bound by EBNA1 protein. Although it is clear that protein binding is required for demethylation of the oriP sites, it is uncertain whether this is a unique feature of the replication origin or whether it is a general phenomenon for all DNA sequences to which sequence-specific proteins are bound. In the present study, we utilize the well-defined Escherichia coli lac repressor/operator system in human cells to determine whether protein binding to methylated DNA, in a region that is neither a replication origin nor a promoter, can also lead to demethylation of the binding sites. We found that demethylation specified by protein binding is not unique to the replication origin or to the promoter. We also found that transcriptional activity does not influence demethylation of the lac operator. Isopropyl-β-d-thiogalactopyranoside (IPTG), an inhibitor of the lac repressor, can prevent demethylation of the lac operator DNA sites and can modulate demethylation of the lac operator by affecting the binding affinity of the lac repressor. Using this system, a titration of protein binding can be done. This titration permits one to infer that protein binding site occupancy is the determinant of demethylation at DNA sites and permits a determination of how this process progresses over time.
CpG methylation is tightly regulated during DNA replication and differentiation of somatic cells. The preexisting DNA methylation pattern is maintained by DNA methyltransferase 1 (DNMT1) during DNA replication (14). Changes in the basic methylation pattern occur throughout development through the dynamic processes of de novo methylation and demethylation. Two gene products, DNMT3A and DNMT3B, have been recently reported to have methyltransferase activity both in vitro and in vivo (11, 17). These de novo methyltransferases are believed to be involved in gene regulation, X-inactivation, genomic imprinting, and methylation of endogenous retroviruses and transposable elements (for a review, see reference 27). Several demethylases have been described previously (2, 13, 21, 24). Unfortunately, the activities of some of these reported demethylases cannot be reproduced in other laboratories (16, 22), and in one case they cannot be reproduced in the laboratory reporting the activity (20). It has been found that genes lose their CpG methylation within the promoter when they become activated, whereas genes acquire CpG methylation after they are no longer transcribed (for reviews, see references 4 and 18). Matsuo et al. (15) suggested that demethylation may involve protein-DNA interactions in Xenopus embryos, based on the specificity of the demethylated sites. We have further demonstrated in an episomal system that binding of proteins specifies sites of demethylation through a two-step process in human cells (10). There is also evidence arguing that loss of DNA methylation may be a consequence of transcriptional activation (23). Taking these results together, one can hypothesize that when a gene is activated, transcription factor binding can initiate demethylation in the promoter and then lead to increased transcription.
Utilizing an episomal system in human cells, we were able to map the sites and dissect the steps of demethylation at oriP sites bound by EBNA1 protein (10). Although it is clear that protein binding is required for demethylation of the oriP sites, it remains uncertain whether this is a unique feature of the replication origin or whether it is a general phenomenon for all DNA sequences to which sequence-specific proteins are bound. In the present study, we utilize the well-defined Escherichia coli lac repressor/operator system to determine whether protein binding to methylated DNA, in a region that is neither a replication origin nor a promoter, can also lead to demethylation of the binding sites. Many reports indicate that the lac repressor system works in both E. coli and mammalian cells (3, 5, 6, 12) and that isopropyl-β-d-thiogalactopyranoside (IPTG) can inhibit lac repressor (LacI) binding to the lac operator (lacO). Therefore, we placed the lacO sequences on the episomes and tested them in cell lines with or without lacI expression. We found that methylated lacO sequences became demethylated in the lacI-expressing cell line but remained methylated in the cell line with no lacI expression. We also found that IPTG inhibition of LacI binding to lacO could modulate demethylation within the lacO sequences. Furthermore, this LacI-specified demethylation was not affected by transcriptional activity. This is the first example in which patterns of demethylation can be modulated by manipulating the binding affinity of a sequence-specific binding protein.
MATERIALS AND METHODS
Plasmids.
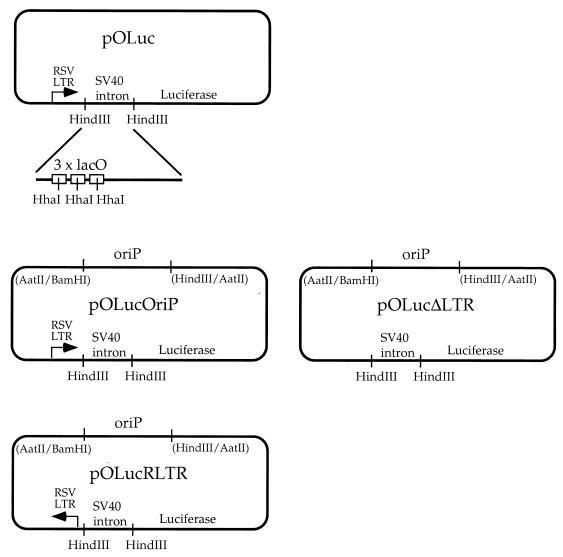
Plasmid pOLuc (Fig. 1) was constructed by replacing the chloramphenicol acetyltransferase gene on pOPI3 (Stratagene) with the luciferase reporter gene. Plasmid pOLucOriP (Fig. 1) has the Epstein-Barr virus EBV latent replication origin, oriP, inserted at the AatII site of pOLuc. Plasmid pOLucOriP was double digested by SacII and BglII to construct pOLucΔLTR and pOLucRLTR (Fig. 1). The DNA ends were blunted after digestion and then recircularized to generate pOLucΔLTR, which does not have the Rous sarcoma virus (RSV) long terminal repeat (LTR) upstream of the intron containing the lacO sequences. The pOLucRLTR was generated by ligating the RSV LTR fragment in the reverse orientation back to the SacII-BglII-double-digested pOLucOriP.
FIG. 1.
Illustration of lacO-bearing plasmids. pOLuc does not have a eukaryotic replication origin. Plasmids pOLucOriP, pOLucΔLTR, and pOLucRLTR have an Epstein-Barr virus latent replication origin inserted at the AatII site of pOLuc.
In vitro methylation.
DNA was methylated with HhaI- or SssI-methylase overnight under conditions recommended by the manufacturer (New England Biolabs). The DNA was phenol-chloroform extracted and ethanol precipitated after in vitro methylation. The status of methylation was confirmed by digestion using methylation-sensitive restriction endonucleases.
Cell lines and transfection.
The 293/EBNA1 cell line has been described previously (9). The 293E/lacI cell line was generated by cotransfection of the linearized pCMVLacI (3) with a puromycin expression vector at a ratio of 10:1 into 293/EBNA1 cells. Twenty independent puromycin-resistant cell clones were isolated and expanded from the transfection. Expression of the LacI protein was examined by immunofluorescence staining and Western blotting using anti-LacI antibody (generous gift of K. Mathews, Rice University, Texas). The amount of LacI protein in the cells was estimated by comparison with a known amount of the purified LacI protein (generous gift of K. Mathews). Throughout this study, the calcium phosphate transfection method (25) was used. All transfections were done in duplicate or triplicate for each experiment, and all experiments were performed multiple times.
Episome recovery and analysis.
Each time the transfected cells reached confluence, 2.5% of the cells were replated into a 100-mm plate and the remaining cells were harvested for plasmid DNA extraction by the Hirt method (7). For the luciferase assay, 2.5% of the cells were harvested for each assay. All the transfection experiments were carried out without any selection for the episomal plasmid. IPTG was added 4 h prior to transfection in all of the IPTG suppression experiments.
DNA harvested from each transfection was digested with restriction enzymes to determine the methylation status. The digested DNA was fractionated on 1% agarose gels, Southern transferred onto nylon membranes, and probed with either the entire plasmid or the HindIII fragment containing the lacO sequence. The Southern blots were analyzed using a phosphorimager (GS525; Bio-Rad).
Luciferase expression analysis.
An aliquot (2.5%) of the transfected cells was harvested when the cells became confluent and lysed for luciferase activity analysis. The luciferase activity was analyzed on a luminometer (Monolight 2020; Analytical Luminescence) as described previously (8). In this study, the measurement of luciferase gene expression is normalized by the amount of plasmid DNA in the cells from each transfection. From the Southern blot described in the previous section, the reading of radioactivity in each lane from the phosphorimager was divided by that of the lane with the lowest reading to normalize for the amount of DNA in each transfection. The luciferase reading was then divided by the normalization factor after subtracting the background luciferase reading to obtain the normalized luciferase activity. Therefore, the levels of gene expression from the same quantity of plasmid DNA with different methylation states were compared in this study.
RESULTS
The lac repressor protein can regulate gene expression in human cells when the lac operator is placed between the promoter and the coding region.
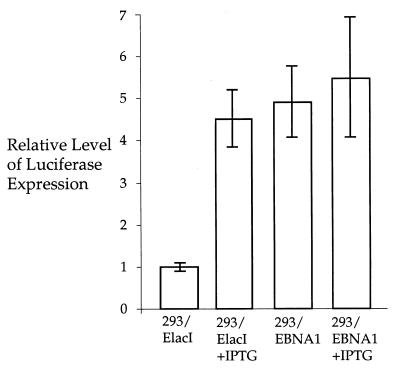
The E. coli lac repressor (LacI) has been demonstrated to bind to the lac operator (lacO) positioned immediately downstream of the simian virus 40 (SV40) promoter on a plasmid, pSVlacO, and inhibit the transcription of downstream sequences in mammalian cells (3). For the studies below, it was necessary to ensure that a stably integrated lacI could function in 293/EBNA1 cells. The plasmid, pOLuc, has an intron between the RSV LTR and the luciferase reporter gene, and there are three copies of the lacO sequence within this intron (Fig. 1). When LacI protein is present in the cells, it is expected to bind to the lacO and inhibit luciferase expression. The pOLuc plasmid was transfected into 293/EBNA1 and 293/ElacI cells to test the transcriptional inhibition of luciferase by LacI. The luciferase expression was reduced approximately fivefold in the 293/ElacI cells compared to that in the 293/EBNA1 cells (Fig. 2). This indicates that the LacI expression in 293/ElacI cells is sufficient to inhibit luciferase expression, most probably by binding to the lacO positioned between the promoter and the luciferase gene.
FIG. 2.
Transcriptional suppression of the luciferase gene on pOLuc by lac repressor. The relative luciferase expression from pOLuc transfected into cell lines with (293/ElacI) and without (293/EBNA1) lac repressor expression is shown. The bars indicate the standard deviations.
If suppression of the luciferase gene expression in 293/ElacI cells is due to LacI binding to lacO, the presence of IPTG, a LacI inhibitor, should reverse the inhibition of transcription. The effect of IPTG on transcription was tested in the 293/ElacI cells. IPTG was added to the tissue culture medium to a final concentration of 5 mM 24 h before transfection. The cells were trypsinized 24 h after transfection and divided equally between two tissue culture plates. One of these plates was continuously treated with IPTG at a final concentration of 5 mM, and the other was cultured without IPTG. The cells were harvested for luciferase expression 72 h after transfection. No significant changes were observed in 293/EBNA1 cells, although approximately a 10% increase in luciferase expression was observed when the cells were treated with IPTG. In contrast, the luciferase expression was 4.5-fold higher in the IPTG-treated 293/ElacI cells compared with that in the untreated cells (Fig. 2). The levels of luciferase expression in the 293/EBNA1 cells and the IPTG-treated 293/ElacI cells were very comparable. This indicates that the suppression of luciferase expression is due to LacI binding to lacO and that this suppression can be reversed by the presence of IPTG.
The lacO sites become demethylated in a LacI-expressing cell line.
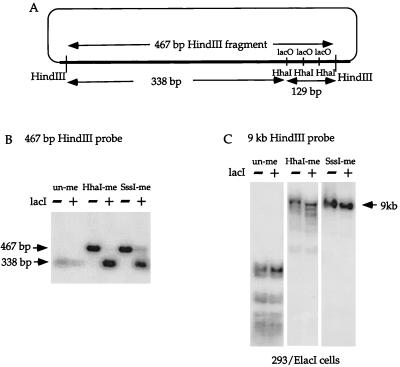
Binding of LacI to lacO should lead to demethylation of lacO if sites of demethylation are specified by protein binding. The plasmid, pOLucOriP, which was generated by inserting oriP into pOLuc and can be stably maintained as an episome in 293/EBNA1 and 293/ElacI cells, was used to test this hypothesis. The unmethylated, HhaI-methylated, or SssI-methylated pOLucOriP was transfected into 293/ElacI cells and into 293/EBNA1 cells. Transfected plasmid DNA was harvested from the cells three times, first at 6 or 7 days after transfection, then at 10 or 11 days after transfection, and finally at 15 or 17 days after transfection. The plasmid DNA was double digested with HindIII and HhaI, fractionated on a 1% agarose gel, Southern transferred, and probed with the 467-bp HindIII fragment (Fig. 3A). There is a single HhaI site at the center of the palindrome in each of the three ideal lacO sites within the HindIII fragment. The unmethylated plasmid can be completely digested by HindIII and HhaI and therefore, should give rise to a 338-bp fragment and three 43-bp fragments (Fig. 3A). The methylated plasmid is resistant to HhaI enzyme digestion and should give rise only to the 467-bp HindIII fragment. If the HhaI sites within lacO become demethylated in 293/ElacI cells, these HhaI sites on the methylated plasmid should become demethylated and show the same band pattern as on the unmethylated plasmid. The unmethylated pOLucOriP harvested from both 293/EBNA1 and 293/ElacI cells showed the same 338-bp fragment after HindIII-HhaI double digestion and probing with the 467-bp HindIII fragment (Fig. 3B). The same result was observed in all three harvests of the same transfections. This indicates that the HhaI sites within the lacO sequence remained unmethylated on the unmethylated pOLucOriP in cell lines with or without LacI expression. A 467-bp fragment was detected in the HhaI-methylated pOLucOriP harvested from 293/EBNA1 cells, while two fragments of 467 bp (very faint) and 338 bp were detected in the HhaI-methylated pOLucOriP harvested from 293/ElacI cells (Fig. 3B). The SssI-methylated plasmid also showed the same results (Fig. 3B). This indicates that lacO remains methylated at the HhaI sites on either the HhaI- or SssI-methylated plasmids when LacI is absent. These results also indicate that a large fraction of the methylated pOLucOriP episomes become demethylated at the HhaI sites within the lacO sequence when LacI is present.
FIG. 3.
Demethylation of lacO sites. (A) Illustration of the SV40 intron harboring the three lacO sites and the sizes of HindIII and HhaI restriction fragments in the region. (B) Southern blot of HindIII-HhaI-double-digested plasmid DNA harvested 15 days after transfection into cell lines with (+ lacI, 293/ElacI) and without (−lacI, 293/EBNA1) lacI expression. The unmethylated plasmid can be digested to completion by HindIII and HhaI; therefore, a 338-bp band is detected. The HhaI- or SssI-methylated plasmids remain methylated at the three HhaI sites within lacO when LacI is absent, so that a 467-bp band is detected. In contrast, these HhaI sites are demethylated when LacI is present, and they are digestable by HhaI to give rise to the 338-bp band. (C) Lack of demethylation in the vector backbone. There is no detectable difference in the HhaI-digested vector backbone with or without lacI.
It is important to confirm that the presence of LacI does not lead to overall demethylation of the plasmids in addition to the lacO sequence. The same blots were stripped and reprobed with the large HindIII fragment of the pOLucOriP that lacks the SV40 intronic sequence and the lacO sequences. The unmethylated plasmid harvested from 293/ElacI cells was digested to completion by HindIII and HhaI (Fig. 3C). Only one fragment, of approximately 9 kb, was detected in HindIII-HhaI-double-digested SssI-methylated plasmid DNA harvested from 293/ElacI cells (Fig. 3C). It has been shown that a small fraction of the HhaI-methylated plasmid demethylates at HhaI sites in the oriP region and in the herpes simplex virus HSV thymidine kinase (tk) promoter region in 293/EBNA1 cells (9). Therefore, multiple bands were detected in the HindIII-HhaI double digestion of HhaI-methylated plasmid DNA harvested from 293/ElacI cells (Fig. 3C). Furthermore, the sizes of the HhaI fragments generated from the vector backbone are not altered when LacI is present in the cells (Fig. 3C). These findings indicate that the presence of LacI specifically leads to demethylation of HhaI sites within lacO.
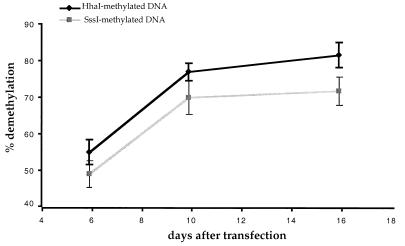
The percentage of methylated plasmids that become demethylated at the HhaI sites in the 293/ElacI cells can be measured as follows. The radioactivity in the 338-bp band is divided by the total radioactivity in the 467- and 338-bp bands after correction for the fraction of the probe that can hybridize to each band. The HhaI-methylated DNA recovered from the first of the three harvests at 6 or 7 days after transfection was approximately 55% demethylated within the lacO sites (Fig. 4). The percentage of HhaI-methylated plasmids demethylated within the lacO sites increased to 77.4% at 10 or 11 days after transfection and to 82.1% at 15 to 16 days after transfection (Fig. 4). The SssI-methylated plasmid showed the same increase in the fraction of plasmids becoming demethylated within the lacO sites as did the HhaI-methylated plasmid in the 293/ElacI cells (Fig. 4). This indicates that a larger number of methylated plasmids become demethylated within the lacO sites over time. In our experience, the amount of episomal DNA in the cell drops significantly between 5 and 8 days after transfection (data not shown). It is possible that not all lacO sites are occupied by LacI during the first few days after transfection due to the abundance of the episome in the cells. Therefore, the fraction of plasmids undergoing demethylation at the lacO sites is limited by ratio of the lacO sites and LacI protein in the cells (the equilibrium for LacI binding to lacO).
FIG. 4.
Quantitation of lacO demethylation observed on pOLucOriP. The fraction of demethylated lacO increases over time. The radioactivity in the 338-bp band is divided by the total radioactivity in the 467- and 338-bp bands after correction for the fraction of the probe that can hybridize to each band. The bars indicate the standard deviations.
IPTG, a LacI binding inhibitor, can prevent lacO demethylation.
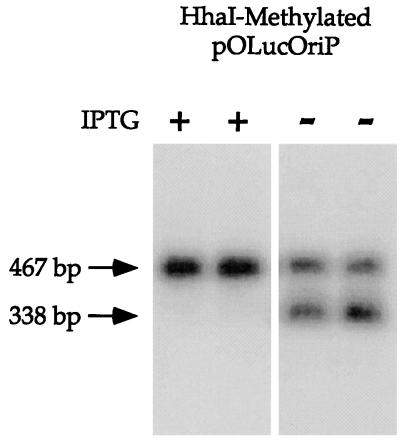
In both E. coli and mammalian cells, IPTG can induce transcription by binding to LacI, thereby decreasing its affinity for lacO (equilibrium dissociation constant, 10−13 M [19]) to the background level of general DNA binding (equilibrium association constant, 3 × 10−10 M [1]). We also confirmed that the presence of 5 mM IPTG in the tissue culture medium can induce luciferase expression from pOLuc in 293/ElacI cells as described above. If the demethylation of lacO sites observed in the 293/ElacI cells is a direct consequence of LacI binding to lacO, the presence of a sufficient concentration of IPTG should prevent LacI binding to lacO and inhibit the demethylation of lacO. Furthermore, the effect of IPTG should be fully reversible. When IPTG is removed from the cells, LacI should be able to bind lacO and lead to demethylation of the lacO sites. The experiment was designed to compare the methylation status within the lacO sites in the presence and absence of IPTG in the cells from the same transfection. IPTG was added to the 293/ElacI cells to a final concentration of 5 mM at least 4 h before transfection. The high-concentration IPTG treatment ensures that most, if not all, of the repressors are fully occupied by the inducer before the lacO-bearing plasmid is transfected into these cells. The cells from each transfection were trypsinized and divided equally between two tissue culture plates 3 days after transfection with HhaI-methylated pOLucOriP. IPTG was added to one of these plates to a final concentration of 10 μM, and no IPTG was added to the other plate. Plasmid DNA was harvested 4 days later, digested with HindIII and HhaI, fractionated on a 1% agarose gel, and analyzed by Southern blotting. When the transfected plasmid DNA was probed with the 467-bp HindIII fragment, only the 467-bp fragment was detected in the DNA with IPTG present the entire time (Fig. 5). In contrast, 467- and 338-bp fragments were detected in the plasmid DNA harvested 7 days after transfection when IPTG was withdrawn from the culture 3 days after transfection (data not shown). This was seen for HhaI-methylated DNA as well as SssI-methylated DNA (data not shown). IPTG was removed from the cells transfected with methylated plasmid DNA 15 days after transfection to confirm that the IPTG effect is reversible after long-term treatment. Approximately 50% of the plasmids were demethylated within the lacO sites 4 days after IPTG withdrawal (Fig. 5). This finding indicates that a high concentration of IPTG can prevent demethylation of the lacO sites on the episome by inhibiting LacI binding to lacO and that the IPTG effect is fully reversible. This suggests that high-affinity protein binding is crucial for demethylation site targeting.
FIG. 5.
The presence of a high concentration of IPTG can prevent lacO demethylation. The 293/ElacI cells were treated with a final IPTG concentration of 5 mM beginning 4 h before transfection. The cells were divided evenly between two plates 3 days after transfection. The cells in one plate were treated with 10 μM IPTG, and the cells in the other plate were not treated with IPTG. Plasmid DNA was harvested for probing multiple times during a 19-day interval for Southern blot analysis using the 467-bp HindIII fragment as a probe. When IPTG was present, the HhaI-methylated plasmid remained methylated at the HhaI sites within the lacO sequence at 19 days after transfection (lanes 1 and 2). In contrast, after IPTG was withdrawn from the tissue culture medium 15 days after transfection, demethylation within the lacO sites could be detected 4 days later (lanes 3 and 4).
The IPTG concentration is inversely proportional to the lacO demethylation.
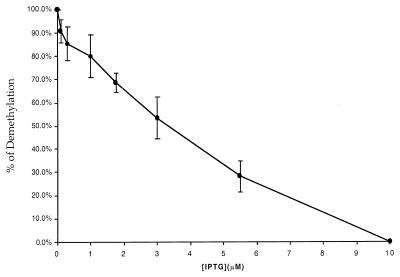
We were interested in determining if the amount of IPTG in the cells correlates with demethylation of the lacO sites. The 293/ElacI cells were treated with a final concentration of 5 mM IPTG 4 h prior to transfection with HhaI-methylated pOLucOriP. The transfected cells were trypsinized and divided among five or six plates as needed at 3 or 4 days after transfection, as the cells first became confluent. Various amounts of IPTG were added to each plate. The goal of the first few sets of experiments was to find the lowest concentration of IPTG that can effectively prevent demethylation of the lacO sites. Various final concentrations of IPTG over the range of 5 mM to 5 nM were tested and compared with no IPTG treatment. A similar fraction of the plasmid became demethylated with an IPTG concentration between 0 and 100 nM (data not shown). No demethylation of the lacO sites was detected from the HhaI-methylated plasmid when the IPTG concentration was above 10 μM (data not shown). It is clear that 100 nM or less IPTG is not sufficient to block LacI-specified demethylation while 10 μM IPTG can effectively prevent demethylation of the lacO sites. Six different intermediate concentrations of IPTG, 0.1, 0.3, 1, 1.75, 3, and 5.5 μM, were further tested with 0 and 10 μM IPTG as controls for inhibition of lacO site demethylation. A higher percentage of plasmid became demethylated with decreasing concentrations of IPTG (Fig. 6). This demonstrates that demethylation of the lacO sites is directly modulated by the concentration of IPTG and the ability of LacI to bind to the lacO sites.
FIG. 6.
Modulation of demethylation by the IPTG concentration. The 293/ElacI cells were treated with a final IPTG concentration of 5 mM for 4 h before transfection with HhaI-methylated pOLucOriP. The transfected cells were divided among several plates and treated with lower concentrations of IPTG. The fraction of plasmids becoming demethylated in the lacO sites was analyzed as described previously. A linear and inversely correlated relationship between the IPTG concentration and demethylation of lacO was observed.
The amount of LacI protein in the 293/ElacI cells was estimated by Western blotting using a known amount of purified LacI protein as a comparison. There was approximately 0.3 μg of LacI in 2 × 106 293/ElacI cells, equivalent to 2.7 × 106 molecules per cell. The minimum concentration of IPTG in the medium needed to inhibit demethylation entirely was 10 μM, equivalent to approximately 4 × 107 molecules per nucleus based on the findings in Wyborski and Short (26). From this, we can calculate that approximately a 20-fold excess of IPTG over LacI may be needed to suppress LacI binding to the lacO sites sufficiently to prevent all demethylation at those sites in this system.
Transcriptional activity does not affect the demethylation of lacO sites.
The lacO sites within an SV40 intron are positioned between the promoter and the luciferase gene on the episome. Transcription through this region may affect demethylation in two possible ways that would lead to opposite effects. It is possible that active transcription can also interfere with maintenance methylation, thereby enhancing demethylation of the lacO. If this were the case, increased transcriptional activity should lead to increased demethylation. Alternatively, it is possible that transcription can displace LacI binding to lacO. If so, increased transcriptional activity should lead to decreased demethylation at lacO sites.
We found that only a slightly smaller fraction of the SssI-methylated plasmid DNA became demethylated compared with the HhaI-methylated plasmid (as described above), although the luciferase activity of the SssI-methylated plasmid was more than 100-fold lower than that of its HhaI-methylated counterpart (data not shown). This suggests that transcription does not enhance demethylation.
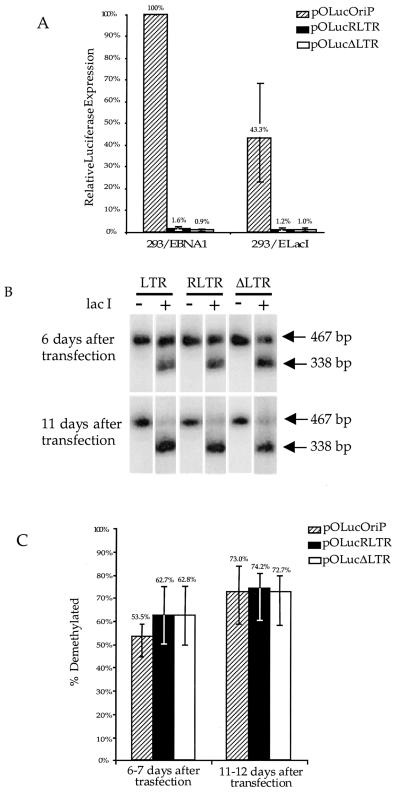
To further test the possible effects of transcription on the demethylation process, plasmids with the same density of methylation and different transcriptional activity could be tested. Plasmid pOLucRLTR was used to vary the transcriptional activity by approximately 40-fold and maintain the same density of methylation. Plasmid pOLucΔLTR was used as the promoterless comparison. HhaI-methylated pOLucΔLTR, pOLucRLTR, or pOLucOriP was transfected into 293/EBNA1 and 293/ElacI cells. The luciferase activity and the fraction of plasmid demethylation were measured 6 or 7 days after transfection and 11 or 12 days after transfection. The luciferase activity was normalized by the amount of plasmid DNA in the cells and was standardized against the luciferase activity of HhaI-methylated pOLucOriP plasmid in 293/EBNA1 cells. The luciferase expression from HhaI-methylated pOLucOriP in 293/ElacI cells was at 43% of the expression level of its counterpart in 293/EBNA1 cells (Fig. 7A). The luciferase expression from HhaI-methylated pOLucΔLTR, which does not have a promoter upstream of the luciferase gene, was 1% of that of the HhaI-methylated pOLucOriP in 293/EBNA1 cells (Fig. 7A). The luciferase expression from HhaI-methylated pOLucRLTR, which has the RSV LTR pointed away from the luciferase gene, was also approximately 1% of the expression level of the HhaI-methylated pOLucOriP in the 293/EBNA1 cells (Fig. 7A). Levels of expression from HhaI-methylated pOLucΔLTR and HhaI-methylated pOLucRLTR were similar in 293/EBNA1 and 293/ElacI cells (Fig. 7A). Southern blot analysis of HindIII-HhaI-double-digested plasmid DNA harvested from 293/ElacI cells showed a similar degree of demethylation for all three plasmids (Fig. 7B). The fraction of demethylated plasmid from each transfection was calculated based on the radioactivity in the 467- and 338-bp bands as described above. Despite these dramatic differences in transcriptional activity, there was only a slight decrease in the fraction of demethylated HhaI-methylated pOLucOriP compared with either HhaI-methylated pOLucΔLTR or HhaI-methylated pOLucRLTR (Fig. 7C). The plasmid DNA harvested 11 or 12 days after transfection showed similar results to the DNA harvested 6 or 7 days after transfection (Fig. 7C). Despite the 40-fold difference in transcriptional activity, demethylation of lacO sites occurred to similar extents on these plasmids. This indicates that the level of transcription through a DNA region does not play a major role in the demethylation process and that the presence of the promoter does not play a role in demethylation at the lacO sites. This also implies that transcriptionally active chromatin is not a requirement for demethylation. LacI is not known to be associated with a histone acetylase complex; therefore, demethylation is not likely to involve the recruitment of a histone acetylase complex.
FIG. 7.
Demethylation of lacO sites is not affected by transcriptional activity through the DNA sequence. (A) The expression of luciferase from three different plasmids was measured and normalized by the amount of plasmid DNA in the transfected cells. The relative luciferase expression was calculated using the luciferase expression of pOLucOriP in 293/EBNA1 cells as 100% expression. (B) Southern blot analysis of lacO demethylation. Three different plasmids showed a similar fraction of lacO demethylation over time. (C) Quantitation of lacO demethylation on three different plasmids. The bars indicate the range of the values. There is 56.9, 60, and 65% demethylated lacO on pOLucOriP, pOLucRLTR, and pOLucΔLTR, respectively, at 6 days after transfection in panel B, and there is 84.9, 81.1, and 75.7% demethylated lacO on pOLucOriP, pOLucRLTR, and pOLucΔLTR, respectively, at 11 days after transfection in panel B.
DISCUSSION
This study utilizes a well-characterized system to demonstrate that (i) lac repressor binding to a lac operator can lead to demethylation of the lac operator, indicating that demethylation specified by protein binding is not unique to the EBV latent replication origin, oriP; (ii) IPTG, an inhibitor of the lac repressor, can prevent demethylation of the lac operator; (iii) demethylation of the lac operator by the lac repressor can be modulated by affecting the binding affinity of the lac repressor; and (iv) transcriptional activity does not influence demethylation of the lac operator.
The demethylation of lacO specified by LacI binding strongly supports our previous finding that protein binding is essential in the demethylation process. Although we have demonstrated previously that the structure of the replication origin alone cannot lead to demethylation (10), it has not been ruled out as a requirement of protein binding-specified demethylation. This possible requirement is clearly ruled out in this study since the binding of LacI can lead to demethylation of the lacO sequence located about 4 kb away from oriP on the opposite side of the episome.
The binding affinity of the protein appears to be important. Although LacI can bind to DNA in general, the presence of LacI does not lead to demethylation of CpG sites on the plasmid outside of the lacO sequences. When a sufficient amount of IPTG is present in the cells, demethylation of lacO can be completely blocked. The equilibrium dissociation constant for the LacI-IPTG complex binding to lacO is 300-fold weaker than that of free LacI binding to lacO (1). These two lines of evidence suggest that a strong binding protein is required to specify sites for demethylation. Different IPTG concentrations in the cells can effectively modulate demethylation of lacO on the plasmid. This modulation appears to be inversely correlated in a linear fashion and indicates that the availability of LacI in the nucleus can directly affect the fraction of plasmids which become demethylated at the lacO sequence. These results strongly suggest that demethylation is determined primarily by occupancy of sites by binding proteins.
In this study, we found that transcriptional activity through the region of demethylation is neither required for nor inhibitory to the DNA demethylation specified by protein binding. A similar degree of demethylation was observed on episomes with a 40-fold difference in transcriptional activity. This clearly indicates that transcription through the protein binding site does not affect demethylation of lacO. The first step of the protein binding-specified demethylation appears to be replication dependent (10). This suggests that transcriptional displacement of LacI binding to lacO does not occur during the time that DNA is remethylated by the maintenance methyltransferase immediately after replication. The promoterless episome also demethylates to the same extent at a similar rate. This rules out the possibility that demethylation occurs through recruitment of a histone acetylase complex as proposed by Matsuo et al. (15). The present study demonstrates that demethylation specified by protein binding is most likely to be a general phenomenon that can occur at sites of strong factor binding. Although this has been found to occur only on plasmids (10, 15), it is conceivable that it also occurs in the mammalian genome. The oriP-based episomes are maintained at a relatively low copy number of 10 to 50 copies in the human cells. These episomes mimic the chromosomal events very closely, as described previously (9). We are currently testing whether protein binding can lead to demethylation in the chromosome and whether protein binding inhibitors can modulate demethylation in the genome.
It has been proposed that DNA methyltransferase can be excluded by proteins that bind cooperatively to CpG islands (28). Another model also proposed that unmethylated DNA regions are either at or near sequences bound by nonhistone proteins (29). This study provides strong support for these early models on methylation-free DNA regions on the chromosome. The fact that affinity of the binding protein is crucial would be consistent with the possibility that cooperative binding of proteins would also result in methylation-free regions on the chromosome.
ACKNOWLEDGMENTS
We thank M. R. Lieber, B. Tracy, D. Van Den Berg, and D. Foti for critical reading of the manuscript.
This work was supported by NIH grant GM54781.
REFERENCES
- 1.Barkley M D, Bourgeois S. Repressor recognition of operator and effectors. In: Miller J H, Reznikoff W S, editors. The operon. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. pp. 177–220. [Google Scholar]
- 2.Bhattacharya S K, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 3.Brown M, Figge J, Hansen U, Wright C, Jeang K-T, Khoury G, Livingston D M, Roberts T M. lac repressor can regulate expression from a hybrid SV40 early promoter containing a lac operator in animal cells. Cell. 1987;49:603–612. doi: 10.1016/0092-8674(87)90536-8. [DOI] [PubMed] [Google Scholar]
- 4.Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 5.Fieck A, Wyborski D L, Short J M. Modifications of the E. coli Lac repressor for expression in eukaryotic cells: effects of nuclear signal sequences on protein activity and nuclear accumulation. Nucleic Acids Res. 1992;20:1785–1791. doi: 10.1093/nar/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figge J, Wright D, Collins C J, Roberts T M, Livingston D M. Stringent regulation of stably integrated chloramphenicol acetyl transferase genes by E. coli lac repressor in monkey cells. Cell. 1988;52:713–722. doi: 10.1016/0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- 7.Hirt B. Selective extraction of polyoma DNA from infected mouse cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh C-L, McCloskey R P, Lieber M R. V(D)J recombination on minichromosomes is not affected by transcription. J Biol Chem. 1992;267:15613–15619. [PubMed] [Google Scholar]
- 9.Hsieh C-L. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh C-L. Evidence that protein binding specifies sites of DNA Demethylation. Mol Cell Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh C-L. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol Cell Biol. 1999;19:8211–8218. doi: 10.1128/mcb.19.12.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu M C, Davidson N. The inducible lac operator-repressor system is functional in mammalian cells. Cell. 1987;48:555–566. doi: 10.1016/0092-8674(87)90234-0. [DOI] [PubMed] [Google Scholar]
- 13.Jost J-P. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc Natl Acad Sci USA. 1993;90:4684–4688. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leohnardt H, Bestor T H. Structure, function and regulation of mammalian DNA methyltransferase. In: Jost J P, Saluz H P, editors. DNA methylation: molecular biology and biological significance. Basel, Switzerland: Birkhauser Verlag; 1993. pp. 109–119. [DOI] [PubMed] [Google Scholar]
- 15.Matsuo K, Silke J, Georgiev O, Marti P, Giovannini N, Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng H-H, Zhang Y, Hendrich B, Johnson C A, Turner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histon deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 17.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 18.Razin A, Kafri T. DNA methylation from embryo to adult. Prog Nucleic Acid Res Mol Biol. 1994;48:53–81. doi: 10.1016/s0079-6603(08)60853-3. [DOI] [PubMed] [Google Scholar]
- 19.Riggs A D, Suzuki H, Bourgeoss S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970;48:67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- 20.Swisher J F, Rand E, Cedar H, Pyle A M. Analysis of putative RNase sensitivity and protease insensitivity of demethylation activity in extracts from rat myoblasts. Nucleic Acids Res. 1998;26:5573–5580. doi: 10.1093/nar/26.24.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vairapandi M, Duker N J. Enzymic removal of 5-methylcytosine from DNA by a human DNA-glycosylase. Nucleic Acids Res. 1993;21:5323–5327. doi: 10.1093/nar/21.23.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 23.Walsh C P, Bestor T H. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: involvement of RNA. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 25.Wigler M, Sweet R, Sim G K, Wold B, Pellicer A, Lacy E, Maniatis T, Silverstein S, Axel R. Transformation of mammalian cells with genes from prokaryotes and eukaryotes. Cell. 1979;16:777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- 26.Wyborski D L, Short J M. Analysis of inducers of the E. coli lac repressor system in mammalian cells and whole animals. Nucleic Acids Res. 1991;19:4647–4653. doi: 10.1093/nar/19.17.4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoder J A, Walsh C P, Bestor T H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 28.Bird A P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 29.Selker E C. DNA methylation and chromatin structure: a view from below. Trends Biochem Sci. 1990;15:103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]