Abstract
Recent work has shown that transcription of the yeast HO gene involves the sequential recruitment of a series of transcription factors. We have performed a functional analysis of HO regulation by determining the ability of mutations in SIN1, SIN3, RPD3, and SIN4 negative regulators to permit HO expression in the absence of certain activators. Mutations in the SIN1 (=SPT2) gene do not affect HO regulation, in contrast to results of other studies using an HO:lacZ reporter, and our data show that the regulatory properties of an HO:lacZ reporter differ from that of the native HO gene. Mutations in SIN3 and RPD3, which encode components of a histone deacetylase complex, show the same pattern of genetic suppression, and this suppression pattern differs from that seen in a sin4 mutant. The Sin4 protein is present in two transcriptional regulatory complexes, the RNA polymerase II holoenzyme/mediator and the SAGA histone acetylase complex. Our genetic analysis allows us to conclude that Swi/Snf chromatin remodeling complex has multiple roles in HO activation, and the data suggest that the ability of the SBF transcription factor to bind to the HO promoter may be affected by the acetylation state of the HO promoter. We also demonstrate that the Nhp6 architectural transcription factor, encoded by the redundant NHP6A and NHP6B genes, is required for HO expression. Suppression analysis with sin3, rpd3, and sin4 mutations suggests that Nhp6 and Gcn5 have similar functions. A gcn5 nhp6a nhp6b triple mutant is extremely sick, suggesting that the SAGA complex and the Nhp6 architectural transcription factors function in parallel pathways to activate transcription. We find that disruption of SIN4 allows this strain to grow at a reasonable rate, indicating a critical role for Sin4 in detecting structural changes in chromatin mediated by Gcn5 and Nhp6. These studies underscore the critical role of chromatin structure in regulating HO gene expression.
The Saccharomyces cerevisiae HO gene encodes an endonuclease that is responsible for initiating mating type switching in yeast. The transcriptional regulation of HO is complex and has been the subject of intensive study (for reviews, see references 22 and 37). Recent studies have shown that transcription of specific genes can be affected by chromatin structure at the promoter (for reviews, see references 25, 28, 54, and 59). Chromatin structure plays an important role in regulation of HO transcription, as HO expression is altered by mutations in a number of important transcriptional regulators, including components of the Swi/Snf chromatin remodeling complex, the SAGA histone acetyltransferase complex, and the Sin3/Rpd3 histone deacetylase complex. GCN5, ADA2, and ADA3, which encode members of the SAGA histone acetyltransferase complex (18), are required for HO:lacZ expression (43), and native HO expression is also reduced in a gcn5 mutant (41). The yeast RPD3 gene encodes a histone deacetylase that is associated with Sin3 (26, 27). SIN3 and RPD3 are negative regulators of transcription, and mutations in SIN3 or RPD3 allow an HO:lacZ reporter to be expressed in the absence of specific activators (41, 53).
HO is cell cycle regulated and is expressed in late G1 (36). Recent work using chromatin immunoprecipitation provides new insights as to changes at the HO promoter during the cell cycle. Cosma et al. (12) showed that activation of HO transcription involves ordered recruitment of transcription factors. Swi5 enters the nucleus at the end of anaphase, binds to the promoter, and then recruits Swi/Snf. Swi/Snf, in turn, recruits SAGA, and Swi/Snf and SAGA are both required for SBF binding. It is believed that SBF, composed of the Swi4 and Swi6 factors, is then directly responsible for HO activation. Krebs et al. (30) showed that a 1-kb region of the HO promoter undergoes histone acetylation in mid-G1 phase of the cell cycle, and these promoter changes require the activity of the Swi5, Swi/Snf, and SAGA transcription factors. Mutations in SIN3 or RPD3 result in acetylation of the HO promoter throughout the cell cycle.
The SIN4 gene was identified as regulator of HO expression (24). A sin4 mutation causes decreased expression of some genes, including HIS4, CTS1, and MATα. However, expression of other genes, including HO:lacZ, IME1, GAL1, SUC2, DIT1, DIT2, and a-specific genes, is increased in a sin4 mutant (11, 14, 16, 24, 50, 56). A sin4 mutation has effects similar to those seen in strains with histone mutations, including changes in linking number of plasmid DNA and sensitivity of chromatin to nucleases, and it has been suggested that these effects on transcription are caused by changes in chromatin structure (23, 24, 33). The Sin4 protein is part of the RNA polymerase II holoenzyme/mediator, in a subcomplex with Rgr1, Gal11, Med2, and Pgd1 (32, 35). Importantly, mutations in other components of the RNA polymerase II mediator complex also have diverse effects on transcriptional regulation (for reviews, see references 5, 9, and 20). It has been recently demonstrated that Sin4 is also part of the SAGA complex (P. Grant and J. Workman, personal communication).
The HO gene promoter is quite large, by yeast standards, with regulatory sites identified nearly 2 kb from the transcription start site. The SWI5 gene encodes a zinc finger DNA binding protein that is required for HO expression. There are two Swi5 binding sites in the HO promoter, at −1800 and at −1300. Genetic analysis demonstrates that both Swi5 binding sites are required for HO expression, suggesting that there is a physical interaction between these two sites separated by 500 bp (34). The term “architectural transcription factor” has been applied to proteins that bend DNA and promote assembly of distantly bound factors into a productive complex (58). It is possible that architectural transcription factors, by promoting DNA bending, could facilitate this proposed interaction between Swi5 molecules bound at these two sites. We decided to examine whether architectural transcription factors contribute to Swi5-dependent activation of HO by determining whether mutations in these factors affect HO expression.
Architectural transcription factors often contain the DNA-binding domain first identified in mammalian high-mobility-group 1 and 2 (HMG1/2) proteins (8). There are a number of yeast genes encoding proteins with homology to the HMG domain, including ABF2, ROX1, SIN1, and the duplicated NHP6A and NHP6B genes. Some HMG proteins, such as Rox1 (15), bind DNA in a sequence-specific manner; other HMG proteins have little specificity in DNA sequence recognition but may recognize structural elements in DNA or chromatin, such as cruciform structures (4). We directed our attention to SIN1 and NHP6A/B because mutations in these genes have been reported to affect transcriptional regulation.
The SIN1 gene was originally identified as SPT2, as sin1/spt2 mutations suppress the transcriptional defects due to insertions of the Ty1 transposable element into the HIS4 and LYS2 promoters (49). SIN1 mutations were also identified as bypass suppressors allowing expression of an HO:lacZ reporter in strains lacking either the Swi1 or Swi5 transcriptional activator (51). As we show below, sin1 mutations do not restore expression of the native HO gene; the original observation appears to be an artifact of the bacterial sequences present in the HO:lacZ reporter. A sin1 mutation can suppress the transcriptional defects at the SUC2 and HIS3 loci caused by mutations in SWI1 and GCN5, respectively (41, 43). Additionally, a sin1 mutation increases expression of the SSA3 gene (1).
The 11-kDa Nhp6 protein of yeast shows 40% identity to the HMG domain of mammalian HMG1/2 proteins (29). There are two highly related genes, NHP6A and NHP6B, that express the Nhp6 protein. These two genes appear to be functionally redundant, as deletion of both genes is required for any observable phenotype (13). The nhp6a nhp6b double mutant is temperature sensitive for growth and shows defects in transcriptional activation of a number of LacZ reporter constructs (13, 39). Finally, in vitro experiments show that Nhp6 protein can promote the assembly of multicomponent protein-DNA complexes (40).
In this report we show that the Gcn5 and Nhp6 proteins are required for expression of HO. Suppressor analysis shows that mutations in the SIN3, RPD3, or SIN4 gene can allow HO expression in the absence of these activators. We also find that gcn5 nhp6a nhp6b triple mutants are very sick, suggesting that Gcn5 and Nhp6 are both required for transcription of important genes. A sin4 mutation suppresses this growth defect, suggesting that Sin4 has a unique role in regulating chromatin structure. The genetic analysis shows differences in the ability of sin3 and sin4 mutations to suppress swi5 and swi6 defects, and these results provide new insights as to regulation of HO expression.
MATERIALS AND METHODS
The yeast strains used in this study are listed in Table 1. Standard genetic methods were used for strain construction (45, 46). W303 strains with SWI5, SIN3, and SIN4 disruptions have been previously described (24, 57). W303 strains with gene disruptions in GCN5, HDA1, and HPR1 were provided by Sharon Roth, Michael Grunstein, and Hannah Klein, respectively. The SIN1 gene was disrupted with plasmid WB39 (31), provided by Ira Herskowitz, and the NHP6A and NHP6B genes were disrupted with plasmids pDK201 and pDK262, respectively, provided by David Kolodrubetz. All gene disruptions were confirmed by Southern analysis. The swi6::TRP1 allele from the closely related K1107 strain background was backcrossed four times into W303. Plasmid M4195 was constructed by inserting a 2.2-kb EcoRI-HindIII fragment with NHP6B from plasmid pDK227 (from David Kolodrubetz) into YEplac195 (17).
TABLE 1.
Yeast strains used
| DY150a | MATa ade2 can1 his3 leu2 trp1 ura3 |
| DY151a | MATα ade2 can1 his3 leu2 trp1 ura3 |
| DY161a | MATa swi5::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY411a | MATa swi5::hisG ade2 can1 his3 leu2 trp1 ura3 |
| DY773a | MATa sin3::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY775a | MATa swi5::hisG sin3::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY984a | MATa sin3::ADE2 ade2 can1 his3 leu2 trp1 ura3 |
| DY1699a | MATa sin4::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY1702a | MATa sin4::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY2133a | MATa swi5::LEU2 sin4::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY2348a | MATa swi2::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY2378a | MATa nhp6a::URA3 ade2 can1 his3 leu2 trp1 ura3 |
| DY2380a | MATa nhp6b::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY2381a | MATα nhp6a::URA3 nhp6b::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY2382a | MATa nhp6a::URA3 nhp6b::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY2389a | MATα ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY2395a | MATa ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY2499a | MATα swi2::ADE2 sin4::TRP1 ade2 can1 leu2 trp1 ura3 |
| DY2763a | MATa sin4::TRP1 ade2 can1 leu2 lys2 trp1 ura3 |
| DY2870a | MATa swi2::ADE2 sin3::LEU2 ade2 can1 leu2 trp1 ura3 |
| DY3944a | MATα swi2::ADE2 ade2 can1 leu2 lys2 trp1 ura3 |
| DY3658a | MATα sin1::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY4548a | MATα rpd3::LEU2 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5068a | MATα hda1::URA3 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5116a | MATa gcn5::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY5153a | MATa nhp6a::URA3 nhp6b::HIS3 sin1::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY5155a | MATa nhp6a::URA3 nhp6b::HIS3 sin4::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY5157a | MATa nhp6a::URA3 nhp6b::HIS3 sin3::ADE2 ade2 can1 his3 leu2 trp1 ura3 |
| DY5168a | MATα gcn5::TRP1 hda1::URA3 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5170a | MATα gcn5::TRP1 rpd3::LEU2 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5199a | MATα gcn5::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5265a | MATa gcn5::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5285a | MATa sin3::ADE2 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5289a | MATa gcn5::TRP1 sin4::LEU2 ade2 can1 his3 leu2 trp1 ura3 |
| DY5294a | MATa sin3::ADE2 sin4::LEU2 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5297a | MATa gcn5::TRP1 sin3::ADE2 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5299a | MATa swi5::hisG-URA3-hisG sin3::ADE2 sin4::LEU2 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5306a | MATa gcn5::TRP1 nhp6a::URA3 nhp6b::HIS3 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5315a | MATa gcn5::TRP1 sin3::ADE2 sin4::LEU2 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5323a | MATa sin1::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5326a | MATa gcn5::hisG-URA3-hisG sin1::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5410a | MATa swi5::LEU2 sin1::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5420a | MATa swi2::HIS3 sin1::TRP1 ade2 can1 his3 leu2 lys2 trp1 ura3 |
| DY5780a | MATa swi6::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY5781a | MATα swi6::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY5820a | MATα sin4::LEU2 gcn5::TRP1 nhp6a::URA3 nhp6b::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY5825a | MATa gcn5::TRP1 nhp6a::URA3 nhp6b::HIS3 ade2 can1 his3 leu2 trp1 ura3 |
| DY5907a | MATa swi6::TRP1 sin4::URA3 ade2 can1 his3 leu2 trp1 ura3 |
| DY5908a | MATα swi6::TRP1 sin4::URA3 ade2 can1 his3 leu2 trp1 ura3 |
| DY5909a | MATa swi5::LEU2 swi6::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY5910a | MATα swi5::LEU2 swi6::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY5911a | MATa sin4::URA3 swi5::LEU2 swi6::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY5912a | MATα sin4::URA3 swi5::LEU2 swi6::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY6103a | MATα sin3::LEU2 swi6::TRP1 ade2 can1 his3 leu2 trp1 ura3 |
| DY881b | MATa ade2 his3 leu2 lys2 trp1 ura3 |
| DY1712b | MATa sin4::URA3 ade2 his3 leu2 lys2 trp1 ura3 |
| DY2532b | MATα nhp6A::URA3 nhp6B::HIS3 ade2 his3 leu2 lys2 trp1 ura3 |
| DY2533b | MATα nhp6A::URA3 nhp6B::HIS3 sin4::TRP1 ade2 his3 leu2 lys2 trp1 ura3 |
Cells were grown at 30°C in standard media (46). YEPD medium was used, except where use of YEP-galactose medium is indicated or when strains had plasmids. In the latter case, cells were grown in synthetic complete medium with 2% glucose supplemented with adenine, uracil, and amino acids, as appropriate, but lacking essential components to select for plasmids.
RNA levels were determined with S1 nuclease protection assays using HO and CMD1 probes as described elsewhere (3). Protein extracts were prepared for quantitative measurement of β-galactosidase activity as described previously (7).
RESULTS
Role of architectural transcription factors in HO transcription.
Genetic studies demonstrated that Swi5 binding at two sites, separated by 500 bp, was required for transcription of the HO gene (34). An architectural transcription factor might promote interaction between Swi5 molecules bound at these sites, and we determined whether mutations in the SIN1 (=SPT2) gene, which encodes an HMG protein (31), affect HO expression. RNA was isolated from isogenic SIN1 and sin1 strains, and HO mRNA levels were measured with an S1 nuclease protection assay. As shown in Fig. 1A, a sin1 mutation does not affect expression of HO (compare lanes 1 and 3). Two groups recently reported that a gcn5 mutation reduced expression of an HO:lacZ reporter (41, 43). It was also reported that a sin1 mutation suppresses the gcn5 mutation, as the HO:lacZ reporter is expressed in the gcn5 sin1 double mutant (41). However, we measured HO mRNA and found that while the gcn5 mutation does reduce HO expression (Fig. 1A, lane 2), this reduction is not reversed in the gcn5 sin1 mutant (Fig. 1A, lane 4). We attribute this difference in results in the gcn5 sin1 double mutant to the use of an HO:lacZ reporter rather than native HO. (The differences between regulation of the native HO gene and the HO:lacZ reporter are considered in Discussion.) As GCN5 encodes a histone acetyltransferase, it seemed possible that a mutation in a histone deacetylase would suppress the gcn5 defect in HO expression. HO is expressed in a gcn5 rpd3 double mutant (Fig. 1B, lane 4), consistent with results with an HO:lacZ reporter (41). In contrast, a mutation in a different histone deacetylase, HDA1, does not suppress the gcn5 mutation (Fig. 1B, lane 6). The S1 protection assay in Fig. 1C shows that HO is not expressed in a swi5 sin1 or swi2 sin1 double mutant. Thus, a sin1 mutation does not suppress defects in HO transcription caused by mutations in GCN5, SWI5, or SWI2. These results suggest that SIN1/SPT2 is not a true negative regulator of native HO expression.
FIG. 1.
HO expression is not altered by a sin1 mutation. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control). HO RNA levels were quantitated by phosphorimager, normalized by dividing by the value for CMD1, and expressed as a percentage of the wild-type (WT) value in lane 1 in each panel. RNAs were prepared from strains DY2395, DY5116, DY5323, and DY5326 (A), DY2389, DY5199, DY4548, DY5170, DY5068, and DY5168 (B), and DY150, DY5323, DY161, DY5410, DY2348, and DY5420 (C).
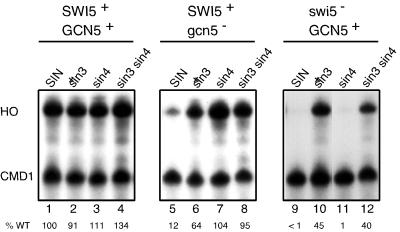
We next evaluated the role of the NHP6A and NHP6B genes, which encode HMG proteins, in activation of HO. The two genes encode nearly identical proteins, and a temperature-sensitive phenotype is seen in the nhp6a nhp6b double mutant but not in either single mutant (13). Expression of a number of lacZ reporters is reduced in a nhp6a nhp6b mutant strain (39). We find that expression of HO is reduced nearly 20-fold in the nhp6a nhp6b double mutant (Fig. 2, lane 2). Sidorova and Breeden (47) recently showed that NHP6A acts as a multicopy suppressor allowing expression of an HO:lacZ reporter at the nonpermissive temperature in a swi6 temperature-sensitive mutant. The YEp-NHP6A plasmid does not suppress a swi6 null mutation, however. They also observed reduced HO expression in a nhp6a nhp6b double mutant; however, the strains were not isogenic and only a modest reduction in HO expression was seen in this study (47).
FIG. 2.
The nhp6 defect in HO transcription can be suppressed by sin3 or sin4 mutations. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control). HO RNA levels were quantitated by phosphorimager, normalized by dividing by the value for CMD1, and expressed as a percentage of the wild-type (WT) value in lane 1. RNAs were prepared from strains DY150, DY2381, DY3658, DY5153, DY984, DY5157, DY1699, and DY5155.
Mutations in sin3 and sin4 suppress nhp6 and gcn5 transcription defects.
Isogenic yeast strains were constructed to test the ability of mutations in regulatory genes to suppress the nhp6 defect in HO transcription. A sin1 mutation does not suppress the defect in HO expression due to the absence of the Nhp6 protein (Fig. 2, lane 4). However, mutations in the SIN3 or SIN4 genes do permit HO expression in the nhp6a nhp6b mutant (Fig. 2, lanes 6 and 8).
As sin3 and sin4 mutations were effective in suppressing defects in nhp6 mutants, we sought to determine whether sin3 or sin4 could suppress defects in other activators of HO expression, such as GCN5 and SWI5. A sin3 mutation suppresses the defects in HO expression caused by a gcn5 mutation (Fig. 3, lane 6) or a swi5 mutation (Fig. 3, lane 10) to 64 or 45%, respectively, of the wild-type level. Similar levels of suppression are seen in an rpd3 mutant (data not shown). This last result is not surprising as mutations in SIN3 and RPD3 cause similar phenotypes (53) and the Sin3 protein physically interacts with the Rpd3 histone deacetylase (26, 27). A sin4 mutation shows striking differences in the ability to suppress gcn5 or swi5 mutations for expression of HO. HO is not expressed in a swi5 sin4 strain (Fig. 3, lane 11), despite the fact a sin4 mutation does suppress the swi5 defect when an HO:lacZ reporter is used (24) (see Discussion). In contrast, HO is expressed in a gcn5 sin4 mutant at 104% of wild-type levels (Fig. 3, lane 7), and thus sin4 is an effective gcn5 suppressor. The combination of the sin3 and sin4 mutations is able to suppress either a gcn5 mutation (Fig. 3, lane 8) or a swi5 mutation (Fig. 3, lane 12). In summary, a sin3 mutation is able to suppress both swi5 and gcn5 defects in HO expression, but a sin4 mutation can suppress only gcn5. Thus, sin3 and sin4 suppress transcriptional defects by different mechanisms.
FIG. 3.
Both sin3 and sin4 mutations suppress the gcn5 defect. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control). HO RNA levels were quantitated by phosphorimager, normalized by dividing by the value for CMD1, and expressed as a percentage of the wild-type (WT) value in lane 1. RNAs were prepared from strains DY150, DY5285, DY2763, DY5294, DY5265, DY5297, DY5289, DY5315, DY411, DY775, DY2133, and DY5299.
Analysis of suppression of swi6 and swi2 transcription defects by sin3 and sin4.
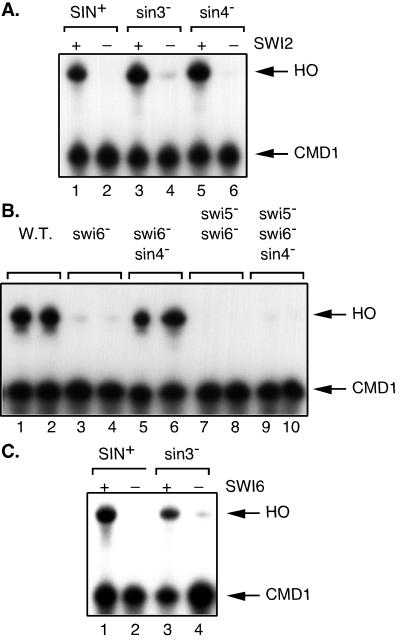
Based on the difference in suppression of a swi5 mutation by sin3 and sin4, we decided to examine suppression of mutations affecting other types of HO transcriptional activators. SWI2 encodes part of the Swi/Snf chromatin remodeling complex, and HO is not expressed in a swi2 mutant. We first examined HO expression in swi2 sin3 and swi2 sin4 mutants to look for suppression of the swi2 transcriptional defect. The results in Fig. 4A show that neither sin3 nor sin4 can suppress the reduced HO expression caused by the swi2 mutation. Thus, the requirement for the Swi/Snf chromatin remodeling complex cannot be suppressed by mutations in either SIN3 or SIN4.
FIG. 4.
A sin4 mutation suppresses swi6 but not swi2. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control). (A) HO is not expressed in swi2 sin3 or swi2 sin4 strains. RNAs were prepared from strains DY150, DY3944, DY773, DY2870, DY1702, and DY2499. (B) HO is expressed in swi6 sin4 strains. RNAs were prepared from strains DY150, DY151, DY5780, DY5781, DY5907, DY5908, DY5909, DY5910, DY5911, and DY5912. (C) HO is not expressed in a swi6 sin3 strains. RNAs were prepared from strains DY150, DY5780, DY773, and DY6103.
The SBF DNA binding factor binds to the HO promoter only after Swi/Snf and SAGA are recruited to the HO promoter (12). The SWI6 gene encodes a subunit of SBF, and HO is not expressed in a swi6 mutant (Fig. 4B, lanes 1 to 4). However, in the swi6 sin4 double mutant, HO is expressed at nearly wild-type levels (Fig. 4B, lanes 5 and 6). Thus, HO can be expressed in a sin4 mutant in the absence of SBF. Importantly, HO is not expressed in the swi5 swi6 sin4 mutant (Fig. 4B, lanes 9 and 10). This suggests that Swi5, or a factor recruited in a SWI5-dependent manner such as Swi/Snf or SAGA, is still required for HO expression in the sin4 mutant. Finally, HO is not expressed in a swi6 sin3 double mutant, and thus a sin3 mutation does not permit HO transcription without SBF (Fig. 4C). Thus there is a striking difference in the ability of sin3 and sin4 mutations to suppress activator mutations. A sin4 mutation allows HO to be expressed in the absence of the SBF factor, while a sin3 mutation does not suppress. The pattern of suppression of a swi5 mutation (Fig. 3) is just the opposite, with a sin3 mutation suppressing but not sin4.
Suppression of gcn5 by Nhp6b overexpression.
We determined whether overexpression of Nhp6b could suppress HO transcriptional defects caused by mutations in transcriptional activators. A YEp multicopy plasmid with the NHP6B gene was transformed into various strains. An S1 nuclease protection assay shows that overexpression of Nhp6b does not suppress swi5, swi2, or swi6 null mutations (Fig. 5). However, Nhp6b overexpression partially suppresses the reduced HO expression caused by a gcn5 mutation (lanes 7 and 8). In the gcn5 mutant, HO is expressed at 6% of the wild-type level, and YEp-NHP6B causes a threefold increase in HO expression.
FIG. 5.
Nhp6b overexpression partially suppresses the gcn5 defect. Strains DY150 (wild type [w.t.]), DY161 (swi5), DY2348 (swi2), DY5116 (gcn5), and DY5780 (swi6) were transformed with either the YEplac195 vector or M1195, a YEp-NHP6B plasmid. S1 nuclease protection assays were performed using probes specific for HO and CMD1 (internal control), using RNA prepared from strains grown under selective conditions to maintain the plasmid. The upper panel was exposed to film for 8 h; the lower panel was exposed for 24 h.
Genetic interactions based on growth phenotypes.
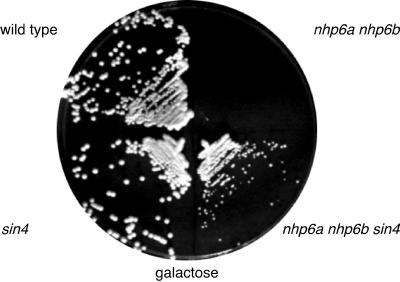
As the sin3 and sin4 mutations suppress the defect in HO transcription caused by the lack of the Nhp6 protein, we investigated whether these mutations would also suppress other nhp6a nhp6b phenotypes. The nhp6a nhp6b double mutant displays a number of phenotypes, including temperature-sensitive growth (13) and inability to grow on galactose medium (Fig. 6). Interestingly, we observed this galactose growth defect for nhp6a nhp6b double mutants only in the S288C background, not in W303 strains. The sin3 mutation was unable to suppress any of the nhp6 defects; in fact, the nhp6a nhp6b sin3 triple mutant grows much more slowly than either the nhp6a nhp6b or sin3 mutant strains. We were unable to demonstrate suppression of the 37°C growth defect, as the sin4 single mutant is also temperature sensitive for growth (24). However, a sin4 mutation is able to suppress one of the nhp6 phenotypes. The nhp6a nhp6b sin4 triple mutant can grow on galactose, whereas the nhp6a nhp6b double mutant cannot (Fig. 6). This suggests that the suppression of nhp6 by sin4 may be more general and not limited to HO transcription.
FIG. 6.
A sin4 mutation suppresses the nhp6 growth defect on galactose. Strains DY881, DY1712, DY2532, and DY2533 were plated on YEP-galactose medium and grown for 4 days at 30°C.
Combining two mutations can sometimes cause a severe additive phenotype, suggesting that these two genes affect the same function but from different pathways (19). For example, Roberts and Winston (44) found that combining a gcn5 mutation with a mutation in the Swi/Snf chromatin remodeling complex causes a severe growth defect. They suggested that either the SAGA histone acetyltransferase complex (which contains Gcn5) or the Swi/Snf chromatin remodeling factor can supply certain critical functions for gene activation, but that the absence of both activities is manifested as the growth defect. We constructed gcn5 nhp6a nhp6b triple mutant strains and found that these strains grew extremely poorly (Fig. 7A). To test whether a sin4 mutation could suppress this growth defect, we crossed a gcn5 nhp6a strain to a nhp6a nhp6b sin4 strain and examined the phenotype of haploid progeny. The experiment in Fig. 7B show that the gcn5 nhp6a nhp6b sin4 quadruple mutant strain grows much better than the gcn5 nhp6a nhp6b triple mutant. Thus, the effect of a sin4 mutation is not limited to allowing HO expression in gcn5 or nhp6 mutants. SIN4 has global effects on transcription, as a sin4 mutation overcomes the marked growth defects in a gcn5 nhp6a nhp6b strain.
FIG. 7.
The severe growth defect of a gcn5 nhp6a nhp6b triple mutant is suppressed by a sin4 mutation. (A) Strains DY150, DY2378, DY2380, DY2382, DY5116, and DY5306 were plated on YEPD medium and grown for 3 days at 30°C. (B) Strains DY5825 and DY5820 were plated on YEPD medium and grown for 5 days at 30°C.
DISCUSSION
The promoter of the yeast HO gene is large and complex, and genetic analysis has shown that chromatin structure plays an important role in transcriptional regulation of this gene. Through studies of HO regulation, we have identified common features between the NHP6A and NHP6B genes, which encode architectural transcription factors, and GCN5, which encodes a hitone acetyltransferase subunit of the SAGA complex. HO expression is reduced in either an nhp6a nhp6b double mutant or a gcn5 mutant. Moreover, these mutants show similar suppression patterns, with either a sin3 or a sin4 mutation restoring HO expression despite mutations in transcriptional activators. We found that a nhp6a nhp6b gcn5 triple mutant grows extremely slowly. One interpretation of this result is that Nhp6 and Gcn5 may provide two distinct mechanisms for transcriptional activation of certain important genes. Disruption of the SIN4 gene suppresses this defect, and thus the nhp6a nhp6b gcn5 sin4 quadruple mutant grows reasonably well. A sin4 mutation also suppresses galactose growth defects of a nhp6a nhp6b mutant.
How do sin3 and sin4 mutations suppress transcriptional defects caused by the absence of Gcn5 or Nhp6? To investigate this further, we determined whether sin3 or sin4 can suppress other mutations in other activators required for HO expression (Table 2). Cosma et al. (12) used chromatin immunoprecipitation to examine transcription factor binding to the HO promoter, and they showed that factors bind sequentially. Their model is that Swi5 enters the nucleus in late anaphase, binds to the promoter, and recruits Swi/Snf. Swi/Snf, in turn, recruits SAGA, and SAGA is required for SBF binding. It is suggested that SBF is responsible for recruiting general transcription factors to the promoter (12). (The term “recruit” means brings to the promoter and does not necessarily imply a direct physical interaction.) Krebs et al. (30) showed that histone acetylation of a 1-kb region of the HO promoter occurs in late G1 phase, and this histone acetylation is dependent on Swi5, Swi/Snf, and the Gcn5 component of SAGA. Importantly, inactivation of the Sin3/Rpd3 histone deacetylase complex causes the promoter to be constitutively acetylated. In light of these findings, we explain our results on suppression by sin3 mutations by suggesting that SBF binds poorly to HO when it is deacetylated, and that either the sin3 mutation or activity of the Gcn5 histone acetyltransferase results in histone acetylation that permits SBF binding. This model explains why a sin3 mutation is able to suppress swi5 and gcn5 mutations (Table 2). The failure of a sin3 mutation to suppress the swi6 defect in HO transcription is also consistent with this model. Why then is HO not expressed in a swi2 sin3 double mutant? We suggest that Swi/Snf has multiple roles in activation of HO expression, with only one being to recruit SAGA. By this model, the second role of Swi/Snf, revealed in the swi5 sin3 mutant, is to assist the TATA-binding protein (TBP), or possibly SBF, to bind the HO promoter. We suggest that Swi/Snf need not be stably bound to the HO promoter to assist TBP to bind. Thus, Swi/Snf is still required for HO activation in a swi5 mutant although it may not be stably bound to the promoter.
TABLE 2.
Suppression of mutations in activators of HO transcription by sin3 and sin4
| Genotype | Function | SIN+ | sin3 (deacetylase) | sin4 (mediator, SAGA) |
|---|---|---|---|---|
| SWI+ | + | + | + | |
| swi5 | DNA-binding factor | − | + | − |
| swi2 | Swi/Snf complex | − | − | − |
| gcn5 | SAGA | − | + | + |
| nhp6ab | Architectural transcription factor | − | + | + |
| swi6 | SBF DNA-binding factor | − | − | + |
| swi5 swi6 | − | ND | − |
ND, not determined.
Suppression of HO activation defects by a sin4 mutation is quite different (Table 2). The Sin4 protein is a component of the RNA polymerase II mediator complex (32), and thus it is possible that the sin4 mutation relaxes the RNA polymerase holoenzyme's specificity, allowing it to activate in the absence of certain factors such as SBF. RNA polymerase binding and transcriptional initiation at HO normally require both SBF and Swi/Snf, and a sin4 mutation could allow polymerase to start in the absence of SBF. According to this scenario, the mediator part of RNA polymerase functions as an “activator checkpoint,” verifying that an activator is at the promoter before RNA polymerase can initiate transcription.
An alternative model for Sin4 function is possible based upon the recent observation that Sin4 is also present in the SAGA complex (Grant and Workman, personal communication). Genetic analysis clearly shows that the SAGA complex has additional roles besides the Gcn5 histone acetyltransferase complex; one of these functions, mediated by the Spt3 and Spt8 proteins, may be to inhibit DNA binding by TBP (2, 52). The model most consistent with the data suggests that in a sin4 mutant the activity of SAGA is altered, with the sin4-mutant SAGA not inhibiting, and thus stimulating, TBP binding. In the wild type, TBP binding requires SBF and Swi/Snf; in the sin4 mutant, TBP binding occurs in the absence of SBF. This model fits the data nicely as HO can be activated in the absence of SBF in a sin4 mutant. Similarly, a sin4 mutation allows HO to be expressed in the absence of Gcn5, normally required for SBF binding. HO is not expressed in a swi2 sin4 double mutant because Swi/Snf is still required, probably to promote binding by TBP.
We first examined the role of Nhp6 in HO expression based on the hypothesis that architectural transcription factors might be involved in bridging the two Swi5 molecules bound at distant sites (34). While we have shown that the Nhp6 protein is required for HO activation, at present we have no evidence that it mediates this long-range interaction in vivo. Instead, our data suggest that Nhp6 functions with the Gcn5 histone acetyltransferase. The nhp6a nhp6b mutant shows the same suppression pattern as the gcn5 strain (Table 2), and thus Nhp6 may work through SAGA. The Nhp6 protein could assist in the recruitment of SAGA to the HO promoter, possibly by stabilizing binding by SAGA. Alternatively, Nhp6 could act downstream of SAGA, by establishing a chromatin structure that facilitates activities of SAGA, or by assisting in DNA binding by SBF. Overexpression of Nhp6 allows HO expression in the absence of Gcn5 (Fig. 5) and suppresses the reduced HO:lacZ expression caused by mutations within the ankyrin repeat region of Swi6 (47). Increased levels of Nhp6 do not suppress swi6 null mutations, however. In contrast, the fact that the nhp6a nhp6b gcn5 mutant grows very slowly suggests that Nhp6 and Gcn5 have independent functions. How does a sin4 mutation suppress the growth defect in the nhp6a nhp6b gcn5 mutant? Further work will be needed to determine whether the absence of Sin4 from the holoenzyme or from SAGA is responsible for suppression of this growth defect. Finally, while we believe that the Nhp6 and Sin4 have direct effects on HO expression, it remains possible that there are indirect effects caused by these mutations altering expression of other genes.
Differences between native HO and an HO:lacZ reporter.
The sin3 and sin4 mutations were identified as suppressor mutations that allow an HO:lacZ reporter to be expressed in the absence of the SWI5 transcriptional activator. As shown in Table 3, a swi5 mutation reduces expression of the HO:lacZ reporter 100-fold. A mutation in either SIN3 or SIN4 restores expression of HO:lacZ, although to different extents. We have found that the regulation of the HO:lacZ reporter can be strikingly different from that of the native HO gene. This HO:lacZ reporter is integrated at the HO locus on chromosome IV, with the entire flanking HO promoter sequences present. HO is expressed in a swi5 sin3 double mutant strain, and thus SIN3 is a bona fide swi5 suppressor. However, a sin4 mutation does not overcome the defect in HO expression due to the mutation in the SWI5 transcription factor. This inability to allow HO expression in a swi5 mutant was described before for the sdi3-1 allele of sin4 (38).
TABLE 3.
Differences in expression of an integrated HO:lacZ reporter and the native HO gene
| Genotype | % Expression
|
|
|---|---|---|
| HO:lacZ reporter | Native HO gene | |
| SWI5 | 100 | 100 |
| swi5 | 1 | 1 |
| swi5 sin3 | 30 | 45 |
| swi5 sin4 | 150 | 1 |
Are there other differences between HO:lacZ and HO in terms of regulatory properties? Although sin1 mutations do suppress the defect in HO:lacZ expression due to the absence of the Swi5 or Swi2 transcriptional activators (data not shown) (31, 41, 43), the same result is not observed when native HO mRNA is measured. The difference between the effects of a sin1 mutation on HO versus HO:lacZ regulation may reflect unique properties of HO, as a sin1 mutation has marked effects on regulation of SUC2, INO1 and SSA3 (1, 42, 43).
A pho2 mutation reduces expression of an HO:lacZ reporter (6) but has no effect on expression of the native HO gene (34). Zhu et al. (60) reported that an hpr1 mutation reduced expression of an HO:lacZ reporter. However, we have determined that expression of the native HO gene is not affected by an hpr1 mutation (data not shown). Chávez and Aguilera (10) have shown that an hpr1 mutation has different effects on lacZ reporters and native genes, and that these effects are transcriptional and not translational.
It is possible that there are sequences present within the bacterial lacZ gene that act in cis to affect regulation of the HO promoter, and that these effects become apparent in a sin4 mutant. Supporting this idea of cis effects from within lacZ, W. Hörz (personal communication) has shown that a sin4 mutation affects expression of a PHO5-lacZ reporter, but a sin4 mutation does not cause derepression of the native PHO5 gene. Additionally, the fact that a sin4 mutation derepresses PHO5 transplaced into the URA3 locus, but not the native PHO5 locus, suggests that effects of a sin4 mutation can be influenced by the chromosomal context (21). Finally, the concept of cis-acting effects of lacZ sequences affecting transcriptional regulation is supported by the work of Chávez and Aguilera (10) showing that an hpr1 mutation affects native genes and lacZ reporters differently.
ACKNOWLEDGMENTS
We thank Leena Bhoite, Brad Cairns, Tim Formosa, and Warren Voth for helpful discussions and comments on the manuscript, Michael Grunstein, Ira Herskowitz, Hannah Klein, David Kolodrubetz, and Sharon Roth for providing strains and plasmids, and Patrick Grant, Wolfram Hörz, and Jerry Workman for communicating unpublished results.
P.E. was supported by a fellowship from the Swedish Foundation for International Cooperation in Research and Higher Education. This work was supported by grants from the NIH awarded to D.J.S.
REFERENCES
- 1.Baxter B K, Craig E A. Suppression of an Hsp70 mutant phenotype in Saccharomyces cerevisiae through loss of function of the chromatin component Sin1p/Spt2p. J Bacteriol. 1998;180:6484–6492. doi: 10.1128/jb.180.24.6484-6492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belotserkovskaya R, Sterner D E, Deng M, Sayre M H, Lieberman P M, Berger S L. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhoite L T, Stillman D J. Residues in the Swi5 zinc finger protein that mediate cooperative DNA binding with the Pho2 homeodomain protein. Mol Cell Biol. 1998;18:6436–6446. doi: 10.1128/mcb.18.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi M E, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 5.Björklund S, Almouzni G, Davidson I, Nightingale K P, Weiss K. Global transcription regulators of eukaryotes. Cell. 1999;96:759–767. doi: 10.1016/s0092-8674(00)80586-3. [DOI] [PubMed] [Google Scholar]
- 6.Brazas R M, Stillman D J. The Swi5 zinc finger and the Grf10 homeodomain proteins bind DNA cooperatively at the yeast HO promoter. Proc Natl Acad Sci USA. 1993;90:11237–11241. doi: 10.1073/pnas.90.23.11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 8.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 9.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Chávez S, Aguilera A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, West R W J, Johnston S L, Gans H, Ma J. TSF3, a global regulatory protein that silences transcription of yeast GAL genes, also mediates repression by α2 repressor and is identical to SIN4. Mol Cell Biol. 1993;13:831–840. doi: 10.1128/mcb.13.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 13.Costigan C, Kolodrubetz D, Snyder M. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol Cell Biol. 1994;14:2391–2403. doi: 10.1128/mcb.14.4.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covitz P A, Song W, Mitchell A P. Requirement for RGR1 and SIN4 in RME1-dependent repression in Saccharomyces cerevisiae. Genetics. 1994;138:577–586. doi: 10.1093/genetics/138.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deckert J, Rodriguez Torres A M, Simon J T, Zitomer R S. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6109–6117. doi: 10.1128/mcb.15.11.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friesen H, Tanny J C, Segall J. Spe3, which encodes spermidine synthase, is required for full repression through NRE(DIT) in Saccharomyces cerevisiae. Genetics. 1998;150:59–73. doi: 10.1093/genetics/150.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 18.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 19.Guarente L. Synthetic enhancement in gene interaction: a tool come of age. Trends Genet. 1993;9:362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- 20.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harashima S, Mizuno T, Mabuchi H, Yoshimitsu S, Ramesh R, Hasebe M, Tanaka A, Oshima Y. Mutations causing high basal level transcription that is independent of transcriptional activators but dependent on chromosomal position in Saccharomyces cerevisiae. Mol Gen Genet. 1995;247:716–725. doi: 10.1007/BF00290403. [DOI] [PubMed] [Google Scholar]
- 22.Herskowitz I, Andrews B, Kruger W, Ogas J, Sil A, Coburn C, Peterson C. Integration of multiple regulatory inputs in the control of HO expression in yeast. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 949–974. [Google Scholar]
- 23.Jiang Y W, Dohrmann P R, Stillman D J. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y W, Stillman D J. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 27.Kasten M M, Dorland S, Stillman D J. A large protein complex containing the Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997;16:4215–4221. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 29.Kolodrubetz D, Burgum A. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J Biol Chem. 1990;265:3234–3239. [PubMed] [Google Scholar]
- 30.Krebs J E, Kuo M H, Allis C D, Peterson C L. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruger W, Herskowitz I. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol Cell Biol. 1991;11:4135–4156. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Bjorklund S, Jiang Y W, Kim Y J, Lane W S, Stillman D J, Kornberg R D. Yeast global transcriptional repressors Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macatee T, Jiang Y W, Stillman D J, Roth S Y. Global alterations in chromatin accessibility associated with loss of SIN4 function. Nucleic Acids Res. 1997;25:1240–1248. doi: 10.1093/nar/25.6.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McBride H J, Brazas R M, Yu Y, Nasmyth K, Stillman D J. Long range interactions at the HO promoter. Mol Cell Biol. 1997;17:2669–2678. doi: 10.1128/mcb.17.5.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasmyth K. Molecular analysis of a cell lineage. Nature. 1983;302:670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- 37.Nasmyth K. Regulating the HO endonuclease in yeast. Curr Opin Genet Dev. 1993;3:286–294. doi: 10.1016/0959-437x(93)90036-o. [DOI] [PubMed] [Google Scholar]
- 38.Nasmyth K, Stillman D, Kipling D. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell. 1987;48:579–587. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- 39.Paull T T, Carey M, Johnson R C. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 40.Paull T T, Johnson R C. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J Biol Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 41.Pérez-Martín J, Johnson A D. Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1049–1054. doi: 10.1128/mcb.18.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peterson C L, Kruger W, Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991;64:1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- 43.Pollard K J, Peterson C L. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–302. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 46.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:1–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 47.Sidorova J, Breeden L. The MSN1 and NHP6A genes suppress SWI6 defects in Saccharomyces cerevisiae. Genetics. 1999;151:45–55. doi: 10.1093/genetics/151.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simchen G, Winston F, Styles C A, Fink G R. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc Natl Acad Sci USA. 1984;81:2431–2434. doi: 10.1073/pnas.81.8.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song W, Treich I, Qian N, Kuchin S, Carlson M. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol Cell Biol. 1996;16:115–120. doi: 10.1128/mcb.16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sternberg P W, Stern M J, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 52.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stillman D J, Dorland S, Yu Y. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SWI5 transcriptional activator. Genetics. 1994;136:781–788. doi: 10.1093/genetics/136.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 55.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 56.Wahi M, Johnson A D. Identification of genes required for α2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H, Clark I, Nicholson P R, Herskowitz I, Stillman D J. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol Cell Biol. 1990;10:5927–5936. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Werner M H, Burley S K. Architectural transcription factors: proteins that remodel DNA. Cell. 1997;88:733–736. doi: 10.1016/s0092-8674(00)81917-0. [DOI] [PubMed] [Google Scholar]
- 59.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 60.Zhu Y, Peterson C L, Christman M F. HPR1 encodes a global positive regulator of transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1698–1708. doi: 10.1128/mcb.15.3.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]