Abstract
Gnomoniopsis (Gnomoniaceae, Diaporthales) is a well-classified genus inhabiting leaves, branches and fruits of the hosts in three plant families, namely Fagaceae, Onagraceae and Rosaceae. In the present study, eighteen Gnomoniopsis isolates were obtained from diseased leaves of Fagaceae hosts collected from Fujian, Guangdong, Hainan, Henan, Jiangxi and Shaanxi provinces in China. Morphology from the cultures and phylogeny based on the 5.8S nuclear ribosomal DNA gene with the two flanking internally transcribed spacer (ITS) regions, the translation elongation factor 1-alpha (tef1) and the beta-tubulin (tub2) genes were employed to identify these isolates. As a result, seven species were revealed, viz. Gnomoniopsis castanopsidis, G. fagacearum, G. guangdongensis, G. hainanensis, G. rossmaniae and G. silvicola spp. nov, as well as a known species G. daii. In addition, G. daii was firstly reported on the host Quercus aliena.
Keywords: Ascomycota, leaf disease, new species, oak, taxonomy
1. Introduction
Diaporthales is a species-rich fungal order usually associated with forest trees as endophytes, pathogens and saprophytes [1,2,3,4,5,6,7,8,9,10]. Amongst the numerous tree pathogens, the most notorious one is Cryphonectria parasitica (Cryphonectriaceae) causing chestnut (Castanea spp.) blight worldwide [11,12,13]. An example for endophytic lifestyle is Diaporthe biconispora (Diaporthaceae) and an additional six Diaporthe species that are endophytic in healthy Citrus tissues in China [14]. As an example of a saprophyte, Apiosporopsis carpinea (Apiosporopsidaceae) occurs on over-wintered leaves of Carpinus betulus [15].
Gnomoniaceae is a large family of the Diaporthales, with currently 38 accepted genera, including Gnomoniopsis [16,17,18,19]. Gnomoniopsis, based on the type species G. chamaemori, is a well-studied genus in regard to morphology, phylogeny and host associations. This genus is characterized by having small, black perithecia immersed in the host tissue and one-septate, oval to fusiform ascospores, and is well-distinguished by phylogenies based on the 5.8S nuclear ribosomal DNA gene with the two flanking internally transcribed spacer (ITS) regions, the translation elongation factor 1-alpha (tef1) and the beta-tubulin (tub2) genes [20,21]. Species of Gnomoniopsis are currently known to inhabit only members of three plant families as hosts, viz. Fagaceae, Onagraceae and Rosaceae [20,21,22,23,24].
Until now, thirty species epithets of Gnomoniopsis have been recorded in Index Fungorum, six of them were reported from fagaceous trees [22]. Two species, Gnomoniopsis clavulata and G. paraclavulata, were firstly discovered on overwintered leaves of Quercus trees in the USA [20,21]. Subsequently, Gnomoniopsis smithogilvyi with its synonym G. castaneae were proposed from rotten fruits of Castanea in Australia and Europe by two independent studies [25,26]. However, these two names were proven to be a single species based on phylogeny and morphological characters [27]. Hence, G. castaneae becomes a synonym of G. smithogilvyi based on priority. In China, G. daii was described from rotten fruits and diseased leaves of Castanea mollissima [23,28]. Meanwhile, a different species named G. chinensis was reported to cause Chinese chestnut branch canker [29]. Later, Yang et al. described G. xunwuensis from leaf spots of Castanopsis fissa in China [24]. Since three Fagaceae-inhabiting species from China are now only known in the asexual morph, it is hard to separate them based on only morphological characters [23,24,29]. Hence, it is necessary to conduct phylogenetic analyses in order to recognize and identify the species [29].
Fagaceae is a common plant family widely distributed in the northern hemisphere, with seven genera namely Castanea, Castanopsis, Cyclobalanopsis, Fagus, Lithocarpus, Quercus and Trigonobalanus [30]. Previously, Gnomoniopsis has been reported from Castanea, Castanopsis and Quercus species [22]. The aims of present study are to investigate fagaceous hosts to collect Gnomoniopsis samples in China, and to identify them to species level based on combined morphology and phylogeny of ITS, tef1 and tub2 loci.
2. Materials and Methods
2.1. Field Sampling and Isolation
In the present study, we investigated leaf diseases of fagaceous trees in Fujian, Guangdong, Hainan, Henan, Jiangxi and Shaanxi provinces of China during 2018 and 2020. The diseased leaf samples were packed in paper bags and transferred to the laboratory for isolation. The infected leaves were firstly surface-sterilized for 1 min in 75% ethanol, 3 min in 1.25% sodium hypochlorite, and 1 min in 75% ethanol, then rinsed for 2 min in distilled water and blotted on dry sterile filter paper. Then samples were cut into 0.5 × 0.5 cm pieces using a double-edge blade, and transferred onto the surface of potato dextrose agar (PDA; 200 g potatoes, 20 g dextrose, 20 g agar per L) and malt extract agar (MEA; 30 g malt extract, 5 g mycological peptone, 15 g agar per L), and incubated at 25 °C to obtain the pure culture. The cultures were deposited in China Forestry Culture Collection Center (CFCC), and the specimens in the herbarium of the Chinese Academy of Forestry (CAF).
2.2. DNA Extraction, Sequencing and Phylogenetic Analyses
Genomic DNA was extracted from mycelia grown on cellophane-covered PDA using a cetyltrimethylammonium bromide (CTAB) method [31]. DNA was checked by electrophoresis in 1% agarose gel, and the quality and quantity were measured using a NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA). Three partial loci, ITS region, tef1 and tub2 genes were amplified by the following primer pairs: ITS1 and ITS4 for ITS [32], EF1-688F and EF2 for tef1 [33], and T1/Bt2a and Bt2b for tub2 [34,35]. The polymerase chain reaction (PCR) conditions were as follows: an initial denaturation step of 5 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 50 s at 48 °C (ITS) or 54 °C (tub2) or 55 °C (tef1), and 1 min at 72 °C, and a final elongation step of 10 min at 72 °C. PCR products were assayed via electrophoresis in 2% agarose gels. DNA sequencing was performed using an ABI PRISM 3730XL DNA Analyser with a BigDye Terminator Kit v.3.1 (Invitrogen, Waltham, MA, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
The sequences obtained in the present study were assembled using SeqMan v.7.1.0, and reference sequences were retrieved from the National Center for Biotechnology Information (NCBI), based on recent publications on the genus Gnomoniopsis [20,21,22,23,24,29]. Sequences of an accession of Apiognomonia errabunda (AR 2813) were added to represent the outgroup. The sequences were aligned using MAFFT v.6 and corrected manually using MEGA 7.0.21 [36].
The phylogenetic analyses of the ITS region and of a combined matrix of the three loci (ITS-tef1-tub2) were performed using Maximum Likelihood (ML) and Bayesian Inference (BI) methods. ML was implemented on the CIPRES Science Gateway portal (https://www.phylo.org) using RAxML-HPC BlackBox 8.2.10 [37,38], employing a GTRGAMMA substitution model with 1000 bootstrap replicates. Bayesian inference was performed using a Markov Chain Monte Carlo (MCMC) algorithm in MrBayes v. 3.0 [39]. Two MCMC chains, starting from random trees for 1,000,000 generations and trees, were sampled every 100th generation, resulting in a total of 10,000 trees. The first 25% of trees were discarded as burn-in of each analysis. Branches with significant Bayesian Posterior Probabilities (BPP > 0.9) were estimated in the remaining 7500 trees. Phylogenetic trees were viewed with FigTree v.1.3.1 and processed by Adobe Illustrator CS5. The nucleotide sequence data of the new taxa were deposited in GenBank, and the GenBank accession numbers of all accessions included in the phylogenetic analyses are listed in Table 1.
Table 1.
Strains and GenBank accession numbers used in this study.
| Species | Country | Host | Host Family | Strain | GenBank Accession Number | ||
|---|---|---|---|---|---|---|---|
| ITS | tef1 | tub2 | |||||
| Apiognomonia errabunda | Switzerland | Fagus sylvatica | Fagaceae | AR 2813 | DQ313525 | DQ313565 | DQ862014 |
| Gnomoniopsis alderdunensis | USA | Rubus pedatus | Rosaeace | CBS 125679 | GU320826 | GU320813 | GU320788 |
| Gnomoniopsis alderdunensis | USA | Rubus parviflorus | Rosaeace | CBS 125680 * | GU320825 | GU320801 | GU320787 |
| Gnomoniopsis alderdunensis | USA | Rubus parviflorus | Rosaeace | CBS 125681 | GU320827 | GU320802 | GU320789 |
| Gnomoniopsis chamaemori | Finland | Rubus chamaemorus | Rosaeace | CBS 804.79 | GU320817 | GU320809 | GU320777 |
| Gnomoniopsis chinensis | China | Castanea mollissima | Fagaceae | CFCC 52286 * | MG866032 | MH545370 | MH545366 |
| Gnomoniopsis chinensis | China | Castanea mollissima | Fagaceae | CFCC 52287 | MG866033 | MH545371 | MH545367 |
| Gnomoniopsis chinensis | China | Castanea mollissima | Fagaceae | CFCC 52288 | MG866034 | MH545372 | MH545368 |
| Gnomoniopsis chinensis | China | Castanea mollissima | Fagaceae | CFCC 52289 | MG866035 | MH545373 | MH545369 |
| Gnomoniopsis clavulata | USA | Quercus falcata | Fagaceae | CBS 121255 | EU254818 | GU320807 | EU219211 |
| Gnomoniopsis castanopsidis | China | Castanopsis hystrix | Fagaceae | CFCC 54437 * | MZ902909 | MZ936385 | NA |
| Gnomoniopsis castanopsidis | China | Castanopsis hystrix | Fagaceae | CFCC 55878 | MZ902910 | MZ936386 | NA |
| Gnomoniopsis comari | Finland | Comarum palustre | Rosaeace | CBS 806.79 | EU254821 | GU320810 | EU219156 |
| Gnomoniopsis comari | Finland | Comarum palustre | Rosaeace | CBS 807.79 | EU254822 | GU320814 | GU320779 |
| Gnomoniopsis comari | Switzerland | Comarum palustre | Rosaeace | CBS 809.79 | EU254823 | GU320794 | GU320778 |
| Gnomoniopsis daii | China | Castanea mollissima | Fagaceae | CFCC 54043 * | MN598671 | MN605517 | MN605519 |
| Gnomoniopsis daii | China | Castanea mollissima | Fagaceae | CMF002B | MN598672 | MN605518 | MN605520 |
| Gnomoniopsis daii | China | Quercus aliena | Fagaceae | CFCC 55517 | MZ902911 | MZ936387 | MZ936403 |
| Gnomoniopsis daii | China | Quercus aliena | Fagaceae | CFCC 55294B | MZ902912 | MZ936388 | MZ936404 |
| Gnomoniopsis fagacearum | China | Castanopsis faberi | Fagaceae | CFCC 54288 | MZ902913 | MZ936389 | MZ936405 |
| Gnomoniopsis fagacearum | China | Quercus variabilis | Fagaceae | CFCC 54439 | MZ902914 | MZ936390 | MZ936406 |
| Gnomoniopsis fagacearum | China | Castanopsis eyrei | Fagaceae | CFCC 54414 | MZ902915 | MZ936391 | MZ936407 |
| Gnomoniopsis fagacearum | China | Lithocarpus glaber | Fagaceae | CFCC 54316 * | MZ902916 | MZ936392 | MZ936408 |
| Gnomoniopsis fagacearum | China | Castanopsis chunii | Fagaceae | CFCC 54412 | MZ902917 | MZ936393 | MZ936409 |
| Gnomoniopsis fragariae = G. fructicola | USA | Fragaria vesca | Rosaeace | CBS 121226 | EU254824 | GU320792 | EU219144 |
| Gnomoniopsis fragariae = G. fructicola | France | Fragaria sp. | Rosaeace | CBS 208.34 | EU254826 | GU320808 | EU219149 |
| Gnomoniopsis fragariae = G. fructicola | USA | Fragaria sp. | Rosaeace | CBS 125671 | GU320816 | GU320793 | GU320776 |
| Gnomoniopsis guangdongensis | China | Castanopsis fargesii | Fagaceae | CFCC 54443 * | MZ902918 | MZ936394 | MZ936410 |
| Gnomoniopsis guangdongensis | China | Castanopsis fargesii | Fagaceae | CFCC 54331 | MZ902919 | MZ936395 | MZ936411 |
| Gnomoniopsis guangdongensis | China | Castanopsis fargesii | Fagaceae | CFCC 54282 | MZ902920 | MZ936396 | MZ936412 |
| Gnomoniopsis guttulata | Bulgaria | Agrimonia eupatoria | Rosaeace | MS 0312 | EU254812 | NA | NA |
| Gnomoniopsis hainanensis | China | Castanopsis hainanensis | Fagaceae | CFCC 54376 * | MZ902921 | MZ936397 | MZ936413 |
| Gnomoniopsis hainanensis | China | Castanopsis hainanensis | Fagaceae | CFCC 55877 | MZ902922 | MZ936398 | MZ936414 |
| Gnomoniopsis idaeicola | USA | Rubus sp. | Rosaeace | CBS 125672 | GU320823 | GU320797 | GU320781 |
| Gnomoniopsis idaeicola | USA | Rubus pedatus | Rosaeace | CBS 125673 | GU320824 | GU320798 | GU320782 |
| Gnomoniopsis idaeicola | France | Rubus sp. | Rosaeace | CBS 125674 | GU320820 | GU320796 | GU320780 |
| Gnomoniopsis idaeicola | USA | Rubus procerus | Rosaeace | CBS 125675 | GU320822 | GU320799 | GU320783 |
| Gnomoniopsis idaeicola | USA | Rubus procerus | Rosaeace | CBS 125676 | GU320821 | GU320811 | GU320784 |
| Gnomoniopsis macounii | USA | Spiraea sp. | Rosaeace | CBS 121468 | EU254762 | GU320804 | EU219126 |
| Gnomoniopsis occulta | USA | Potentilla sp. | Rosaeace | CBS 125677 | GU320828 | GU320812 | GU320785 |
| Gnomoniopsis occulta | USA | Potentilla sp. | Rosaeace | CBS 125678 | GU320829 | GU320800 | GU320786 |
| Gnomoniopsis paraclavulata | USA | Quercus alba | Fagaceae | CBS 123202 | GU320830 | GU320815 | GU320775 |
| Gnomoniopsis racemula | USA | Chamerion angustifolium | Onagraceae | CBS 121469 * | EU254841 | GU320803 | EU219125 |
| Gnomoniopsis rossmaniae | China | Castanopsis hainanensis | Fagaceae | CFCC 54307 * | MZ902923 | MZ936399 | MZ936415 |
| Gnomoniopsis rossmaniae | China | Castanopsis hainanensis | Fagaceae | CFCC 55876 | MZ902924 | MZ936400 | MZ936416 |
| Gnomoniopsis sanguisorbae | Switzerland | Sanguisorba minor | Rosaeace | CBS 858.79 | GU320818 | GU320805 | GU320790 |
| Gnomoniopsis silvicola | China | Castanopsis hystrix | Fagaceae | CFCC 54304 | MZ902925 | MZ936401 | MZ936417 |
| Gnomoniopsis silvicola | China | Quercus serrata | Fagaceae | CFCC 54418 * | MZ902926 | MZ936402 | MZ936418 |
| Gnomoniopsis smithogilvyi | Australia | Castanea sp. | Fagaceae | CBS 130190 * | JQ910642 | KR072534 | JQ910639 |
| Gnomoniopsis smithogilvyi | Australia | Castanea sp. | Fagaceae | CBS 130189 | JQ910644 | KR072535 | JQ910641 |
| Gnomoniopsis smithogilvyi | Australia | Castanea sp. | Fagaceae | CBS 130188 | JQ910643 | KR072536 | JQ910640 |
| Gnomoniopsis smithogilvyi | Italy | Castanea sativa | Fagaceae | MUT 401 | HM142946 | KR072537 | KR072532 |
| Gnomoniopsis smithogilvyi | New Zealand | Castanea sativa | Fagaceae | MUT 411 | HM142948 | KR072538 | KR072533 |
| Gnomoniopsis tormentillae | Switzerland | Potentilla sp. | Rosaeace | CBS 904.79 | EU254856 | GU320795 | EU219165 |
| Gnomoniopsis xunwuensis | China | Castanopsis fissa | Fagaceae | CFCC 53115 * | MK432667 | MK578141 | MK578067 |
| Gnomoniopsis xunwuensis | China | Castanopsis fissa | Fagaceae | CFCC 53116 | MK432668 | MK578142 | MK578068 |
Note: NA, not applicable. Ex-type strains are marked with *, and strains from present study are in black bold.
2.3. Morphological Identification and Characterization
The morphological data of the isolates collected in the present study were based on the cultures sporulating on PDA in the dark at 25 °C. The conidiomata were observed and photographed under a dissecting microscope (M205 C, Leica, Wetzlar, Germany). The conidiogenous cells and conidia were immersed in tap water, then the microscopic photographs were captured with an Axio Imager 2 microscope (Zeiss, Oberkochen, Germany) equipped with an Axiocam 506 color camera, using differential interference contrast (DIC) illumination. More than 50 conidia were randomly selected for measurement. Culture characteristics were recorded from PDA and MEA after 10 days incubation at 25 °C in the dark.
3. Results
3.1. Phylogeny
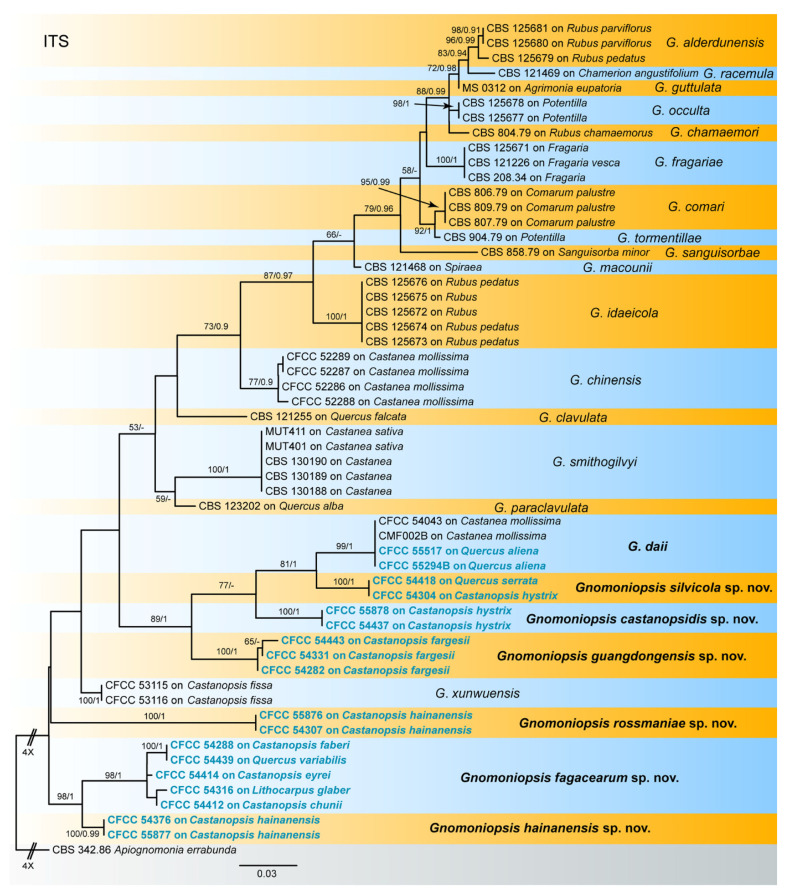
The sequence dataset of the ITS gene matrix was analysed to infer the interspecific relationships within Gnomoniopsis. The dataset consisted of 56 sequences including one outgroup taxon, Apiognomonia errabunda (CBS 342.86). A total of 538 characters including gaps were included in the phylogenetic analysis. The topologies resulting from ML and BI analyses of the concatenated dataset were congruent (Figure 1). Isolates from the present study formed seven individual clades representing seven species of Gnomoniopsis, including six new species and one known species.
Figure 1.
Phylogram of Gnomoniopsis resulting from a maximum likelihood analysis based on the ITS gene. Numbers above the branches indicate ML bootstrap values (left, ML BS ≥ 50%) and Bayesian Posterior Probabilities (right, BPP ≥ 0.9). The tree is rooted with Apiognomonia errabunda (CBS 342.86). Isolates from the present study are marked in blue, and taxa in bold face are studied in the present study.
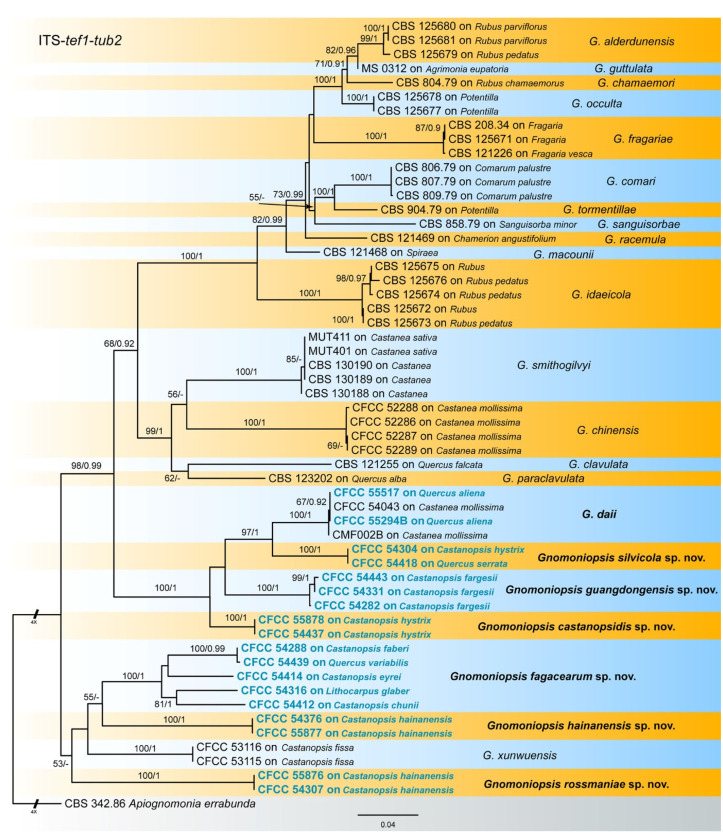
The combined three-gene sequence dataset (ITS, tef1 and tub2) was further analysed to compare with results of the phylogenetic analyses of the ITS gene. The dataset consisted of 56 sequences including one outgroup taxon, Apiognomonia errabunda (CBS 342.86). A total of 1426 characters including gaps (538 for ITS, 348 for tef1 and 540 for tub2) were included in the phylogenetic analysis. The topologies resulting from ML and BI analyses of the concatenated combined dataset were congruent (Figure 2). Isolates from the present study formed seven individual clades which were congruent with those in Figure 1.
Figure 2.
Phylogram of Gnomoniopsis resulting from a maximum likelihood analysis based on a combined matrix of ITS, tef1 and tub2. Numbers above the branches indicate ML bootstrap values (left, ML BS ≥ 50%) and Bayesian Posterior Probabilities (right, BPP ≥ 0.9). The tree is rooted with Apiognomonia errabunda (CBS 342.86). Isolates from present study are marked in blue, and taxa in bold face are studied in the present study.
3.2. Taxonomy
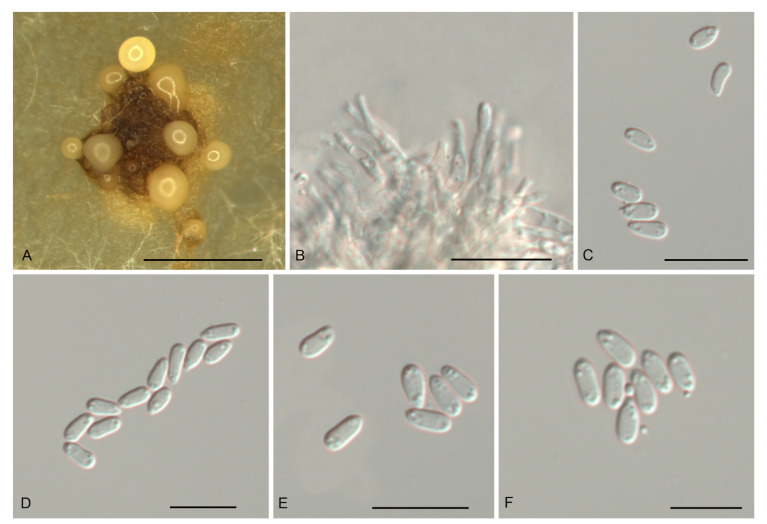
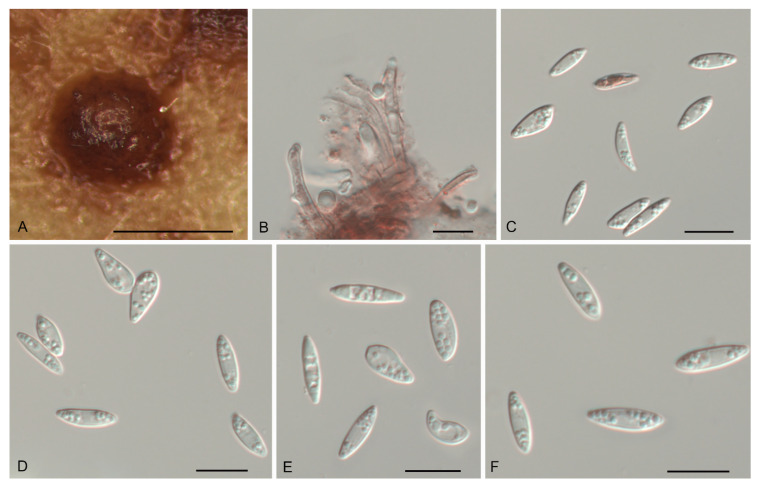
Gnomoniopsis castanopsidis N. Jiang, sp. nov. Figure 3.
Figure 3.
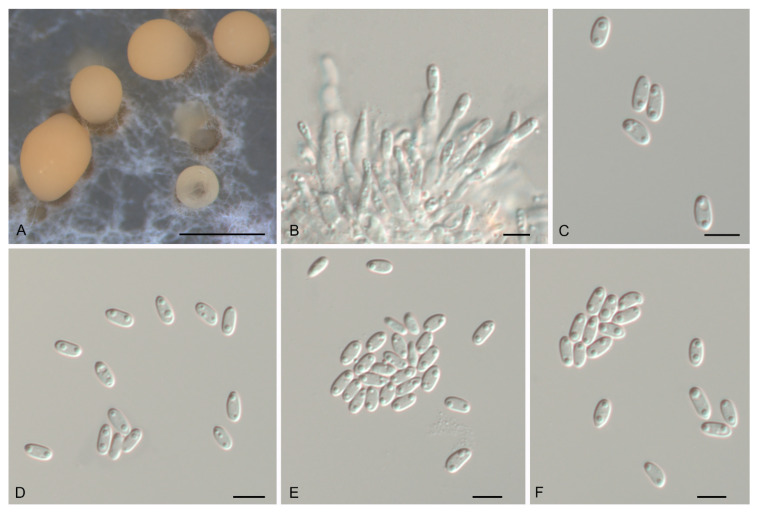
Morphology of Gnomoniopsis castanopsidis (CFCC 54437). (A) Conidiomata formed on PDA; (B) Conidiogenous cells giving rise to conidia; (C–F) Conidia. Scale bars: A = 500 μm; (B–F) = 10 μm.
Mycobank No.: 840969.
Etymology—Named after the host genus, Castanopsis.
Description—Conidiomata pycnidial, aggregated or solitary, erumpent, globose to pulvinate, brown, 300–700 μm diam., exuding a creamy conidial mass. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, multi-guttulate, cylindrical to ampulliform, attenuate towards apex, phialidic, 6.5–13 × 1.5–3 μm. Conidia aseptate, hyaline, smooth, multi-guttulate, oval to fusoid, straight or slightly curved, base truncate, (4.3–) 4.6–5.1 (–5.4) × (1.8–) 2.1–2.5 (–2.6) μm (n = 50), L/W = 1.8–2.6.
Culture characteristics—Colonies flat, spreading, with moderate aerial mycelium and undulate margin, fawn on MEA, dirty-white to fawn on PDA, forming abundant brown conidiomata with creamy conidial masses.
Material examined—CHINA, Hainan Province, Changjiang Li Autonomous County, on diseased leaves of Castanopsis hystrix, 16 November 2018, Yong Li (JNH0003 holotype; ex-type living culture, CFCC 54437); Ibid. (living culture CFCC 55878).
Notes—Two isolates from leaf spots of Castanopsis hystrix clustered into a well-supported clade named Gnomoniopsis castanopsidis, which is distinct from any known species phylogenetically (Figure 1 and Figure 2). Morphologically, G. castanopsidis is similar to G. silvicola in conidial size and shape. However, G. castanopsidis is separated from G. silvicola in 36 bp differences in ITS.
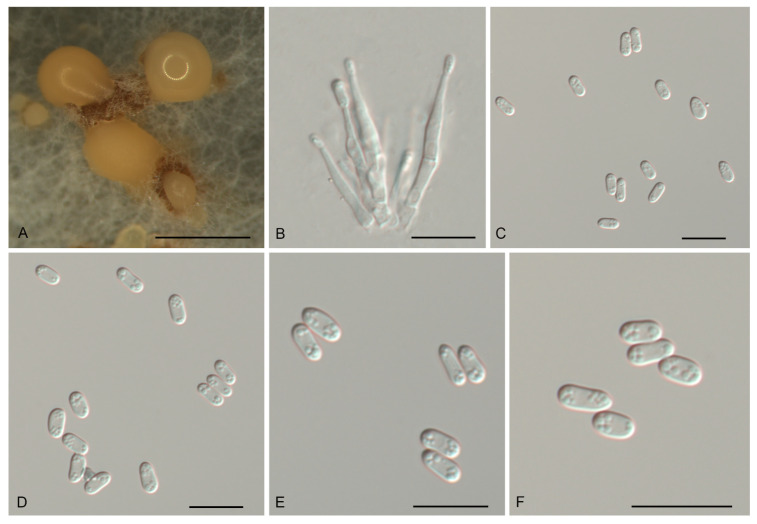
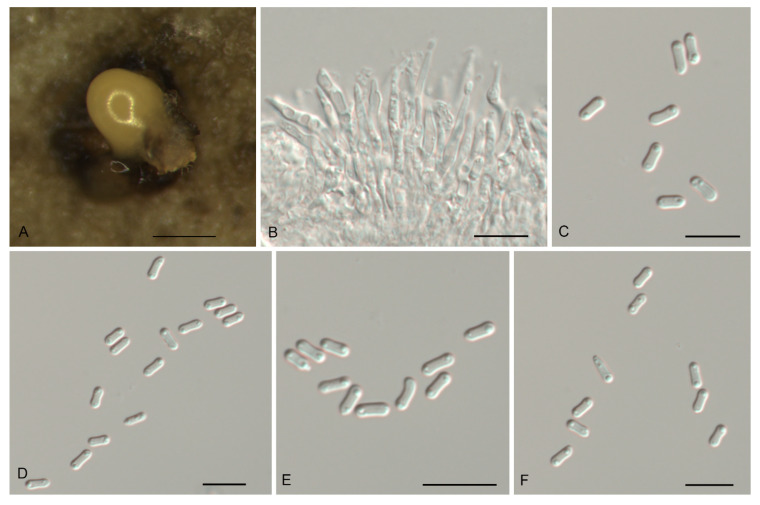
Gnomoniopsis daii C.M. Tian & N. Jiang, Forests 10(11/1016): 6 (2019). Figure 4.
Figure 4.
Morphology of Gnomoniopsis daii (CFCC 55517). (A) Conidiomata formed on PDA; (B) Conidiogenous cells giving rise to conidia; (C–F) Conidia. Scale bars: A = 500 μm; (B–F) = 10 μm.
Description—Conidiomata pycnidial, aggregated or solitary, erumpent, globose to pulvinate, brown, 200–600 μm diam., exuding a creamy conidial mass. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, multi-guttulate, cylindrical, attenuate towards apex, phialidic, 7.5–19.5 × 2–3.5 μm. Conidia aseptate, hyaline, smooth, multi-guttulate, oval to fusoid, straight or slightly curved, base truncate, (5.1–) 5.6–6.1 (–6.3) × (2.3–) 2.8–3.2 (–3.6) μm (n = 50), L/W = 1.4–2.5.
Culture characteristics—Colonies flat, spreading, with moderate aerial mycelium and undulate margin, dirty-white to sienna on MEA, dirty-white to fawn on PDA, forming abundant brown conidiomata with creamy conidial masses.
Material examined—CHINA, Henan Province, Xinyang City, Shihe District, on diseased leaves of Quercus aliena, 7 August 2019, Yong Li (JNH0004; living culture, CFCC 55517); Ibid. (living culture CFCC 55294B).
Notes—Gnomoniopsis daii was initially described as the pathogen of Chinese chestnut (Castanea mollissima) fruit rot [23], and subsequently discovered to be the leaf spot pathogen of Chinese chestnut [28]. In the present study, two isolates from diseased leaves of Quercus aliena formed a well-supported clade with the ex-type strain of G. daii (Figure 1 and Figure 2). Hence, Gnomoniopsis daii is for the first time reported on the host genus Quercus.
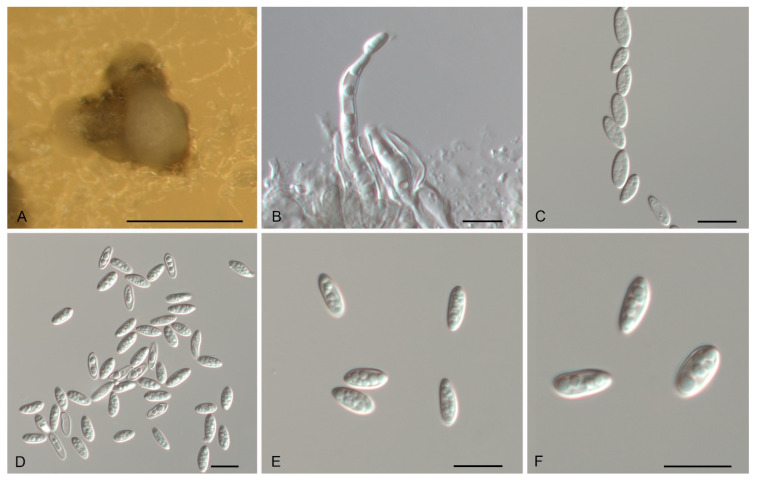
Gnomoniopsis fagacearum N. Jiang, sp. nov. Figure 5.
Figure 5.
Morphology of Gnomoniopsis fagacearum (CFCC 54316). (A) Conidioma formed on PDA; (B) Conidiogenous cells giving rise to conidia; (C–F) Conidia. Scale bars: A = 300 μm; (B–F) = 10 μm.
Mycobank No.: 840970.
Etymology—Named after the host family, Fagaceae.
Description—Conidiomata acervular, solitary, erumpent, pulvinate, red-brown, 250–450 μm diam. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells red-brown, smooth, multi-guttulate, cylindrical, slightly curved, attenuate towards apex, phialidic, 16–33.5 × 2–5 μm. Conidia aseptate, hyaline or seldom red-brown, smooth, multi-guttulate, fusoid, straight or curved, base truncate, (9–) 9.6–11.4 (–12.6) × (2.8–) 3.1–4 (–4.5) μm (n = 50), L/W = 2.1–4.2.
Culture characteristics—Colonies flat, spreading, with moderate aerial mycelium, folded surface and lobate margin, sienna to red-brown on MEA, dirty-white to slightly red-brown on PDA, occasionally forming red-brown conidiomata.
Material examined—CHINA, Guangdong Province, Qingyuan City, Yangshan County, on diseased leaves of Lithocarpus glaber, 26 November 2019, Dan-Ran Bian (JNH0005 holotype; ex-type living culture, CFCC 54316); Jiangxi Province, Xinyu City, Fenyi County, on diseased leaves of Castanopsis faberi, 20 October 2019, Yong Li (living culture, CFCC 54288); Shaanxi Province, Hanzhong City, Foping County, on diseased leaves of Quercus variabilis, 13 August 2019, Yong Li (living culture, CFCC 54439); Fujian Province, Nanping City, Yanping County, on diseased leaves of Castanopsis eyrei, 13 July 2019, Dan-Ran Bian (living culture, CFCC 54414); Guangdong Province, Qingyuan City, Yangshan County, on diseased leaves of Castanopsis chunii, 26 November 2019, Dan-Ran Bian (living culture, CFCC 54412).
Notes—Five isolates from leaf spots of Castanopsis chunii, C. eryei, C. faberi, Lithocarpus glaber and Quercus variabilis clustered into a well-supported clade here newly described as Gnomoniopsis fagacearum, which is distinct from any known species phylogenetically (Figure 1 and Figure 2). Morphologically, G. guangdongensis can be distinguished from the other Gnomoniopsis species by red-brown conidiogenous cells.
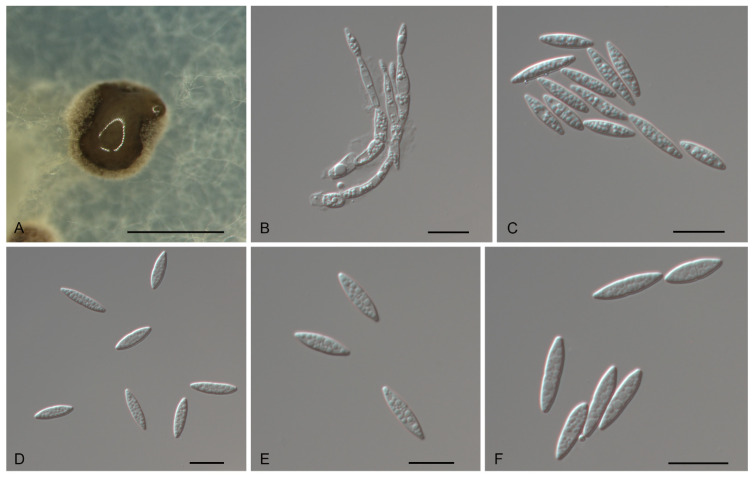
Gnomoniopsis guangdongensis N. Jiang, sp. nov. Figure 6.
Figure 6.
Morphology of Gnomoniopsis guangdongensis (CFCC 54443). (A) Conidioma formed on PDA; (B) Conidiogenous cells giving rise to conidia; (C–F) Conidia. Scale bars: A = 300 μm; (B–F) = 10 μm.
Mycobank No.: 840971.
Etymology—Named after the collection site, Guangdong Province.
Description—Conidiomata pycnidial, aggregated or solitary, erumpent, globose to pulvinate, dark brown, 150–600 μm diam., exuding a creamy conidial mass. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, multi-guttulate, cylindrical to ampulliform, attenuate towards apex, phialidic, 12.5–24 × 1.5–3 μm. Conidia aseptate, hyaline, smooth, multi-guttulate, cylindrical, constricted at the middle, straight or slightly curved, base truncate, (4.3–) 4.6–5 (–5.2) × (1.4–) 1.6–1.8 (–2) μm (n = 50), L/W = 2.4–3.3.
Culture characteristics—Colonies flat, spreading, with sparse to moderate aerial mycelium and diffuse margin, buff to fawn on MEA, dirty-white on PDA, with age forming narrow concentric zones, forming abundant dark brown conidiomata with creamy conidial masses.
Material examined—CHINA, Guangdong Province, Qingyuan City, Yangshan County, on diseased leaves of Castanopsis fargesii, 26 November 2019, Dan-Ran Bian (JNH0006 holotype; ex-type living culture, CFCC 54443); Ibid. (living cultures CFCC 54331 and CFCC 54282).
Notes—Three isolates from leaf spots of Castanopsis fargesii clustered into a well-supported clade named Gnomoniopsis guangdongensis, which is distinct from any known species phylogenetically (Figure 1 and Figure 2). Morphologically, G. guangdongensis can be distinguished from the other Gnomoniopsis species by its conidia constricted at the middle.
Gnomoniopsis hainanensis N. Jiang, sp. nov. Figure 7.
Figure 7.
Morphology of Gnomoniopsis hainanensis (CFCC 54376). (A) Conidioma formed on PDA; (B) Conidiogenous cells giving rise to conidia; (C–F) Conidia. Scale bars: A = 300 μm; (B–F) = 10 μm.
Mycobank No.: 840972.
Etymology—Named after the collection site, Hainan Province.
Description—Conidiomata pycnidial, solitary, erumpent, globose to pulvinate, light brown, 100–300 μm diam., exuding a creamy conidial mass. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, multi-guttulate, cylindrical, attenuate towards apex, phialidic, 16.5–26 × 2.5–4.5 μm. Conidia aseptate, hyaline, smooth, multi-guttulate, fusoid, straight, base truncate, (7.3–) 8–10 (–12.2) × (3.3–) 3.4–3.9 (–4.2) μm (n = 50), L/W = 1.9–3.3.
Culture characteristics—Colonies flat, spreading, with sparse aerial mycelium and lobate to undulate margin, sienna to luteous on MEA, luteous on PDA, with age forming narrow concentric zones, forming abundant light brown conidiomata with creamy conidial masses.
Material examined—CHINA, Hainan Province, Changjiang Li Autonomous County, on diseased leaves of Castanopsis hainanensis, 16 November 2018, Yong Li (JNH0007 holotype; ex-type living culture, CFCC 54376); Ibid. (living culture CFCC 55877).
Notes—Two isolates from leaf spots of Castanopsis hainanensis clustered into a well-supported clade here newly described as Gnomoniopsis hainanensis, which is distinct from any known species phylogenetically (Figure 1 and Figure 2). G. guangdongensis is different from the phylogenetically close species G. fagacearum by its conidial size and length-width ratio (7.3–12.2 × 3.3–4.2 μm, L/W = 1.9–3.3 in G. guangdongensis vs. 9–12.6 × 2.8–4.5 μm, L/W = 2.1–4.2 in G. fagacearum).
Gnomoniopsis rossmaniae N. Jiang, sp. nov. Figure 8.
Figure 8.
Morphology of Gnomoniopsis rossmaniae (CFCC 54307). (A) Conidioma formed on PDA; (B) Conidiogenous cells giving rise to conidia; (C–F) Conidia. Scale bars: A = 300 μm; (B–F) = 10 μm.
Mycobank No.: 840973.
Etymology—In honor of Amy Y. Rossman for her contributions to the study of the fungal order Diaporthales.
Description—Conidiomata pycnidial, solitary, erumpent, pulvinate, dark brown, 250–650 μm diam., exuding a brown conidial mass. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, multi-guttulate, cylindrical to ampulliform, attenuate towards apex, phialidic, 9–19 × 2–3 μm. Conidia aseptate to 1-septate, slightly constricted at septum, hyaline, smooth, multi-guttulate, elongate-fusoid, straight, base truncate, (10–) 11.6–14.6 (–16.1) × (3.1–) 3.3–3.9 (–4.1) μm (n = 50), L/W = 2.8–4.5.
Culture characteristics—Colonies flat, spreading, with sparse aerial mycelium and lobate to undulate margin, hazel on MEA, dirty-white on PDA, seldom forming dark brown conidiomata with brown conidial masses.
Material examined—CHINA, Hainan Province, Changjiang Li Autonomous County, on diseased leaves of Castanopsis hainanensis, 16 November 2018, Yong Li (JNH0008 holotype; ex-type living culture, CFCC 54307); Ibid. (living culture CFCC 55876).
Notes—Two isolates from leaf spots of Castanopsis hainanensis clustered into a well-supported clade here newly described as Gnomoniopsis rossmaniae, which is distinct from any known species phylogenetically (Figure 1 and Figure 2). Morphologically, G. rossmaniae can be distinguished from the other Gnomoniopsis species by its aseptate to 1-septate, elongate-fusoid conidia.
Gnomoniopsis silvicola N. Jiang, sp. nov. Figure 9.
Figure 9.
Morphology of Gnomoniopsis silvicola (CFCC 54418). (A) Conidiomata formed on PDA; (B) Conidiogenous cells giving rise to conidia; (C–F) Conidia. Scale bars: A = 500 μm; (B–F) = 5 μm.
Mycobank No.: 840974.
Etymology—Name from “silva” = forest and “-cola” = inhabiting; with reference to its woody host.
Description—Conidiomata pycnidial, aggregated or solitary, erumpent, globose to pulvinate, brown, 250–650 μm diam., exuding a creamy conidial mass. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, multi-guttulate, cylindrical to ampulliform, attenuate towards apex, phialidic, 7–15 × 1.5–2.5 μm. Conidia aseptate, hyaline, smooth, multi-guttulate, oval to fusoid, straight or slightly curved, base truncate, (4.3–) 4.5–5.3 (–5.9) × (1.9–) 2.2–2.6 (–2.7) μm (n = 50), L/W = 1.7–2.5.
Culture characteristics—Colonies flat, spreading, with moderate aerial mycelium and undulate margin, luteous to brown on MEA, dirty-white on PDA, forming abundant brown conidiomata with creamy conidial masses.
Material examined—CHINA, Shaanxi Province, Hanzhong City, Foping County, on diseased leaves of Quercus serrata, 13 August 2019, Yong Li (JNH0009 holotype; ex-type living culture, CFCC 54418); Guangdong Province, Shaoguan City, Lechang County, on diseased leaves of Castanopsis hystrix, 4 December 2019, Dan-Ran Bian (living culture, CFCC 54304).
Notes—Two isolates from leaf spots of Castanopsis hystrix and Quercus serrata clustered into a well-supported clade here described as the new species Gnomoniopsis silvicola, which is distinct from any known species phylogenetically (Figure 1 and Figure 2). Morphologically, G. silvicola has a bit smaller conidia than its phylogenetically close species G. daii (4.3–5.9 × 1.9–2.7 μm in G. silvicola vs. 5.1–6.3 × 2.3–3.6 μm in G. daii). In addition, G. silvicola is separated from G. daii in 34 bp differences in ITS.
4. Discussion
In the present study, six new Gnomoniopsis species (viz. G. castanopsidis, G. fagacearum, G. guangdongensis, G. hainanensis, G. rossmaniae and G. silvicola) are described and illustrated (Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, Figure 9 and Figure 10), and a new host, Quercus aliena, is reported for the known species G. daii. As noted in previous studies, the fungal genus Gnomoniopsis is so far only known from hosts of three plant families, Fagaceae, Onagraceae and Rosaceae [20,21,40], of which only one species, G. racemula was described from the family Onagraceae [20]. Hence, Fagaceae and Rosaceae are the main hosts for Gnomoniopsis species. Although several new species and host records are reported from Fagaceae in China herein, numerous additional hidden species might remain to be revealed from the widely spread fagaceous species in China.
Figure 10.
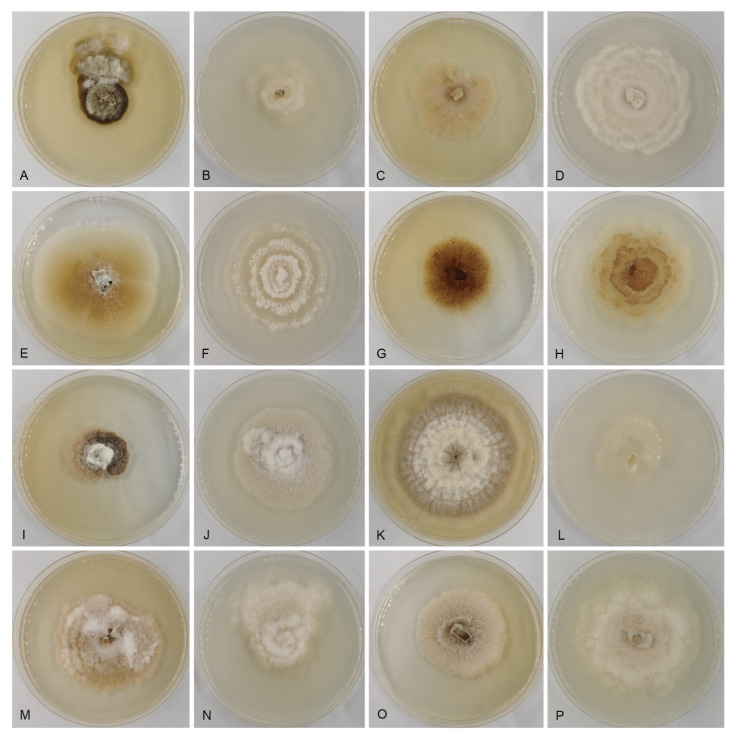
Gnomoniopsis cultures at 10 days. (A) G. silvicola (CFCC 54304) on MEA; (B) G. silvicola (CFCC 54304) on PDA; (C) G. rossmaniae (CFCC 54307) on MEA; (D) G. rossmaniae (CFCC 54307) on PDA; (E) G. guangdongensis (CFCC 54443) on MEA; (F) G. guangdongensis (CFCC 54443) on PDA; (G) G. hainanensis (CFCC 54376) on MEA; (H) G. hainanensis (CFCC 54376) on PDA; (I) G. fagaceaerum (CFCC 54316) on MEA; (J) G. fagaceaerum (CFCC 54316) on PDA; (K) G. silvicola (CFCC 54418) on MEA; (L) G. silvicola (CFCC 54418) on PDA; (M) G. castanopsidis (CFCC 54437) on MEA; (N) G. castanopsidis (CFCC 54437) on PDA; (O) G. daii (CFCC 55517) on MEA; (P) G. daii (CFCC 55517) on PDA.
So far, eleven Gnomoniopsis species were reported from fagaceous hosts, of which G. clavulata and G. paraclavulata were described from Quercus in the USA [20]. Gnomoniopsis smithogilvyi was reported as causal agent of sweet chestnut fruit rot in Australia, Europe and North America [25,26,41,42,43,44,45]. The remaining eight species are only known from China. They were well distinguished in phylogenetic analyses of the ITS gene and of combined matrices of ITS, tef1 and tub2 genes (Figure 1 and Figure 2). The conidial characters as well as the hosts and distribution provide useful information for species delimitation (Table 2).
Table 2.
Comparison of Gnomoniopsis species on hosts belonging to Fagaceae.
| Species | Host | Conidial Length (µm) | Conidial Width (µm) | L/W Ratio | Reference |
|---|---|---|---|---|---|
| G. castanopsidis | Castanopsis hystrix | (4.3–) 4.6–5.1 (–5.4) | (1.8–) 2.1–2.5 (–2.6) | 1.8–2.6 | This study |
| G. chinensis | Castanea mollissima | (6.0–) 6.5–8.5 (–9.0) | (2.2–) 2.7–3 (–3.5) | NA | [29] |
| G. clavulata | Quercus falcata | (5–) 6–6.5 (–8) | (2–) 2.5–3 (–4) | 1.4–3.7 | [20] |
| G. daii | Castanea mollissima | (5.0–) 5.5–7.0 (–8.0) | 2.0–3.5 | NA | [23,28] |
| G. daii | Quercus aliena | (5.1–) 5.6–6.1 (–6.3) | (2.3–) 2.8–3.2 (–3.6) | 1.4–2.5 | This study |
| G. fagacearum | Castanopsis chunii, C. eryei, C. faberi, Lithocarpus glaber and Quercus variabilis | (9–) 9.6–11.4 (–12.6) | (2.8–) 3.1–4 (–4.5) | 2.1–4.2 | This study |
| G. guangdongensis | Castanopsis fargesii | (4.3–) 4.6–5 (–5.2) | (1.4–) 1.6–1.8 (–2) | 2.4–3.3 | This study |
| G. hainanensis | Castanopsis hainanensis | (7.3–) 8–10 (–12.2) | (3.3–) 3.4–3.9 (–4.2) | 1.9–3.3 | This study |
| G. paraclavulata | Quercus alba | (6–) 7.5–8 (–9.5) | (2–) 3–3 (–3.5) | 1.6–4.2 | [20] |
| G. rossmaniae | Castanopsis hainanensis | (10–) 11.6–14.6 (–16.1) | (3.1–) 3.3–3.9 (–4.1) | 2.8–4.5 | This study |
| G. silvicola | Castanopsis hystrix and Quercus serrata | (4.3–) 4.5–5.3 (–5.9) | (1.9–) 2.2–2.6 (–2.7) | 1.7–2.5 | This study |
| G. smithogilvyi | Castanea sativa | (6.0–) 8 (–9.5) | (2.0–) 2.5 (–4.0) | 2.5–3.5 | [25] |
Several Gnomoniopsis species are pathogens of leaves, branches or fruits [29,46]. For example, G. smithogilvyi causes sweet chestnut branch canker and fruit rot in in Australia, Europe and the USA [26,42,45], whereas in China G. daii is one of the main pathogens of Chinese chestnut causing fruit rot and leaf spot diseases [23,28]. In addition, G. chinensis causes branch canker of Chinese chestnut in China [29]. The newly described species of the present study were isolated from diseased leaves; however, additional studies are required to confirm their pathogenicity.
5. Conclusions
Eight Gnomoniopsis species are known from fagaceous hosts in China based on morphology and phylogeny, viz. G. chinensis on Castanea mollissima, G. castanopsidis on Castanopsis hystrix, G. daii on Castanea mollissima and Quercus aliena, G. fagacearum on Castanopsis chunii, Castanopsis eyrei, Castanopsis faberi, Lithocarpus glaber and Quercus variabilis, G. guangdongensis on Castanopsis fargesii, G. hainanensis on Castanopsis hainanensis, G. rossmaniae on Castanopsis hainanensis and G. silvicola on Castanopsis hystrix and Quercus serrata. They can be well distinguished by the combined approaches of morphology and phylogeny based on ITS, tef1 and tub2 genes.
Author Contributions
Conceptualization, Y.L. and N.J.; methodology, N.J. and D.-R.B.; software, N.J.; validation, N.J., S.-K.W., Y.L. and C.-G.P.; formal analysis, N.J.; investigation, Y.L.; resources, Y.L.; data curation, N.J.; writing—original draft preparation, N.J.; writing—review and editing, H.V.; visualization, N.J.; supervision, C.-G.P.; project administration, C.-G.P.; funding acquisition, C.-G.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundamental Research Funds for the Central Non-profit Research Institution of CAF (CAFYBB2018ZB001), and National Microbial Resource Center of the Ministry of Science and Technology of the People’s Republic of China (NMRC-2021-7).
Institutional Review Board Statement
Not applicable for studies involving humans or animals.
Informed Consent Statement
Not applicable for studies involving humans.
Data Availability Statement
The sequences from the present study were submitted to the NCBI website (https://www.ncbi.nlm.nih.gov/) and the accession numbers were listed in Table 1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barr M.E. The Diaporthales in North America with emphasis on Gnomonia and its segregates. Mycol. Mem. 1978;7:1–232. [Google Scholar]
- 2.Rossman A.Y., Farr D.F., Castlebury L.A. A review of the phylogeny and biology of the Diaporthales. Mycoscience. 2007;48:135–144. doi: 10.1007/S10267-007-0347-7. [DOI] [Google Scholar]
- 3.Voglmayr H., Castlebury L.A., Jaklitsch W.M. Juglanconis gen. nov. on Juglandaceae, and the new family Juglanconidaceae (Diaporthales) Persoonia. 2017;38:136–155. doi: 10.3767/003158517X694768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voglmayr H., Rossman A.Y., Castlebury L.A., Jaklitsch W.M. Multigene phylogeny and taxonomy of the genus Melanconiella (Diaporthales) Fungal Divers. 2012;57:1–44. doi: 10.1007/s13225-012-0175-8. [DOI] [Google Scholar]
- 5.Fan X.L., Bezerra J.D., Tian C.M., Crous P.W. Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia. 2018;40:119–134. doi: 10.3767/persoonia.2018.40.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senanayake I.C., Jeewon R., Chomnunti P., Wanasinghe D.N., Norphanphoun C., Karunarathna A., Pem D., Perera R.H., Camporesi E., Eric H.C., et al. Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Divers. 2018;93:241–443. doi: 10.1007/s13225-018-0410-z. [DOI] [Google Scholar]
- 7.Jiang N., Voglmayr H., Tian C.M. New species and records of Coryneum from China. Mycologia. 2018;110:1172–1188. doi: 10.1080/00275514.2018.1516969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang N., Fan X.L., Crous P.W., Tian C.M. Species of Dendrostoma (Erythrogloeaceae, Diaporthales) associated with chestnut and oak canker diseases in China. MycoKeys. 2019;48:67–96. doi: 10.3897/mycokeys.48.31715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaklitsch W.M., Voglmayr H. European species of Dendrostoma (Diaporthales) MycoKeys. 2019;59:1–26. doi: 10.3897/mycokeys.59.37966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaklitsch W.M., Voglmayr H. The genus Melanconis (Diaporthales) MycoKeys. 2020;63:69–117. doi: 10.3897/mycokeys.63.49054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rigling D., Prospero S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018;19:7–20. doi: 10.1111/mpp.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang N., Fan X.L., Tian C.M. Identification and pathogenicity of Cryphonectriaceae species associated with chestnut canker in China. Plant Pathol. 2019;68:1132–1145. doi: 10.1111/ppa.13033. [DOI] [Google Scholar]
- 13.Jiang N., Fan X.L., Tian C.M., Crous P.W. Reevaluating Cryphonectriaceae and allied families in Diaporthales. Mycologia. 2020;112:267–292. doi: 10.1080/00275514.2019.1698925. [DOI] [PubMed] [Google Scholar]
- 14.Huang F., Udayanga D., Wang X., Hou X., Mei X., Fu Y., Hyde K.D., Li H. Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biol. 2015;119:331–347. doi: 10.1016/j.funbio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Senanayake I.C., Crous P.W., Groenewald J.Z., Maharachchikumbura S.S., Jeewon R., Phillips A.J., Bhat J.D., Perera R.H., Li Q.R., Li W.J., et al. Families of Diaporthales based on morphological and phylogenetic evidence. Stud. Mycol. 2017;86:217–296. doi: 10.1016/j.simyco.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mejía L.C., Castlebury L.A., Rossman A.Y., Sogonov M.V., White J.F. A systematic account of the genus Plagiostoma (Gnomoniaceae, Diaporthales) based on morphology, host-associations, and a four-gene phylogeny. Stud. Mycol. 2011;68:211–235. doi: 10.3114/sim.2011.68.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mejía L.C., Rossman A.Y., Castlebury L.A., Yang Z.L., White J.F. Occultocarpon, a new monotypic genus of Gnomoniaceae on Alnus nepalensis from China. Fungal Divers. 2012;52:99–105. doi: 10.1007/s13225-011-0108-y. [DOI] [Google Scholar]
- 18.Walker D.M., Castlebury L.A., Rossman A.Y., Mejía L.C., White J.F. Phylogeny and taxonomy of Ophiognomonia (Gnomoniaceae, Diaporthales), including twenty-five new species in this highly diverse genus. Fungal Divers. 2012;57:85–147. doi: 10.1007/s13225-012-0200-y. [DOI] [Google Scholar]
- 19.Jiang N., Yang Q., Liang Y.M., Tian C.M. Taxonomy of two synnematal fungal species from Rhus chinensis, with Flavignomonia gen. nov. described. MycoKeys. 2019;60:17–29. doi: 10.3897/mycokeys.60.46395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sogonov M.V., Castlebury L.A., Rossman A.Y., Mejía L.C., White J.F. Leaf-inhabiting genera of the Gnomoniaceae, Diaporthales. Stud. Mycol. 2008;62:1–77. doi: 10.3114/sim.2008.62.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker D.M., Castlebury L.A., Rossman A.Y., Sogonov M.V., White J.F. Systematics of genus Gnomoniopsis (Gnomoniaceae, Diaporthales) based on a three gene phylogeny, host associations and morphology. Mycologia. 2010;102:1479–1496. doi: 10.3852/10-002. [DOI] [PubMed] [Google Scholar]
- 22.Farr D.F., Rossman A.Y., Fungal Databases, U.S National Fungus Collections, ARS, USDA. [(accessed on 13 September 2021)]; Available online: https://nt.ars-grin.gov/fungaldatabases/
- 23.Jiang N., Tian C.M. An emerging pathogen from rotted chestnut in China: Gnomoniopsis daii sp. nov. Forests. 2019;10:1016. doi: 10.3390/f10111016. [DOI] [Google Scholar]
- 24.Yang Q., Jiang N., Tian C.M. Tree inhabiting gnomoniaceous species from China, with Cryphogonomonia gen. nov. proposed. MycoKeys. 2020;69:71–89. doi: 10.3897/mycokeys.69.54012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crous P.W., Summerell B.A., Shivas R.G., Burgess T.I., Decock C.A., Dreyer L.L., Granke L.L., Guest D.I., Hardy G.E.S.J., Hausbeck M.K., et al. Fungal Planet Description Sheets: 107–127. Persoonia. 2012;28:138–182. doi: 10.3767/003158512X652633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visentin I., Gentile S., Valentino D., Gonthier P., Tamietti G., Cardinale F. Gnomoniopsis castanea sp. nov. (Gnomoniaceae, Diaporthales) as the causal agent of nut rot in sweet chestnut. J. Plant Pathol. 2012;94:411–419. [Google Scholar]
- 27.Shuttleworth L.A., Walker D.M., Guest D.I. The chestnut pathogen Gnomoniopsis smithogilvyi (Gnomoniaceae, Diaporthales) and its synonyms. Mycotaxon. 2016;130:929–940. doi: 10.5248/130.929. [DOI] [Google Scholar]
- 28.Jiang N., Fan X.L., Tian C.M. Identification and characterization of leaf-inhabiting Fungi from Castanea plantations in China. J. Fungi. 2021;7:64. doi: 10.3390/jof7010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang N., Liang L.Y., Tian C.M. Gnomoniopsis chinensis (Gnomoniaceae, Diaporthales), a new fungus causing canker of Chinese chestnut in Hebei Province, China. MycoKeys. 2020;67:19–32. doi: 10.3897/mycokeys.67.51133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson M.G. Diversity and Classification of Flowering Plants: Eudicots. Academic Press; San Diego, CA, USA: 2010. [Google Scholar]
- 31.Doyle J.J., Doyle J.L. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 32.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 33.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- 34.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Donnell K., Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 36.Katoh K., Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. Institute of Electrical and Electronics Engineers; New Orleans, LA, USA: 2010. [Google Scholar]
- 38.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 40.Udayanga D., Miriyagalla S.D., Manamgoda D.S., Lewers K.S., Gardiennet A., Castlebury L.A. Molecular reassessment of diaporthalean fungi associated with strawberry, including the leaf blight fungus, Paraphomopsis obscurans gen. et comb. nov.(Melanconiellaceae) IMA Fungus. 2021;12:1–21. doi: 10.1186/s43008-021-00069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shuttleworth L.A., Liew E.C.Y., Guest D.I. Survey of the incidence of chestnut rot in south-eastern Australia. Australas. Plant Pathol. 2013;42:63–72. doi: 10.1007/s13313-012-0170-2. [DOI] [Google Scholar]
- 42.Shuttleworth L.A., Guest D.I. The infection process of chestnut rot, an important disease caused by Gnomoniopsis smithogilvyi (Gnomoniaceae, Diaporthales) in Oceania and Europe. Australas. Plant Pathol. 2017;46:397–405. [Google Scholar]
- 43.Dennert F.G., Broggini G.A., Gessler C., Storari M. Gnomoniopsis castanea is the main agent of chestnut nut rot in Switzerland. Phytopathol. Mediterr. 2015;54:199–211. [Google Scholar]
- 44.Pasche S., Calmin G., Auderset G., Crovadore J., Pelleteret P., Mauch-Mani B., Barja F., Paul B., Jermini M., Lefort F. Gnomoniopsis smithogilvyi causes chestnut canker symptoms in Castanea sativa shoots in Switzerland. Fungal Genet. Biol. 2016;87:9–21. doi: 10.1016/j.fgb.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Sakalidis M.L., Medina-Mora C.M., Kolp M., Fulbright D.W. First report of Gnomoniopsis smithogilvyi causing chestnut brown rot on chestnut fruit in Michigan. Plant Dis. 2019;103:2134. doi: 10.1094/PDIS-03-19-0562-PDN. [DOI] [Google Scholar]
- 46.Linaldeddu B.T., Deidda A., Scanu B., Franceschini A., Alves A., Abdollahzadeh J., Phillips A.J.L. Phylogeny, morphology and pathogenicity of Botryosphaeriaceae, Diatrypaceae and Gnomoniaceae associated with branch diseases of hazelnut in Sardinia (Italy) Eur. J. Plant Pathol. 2016;146:259–279. doi: 10.1007/s10658-016-0912-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences from the present study were submitted to the NCBI website (https://www.ncbi.nlm.nih.gov/) and the accession numbers were listed in Table 1.