Abstract
Yeast Smt3 and its vertebrate homolog SUMO-1 are ubiquitin-like proteins (Ubls) that are reversibly ligated to other proteins. Like SMT3, SMT4 was first isolated as a high-copy-number suppressor of a defective centromere-binding protein. We show here that SMT4 encodes an Smt3-deconjugating enzyme, Ulp2. In cells lacking Ulp2, specific Smt3-protein conjugates accumulate, and the conjugate pattern is distinct from that observed in a ulp1ts strain, which is defective for a distantly related Smt3-specific protease, Ulp1. The ulp2Δ mutant exhibits a pleiotropic phenotype that includes temperature-sensitive growth, abnormal cell morphology, decreased plasmid and chromosome stability, and a severe sporulation defect. The mutant is also hypersensitive to DNA-damaging agents, hydroxyurea, and benomyl. Although cell cycle checkpoint arrest in response to DNA damage, replication inhibition, or spindle defects occurs with normal kinetics, recovery from arrest is impaired. Surprisingly, either introduction of a ulp1ts mutation or overproduction of catalytically inactive Ulp1 can substantially overcome the ulp2Δ defects. Inactivation of Ulp2 also suppresses several ulp1ts defects, and the double mutant accumulates far fewer Smt3-protein conjugates than either single mutant. Our data suggest the existence of a feedback mechanism that limits Smt3-protein ligation when Smt3 deconjugation by both Ulp1 and Ulp2 is compromised, allowing a partial recovery of cell function.
The ubiquitin system is central to many biological regulatory mechanisms, including aspects of signal transduction, cell cycle progression, differentiation, and the stress response (reviewed in references 12, 13, 19, and 38). Covalent attachment of the ubiquitin polypeptide to cellular proteins is achieved through a highly conserved enzymatic pathway. In an ATP-consuming reaction, the C terminus of ubiquitin is first activated by an enzyme called E1, to which it becomes attached by a thiolester bond. The ubiquitin is then transferred to an E2 ubiquitin-conjugating enzyme. Together with an additional factor called E3, or ubiquitin-protein ligase, E2 enzymes catalyze formation of an amide (isopeptide) bond between the C-terminal carboxyl group of ubiquitin and a lysine side chain(s) of the acceptor protein. Most frequently, the modified protein is targeted to the 26S proteasome, a protease that degrades the substrate into small peptides but allows recycling of ubiquitin.
Eukaryotes express a set of ubiquitin-like proteins (Ubls) that diverge significantly from ubiquitin yet in some cases are also ligated to other proteins (reviewed in references 10 and 14). A recently discovered Ubl that is ligated to cellular proteins is the vertebrate SUMO-1 protein (also called UBL1, PIC1, sentrin, SMT3C, or GMP1) (17, 30). Human SUMO-1 is only 18% identical to ubiquitin but is 48% identical to a yeast protein called Smt3 (25), and the human and yeast proteins are functional homologs. Despite its limited sequence similarity to ubiquitin, SUMO-1 shares the ubiquitin superfold, although it has a flexible N-terminal extension that is found only in the SUMO family (3). The reactions involving SUMO-1 and Smt3 have much in common with those of ubiquitin (reviewed in reference 14). For example, conjugation of substrates to Smt3 or SUMO-1 has been shown to depend on an E1-related heterodimeric activating enzyme, Uba2-Aos1, and an E2-like enzyme, Ubc9 (5, 16).
The SUMO conjugation system has been implicated in multiple physiological pathways. SMT3, UBA2, AOS1, and UBC9 are all essential for viability in the yeast Saccharomyces cerevisiae, and conditional ubc9 mutants are defective in cell cycle progression (32). However, little else is known about the functions of the yeast pathway. The only known target for Smt3 in yeast is the septin Cdc3, and the functional consequences of this modification are not yet clear (36). In vertebrates, RanGAP1, a protein required for nucleocytoplasmic trafficking, must be sumoylated for localization to the nuclear pore complex (23, 24). Other targets of SUMO-1 are RanBP2, another nuclear pore component (31); PML, a protein encoded by a gene involved in chromosomal translocations that are responsible for certain leukemias (26); Sp100, a protein which, like PML, is found in nuclear substructures called PML nuclear bodies (35); and IκBα, the NF-κB inhibitor (6).
Modification of proteins by ubiquitin and Ubls is reversible. In addition, both ubiquitin and Ubls are synthesized in precursor form, with one or more amino acids following a Gly-Gly dipeptide that will form the C terminus of the mature protein. Ubiquitin-substrate deconjugation and precursor processing are performed by members of a diverse group of specialized cysteine proteases called deubiquitinating enzymes, or Dubs (39). Analyses of these enzymes indicate that they have diverse regulatory roles in the ubiquitin system. Much less is known about the analogous reactions involving Ubls, so we recently initiated a search for SUMO-specific proteases. We found a yeast enzyme, Ulp1, that specifically removes Smt3 and SUMO-1 from proteins and is required for progression through the G2/M phase of the cell cycle (22). Ulp1 (Ubl-specific protease 1) lacks sequence similarity to any Dub and is unable to process ubiquitin-linked substrates. However, Ulp1 has weak sequence similarity to the protein encoded by the yeast SMT4 gene.
Here we show that Ulp2 (Smt4) is also an Smt3-specific protease. (For the purpose of cataloging the yeast Ubl-specific processing enzymes, we propose calling Smt4 by the alternative name, Ulp2.) This has allowed a detailed comparison of the two Smt3 proteases, leading to several surprising observations. Yeast cells lacking ULP2 have multiple defects and accumulate a specific set of Smt3-protein conjugates that are distinguishable from those that accumulate in ulp1 cells. Interestingly, both catalytically active and inactive forms of overexpressed Ulp1 can suppress the temperature-sensitive growth of ulp2Δ cells, and mutation of either of the Smt3 proteases suppresses defects associated with a mutation of the other. These results could be explained by functional antagonism between the two enzymes and/or by both enzymes acting as positive regulators of Smt3-protein ligation. In fact, a strong reduction in Smt3 conjugates is observed in the double mutant, and mutation of the Smt3-conjugating enzyme Ubc9 suppresses the ulp2Δ defect as well. Our data also suggest an important role for Ulp2 in the recovery of cells from checkpoint arrest induced by DNA damage, inhibition of DNA replication, or defects in spindle assembly. A connection between Ulp2 action and mitotic spindle dynamics in particular is reinforced by the finding that ulp2Δ cells lose chromosomes at high rates, have structurally abnormal mitotic spindles, and are hypersensitive to microtubule-depolymerizing agents.
MATERIALS AND METHODS
Plasmid and strain construction.
Standard methods were used for the growth of yeast and bacteria and for recombinant DNA work (2). The yeast strains used in the present work are listed in Table 1. To generate yeast strains with a null allele of ULP2, we amplified the yeast HIS3 gene by PCR using primers with 5′ sequence segments matching the beginning and end of the ULP2 open reading frame sequences; the amplified fragments were transformed into diploid MHY606 cells (27), and the resulting histidine prototrophs were checked by colony PCR for the correct gene replacements. Tetrad analysis of the heterozygotes was used to verify single-site insertion of the HIS3 gene and to evaluate the phenotype of haploid strains carrying the deletion allele. MHY1628 was made by transformation of MHY501 with HindIII/XhoI-digested pRC10.1, which carries a mad2Δ::URA3 allele (from S. Kron). Fully functional, C-terminal myc9-tagged versions of Ulp1 and Ulp2 were created by amplifying a myc9-his5+ DNA segment (from R. Deshaies) with the appropriate primers and single-step integration at the corresponding ULP loci. His+ colonies were screened by anti-myc immunoblotting.
TABLE 1.
Yeast strains
| BWG9a-1 | MATα his4 ade6 ura3 |
| W303a | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
| MHY500a | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 |
| MHY501a | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 |
| MHY606b | MHY500 × MHY501 |
| MHY612a | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 rad6Δ::LEU2 |
| MHY1380 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 ulp2-Δ1::HIS3 |
| MHY1381 | MATa his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 ulp2-Δ1::HIS3 |
| MHY1569 | MATa/MATα his3-Δ200/his3-Δ200 leu2-3,112/ leu2-3,112 ura3-52/ura3-52 lys2-801/lys2-801 trp1-1/trp1-1 ulp2-Δ1::HIS3/ulp2-Δ1::HIS3 |
| MHY1614 | MATa his3-Δ200 leu2-3,112::LEU2::ulp1-333 ura3-52 lys2-801 trp1-1 ulp2-1::HIS3 ulp1-Δ1::his3::URA3 |
| MHY1615 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 ulp2-Δ1::HIS3 |
| MHY1616 | MATa his3-Δ200 leu2-3,112::LEU2::ulp1-333 ura3-52 lys2-801 trp1-1 ulp1-Δ1::his3::URA3 |
| MHY1617 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 |
| MHY1620 | MATa his3-Δ200 leu2-3,112::LEU2::ubc9-1 ura3-52 lys2-801 trp1-1 ubc9Δ::TRP1 |
| MHY1621c | MATa leu2 trp1 his3 ura3 mec1 |
| MHY1624 | MATα his3-Δ200 leu2-3,112::LEU2::ubc9-1 ura3-52 lys2-801 trp1-1 ubc9Δ::TRP1 ulp2-Δ1::HIS3 |
| MHY1628 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 mad2Δ::URA3 |
| MHY1678d | MATa leu2-3,112 trp1Δ1 his3-Δ200 ade2-1 can1 ura3-52::URA3-GAL-myc-MPS1 |
| MHY1705 | MATa leu2-3,112 trp1Δ1 his3-Δ200 ade2-1 can1 ura3-52::URA3-GAL-myc-MPS1 ulp2Δ::HIS3 |
For bacterial expression of Ulp2, the ULP2 coding sequence was amplified from yeast genomic DNA, and the PCR fragment was digested with SmaI and SalI and ligated into SmaI/XhoI-digested pGEX-KG, yielding pGEX-ULP2. The plasmid YRTAG310-cup1Δ-ULP2, which allows ULP2 expression from its own promoter, was made as follows. ULP2 genomic sequence, including ∼500 bp of DNA both upstream and downstream of the ULP2 open reading frame, was amplified by PCR, and the PCR product was sequentially treated with PstI, T4 DNA polymerase, and XhoI. YRTAG310 was incubated sequentially with BamHI, T4 DNA polymerase, and XhoI, the larger vector DNA fragment lacking the CUP1 promoter was purified, and the vector and PCR DNA fragments were ligated together.
Protein purification and enzyme assays.
Recombinant protein expression in bacterial cells harboring the appropriate expression plasmids was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). For radiolabeling substrates, induced cells were incubated with 35S-Translabel (ICN) for 20 min prior to being harvested. Glutathione S-transferase (GST)–Ulp1 and GST-Ulp2 were purified on glutathione-agarose (Sigma). His6-tagged substrates were purified on the Talon affinity matrix (Clontech). Smt3 cleavage assays were done at 30°C with different concentrations of substrate and GST-Ulp1 or GST-Ulp2 in a reaction mixture containing 150 mM NaCl, 1 mM dithiothreitol, 10 mM Tris-HCl (pH 8.0), and 0.2% Triton X-100 (22).
For cleavage of yeast Smt3 conjugates, MHY1380 cells were grown to log phase (total, ∼109 cells) in rich medium (yeast extract-peptone-dextrose [YPD]) at 30°C. The cells were pelleted, washed once with buffer A (1.2 M sorbitol, 50 mM Tris-HCl, pH 7.5), and incubated in 1 ml of buffer A containing 0.5 mg of Zymolyase 100T/ml at 30°C for 30 min. After cell wall digestion, the cells were washed once in cold buffer A and lysed by sonication in 0.5 ml of buffer B (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.2% Triton X-100, 2 mM N-ethylmaleimide [NEM], 2 mM phenylmethylsulfonyl fluoride, and 20 μg each of leupeptin, pepstatin, and antipain per ml) on ice. The lysates were centrifuged at 14,000 × g for 10 min to clear cell debris, and soluble protein concentrations were determined by the bicinchoninic acid protein assay (Pierce). Prior to the cleavage reactions, l-cysteine and β-mercaptoethanol were added to 2 mM each and incubated at room temperature for 10 min to consume any remaining unreacted NEM. Reactions were started by adding 50 ng of purified GST-Ulp2 to a 20-μl reaction mixture containing 25 μg of soluble yeast proteins in buffer C (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM dithiothreitol, 0.1% Triton X-100) and stopped after 2 to 3 h by boiling in sodium dodecyl sulfate (SDS) sample buffer (22).
Immunofluorescence microscopy.
For immunofluorescent staining of yeast cells, the cells were fixed by addition to the growth medium of formaldehyde to 4% (wt/vol) for 2 h at room temperature. The fixed cells were washed once and resuspended in 0.5 ml of buffer D (50 mM Tris-HCl [pH 8.0], 1.2 M sorbitol, 1 mM MgCl2). Zymolyase 100T (ICN) was added to 0.5 mg/ml, and the mixture was incubated at room temperature for 30 min. The cells were again washed, resuspended, and spotted on polylysine-coated microscope slides. Excess cells were removed by aspiration, and the cells retained on the slide were permeabilized in phosphate-buffered saline (PBS) containing 0.1% Triton X-100. After washes with PBS and PBS–1% bovine serum albumin (BSA) (Sigma; immunoglobulin G free), the cells were incubated with anti-β-tubulin (Boehringer Mannheim) or anti-myc (9E10; Santa Cruz Biotechnology) monoclonal antibodies diluted 100-fold and 500-fold, respectively, in PBS–1% BSA for at least 2 h at room temperature in a moist chamber. Primary antibody was removed by aspiration, followed by four PBS washes. Secondary Oregon Green-conjugated anti-mouse immunoglobulin G (Molecular Probes) diluted 100-fold in PBS–1% BSA was added and incubated for 1 h at room temperature, followed by four washes in PBS. A drop of 25-ng/ml 4,6-diamidino-2-phenylindole (DAPI)-containing mounting solution was applied to the air-dried cells, and the samples were sealed under a cover slide.
Microcolony assays.
Logarithmically growing MHY1381 or MHY500 yeast cells (2.5 μl) were placed on YPD plus methyl methanesulfonate (MMS) (0.01%), 0.1 M hydroxyurea, or 15 μg of benomyl sulfate/ml. For the GAL-MPS1 experiments, cells were synchronized in G1 with α-factor and placed on YPGal (2%) plates. Fifty to 60 isolated small, unbudded cells were identified and marked by punctures in the agar with a dissection needle (21). The cells or buds in each position were counted every 3 h. Cell viability was scored after 24 h at 30°C. At that point, viable cells usually formed colonies with >100 cells whereas growth-arrested colonies usually had just one to four large-budded cells.
For serial-dilution growth assays, log-phase cultures were normalized based on optical density at 600 nm to 1 optical density unit/ml and serially diluted in 10-fold steps. A 2.5-μl sample from each dilution was spotted on YPD plates containing the following: benomyl sulfate (Sigma), 15 μg/ml; hydroxyurea (Molecular Probes), 0.1 M; and MMS (Sigma), 0.005 to 0.01% (wt/vol). The radiation exposures used were 200 kilorads of γ rays or 1.3 J/m2 of UV (Bio-Rad UV cross-linker).
Plasmid and chromosome loss measurements.
To measure plasmid loss, log-phase cultures of wild-type (MHY500) or ulp2Δ (MHY1381) cells harboring YCplac22 (TRP1 CEN) (9) or pHRΔUb (TRP1 ARS) (M. Hochstrasser, unpublished plasmid) grown in minimal medium lacking tryptophan were diluted 100-fold with YPD and grown at 30°C. At 4-h intervals, aliquots were taken, diluted, and plated in triplicate on minimal medium (SD/complete). The cultures were monitored for at least 48 h, and fresh dilutions in YPD were made every 12 h to maintain the cultures in exponential growth. Colonies grown on SD/complete plates were replica plated to SD lacking tryptophan (SD-trp) dropout plates. Percentages of colonies that grew on SD/complete plates but failed to grow on SD-trp plates were counted and divided by the number of generations in YPD (8).
Chromosome loss rates were measured by a quantitative mating assay (21). Haploid MATα lys2 HIS4 strains and the tester strain BWG9a-1 (MATα his4 ade6) were mated by spotting ∼3 × 106 log-phase cells of each partner on a YPD plate and incubating them at 30°C for 12 h. After being washed and resuspended in water, the cells were diluted and plated on medium lacking both adenine and lysine to select diploid cells or on SD/complete plates to determine total viable cells. Mating can occur only if one of the two haploid strains loses its copy of MATα, allowing it to mate as an a cell. Ade+ Lys+ colonies were replica plated to medium lacking histidine. MATα and HIS4 are on opposite arms of chromosome III. Complete loss of chromosome III from the HIS4 strain would result in Ade+ Lys+ His− colonies following mating. Loss of just part of chromosome III from the HIS4 strain, mutation of MATα, gene conversion, or loss of all or part of chromosome III from the his4 tester strain gives rise to Ade+ Lys+ His+ cells. Chromosome loss frequencies were expressed as the number of Ade+ Lys+ His− colonies divided by the number of viable colonies.
RESULTS
ULP2 encodes an Smt3-specific protease.
Our recent biochemical screen for yeast Smt3-specific processing proteases yielded Ulp1 but no other Smt3-cleaving enzymes (22). Deletion of the ULP1 gene in yeast is lethal, and partial-loss-of-function alleles result in the accumulation of the Smt3 precursor as well as Smt3-protein conjugates. Hence, it was possible that Ulp1 would be the only yeast Smt3-processing enzyme. However, sequence database searches with Ulp1 identified a number of related sequences, including a weakly similar S. cerevisiae protein, Ulp2 (Fig. 1). The ULP2 gene was identified in the same high-copy-number suppressor screen that yielded SMT3 (25), although no characterization of ULP2 has been published.
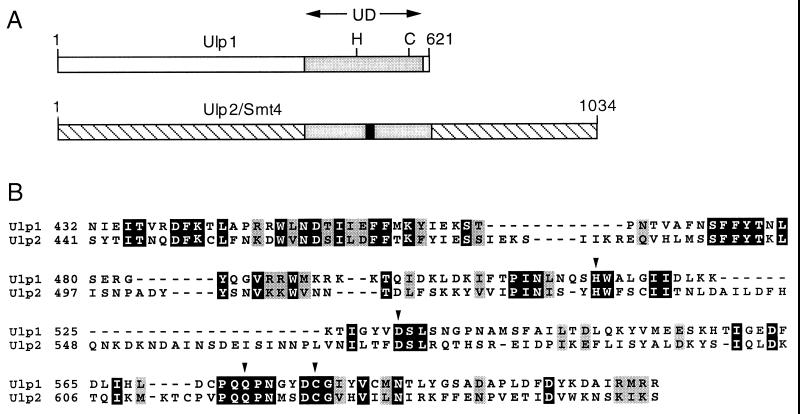
FIG. 1.
Yeast Ulp enzymes. (A) UD in S. cerevisiae Ulp1 and Ulp2. Positions of the catalytic His (H) and Cys (C) residues and of the Ulp2-specific insertion in the UD (solid bar) are indicated. (B) Sequence alignment of the UDs of yeast Ulp1 and Ulp2. The arrowheads mark presumptive catalytic residues; black and grey boxes mark identical and similar residues, respectively.
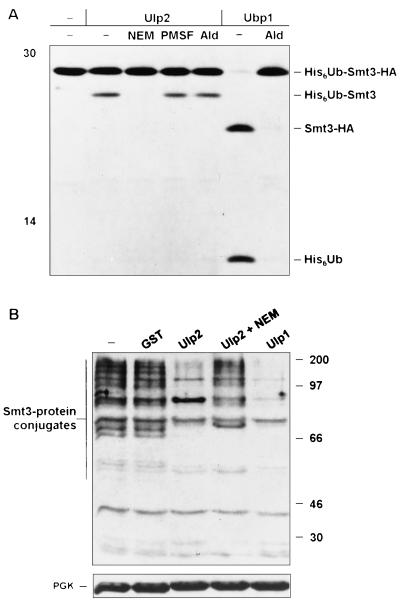
The similarity between the 621-residue Ulp1 and the 1,034-residue Ulp2 proteins is confined primarily to a region of ∼200 amino acids, and this region is also the part most readily aligned with other Ulp1-related proteins from other organisms (Fig. 1A). This region has been dubbed the Ulp domain (UD), and it includes all the residues implicated in the formation of the cysteine protease-like catalytic site (22). These residues are all conserved in Ulp2 (Fig. 1B), but because the overall conservation even in the UD is low, it was uncertain whether Ulp2 would have Smt3-cleaving activity. We purified recombinant GST-Ulp2 from Escherichia coli and found that it was indeed able to cleave after the Smt3 moiety in both His6-ubiquitin-Smt3-hemagglutinin (HA) and Smt3–β-galactosidase protein fusions, yielding the same cleavage products observed with Ulp1 (Fig. 2A and not shown). As expected for a cysteine protease distinct from Dubs, Ulp2 did not cleave after the ubiquitin in His6-ubiquitin-Smt3-HA and was inhibited by NEM but not by phenylmethylsulfonyl fluoride or the Dub-specific inhibitor ubiquitin aldehyde (Fig. 2A). The ubiquitin-specific protease Ubp1 was used as a control for cleavage after ubiquitin in the chimeric substrate and for verifying the inhibitory activity of the ubiquitin aldehyde. Incubation of the Ulp2 protein with Smt3-protein conjugates from yeast also resulted in a strong reduction in the levels of the conjugates (Fig. 2B), indicating that Ulp2 was also able to cleave isopeptide-linked Smt3 molecules from natural substrates.
FIG. 2.
S. cerevisiae Ulp2 is an Smt3-cleaving enzyme. (A) Cleavage of radiolabeled His6-ubiquitin-Smt3-HA by GST-Ulp2 and yeast Ubp1 analyzed after 2 h at 30°C by SDS-polyacrylamide gel electrophoresis (12.5% gel). Left lane, no added enzyme. Inhibitor preincubations were done for 15 min at room temperature. Ald, ubiquitin aldehyde; −, absent. (B) In vitro cleavage by purified GST-Ulp2 or GST-Ulp1 of yeast Smt3-protein conjugates. The blotting conditions did not allow visualization of free Smt3. NEM-treated extracts (25 μg) from ulp2Δ cells grown at 23°C (two left lanes) or 37°C (three right lanes) are shown. A species of ∼85 kDa was reproducibly enhanced following Ulp2 digestion in vitro; this might represent a multiply modified protein from which not all Smt3 molecules were cleaved. A rabbit anti-Smt3 antibody was used for immunoblot analysis. In the lower gel, filters were reprobed with anti-PGK (3-phosphoglycerate kinase) to compare protein loadings. The positions of molecular mass markers are indicated in each panel (in kilodaltons).
Characterization of a ulp2 null mutant.
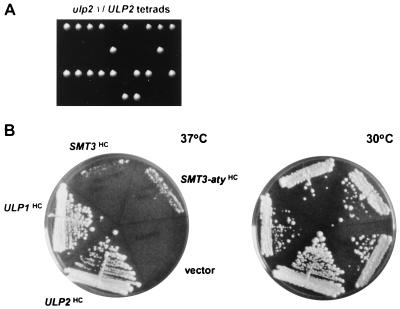
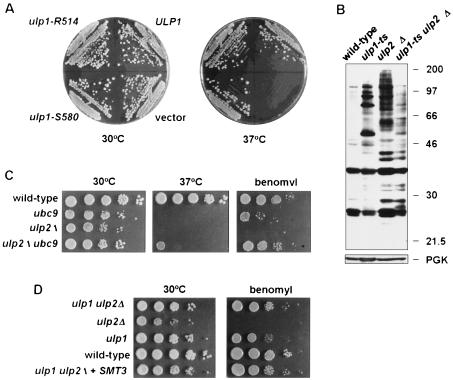
A diploid strain heterozygous for a null allele of ULP2, ulp2Δ::HIS3, was constructed. Tetrad dissection of sporulated cells yielded primarily tetrads with two viable and two inviable segregants, and the fast-growing segregants were all histidine auxotrophs (Fig. 3A). Microscopic examination of the inviable segregants indicated that some segregants arrested after one or two divisions and others proceeded through at least five or six divisions. However, prolonged incubation on rich medium (YPD) revealed an occasional slow-growing His+ segregant. The heterozygote was transformed with YCplac33-ULP2, and haploid ulp2Δ segregants carrying the URA3-marked plasmid were obtained. Plating on 5-fluoroorotic acid, which is toxic to cells expressing URA3, resulted in viable but slow-growing cells that had lost the plasmid. In liquid YPD cultures at 30°C, ulp2Δ cells had a 143-min doubling time compared to 91 min for the congenic wild type. Mutant cells were unable to form colonies at 37°C (Fig. 3B, vector; also see Fig. 5B). Provision of high levels of mature Smt3 very weakly suppressed the temperature sensitivity of ulp2Δ cells, but overexpression of the precursor did as well. Therefore, the ulp2Δ growth defects are unlikely to be due to impaired processing of proSmt3 (see below). The basis of the weak suppression by Smt3 is unknown. Association of some Smt3-protein conjugates with cellular targets may be growth inhibitory, so excess Smt3 might be able to compete for binding to these sites. In summary, Ulp2 is essential for viability at high temperature, but loss of ULP2 also leads to poor growth at lower temperatures and appears to be important for the return to growth of spores.
FIG. 3.
Growth of ulp2Δ cells. (A) Tetrad analysis of an ulp2Δ/ULP2 heterozygote. One His+ segregant from the 10 tetrads shown eventually grew into a small colony. (B) Growth on rich medium at 37°C for 5 days or 30°C for 4 days of ulp2Δ cells carrying high-copy-number (HC) plasmids with the indicated genes. The Smt3 proteins were tagged with HA.
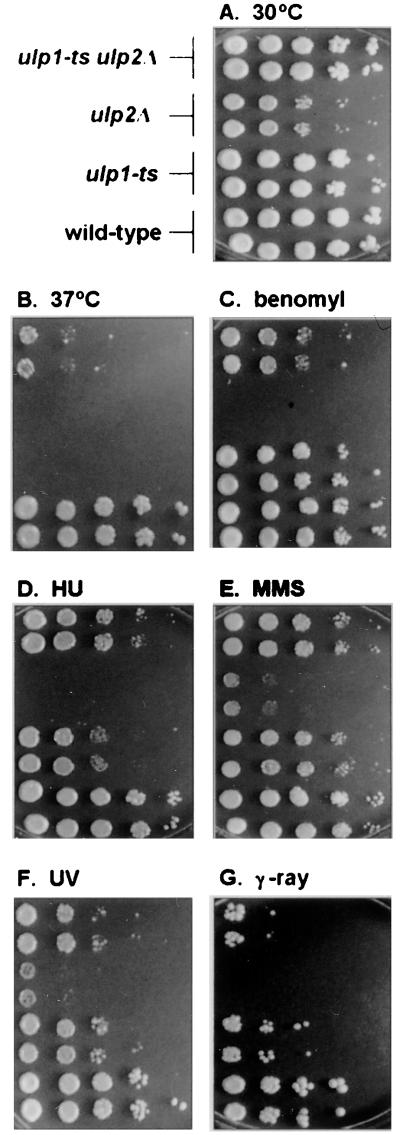
FIG. 5.
Comparison of ulp2Δ, ulp1ts, and ulp2Δ ulp1ts mutants. Tenfold serial dilutions of logarithmically growing cultures were spotted onto YPD plates that were placed at 30°C (A) or 37°C (B). Alternatively, cells were incubated at 30°C in the presence of 15 μg of benomyl sulfate/ml (C), 0.1 M hydroxyurea (D), or 0.005% MMS (E) or they were exposed to 1.3 J/m2 of UV radiation (F) or 200 kilorads of γ rays (G).
The deficiency of ulp2Δ cells in growth from spores led us to test the ability of homozygous ulp2Δ/ulp2Δ diploids to sporulate. The mutant cells were severely defective for sporulation at both 23 and 30°C (<0.001% asci versus 28% for the congenic wild type). Notably, transcript levels of ULP2 increase over 10-fold early in meiosis (http://cmgm.stanford.edu/pbrown /sporulation). We have also examined Smt3-protein conjugate profiles as a function of meiotic progression by anti-Smt3 immunoblot analysis. The pattern of Smt3-linked proteins changed strikingly as the cells progressed through meiosis (not shown). These data suggest that Smt3-protein deconjugation by Ulp2 is important for normal meiotic development.
Enhanced chromosome and plasmid loss rates in ulp2Δ.
We noticed that ulp2Δ strains formed colonies of irregular size and with uneven borders (Fig. 3B). If either small or large colonies were restreaked on plates, they again gave rise to both small and large colonies. Moreover, plasmids were difficult to maintain in the mutant without continuous selection for a plasmid-borne marker. These observations suggested that ulp2− cells might lose chromosomes and plasmids at abnormally high rates. We compared loss rates for both CEN/ARS and ARS plasmids in wild-type and ulp2− cells under logarithmic growth conditions in rich medium, which allowed plasmids to be lost during cell division. Whereas the ARS plasmid was lost only slightly more rapidly in the mutant than in the wild-type control, the centromeric plasmid was lost far faster than normal, with 50% loss within ∼4 generations versus ∼25 generations for wild-type strains. Specifically, the YCplac22 plasmid was lost at a rate of 2% (±0.7%) per generation in MHY500 wild-type cells and 13% (±0.8%) per generation in MHY1380 (ulp2Δ) cells. To measure rates of chromosome loss, we used a variation of a quantitative mating assay described previously (21). Chromosome III loss rates were nearly 10-fold higher in ulp2Δ than in wild-type cells [(2.2 ± 0.6) × 10−7 versus (1.5 ± 0.6) × 10−6 chromosome loss events/viable cell]. The magnitude of this effect was comparable to that observed with the mad checkpoint mutants (21). Thus, stable maintenance of chromosomes and centromeric plasmids in yeast depends on Smt3 deconjugation by Ulp2.
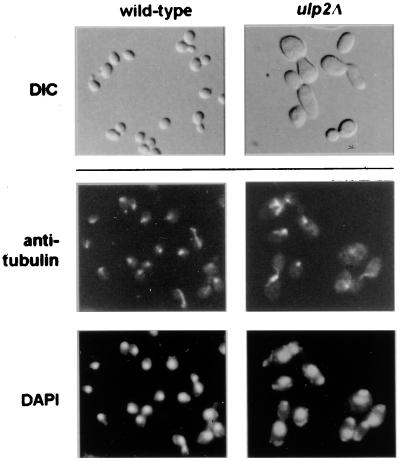
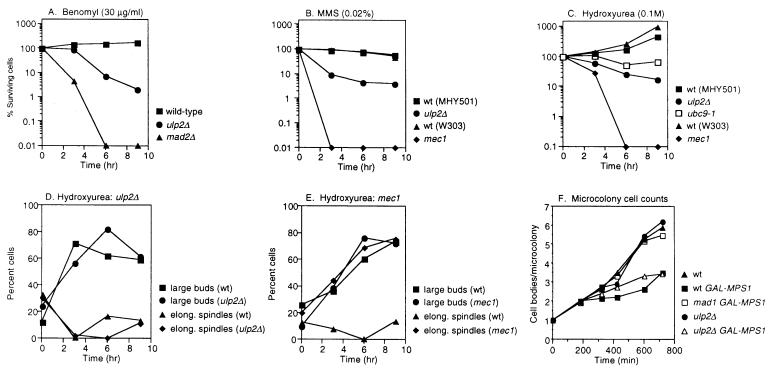
The high rates of chromosome loss in ulp2Δ cells correlated with aberrant mitotic spindle morphology. We examined microtubule organization by antitubulin immunofluorescent staining (Fig. 4). The spindles seen in ulp2Δ cells frequently had thick bars of staining or brightly staining microtubule clusters. Consistent with these data, we found that the ulp2Δ mutant failed to grow on low concentrations of the microtubule-depolymerizing drug benomyl (Fig. 5C). If the mutant cells were incubated with 15 μg of benomyl/ml, very few spindles could be detected, unlike similarly treated wild-type cells (not shown). These results suggest that the benomyl sensitivity of the ulp2Δ strain is due to a defect in microtubule or spindle assembly. This defect might underlie the enhanced rates of chromosome loss in the mutant.
FIG. 4.
Cell morphology of ulp2Δ cells. Wild-type and congenic ulp2Δ cells grown at 30°C were viewed with Nomarski optics (DIC). Mutant cells were enlarged relative to the wild type, frequently had elongated buds, and occasionally formed daisy chains. Microtubules were visualized by antitubulin immunofluorescence, and nuclei were visualized by DAPI fluorescence.
Sensitivity of ulp2Δ cells to hydroxyurea and to DNA damage.
The ulp2Δ strain grew poorly at 30°C and could not form colonies at 37°C (Fig. 4B and 5B). Flow cytometric analysis of logarithmic cultures grown at 30°C or shifted for 3 h to 37°C (or of α-factor-synchronized cultures released at 37°C and assayed at various times thereafter) did not suggest a unique cell cycle arrest point. However, the profiles were much flatter and broader than those of wild-type cells, which could reflect both chromosome segregation and replication defects (not shown). The mutant cells also frequently displayed elongated buds (Fig. 4). Buds can become elongated when cells fail to switch from apical to isotropic bud growth, a switch that requires activation of the Cdc28 cell cycle kinase by G2/M cyclins (20). A delay in S-phase entry, for example, can lead to elongated buds. We assayed the sensitivity of ulp2Δ cells to hydroxyurea, an inhibitor of DNA replication, at a concentration that slows but does not completely block replication. The mutant strain was found to be strongly sensitive to the drug (Fig. 5D).
Human Ubc9 and SUMO-1 have been isolated in two-hybrid interaction screens by their ability to bind to the RAD51 and RAD52 DNA repair proteins (18, 34). Moreover, deletion of the fission yeast homologs of Ubc9 and Aos1, which are expected to be required for Smt3-protein ligation, results in much greater sensitivity of cells to DNA-damaging agents (1, 33). Therefore, we asked whether impaired Smt3-protein deconjugation by either Ulp1 or Ulp2 might also result in hypersensitivity to different types of DNA damage. The ulp1ts strain was slightly less resistant than wild-type cells to UV or γ radiation or to DNA alkylation by MMS; the ulp2Δ mutant was sensitive to γ rays and, to a lesser extent, to UV and MMS (Fig. 5E to G). For comparison, we also exposed a ubc9-1 mutant (32) to benomyl, hydroxyurea, UV, and MMS. This Smt3-conjugating enzyme mutant grew poorly on hydroxyurea and was moderately sensitive to UV and benomyl but was not strongly sensitive to the concentrations of MMS tested (not shown [but see Fig. 9C]). Thus, both Smt3-protein conjugation and Smt3-protein deconjugation are required for full resistance of cells to DNA damage, hydroxyurea, and benomyl.
FIG. 9.
Smt3 processing and Smt3-protein conjugates in ulp mutants. (A) Suppression of ulp2Δ temperature sensitivity by catalytically defective Ulp1 mutants. (B) Smt3-protein conjugates in ulp mutants at 30°C. The strains used were MHY1614 to MHY1617. (C) Reciprocal suppression of ulp2Δ and ubc9-1 growth defects. (D) Provision of additional mature Smt3 does not impair the suppression of benomyl sensitivity in the ulp2Δ ulp1ts mutant. The YPD plates included 100 μM CuSO4 to induce His6-Smt3 expression from the YRTAG310-His6-SMT3-gg plasmid.
Analysis of checkpoint function in ulp2Δ mutants.
Cell cycle progression involves an interdependent series of steps. For instance, chromosome segregation normally does not occur until the completion of DNA replication. The proper order of these events requires cellular surveillance or checkpoint functions (7, 29). DNA damage, incomplete DNA replication, or an incompletely assembled mitotic spindle all normally cause a delay in chromosome separation and a transient arrest as large-budded cells, which allows time for the defects to be corrected. In checkpoint mutants, failure to arrest results in rapid loss of viability when the cells divide. DNA replication, DNA damage, and spindle assembly checkpoint mutants are hypersensitive to hydroxyurea, DNA-damaging agents, and benomyl, respectively. Hence, from the analysis described above, it was possible that the ulp2Δ mutant was defective in one or more of these checkpoints.
We assessed the functions of all of these checkpoints in ulp2Δ cells, and in each case it appeared that checkpoint arrest occurred normally but that the cells were impaired for recovery from the arrest. Using a visual assay to follow multiplication of single cells into microcolonies in the presence of 15 μg of benomyl/ml, we found that ulp2Δ cells arrested with large buds with the same kinetics as wild-type cells, but unlike the wild type, most of the mutant cells (92%) never resumed division and remained arrested as large-budded cells. Consistent with this, exposure of the ulp2Δ strain to benomyl for up to 9 h caused a substantial but relatively moderate loss of viability (Fig. 6A) in comparison to a mad2Δ spindle checkpoint mutant (29), which already showed severely reduced viability by 6 h. Hence, mitotic arrest appears to occur normally in ulp2Δ cells but recovery from arrest is defective either because the mutant is impaired in the checkpoint recovery process per se or because its microtubules are hypersensitive to the drug (or both).
FIG. 6.
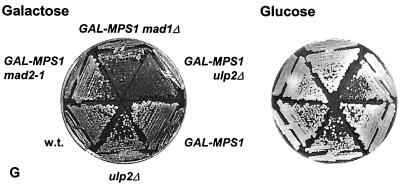
Evaluation of checkpoint function in the ulp2Δ mutant. (A) Growth of cells after incubation for the indicated times in benomyl. The treated cells were spread on YPD plates and incubated at 30°C. Survival was calculated as the percentage of colonies formed from each time point sample relative to the number counted at the outset of the treatment for each strain. The strains used were MHY501, MHY1380, and MHY1628. (B) Growth of cells after incubation in MMS. The ulp2Δ and ubc9-1 mutants are congenic with MHY501, and the mec1 mutant MHY1621 is congenic with W303a. wt, wild type. (C) Growth of cells after incubation in hydroxyurea. (D) Normal block to spindle elongation in ulp2Δ cells exposed to 0.1 M hydroxyurea. Cell budding and spindles were evaluated from micrographs of anti-tubulin-stained cells. The mean numbers of cells counted for each time point were 51 and 68 for MHY501 (wild type) and MHY1380 (ulp2Δ), respectively. (E) Abnormal spindle elongation in the mec1 checkpoint mutant evaluated as for panel D. The mean numbers of cells counted for each time point were 56 and 44 for W303a (wild type) and MHY1621 (mec1), respectively. (F) Average number of cell bodies (cells plus buds) observed in microcolonies that initiated from single cells of the indicated genotype, which were isolated from an α-factor synchronized cell population. Strains were isogenic except as indicated (11). Plates contained 2% galactose. (G) Inhibition of colony formation of ulp2Δ cells overexpressing MPS1.
If ulp2Δ cells were unable to recover properly from activation of the spindle checkpoint, rather than simply being impaired for spindle assembly under conditions that enhance the depolymerization of microtubules, then the mutant should also be sensitive to activation of the checkpoint when spindle assembly is not perturbed. This can be achieved by overproduction of the Mps1 kinase, which phosphorylates the Mad1 checkpoint protein and temporarily arrests wild-type cells in mitosis with morphologically normal spindles (11). Deletion of ULP2 in cells overproducing Mps1 resulted in severe growth impairment similar to that seen with mad1Δ cells but unlike congenic ulp2Δ cells with normal levels of Mps1 or wild-type cells overproducing Mps1 (Fig. 6G). Unlike mad mutants, however, which are defective for activating checkpoint arrest, ulp2Δ cells arrested cell division in response to overexpressed Mps1 as large-budded cells in a manner similar to wild-type cells, based on microcolony assays (Fig. 6F). These data are consistent with the possibility that the Ulp2 enzyme is required for normal recovery from or adaptation to activation of the spindle checkpoint arrest.
We evaluated the DNA damage checkpoint in ulp2Δ cells exposed to MMS using a microcolony assay (not shown) or plating on YPD following different times of exposure to MMS (Fig. 6B). The kinetics of arrest as single, large-budded cells were similar to those of the wild type, but the mutant cells failed to resume cell division. Transient exposure of a ulp2Δ (or ubc9) strain to MMS led to a significant but relatively modest drop in colony-forming ability compared to that of a rad6Δ DNA repair mutant (not shown) or a mec1 checkpoint mutant (Fig. 6B). We conclude that loss of Ulp2 does not prevent DNA damage-induced cell cycle arrest.
Finally, when ulp2Δ cells were exposed to 0.1 M hydroxyurea for as long as 9 h, relatively little loss of viability was observed, unlike mec1 cells, which are also defective for the DNA replication checkpoint (Fig. 6C). As with wild-type cells, the mutant accumulated as large-budded cells, and few elongated spindles were detected by quantitative analysis of cells by indirect immunofluorescent detection of microtubules (Fig. 6D). By contrast, the mec1 mutant underwent spindle elongation even without full DNA replication (Fig. 6E). These results argue against a requirement for Ulp2 in DNA replication checkpoint arrest, but the enzyme may be important for recovery after the checkpoint is activated. The defect in the resumption of cell division for ulp2Δ cells exposed to agents that activate distinct cell cycle checkpoints suggests that adaptation to or recovery from these different checkpoints may involve a common desumoylation-dependent step(s).
Smt3-protein conjugates in ulp2Δ cells.
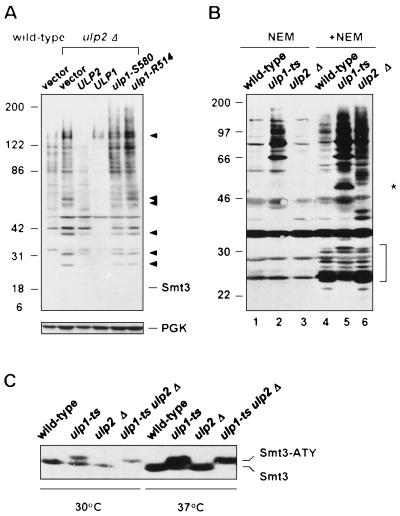
To begin to examine what biochemical changes underlie the phenotypic differences between the ulp1 and ulp2 mutants, we assessed the accumulation of Smt3-protein conjugates as well as Smt3 precursor processing. Mutant ulp2Δ cells amassed specific, high-molecular-mass Smt3-protein conjugates (Fig. 7A). The pattern of conjugates was distinct from that which accumulated in ulp1ts cells (Fig. 7B, lanes 4 to 6, and Fig. 8B). This supported the inference of distinct substrate specificities for these enzymes suggested by the disparate phenotypes of the two mutants. Nevertheless, when Ulp1 was overproduced in an ulp2Δ strain, the levels of most of the Smt3 conjugates were strongly reduced, indicating that Ulp1 at high concentration can cleave conjugates that are normally acted on primarily or exclusively by Ulp2. Overproduced, catalytically impaired Ulp1 proteins did not cause the same reduction in ulp2Δ cell-specific Smt3 species (Fig. 7A).
FIG. 7.
Smt3-protein conjugates in ulp2Δ cells. (A) Effect on Smt3-protein conjugates of overproduction of Ulp2 or Ulp1. vector, pRS424; all alleles are in pRS424. The arrowheads indicate prominent Smt3 conjugates that accumulate in ulp2Δ cells. Samples were run on an SDS–7.5 to 18% polyacrylamide gradient gel followed by anti-Smt3 immunoblotting. Free Smt3 was not detected under the blotting conditions used. (B) Cleavage of Smt3-protein conjugates in yeast extracts. Spheroplasts were incubated at 37°C for 30 min and then lysed with (+) or without (−) NEM. After 30 min at 30°C, 90 μg of protein from each sample was loaded on a 10 to 15% gradient gel, followed by immunoblot analysis. The asterisk and bracket denote Smt3 conjugates from ulp1ts cells that, unlike most of the conjugates, disappeared during in vitro incubation without NEM. (C) Smt3-precursor processing analyzed by anti-Smt3 immunoblotting with enhanced chemiluminescent detection. The strains used were MHY1614 to MHY1617 that expressed SMT3 from a plasmid-borne CUP1 promoter.
FIG. 8.
Localization of Ulp1 and Ulp2. (A) Anti-myc immunoblot analysis of myc9-tagged Ulps. Lane 1, untagged control MHY500 cells. (B) Indirect immunofluorescence localization of tagged Ulps. The nuclei were visualized with DAPI.
Interestingly, when whole-cell lysates from either ulp2Δ or ulp1ts cells were incubated at 30°C for 30 min to allow cleavage of the mutant cell-specific Smt3-protein conjugates by the remaining endogenous Ulp, very different results were obtained with the two mutants (Fig. 7B). If ulp1ts extracts were incubated in the absence of NEM, only limited changes in the pattern or intensity of Smt3-protein conjugates were observed compared to what occurred with the same extracts treated with NEM to inactivate all Ulps (Fig. 7B, compare lanes 2 and 5). In contrast, incubation of an ulp2Δ lysate without NEM led to nearly complete elimination of most of the ulp2Δ cell-specific Smt3 conjugates (lane 3). Therefore, Ulp1 could cleave ulp2Δ cell-specific Smt3 conjugates when Ulp1 was present at endogenous levels in a cell lysate. This was also true when Ulp1 was overproduced in vivo (Fig. 7A) or was added to an NEM-treated ulp2Δ lysate (Fig. 2B). Ulp2, on the other hand, showed a much more modest ability to cleave ulp1 cell-specific Smt3 conjugates in analogous experiments.
Marked differences between the two mutants were also seen for the processing of the Smt3 precursor, Smt3-ATY (Fig. 7C). Smt3-ATY was expressed from a high-copy-number expression vector to supplement endogenous levels of unconjugated protein because very little free Smt3 is normally detectable (16, 22). Whereas all detectable free Smt3 was found in the mature form in both wild-type and ulp2Δ cells, much of the free Smt3 in ulp1ts cells still retained the C-terminal tripeptide, particularly at 37°C. However, Ulp2 is able to process the Smt3 precursor in vivo, albeit inefficiently relative to Ulp1. This was inferred from a comparison between the ulp1ts strain and an ulp2Δ ulp1ts double mutant (Fig. 7C): the residual activity in the ulp1ts single mutant at restrictive temperature was no longer detectable in the double mutant (a small amount of mature Smt3 is seen at 30°C in longer exposures). Measurements of in vitro processing of a purified 35S-labeled Smt3-HA fusion in extracts from these same mutants were consistent with these results; processing was rapid in both wild-type and ulp2Δ lysates but was not detectable over a 30-min chase in ulp1ts extracts (not shown).
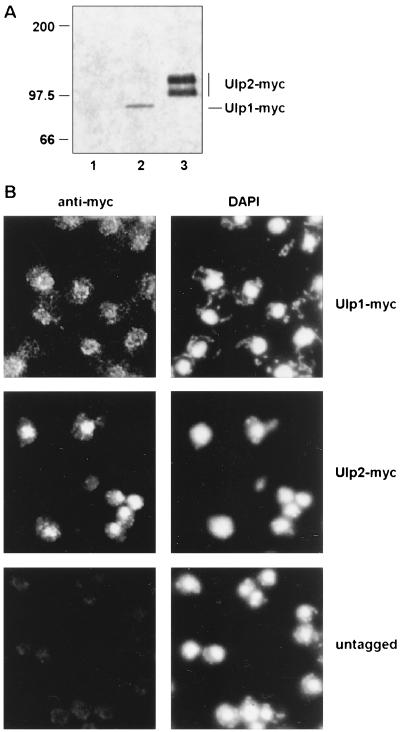
Based on immunoblot analysis of identically epitope-tagged proteins expressed from chromosomally integrated alleles (Fig. 8A), Ulp1 appeared to be present at much lower levels than Ulp2. Moreover, Ulp1 was concentrated in the nuclear periphery-nuclear envelope region, based on immunofluorescent staining (or GFP fluorescence), while Ulp2 localized throughout the nucleus (Fig. 8B). These differences in level and localization of the two yeast Ulps could account at least in part for their distinct in vivo substrate selectivities.
Suppression of ulp2Δ by mutation of ULP1.
ULP1 and ULP2 have nonredundant functions, based on the strong phenotypic defects of the corresponding single mutants. We tested whether either gene, when placed on a high-copy-number plasmid, could compensate for loss of the other gene. No growth of ulp1Δ cells carrying a high-copy-number ULP2 plasmid was detected (22). In contrast, when ulp2Δ cells carrying the high-copy-number pRS424-ULP1 plasmid were placed at 37°C, a temperature that is normally lethal to the mutant, colonies were able to form, albeit at a lower rate than with wild-type cells (Fig. 3B). This finding is in accord with the ability of high-copy-number ULP1 to suppress the accumulation of Smt3-protein conjugates in ulp2Δ cells (Fig. 7A). Little or no suppression of the slow growth or irregular colony size at 30°C of the ulp2Δ mutant was observed, however. Surprisingly, equal or better suppression of ulp2Δ temperature, benomyl, and hydroxyurea sensitivities was also seen with high-copy-number ulp1 alleles encoding catalytically defective Ulp1 proteins (Fig. 9A and not shown) despite their failure to reduce bulk ulp2Δ cell-specific Smt3 conjugates (Fig. 7A).
The suppression of the ulp2Δ growth defect at 37°C by overproduced, inactive Ulp1 might be due to the Ulp1 protein binding to and thereby inactivating a growth-inhibitory Smt3-protein conjugate(s) that accumulates in the ulp2Δ mutant. An analogous suppression of E. coli lon mutant defects by high levels of catalytically inactive derivatives of the Lon protease has been documented (37). However, an alternative explanation is suggested by results with an ulp1ts ulp2Δ double mutant. We anticipated that loss or reduced activity of both Smt3-cleaving enzymes from the same cell would cause more severe phenotypic abnormalities than were observed for either single mutant. Remarkably, the opposite was observed: each mutation suppressed defects associated with the other (Fig. 4). For example, neither single mutant was able to form colonies at 37°C, but weak growth of the double mutant was observed at that temperature (Fig. 4B), indicating reciprocal suppression. For all conditions tested, the ulp1ts ulp2Δ mutant grew better than the ulp2Δ single mutant, and in the presence of hydroxyurea, the double mutant also grew better than the ulp1ts mutant. In this light, the suppression of ulp2Δ by high levels of catalytically defective Ulp1 protein may work by dominant-negative interference with the endogenous wild-type Ulp1 enzyme.
Paralleling the reciprocal phenotypic suppression observed with the ulp1ts ulp2Δ strain was a marked drop in the levels of Smt3-protein conjugates in the double mutant relative to either single mutant (Fig. 9B). These results suggested that cells might require a balance between Smt3-conjugating and -deconjugating activities and that there is a feedback mechanism that limits Smt3 conjugation when Smt3 cleavage rates are severely impaired. Consistent with this possibility, when UBC9, the gene encoding the Smt3-conjugating enzyme, was mutated in ulp2Δ cells, the benomyl sensitivity of the single mutants was strongly suppressed (Fig. 9C). The ulp2Δ ubc9-1 double mutant also grew slightly better than either single mutant at 37°C and on hydroxyurea (Fig. 9C and not shown), indicating reciprocal suppression similar to that observed with ulp2Δ ulp1ts cells.
How might reduced Ulp function inhibit Smt3-protein ligation? Reduced Ulp activity might somehow limit synthesis of Smt3 precursor. Pulse-labeling experiments suggested that there was little or no difference in the rate of Smt3 synthesis in wild-type, ulp2Δ, ulp1ts, and ulp2Δ ulp1ts cells, at least when SMT3 was expressed from the CUP1 promoter (which was necessary to detect free Smt3 reliably) (not shown). A more obvious possibility is that Ulp activity in the double mutant reduces the rate of Smt3 precursor processing to the point where mature Smt3 levels become limiting for conjugation. As discussed earlier, precursor processing was indeed more severely impaired in the double mutant (Fig. 7C). If reduced Smt3 precursor processing fully accounted for the phenotypic recovery of the ulp2Δ ulp1ts mutant, provision of additional mature Smt3 to the double mutant should again lead to an imbalance between Smt3-protein conjugation and deconjugation, with a concomitant loss in cell function. This was not observed. Overexpression of mature Smt3, which was verified by immunoblot analysis, did not make the double mutant grow more poorly on benomyl (Fig. 9D).
Another possibility is that a component(s) of the Smt3 ligation machinery, such as Ubc9 or an E3-like factor, is positively regulated by Ulps. For example, such factors may be susceptible to inhibitory Smt3 automodification reactions that are reversed by Ulp action. Alternatively, active Smt3 might be depleted from the cell by reaction with abundant cellular nucleophiles, such as glutathione (although no increase in Smt3 species close to the size of free Smt3 was detected). Analogous reactions occur in vitro with ubiquitin and can be reversed by Dubs (28). To begin to address these ideas, we have examined Smt3-protein conjugates in cells overproducing mature Smt3 (not shown). The ulp2Δ ulp1ts double mutant actually accumulated significantly more bulk Smt3-protein conjugates under these conditions than did wild-type cells, but because of the severely impaired deconjugation capacity of the double mutant, more Smt3 might get trapped in conjugates even if conjugation rates were strongly reduced. The exact pattern of conjugates was different from that of the ulp single mutants, so the level of a specific growth-inhibitory conjugate(s) might remain suppressed under these conditions. Because these results would be most easily reconciled with inhibition of specific E3-like factors, we currently favor the autoinhibition model, but the data do not rule out Smt3 depletion by small nucleophiles or other more complicated models.
DISCUSSION
Ulp1 and Ulp2 specifically hydrolyze peptide and isopeptide bonds between the ubiquitin-like Smt3 and SUMO-1 modifiers and their substrates. These enzymes represent a novel class of cysteine proteases that is unrelated to any known deubiquitinating enzyme. While both Ulp1 and Ulp2 are Smt3-cleaving enzymes, the phenotypic consequences of deleting either of them are different. Ulp1 has an essential role in the cell cycle and is the major Smt3 precursor-processing enzyme in the cell. Deletion of ULP2 results in a diverse set of aberrations, but the gene is not essential. Striking differences in Smt3-protein profiles correlate with these differences in cellular phenotype. Most remarkably, simultaneous mutation of both Smt3-cleaving enzymes suppresses defects associated with either single mutant, and this appears to be due to inhibition of Smt3-protein ligation in the double mutant. These results point to the importance of a dynamic balance between Smt3 addition to and removal from substrate proteins for multiple cellular processes.
The Ulps and related cysteine proteases.
The enzymes responsible for ubiquitin activation (E1) and conjugation (E2) and the analogous enzymes for Smt3 and SUMO-1 and for the Ubl called Rub1 or NEDD8 are clearly related in primary sequence (14). The ubiquitin and Ubl conjugation systems, like the modifier proteins themselves, presumably evolved from a set of common ancestral proteins. Thus, it was surprising to find that the desumoylating enzymes are unrelated to any Dubs but instead are distantly related to the adenovirus processing protease (reference 22 and the present study). All the key catalytic residues of the viral protease are conserved in the Ulps. The finding that Ulp2, which shows minimal similarity to Ulp1 outside the UD, is also an Smt3-specific protease strongly supports the idea that Ulps utilize a catalytic mechanism very similar to that of the adenovirus proteases and suggests that the UD is largely, if not wholly, responsible for Smt3 recognition.
Several eubacterial proteins have limited similarity to the core region of the UD. In E. coli, the product of an uncharacterized cistron called elaD (GenBank no. 1381662) is related to this protease region. The genome of the eubacterium Chlamydia trachomatis, a sexually transmitted intracellular pathogen, is predicted to encode a pair of proteases (GenBank no. AE001359 and AE001360) distantly related to yeast Ulps. These observations suggest that proteins with the proposed cysteine protease core found in the Ulps existed prior to the divergence of eubacteria from eukaryotes or were acquired by horizontal gene transfer. We have not yet found any archaeal gene products bearing the UD core sequence signature. In contrast to the Ulp-related viral and eubacterial proteins, no viral or prokaryotic gene product with clear primary sequence similarity to any known Dub has yet been detected.
Are there enzymes without a UD that are capable of cleaving Smt3-linked substrates at physiologically significant rates? For yeast, at least, the answer is likely to be no. We infer this primarily from the fact that ulp2Δ ulp1ts double mutants exhibit almost no detectable Smt3 precursor processing at the restrictive temperature. In our original screen, we also did not isolate any other Smt3-cleaving enzymes. It remains conceivable, however, that an enzyme with a very restricted substrate specificity exists that was not detectable under any of our experimental conditions.
Substrate specificity and regulation of Ulps.
Despite the broad specificity of Ulp1 in vitro, the enzyme expressed at its normal levels in vivo is unable to compensate for loss of Ulp2, and Ulp2, even in high copy numbers, cannot suppress the lethality of the ULP1 deletion. Loss of either Ulp in vivo results in the accumulation of distinct Smt3-containing species. This indicates that in the cell, Ulps are regulated in ways that limit their activity toward particular Smt3-protein conjugates.
What determines the distinct substrate selectivities of the two yeast Ulps in vivo? Our data indicate that Ulp1 and Ulp2 localize differently and are probably also both posttranslationally modified or processed (Fig. 8A and not shown); these features and/or binding to other cellular factors may contribute to their in vivo specificity. Interestingly, we find that low levels of detergent stimulate Ulp2 activity. This treatment might induce conformational changes that permit substrate attack; analogous changes in the Ulp2 enzyme might occur in a regulated fashion in the cell. Ulp1, which appears to be present at much lower concentrations than Ulp2 in vivo, has a broad and robust ability to cleave Smt3 and SUMO-1-linked substrates in vitro (22), whereas Ulp2 cleavage of several tested substrates, including the natural Smt3 precursor, was slow. It is therefore possible that the Ulp2 enzyme is more substrate selective and we have not yet found its preferred in vivo targets. A caveat is that the recombinant Ulp2 may be defective in some way, e.g., it fails to fold properly, or our in vitro conditions are not ideal. However, the analysis of Smt3 precursor processing in yeast suggests that at least for this simple substrate, Ulp1 has much greater activity than Ulp2 in vivo as well.
Functions of the Smt3-cleaving enzymes.
It is not yet known how sumoylation of a protein changes its functional properties. One possibility is that the ability of the modified protein to interact with other proteins is altered. The correlation between SUMO-1 attachment to RanGAP1 or PML and particular subcellular distributions of these proteins (see the introduction) supports this idea. Sumoylation could also alter a protein's susceptibility to other types of modification. For instance, IκBα becomes resistant to ubiquitination when it is linked to SUMO-1. A striking finding of the present work which has significant implications for our understanding of SUMO function is that a balance between SUMO modification and demodification is critical for normal cell function. This may be so because the ratio of modified to unmodified forms of one or more target proteins is an important parameter determining a particular functional state. It is also possible that different sites of Smt3 modification on a protein have different effects, analogous to the activating and inhibitory phosphate additions to targets such as the CDK1 kinase. Alternatively, the timing of Smt3 attachment to and removal from proteins may be coupled to other dynamic cellular events.
Deletion of ULP2 results in a variety of phenotypic abnormalities. This probably reflects the loss of Ulp2 activity toward a number of different Smt3-modified substrates, based on the array of Smt3 conjugates observed in the mutant; however, some of these traits may trace to the failure to deconjugate Smt3 from a single protein that regulates or is regulated by several cellular processes. It is possible, for instance, that multiple defects result from abnormal regulation of mitotic spindle assembly or kinetochore attachment. The ulp2Δ mutant has pronounced defects in maintaining chromosomes, frequently has morphologically abnormal spindles, and is sensitive to microtubule-depolymerizing drugs. A defect in the CENP-C-related centromere-binding protein called Mif2 can be suppressed by overexpression of Smt3 or Ulp2 (25), and the Smt3-conjugating enzyme Ubc9 can bind yeast centromere proteins (15). It is also possible that a common step in the resumption of cell division following a transient cell cycle arrest in response to DNA damage, replication inhibition, or spindle disassembly might involve Smt3 deconjugation from a spindle or kinetochore component or from a mitotic regulator.
Control of Smt3 ligation by Smt3-cleaving enzymes.
The reciprocal suppression of the ulp1ts and ulp2Δ phenotypic defects was unexpected. We traced at least a component of this unusual genetic phenomenon to suppression of Smt3-protein ligation in the double mutant. Even in the presence of excess mature Smt3, reciprocal suppression continues to be observed, indicating that impaired processing of Smt3 precursor cannot fully account for the effect. While a fraction of this extra Smt3 becomes ligated to other proteins in the double mutant, the pattern of conjugates is distinguishable from that of the ulp1ts and ulp2Δ single mutants. The most straightforward interpretation is that a reduction in the levels of one or more specific Smt3-protein conjugates is necessary to relieve the growth defects of the single mutants, and Ulp1 and Ulp2 may both positively regulate a component(s) of the Smt3 ligation pathway (perhaps including Smt3 itself, by reversal of side reactions). Another possibility is that the growth enhancement of the ulp2Δ ulp1ts strain is due at least in part to the two yeast Ulps working in opposing physiological pathways, e.g., in nuclear import versus nuclear export. The subcellular localization of the two Ulps is not inconsistent with roles in nuclear transport.
A major challenge in understanding the functions of the SUMO system will be to identify the targets of this dynamic modification and to determine how sumoylation affects their functions. The present work indicates that protein sumoylation affects a broad range of cellular processes and is subject to regulation by Ulps. Use of the ulp mutants should greatly facilitate the identification of Smt3-modified proteins, a prerequisite to a full mechanistic understanding of the SUMO system.
ACKNOWLEDGMENTS
We thank Robert Cohen for ubiquitin aldehyde, Mark Winey for GAL-MPS1 strains and helpful advice, Steve Kron and Ray Deshaies for strains and plasmids, Nels Elde for constructing the ULP2-myc9 strain, and Jeff Laney and Nels Elde for critical reading of the manuscript.
This work was supported by NIH grant GM53756.
REFERENCES
- 1.al-Khodairy F, Enoch T, Hagan I M, Carr A M. The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J Cell Sci. 1995;108:475–486. doi: 10.1242/jcs.108.2.475. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989. [Google Scholar]
- 3.Bayer P, Arndt A, Metzger S, Mahajan R, Mechior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 4.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 5.Desterro J M, Rodriguez M S, Kemp G D, Hay R T. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 6.Desterro J M P, Rodriguez M S, Hay R T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 7.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 9.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 10.Haas A L, Siepmann T J. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 11.Hardwick K G, Weiss E, Luca F C, Winey M, Murray A W. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 12.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 13.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser M. There's the Rub: a novel ubiquitin-like modification involved in cell cycle regulation. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 15.Jiang W, Koltin Y. Two-hybrid interaction of a human UBC9 homolog with centromere proteins of Saccharomyces cerevisiae. Mol Gen Genet. 1996;251:153–160. doi: 10.1007/BF02172913. [DOI] [PubMed] [Google Scholar]
- 16.Johnson E S, Schwienhorst I, Dohmen R J, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson P R, Hochstrasser M. SUMO-1: ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- 18.Kovalenko O V, Plug A W, Haaf T, Gonda D K, Ashley T, Ward D C, Radding C M, Golub E I. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. Proc Natl Acad Sci USA. 1996;93:2958–2963. doi: 10.1073/pnas.93.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laney J D, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 20.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, Murray A W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 22.Li S-J, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 23.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 24.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meluh P B, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papa F, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 28.Rose I A, Warms J V. An enzyme with ubiquitin carboxy-terminal esterase activity from reticulocytes. Biochemistry. 1983;22:4234–4237. doi: 10.1021/bi00287a012. [DOI] [PubMed] [Google Scholar]
- 29.Rudner A D, Murray A W. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- 30.Saitoh H, Pu R T, Dasso M. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh H, Sparrow D B, Shiomi T, Pu R T, Nishimoto T, Mohun T J, Dasso M. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr Biol. 1998;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 32.Seufert W, Futcher B, Jentsch S. A ubiquitin-conjugating enzyme involved in the degradation of both S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 33.Shayeghi M, Doe C L, Tavassoli M, Watts F Z. Characterisation of Schizosaccharomyces pombe rad31, a UBA-related gene required for DNA damage tolerance. Nucleic Acids Res. 1997;25:1162–1169. doi: 10.1093/nar/25.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Z, Pardington-Purtymun P E, Comeaux J C, Moyzis R K, Chen D J. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics. 1996;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- 35.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi Y, Iwase M, Konishi M, Tanaka M, Toh-e A, Kikuchi Y. Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem Biophys Res Commun. 1999;259:582–587. doi: 10.1006/bbrc.1999.0821. [DOI] [PubMed] [Google Scholar]
- 37.Van Melderen L, Gottesman S. Substrate sequestration by a proteolytically inactive Lon mutant. Proc Natl Acad Sci USA. 1999;96:6064–6071. doi: 10.1073/pnas.96.11.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson K D, Hochstrasser M. The deubiquitinating enzymes. In: Peters J-M, Horris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 99–125. [Google Scholar]