Abstract
The aim of this study was to apply molecular epidemiology, antimicrobial surveillance, and PK/PD analysis to guide the antimicrobial treatment of gonococci infections in a region of the north of Spain. Antibiotic susceptibility testing was performed on all isolates (2017 to 2019, n = 202). A subset of 35 isolates intermediate or resistant to at least two antimicrobials were selected to search for resistance genes and genotyping through WGS. By Monte Carlo simulation, we estimated the probability of target attainment (PTA) and the cumulative fraction of response (CFR) of the antimicrobials used to treat gonorrhea, both indicative of the probability of treatment success. In total, 2.0%, 6.4%, 5.4%, and 48.2% of the isolates were resistant to ceftriaxone, cefixime, azithromycin, and ciprofloxacin, respectively. Twenty sequence types were identified. Detected mutations were related to antibiotic resistance. PK/PD analysis showed high probability of treatment success of the cephalosporins. In conclusion, multiple populations of N. gonorrhoeae were identified. We can confirm that ceftriaxone (even at the lowest dose: 250 mg) and oral cefixime are good candidates to treat gonorrhea. For patients allergic to cephalosporins, ciprofloxacin should be only used if the MIC is known and ≤0.125 mg/L; this antimicrobial is not recommended for empirical treatment.
Keywords: N. gonorrhoeae, antibiotic resistance, cephalosporins, azithromycin, pharmacokinetic/pharmacodynamic (PK/PD) analysis, whole-genome sequencing (WGS)
1. Introduction
Sexually transmitted infections (STIs) are a serious global health problem [1]. The prevalence of gonorrhea has increased significantly in recent years. The World Health Organization (WHO) estimates that there are 87 million new cases of gonorrhea each year [2].
In recent years, the most frequently prescribed antibiotics for the treatment of gonorrhea have become less effective, in part due to the inexorable progression of gonococcal antimicrobial resistance [3,4]. In 2018, the WHO reported the expansion of multidrug-resistant strains of N. gonorrhoeae worldwide [5]. In Europe, the Euro-GASP (depending on the European Centre for Disease Prevention and Control) provides important data at the European level on antimicrobial resistance, which are used to inform treatment guidelines [6]. Thus, the determination of antimicrobial susceptibility to identify isolates with less susceptibility and resistance to antimicrobial agents, and monitoring of strain populations via molecular techniques to identify gonococcal clones that are important for driving the transmission of multidrug-resistant gonococci, become crucial. Additionally, ensuring effective empirical therapy is also essential.
Current guidelines—such as the European guidelines for the diagnosis and treatment of gonorrhea in adults [7], and the Center for Disease Control and Prevention (CDC) Treatment Guidelines for Gonococcal Infection [8], both from 2020—recommend a dual therapy with extended-spectrum cephalosporins (ESCs, such as ceftriaxone or cefixime) and azithromycin for the treatment of uncomplicated gonorrhea when the antimicrobial susceptibility is unknown. Some guidelines recommend fluoroquinolones as an alternative treatment of pharyngeal infections if the isolate is known to be fluoroquinolone-susceptible and there are indications against using ceftriaxone—for instance, history of severe hypersensitivity to cephalosporins [7,9].
Recently, improved methods for the evaluation of the effects of pharmacokinetics (PK) and pharmacodynamics (PD) on treatment outcomes have become available for a number of other infections. In this regard, better data and new research on PK contributors to gonorrhea treatment outcomes are needed [10]. PK/PD analysis integrates information about the concentration of the drug that reaches the infection site and induces the therapeutic response, and the susceptibility of the pathogen to the antibiotic, expressed as the minimum inhibitory concentration (MIC). This allows researchers or clinicians to select the optimal antibiotic and dosing regimen for each infectious process and patient in order to enhance the effect of the antibiotic, minimizing the incidence of side effects and the emergence of resistance [11].
The aim of this work was to apply molecular epidemiology—including whole-genome sequencing (WGS) information, antimicrobial surveillance data, and PK/PD analysis—to guide the antimicrobial treatment of gonococci infections.
2. Materials and Methods
2.1. N. gonorrhoeae Isolates
All isolates from 2017 to 2019 were collected at the Microbiology Service of the University Hospital of Araba (HUA). The hospital, located in the Basque Country (Spain), covers a population of ~400,000 inhabitants, and has 780 beds.
Non-duplicated N. gonorrhoeae isolates from patients attending to the Emergency, Infectious Disease, and Primary Care services were included in the study. Gonococcal isolation was performed on chocolate agar PolyViteX VCA and Chocolate agar PVX plates (bioMérieux, Marcy-l’Étoile, France). The suspected colonies were then identified by mass spectrometry using the MALDI-TOF MS (Microflex LT, Bruker-Daltonics, Bremen, Germany) methodology [12].
This study met the exemption criteria of the ethics committee of clinical research because the isolates analyzed were collected in routine practice, and did not allow the identification of patients.
2.2. Antibiotic Susceptibility Testing
Isolated strains were subjected to antibiotic susceptibility testing using the MIC gradient strip tests (Liofilchem, Roseto degli Abruzzi, Italy) according to the manufacturer’s instructions. The minimum inhibitory concentrations (MICs) of tetracycline, penicillin, cefixime, ceftriaxone, ciprofloxacin, and azithromycin were determined. From the MIC distribution data of all of the isolates, the calculation of MIC50 and MIC90—i.e., the lowest concentration of the antimicrobial capable of inhibiting 50% and 90% of the isolates, respectively—was performed. Resistance rates were calculated with reference to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [13] and the Clinical and Laboratory Standards Institute (CLSI) [14]. The current EUCAST MIC breakpoints for penicillin are ≤0.06 mg/L (susceptible) and >1 mg/L (resistant); for ceftriaxone and cefixime, ≤0.125 mg/L (susceptible) and >0.125 mg/L (resistant); for ciprofloxacin, ≤0.03 mg/L (susceptible) and >0.06 mg/L (resistant); and for tetracycline, ≤0.5 mg/L (susceptible) and >1 mg/L (resistant). For azithromycin, the EUCAST epidemiological cutoff (ECOFF) of 1 mg/L was used to identify non-wild-type (WT) isolates. CLSI breakpoints were the following: for penicillin, ≤0.06 mg/L (susceptible), 0.12–1 mg/L (intermediate), and ≥2 mg/L (resistant); for ceftriaxone and cefixime, ≤0.25 mg/L (susceptible); for azithromycin, ≤1 mg/L (susceptible); for ciprofloxacin, ≤0.06 mg/L (susceptible), 0.12–0.5 mg/L (intermediate), and ≥1 mg/L (resistant); and for tetracycline, ≤0.25 mg/L (susceptible), 0.5–1 mg/L (intermediate), and ≥2 mg/L (resistant).
2.3. WGS Molecular Epidemiology and Antimicrobial Resistance Determinants
A subset of 35 strains intermediate or resistant to at least two of the antimicrobials tested were selected to search for resistance genes and genotyping through WGS. The DNA of the strains was extracted using the NucleoSpin Tissue Kit (Macherey-Nagel, Duren, Germany) and sequenced using the Illumina MiSeq platform with the Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA, USA). The genomic information was received as small nucleotide sequence reads, and longer sequences as contigs. These sequence data were submitted to GenBank under the BioProject accession number PRJNA684048 (https://www.ncbi.nlm.nih.gov/sra/PRJNA684048, accessed on 10 December 2020). Subsequently, the assembled genomes of the isolates were input to the ResFinder 3.2 tool on the CGE website (https://cge.cbs.dtu.dk, accessed on 10 December 2020) for the identification of acquired antimicrobial resistance genes and chromosomal mutations. Default thresholds of 90% identity and 60% gene coverage were employed. Additionally, the PubMLST tool was employed to detect antimicrobial resistance determinants (https://pubmlst.org/neisseria/, accessed on 10 December 2020). Multilocus sequence typing (MLST) and N. gonorrhoeae multi-antigen sequence typing (NG-MAST) were performed on the sequences produced by WGS, and they were compared with existing alleles on the Neisseria MLST website (http://pubmlst.org/neisseria/, accessed on 10 December 2020) and NG-MAST (http://www.ng-mast.net/, accessed on 10 December 2020) schemes for the determination of allele numbers and sequence types (STs). All new alleles or STs were submitted to the NG-MAST or MLST website curators to be assigned an allelic number and ST. The genomic epidemiology of the isolates was determined based on whole-genome analysis. An SNP phylogenetic tree, based on FASTQ files, was generated by using the default settings of the MINTyper 1.0 server, available at https://cge.cbs.dtu.dk/services/MINTyper/ (accessed on 10 December 2020). The complete genome of isolate FA1090 (NC_002946.2) was used as a reference. Furthermore, a genome-based clustering was also performed using the TYGS platform, accessible at https://tygs.dsmz.de/ (accessed on 10 December 2020).
2.4. Pharmacokinetic/Pharmacodynamic (PK/PD) Analysis
In order to predict the probability of PK/PD target attainment, different antibiotics and dosing regimens were evaluated (Table 1).
Table 1.
Dose regimen, PK/PD index, and pharmacokinetic parameters used for simulations.
| Ciprofloxacin | Azithromycin | Ceftriaxone | Cefixime | |
|---|---|---|---|---|
| Dose regimen | 500 mg single dose, PO |
1 g, 2 g single dose, PO |
0.25, 0.5, 1 g single dose, IM 2 g/day, IV |
400 mg/day, PO |
| PK/PD index | AUC0–∞/MIC ≥125 |
AUC0–∞/MIC ≥59.5 |
fT>MIC ≥ 20 h (IM) fT>MIC ≥ 60% (IV) |
fT>MIC ≥ 60% |
| AUC0–∞ (mg h/L) | 10.7 ± 2.6 | |||
| CL/F (L/h) | 144 ± 39.5 | |||
| Ke (h−1) | 0.082 ± 0.029 | 0.204 ± 0.02 | ||
| Vd (L) | 14.70 ± 4.93 | 19 ± 0.03 | ||
| Fu | 0.05 | 0.35 | ||
| F | 1 | 0.42 ± 0.045 | ||
| Ka (h−1) | 1 | 0.55 | ||
| References | [15,16] | [17,18] | [19] | [19] |
IM: intramuscular; IV: intravenous; PO: oral; AUC0–∞: area under the plasma concentration vs. time curve from 0 to infinity; CL: clearance; F: bioavailability; Fu: unbound fraction; Ka: absorption constant rate; Ke: elimination constant rate; Vd: volume of distribution.
From the published pharmacokinetic parameters listed in Table 1 [15,16,17,18,19], and from the study sample MIC distribution, we estimated the probability of target attainment (PTA, defined as the probability that at least a specific value of a PK/PD index is achieved at a certain minimum inhibitory concentration) and calculated the cumulative fraction of response (CFR, defined as the expected population probability of target attainment for a specific drug dose and a specific population of microorganisms). PK/PD indices and values related to efficacy are also listed in Table 1. Five-thousand-subject Monte Carlo simulations with Oracle® Crystal Ball Fusion Edition v.11.1.2.3.500 (Oracle USA Inc., Redwood City, CA, USA) were used to estimate the PTA and CRF values, indicative of the probability of target success. PTA and CFR ≥ 80% but < 90% were associated with moderate probabilities of success, whereas a CFR ≥ 90% was considered optimal against that bacterial population. More detailed information about the PTA and CFR calculations is presented in the Supplementary Materials.
3. Results
3.1. N. gonorrhoeae Isolates and Antibiotic Susceptibility Testing
A total of 202 isolates of N. gonorrhoeae were collected in the study period (2017–2019). Of these, 85% were collected from men, and 15% from women. In men, the urethral exudate was the most common sample (87%); other locations were anal exudate (11%) and pharyngeal exudate (2%). Endocervical (64%) and vaginal exudate (33%) were the most common among women, and anal exudate (1%) was also detected. The average age was 31 years (range 14–67) and 33 years (age range 16–61) for men and women, respectively. A total of 10 patients had N. gonorrhoeae-positive cultures in two anatomical sites (7 pharynx and rectum, 3 pharynx and urethra), but only one isolate per patient was included in the susceptibility study.
Table 2 shows the antimicrobial susceptibility of N. gonorrhoeae isolates. The higher resistance rates were obtained for ciprofloxacin and tetracycline. Eleven isolates were resistant to azithromycin (5.4% of the total), all of which were collected in 2019. Resistant isolates had MICs of 1.5 mg/L (n = 5), 2 mg/L (n = 5), and 16 mg/L (n = 1). The percentage of isolates resistant to cephalosporins varied depending on the EUCAST and CLSI clinical breakpoints: for cefixime, resistance rates were 6.4% (EUCAST) and 1.6% (CLSI), and for ceftriaxone they were 2.0% (EUCAST) and 0.5% (CLSI). The MIC distribution of the six antibiotics is presented in Figure S1.
Table 2.
Antimicrobial susceptibility of 202 N. gonorrhoeae isolates collected in 2017–2019 (n = 202).
| Antimicrobial | MIC Range (mg/L) | MIC50 (mg/L) |
MIC90 (mg/L) |
R (%) EUCAST |
R (%) CLSI |
|---|---|---|---|---|---|
| Penicillin | 0.002–64 | 0.19 | 3 | 12.6 | 12.6 |
| Cefixime | 0.008–0.75 | 0.008 | 0.047 | 6.4 | 1.6 |
| Ceftriaxone | 0.002–0.5 | 0.008 | 0.094 | 2.0 | 0.5 |
| Azithromycin | 0.016–16 | 0.19 | 0.75 | 5.4 a | 5.4 |
| Ciprofloxacin | 0.015–64 | 0.012 | 8 | 48.2 | 45.2 |
| Tetracycline | 0.9->256 | 1 | 32 | 34.2 | 34.2 |
MIC: minimum inhibitory concentration; MIC50 and MIC90: minimum inhibitory concentration at which 50% and 90% of the isolates were inhibited, respectively; R: percentage of resistant isolates. a: According to the epidemiological cutoff (ECOFF) value.
3.2. Genotyping: MLST and NG-MAST
MLST genotyping identified multiple distinct populations of N. gonorrhoeae in the isolates of HUA in 2017–2019 (Table 3). Overall, a total of 20 MLST STs were identified among the 35 isolates analyzed, with ST-9363 being the most common (n = 8), followed by ST-7363 (n = 4) and ST-7822 (n = 3), while 14 STs were represented by a single isolate. Two new MLST STs (ST-14274 and ST-14304) were previously unreported, and resulted from combinations of known alleles. NG-MAST further discriminated between strains with identical MLST-STs, and 27 different NG-MAST STs were observed among the 35 isolates analyzed. ST-6765 predominated (n = 7), ST-470 and ST-13070 were observed in two isolates, and the remaining STs were represented by single isolates. Additionally, six NG-MAST STs (17.1%) were previously unreported, and resulted from new combinations of known alleles.
Table 3.
Genotypes, MICs, and antimicrobial resistance determinants for the 35 Neisseria gonorrhoeae isolates sequenced by WGS.
| Genotype | Susceptibility (mg/L) | Chromosomal Mutations | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Year | MLST | NG-MAST | P | CRO | CFM | TET | CIP | AZM | Adquired Gene | PBP2 | PBP1 | PorB | GyrA | ParC | S10 | mtrR Promoter a | MtrR | 23S rRNA |
| 2 | 2017 | 10,314 | 12,547 | 0.25 | 0.016 | nd | 0.5 | 3 | 0.125 | - | Type V non-mosaic | L421P | - | S91F, D95A | S87R | V57M | - | A39T | - |
| 3 | 2017 | 1588 | 3750 | 0.38 | 0.023 | nd | 64 | 8 | 0.125 | tet(M) | Type XIX non-mosaic | L421P | G120K A121G |
S91F, D95A | S87R | V57M | - | A39T | - |
| 6 | 2017 | 1596 | 19,728 | 0.125 | 0.008 | nd | 0.125 | 0.006 | 0.25 | - | Type 81 semi Mosaic | - | - | - | - | - | - | - | - |
| 9 | 2017 | 1583 | 19,729 | 0.125 | 0.006 | nd | 0.25 | 0.004 | 0.125 | - | Type II non-mosaic | L421P | - | - | - | - | -35A Del | - | - |
| 11 | 2017 | 14,274 | 587 | 0.19 | 0.008 | nd | 0.5 | 0.006 | 0.75 | - | Type II non-mosaic | - | - | - | - | V57M | G | - | - |
| 17 | 2017 | 1587 | 13,971 | >32 | 0.012 | nd | 24 | 1.5 | 0.094 | blaTEM-1B, tet(M) | Type II non-mosaic | - | - | S91F, D95A | D86N | V57M | - | - | - |
| 19 | 2017 | 10,935 | 15,728 | 2 | 0.006 | nd | 24 | 0.003 | 0.032 | blaTEM-1B, tet(M) | Type XIV non-mosaic | - | - | - | - | V57M | - | - | - |
| 24 | 2017 | 8145 | 19,730 | 2 | 0.004 | nd | 0.5 | 0.004 | 0.125 | bla TEM-1B | Type XIV non-mosaic | - | - | - | - | V57M | - | - | - |
| 31 | 2017 | 7363 | 11,547 | 0.5 | 0.094 | nd | 1 | >32 | 0.094 | - | Type X mosaic | L421P | G120N A121G |
S91F, D95N | S87R, S88P | V57M | - | - | - |
| 33 | 2017 | 7363 | 13,070 | 0.75 | 0.125 | nd | 1 | >32 | 0.25 | - | Type X mosaic | L421P | G120N A121G |
S91F, D95N | S87R, S88P | V57M | - | - | - |
| 35 | 2017 | 8143 | 14,306 | 0.125 | 0.008 | nd | 1 | 8 | 0.125 | - | Type II non-mosaic | - | S91F, D95A | S87R | V57M | - | A39T | - | |
| 50 | 2018 | 14,304 | 13,070 | 0.5 | 0.125 | nd | 1 | >32 | 0.125 | - | Type X mosaic | L421P | G120N A121G |
S91F, D95N | S87R, S88P | V57M | - | - | - |
| 51 | 2018 | 7363 | 9184 | 0.38 | 0.064 | nd | 1.5 | 8 | 0.25 | - | Type IX non-mosaic | L421P | G120K A121D |
S91F, D95G | E91G | V57M | -35A Del | - | - |
| 52 | 2018 | 1901 | 1407 | 0.75 | 0.25 | nd | 3 | >32 | 0.5 | - | Type XXXIV mosaic | L421P | G120K A121N |
S91F, D95G | S87R | V57M | -35A Del | - | - |
| 56 | 2018 | 10,890 | 1407 | 0.5 | 0.125 | nd | 4 | >32 | 0.5 | - | Type XXXIV mosaic | L421P | G120K, A121N | S91F, D95G | S87R | V57M | -35A Del | - | - |
| 58 | 2018 | 1901 | 19,111 | 0.5 | 0.125 | nd | 2 | >32 | 0.5 | - | Type XXXIV mosaic | L421P | G120K, A121N | S91F, D95G | S87R | V57M | -35A Del | - | - |
| 59 | 2018 | 1583 | 217 | >32 | 0.006 | nd | 64 | 16 | 0.064 | blaTEM-1B, tet(M) | Type II non-mosaic | - | G120K, A121D | S91F, D95G | D86N | V57M | - | G45D | - |
| 61 | 2018 | 7363 | 15,198 | 0.25 | 0.064 | nd | 2 | 8 | 0.25 | - | Type IX non-mosaic | L421P | - | S91F, D95G | E91G | V57M | -35A Del | - | - |
| 7244 | 2018 | 11428 | 2992 | 0.025 | 0.008 | <0.016 | 0.25 | 0.016 | 0.75 | - | Type II non-mosaic | - | - | - | - | V57M | - | A39T | - |
| 8661 | 2018 | 15,573 | 17,371 | 0.125 | 0.012 | 0.016 | 2 | 0.016 | 1 | - | Type II non-mosaic | - | G120K, A121N | - | - | V57M | G | - | - |
| 293 | 2019 | 9363 | 6765 | 0.19 | 0.012 | <0.016 | 2 | 0.012 | 1 | - | Type II non-mosaic | - | G120K, A121N | - | - | V57M | G | - | - |
| 1941 | 2019 | 1580 | 470 | 0.125 | 0.03 | 0.016 | 2 | 0.002 | 1.5 | - | Type 93 semi Mosaic | - | A121S | - | - | V57M | - | G45D | C2599T |
| 1943 | 2019 | 1580 | 470 | 0.125 | 0.008 | <0.016 | 2 | 0.002 | 1.5 | - | Type 93 semi Mosaic | - | A121S | - | - | V57M | - | G45D | C2599T |
| 3023 | 2019 | 9363 | 6765 | 0.25 | 0.016 | <0.016 | 1.5 | 0.008 | 2 | - | Type II non-mosaic | - | G120K, A121N | - | - | V57M | G | - | - |
| 3526 | 2019 | 9363 | 6765 | 0.19 | 0.023 | <0.016 | 3 | 0.008 | 0.75 | - | Type II non-mosaic | - | G120K, A121N | - | - | V57M | G | - | - |
| 3569 | 2019 | 9363 | 6765 | 0.38 | 0.012 | <0.016 | 2 | 0.016 | 2 | - | Type II non-mosaic | - | G120K, A121N | - | - | V57M | G | - | - |
| 3700 | 2019 | 11,706 | 17,972 | 0.19 | 0.003 | <0.016 | 1.5 | 2 | 0.75 | - | Type V non-mosaic | L421P | - | S91FD95A | S87R | V57M | - | A39T | - |
| 4458 | 2019 | 13,292 | 9208 | 0.094 | 0.004 | nd | 2 | 0.003 | 0.75 | - | Type II non-mosaic | - | - | - | - | V57M | - | A39T | C2611T |
| 4515 | 2019 | 9363 | 6765 | 0.38 | 0.016 | 0.032 | 1 | 0.008 | 1.5 | - | Type II non-mosaic | - | G120K, A121N | - | - | V57M | G | - | - |
| 4726 | 2019 | 9363 | 6765 | 0.19 | 0.016 | <0.016 | 3 | 0.008 | 1 | - | Type II non-mosaic | - | G120K, A121N | - | - | V57M | G | - | - |
| 5903 | 2019 | 7822 | 14,994 | 0.25 | 0.023 | <0.016 | 1 | 4 | 1 | - | Type V non-mosaic | L421P | - | S91F, D95A | S87R | V57M | G | A39T | - |
| 7079 | 2019 | 9363 | 6765 | 0.125 | 0.008 | <0.016 | 1.5 | 0.012 | 1.5 | - | Type II non-mosaic | - | G120K, A121N | - | - | V57M | G | - | - |
| 7181 | 2019 | 9363 | 19,731 | 0.19 | 0.016 | <0.016 | 3 | 0.004 | 1.5 | - | Type II non-mosaic | - | G120K, A121N | - | - | V57M | G | - | - |
| 7789 | 2019 | 7822 | 14,994 | 0.125 | 0.012 | <0.016 | 0.75 | 2 | 1 | - | Type V non-mosaic | L421P | S91F, D95A | S87R | V57M | G | A39T | - | |
| 9051 | 2019 | 7822 | 14,994 | 0.38 | 0.016 | <0.016 | 1.5 | 6 | 1.5 | - | Type V non-mosaic | L421P | - | S91F, D95A | S87R | V57M | G | A39T | - |
P: penicillin; CRO: ceftriaxone; CFM: cefixime; TET: tetracycline; CIP: ciprofloxacin; AZ: azithromycin; nd: no data available; -: wild type. a: G, deletion of one G (guanine), 20 nucleotides downstream of where the -35A deletion occurs.
3.3. Antimicrobial Resistance Determinants
Genomic markers associated with resistance to ciprofloxacin, tetracycline, β-lactams, and azithromycin were found (Table 3). All of the ciprofloxacin-resistant strains investigated presented two substitutions in GyrA and one or two substitutions in ParC. At GyrA, they presented substitutions in positions 91 (all presented an S91F) and 95 (D95A/G/N). The main amino acid substitution observed in ParC was S87R (n = 13), either alone, or associated with S88P. Other substitutions included D86N and E91G.
The tetM gene was detected in the four highly tetracycline-resistant strains (MIC ≥ 24 mg/L). The V57M substitution in the ribosomal protein S10 subunit encoded by the rpsJ gene was detected in 33/35 (94.3%) of the strains investigated, irrespective of their tetracycline susceptibility. Chromosomal mutations in the porB gene were also found in tetracycline-resistant strains. The G120K substitution was detected in 14 isolates, and was associated with A121G/N/D.
The only sequenced isolate resistant to ceftriaxone (MIC: 0.25 mg/L) presented the penA mosaic allele XXXIV associated with an L421P substitution in PBP1, a G120K/A121N in the PorB, and an adenine deletion (-35A Del) in the mtrR promoter.
Twenty-five isolates showed different degrees of penicillin resistance. The four high-level penicillin-resistant strains (n = 4; MIC ≥ 2 mg/L) carried the blaTEM-1B gene, which encodes the TEM-1 β-lactamase; these four strains were β-lactamase producers, as determined by the nitrocefin test. The remaining isolates (n = 21) presented an MIC range between 0.19 and 0.75 mg/L. Nineteen isolates were associated with non-mosaic penA alleles, with non-mosaic II being the most common (n = 10); the remaining six isolates were associated with mosaic penA alleles (mosaic X and XXXIV, n = 3 each). The major amino acid substitution observed in the PBP1 protein was Leu421Pro (n = 13), which was correlated with a degree of intermediate resistance. Simultaneous substitutions at amino acids G120 and A121 in the PorB porin—associated with decreased intake of several antimicrobials—were observed in 16 penicillin-resistant isolates, with the substitutions of G120K/A121N being the most common (n = 10).
The 13 isolates with an MIC of 1–2 mg/L against azithromycin were associated with mutations in the promoter region and coding sequence of the mtrR genes, and with mutations in the gene encoding the 23S rRNA. Two isolates presented the C2599T mutation in the 23S rRNA gene associated with a G45D substitution in the MtrR protein. A deletion of one guanine (G), 20 nucleotides downstream of the -35A deletion, was observed in the mtrR promoter of 11 isolates; their sequences showed a 98.48% identity with that of strain 38,194 (GenBank accession number KT954125), and 86.36% with strain FA1090 (GenBank accession number NC_002946). The -35A deletion in the repeated sequence of the mtrR promoter was not observed in the resistant isolates; however, it was observed in six susceptible isolates (MIC, <0.5 mg/L) as the only mechanism related to azithromycin resistance. In the MtrR protein, the amino acid substitutions of A39T (n = 3) and G45D (n = 2) were detected in azithromycin-resistant strains. The alignment of partial sequences is presented in Supplementary Figure S2.
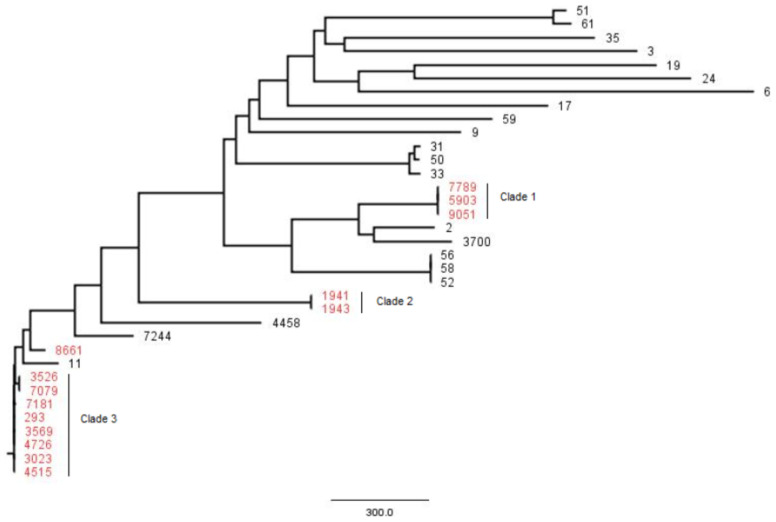
Whole-genome SNP-based phylogenetic analysis of the 35 sequenced isolates revealed a great genetic dissimilarity between isolates, with differences ranging from 0 to 2378 SNPs; however, three clades involving non-azithromycin-susceptible isolates were identified (Figure 1). Clade 1 consisted of three isolates belonging to MLST ST-7822, with an Ala39Thr substitution in the MtrR gene and a deletion of one G, 20 nucleotides downstream of where the -35A deletion occurs; no SNP differences were observed. Clade 2 consisted of two isolates belonging to MLST ST-1580, with the C2599T mutation in the 23S rRNA gene and a Gly45Thr substitution in the MtrR gene; no SNP differences were observed. Clade 3 consisted of eight closely related isolates belonging to MLST ST-9363, with a deletion of one G, 20 nucleotides downstream of where the -35A deletion occurs; differences between these isolates ranged from 0 to 20 SNPs. The same three clusters were obtained when the phylogenetic tree was constructed from the whole genome (data not shown).
Figure 1.
Clustering of the 35 N. gonorrhoeae sequences obtained by WGS, based on genome SNPs; red indicates non-azithromycin-susceptible isolates.
3.4. Pharmacokinetic/Pharmacodynamic (PK/PD) Analysis
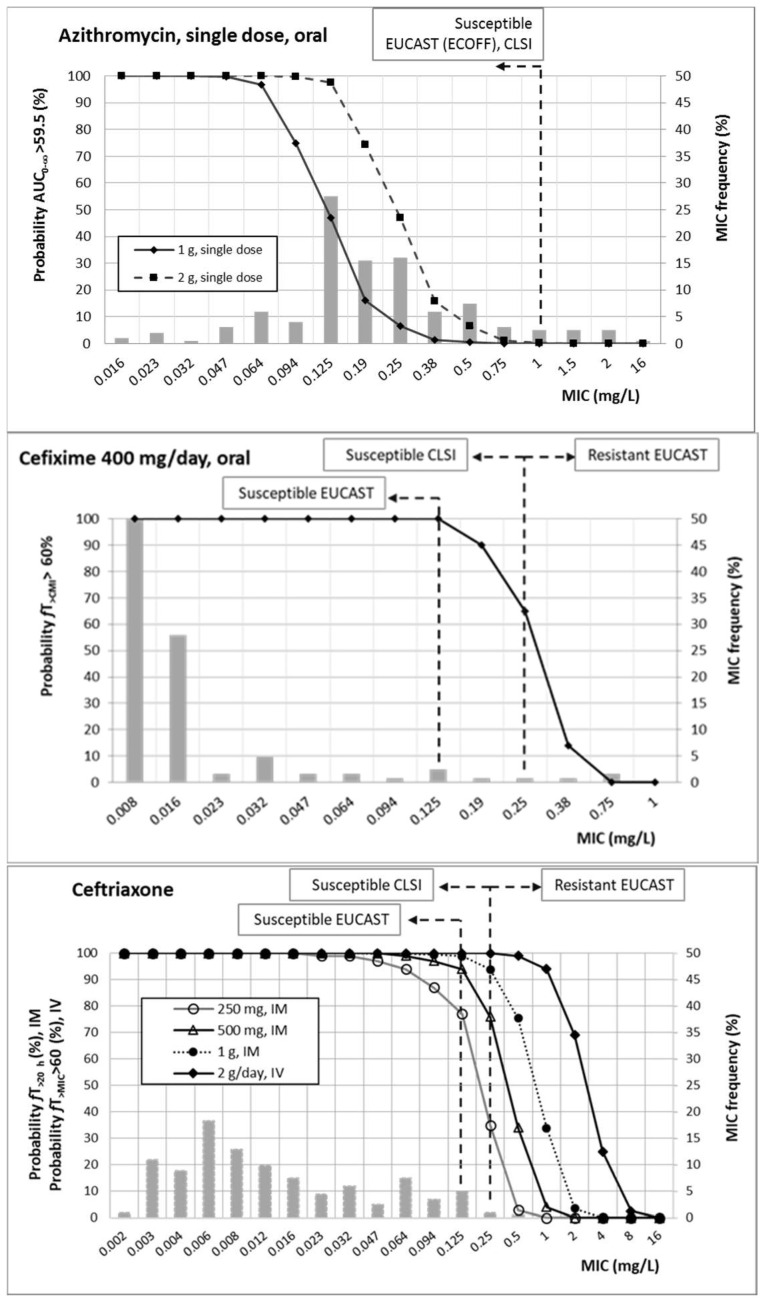
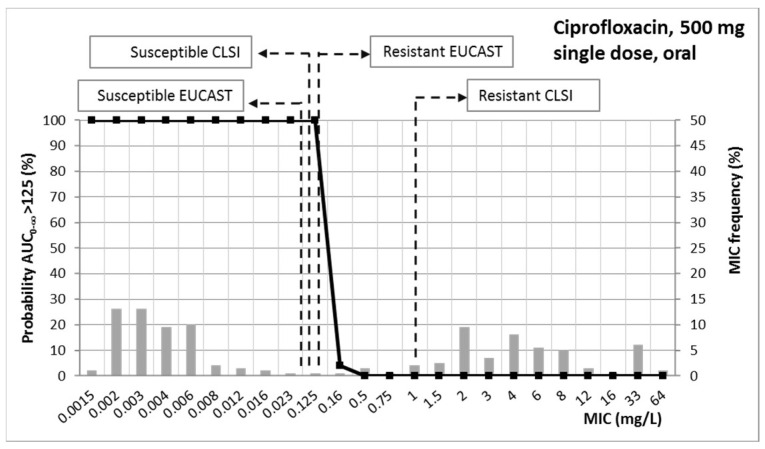
Figure 2 shows the probability of target attainment (PTA) values of all of the antimicrobial agents studied, as well as their MIC distribution. On the basis of the simulation results, and taking into account the targets for the different antimicrobial agents, PTA values equal to or higher than 90% were achieved with (1) azithromycin at single doses of 1 g and 2 g, for MICs up to 0.064 and 0.125 mg/L, respectively; (2) ceftriaxone at 250, 500, and 1000 g, administered intramuscularly as single doses, for MICs up to 0.064, 0.125, and 0.25 mg/L, respectively, and for MICs equal to or lower than 1 mg/L for 2 g/day via the intravenous route; (3) cefixime at 400 mg/day by oral route, for MICs up to 0.125 mg/L; and (4) single oral doses of ciprofloxacin at 500 mg, for MICs up to 0.125 mg/L.
Figure 2.
Probability of target attainment (PTA) of azithromycin, ceftriaxone, cefixime, and ciprofloxacin. Grey bars: MIC frequency.
Table 4 features the CFR values obtained after the PK/PD analysis and Monte Carlo simulations. Values higher than 90% were only achieved with the two cephalosporins.
Table 4.
Cumulative fraction of response (CFR) calculated for all antibiotics and dosing regimens.
| Antibiotic | Dosing Regimen | CFR (%) |
|---|---|---|
| Ciprofloxacin | 500 mg, single dose, PO | 51 |
| Azithromycin | 1 g single dose, PO | 33 |
| 2 g single dose, PO | 64 | |
| Ceftriaxone | 1 g, single dose, IM | 100 |
| 500 mg, single dose, IM | 98 | |
| 250 mg, single dose, IM | 96 | |
| 2 g/day, IV | 100 | |
| Cefixime | 400 mg/day, PO | 97 |
PO: oral administration; IM: intramuscular; IV: intravenous.
In supplementary Tables S2–S9, the mean, median, and 2.5% and 97.5% confidence intervals of PTA and CFR for every antimicrobial and dosing regimen are presented.
4. Discussion
To address the antimicrobial resistance problem, more efficient means of evaluating therapies for gonorrhea are needed. Our purpose in this study was to apply molecular epidemiology—including whole-genome sequencing (WGS) information, antimicrobial surveillance data, and PK/PD analysis—to guide the antimicrobial treatment of gonococci infections in the Basque Country (Spain).
In this region of the north of Spain, the percentage of resistance was of the same order as recent data obtained from Madrid [20] and Barcelona [21,22], and also of those reported in other European countries [23]. As expected, the highest percentage of resistance was detected for ciprofloxacin (>45%). The historical resistance of gonococci to ciprofloxacin is very well known, and it has been related to the incidence of gonorrhea at the population level. In fact, ciprofloxacin is no longer recommended for treatment of this STI, except for pharyngeal infections if susceptibility has been proven or if the patient is allergic to β-lactams [9].
In spite of the differences in the resistance breakpoint between EUCAST [13] and CLSI [14] for ciprofloxacin (0.125 and 1 mg/L, respectively), the percentage resistance we obtained was similar, since very few isolates are affected by the discrepancies in the breakpoints (Figure 1). However, discrepancies in the clinical breakpoint for cephalosporins (0.25 mg/L from EUCAST and 0.5 mg/L from CLSI) justify the difference in the percentage resistance for cefixime (6.5% vs. 1.6%) and ceftriaxone (2.0% vs. 0.5%). Unlike the other antibiotics, all isolates resistant to azithromycin were collected in 2019 (n = 11). These data are consistent with the recent increase in the resistance of N. gonorrhoeae to azithromycin in Spain and worldwide [22,23]. Some studies support the notion that the selection/induction of azithromycin resistance in N. gonorrhoeae may be associated with the general use of azithromycin for the treatment of respiratory infections or non-gonococcal urethritis [24]. According to the susceptibility profile, azithromycin and cephalosporins are the most active antimicrobials. These results are in agreement with the European guidelines, which recommend the use of a dual therapy with ceftriaxone and azithromycin for all gonorrhea cases [7].
Antibiotic susceptibility data were consistent with those obtained through WGS, and the mutations detected were related to resistance to the studied antibiotics. The genomic study detected the main antimicrobial resistance determinants associated with resistance to β-lactams (penA, porB, ponA, mtrR, and blaTEM1-B), ciprofloxacin (gyrA and parC), and tetracycline (tetM and rpsJ). All ciprofloxacin-resistant strains presented two substitutions in GyrA, together with substitutions in the ParC protein, while among the ciprofloxacin-susceptible isolates, no mutations in the gyrA and parC genes were detected, as previously reported [25]. Regarding ceftriaxone—and as expected—the mutations found in the analyzed resistant isolates were associated with reduced susceptibility or resistance to ESC [26]. Tetracycline resistance in N. gonorrhoeae may be related to the presence of the plasmid-mediated TetM protein (MIC, 16–64 mg/L), or to mutations in some chromosomal genes (MIC, 2–4 mg/L) [27,28]. In our study, the tetM gene was detected only in high-level resistant isolates; meanwhile, low-level resistant isolates only presented mutations in the chromosomal genes penB (PorB), rpsJ (S10), and mtrR and its promoter. These isolates presented at least two or three mutated genes, suggesting that more than one antimicrobial resistance determinant is needed to confer resistance.
Resistance to β-lactams, macrolides, and tetracycline may result from overexpression of the MtrCDE efflux pump related to mutations in the promoter and/or coding regions of the mtrR repressor, which causes not only resistance, but also an increase in the MIC [25]. The -35A deletion in the promoter sequence—the most common mutation—was detected only in isolates recovered in 2017 and 2018, but it was not observed in those from 2019. Several mutations—including insertions, deletions, or transversions—have been described in this promoter region [29]. Related to azithromycin resistance, a guanine (G) deletion in this promoter was observed in most of the azithromycin-resistant isolates, either alone or associated with the A39T substitution in MtrR repressor. In some isolates, this mutation was associated with substitutions in the MtrD protein (data not shown), so the exact contribution of this G deletion to antimicrobial resistance should be determined.
The majority of genotyped azithromycin-resistant isolates were MLST ST-9363. In a recent study carried out in Barcelona (Spain) [22], this genotype was associated with outbreaks of N. gonorrhoeae with high-level azithromycin resistance (MIC ≥ 256 mg/L) between 2016 and 2018. In that study, this genotype was found mainly in men who have sex with men. Contrary to Barcelona isolates, our ST-9363 isolates were not high-level azithromycin resistant, and their MIC values were 1 and 2 mg/L. The phylogenetic analysis of both the SNPs and the complete genome using the WGS showed similar results and, together with the application of the other epidemiological markers and antibiotic resistance, allowed the epidemiological characterization of the strains analyzed. The joint analysis of the data obtained from the WGS allowed the detection of three clusters that involved eight, three, and two non-azithromycin-susceptible isolates, respectively, all of them isolated in 2019 and with an MIC ≥ 1 mg/L to azithromycin.
Among the interventions to mitigate the current and future impact of antimicrobial resistance, the development of new generations of antimicrobials is one of the most accepted. Another recognized strategy to diminish antibiotic resistance is the optimization of the dosing regimen of available antimicrobials. PK/PD analysis and Monte Carlo simulation have shown to be very useful tools to select adequate antibiotic dosages, with the goal of increasing treatment efficacy and reducing the risk of multidrug-resistant pathogens [30]. Although integrated PK/PD analysis has been applied to identify changes in the antimicrobial activity of antibiotics [31,32], until now it has not been applied to gonorrhea for optimizing dosage and correlating exposure with clinical outcomes [33]. Therefore, by using Monte Carlo simulation, we evaluated the probability of PK/PD target attainment by MIC (PTA), as well as taking into account the MIC distribution (CFR), which is indicative of treatment success.
At EUCAST susceptibility breakpoints, all antibiotics except for azithromycin and the lowest dose of ceftriaxone provided high (>90%) probability of treatment success (Figure 2). Moreover, the highest doses of ceftriaxone also showed adequate results for higher MICs (1 g IM would cover an MIC of 0.25 mg/L, and 2 g/day IV would cover an MIC of 1 mg/L). Regarding azithromycin in monotherapy, 1 g would be an option only when the MIC is 0.064 mg/L or lower, and 2 g if the MIC is equal to or lower than 0.125 mg/L.
Empirical treatments with antimicrobial drugs (i.e., without susceptibility testing) provide progressive development of decreasing susceptibility, which threatens personal and public health. The estimation of CFR is a very useful tool to guide empirical treatments, since it allows us to know which antimicrobial and dose regimen would have the best likelihood of success to treat bacterial isolates from a particular hospital or geographical area. Considering the MIC distribution of our isolates, CFR values—indicative of the probability of empirical treatment success (Table 4)—allow us to confirm that ceftriaxone (even at the lowest dose: 250 mg) and oral cefixime are good candidates to empirically treat gonorrhea (CFR around 100%) in our geographical area. On the other hand, ciprofloxacin should not be used empirically. For both ESCs and ciprofloxacin, the probability of treatment success was correlated with the susceptibility rate. However, the susceptibility rate of azithromycin was much higher than the estimated CFR. This discrepancy may be explained in part by the fact that, in contrast to β-lactams—which do not accumulate within the cell (extracelluar: intracellular ratio of 1)—azithromycin achieves intracellular concentrations several-fold higher than in plasma and extracellular fluid [34]. Taking into account that N. gonorrhoeae presents intracellular localization, the treatment success of azithromycin may be underestimated.
Our results are consistent with the European guidelines for the diagnosis and treatment of gonorrhea in adults [7], and with the CDC [8], both of which recommend ceftriaxone monotherapy for the treatment of uncomplicated gonorrhoeae. Because N. gonorrhoeae remains highly susceptible to ceftriaxone, and azithromycin resistance is increasing, prudent use of antimicrobial agents supports limiting their use. In spite of that, continuous monitoring for the emergence of resistance to this cephalosporin is necessary in order to ensure the efficacy of the recommended regimens. In this sense, surveillance programs and health care provider reports of treatment failures are essential.
For empirical treatment, the WHO recommends that the selected gonorrhea treatment should have ≥95% probability of being effective—that is, 5% or fewer of gonococcal isolates are likely to be resistant to the antimicrobial used for first-line empirical treatment [35]. In our opinion, PK/PD analysis must be also considered; in this sense, the selection of the most adequate antibiotic and dosing regimen should be based on local epidemiology. Moreover, PK/PD analysis results are also beneficial when the MIC value is known, and in the case of ceftriaxone, would allow the selection of lower doses. In previous studies, PK/PD analysis has been shown to be a useful tool for the surveillance of antimicrobial activity, as a complement to the simple assessment of MIC values [32,36,37,38]. Thus, it could be applied to guide therapy along with surveillance programs of gonococcal resistance. In this sense, the surveillance of gonococcal antimicrobial resistance could be implemented with PK/PD analysis by using the susceptibility data collected by the Euro-GASP.
This study presents some limitations. First, PK/PD analysis was performed by using the mean and standard deviation of the PK parameters, without considering the potential influence of covariates on the PK profile; second, our simulations are based on plasma concentrations and, therefore, dosing recommendations may be more beneficial for gonococcal bacteremia than for extragenital infection.
In spite of these limitations, our study represents a good example of how to combine molecular epidemiology, antimicrobial surveillance, and PK/PD analysis to provide quality data to support treatment guidelines—especially empirical treatments.
5. Conclusions
In our geographical area, multiple distinct populations of N. gonorrhoeae were identified. We can confirm that ceftriaxone (even at the lowest dose: 250 mg) and oral cefixime are good candidates to treat gonorrhea. For patients allergic to β-lactam antibiotics, or with pharyngeal infections, ciprofloxacin should be only used if the MIC is known and is equal to or lower than 0.125 mg/L; this antimicrobial is not recommended to be used empirically.
Acknowledgments
We thank Irati Miguel from SGIKER, UPV/EHU, Spain, and Javier Gamboa from Biogenetics, Spain, for their technical advice and the performance of the sequencing procedure.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pharmaceutics13101699/s1, Figure S1: MIC frequency of all antimicrobials studie. Figure S2: Alignment of partial sequences of the mtrR promoter from Neisseria gonorrhoeae FA1090 (GenBank accession number NC_002946), Neisseria gonorrhoeae 38197 (GenBank accession number KT954125), and some N. gonorrhoeae isolates from this study. The -10 and -35 promoter sequences for the mtrR gene are shown in overlines. The 13-bp inverted repeat (IR) is underlined. Lowercase letters represent the single-base-pair deletion (-35A Del) within the IR. *—location of the single base-pair deletion (G) 20 nucleotides downstream of where the -35A deletion occurs; Table S1: Dose regimen, PK/PD index, and pharmacokinetic parameters used for simulations; Table S2: Probability of target attainment (PTA) and cumulative fraction of response (CFR) of ciprofloxacin, 500 mg, single dose, oral administration; Table S3: Probability of target attainment (PTA) and cumulative fraction of response (CFR) of azithromycin, 1 g, single dose, oral administration; Table S4: Probability of target attainment (PTA) and cumulative fraction of response (CFR) of azithromycin, 2 g, single dose, oral administration; Table S5: Probability of target attainment (PTA) and cumulative fraction of response (CFR) of ceftriaxone, 250 mg, single dose, intramuscular administration; Table S6: Probability of target attainment (PTA) and cumulative fraction of response (CFR) of ceftriaxone, 500 mg, single dose, intramuscular administration; Table S7: Probability of target attainment (PTA) and cumulative fraction of response (CFR) of ceftriaxone, 1 g, single dose, intramuscular administration; Table S8: Probability of target attainment (PTA) and cumulative fraction of response (CFR) of ceftriaxone, 2 g/day, intravenous administration; Table S9: Probability of target attainment (PTA) and cumulative fraction of response (CFR) of cefixime, 400 mg/day, oral administration.
Author Contributions
Conceptualization, R.A., A.R.-G., J.G. and A.C.; data curation, A.R.-A., A.A.-Q., A.A. and I.M.-B.; formal analysis, R.A., I.M.-B. and A.R.-G.; funding acquisition, A.R.-G. and J.G.; investigation, R.A., J.G. and A.C.; methodology, R.A., A.A.-Q., A.A. and I.M.-B.; project administration, A.C.; supervision, A.R.-G. and J.G.; validation, A.C.; writing—original draft, R.A., A.R.-G. and A.C.; writing—review and editing, R.A., A.R.-A., A.A.-Q., A.A., A.R.-G., I.M.-B., J.G. and A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of the Basque Country UPV/EHU (GIU20/048; PA20/03), Spain.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest in the interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fasciana T., Capra G., Di Carlo P., Calà C., Vella M., Pistone G., Colomba C., Giammanco A. Socio-Demographic Characteristics and Sexual Behavioral Factors of Patients with Sexually Transmitted Infections Attending a Hospital in Southern Italy. Int. J. Environ. Res. Public Health. 2021;18:4722. doi: 10.3390/ijerph18094722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization, WHO . Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- 3.Chesson H.W., Kirkcaldy R.D., Gift T.L., Owusu-Edusei K., Jr., Weinstock H.S. Ciprofloxacin resistance and gonorrhea incidence rates in 17 cities, United States, 1991–2006. Emerg. Infect. Dis. 2014;20:612–619. doi: 10.3201/eid2004.131288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hook E.W., 3rd, Kirkcaldy R.D. A Brief History of Evolving Diagnostics and Therapy for Gonorrhea: Lessons Learned. Clin. Infect. Dis. 2018;67:1294–1299. doi: 10.1093/cid/ciy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization, WHO . Report on Global Sexually Transmitted Infection Surveillance. World Health Organization; Geneva, Switzerland: 2018. [(accessed on 3 August 2021)]. Available online: https://www.who.int/reproductivehealth/publications/stis-surveillance-2018/en/ [Google Scholar]
- 6.Cole M.J., Quinten C., Jacobsson S., Amato-Gauci A.J., Woodford N., Spiteri G., Unemo M., Euro-GASP Network The European gonococcal antimicrobial surveillance programme (Euro-GASP) appropriately reflects the antimicrobial resistance situation for Neisseria gonorrhoeae in the European Union/European Economic Area. BMC Infect. Dis. 2019;19:1040. doi: 10.1186/s12879-019-4631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unemo M., Ross J., Serwin A.B., Gomberg M., Cusini M., Jensen J.S. Background review for the ‘2020 European guideline for the diagnosis and treatment of gonorrhoea in adults’. Int. J. STD AIDS. 2021;32:108–126. doi: 10.1177/0956462420948739. [DOI] [PubMed] [Google Scholar]
- 8.St Cyr S., Barbee L., Workowski K.A., Bachmann L.H., Pham C., Schlanger K., Torrone E., Weinstock H., Kersh E.N., Thorpe P. Update to CDC’s Treatment Guidelines for Gonococcal Infection, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1911–1916. doi: 10.15585/mmwr.mm6950a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unemo M. Current and future antimicrobial treatment of gonorrhoea—The rapidly evolving Neisseria gonorrhoeae continues to challenge. BMC Infect. Dis. 2015;15:364. doi: 10.1186/s12879-015-1029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hook E.W., Newman L., Drusano G., Evans S., Handsfield H.H., Jerse A.E., Kong F.Y.S., Lee J.Y., Taylor S.N., Deal C. Development of New Antimicrobials for Urogenital Gonorrhea Therapy: Clinical Trial Design Considerations. Clin. Infect Dis. 2020;70:1495–1500. doi: 10.1093/cid/ciz899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asín-Prieto E., Rodríguez-Gascón A., Isla A. Applications of the pharmacokinetic/pharmacodynamic (PK/PD) analysis of antimicrobial agents. J. Infect. Chemother. 2015;21:319–329. doi: 10.1016/j.jiac.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Caruso G., Giammanco A., Virruso R., Fasciana T. Current and Future Trends in the Laboratory Diagnosis of Sexually Transmitted Infections. Int. J. Environ. Res. Public Health. 2021;18:1038. doi: 10.3390/ijerph18031038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The European Committee on Antimicrobial Susceptibility Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. [(accessed on 3 August 2021)]. Available online: http://www.eucast.org.
- 14.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2020. CLSI Supplement M100. [Google Scholar]
- 15.Lettieri J.T., Rogge M.C., Kaiser L., Echols R.M., Heller A.H. Pharmacokinetic profiles of ciprofloxacin after single intravenous and oral doses. Antimicrob. Agents Chemother. 1992;36:993–996. doi: 10.1128/AAC.36.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koomanachai P., Bulik C.C., Kuti J.L., Nicolau D.P. Pharmacodynamic modeling of intravenous antibiotics against gram-negative bacteria collected in the United States. Clin. Ther. 2010;32:766–779. doi: 10.1016/j.clinthera.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Dumitrescu T.P., Anic-Milic T., Oreskovic K., Padovan J., Brouwer K.L.R., Zuo P., Schmith V.D. Development of a population pharmacokinetic model to describe azithromycin whole-blood and plasma concentrations over time in healthy subjects. Antimicrob. Agents Chemother. 2013;57:3194–3201. doi: 10.1128/AAC.02430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soda M., Ito S., Matsumaru N., Matsumaru N., Nakamura S., Nagase I., Takahashi H., Ohno Y., Yasuda M., Yamamoto M., et al. Evaluation of the microbiological efficacy of a single 2-gram dose of extended-release azithromycin by population pharmacokinetics and simulation in japanese patients with gonococcal urethritis. Antimicrob. Agents Chemother. 2017;62:e01409-17. doi: 10.1128/AAC.01409-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chisholm S.A., Mouton J.W., Lewis D.A., Nichols T., Ison C.A., Livermore D.M. Cephalosporin MIC creep among gonococci: Time for a pharmacodynamic rethink? J. Antimicrob. Chemother. 2010;65:2141–2148. doi: 10.1093/jac/dkq289. [DOI] [PubMed] [Google Scholar]
- 20.Guerrero-Torres M.D., Menéndez M.B., Guerras C.S., Tello E., Ballesteros J., Clavo P., Puerta T., Vera M., Ayerdi O., Carrio J.C., et al. Epidemiology, molecular characterisation and antimicrobial susceptibility of Neisseria gonorrhoeae isolates in Madrid, Spain, in 2016. Epidemiol. Infect. 2019;147:e274. doi: 10.1017/S095026881900150X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmerón P., Viñado B., El Ouazzani R., Hernández M., Barbera M.J., Alberny M., Jané M., Larrosa N., Pumarola T., Hoyos-Mallecot Y. Antimicrobial susceptibility of Neisseria gonorrhoeae in Barcelona during a five-year period, 2013 to 2017. Eur. Surveill. 2020;25:1900576. doi: 10.2807/1560-7917.ES.2020.25.42.1900576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmerón P., Moreno-Mingorance A., Trejo J., Amado R., Viñado B., Cornejo-Sanchez T., Alberny M., Barbera M.J., Arando M., Pumarola T., et al. Emergence and dissemination of three mild outbreaks of Neisseria gonorrhoeae with high-level resistance to azithromycin in Barcelona, 2016–2018. J. Antimicrob. Chemother. 2021;76:930–935. doi: 10.1093/jac/dkaa536. [DOI] [PubMed] [Google Scholar]
- 23.Unemo M., Lahra M.M., Cole M.M., Galarza P., Ndowa F., Martin I., Dillon I.A.R., Ramon-Pardo P., Bolan G., Ti T.W. World Health Organization Global Gonococcal Antimicrobial Surveillance Program (WHO GASP): Review of new data and evidence to inform international collaborative actions and research efforts. Sex Health. 2019;16:412–425. doi: 10.1071/SH19023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unemo M., Workowski K. Dual antimicrobial therapy for gonorrhoea: What is the role of azithromycin? Lancet Infect. Dis. 2018;18:486–488. doi: 10.1016/S1473-3099(18)30162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calado J., Castro R., Lopes Â., Campos M.J., Rocha M., Pereira F. Antimicrobial resistance and molecular characteristics of Neisseria gonorrhoeae isolates from men who have sex with men. Int. J. Infect. Dis. 2019;79:116–122. doi: 10.1016/j.ijid.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 26.Ryan L., Golparian D., Fennelly N., Rose L., Walsh P., Lawlor B., Mac Aogáin M., Unemo M., Crowley B. Antimicrobial resistance and molecular epidemiology using whole-genome sequencing of Neisseria gonorrhoeae in Ireland, 2014–2016: Focus on extended-spectrum cephalosporins and azithromycin. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:1661–1672. doi: 10.1007/s10096-018-3296-5. [DOI] [PubMed] [Google Scholar]
- 27.Młynarczyk-Bonikowska B., Majewska A., Malejczyk M., Młynarczyk G., Majewski S. Multiresistant Neisseria gonorrhoeae: A new threat in second decade of the XXI century. Med. Microbiol. Immunol. 2020;209:95–108. doi: 10.1007/s00430-019-00651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Low N., Unemo M. Molecular tests for the detection of antimicrobial resistant Neisseria gonorrhoeae: When, where, and how to use? Curr. Opin. Infect. Dis. 2016;29:45–51. doi: 10.1097/QCO.0000000000000230. [DOI] [PubMed] [Google Scholar]
- 29.Ohneck E.A., Zalucki Y.M., Johnson P.J., Dhulipala V., Golparian D., Unemo M., Jerse A.E., Shafer W.M. A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. mBio. 2011;2:e00187-11. doi: 10.1128/mBio.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asín E., Isla A., Canut A., Rodríguez Gascón A. Comparison of antimicrobial pharmacokinetic/pharmacodynamic breakpoints with EUCAST and CLSI clinical breakpoints for Gram-positive bacteria. Int. J. Antimicrob. Agents. 2012;40:313–322. doi: 10.1016/j.ijantimicag.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Valero A., Isla A., Rodríguez-Gascón A., Canut A., Solinís M.A. Susceptibility of Pseudomonas aeruginosa and antimicrobial activity using PK/PD analysis: An 18-year surveillance study. Enferm. Infecc. Microbiol. Clin. 2019;37:626–633. doi: 10.1016/j.eimc.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Zelenitsky S.A., Rubinstein E., Ariano R.E., Zhanel G.G., Canadian Antimicrobial Resistance Alliance Integrating pharmacokinetics, pharmacodynamics and MIC distributions to assess changing antimicrobial activity against clinical isolates of Pseudomonas aeruginosa causing infections in Canadian hospitals (CANWARD) J. Antimicrob. Chemother. 2013;68((Suppl. S1)):i67–i72. doi: 10.1093/jac/dkt028. [DOI] [PubMed] [Google Scholar]
- 33.Theuretzbacher U., Barbee L., Connolly K., Drusano G., Fernandes P., Hook E., Jerse A., O’Donnell J., Unemo M., Van Bambeke F., et al. Pharmacokinetic/pharmacodynamic considerations for new and current therapeutic drugs for uncomplicated gonorrhoea-challenges and opportunities. Clin. Microbiol. Infect. 2020;26:1630–1635. doi: 10.1016/j.cmi.2020.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Matzneller P., Krasniqi S., Kinzig M., Sörgel F., Hüttner S., Lackner E., Müller M., Zeitlinger M. Blood, tissue, and intracellular concentrations of azithromycin during and after end of therapy. Antimicrob. Agents Chemother. 2013;57:1736–1742. doi: 10.1128/AAC.02011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tapsall J. Antimicrobial Resistance in Neisseria Gonorrhoeae. World Health Organization; Geneva, Switzerland: 2001. [(accessed on 3 August 2021)]. Available online: https://apps.who.int/iris/bitstream/handle/10665/66963/WHO_CDS_CSR_DRS_2001.3.pdf. [Google Scholar]
- 36.Ibar-Bariain M., Isla A., Solinís M.Á., Sanz-Moreno J.C., Canut A., Rodríguez-Gascón A. Pharmacokinetic/pharmacodynamic evaluation of the antimicrobial therapy of pneumococcal invasive disease in adults in post-PCV13 vaccine period in Madrid, Spain. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:2145–2152. doi: 10.1007/s10096-021-04255-w. [DOI] [PubMed] [Google Scholar]
- 37.Valero A., Isla A., Rodríguez-Gascón A., Calvo B., Canut A., Solinís M.Á. Pharmacokinetic/pharmacodynamic analysis as a tool for surveillance of the activity of antimicrobials against Pseudomonas aeruginosa strains isolated in critically ill patients. Enferm. Infecc. Microbiol. Clin. 2019;37:380–386. doi: 10.1016/j.eimc.2018.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez-Gascón A., Solinís M.Á., Isla A. The Role of PK/PD Analysis in the Development and Evaluation of Antimicrobials. Pharmaceutics. 2021;13:833. doi: 10.3390/pharmaceutics13060833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in Supplementary Materials.