Abstract
A new series of quinoline-based benzenesulfonamides (QBS) were developed as potential carbonic anhydrase inhibitors (CAIs). The target QBS CAIs is based on the 4-anilinoquinoline scaffold where the primary sulphonamide functionality was grafted at C4 of the anilino moiety as a zinc anchoring group (QBS 13a–c); thereafter, the sulphonamide group was switched to ortho- and meta-positions to afford regioisomers 9a–d and 11a–g. Moreover, a linker elongation approach was adopted where the amino linker was replaced by a hydrazide one to afford QBS 16. All the described QBS have been synthesized and investigated for their CA inhibitory action against hCA I, II, IX and XII. In general, para-sulphonamide derivatives 13a–c displayed the best inhibitory activity against both cancer-related isoforms hCA IX (KIs = 25.8, 5.5 and 18.6 nM, respectively) and hCA XII (KIs = 9.8, 13.2 and 8.7 nM, respectively), beside the excellent hCA IX inhibitory activity exerted by meta-sulphonamide derivative 11c (KI = 8.4 nM). The most promising QBS were further evaluated for their anticancer and pro-apoptotic activities on two cancer cell lines (MDA-MB-231 and MCF-7). In addition, molecular docking simulation studies were applied to justify the acquired CA inhibitory action of the target QBS.
Keywords: sulfonamides, 4-anilinoquinoline, carbonic anhydrase IX inhibitors, anticancer agents, hypoxic tumors
1. Introduction
Carbonic anhydrases (CA) are metalloenzymes that catalyze the reversable interconversion between carbon dioxide and bicarbonate ion reaction [1]. Human carbon anhydrases (hCAs) belong to the α-CA family, one of the eight discovered families of carbon anhydrases [2]. Only twelve hCAs (hCAs I–VII, hCA IX and hCAs XII–XIV) out of the sixteen isozymes discovered exhibit catalytic action [3]. CAs are involved in both biological and pathological processes such as homeostasis of pH, respiration, bone resorption, epilepsy, tumorigenicity and obesity [4,5]. The role of CAs in various diseases has been confirmed and several hCAs isoforms are, therefore, valuable targets for designing inhibitors with clinical applications, such as anti-glaucoma, antiepileptic, anti-obesity, anticancer, etc. [6,7,8,9,10,11].
Tumor progression induces a hypoxic environment that triggers extracellular acidosis as a result of anaerobic glycolysis in the tumor cell and this drop in pH further stimulates the tumor growth [12,13]. hCA IX and hCA XII isoforms are generally called “cancer-associated” CA isoforms. The hCA IX isoform is upregulated in nearly all hypoxic tumors so that it can maintain the intracellular pH and promote the acidic extracellular environment required for promoting tumor growth and metastasis [13,14]. In addition, hCA IX is involved in cell proliferation as well as cell to cell communication. Overexpression of hCA IX isoform is strongly correlated with poor prognosis in many cancers [15]. Moreover, hCA XII is also accompanied with many tumor types, but it is less associated to a hypoxic tumor when compared with hCA IX [16].
Quinoline-based small molecules have been reported to exhibit diverse biological activities including anticancer activity [17]. Bosutinib and Lenvatinib (Figure 1) are quinoline-based kinase inhibitors approved for chronic myelogenous leukemia and thyroid cancer, respectively [18]. Moreover, Bosutinib has been investigated in clinical trials for the treatment of breast cancer [19,20,21]. In addition, Neratinib (Figure 1), a tyrosine kinase inhibitor, is FDA approved for metastatic HER2-positive breast cancer [22], whereas, Pelitinib (Figure 1) is a second generation irreversible epidermal growth factor receptor tyrosine kinase (EGFR TK) inhibitor that is currently examined in phase II clinical trials for the treatment of non-small cell lung cancer and colorectal cancer [23].
Figure 1.
Chemical structure of clinically approved quinoline-based drugs; Bosutinib, Lenvatinib and Neratinib, and the investigational drug Pelitinib, as well as CAIs quinolines I and II.
To the best of our knowledge, few studies have reported on the development of quinoline-based sulfonamides as carbonic anhydrase inhibitors. In 2019, a novel series of 3-(quinolin-4-ylamino)benzenesulfonamides was reported as hCA I and II inhibitors. These derivatives, with general structure I (Figure 1), displayed weak inhibitory activity against the hCA I isoform (KI range: 0.96 μM–9.09 μM) and moderate activity toward the hCA II isoform (KI range: 83.3 nM–3.59 μM) [24]. In the same year, another series of quinoline-2-carboxamides (general structure II, Figure 1) was reported as a novel hCA inhibitor [25]. Quinolines II exerted moderate inhibitory activity against hCA II and IV isoforms, whereas they did not display any significant activity against the cancer-associated hCA IX isoform.
Resuming the efforts to create effective hCA IX and hCA XII inhibitors, here we describe the design and synthesis of novel 4-aminoquinoline-based sulfonamides (Figure 2). The design of the herein reported target QBS CAIs is based on the incorporation of 6-substituted quinoline as a lipophilic tail. The fused lipophilic quinoline tails are anticipated to achieve significant hydrophobic interactions within the roomier hCA IX and XII binding sites. The substitutions on a quinoline ring including -CH3, -OCH3, -Cl and di-CF3 span different electronic properties. Furthermore, different positional isomers “ortho, meta and para” of aminobenzenesulfonamide moieties were incorporated to provide the target series QBS 9a–d, 11a–g and 13a–c, Figure 2. In addition, a linker elongation approach was adopted where the amino linker was replaced by a hydrazide one to afford QBS 16.
Figure 2.
Design of target QBS (9a–d, 11a–g, 13a–c and 16).
All the herein designed quinolines were synthesized, characterized and explored for their CA inhibitory action against hCA I, II, IX and XII. Then, the anti-proliferation and the apoptosis induction effects of the most efficient hCA IX inhibitors were in vitro investigated. Molecular docking simulation studies were applied to justify the CA inhibitory action of the target quinolines.
2. Results and Discussion
2.1. Chemistry
The preparation of QBS (9a–d, 11a–g, 13a–c and 16) is illustrated in Scheme 1, Scheme 2 and Scheme 3.
Scheme 1.
Reagent and conditions: (i) Acetic anhydride, conc. H2SO4, acetone RT; (ii) Triethyl orthoforamte, reflux 3 h; (iii) DMF, reflux 2 h; (iv) Diphenyl ether, Microwave at 250 °C, 10–15 min; (V) POCl3, 90 °C, 4 h.
Scheme 2.
Reagent and conditions: (i) Isopropanol, catalytic HCl, reflux 2 h.
Scheme 3.
Reagent and conditions: (i) Isopropyl alcohol, NH2NH2.H2O, reflux 3 h; (ii) Isopropanol, catalytic HCl, reflux 2 h.
Meldrum’s acid 2 was prepared from malonic acid and acetone according to the reported procedures [26]. Then, heating 2 with triethyl orthoformate under gentile reflux followed by the addition of substituted anilines 4a–d, afforded 5-(phenylaminomethylene) meldrum’s acids 5a–d. The cyclization of 5a–d was achieved via microwave irradiation for 10–15 min at 250 °C to produce 6-substiuted 4-hydroxyquinolines 6a–d in 66–70% yield [27]. Heating Meldrum’s acid at temperatures greater than 200 °C leads to a pericyclic reaction that produces the highly reactive ketene, which was subjected to nucleophilic addition by the phenyl ring [28,29]. Thereafter, 6-substiuted 4-hydroxyquinolines 6a–d were converted to the corresponding 4-chloroquinoline derivatives 7a–d in 70–75% yield by heating with an excess of phosphorus oxychloride for 4 h. (Scheme 1).
In Scheme 2, the 4-chloroquinoline derivatives 7a–d were reacted with different aminobenzenesulfonamide derivatives (8, 10a–b and 12) in refluxing isopropanol in the presence of a catalytic amount of HCl to furnish the corresponding target QBS derivatives (9a–d, 11a–g and 13a–c) with a 65–78% yield.
The methyl 4-sulfamoylbenzoate 14 was subjected to hydrazinolysis to afford hydrazide 15, through refluxing with hydrazine hydrate in isopropyl alcohol. Thereafter, 6-methoxy-4-chloroquinoline 7b was reacted with 4-(hydrazinecarbonyl)benzenesulfonamide 15 in refluxing isopropanol to furnish the corresponding target QBS 16 with a 69% yield (Scheme 3).
Postulated structures for the newly synthesized QBS compounds were in full agreement with their spectral analyses data (Supporting Information).
2.2. Biological Evaluation
2.2.1. Carbonic Anhydrases Inhibition
The potential CA inhibition activity of the newly synthesized QBS (9a–d, 11a–g, 13a–c and 16) were assessed applying the stopped flow carbon dioxide hydrase assay [30] for both the ubiquitous CA isoforms hCA I and II, and the cancer-related isoforms hCA IX and XII. The tested CA isoforms were suppressed to varying degrees by the herein reported QBS, and the inhibition data are displayed in Table 1.
Table 1.
Inhibition data for the target sulfonamides on human CA isoforms hCA I, II, IX and XII using AAZ as a reference drug.
| cmpd | o-/m-/p- | R1 | R2 | KI (nM) 1 | |||
|---|---|---|---|---|---|---|---|
| CA I | CA II | CA IX | CA XII | ||||
| 9a | o- | 6-CH3 | H | 558.3 | 78.4 | 36.4 | 22.8 |
| 9b | o- | 6-OCH3 | H | 864.4 | 49.7 | 39.2 | 42.1 |
| 9c | o- | 6-Cl | H | 1239 | 86.8 | 65.6 | 38.4 |
| 9d | o- | 5,7-diCF3 | H | 1562 | 112.4 | 25.9 | 26.5 |
| 11a | m- | 6-CH3 | H | 663.1 | 265.2 | 70.1 | 41.2 |
| 11b | m- | 6-OCH3 | H | 258.4 | 365.7 | 86.5 | 73.1 |
| 11c | m- | 6-Cl | H | 442.3 | 154.8 | 8.4 | 55.3 |
| 11d | m- | 5,7-diCF3 | H | 105.3 | 221.0 | 52.9 | 34.0 |
| 11e | m- | 6-CH3 | CH3 | 3498 | 1503 | 103.5 | 88.1 |
| 11f | m- | 6-OCH3 | CH3 | 2256 | 998.2 | 853.4 | 152.2 |
| 11g | m- | 6-Cl | CH3 | 4521 | 2356 | 116.2 | 92.7 |
| 13a | p- | 6-CH3 | - | 78.4 | 36.5 | 25.8 | 9.8 |
| 13b | p- | 6-OCH3 | - | 92.1 | 58.4 | 5.5 | 13.2 |
| 13c | p- | 6-Cl | - | 55.4 | 7.3 | 18.6 | 8.7 |
| 16 | - | - | - | 81.4 | 31.1 | 21.7 | 25.4 |
| AAZ | - | - | - | 250 | 12.5 | 25.0 | 5.7 |
1 Mean from 3 different assays, using a stopped flow technique (errors were in the range of ±5–10% of the reported values).
The cytosolic hCA I isoform was inhibited by all the quinoline-based sulfonamides reported in this study with inhibition constants (KIs) ranging from low nanomolar to low micromolar concentrations, between 55.4 nM and 1.56 μM, apart from 4-methyl-3-sulfonamide bearing counterparts 11e–g, which exerted lower inhibitory actions (KI = 3.498, 2.256 and 4.521 μM, respectively). In particular, all the para-sulphonamide analogous 13a–c and 16 were found to be the most effective hCA I inhibitors with two-digit nanomolar inhibition constants (KIs = 78.4, 92.1, 55.4 and 81.4 nM, respectively). Moreover, meta-sulphonamide analogous 11a–d and ortho-sulphonamide analogous 9a–b displayed moderate submiclomolar hCA I inhibitory activity with KIs spanning in the range 105.3–864.4 nM.
It is worth stressing that shifting the primary sulphonamide functionality from the ortho-position (9a–d; KIs: 558.3–1562.0 nM) to the meta-position (11a–d; KIs: 105.3–663.1 nM), as well as shifting from the meta- to para-position (13a–c; KIs: 55.4–92.1 nM) dramatically enhanced the inhibitory activity towards hCA I. In addition, incorporation of the hydrazide linker in compound 16 (KI = 81.4 nM) slightly improved hCA I inhibitory activity in comparison to its analogue 13b (KI = 92.1 nM). In contrast, grafting a 4-methyl group within the meta-sulphonamide derivatives 11a–c led to compounds 11e–g with much lower inhibitory activity (KIs: 2.256–4.521 μM, respectively).
The in vitro kinetic data demonstrated in Table 1 revealed that the physiologically dominant off-target hCA II isoform has been inhibited by the herein prepared QBS (9a–d, 11a–g, 13a–c and 16) with KIs in the nanomolar range (from 7.3 to 998.2 nM), except compounds 11e and 11g, which displayed inhibitory activity in the low micromolar concentration (KIs = 1.503 and 2.356 μM, respectively). The para-sulphonamide derivative 13c emerged as the most efficient hCA II inhibitor, in this study, with a single-digit nanomolar inhibition constant (KI = 7.3 nM). Additionally, compounds 9a–c, 13a, 13b and 16 exerted potent activities with KI values equal 78.4, 49.7, 86.8, 36.5, 58.4 and 31.1 nM, respectively.
It is noteworthy that switching the sulphonamide functionality from the ortho-position (9a–d; KIs: 49.7–112.4 nM) to the meta-position (11a–d; KIs: 154.8–365.7 nM) elicited a worsening of effectiveness toward the hCA II isoform, whereas shifting to the para-position led to compounds 13a–c with an enhanced hCA II inhibitory activity. Moreover, incorporation of the 4-methyl group in compounds 11e–g resulted in a decrease in the inhibitory activity (KIs: 998.2 nM–2.356 μM) in comparison to their analogues 11a–c (KIs: 154.8 nM–365.7 nM), whereas the linker elongation approach (compound 16) improved the inhibitory activity from 58.4 nM (for anilino derivative 13b) to 31.1 nM, Table 1.
The examined quinoline-based sulfonamides displayed potent to moderate inhibitory activity towards the target tumor-associated hCA IX isoform (KI values spanning between 5.5 and 116.2 nM, Table 1), except compound 11f (KI = 853.4 nM). In particular, QBS 11c and 13b emerged as excellent single-digit nanomolar hCA IX inhibitors (KIs = 8.4 and 5.5 nM, respectively). Additionally, sulfonamides 9d, 13a, 13c and 16 exerted better or equipotent inhibitory activity (KIs = 25.9, 25.8, 18.6 and 21.7 nM, respectively) compared to the reference AAZ (KI = 25 nM).
Further analysis of the obtained results showed that grafting the sulfamoyl group at the para-position (13a–c; KIs: 5.5–25.8 nM) was more beneficial for hCA IX inhibitory activity than the ortho-substitution (9a–d; KIs: 25.9–65.6 nM) and the meta-substitution (11a–d; KIs: 8.4–86.5 nM), except for 6-chloro bearing derivative 11b. Dissimilar to the inhibitory profile of target sulfonamides toward hCA I and II isoforms, the linker elongation approach failed to improve the hCA IX inhibitory activity (anilino derivative 13b; KI = 5.5 nM vs. hydrazido derivative 16; KI = 21.7 nM). It is worth noting that, appending a methyl group at C4 within the benzenesulfonamide moiety of QBS 11a–c resulted in QBS 11e–g analogues with about a 1.5- to 13.8-fold decreased potency, Table 1.
As shown in Table 1, the second cancer-related isoform here examined hCA XII has been potently inhibited by all the newly prepared quinoline-based sulfonamides 9a–d, 11a–g, 13a–c and 16 (KIs range: 8.7–88.12 nM), except compound 11f, which moderately affected the hCA XII isoform (KI = 152.2 nM). Interestingly, grafting the zinc anchoring sulfamoyl group at the para-position achieved the best hCA XII inhibitory action in this study (QBS 13a–c; KIs = 9.8, 13.2 and 8.7 nM, respectively). Moreover, ortho-sulphonamide derivatives 9a and 9d, as well as the hydrazido derivative 16 possessed efficient inhibitory activity against the hCA XII isoform (KIs = 22.8, 26.5 and 25.4 nM, respectively).
In conclusion, for both cancer-related isoforms hCA IX and hCA XII, grafting the sulfamoyl functionality at the para-position was more advantageous for inhibitory activity than the ortho-position, which, in turn, was more advantageous than meta-substitution. Furthermore, C4 substitution of the benzenesulfonamide moiety by a methyl group, as well as the incorporation of the hydrazide linker, slightly decreased the inhibitory activities toward both isoforms.
The selectivity index (SI) presented in Table 2 obviously displayed a good selectivity profile for target QBS toward hCA IX over hCA I with 9d, 11c, 11e and 11g having the highest SIs (60.3, 52.7, 33.8 and 38.9, respectively). In addition, the QBS showed good selectivity toward hCA IX over hCA II, with 11c, 11e, 11g and 13b exhibiting the best SIs (18.4, 14.4, 20.3 and 10.6, respectively). Similarly, excellent selectivity toward hCA XII over hCA I was demonstrated by all the QBS except 11b, 11d and 16; also, QBS showed a good selectivity towards hCA XII over hCA II with 11e and 11g being the highest (SIs: 17.1 and 25.4, respectively).
Table 2.
Selectivity ratios for the inhibition of hCA IX and XII over hCA I and II for target compounds 9a–d, 11a–g, 13a–c and 16.
| cmpd | I/IX | II/IX | I/XII | II/XII |
|---|---|---|---|---|
| 9a | 15.3 | 2.2 | 24.5 | 3.4 |
| 9b | 22.1 | 1.3 | 20.5 | 1.2 |
| 9c | 18.9 | 1.3 | 32.3 | 2.3 |
| 9d | 60.3 | 4.3 | 58.9 | 4.2 |
| 11a | 9.5 | 3.8 | 16.1 | 6.4 |
| 11b | 3.0 | 4.2 | 3.5 | 5.0 |
| 11c | 52.7 | 18.4 | 8.0 | 2.8 |
| 11d | 2.0 | 4.2 | 3.1 | 6.5 |
| 11e | 33.8 | 14.5 | 39.7 | 17.1 |
| 11f | 2.6 | 1.2 | 14.8 | 6.6 |
| 11g | 38.9 | 20.3 | 48.8 | 25.4 |
| 13a | 3.0 | 1.4 | 8.0 | 3.7 |
| 13b | 16.7 | 10.6 | 7.0 | 4.4 |
| 13c | 3.0 | 0.4 | 6.4 | 0.8 |
| 16 | 3.8 | 1.4 | 3.2 | 1.2 |
| AAZ | 10.0 | 0.5 | 43.9 | 2.2 |
2.2.2. Anticancer Activity
In Vitro Anti-Proliferative Activity
The CA inhibition data presented in Table 1 revealed that not only was an efficient single-digit nanomolar inhibition of CA IX isoform exerted by QBS 11c and 13b (KIs = 8.4 and 5.5 nM, respectively), but also, both compounds demonstrated good selectivity toward the hCA IX isoform over the off-target isoforms hCA I (S.I. = 52.7 and 16.7, respectively) and hCA II (S.I. = 18.4 and 10.6, respectively), Table 2. Therefore, QBS 11c and 13b were further screened for their potential in vitro anti-proliferative action against two breast cancer cell lines (MDA-MB-231 and MCF-7) under hypoxic conditions, exploiting a 72 h MTT assay protocol [31] and using Doxorubicin as a positive control drug. The IC50 values for the examined derivatives are listed in Table 3.
Table 3.
Anti-proliferative action of QBS 11c and 13b against breast MDA-MB-231 and MCF-7 cancer cell lines.
| Compound | IC50 (μM) 1 | |
|---|---|---|
| MDA-MB-231 | MCF-7 | |
| 11c | 1.03 ± 0.05 | 0.43 ± 0.02 |
| 13b | 2.24 ± 0.1 | 3.69 ± 0.17 |
| Doxorubicin | 1.67 ± 0.08 | 3.04 ± 0.14 |
1 IC50 values are the mean ± S.D. of three separate experiments.
The results of the MTT assay are ascribed to both QBS 11c and 13b potent anti-proliferative action toward the tested MDA-MB-231 and MCF-7 cell lines (IC50 range: from 0.43 ± 0.02 to 3.69 ± 0.17). Interestingly, QBS 11c showed submicro-molar activity against the MCF-7 cancer cell line (IC50 = 0.43 ± 0.02 μM). It is worth noting that QBS 11c displayed slight better activity (IC50 = 1.03 ± 0.05 μM and 0.43 ± 0.02 μM) than QBS 13b (IC50 = 2.24 ± 0.1 μM and 3.69 ± 0.17 μM) against MDA-MB-231 and MCF-7 cell lines, respectively (Table 3).
Effect of QBS 11c and 13b on Apoptotic Markers Bax, Bcl-2, and Active Caspase-3
The levels of two members of Bcl-2 family proteins, the anti-apoptotic Bcl-2 protein, and the counteracting pro-apoptotic Bax protein, as well as the level of active caspase-3 (a key executioner protease) in MDA-MB-231 and MCF-7 cells have been assessed after incubation with QBS 11c and 13b for 24 h. The obtained results (Table 4 and Table 5) revealed that the expression levels of the examined proteins (Bax, Bcl-2, and active caspase-3) have been significantly affected upon treatment with QBS 11c and 13b.
Table 4.
Effect of QBS 11c and 13b on the expression levels of Bax, Bcl-2 and active Caspase-3 in breast cancer MDA-MB-231 cells.
| Compound | MDA-MB-231 | ||
|---|---|---|---|
| BAX (Pg/mg of Total Protein) |
Bcl-2 (Pg/mg of Total Protein) |
Active Caspase-3 (Pg/mg of Total Protein) |
|
| 11c | 420.9 ± 8.24 (7.1) 1 |
6.45 ± 0.17 (0.45) 1 |
12.69 ± 1.14 (4.93) 1 |
| 13b | 373.5 ± 10.4 (6.3) 1 |
5.304 ± 0.13 (0.37) 1 |
9.32 ± 0.73 (3.62) 1 |
| Control | 59.43 ± 24.8 | 14.29 ± 0.10 | 2.57 ± 0.35 |
1 Numbers given between parentheses are the numbers of folds of control.
Table 5.
Effect of QBS 11c and 13b on the expression levels of Bax, Bcl-2 and active Caspase-3 in breast cancer MCF-7 cells.
| Compound | MCF-7 | ||
|---|---|---|---|
| BAX (Pg/mg of Total Protein) |
Bcl-2 (Pg/mg of Total Protein) |
Active Caspase-3 (Pg/mg of Total Protein) |
|
| 11c | 512.9 ± 11.7 (5.36) 1 |
7.29 ± 0.15 (0.79) 1 |
17.83 ± 0.62 (3.64) 1 |
| 13b | 443.9 ± 10.2 (4.64) 1 |
5.72 ± 0.13 (0.62) 1 |
14.82 ± 0.43 (3.02) 1 |
| Control | 95.6 ± 11.6 | 9.14 ± 0.21 | 4.895 ± 0.66 |
1 Numbers given between parentheses are the numbers of folds of control.
Regarding MDA-MB-231 cells, their treatment with QBS 11c and 13b resulted in significant elevation in the expression levels for Bax protein (by 7.1-fold and 6.3-fold, respectively) and active caspase-3 (by 4.93-fold and 3.62-fold, respectively), whereas such treatment led to a decrease in the expression levels for the anti-apoptotic Bcl-2 protein (by 55 and 63%, respectively) in comparison to the untreated control (Table 4).
On the other hand, treatment of MCF-7 cells with QBS 11c and 13b led to an increase in the expression levels for Bax protein (by 5.36-fold and 4.64-fold, respectively) and active caspase-3 (by 3.64-fold and 3.02-fold, respectively); in addition, it resulted in a suppression of the expression levels for Bcl-2 protein (by 21 and 38%, respectively) compared to the untreated control (Table 5).
2.3. Molecular Modeling Studies
Molecular docking and MM-GBSA-based refinements inside hCA isozymes IX and XII (PDB 5FL4 [32] and 4WW8 [33]) were employed to explore the binding modalities of the synthesized compounds and to correlate their structural characteristics with the inhibition activity. The co-crystalized ligands in both hCA isoforms IX and XII showed the usual pattern of the sulfonamide moiety where it binds to the zinc(II) ion after the displacement of the metal-bound water molecule to form the tetrahedral adduct with the zinc atom. Both ligands are positioned toward the hydrophobic half of the active site and the ligand binding is achieved mostly by van der Waals and hydrophobic interactions [32,33].
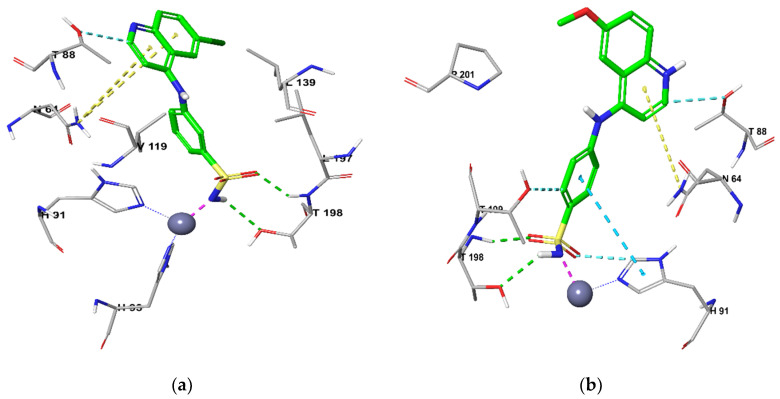
The benzenesulfonamide ring fit deeply inside the shallow CA active site for both isoforms anchoring the zinc atom in a typical manner for sulfonamide CAIs through an NH−---Zn2+ bond. In addition, in the active site of CA IX, two hydrogen bonds occurred between the sulfamoyl NH---O (T200) and sulfamoyl S = O---HN (T201) for 11c, while 13b sulfamoyl S = O made two hydrogen bonds with T200 (NH) and T201(OH) alongside the π−π stacking between H94 and the sulfonamide benzene ring in both 11c and 13b (Figure 3).Furthermore, 11c was a correct distance from the additional three hydrogen bonds by its linker NH with Q71 (C = O) and by its halogen that made two interactions with W9 (NH) and T201 (OH). On the other hand, 13b had only two additional hydrogen bonds (linker NH---C = O of Q71 and CH3O---NH of W9) in concurrence with the hydrophobic interaction of its quinoline ring with Q71, Q92 and P202.
Figure 3.
Docking CA IX for (a) 11c and (b) 13b.
Similarly, docking of QBS into CAXII revealed two hydrogen bonds (sulfamoyl NH---O (T198) and sulfamoyl S = O---HN (T198) for both 11b and 13c (Figure 4). Moreover, the quinoline ring in both QBS engaged in a N-π interaction with N64 and hydrophobic interaction with T88. Additionally, the sulfonamide benzene ring in 13b was observed to make a π−π stacking interaction with H94.
Figure 4.
Docking CA XII for (a) 11c and (b) 13b.
The target QBS showed a similar interaction to the co-crystalized ligand maintaining the sulfonamide moiety interactions; in addition, the quinoline ring was similarly positioned toward the hydrophobic half of the active site of hCA IX and XII, similar to the co-crystalized ligands interaction.
The narrow range of docking scores and prime MMGBSA binding energy was in accordance with the observed in vitro inhibitory activity of 11c and 13b on each isozyme, as shown in Table 6.
Table 6.
The docking scores and binding energy from MMGBSA for QBS 11c and 13b on hCA IX and XII.
| Comp. | hCA IX | hCA XII | ||
|---|---|---|---|---|
| Docking Score | MMGBSA dG Bind | Docking Score | MMGBSA dG Bind | |
| 11c | −6.029 | −34.30 | −6.029 | −46.69 |
| 13b | −6.046 | −42.14 | −6.441 | −49.81 |
3. Materials and Methods
3.1. Chemistry
General
All reaction solvents and reagents were purchased from commercial suppliers; Sigma-Aldrich (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) and Alfa Aesar(Thermo Fisher GmbH, Kandel, Germany) and used without further purification. Melting points were measured with a Stuart melting point apparatus and were uncorrected. The NMR spectra were obtained on Bruker Avance 400 (400 MHz 1H and 100 MHz 13C NMR). 1H NMR spectra were referenced to tetramethylsilane (δ = 0.00 ppm) as an internal standard and were reported as follows: chemical shift, multiplicity (b = broad, s = singlet, d = doublet, t = triplet, dd = doublet of doublet, m = multiplet). IR spectra were recorded with a Bruker FT-IR spectrophotometer. Reaction courses and product mixtures were routinely monitored using thin layer chromatography (TLC) that was carried out using aluminum sheets pre-coated with silica gel 60 F254 purchased from Merk (Merck Group, Darmstadt, Germany) Compounds 5–7 and 15 were prepared according to the reported methods [27,34,35].
General Procedures for Preparation of Target QBS (9a–d, 11a–g, 13a–c and 16)
To a heated solution of the appropriate 4-chloroquinoline derivative 7a–d (0.5 mmol) in dry isopropanol (3 mL) in a round-bottom flask, a catalytic amount of HCl, then the appropriate benzenesulfonamide derivatives 8, 10a–b, 12 and 15 (0.5 mmol) were added. The reaction mixture was stirred under reflux temperature for 2 h. The precipitated solid was collected by filtration while hot, washed with cold water, cold ethanol and petroleum ether then recrystallized from methanol to produce the target QBS derivatives 9a–d, 11a–g, 13a–c and 16.
Synthesis of 2-((6-methylquinolin-4-yl)amino)benzenesulfonamide (9a). QBS 9a was obtained following the general procedure mentioned above using 7a (0.09 g, 0.50 mmol) and 2-aminobenzenesulfonamide 8 (0.086 g, 0.50 mmol). A 78% yield; m.p. 264–265 °C; IR (KBr, ν cm−1): 3369, 3249, 3146 (NH, NH2), and 1338, 1163 (SO2); 1H NMR (DMSO-d6) δ ppm: 2.58 (s, 3H, CH3), 6.2 (d, 1H, Ar-H, H3 of quinoline, J = 6.4 Hz), 7.62 (d, 1H, Ar-H, H3 of C6H4SO2NH2, J = 7.6 Hz), 7.73 (t, 1H, Ar-H, H5 of C6H4SO2NH2, J = 7.6 Hz), 7.78 (s, 2H, NH2, D2O exchangeable of -SO2NH2), 7.84 (t, 1H, Ar-H, H4 of C6H4SO2NH2, J = 7.6, Hz), 7.89 (d, 1H, Ar-H, H7 of quinoline, J = 8.8 Hz), 8.02 (d, 1H, Ar-H, H8 of quinoline, J = 8.8 Hz), 8.11 (d, 1H, Ar-H, H6 of C6H4SO2NH2, J = 7.6 Hz), 8.43 (d, 1H, Ar-H, H2 of quinoline, J = 6.4 Hz), 8.98 (s, 1H, Ar-H, H5 of quinoline), 11.00 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 21.60, 101.02, 117.60, 120.23, 123.80, 129.58, 129.69, 131.11, 134.46, 134.69, 135.98, 136.81, 137.40, 141.44, 142.01, 156.46; Anal. Calcd. for C16H15N3O2S: C, 61.32; H, 4.82; N, 13.41; found C, 61.44; H, 4.85; N, 13.32.
Synthesis of 2-((6-methoxyquinolin-4-yl)amino)benzenesulfonamide (9b). QBS 9b was obtained following the general procedure mentioned above using 7b (0.1 g, 0.50 mmol) and 2-aminobenzenesulfonamide 8 (0.086 g, 0.50 mmol). A 65% yield; m.p. 275–277 °C; IR (KBr, ν cm−1): 3457, 3218, 3124 (NH, NH2) and 1341, 1167 (SO2); 1H NMR (DMSO-d6) δ ppm: 4.03 (s, 3H, -OCH3), 6.18 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.63 (d, 1H, Ar-H, H3 of C6H4SO2NH2, J = 7.6 Hz), 7.66–7.75 (m, 2H, Ar-H, H7 of quinoline and H5 of C6H4SO2NH2), 7.76 (s, 2H, NH2, D2O exchangeable of -SO2NH2), 7.85 (t, 1H, Ar-H, H4 of C6H4SO2NH2, J = 7.0 Hz), 8.02 (d, 1H, Ar-H, H8 of quinoline J = 8.8 Hz), 8.11 (d, 1H, Ar-H, H6 of C6H4SO2NH2, J = 7.0 Hz), 8.38 (s, 1H, Ar-H, H5 of quinoline), 8.49 (d, 1H, Ar-H, H2 of quinoline J = 6.8 Hz), 11.01 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 57.26, 101.03, 104.21, 118.90, 122.18, 125.90, 129.57, 129.70, 131.32, 133.72, 134.49, 134.84, 140.72, 141.65, 156.10, 158.43; Anal. Calcd. for C16H15N3O3S: C, 58.35; H, 4.59; N, 12.76; found C, 58.67; H, 4.62; N, 12.68.
Synthesis of 2-((6-chloroquinolin-4-yl)amino)benzenesulfonamide (9c). QBS 9c was obtained following the general procedure mentioned above using 7c (0.1 g, 0.50 mmol) and 2-aminobenzenesulfonamide 8 (0.086 g, 0.50 mmol). A 71% yield; m.p. 258–260 °C; IR (KBr, ν cm−1): 3257, 3144, 3102 (NH, NH2) and 1335, 1163 (SO2); 1H NMR (DMSO-d6) δ ppm: 6.22 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.63 (d, 1H, Ar-H, H3 of C6H4SO2NH2, J = 7.6 Hz), 7.68–7.82 (m, 3H, Ar-H, H5 of C6H4SO2NH2, NH2, D2O exchangeable of -SO2NH2), 7.85 (t, 1H, Ar-H, H4 of C6H4SO2NH2, J = 6.8, 7.6 Hz), 8.02–8.19 (m, 3H, Ar-H, H2, H7 and H8 of quinoline), 8.5 (d, 1H, Ar-H, H6 of C6H4SO2NH2, J = 7.2 Hz), 9.31 (s, 1H, Ar-H, H5 of quinoline), 11.01 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 101.84, 118.59, 122.67, 124.38, 129.59, 129.98, 131.15, 131.99, 134.35, 134.57, 134.57, 137.29, 141.57, 143.01, 156.39; Anal. Calcd. for C15H12ClN3O2S: C, 53.98; H, 3.62; N, 12.59; found C, 54.21; H, 3.58; N, 12.51.
Synthesis of 2-((5,7-bis(trifluoromethyl)quinolin-4-yl)amino)benzenesulfonamide (9d). QBS 9d was obtained following the general procedure mentioned above using 7d (0.15 g, 0.50 mmol) and 2-aminobenzenesulfonamide 8 (0.086 g, 0.50 mmol). A 78% yield; m.p. 240–242 °C; IR (KBr, ν cm−1): 3331, 3159, 3115 (NH, NH2), 1583 (C = N) and 1342, 1165 (SO2); 1H NMR (DMSO-d6) δ ppm: 6.84 (t, 1H, Ar-H, H5 of C6H4SO2NH2, J = 7.2, 7.6 Hz ), 7.01 (d, 1H, Ar-H, H3 of quinoline, J = 8.0 Hz), 7.33–7.47 (m, 7H, Ar-H, NH2, D2O exchangeable of -SO2NH2), 7.63 (d, 1H, Ar-H, H2 of quinoline, J = 8.0 Hz); Anal. Calcd. for C17H11F6N3O2S: C, 46.90; H, 2.55; N, 9.65; found C, 47.05; H, 2.53; N, 9.73.
Synthesis of 3-((6-methylquinolin-4-yl)amino)benzenesulfonamide (11a). QBS 11a was obtained following the general procedure mentioned above using 7a (0.09 g, 0.50 mmol) and 3-aminobenzenesulfonamide 10a (0.086 g, 0.50 mmol). A 75% yield; m.p. 272–274 °C; IR (KBr, ν cm−1): 3286, 3148, 3107 (NH, NH2) and 1348, 1156 (SO2); 1H NMR (DMSO-d6) δ ppm: 2.58 (s, 3H, CH3), 6.91 (d, 1H, Ar-H, H3 of quinoline, J = 4.8 Hz), 7.61 (s, 2H, NH2, D2O exchangeable of -SO2NH2), 7.77 (d, 2H, Ar-H, H4 and H6 of C6H4SO2NH2, J = 8.4 Hz), 7.84–7.96 (m, 3H, Ar-H, H7 of quinoline, H3 and H5 of C6H4SO2NH2), 8.08 (d, 1H, Ar-H, H8 of quinoline, J = 8.4, Hz), 8.55 (d, 1H, Ar-H, H2 of quinoline, J = 6.8 Hz), 8.74 (s, 1H, Ar-H, H5 of quinoline), 11.14 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 21.69, 100.27, 117.87, 120.50, 122.51, 123.20, 124.51, 128.79, 131.18, 136.11, 137.01, 137.77, 138.42, 142.67, 146.11, 154.49; Anal. Calcd. for C16H15N3O2S: C, 61.32; H, 4.82; N, 13.41; found C, 61.07; H, 4.86; N, 13.33.
Synthesis of 3-((6-methoxyquinolin-4-yl)amino)benzenesulfonamide (11b). QBS 11b was obtained following the general procedure mentioned above using 7b (0.1 g, 0.50 mmol) and 3-aminobenzenesulfonamide 10a (0.086 g, 0.50 mmol). A 69% yield; m.p. 244–246 °C; IR (KBr, ν cm−1): 3349, 3152, 3111 (NH, NH2) and 1341, 1155 (SO2); 1H NMR (DMSO-d6) δ ppm: 3.99 (s, 3H,-OCH3), 6.90 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.60 (s, 2H, NH2, D2O exchangeable of -SO2NH2), 7.69 (d, 1H, Ar-H, H7 of quinoline, J = 9.2 Hz), 7.76–7.88 (m, 3H, Ar-H, H4, H5 and H6 of C6H4SO2NH2,), 7.94 (s, 1H, Ar-H, H3 of C6H4SO2NH2), 8.11 (d, 1H, Ar-H, H8 of quinoline, J = 9.2 Hz), 8.33 (s, 1H, Ar-H, H5 of quinoline), 8.50 (d, 1H, Ar-H, H2 of quinoline, J = 6.8 Hz), 11.18 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 57.11, 100.22, 103.48, 119.20, 122.42, 122.60, 124.43, 126.10, 128.95, 131.18, 133.97, 138.55, 141.22, 146.11, 153.97, 158.60; Anal. Calcd. for C16H15N3O3S: C, 58.35; H, 4.59; N, 12.76; found C, 58.59; H, 4.63; N, 12.70.
Synthesis of 3-((6-chloroquinolin-4-yl)amino)benzenesulfonamide (11c). QBS 11c was obtained following the general procedure mentioned above using 7c (0.1 g, 0.50 mmol) and 3-aminobenzenesulfonamide 10a (0.086 g, 0.50 mmol). A 70% yield; m.p. 272–274 °C; IR (KBr, ν cm−1): 3291, 3159, 3100 (NH, NH2) and 1348, 1156 (SO2); 1H NMR (DMSO-d6) δ ppm: 6.96 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.63 (s, 2H, NH2, D2O exchangeable of -SO2NH2) 7.74–7.81 (m, 2H, Ar-H, H4 and H5 of C6H4SO2NH2), 7.87 (d, 1H, Ar-H, H6 of C6H4SO2NH2, J = 4.4, Hz), 7.93 (s, 1H, Ar-H, H3 of C6H4SO2NH2), 8.10 (d, 1H, Ar-H, H7 of quinoline, J = 9.2 Hz), 8.23 (d, 1H, Ar-H, H8 of quinoline, J = 9.2 Hz), 8.61 (d, 1H, Ar-H, H2 of quinoline, J = 6.8 Hz), 9.12 (s, 1H, Ar-H, H5 of quinoline), 11.37 (s, 1H, NH, D2O exchangeable);13C NMR (DMSO-d6) δ ppm: 101.00, 118.92, 122.54, 122.91, 123.79, 124.79, 128.79, 130.82, 131.27, 134.66, 137.50, 138.08, 143.71, 146.15, 154.39; Anal. Calcd. for C15H12ClN3O2S: C, 53.98; H, 3.62; N, 12.59; found C, 54.20; H, 3.59; N, 12.66.
Synthesis of 3-((5,7-bis(trifluoromethyl)quinolin-4-yl)amino)benzenesulfonamide (11d). QBS 11d was obtained following the general procedure mentioned above using 7d (0.15 g, 0.50 mmol) and 3-aminobenzenesulfonamide 10a (0.086 g, 0.50 mmol). A 78% yield; m.p. 172–174 °C; IR (KBr, ν cm−1): 3331, 3159, 3115 (NH, NH2), 1583 (C = N) and 1342, 1165 (SO2); 1H NMR (DMSO-d6) δ ppm: 7.49 (d, 2H, Ar-H, H4 and H6 of C6H4SO2NH2, J = 7.6 Hz), 7.50 - 7.68 (m, 7H, Ar-H, NH2, D2O exchangeable of -SO2NH2), 7.71 (s, 1H, Ar-H, H6 of quinoline), 7.73 (s, 1H, NH, D2O exchangeable); Anal. Calcd. for C17H11F6N3O2S: C, 46.90; H, 2.55; N, 9.65; found C, 46.69; H, 2.60; N, 9.57.
Synthesis of 2-methyl-5-((6-methylquinolin-4-yl)amino)benzenesulfonamide (11e). QBS 11e was obtained following the general procedure mentioned above using 7a (0.09 g, 0.50 mmol) and 5-amino-2-methylbenzenesulfonamide 10b (0.093 g, 0.50 mmol). A 66% yield; m.p. 280–282 °C; IR (KBr, ν cm−1): 3324, 3205, 3144 (NH, NH2) and 1333, 1163 (SO2); 1H NMR (DMSO-d6) δ ppm: 2.57 (s, 3H, CH3 of quinoline), 2.66 (s, 3H, CH3 of C6H4SO2NH2), 6.84 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.52–7.69 (m, 4H, Ar-H, H3 and H4 of C6H4SO2NH2, NH2, D2O exchangeable of -SO2NH2), 7.88 (d, 1H, Ar-H, H7 of quinoline, J = 8.8, Hz), 7.94 (s, 1H, Ar-H, H6 of C6H4SO2NH2), 8.06(d, 1H, Ar-H, H8 of quinoline, J = 8.8, Hz), 8.50 (d, 1H, Ar-H, H2 of quinoline, J = 6.8, Hz), 8.73 (s, 1H, Ar-H H5 of quinoline), 11.07 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 19.94, 21.67, 100.12, 117.73, 120.47, 123.18, 124.25, 128.89, 134.20, 135.20, 135.73, 136.03, 136.98, 137.66, 142.51, 143.80, 154.70; Anal. Calcd. for C17H17N3O2S: C, 62.37; H, 5.23; N, 12.83; found C, 62.54; H, 5.26; N, 12.91.
Synthesis of 5-((6-methoxyquinolin-4-yl)amino)-2-methylbenzenesulfonamide (11f). QBS 11f was obtained following the general procedure mentioned above using 7b (0.1 g, 0.50 mmol) and 5-amino-2-methylbenzenesulfonamide 10b (0.093 g, 0.50 mmol). A 68% yield; m.p. 248–250 °C; IR (KBr, ν cm−1): 3378, 3203, 3146 (NH, NH2) and 1331, 1155 (SO2); 1H NMR (DMSO-d6) δ ppm: 2.67 (s, 3H, CH3), 4.01 (s, 3H, -OCH3), 6.84 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.49–7.74 (m, 5H, Ar-H, H7 of quinoline, H3 and H4 of C6H4SO2NH2,NH2, D2O exchangeable of -SO2NH2), 7.94 (s, 1H, Ar-H, H6 of C6H4SO2NH2), 8.09 (d, 1H, Ar-H, H8 of quinoline, J = 9.2, Hz), 8.31 (s, 1H, Ar-H, H5 of quinoline), 8.45 (d, 1H, Ar-H, H2 of quinoline, J = 6.8, Hz), 11.10 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 19.94, 57.09, 100.05, 103.51, 119.05, 122.38, 124.35, 125.98, 129.08, 133.91, 134.22, 135.14, 135.83, 141.09, 143.80, 154.21, 158.55; Anal. Calcd. for C17H17N3O3S: C, 59.46; H, 4.99; N, 12.24; found C, 59.23; H, 5.03; N, 12.32.
Synthesis of 5-((6-chloroquinolin-4-yl)amino)-2-methylbenzenesulfonamide (11g). QBS 11g was obtained following the general procedure mentioned above using 7c (0.1 g, 0.50 mmol) and 5-amino-2-methylbenzenesulfonamide 10b (0.093 g, 0.50 mmol). A 77% yield; m.p. 282–285 °C; IR (KBr, ν cm−1): 3277, 3204, 3174 (NH, NH2) and 1326, 1156 (SO2); 1H NMR (DMSO-d6) δ ppm: 2.66 (s, 3H, CH3), 6.89 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.54–7.69 (m, 4H, Ar-H, H3 and H4 of C6H4SO2NH2, NH2, D2O exchangeable of -SO2NH2), 7.94 (s, 1H, Ar-H, H6 of C6H4SO2NH2), 8.08 (d, 1H, Ar-H, H7 of quinoline, J = 9.2, Hz), 8.20 (d, 1H, Ar-H, H8 of quinoline, J = 9.2, Hz), 8.57 (d, 1H, Ar-H, H2 of quinoline, J = 6.8, Hz), 9.09 (s, 1H, Ar-H, H5 of quinoline), 11.28 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 19.94, 100.87, 118.81, 122.94, 123.69, 124.25, 128.83, 132.22, 134.28, 134.59, 135.40, 135.51, 137.51, 143.61, 143.85, 154.56; Anal. Calcd. for C16H14ClN3O2S: C, 55.25; H, 4.06; N, 12.08; found C, 54.96; H, 4.08; N, 11.99.
Synthesis of 4-((6-methylquinolin-4-yl)amino)benzenesulfonamide (13a). QBS 13a was obtained following the general procedure mentioned above using 7a (0.09 g, 0.50 mmol) and 4-aminobenzenesulfonamide 12 (0.086 g, 0.50 mmol). A 70% yield; m.p. 270–272°C; IR (KBr, ν cm−1): 3335, 3216, 3159 (NH, NH2) and 1324, 1156 (SO2); 1H NMR (DMSO-d6) δ ppm: 2.57 (s, 3H, CH3), 7.01 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.53 (s, 2H, NH2, D2O exchangeable of -SO2NH2), 7.73 (d, 2H, Ar-H, H3 and H5 of C6H4SO2NH2, J = 8.4, Hz), 7.89 (d, 1H, Ar-H, H7 of quinoline, J = 8.8, Hz), 7.99 (d, 2H, Ar-H, H2 and H6 of C6H4SO2NH2, J = 8.4, Hz), 8.07 (d, 1H, Ar-H, H8 of quinoline, J = 8.8 Hz), 8.55 (d, 1H, Ar-H, H2 of quinoline, J = 6.8, Hz), 8.78 (s, 1H, Ar-H, H5 of quinoline), 11.18 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 21.67, 100.80, 118.07, 120.50, 123.30, 125.33, 125.33, 127.85, 127.85, 136.14, 137.04, 137.82, 141.09, 142.42, 142.69, 154.23; Anal. Calcd. for C16H15N3O2S: C, 61.32; H, 4.82; N, 13.81; found C, 61.49; H, 4.86; N, 13.89.
Synthesis of 4-((6-methoxyquinolin-4-yl)amino)benzenesulfonamide (13b). QBS 13b was obtained following the general procedure mentioned above using 7b (0.1 g, 0.50 mmol) and 4-aminobenzenesulfonamide 12 (0.086 g, 0.50 mmol). A 67% yield; m.p. 250–252 °C; IR (KBr, ν cm−1): 3333, 3192, 3107 (NH, NH2) and 1338, 1162 (SO2); 1H NMR (DMSO-d6) δ ppm: 4.03 (s, 3H, -OCH3), 7.01 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.53 (s, 2H, NH2, D2O exchangeable of -SO2NH2), 7.69 (dd, 1H, Ar-H, H7 of quinoline, J = 9.2 Hz), 7.75 (d, 2H, Ar-H, H3 and H5 of C6H4SO2NH2, J = 8.4, Hz), 7.99 (d, 2H, Ar-H, H2 and H6 of C6H4SO2NH2, J = 8.4, Hz), 8.10 (d, 1H, Ar-H, H8 of quinoline, J = 9.2 Hz), 8.37 (s, 1H, Ar-H, H5 of quinoline), 8.50 (d, 1H, Ar-H, H2 of quinoline, J = 6.8, Hz), 11.24 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 57.18, 100.77, 103.63, 119.44, 122.41, 125.44, 125.44, 126.16, 127.82, 127.82, 134.01, 141.21, 141.23, 142.34, 153.70, 158.61; Anal. Calcd. for C16H15N3O3S: C, 58.35; H, 4.59; N, 12.76; found C, 58.61; H, 4.55; N, 12.83.
Synthesis of 4-((6-chloroquinolin-4-yl)amino)benzenesulfonamide (13c). QBS 13c was obtained following the general procedure mentioned above using 7c (0.1 g, 0.50 mmol) and 4-aminobenzenesulfonamide 12 (0.086 g, 0.50 mmol). A 72% yield; m.p. 284–286 °C; IR (KBr, ν cm−1): 3281, 3208, 3148 (NH, NH2) and 1330, 1166 (SO2); 1H NMR (DMSO-d6) δ ppm: 7.04 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.54 (s, 2H, NH2, D2O exchangeable of -SO2NH2), 7.73 (d, 2H, Ar-H, H3 and H5 of C6H4SO2NH2, J = 8.0, Hz), 8.00 (d, 2H, Ar-H, H2 and H6 of C6H4SO2NH2, J = 8.0, Hz), 8.09 (d, 1H, Ar-H, H7 of quinoline, J = 8.8 Hz), 8.21 (d, 1H, Ar-H, H8 of quinoline, J = 8.8, Hz), 8.62 (d, 1H, Ar-H, H2 of quinoline, J = 6.8, Hz), 9.16 (s, 1H, Ar-H, H5 of quinoline), 11.18 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 101.50, 119.07, 122.91, 123.85, 125.42, 125.42, 127.89, 127.89, 132.35, 134.67, 137.53, 140.73, 142.70, 143.73, 154.11; Anal. Calcd. for C15H12ClN3O2S: C, 53.98; H, 3.62; N, 12.59; found C, 54.17; H, 3.66; N, 12.53.
Synthesis of 4-(2-(6-Methoxyquinolin-4-yl)hydrazine-1-carbonyl)benzenesulfonamide (16). QBS 16 was obtained following the general procedure mentioned above using 7b (0.1 g, 0.50 mmol) and 4-(hydrazinecarbonyl)benzenesulfonamide 15 (0.108 g, 0.50 mmol). A 69% yield; m.p. 228–230 °C; IR (KBr, ν cm−1): 3345, 3208, 3119, 3066 (NH, NH2), 1685 (C = O) and 1333, 1162 (SO2); 1H NMR (DMSO-d6) δ ppm: 3.99 (s, 3H, -OCH3), 7.01 (d, 1H, Ar-H, H3 of quinoline, J = 6.8 Hz), 7.56 (s, 1H, Ar-H, H5 of quinoline), 7.63 (s, 2H, NH2, D2O exchangeable of -SO2NH2), 7.72 (dd, 1H, Ar-H, H7 of quinoline, J = 8.4, Hz), 7.94 (d, 1H, Ar-H, H3 of C6H4SO2NH2, J = 8.8, Hz), 8.01–8.11 (m, 3H, Ar-H, H2, H5 and H6 of C6H4SO2NH2,), 8.23 (d, 1H, Ar-H, H8 of quinoline, J = 8.4 Hz), 8.55 (d, 1H, Ar-H, H2 of quinoline, J = 6.8 Hz), 11.23 (s, 1H, NH, D2O exchangeable), 11.66 (s, 1H, NHC=O, D2O exchangeable); 13C NMR (DMSO-d6) δ ppm: 56.87, 98.96, 102.66, 116.73, 122.60, 126.28, 126.44, 128.54, 129.05, 133.52, 135.08, 147.25, 147.83, 155.92, 158.51, 165.27, 165.59; Anal. Calcd. for C17H16N4O4S: C, 54.83; H, 4.33; N, 15.05; found C, 55.06; H, 4.31; N, 14.96.
3.2. Biological Evaluation
All adopted procedures for the conducted in vitro biological assays were performed as described earlier; CA (stopped-flow [4,30,36]), cytotoxicity (MTT [37,38]), and assessment of apoptotic markers [39,40] assays, as well as induction of hypoxia with cobalt chloride [41,42], and were mentioned in the Supporting Materials.
3.3. Molecular Modelling
The utilized procedures within the docking experiments for QBS 11c and 13b in hCA IX (pdb: 5FL4, [32]) and hCA XII (pdb: 4WW8, [33]) active sites are provided in the Supplementary Materials.
4. Conclusions
In this work, the design, synthesis and characterization of different novel series of quinoline-based sulfonamides (QBS; 9a–c, 11a–h, 13a–c and 16) were reported, afterwards their CA inhibition activity were examined against hCA I, II, IX and XII. Most of the newly reported QBS efficiently inhibited the herein investigated hCA IX and XII (tumor-related isoforms) with KIs in the ranges 5.5–853.4 nM and 8.7–152.2 nM, respectively. Furthermore, QBS 9a, 9b, 9d, 13a, 13c and 16 showed KIs values in the nanomolar range from 18.6–39.2 nM and compounds 11c and 13b were shown to be the most effective hCA IX inhibitor in this investigation (KIs = 8.4 and 5.5 nM, respectively). Similarly, 13a and 13c showed one-digit nanomolar inhibition activity against hCA XII (KIs = 9.8 and 8.7 nM, respectively). In addition, 11c and 13b showed good selectivity towards hCA IX over the physiological isomer hCAI (SI = 52.7 and 16.7) and hCAII (SI = 18.4 and 10.6). SAR analysis pointed out that grafting the sulfamoyl functionality at the para-position was more advantageous for hCA IX and hCA XII inhibitory activities than the ortho-position, which, in turn, was more advantageous than meta-substitution. Additionally, the C4 substitution of the benzenesulfonamide moiety by a methyl group, as well as the incorporation of the hydrazide linker, slightly reduced the inhibitory activities toward both the hCA IX and hCA XII isoforms. Thereafter, a utilized MTT assay revealed that QBS 11c (IC50 = 1.03 ± 0.05 μM and 0.43 ± 0.02 μM) and QBS 13b (IC50 = 2.24 ± 0.1 μM and 3.69 ± 0.17 μM) possessed potent activity against MDA-MB-231 and MCF-7 cell lines, respectively, under hypoxic conditions. Moreover, the incubation of MDA-MB-231 and MCF-7 cells with QBS 11b and 13b enhanced the expression levels for pro-apoptotic markers Bax and active Caspase-3 proteins, while the level of anti-apoptotic Bcl-2 protein was suppressed. Finally, the molecular docking simulations have provided insights for the binding interactions of QBS 11b and 13b within hCA IX (pdb: 5FL4) and hCA XII (pdb: 4WW8) binding sites.
Acknowledgments
This work was supported by Bilateral Project (2019–2020) Agreement between CNR, Italy and ASRT, Egypt.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222011119/s1.
Author Contributions
M.S.: Investigation, data curation, formal analysis and writing—original draft. A.N.: Investigation and data curation. Z.M.E.: Investigation. T.M.I.: Formal analysis, visualization and writing—review and editing. R.S.: Investigation, visualization and writing—review and editing. R.A.E.-D.: Supervision. C.C.: Investigation, data curation and writing—review and editing. C.T.S.: Conceptualization, resources and writing—review and editing. W.M.E.: Methodology, conceptualization, resources, data curation, visualization and writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Bilateral Project (2019–2020) Agreement between CNR, Italy and ASRT, Egypt.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Angeli A., Carta F., Supuran C.T. Carbonic Anhydrases: Versatile and Useful Biocatalysts in Chemistry and Biochemistry. Catalysts. 2020;10:1008. doi: 10.3390/catal10091008. [DOI] [Google Scholar]
- 2.Mishra C.B., Tiwari M., Supuran C.T. Progress in the development of human carbonic anhydrase inhibitors and their pharmacological applications: Where are we today? Med. Res. Rev. 2020;40:2485–2565. doi: 10.1002/med.21713. [DOI] [PubMed] [Google Scholar]
- 3.Taslimi P., Gulcin I., Ozgeris B., Goksu S., Tumer F., Alwasel S.H., Supuran C.T. The human carbonic anhydrase isoenzymes I and II (hCA I and II) inhibition effects of trimethoxyindane derivatives. J. Enzym. Inhib. Med. Chem. 2016;31:152–157. doi: 10.3109/14756366.2015.1014476. [DOI] [PubMed] [Google Scholar]
- 4.Eldehna W.M., Abdelrahman M.A., Nocentini A., Bua S., Al-Rashood S.T., Hassan G.S., Bonardi A., Almehizia A.A., Alkahtani H.M., Alharbi A., et al. Synthesis, biological evaluation and in silico studies with 4-benzylidene-2-phenyl-5(4H)-imidazolone-based benzenesulfonamides as novel selective carbonic anhydrase IX inhibitors endowed with anticancer activity. Bioorganic Chem. 2019;90:103102. doi: 10.1016/j.bioorg.2019.103102. [DOI] [PubMed] [Google Scholar]
- 5.Aggarwal M., Boone C.D., Kondeti B., McKenna R. Structural annotation of human carbonic anhydrases. J. Enzym. Inhib. Med. Chem. 2013;28:267–277. doi: 10.3109/14756366.2012.737323. [DOI] [PubMed] [Google Scholar]
- 6.Waheed A., Sly W.S. Carbonic anhydrase XII functions in health and disease. Gene. 2017;623:33–40. doi: 10.1016/j.gene.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Occhipinti R., Boron W.F. Role of Carbonic Anhydrases and Inhibitors in Acid-Base Physiology: Insights from Mathematical Modeling. Int. J. Mol. Sci. 2019;20:3841. doi: 10.3390/ijms20153841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Provensi G., Carta F., Nocentini A., Supuran C.T., Casamenti F., Passani M.B., Fossati S. A New Kid on the Block? Carbonic Anhydrases as Possible New Targets in Alzheimer’s Disease. Int. J. Mol. Sci. 2019;20:4724. doi: 10.3390/ijms20194724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Supuran C.T. Carbonic Anhydrases and Metabolism. Metabolites. 2018;8:25. doi: 10.3390/metabo8020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabrizi F., Mincione F., Somma T., Scozzafava G., Galassi F., Masini E., Impagnatiello F., Supuran C.T. A new approach to antiglaucoma drugs: Carbonic anhydrase inhibitors with or without NO donating moieties. Mechanism of action and preliminary pharmacology. J. Enzyme Inhib. Med. Chem. 2012;27:138–147. doi: 10.3109/14756366.2011.597749. [DOI] [PubMed] [Google Scholar]
- 11.Aggarwal M., Kondeti B., McKenna R. Anticonvulsant/antiepileptic carbonic anhydrase inhibitors: A patent review. Expert Opin. Ther. Pat. 2013;23:717–724. doi: 10.1517/13543776.2013.782394. [DOI] [PubMed] [Google Scholar]
- 12.Pastorekova S., Gillies R.J. The role of carbonic anhydrase IX in cancer development: Links to hypoxia, acidosis, and beyond. Cancer Metastasis Rev. 2019;38:65–77. doi: 10.1007/s10555-019-09799-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiche J., Ilc K., Laferrière J., Trottier E., Dayan F., Mazure N.M., Brahimi-Horn M.C., Pouysségur J. Hypoxia-Inducible Carbonic Anhydrase IX and XII Promote Tumor Cell Growth by Counteracting Acidosis through the Regulation of the Intracellular pH. Cancer Res. 2009;69:358–368. doi: 10.1158/0008-5472.CAN-08-2470. [DOI] [PubMed] [Google Scholar]
- 14.Chafe S.C., Vizeacoumar F.S., Venkateswaran G., Nemirovsky O., Awrey S., Brown W.S., McDonald P.C., Carta F., Metcalfe A., Karasinska J.M., et al. Genome-wide synthetic lethal screen unveils novel CAIX-NFS1/xCT axis as a targetable vulnerability in hypoxic solid tumors. Sci. Adv. 2021;7:eabj0364. doi: 10.1126/sciadv.abj0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuno K., Matsubara T., Nakamura T., Iino T., Kakimoto T., Asanuma K., Matsumine A., Sudo A. Carbonic anhydrase IX enhances tumor cell proliferation and tumor progression in osteosarcoma. Onco Targets Ther. 2018;11:6879–6886. doi: 10.2147/OTT.S177605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tafreshi N.K., Lloyd M.C., Proemsey J.B., Bui M.M., Kim J., Gillies R.J., Morse D.L. Evaluation of CAIX and CAXII Expression in Breast Cancer at Varied O2 Levels: CAIX is the Superior Surrogate Imaging Biomarker of Tumor Hypoxia. Mol. Imaging Biol. 2016;18:219–231. doi: 10.1007/s11307-015-0885-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhat H.R., Masih A., Shakya A., Ghosh S.K., Singh U.P. Design, synthesis, anticancer, antibacterial, and antifungal evaluation of 4-aminoquinoline-1,3,5-triazine derivatives. J. Heterocycl. Chem. 2020;57:390–399. doi: 10.1002/jhet.3791. [DOI] [Google Scholar]
- 18.Musiol R. An overview of quinoline as a privileged scaffold in cancer drug discovery. Expert Opin. Drug Discov. 2017;12:583–597. doi: 10.1080/17460441.2017.1319357. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy L.C., Mayer I.A. New targets in endocrine-resistant hormone receptor-positive breast cancer. Clin. Adv. Hematol. Oncol. HO. 2021;19:511–521. [PubMed] [Google Scholar]
- 20.ClinicalTrials WI231696: Bosutinib, Palbocicilib and Fulvestrant for HR+HER2- Advanced Breast Cancer Refractory to a CDK4/6 Inhibitor. [(accessed on 7 September 2021)]; Available online: https://clinicaltrials.gov/ct2/show/NCT03854903.
- 21.Campone M., Bondarenko I., Brincat S., Hotko Y., Munster P.N., Chmielowska E., Fumoleau P., Ward R., Bardy-Bouxin N., Leip E., et al. Phase II study of single-agent bosutinib, a Src/Abl tyrosine kinase inhibitor, in patients with locally advanced or metastatic breast cancer pretreated with chemotherapy. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012;23:610–617. doi: 10.1093/annonc/mdr261. [DOI] [PubMed] [Google Scholar]
- 22.Chan A., Moy B., Mansi J., Ejlertsen B., Holmes F.A., Chia S., Iwata H., Gnant M., Loibl S., Barrios C.H., et al. Final Efficacy Results of Neratinib in HER2-positive Hormone Receptor-positive Early-stage Breast Cancer from the Phase III ExteNET Trial. Clin. Breast Cancer. 2021;21:80–91.e87. doi: 10.1016/j.clbc.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 23.To K.K.W., Poon D.C., Wei Y., Wang F., Lin G., Fu L. Pelitinib (EKB-569) targets the up-regulation of ABCB1 and ABCG2 induced by hyperthermia to eradicate lung cancer. Br. J. Pharmacol. 2015;172:4089–4106. doi: 10.1111/bph.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Sanea M.M., Elkamhawy A., Paik S., Bua S., Ha Lee S., Abdelgawad M.A., Roh E.J., Eldehna W.M., Supuran C.T. Synthesis and biological evaluation of novel 3-(quinolin-4-ylamino)benzenesulfonamidesAQ3 as carbonic anhydrase isoforms I and II inhibitors. J. Enzym. Inhib. Med. Chem. 2019;34:1457–1464. doi: 10.1080/14756366.2019.1652282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thacker P.S., Shaikh P., Angeli A., Arifuddin M., Supuran C.T. Synthesis and biological evaluation of novel 8-substituted quinoline-2-carboxamides as carbonic anhydrase inhibitors. J. Enzym. Inhib. Med. Chem. 2019;34:1172–1177. doi: 10.1080/14756366.2019.1626376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo S.-E., Lee S.-H., Kim S.-K., Lee S.-H. The conformation and activity relationship of benzofuran derivatives as angiotensin II receptor antagonists. Bioorganic Med. Chem. 1997;5:445–459. doi: 10.1016/S0968-0896(96)00244-1. [DOI] [PubMed] [Google Scholar]
- 27.Madrid P.B., Sherrill J., Liou A.P., Weisman J.L., DeRisi J.L., Guy R.K. Synthesis of ring-substituted 4-aminoquinolines and evaluation of their antimalarial activities. Bioorganic Med. Chem. Lett. 2005;15:1015–1018. doi: 10.1016/j.bmcl.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 28.Gaber A.E.-A.M., McNab H. Synthetic Applications of the Pyrolysis of Meldrum’s Acid Derivatives. Synthesis. 2001;2001:2059–2074. doi: 10.1055/s-2001-18057. [DOI] [Google Scholar]
- 29.Dumas A.M., Fillion E. Meldrum’s Acids and 5-Alkylidene Meldrum’s Acids in Catalytic Carbon−Carbon Bond-Forming Processes. Acc. Chem. Res. 2010;43:440–454. doi: 10.1021/ar900229z. [DOI] [PubMed] [Google Scholar]
- 30.Khalifah R.G. The Carbon Dioxide Hydration Activity of Carbonic Anhydrase: I. stop-flow kinetic studies on the native human isoenzymes B and C. J. Biol. Chem. 1971;246:2561–2573. doi: 10.1016/S0021-9258(18)62326-9. [DOI] [PubMed] [Google Scholar]
- 31.Van Meerloo J., Kaspers G.J.L., Cloos J. Cell Sensitivity Assays: The MTT Assay. In: Cree I.A., editor. Cancer Cell Culture: Methods and Protocols. Humana Press; Totowa, NJ, USA: 2011. pp. 237–245. [DOI] [PubMed] [Google Scholar]
- 32.Leitans J., Kazaks A., Balode A., Ivanova J., Zalubovskis R., Supuran C.T., Tars K. Efficient Expression and Crystallization System of Cancer-Associated Carbonic Anhydrase Isoform IX. J. Med. Chem. 2015;58:9004–9009. doi: 10.1021/acs.jmedchem.5b01343. [DOI] [PubMed] [Google Scholar]
- 33.Zubrienė A., Smirnovienė J., Smirnov A., Morkūnaitė V., Michailovienė V., Jachno J., Juozapaitienė V., Norvaišas P., Manakova E., Gražulis S., et al. Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry. Biophys. Chem. 2015;205:51–65. doi: 10.1016/j.bpc.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Cassis R., Tapia R., Valderrama J.A. Synthesis of 4(1H)-Quinolones by Thermolysis of Arylaminomethylene Meldrum’s Acid Derivatives. Synth. Commun. 1985;15:125–133. doi: 10.1080/00397918508076818. [DOI] [Google Scholar]
- 35.Takaoka Y., Tsutsumi H., Kasagi N., Nakata E., Hamachi I. One-Pot and Sequential Organic Chemistry on an Enzyme Surface to Tether a Fluorescent Probe at the Proximity of the Active Site with Restoring Enzyme Activity. J. Am. Chem. Soc. 2006;128:3273–3280. doi: 10.1021/ja057926x. [DOI] [PubMed] [Google Scholar]
- 36.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 37.Elmetwally S.A., Saied K.F., Eissa I.H., Elkaeed E.B. Design, synthesis and anticancer evaluation of thieno[2,3-d]pyrimidine derivatives as dual EGFR/HER2 inhibitors and apoptosis inducers. Bioorganic Chem. 2019;88:102944. doi: 10.1016/j.bioorg.2019.102944. [DOI] [PubMed] [Google Scholar]
- 38.Eldehna W.M., Al-Rashood S.T., Al-Warhi T., Eskandrani R.O., Alharbi A., El Kerdawy A.M. Novel oxindole/benzofuran hybrids as potential dual CDK2/GSK-3β inhibitors targeting breast cancer: Design, synthesis, biological evaluation, and in silico studies. J. Enzym. Inhib. Med. Chem. 2021;36:270–285. doi: 10.1080/14756366.2020.1862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Naggar A.M., Eissa I.H., Belal A., El-Sayed A.A. Design, eco-friendly synthesis, molecular modeling and anticancer evaluation of thiazol-5(4H)-ones as potential tubulin polymerization inhibitors targeting the colchicine binding site. RSC Adv. 2020;10:2791–2811. doi: 10.1039/C9RA10094F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Rashood S.T., Hamed A.R., Hassan G.S., Alkahtani H.M., Almehizia A.A., Alharbi A., Al-Sanea M.M., Eldehna W.M. Antitumor properties of certain spirooxindoles towards hepatocellular carcinoma endowed with antioxidant activity. J. Enzym. Inhib. Med. Chem. 2020;35:831–839. doi: 10.1080/14756366.2020.1743281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rana N.K., Singh P., Koch B. CoCl2 simulated hypoxia induce cell proliferation and alter the expression pattern of hypoxia associated genes involved in angiogenesis and apoptosis. Biol. Res. 2019;52:12. doi: 10.1186/s40659-019-0221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muñoz-Sánchez J., Chánez-Cárdenas M.E. The use of cobalt chloride as a chemical hypoxia model. J. Appl. Toxicol. 2019;39:556–570. doi: 10.1002/jat.3749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.