Abstract
Patients with advanced ovarian cancer usually exhibit high mortality rates, thus more efficient therapeutic strategies are expected to be developed. Recent transcriptomic studies revealed that long intergenic noncoding RNAs (lincRNAs) can be a new class of molecular targets for cancer management, because lincRNAs likely exert tissue-specific activities compared with protein-coding genes or other noncoding RNAs. We here show that an unannotated lincRNA originated from chromosome 10q21 and designated as ovarian cancer long intergenic noncoding RNA 1 (OIN1), is often overexpressed in ovarian cancer tissues compared with normal ovaries as analyzed by RNA sequencing. OIN1 silencing by specific siRNAs significantly exerted proliferation inhibition and enhanced apoptosis in ovarian cancer cells. Notably, RNA sequencing showed that OIN1 expression was negatively correlated with the expression of apoptosis-related genes ras association domain family member 5 (RASSF5) and adenosine A1 receptor (ADORA1), which were upregulated by OIN1 knockdown in ovarian cancer cells. OIN1-specifc siRNA injection was effective to suppress in vivo tumor growth of ovarian cancer cells inoculated in immunodeficient mice. Taken together, OIN1 could function as a tumor-promoting lincRNA in ovarian cancer through modulating apoptosis and will be a potential molecular target for ovarian cancer management.
Keywords: long intergenic noncoding RNA (lincRNA), ovarian cancer, RNA sequencing, small interfering RNA (siRNA), xenograft
1. Introduction
Ovarian cancer is one of the most common cancers in women [1]. Despite efforts to develop effective therapeutic strategies for ovarian cancer, the mortality rate remains the highest among gynecological cancers [2]. Because of the absence of symptoms in early stages, ~60% of ovarian cancers are diagnosed as advanced disease. The 5-year overall survival rate of ovarian cancer patients remains <50% [3]. The development of new biomarkers and therapeutic targets is expected to improve current diagnosis and treatment of ovarian cancer.
Advancements in cloning and sequencing technologies have revealed that 70–90% of mammalian genomes are transcribed into a variety of noncoding RNAs (ncRNAs) that do not encode proteins [4]. ncRNAs with more than 200 bases are classified as long noncoding RNAs (lncRNAs), while others are classified as small noncoding RNAs (sncRNAs). MicroRNAs (miRNAs) consist of a class of sncRNAs, and some miRNAs, including miR-200c-3p, have been indicated to be key regulators of ovarian cancer pathophysiology [5,6]. Moreover, several lncRNAs have been characterized as playing oncogenic or tumor-suppressive roles in cancers including ovarian cancer [7,8,9,10,11,12,13]. For example, metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is an lncRNA which is upregulated in ovarian cancer and associated with poor prognosis in ovarian cancer patients. Previous studies demonstrated that MALAT1 promotes proliferation and suppresses apoptosis in ovarian cancer by sponging miRNAs such as miR-211 [12]. Other reports have also indicated that MALAT1 regulates the migration and invasion of ovarian cancer cells through controlling the expression of extracellular matrix genes and cell signaling pathways such as phosphoinositide 3-kinase (PI3K)/AKT and Wnt/β-catenin [12]. In contrast, growth arrest-specific 5 (GAS5) is an lncRNA that exerts a tumor-suppressive function in ovarian cancer through sponging miRNAs such as miR-21 [12], and by facilitating the E2F4-mediated transcriptional repression of poly (ADP-ribose) polymerase 1 (PARP1) [14]. Nevertheless, the majority of lncRNAs remain to be characterized in ovarian cancer, and their functional annotation may provide useful information for potential diagnostic and therapeutic targets for the disease.
We previously performed RNA-sequencing (RNA-seq) analysis using clinical ovarian tissues and cancer specimens and identified candidate genes associated with the pathophysiology of ovarian cancer [15,16]. We also identified novel mutations in known ovarian cancer-associated genes, such as TP53, breast cancer 2 (BRCA2), and AT-rich interaction domain 1A (ARID1A) [16]. These results indicate that RNA-seq analysis using clinical samples is useful for the characterization of ovarian cancer gene signature and the screen of new therapeutic targets. Based on the RNA-seq, we here focused on functional lncRNAs, particularly long intergenic noncoding RNAs (lincRNAs) predominantly expressed in ovarian cancer tissues. Apart from a genic lncRNA subclass that shares sequence with protein-coding transcript, the lincRNA subclass may exert tissue-specific activities, as lincRNAs derived from intergenic “gene deserts” generally show less conservation than protein-coding genes or other noncoding RNAs across species [17]. We identified an uncharacterized lincRNA originated from chromosome 10q21, whose expression was substantially higher in clinical ovarian cancer specimens compared with normal ovarian tissues. We designated the lincRNA as ovarian cancer long intergenic noncoding RNA 1 (OIN1). Loss-of-function study using OIN1-specific siRNAs showed significant proliferation inhibition and enhanced apoptosis in A2780 and SKOV3 ovarian cancer cells. Moreover, we found that the expression of apoptosis-related genes ras association domain family member 5 (RASSF5) and adenosine A1 receptor (ADORA1) could be modulated by OIN1 in ovarian cancer. We propose that lincRNA OIN1 could contribute to ovarian cancer progression by fine-tuning gene expression, leading to the suppression of apoptosis.
2. Results
2.1. OIN1 Is Highly Expressed in Ovarian Cancer Tissues and Cells
In the present study, we aimed to identify functional lincRNAs predominantly expressed in ovarian cancer. We screened our RNA-seq data analyzed by NONCODE database [18] obtained from clinical specimens of normal ovarian tissues (n = 6) and ovarian cancers (n = 15) [15,16]. Screening NONCODE v4 transcripts abundantly expressed in ovarian cancer compared with normal ovarian tissues by ≥10-folds at an FDR q-value threshold <0.05, we identified 10 particular lincRNAs (Table 1), including known oncogenic RNAs: competing endogenous lncRNA 2 for microRNA let-7b (CERNA2)/human ovarian cancer-specific transcript 2 (HOST2) [19], urothelial cancer associated 1 (UCA1) [14,20,21,22], and long intergenic non-protein coding RNA 958 (LINC00958) [23]. CERNA2/HOST2 was shown to promote ovarian cancer cell proliferation, partly by sponging the tumor suppressor let-7b [19]. In terms of UCA1, the elevated expression was shown to enhance cell migration, invasion and cisplatin resistance of ovarian cancer [20,21], and the lincRNA activates Hippo-Yes-associated protein (YAP) signaling in ovarian cancer [22].
Table 1.
Differentially upregulated lincRNAs in ovarian cancer versus normal ovary tissues a.
| NONCODE ID | Chr | Start | End | Strand | Alias | Normal Ovary | Ovarian Cancer | Fold Change b | q-Value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean RPKM | SEM RPKM | Mean RPKM | SEM RPKM | ||||||||
| NONHSAT013448 | 10 | 54,785,023 | 54,789,855 | - | OIN1 | 3.0 | 2.6 | 79.2 | 24.8 | 26.1 | 5.6 × 10−3 |
| NONHSAT099419 | 4 | 182,443,812 | 182,444,154 | + | 3.2 | 2.7 | 40.4 | 5.5 | 12.7 | 2.9 × 10−4 | |
| NONHSAT017219 | 11 | 287,304 | 288,298 | + | 0.6 | 0.2 | 38.6 | 18.3 | 62.9 | 1.0 × 10−2 | |
| NONHSAT027397 | 12 | 26,383,751 | 26,472,653 | - | 0.5 | 0.2 | 27.4 | 11.7 | 51.5 | 1.6 × 10−3 | |
| NONHSAT015316 | 10 | 85,926,985 | 85,931,832 | - | CERNA2/HOST2 | 0.8 | 0.7 | 17.2 | 5.9 | 20.2 | 7.4 × 10−4 |
| NONHSAT032437 | 13 | 23,477,401 | 23,493,348 | + | 0.3 | 0.1 | 9.2 | 3.0 | 31.0 | 1.5 × 10−4 | |
| NONHSAT080725 | 20 | 60,880,487 | 60,881,452 | - | 0.5 | 0.2 | 8.8 | 3.8 | 19.2 | 1.3 × 10−3 | |
| NONHSAT122583 | 7 | 104,581,509 | 104,602,507 | + | 0.3 | 0.2 | 8.2 | 3.7 | 27.0 | 3.0 × 10−2 | |
| NONHSAT061517 | 19 | 15,939,789 | 15,947,064 | + | UCA1 | 0.6 | 0.5 | 6.7 | 2.0 | 11.3 | 8.1 × 10−3 |
| NONHSAT018088 | 11 | 13,002,033 | 13,005,839 | - | LINC00958 | 0.6 | 0.5 | 5.7 | 0.9 | 10.3 | 2.1 × 10−2 |
a Differentially upregulated lincRNAs in ovarian cancer were selected by the criterion described below: among transcripts mapped to NONCODE v4 gene sets, we selected 10 putative differentially expressed lincRNAs, which were particularly upregulated in ovarian cancer compared with normal tissues by ≥10-folds at an FDR q-value threshold <0.05. b Fold change of RPKM values in ovarian cancer versus normal ovary tissues. RPKM: reads per kilobase of transcript length per million mapped reads.
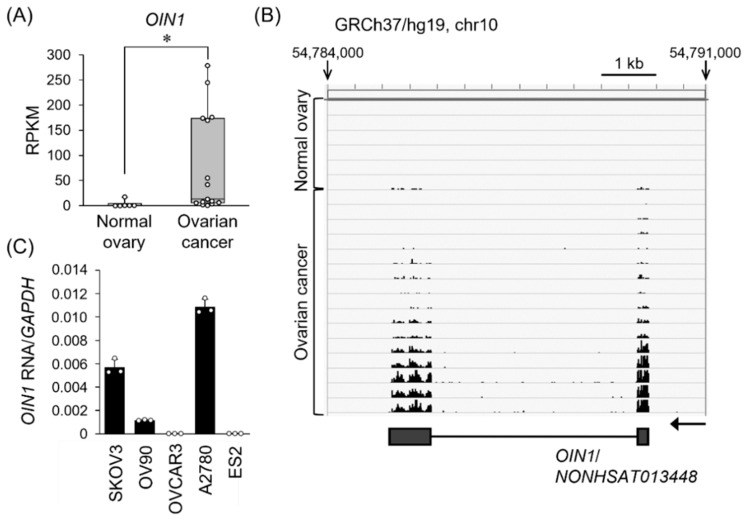
Since we noticed functional ovarian cancer-related lincRNAs among the top 10 RNAs, we next characterized the most abundantly expressed lincRNA in ovarian cancer. The sequence of the top lincRNA corresponds to the NONHSAT013448 in NONCODE database, with the mean RPKM (reads per kilobase of transcript length per million mapped reads) values as 79.2 ± 24.8 and 3.0 ± 2.6 in ovarian cancer and normal tissues, respectively (q = 0.006) (Figure 1A,B; Table 1). Coding potential calculator (CPC) algorithm showed that the coding potential score of NONHSAT013448 was −0.62195, suggesting that it is a putative noncoding RNA. NONHSAT013448 is transcribed from a gene NONHSAG005930 at chromosome 10q21.1, whose location is ~0.78 and ~0.20 Mb apart from the neighboring protein-coding genes protocadherin-related 15 (PCDH15) and mannose binding lectin 2 (MBL2), respectively. We designated the lincRNA as ovarian cancer long intergenic noncoding RNA 1, or OIN1. Based on quantitative real-time PCR (qRT-PCR) analysis in ovarian cancer cell lines, we found that high expression of OIN1 was observed in A2780 and SKOV3 cells, and moderate expression in OV90 cells (Figure 1C).
Figure 1.
Overexpression of lincRNA OIN1 in ovarian cancer. (A) OIN1 RNA (registered as NONHSAT013448 in NONCODE) is overexpressed in ovarian cancer. RPKM (reads per kilobase of transcript length per million mapped reads) values of OIN1 RNA were estimated by RNA-seq analysis using ovarian cancer (n = 15) and normal tissues (n = 6) [15,16]. Data are presented by box plots. *, q < 0.01. (B) The mapping data of RNA-seq reads derived from normal ovary tissues and ovarian cancer on the OIN1 locus. A genome browser snapshot of the OIN1 region (chr10:54,784,000–54,791,000 in hg19) was shown with the schematic representation of OIN1 gene. The grey boxes indicated the exons of OIN1. The arrow shows the direction of OIN1 gene. (C) OIN1 RNA expression levels in ovarian cancer cell lines. qRT-PCR was performed to quantify the expression levels of OIN1 RNA normalized to GAPDH mRNA levels. Data are presented as mean ± SD (n = 3).
2.2. OIN1 Promotes Proliferation and Suppresses Apoptosis of Ovarian Cancer Cells
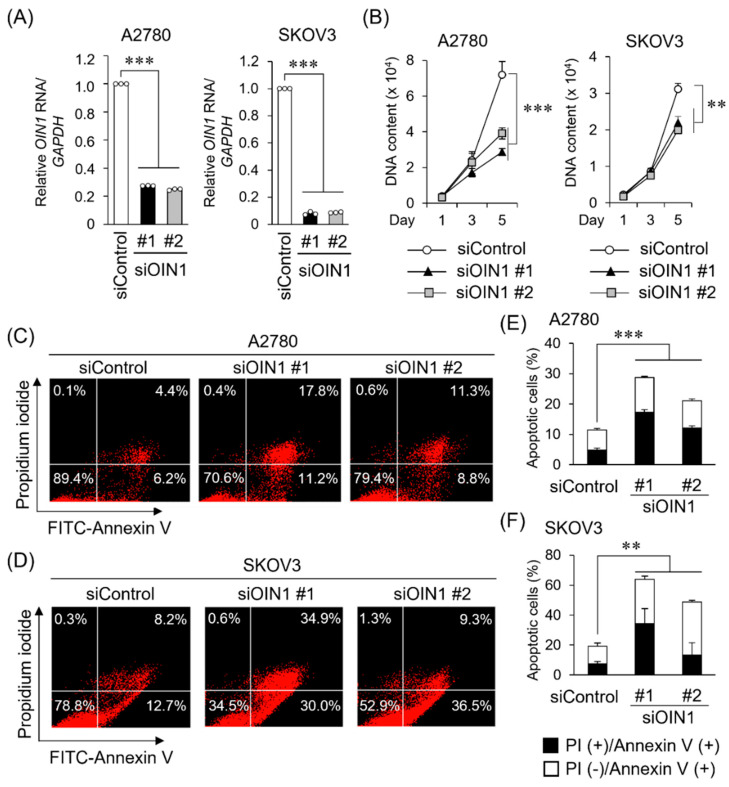
To examine the significance of OIN1 in ovarian cancer, we performed knockdown experiments using OIN1-specific siRNAs (siOIN1 #1 and #2). OIN1 expression was substantially decreased by these siRNAs in A2780 and SKOV3 cells as analyzed by qRT-PCR (Figure 2A). We showed that the siRNA-mediated OIN1 knockdown significantly suppressed the proliferation of these A2780 and SKOV3 cells (Figure 2B). Conversely, the exogenous expression of OIN1 promoted the proliferation of A2780 and SKOV3 cells (Figure S1A–C,F). We next examined whether OIN1 knockdown modulates the apoptosis of ovarian cancer cells. As analyzed by flow cytometry, OIN1 knockdown increased annexin V-positive fractions in A2780 and SKOV3 cells (Figure 2C–F). The expression levels of an apoptosis marker, cleaved PARP1 protein, were increased in OIN1-silenced A2780 and SKOV3 cells (Figure S2A,B). In addition, the mRNA expression of anti-apoptotic B-cell lymphoma 2 (BCL2) was downregulated, and the ratio of the expression of proapoptotic BCL2-associated X, apoptosis regulator (BAX) to BCL2 was increased in OIN1-silenced cells (Figure S2C–H). These results suggest that OIN1 promotes proliferation and suppresses apoptosis of ovarian cancer cells.
Figure 2.
OIN1 regulates proliferation and apoptosis in ovarian cancer cells. (A) Knockdown efficiencies of siRNAs targeting OIN1 (siOIN1 #1 and #2) in A2780 and SKOV3 cells 48 h after siRNA transfection were analyzed by qRT-PCR. Relative OIN1 RNA expression levels were normalized to GAPDH mRNA levels and presented as mean fold change ± SD compared with control siRNA (siControl) in each cell type (n = 3). (B) Inhibitory effects of OIN1 knockdown in A2780 and SKOV3 cell proliferation were analyzed by DNA assay. Data are presented as mean ± SD (A2780, n = 5; SKOV3, n = 3). (C–F) Promoting effects of OIN1 knockdown in apoptosis of A2780 (C) and SKOV3 (D) cells 72 h after siRNA transfection were analyzed by flow cytometry with propidium iodide and annexin V. Percentages of annexin V-positive populations of A2780 (E) and SKOV3 (F) cells treated with indicated siRNAs were quantified (n = 3). **, p < 0.001; ***, p < 0.0001, two-way ANOVA.
2.3. OIN1 Modulates the Expression of Apoptosis- or Cell Proliferation-Related Genes
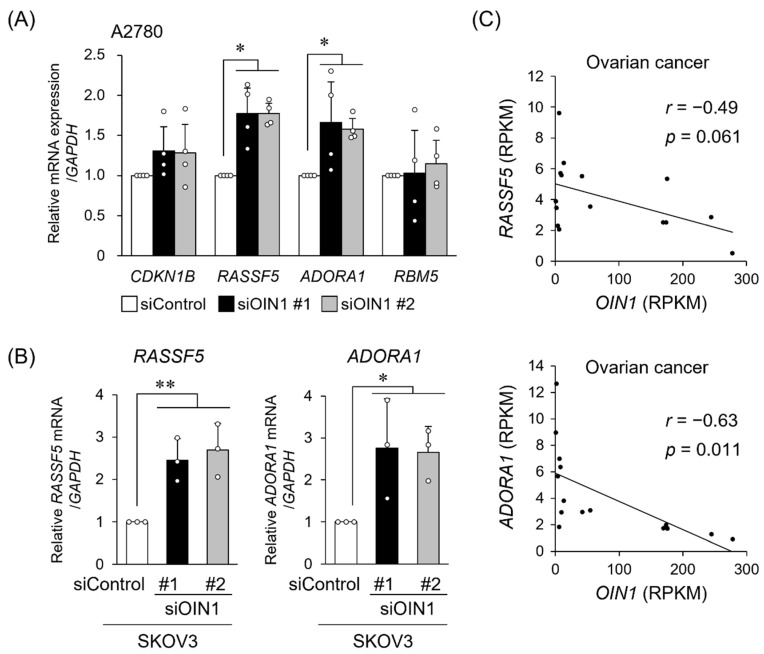
We next screened OIN1-associated genes whose expression levels exhibited positive or negative correlations with OIN1 in the RNA-seq data from ovarian cancer specimens [15,16]. We found that 323 and 312 genes had correlation coefficients of ≥0.6 and ≤−0.45 with OIN1, respectively, and analyzed enriched biological pathways in these identified genes using the DAVID Bioinformatics Resources 6.8 [24] (Tables S1 and S2). We noted that the apoptosis- and cell proliferation-related pathways were enriched in the genes showing negative correlation coefficient values (≤−0.45) (Table S2). Among the selected genes, we selected 4 genes including cyclin-dependent kinase inhibitor 1B (CDKN1B), RASSF5, ADORA1, and RNA-binding motif protein 5 (RBM5), which were previously characterized as those involved in apoptosis and cell proliferation (Figure 3A). Of the 4 genes, we found that OIN1 knockdown significantly upregulated RASSF5 and ADORA1 mRNA levels in both A2780 and SKOV3 cells (Figure 3A,B). In contrast, OIN1 overexpression substantially downregulated RASSF5 and ADORA1 mRNA levels in A2780 and SKOV3 cells (Figure S1D,E,G,H). Scattered plots of RASSF5 or ADORA1 mRNA levels versus OIN1 levels, shown as RPKM values of our RNA-seq data of clinical ovarian cancer tissues, revealed that OIN1 had a tendency of negative correlation with RASSF5 (r = −0.49 and p = 0.06) and a significant negative correlation with ADORA1 (r = −0.63 and p = 0.01) (Figure 3C). Taken together, we proposed that OIN1 negatively modulates expression of RASSF5 or ADORA1, which may contribute to the alteration of ovarian cancer cell proliferation.
Figure 3.
OIN1 regulates expression levels of RASSF5 and ADORA1. (A) Increased expression of RASSF5 and ADORA1 in OIN1-downregulated A2780 cells. A2780 cells were transfected with OIN1 siRNAs (siOIN1 #1 and #2) or siControl. Relative expression levels of selected genes 48 h after siRNA transfection were quantified by qRT-PCR and normalized to GAPDH mRNA levels. Data are presented as mean fold change ± SD in each gene group (n = 4). (B) Increased expression of RASSF5 and ADORA1 in OIN1-downregulated SKOV3 cells. SKOV3 cells were transfected as above, and expression levels of RASSF5 and ADORA1 mRNAs 72 h after siRNA transfection were quantified by qRT-PCR. Data are normalized to GAPDH mRNA levels and presented as mean fold change ± SD in each gene group (n = 3). *, p < 0.05; **, p < 0.01; two-way ANOVA. (C) Relationship between expression levels of RASSF5 or ADORA1 mRNA and OIN1 RNA analyzed using our RNA-seq data for clinical ovarian cancer specimens (n = 15) [15,16].
2.4. OIN1 Silencing Suppresses In Vivo Tumor Growth of Ovarian Cancer Cells
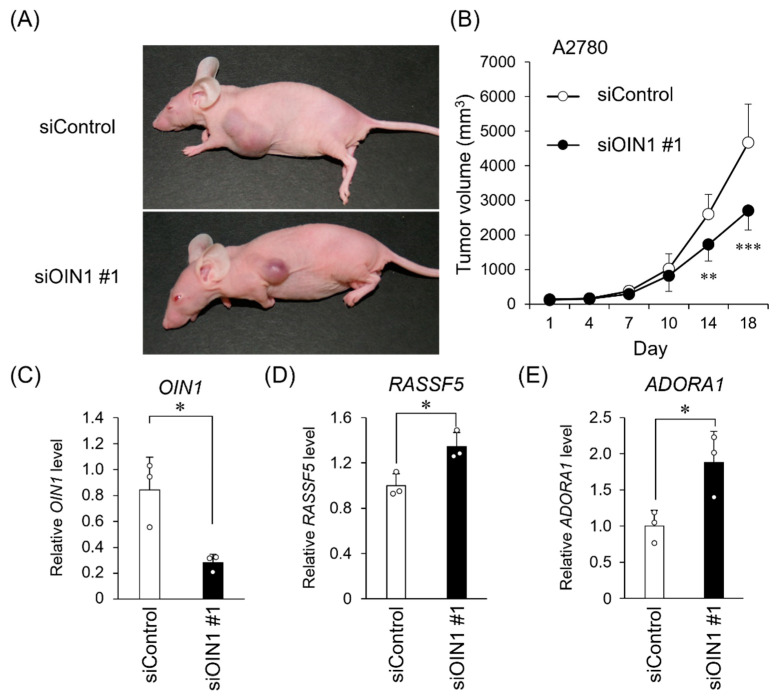
We further examined the role of OIN1 in in vivo tumor formation of ovarian cancer cells. We generated a A2780 cell-derived xenograft model by the subcutaneous administration of cells with Matrigel into female athymic mice, followed by the intratumoral injection of siRNAs twice a week. We showed that the OIN1-specific siRNA injection significantly suppressed tumor formation in the xenografted mice (Figure 4A,B and Figure S3). Notably, OIN1 expression was downregulated (Figure 4C), whereas RASSF5 and ADORA1 expression was increased (Figure 4D,E) in dissected xenograft tumors injected with OIN1-specific siRNA. Overall, these results suggest that OIN1 is a functional lincRNA that may contribute to ovarian cancer progression by modulating apoptosis-related gene expression.
Figure 4.
OIN1 siRNA inhibits tumor formation of ovarian cancer xenografts. (A) Athymic mice were subcutaneously xenografted with A2780 cells and, then, siOIN1 #1 (n = 8) or siControl (n = 7) was injected into the tumors twice a week. Representative images of generated tumors after 18 days are shown. (B) Tumor volumes are presented as mean ± SD. (C–E) Relative expression levels of OIN1 RNA (C), RASSF5 (D), and ADORA1 (E) mRNAs in the tumors dissected 18 days after the start of siRNA injection were quantified by qRT-PCR and normalized to GAPDH mRNA levels. Data are presented as mean fold change ± SD versus siControl in tumors (siControl, n = 3; siOIN1 #1, n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001; Student’s t-test.
3. Discussion
In the present study, we identified the novel ovarian cancer-related lincRNA OIN1, which is highly expressed in ovarian cancer. Functional analyses revealed that OIN1 substantially suppresses apoptosis and promotes the proliferation of ovarian cancer cells. Moreover, candidate apoptosis- or cell proliferation-related genes RASSF5 and ADORA1 were identified as OIN1 downstream genes in ovarian cancer. siRNA-mediated OIN1 silencing significantly decreased the in vivo tumor formation of ovarian cancer cells, along with the upregulation of RASSF5 and ADORA1. Although there was a difference in the experiment conditions of in vitro and in vivo OIN1 silencing experiments, these results suggest that OIN1 plays a crucial role in ovarian cancer progression.
Among the ovarian cancer cells used in the present study, OIN1 was highly expressed in A2780 and SKOV3 cells, and was moderately expressed in OV90 cells. Ovarian cancer is a heterogeneous disease and is classified into multiple histological subtypes. Regarding the cells used in this study, OV90 and OVCAR3 cells are derived from high-grade serous carcinoma while ES2 cells are derived from clear cell carcinoma, and the histological subtypes from which A2780 and SKOV3 originate remain controversial [25,26]. All the cells used in this study possess wild-type breast cancer 1/2 (BRCA1/2) [25,26,27]. Meanwhile, TP53 mutations have been reported in SKOV3, OV90, OVCAR3, and ES2 cells, while A2780 cells have wild-type TP53 [26]. Since there is no correlation between TP53 mutation status and OIN1 expression levels in these cells, we assume that TP53 mutation status may not be a determinant of OIN1 expression levels in ovarian cancer cells.
RASSF5 belongs to the ras-association domain family, which is generally known as a tumor suppressor [28]. Through the conformational change in RASSF5 upon ras association, RASSF5 modulates the Hippo pathway by phosphorylating its component pro-apoptotic mammalian STE20-like protein kinase 1/2 (MST1/2), leading to the activation of large tumor suppressor kinase 1/2 (LATS1/2) and the protein degradation of YAP1, controlling the expression of genes involved in proliferation (e.g., proliferating cell nuclear antigen; PCNA), invasion (e.g., matrix metallopeptidase 9; MMP9), and apoptosis (e.g., p53) [29]. Besides MST1/2-mediated RASSF5 functions on cell proliferation and apoptosis, RASSF5 was also reported to function independently of ras or MST1/2 in lung cancer cells [30]. RASSF5 was also shown to be involved in apoptosis, mediated by the tumor necrosis factor α (TNF-α), TNF-related apoptosis-inducing ligand (TRAIL), and CD40 ligand [31,32]. Overall, RASSF5 may exert tumor-suppressive functions through multiple context-dependent mechanisms. In ovarian cancer cells, RASSF5 downregulation was shown due to CpG hypermethylation in the RASSF5 promoter. In contrast, RASSF5 upregulation by ten-eleven translocation 1 (TET1)-mediated demethylation may result in the suppression of ovarian cancer cell proliferation [33].
ADORA1 is a G-protein coupled receptor (GPCR) for adenosine, typically mediated by the G-proteins Gi and Go [34]. The role of ADORA1 in cancer remains controversial. ADORA1 was shown to suppress proliferation and induce apoptosis in CW2 colon cancer and MCF-7 breast cancer cells [34]. Conversely, ADORA1 was reported to promote the growth and survival of 786-O and ACHN kidney cancer cells, or MDA-MB-468 breast cancer cells [34,35]. Notably, treatment with the ADORA1 antagonist SLV320 partially rescued the adenosine-mediated decrease in A2780 cell survival, suggesting that ADORA1 may play a tumor-suppressive role in ovarian cancer [36].
A question remains how OIN1 modulates the expression of RASSF5 and ADORA1 in ovarian cancer. Previous literature showed that LUCAT1 and TUG1 are examples of oncogenic lncRNAs that suppress apoptosis in ovarian cancer: the former promotes the proliferation and migration of cancer cells through the miR-612/homeobox A13 (HOXA13) axis [37], and the latter decreases apoptosis by targeting miR-29b-3p, leading to paclitaxel resistance [38]. Similarly, OIN1 may also have a possibility to promote ovarian cancer progression by targeting particular miRNAs. In terms of sequence similarity, we did not observe substantial homology between OIN1 and RASSF5 or ADORA1 mRNAs, suggesting that OIN1 may interact with RASSF5 and ADORA1 mRNAs through a mechanism other than direct RNA-RNA binding. Recent study of long terminal repeat (LTR) retrotransposon-derived lncRNA p53-regulated lncRNA for homologous recombination repair 1 (PRLH1) in p53-mutated hepatocellular carcinoma showed that the lncRNA functions as a homologous recombination-promoting factor [39]. Interestingly, we found that OIN1 may have a similarity with human endogenous retrovirus (HERV)-like long terminal repeat (LTR) retrotransposon ERV1 clade [40] as shown by the UCSC Genome Browser. The oncogenic role of ERV1 has been shown as high ERV1 expression, and in kidney cancer was shown to be associated with worse patient survival outcomes [40]. Given that OIN1 was originated from an endogenous retrovirus-like sequence, future study may reveal whether the lincRNA may affect the homologous recombination or immune reactions contributing to the pathophysiology of ovarian cancer.
In the present study, we examined the role of OIN1 in ovarian cancer mainly using A2780 and SKOV3 cells. Considering the heterogeneity of ovarian cancer, studies with other ovarian cancer cells may also provide useful information to understand the oncogenic role of OIN1 (e.g., OIN1 overexpression in ovarian cancer cells with low expression of OIN1, such as OVCAR3 and ES2). Analyzing the role of OIN1 in xenograft models using ovarian cancer cells other than A2780 will provide useful information to characterize the in vivo role of OIN1. Recently, three-dimensional cultures of patient-derived cancer cells (PDCs) and patient-derived xenograft (PDX) models have been applied to preclinical studies because they usually retain the properties of original tumors [41,42,43,44,45]. Characterizing OIN1 in in vitro experiments with PDCs and their xenograft models or PDX models will be useful to elucidate the precise role of this lincRNA. For clinical applications of OIN1 to ovarian cancer treatment, it will be important to examine whether intravenous siOIN1 injections substantially repress tumor formation of ovarian cancer models. As we recently demonstrated that intravenous injections of an siRNA targeting an oncogenic lncRNA thymopoietin antisense transcript 1 (TMPO-AS1) efficiently suppressed the xenograft tumor growth and lung metastasis derived from breast cancer cells [46], the therapeutic efficacy of siOIN1 can be evaluated in similar ovarian cancer xenograft models or patient-derived cancer models.
4. Materials and Methods
4.1. RNA-Seq Analysis of Clinical Specimens from Normal and Ovarian Cancer Tissues
Study protocols and patient consent were approved by the Institutional Review Board of Saitama Medical University International Medical Center (#12-096, #13-165). Ovarian cancer and normal tissue specimens were obtained from patients who underwent surgery for primary ovarian tumor. Detailed information for the clinical specimens and RNA-seq methods were described previously [15,16]. Expression values of RNA-seq were quantified as RPKM based on RefSeq and NONCODE (http://www.noncode.org/ [accessed on 22 January 2021]) gene models [18]. Among transcripts mapped to NONCODE v4 gene sets, we selected 10 putative differentially expressed lincRNAs, which were particularly upregulated in ovarian cancer compared with normal tissues by ≥10-folds at an FDR adjusted p-value, or q-value threshold <0.05 (Table 1). Among the 10 lincRNAs, NONHSAT013448 designated as OIN1 in this study exhibited the highest expression in ovarian cancer tissues. We screened positively or negatively OIN1-correlated Refseq genes showing correlation coefficients (r) between OIN1 as ≥0.6 or ≤−0.45. Biological pathways enriched among the OIN1-associated genes were analyzed using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/summary.jsp [accessed on 22 January 2021]) [24].
4.2. Human Ovarian Cancer Cell Culture
OV90, OVCAR3, and SKOV3 ovarian cancer cells, A2780 ovarian cancer cells, and ES2 ovarian cancer cells were grown in Dulbecco’s modified Eagle’s medium (DMEM), in RPMI 1640 and DMEM/F12, respectively, supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a 5% CO2 atmosphere. All cell lines were authenticated using short-tandem-repeat (STR) analysis (BEX, Tokyo, Japan).
4.3. siRNA and Plasmid Transfection
The custom small interfering RNAs (siRNAs) against OIN1 synthesized by Sigma-Aldrich (St Louis, MO, USA) were as follows: siOIN1 #1, 5’-GCUCAGCUCACGGCUUCUACC-3’ (sense) and 5’-UAGAAGCCGUGAGCUGAGCUC-3’ (antisense); siOIN1 #2, 5’-GACAGGAGACUCCAGAAAAGG-3’ (sense) and 5’-UUUUCUGGAGUCUCCUGUCUG-3’ (antisense). The control siRNA (siControl) was synthesized at RNAi Inc. (Tokyo, Japan) [47]. Cells were transfected with siRNAs (10 nM) using Lipofectamine RNAiMax (Thermo Fisher Scientific, Waltham, MA, USA) or transfected with the indicated plasmids using Lipofectamine 3000 reagent (Thermo Fisher Scientific) or FuGene HD transfection reagent (Promega, Madison, WI, USA). After 24, 48 or 72 h incubation, cells were harvested for qRT-PCR.
4.4. RNA Extraction and qRT-PCR
Total RNA was extracted from the ovarian cancer cells and A2780-derived xenografted tumors using ISOGEN reagent (Nippon Gene Co., Toyama, Japan) or Sepasol-RNA I Super G (Nacalai Tesque, Kyoto, Japan). One microgram of total RNA was reverse-transcribed to single-stranded cDNAs using SuperScript III (Thermo Fisher Scientific) or PrimeScript™ RT reagent kit (perfect real time) (Takara Bio Inc., Shiga, Japan). qRT-PCR was carried out on a StepOnePlus™ real-time PCR System (Thermo Fisher Scientific) using KAPA SYBR FAST qPCR Kit (KAPA Biosystems, Wilmington, MA, USA) with gene-specific primers. Relative RNA levels were analyzed using the ΔΔCt method according to the manufacturer’s protocol and normalized to GAPDH. Primers used for qPCR are listed in Table S3.
4.5. Cell Proliferation Assay (DNA Assay)
A2780 and SKOV3 cells were plated at 3000 or 1000 cells/well, respectively, in 96-well plates. After 24 h, these cells were transfected with the indicated siRNAs or plasmids as described above. Cells were collected at the indicated days after cell plating. To evaluate cell proliferation ability, the extracted DNA was stained with Hoechst 33258 pentahydrate (Thermo Fisher Scientific) at a final concentration of 5 μg/mL. DNA content in each well was measured using a 2030 ARVO X5 multilabel plate reader or VICTOR Nivo multimode microplate reader (Perkin Elmer, Foster City, CA, USA) [48].
4.6. Apoptosis Assay
A2780 and SKOV3 cells were plated at 3 × 105 or 1 × 105 cells/well, respectively, in 6-well plates. After 24 h, the cells were transfected with the indicated siRNAs at a final concentration of 10 nM and collected 72 h after transfection. Apoptotic cells were stained with a FITC Annexin V Apoptosis Detection Kit I (BD Biosciences, San Jose, CA, USA) following the manufacturer’s instructions. Annexin V- and propidium iodide (PI)-positive cells were analyzed with BD FACSCalibur (BD Biosciences) [48].
4.7. Western Blotting
Whole cell lysates were prepared from A2780 or SKOV3 cells transfected with indicated siRNAs for 48 or 72 h. Western blotting using anti-cleaved PARP1 (Abcam, Cambridge, UK) and β-actin (Sigma–Aldrich) antibodies and subsequent detection were performed as described [46].
4.8. In Vivo Tumor Formation and siRNA Treatment
All animal experiments were approved by the Animal Care and Use Committee of Saitama Medical University, and performed following the institutional Guidelines and Regulations. Female athymic mice (BALB/cAJcl-nu/nu) were purchased from CREA Japan Inc (Tokyo, Japan). A2780 cells (5 million cells per mouse) were mixed with an equal volume of Matrigel matrix (Corning, Corning, NY, USA) and injected subcutaneously into the side flank of 10-week-old female athymic mice. We inoculated A2780 cells subcutaneously into 20 female athymic mice, and tumors were generated in 15 mice. Then, we randomly assigned these 15 mice to the siControl (n = 7) or siOIN1 #1 groups (n = 8). siControl or siOIN1 #1 (5 μg each) was prepared with the GeneSilencer reagent (Gene Therapy System, San Diego, CA, USA) as described previously [49] and injected into the generated tumors twice a week. Three dimensions of tumor were measured twice a week, and tumor volumes were calculated using the following formula: 0.5 × largest dimension × intermediate dimension × shortest dimension.
4.9. Statistical Analysis
Statistical analysis was performed using two-way analysis of variance (ANOVA), or unpaired two-tailed Student’s t-test, as indicated. JMP 9.0.0 (SAS Institute) was used for statistical analysis.
5. Conclusions
In summary, we showed that OIN1 is a functional lincRNA abundantly expressed in ovarian cancer and functions as a tumor-promoting molecule by suppressing apoptosis, suggesting that OIN1 may be served as a potential diagnostic and therapeutic target for ovarian cancer.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222011242/s1.
Author Contributions
Conceptualization, T.T., K.H. (Kuniko Horie) and S.I.; methodology, T.T., Y.M. and K.I.; software, T.T. and Y.M.; validation, T.T., Y.M. and K.I.; formal analysis, K.H. (Kuniko Horie); investigation, T.T. and Y.M.; resources, K.H. (Kosei Hasegawa) and K.H. (Kuniko Horie); data curation, K.I., K.H. (Kuniko Horie) and S.I.; writing—original draft preparation, T.T.; writing—review and editing, Y.M., K.I., K.H. (Kosei Hasegawa), K.H. (Kuniko Horie) and S.I.; visualization, T.T.; supervision, K.H. (Kuniko Horie) and S.I.; project administration, K.H. (Kuniko Horie) and S.I.; funding acquisition, T.T., K.I., K.H. (Kuniko Horie) and S.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by grants from the Japan Society for the Promotion of Science, Japan [20K08916 and 20K21636 (to K.I.); 20H03734 (to K.H.); 20K21667 (to S.I.); 21K16798 (to T.T.)]; by the Takeda Science Foundation (to K.I. and S.I.); by the Vehicle Racing Commemorative Foundation (to K.H.); and by the Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan (to T.T.).
Institutional Review Board Statement
The study using clinical samples abided by the Declaration of Helsinki principles and was approved by the Institutional Review Board of the Saitama Medical University International Medical Center (#13-165, 7 February 2014).
Informed Consent Statement
Ovarian cancer and normal tissue specimens were obtained from patients who underwent surgery for primary ovarian tumor with their informed consent in a previous study (#12-096, 5 September 2012).
Data Availability Statement
The data used to support the findings of the present study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Momenimovahed Z., Tiznobaik A., Taheri S., Salehiniya H. Ovarian cancer in the world: Epidemiology and risk factors. Int. J. Women’s Health. 2019;11:287–299. doi: 10.2147/IJWH.S197604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coburn S.B., Bray F., Sherman M.E., Trabert B. International patterns and trends in ovarian cancer incidence, overall and by histologic subtype. Int. J. Cancer. 2017;140:2451–2460. doi: 10.1002/ijc.30676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torre L.A., Trabert B., DeSantis C.E., Miller K.D., Samimi G., Runowicz C.D., Gaudet M.M., Jemal A., Siegel R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kashi K., Henderson L., Bonetti A., Carninci P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochim. Biophys. Acta. 2016;1859:3–15. doi: 10.1016/j.bbagrm.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Anastasiadou E., Messina E., Sanavia T., Labruna V., Ceccarelli S., Megiorni F., Gerini G., Pontecorvi P., Camero S., Perniola G., et al. Calcineurin gamma catalytic subunit PPP3CC inhibition by miR-200c-3p affects apoptosis in epithelial ovarian cancer. Genes. 2021;12:1400. doi: 10.3390/genes12091400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alshamrani A.A. Roles of microRNAs in ovarian cancer tumorigenesis: Two decades later, what have we learned. Front. Oncol. 2020;10:1084. doi: 10.3389/fonc.2020.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takayama K.I., Inoue S. The emerging role of noncoding RNA in prostate cancer progression and its implication on diagnosis and treatment. Brief. Funct. Genom. 2016;15:257–265. doi: 10.1093/bfgp/elv057. [DOI] [PubMed] [Google Scholar]
- 8.Misawa A., Takayama K.I., Inoue S. Long non-coding RNAs and prostate cancer. Cancer Sci. 2017;108:2107–2114. doi: 10.1111/cas.13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitobe Y., Takayama K.I., Horie-Inoue K., Inoue S. Prostate cancer-associated lncRNAs. Cancer Lett. 2018;418:159–166. doi: 10.1016/j.canlet.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Arun G., Diermeier S.D., Spector D.L. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeiwa T., Ikeda K., Mitobe Y., Horie-Inoue K., Inoue S. Long noncoding RNAs involved in the endocrine therapy resistance of breast cancer. Cancers. 2020;12:1424. doi: 10.3390/cancers12061424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J.Y., Lu A.Q., Chen L.J. LncRNAs in ovarian cancer. Clin. Chim. Acta. 2019;490:17–27. doi: 10.1016/j.cca.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Zheng J., Guo J., Zhang H., Cao B., Xu G., Zhang Z., Tong J. Four prognosis-associated lncRNAs serve as biomarkers in ovarian cancer. Front. Genet. 2021;12:672674. doi: 10.3389/fgene.2021.672674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeiwa T., Ikeda K., Horie-Inoue K., Inoue S. Mechanisms of apoptosis-related long non-coding RNAs in ovarian cancer. Front. Cell Dev. Biol. 2021;9:641963. doi: 10.3389/fcell.2021.641963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagasawa S., Ikeda K., Horie-Inoue K., Sato S., Itakura A., Takeda S., Hasegawa K., Inoue S. Systematic identification of characteristic genes of ovarian clear cell carcinoma compared with high-grade serous carcinoma based on RNA-sequencing. Int. J. Mol. Sci. 2019;20:4330. doi: 10.3390/ijms20184330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagasawa S., Ikeda K., Horie-Inoue K., Sato S., Takeda S., Hasegawa K., Inoue S. Identification of novel mutations of ovarian cancer-related genes from RNA-sequencing data for Japanese epithelial ovarian cancer patients. Endocr. J. 2020;67:219–229. doi: 10.1507/endocrj.EJ19-0283. [DOI] [PubMed] [Google Scholar]
- 17.Ransohoff J.D., Wei Y., Khavari P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie C., Yuan J., Li H., Li M., Zhao G., Bu D., Zhu W., Wu W., Chen R., Zhao Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y., Meng H., Liu S., Hu J., Zhang Y., Jiao T., Liu Y., Ou J., Wang D., Yao L., et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum. Mol. Genet. 2015;24:841–852. doi: 10.1093/hmg/ddu502. [DOI] [PubMed] [Google Scholar]
- 20.Wang F., Zhou J., Xie X., Hu J., Chen L., Hu Q., Guo H., Yu C. Involvement of SRPK1 in cisplatin resistance related to long non-coding RNA UCA1 in human ovarian cancer cells. Neoplasma. 2015;62:432–438. doi: 10.4149/neo_2015_051. [DOI] [PubMed] [Google Scholar]
- 21.Li Z., Niu H., Qin Q., Yang S., Wang Q., Yu C., Wei Z., Jin Z., Wang X., Yang A., et al. lncRNA UCA1 mediates resistance to cisplatin by regulating the miR-143/FOSL2-signaling pathway in ovarian cancer. Mol. Ther. Nucleic Acids. 2019;17:92–101. doi: 10.1016/j.omtn.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X., Spindler T.J., de Souza Fonseca M.A., Corona R.I., Seo J.H., Dezem F.S., Li L., Lee J.M., Long H.W., Sellers T.A., et al. Super-enhancer-associated LncRNA UCA1 interacts directly with AMOT to activate YAP target genes in epithelial ovarian cancer. iScience. 2019;17:242–255. doi: 10.1016/j.isci.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y., Liu Y., Cai Y., Han P., Wang R., Cao L., He S. Downregulation of LINC00958 inhibits proliferation, invasion and migration, and promotes apoptosis of colorectal cancer cells by targeting miR-3619-5p. Oncol. Rep. 2020;44:1574–1582. doi: 10.3892/or.2020.7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domcke S., Sinha R., Levine D.A., Sander C., Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaufort C.M., Helmijr J.C., Piskorz A.M., Hoogstraat M., Ruigrok-Ritstier K., Besselink N., Murtaza M., van IJcken W.F., Heine A.A., Smid M., et al. Ovarian cancer cell line panel (OCCP): Clinical importance of in vitro morphological subtypes. PLoS ONE. 2014;9:e103988. doi: 10.1371/journal.pone.0103988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stordal B., Timms K., Farrelly A., Gallagher D., Busschots S., Renaud M., Thery J., Williams D., Potter J., Tran T., et al. BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Mol. Oncol. 2013;7:567–579. doi: 10.1016/j.molonc.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao T.J., Tsai C.J., Jang H., Fushman D., Nussinov R. RASSF5: An MST activator and tumor suppressor in vivo but opposite in vitro. Curr. Opin. Struct. Biol. 2016;41:217–224. doi: 10.1016/j.sbi.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X.H., Yang C.Q., Zhang C.L., Gao Y., Yuan H.B., Wang C. RASSF5 inhibits growth and invasion and induces apoptosis in osteosarcoma cells through activation of MST1/LATS1 signaling. Oncol. Rep. 2014;32:1505–1512. doi: 10.3892/or.2014.3387. [DOI] [PubMed] [Google Scholar]
- 30.Aoyama Y., Avruch J., Zhang X.F. Nore1 inhibits tumor cell growth independent of Ras or the MST1/2 kinases. Oncogene. 2004;23:3426–3433. doi: 10.1038/sj.onc.1207486. [DOI] [PubMed] [Google Scholar]
- 31.Park J., Kang S.I., Lee S.Y., Zhang X.F., Kim M.S., Beers L.F., Lim D.S., Avruch J., Kim H.S., Lee S.B. Tumor suppressor ras association domain family 5 (RASSF5/NORE1) mediates death receptor ligand-induced apoptosis. J. Biol. Chem. 2010;285:35029–35038. doi: 10.1074/jbc.M110.165506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elmetwali T., Salman A., Palmer D.H. NORE1A induction by membrane-bound CD40L (mCD40L) contributes to CD40L-induced cell death and G1 growth arrest in p21-mediated mechanism. Cell Death Dis. 2016;7:e2146. doi: 10.1038/cddis.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B.T., Yu C., Xu Y., Liu S.B., Fan H.Y., Pan W.W. TET1 inhibits cell proliferation by inducing RASSF5 expression. Oncotarget. 2017;8:86395–86409. doi: 10.18632/oncotarget.21189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazemi M.H., Raoofi Mohseni S., Hojjat-Farsangi M., Anvari E., Ghalamfarsa G., Mohammadi H., Jadidi-Niaragh F. Adenosine and adenosine receptors in the immunopathogenesis and treatment of cancer. J. Cell Physiol. 2018;233:2032–2057. doi: 10.1002/jcp.25873. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Y., Tong L., Chu X., Deng F., Tang J., Tang Y., Dai Y. The adenosine A1 receptor antagonist DPCPX inhibits tumor progression via the ERK/JNK pathway in renal cell carcinoma. Cell. Physiol. Biochem. 2017;43:733–742. doi: 10.1159/000481557. [DOI] [PubMed] [Google Scholar]
- 36.Sureechatchaiyan P., Hamacher A., Brockmann N., Stork B., Kassack M.U. Adenosine enhances cisplatin sensitivity in human ovarian cancer cells. Purinergic Signal. 2018;14:395–408. doi: 10.1007/s11302-018-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H., Xu Y., Zhang D., Liu G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem. Biophys. Res. Commun. 2018;503:2095–2100. doi: 10.1016/j.bbrc.2018.07.165. [DOI] [PubMed] [Google Scholar]
- 38.Gu L., Li Q., Liu H., Lu X., Zhu M. Long noncoding RNA TUG1 promotes autophagy-associated paclitaxel resistance by sponging miR-29b-3p in ovarian cancer cells. Onco Targets Ther. 2020;13:2007–2019. doi: 10.2147/OTT.S240434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng B., Xu W., Wang Z., Liu C., Lin P., Li B., Huang Q., Yang J., Zhou H., Qu L. An LTR retrotransposon-derived lncRNA interacts with RNF169 to promote homologous recombination. EMBO Rep. 2019;20:e47650. doi: 10.15252/embr.201847650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zapatka M., Borozan I., Brewer D.S., Iskar M., Grundhoff A., Alawi M., Desai N., Sültmann H., Moch H., Cooper C.S., et al. The landscape of viral associations in human cancers. Nat. Genet. 2020;52:320–330. doi: 10.1038/s41588-019-0558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishiguro T., Sato A., Ohata H., Ikarashi Y., Takahashi R.U., Ochiya T., Yoshida M., Tsuda H., Onda T., Kato T., et al. Establishment and characterization of an in vitro model of ovarian cancer stem-like cells with an enhanced proliferative capacity. Cancer Res. 2016;76:150–160. doi: 10.1158/0008-5472.CAN-15-0361. [DOI] [PubMed] [Google Scholar]
- 42.Namekawa T., Ikeda K., Horie-Inoue K., Inoue S. Application of prostate cancer models for preclinical study: Advantages and limitations of cell lines, patient-derived xenografts, and three-dimensional culture of patient-derived cells. Cells. 2019;8:74. doi: 10.3390/cells8010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maru Y., Hippo Y. Current status of patient-derived ovarian cancer models. Cells. 2019;8:505. doi: 10.3390/cells8050505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiba S., Ikeda K., Suzuki T., Shintani D., Okamoto K., Horie-Inoue K., Hasegawa K., Inoue S. Hormonal regulation of patient-derived endometrial cancer stem-like cells generated by three-dimensional culture. Endocrinology. 2019;160:1895–1906. doi: 10.1210/en.2019-00362. [DOI] [PubMed] [Google Scholar]
- 45.Kamada S., Namekawa T., Ikeda K., Suzuki T., Kagawa M., Takeshita H., Yano A., Okamoto K., Ichikawa T., Horie-Inoue K., et al. Functional inhibition of cancer stemness-related protein DPP4 rescues tyrosine kinase inhibitor resistance in renal cell carcinoma. Oncogene. 2021;40:3899–3913. doi: 10.1038/s41388-021-01822-5. [DOI] [PubMed] [Google Scholar]
- 46.Mitobe Y., Ikeda K., Sato W., Kodama Y., Naito M., Gotoh N., Miyata K., Kataoka K., Sasaki H., Horie-Inoue K., et al. Proliferation-associated long noncoding RNA, TMPO-AS1, is a potential therapeutic target for triple-negative breast cancer. Cancer Sci. 2020;111:2440–2450. doi: 10.1111/cas.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato W., Ikeda K., Urano T., Abe Y., Nakasato N., Horie-Inoue K., Takeda S., Inoue S. Efp promotes in vitro and in vivo growth of endometrial cancer cells along with the activation of nuclear factor-κB signaling. PLoS ONE. 2018;13:e0208351. doi: 10.1371/journal.pone.0208351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitobe Y., Ikeda K., Suzuki T., Takagi K., Kawabata H., Horie-Inoue K., Inoue S. ESR1-stabilizing long noncoding RNA TMPO-AS1 promotes hormone-refractory breast cancer progression. Mol. Cell. Biol. 2019;39:e00261-19. doi: 10.1128/MCB.00261-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ueyama K., Ikeda K., Sato W., Nakasato N., Horie-Inoue K., Takeda S., Inoue S. Knockdown of Efp by DNA-modified small interfering RNA inhibits breast cancer cell proliferation and in vivo tumor growth. Cancer Gene Ther. 2010;17:624–632. doi: 10.1038/cgt.2010.19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of the present study are available from the corresponding author upon request.