ABSTRACT
An unavoidable consequence of aerobic metabolism is the production of reactive oxygen species (ROS). Mitochondria have historically been considered the primary source of ROS; however, recent literature has highlighted the uncertainty in primary ROS production sites and it is unclear how variation in mitochondrial density influences ROS-induced damage and protein turnover. Fish skeletal muscle is composed of distinct, highly aerobic red muscle and anaerobic white muscle, offering an excellent model system in which to evaluate the relationship of tissue aerobic capacity and ROS-induced damage under baseline conditions. The present study used a suite of indices to better understand potential consequences of aerobic tissue capacity in red and white muscle of the pinfish, Lagodon rhomboides. Red muscle had a 7-fold greater mitochondrial volume density than white muscle, and more oxidative damage despite also having higher activity of the antioxidant enzymes superoxide dismutase and catalase. The dominant protein degradation system appears to be tissue dependent. Lysosomal degradation markers and autophagosome volume density were greater in white muscle, while ubiquitin expression and 20S proteasome activity were significantly greater in red muscle. However, ubiquitin ligase expression was significantly higher in white muscle. Red muscle had a more than 2-fold greater rate of translation and total ATP turnover than white muscle, results that may be due in part to the higher mitochondrial density and the associated increase in oxidative damage. Together, these results support the concept that an elevated aerobic capacity is associated with greater oxidative damage and higher costs of protein turnover.
KEY WORDS: Oxidative stress, Protein turnover, Autophagy, Ubiquitin proteasome system
Highlighted Article: High aerobic activity is linked to an accrual of oxidative damage and greater costs associated with protein turnover.
INTRODUCTION
It has long been recognized that reactive oxygen species (ROS) inflict damage on cells, organelles and macromolecules such as proteins, lipids and nucleic acids when there is a mismatch between ROS production and antioxidant activity. However, low levels of intracellular ROS in skeletal muscle (e.g. 1–15 µmol l−1 H2O2) are necessary as they act as secondary messengers for transcription factors, regulators of apoptosis and autophagy, are vital for normal force production and thus are essential for maintaining homeostasis via signaling events (Picard et al., 2012; Rani et al., 2015; Reid, 2001; Scherz-Shouval et al., 2007; Triolo and Hood, 2021).
The electron transport chain (ETC) has historically been considered the main source of ROS in skeletal muscle (Banh et al., 2016; Filho, 2007; Jackson, 2011; Rani et al., 2015; St-Pierre et al., 2002). In the ETC, ROS are a product of molecular oxygen (O2) reduction by electrons that primarily arise from complexes I and III of the ETC (Banh et al., 2016; St-Pierre et al., 2002; Wong et al., 2019). The addition of an electron to O2 yields a superoxide radical (O−2) that is quickly converted into hydrogen peroxide (H2O2) by the antioxidant enzyme superoxide dismutase (SOD). H2O2 is then scavenged by the enzyme catalase (CAT) to form water and O2 (Barreiro and Hussain, 2010).
While the ETC has received the bulk of the attention, ROS have other sources that are both mitochondrial and non-mitochondrial in origin. For instance, NADPH oxidases, specifically NOX4, which localizes to the inner mitochondrial membrane, is known to be an important ROS producer (Ferreira and Laitano, 2016; Zhang and Wong, 2021). NADPH oxidases are also found at the sarcoplasmic reticulum, sarcolemma and cytosol, while xanthine oxidase, another ROS-producing enzyme, is located in the extracellular space (Barreiro and Hussain, 2010; Brown and Borutaite, 2012; Ferreira and Laitano, 2016). Although it is commonly thought that mitochondria are the principle producers of ROS, others have questioned this view (Brown and Borutaite, 2012; Zhang and Wong, 2021). Recently, Zhang and Wong (2021) reviewed the case for mitochondria acting as the primary source of ROS and found a lack of evidence in prior studies that support this idea, particularly under baseline conditions. Further, these authors demonstrated that while mitochondrial ROS production was variable across a range of different cell types, in no cases were mitochondria the major source of ROS and they typically accounted for a third or less of the total production. Rather, ROS production was shared by mitochondria, NADPH oxidases and other unidentified sources in vitro, and this pattern extended to cases of hypoxia–reperfusion, where global ROS production from all sources was increased, but mitochondria remained a minority contributor (Zhang and Wong, 2021). While some evidence points to NADPH oxidases as the major contributors of ROS in isolated single skeletal muscle fibers (Sakellariou et al., 2013), other evidence suggests that ∼45% of ROS stems from mitochondria and ∼40% originates from NADPH oxidases in C2C12 myoblasts (Wong et al., 2019). Thus, the role that tissue aerobic capacity plays in ROS production, ROS-induced cellular damage and tissue maintenance costs is clouded by uncertainties about the sources of ROS, especially in vivo where ROS production is difficult to measure.
ROS levels are balanced by antioxidant defenses to prevent excessive damage but to permit vital functional roles of ROS. This inherently leads to a modest basal level of oxidative damage. ROS that are not scavenged by antioxidants can cause excessive oxidation of proteins, lipids and nucleic acids. Protein carbonylation, the addition of carbonyl groups to a protein by the direct interaction with ROS, and lipid peroxidation, the production of reactive aldehydes as a result of ROS (primarily the hydroxyl radical, HO•, and hydroperoxyl radical, HO•2) attacking poly-unsaturated fatty acids, are commonly measured oxidative stress markers (Barreiro and Hussain, 2010; Fedorova et al., 2014; Fritz and Petersen, 2011). Additionally, reactive aldehydes such as 4-hydroxynonenal (4-HNE) produced during lipid peroxidation are persistent, lasting up to 2 min, and have the potential of reacting with cysteine, histidine and lysine residues of proteins, serving as a secondary source of protein carbonylation (Barreiro and Hussain, 2010; Carrard et al., 2002). Conformational changes in protein structure resulting from oxidative damage can lead to the exposure of hydrophobic residues, altering the function of the protein and potentially promoting the formation of protein aggregates (Carrard et al., 2002). Furthermore, the hydroxyl radical is especially damaging to DNA, particularly to the base guanine, which can be oxidized to form 8-hydroxy-2′-deoxyguanosine (8-OHdG; Valavanidis et al., 2009). 8-OHdG is the most common DNA lesion and is widely used as a cellular oxidative stress marker.
Protein turnover is essential to replace damaged proteins and to permit changes in gene expression. Protein degradation primarily occurs via two pathways: the autophagic–lysosomal system (ALS) or the ubiquitin–proteasome system (UPS). Autophagy is essential to cell survival and muscle homeostasis, and it entails engulfment of portions of the cytoplasm, misfolded and/or oxidized proteins, large protein aggregates (formed by the hydrophobic nature of the oxidized residues) and entire organelles. A double membrane phagophore, now thought to originate from the endoplasmic reticulum in mammalian cells, sequesters the damaged macromolecules and/or organelles, forming an autophagosome (Grumati and Bonaldo, 2012; Lamb et al., 2013). The autophagosome then fuses with a lysosome in which the cellular debris is degraded by lysosomal cathepsins (de Duve and Wattiaux, 1966). ATP is indirectly used in autophagy, controlling autophagy rate and autophagosome volume density (AVD; Plomp et al., 1987; Schellens et al., 1988). The production of ROS is an upstream initiator of autophagy, particularly during cell stress, inducing damage to mitochondrial proteins, resulting in mitophagy or the removal of damaged proteins via mitochondrial derived vesicles (Romanello and Sandri, 2021).
In contrast to the ALS, the UPS is relatively rapid, dominating degradation of short-lived proteins, including single proteins and small protein aggregates. The UPS has been shown to operate in both ubiquitin- and ATP-dependent and ubiquitin- and ATP-independent ways (Davies, 2001; Shringarpure et al., 2003). In the former case, ubiquitin is attached to misfolded proteins by a set of enzymes: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases. The E3 ligases MAFbx (atrogin-1) and MuRF1 (TRIM63) are highly expressed in skeletal muscle, are vital for proper placement of ubiquitin molecules on misfolded proteins, and serve as markers of muscle atrophy (Bodine and Baehr, 2014; Bodine et al., 2001; Tacchi et al., 2012). The 26S proteasome targets proteins with poly-ubiquitin chains of four or more ubiquitin molecules (Passmore and Barford, 2004). The 26S proteasome is composed of two large subunits, the 19S regulatory particle, which unfolds and deubiquitinylates proteins, and the 20S catalytic core particle, which hosts protease sites that break proteins via hydrolysis of peptide bonds (Navon and Ciechanover, 2009). In addition, ubiquitin- and ATP-independent proteasome activity occurs when hydrophobic regions of oxidized proteins bind directly to the hydrophobic binding domain on the alpha subunits at each end of the catalytic core, thereby not requiring ubiquitin for recognition (Davies, 2001).
Prior studies in mice evaluated differences in the UPS between predominantly fast- and slow-twitch skeletal muscles (Okamoto et al., 2011), and differences in rates of protein synthesis in specific muscle fiber types (Goodman et al., 2011). In fishes, previous work has examined the effects of fasting on the contributions of the ALS and UPS to protein turnover in aerobic red muscle and anaerobic white muscle (Cassidy et al., 2016; Goodman et al., 2011; Loughna and Goldspink, 1984; Okamoto et al., 2011), and studies using murine models have evaluated the impact of hindleg suspension and denervation on protein degradation (Moriscot et al., 2010; Okamoto et al., 2011). However, while it has been suggested in the literature that there is a relationship between mitochondrial content and protein turnover, to the best of our knowledge, no study has assessed baseline muscle tissue-specific differences in mitochondrial volume density (MVD), ROS-induced damage to macromolecules, antioxidant defenses and rates of protein/ATP turnover.
Fish muscle has three major advantages for addressing these relationships. First, red and white muscle fibers of fish have a dramatic difference in aerobic capacity, where a high MVD is necessary in red muscle to power sustained swimming (Mosse and Hudson, 1977), while white muscle, which is used for burst swimming, often has some of the lowest mitochondrial densities recorded (Johnston, 2001; Burpee et al., 2010). MVD is an important indicator of the oxidative capacity of a tissue (Eya et al., 2017; Johnston and Moon, 1981; Orczewska et al., 2010). In fishes, MVD varies by about 10-fold between red and white muscle, while it varies by <2-fold among the four major fiber types in mouse skeletal muscle (types 1a, 2a, 2x and 2b; Johnston, 2001; Pathi et al., 2012). This makes the fish muscle model an excellent system for evaluating potential effects of MVD. Second, the variation in MVD occurs within discrete regions of the same tissue, allowing comparison of cells with the same basic function (locomotion). Third, fish red and white muscle are easily delineated and tissue samples can be collected from each that have a homogeneous fiber type composition (as we avoided the thin layer of intermediate ‘pink’ fibers), in contrast to the primarily mixed fiber composition of mammalian muscle (Kiessling et al., 2006; Staron et al., 1999; Soukup et al., 2002). The homogeneous composition of fiber types in fish red and white muscle removes a confounding variable when trying to interpret tissue-level properties such as macromolecule oxidative damage and ATP turnover rate. Fish muscle fiber types are consistent with their function in supporting different swimming modes, where red muscle has fibers that are analogs to slow-twitch (type 1) fibers that have been extensively characterized in mammals, while white muscle fibers are analogs of fast-twitch (type IIb) fibers (Carpené et al., 1982; Johnston et al., 1977; Ramírez-Zarsosa et al., 1998).
The pinfish, Lagodon rhomboides, is a saltwater fish inhabiting shallow coastal waters along the Atlantic and Gulf Coasts of the USA and Mexico with distinct division of red and white muscle fibers (Hoese and Moore, 1998). The present study utilized a suite of indices to better understand the association of elevated tissue aerobic capacity with oxidative damage, oxidative defenses, protein turnover and resting ATP demand. We hypothesized that highly aerobic red muscle would accrue greater oxidative damage and have higher protein/ATP turnover rates, which would be consistent with the notion that red muscle is costlier to maintain at least in part as a consequence of its high mitochondrial content.
MATERIALS AND METHODS
Animal maintenance
Adult male and female L. rhomboides (Linnaeus 1766) were collected from the Intracoastal Waterway in Wrightsville Beach, NC, USA. Fish were maintained and processed according to UNCW Institutional Animal Care and Use Committee standards. Animals were immediately transported to the laboratory at the University of North Carolina Wilmington where they were held in filtered seawater in 570 l tanks on a 14 h:10 h light:dark regime. Salinity and temperature were monitored daily; water was held constant at 30±1 ppt and 21±1°C. Animals were fed raw shrimp daily.
Tissue collection
Fish were chosen at random and isolated in the dark in 1 liter of seawater for 1 h prior to tissue collection. Tricaine methanesulfonate (MS-222; Sigma-Aldrich, St Louis, MO, USA) was introduced into the water to a final concentration of 500 mg l−1 to anesthetize the fish. This process was carried out slowly to minimize the stress placed on the fish. Fish were then killed via cervical dislocation and scales and skin were removed quickly. White epaxial muscle and lateral red muscle were extracted parallel to the fiber direction. Muscle samples used for transmission electron microscopy (TEM) were cut into 1 mm3 pieces and immediately placed in 2.5% glutaraldehyde/2% paraformaldehyde in Sörensen's phosphate buffer (pH 7.38; Karnovsky, 1965; Presnell and Schreibman, 1997) and stored at room temperature for >24 h. Samples to be used for immunoassays, protein synthesis measurements and antibody assays were immediately flash-frozen in liquid N2 and stored at −80°C until further use. Samples to be used in 31P nuclear magnetic resonance (NMR) spectroscopy experiments were used immediately as described below.
Volume density of mitochondria (MVD)
TEM was used to examine MVD to verify aerobic capacity. Red and white muscle were analyzed from 3 fish samples. Images were taken from 3–5 grids per sample (n=9 per red muscle, n=11 per white muscle). Briefly, muscle fibers were teased apart into bundles of 5–8 fibers. The tissue was washed 2× in Sörensen's phosphate buffer for 15 min before adding a secondary fixative (1.0% osmium tetroxide/0.8% potassium ferricyanide in Sörensen's phosphate buffer, pH 7.38; Reynolds, 1963). Following secondary fixation, the tissue was washed 2× in Sörensen's phosphate buffer followed by two 15 min washes in distilled water. An ascending ethanol dehydration series (50%, 70%, 95%, 100%, 100%) was then applied for 15 min each. The tissue was infiltrated with a 1:1 mixture of 100% Spurr's low viscosity epoxy resin plus 100% ethanol for 1 h, refreshed with 100% Spurr's low viscosity epoxy resin, and infiltrated overnight (Spurr, 1969). Tissue was placed in fresh 100% Spurr epoxy resin and dried at 70°C for 8 h. Sections (90 nm thick) were cut (Leica UC7 ultramicrotome), stained with uranyl acetate and lead citrate, imaged in brightfield mode at 80 kV (Tecnai 12 Spirit) at a magnification of 6800×, and captured using an Eagle 2K HR camera.
Images were analyzed in Fiji (Schindelin et al., 2012). A 0.6 µm square grid was laid over the image and points where grid cells intersected over mitochondria were counted. The ratio of mitochondria points to total intracellular points was calculated. Images were processed with Adobe Photoshop Express.
Western and dot blotting
For all immunoblotting experiments, previously frozen muscle tissue (n=8 per muscle type) was thoroughly homogenized under liquid N2 with mortar and pestle then combined with RIPA buffer (Santa Cruz Biotechnology, Dallas, TX, USA) in a 1:4 ratio. Samples were sonicated for 30 s (10 s intervals) on ice then centrifuged at 16,000 g for 20 min at 4°C. Supernatant was collected and total protein was determined via a standard Bradford assay.
For western blotting, 2× Laemmli SDS sample buffer was added in a 1:1 ratio to each sample. Samples were heated to 100°C for 5 min; 15 μg μl−1 of protein per sample was loaded into each gel lane and gels were run at 80 V for approximately 30 min. Voltage was then increased to 120 V for approximately 80 min or until migration was complete. PVDF membranes were soaked in methanol for 10 min followed by transfer buffer for 5 min. Filter paper and sponges were soaked in transfer buffer for 5 min. Protein was transferred to the membrane for 60 min at 230 mA at 4°C. Once transfer was complete, membranes were washed 5× with TBST for 5 min (all washes are 5 min unless otherwise noted). Membranes were blocked with 3% low-fat milk/TBST (3% milk) or 3% bovine serum albumin (BSA)/TBST (3% BSA, when noted) for 1 h at room temperature followed by 3 washes with TBST. Membranes were incubated overnight in primary antibody with 3% milk at 4°C. Membranes were washed 5× in TBST and then incubated in secondary antibody with 3% milk. Membranes were again washed 5× in TBST then incubated in enhanced chemiluminescent substrate (ECL; Bio-Rad, Hercules, CA, USA) for 5 min. Clear plastic wrap was used to laminate the membranes and protein was detected with the Bio-Rad Molecular Imager® Gel Doc™ XR+. Both western and dot blot membranes were stained with Coomassie Brilliant Blue as a loading control, and total protein densitometry values were used as a normalization standard. Digital images were analyzed using ImageJ 1.52d (National Institutes of Health).
For dot blotting, PVDF membranes were soaked in methanol for 10 min then air dried (approximately 60 min). Samples were heated to 100°C for 5 min; 15 μg μl−1 of protein per sample was spotted onto the membrane and allowed to dry for 30 min. The membrane was blocked in 3% milk for 1 h, and then washed 3× with TBST and incubated in primary antibody with 3% milk at 4°C overnight. The membrane was then washed 5× with TBST, incubated in secondary antibody with 3% milk for 1 h, washed again 5× with TBST, incubated in ECL substrate for 5 min, laminated in clear plastic wrap and imaged. Total protein and image analysis were performed as described above.
Oxidative damage to proteins, lipids and DNA
A standard dot blot as described above with modifications from Robinson et al. (1999) and Wilson et al. (2015) was used to measure protein carbonylation. An initial incubation used a concentration of 100 mg ml−1 2,4-dinitrophenylhydrazine (DNPH, Sigma-Aldrich), which reacts with protein carbonyl groups. A primary antibody (Invitrogen Molecular Probes: A11097, anti-2,4-dinitrophenol) at a dilution of 1:2500 and a secondary antibody (Santa Cruz Biotechnology: sc-2357, mouse-anti-rabbit) at a dilution of 1:5000 were used to determine levels of 2,4-DNPH derivatives in samples.
Dot blots were used to measure the levels of 4-HNE, a product of lipid peroxidation found in higher concentrations when cells are experiencing oxidative stress, and the levels of 8-OHdG, the predominant form of oxidative DNA base lesions due to ROS and reactive nitrogen species. A primary antibody (Cell BioLabs: STA-035, anti-4-hydroxynonenal; and Santa Cruz Biotechnology: sc-393871, 8-OHdG, respectively) in a dilution of 1:1000 and a secondary antibody (Santa Cruz Biotechnology: sc-2357, mouse-anti-rabbit; and Sigma-Aldrich: NA931V/AH, sheep-anti-mouse, respectively) at a dilution of 1:2000 were used to determine expression of 4-HNE and 8-OHdG.
Antioxidant activity
Antioxidant capacity was assessed by measuring SOD and catalase activity. SOD activity was measured using the superoxide dismutase assay kit (Cayman Chemical: 706002) according to the manufacturer’s instructions with slight modifications. Briefly, tissue was lightly homogenized under liquid N2 and added to 20 mmol l−1 Hepes buffer, pH 7.2 in a 1:5 ratio. Samples were sonicated on ice for 15 s (5 s intervals) and centrifuged at 1500 g for 5 min at 4°C. The supernatant was collected and considered to contain Cu,Zn-SOD (cytosolic fraction) as per the manufacturer’s protocol. The pellet was well mixed with 1 ml 20 mmol l−1 Hepes buffer and centrifuged at 10,000 g for 15 min at 4°C. The supernatant was collected and considered to contain Mn-SOD (mitochondrial fraction) as per the manufacturer’s protocol. Potassium cyanide (3 mmol l−1) was added to the Mn-SOD fraction to inhibit Cu,Zn-SOD and extracellular SOD, ensuring only Mn-SOD was present from the mitochondrial fraction. However, previous studies have found a small amount of Cu,Zn-SOD in the intermembrane space in addition to the cytosol, providing protection to mitochondrial proteins along with cytosolic proteins (Okado-Matsumoto and Fridovich, 2001; Sturtz et al., 2001). Therefore, we analyzed Cu,Zn-SOD as the cytosolic fraction but recognize that it is dual-localized, with the majority of Cu,Zn-SOD activity occurring in the cytosol (Okado-Matsumoto and Fridovich, 2001). A 10 µg µl−1 sample was utilized in the assay. The addition of xanthine oxidase produced H2O2 and the remaining O2•− reacted with tetrazolium salt to produce a formazan dye. Absorbance was read at 440 nm, 30 min after reaction initiation. SOD activity (U ml−1) was calculated based on the standard curve and converted to U g−1 tissue assuming 70% of tissue mass was intracellular. SOD activity in relation to cytosolic density was calculated by dividing Cu,Zn-SOD activity (U ml−1 g−1) by the cytosolic volume density from the TEM analysis to yield Cu,Zn-SOD activity per gram of cytosol (U g−1 cytosol).
CAT activity was measured using a catalase activity kit (Abcam: ab83464) according to the manufacturer's protocol. Briefly, tissue was homogenized under liquid N2 and 100 mg of tissue was added to 200 µl of the provided buffer. Samples were centrifuged at 10,000 g for 15 min at 4°C. The supernatant was combined with the CAT assay buffer provided in the kit in a 1:1 ratio (39 µl:39 µl) and added to the sample wells. Control wells were designated for each sample in which the reaction was stopped prior to 1 mmol l−1 H2O2 incubation. All control and sample wells were incubated in 1 mmol l−1 H2O2 for 30 min at 25°C. The reaction was stopped and developer mix (containing catalase assay buffer, OxiRed and HRP solution) was added to each well, including standard curve wells, and incubated for 10 min at 25°C. The unconverted H2O2 reacts with the OxiRed probe and the resulting product was measured colorimetrically at 570 nm. To calculate catalase activity, the optical density of the sample well was subtracted from the optical density of the control well. This value was then used in the H2O2 standard curve to find the amount of H2O2 (nmol) decomposed by catalase in 30 min. CAT activity (nmol min−1 ml−1) was then calculated as:
| (1) |
where B is the amount of H2O2 calculated from the standard curve, V is the volume of sample used and D is the sample dilution factor.
Autophagic–lysosomal system (ALS)
The ALS was assessed by examining LC3B-II (the lipid-conjugated form of microtubule-associated protein 1A/1B-light chain 3) expression via western blot. LC3B-II is recruited to autophagosomes, and high expression of LC3B-II indicates greater autophagic activity. A primary antibody (Cell Signaling: 2775S, anti-LC3B) at a dilution of 1:1000 and a secondary antibody (Santa Cruz Biotechnology: sc-2357, mouse anti-rabbit) at a dilution of 1:2000 were used.
In addition, TEM analysis of AVD was conducted as described above for the analysis of MVD using the same magnification and grid size.
Ubiquitin–proteasome system (UPS)
The UPS was assessed by examining ubiquitin and ubiquitin ligase (MAFbx and MuRF1) expression. Standard dot blotting was used to measure the amount of protein ubiquitinylation. A primary antibody (ENZO Life Sciences: BML-PW8810-0100, anti-polyubiquitin) was used at a dilution of 1:400 and a secondary antibody (Sigma-Aldrich: NA931V/AH, sheep-anti-mouse) was used at a dilution of 1:4000. Standard western blotting was used to measure the expression of MAFbx and MuRF1. For MAFbx detection, a primary antibody (Santa Cruz Biotechnology: sc-33782, MAFbx antibody) at a dilution of 1:1000 and a secondary antibody (Santa Cruz Biotechnology: sc-2357, mouse anti-rabbit) at a dilution of 1:2000 were used. For MuRF1 detection, a primary antibody (R&D Systems: AF5366, MuRF1/TRIM63 antibody) was used at a dilution of 1:1000 in 3% BSA and a secondary antibody (Santa Cruz Biotechnology: sc-2768, rabbit anti-goat) was used at a dilution of 1:2000 in 3% BSA.
Activity of the 20S proteasome, the core of the 26S proteasome, was evaluated as another indicator of the UPS. Muscle was extracted from 12 individuals, stored at −80°C, and homogenized as described above for immunoblotting with the modification of phosphate-buffered saline (PBS: 8 g l−1 NaCl, 0.2 g l−1 KCl, 1.44 g l−1 Na2HPO4, 0.24 g l−1 KH2PO4, pH 7.4) in place of RIPA buffer. Activity was assessed using a 20S proteasome activity assay kit (Millipore Sigma: APT280) as described by Hsu et al. (2014) and Lu and Hsu (2015).
Relative rate of translation
Relative rates of translation were determined via in vivo SUnSET (surface sensing of translation) using a protocol modified from Goodman et al. (2011). The introduction of puromycin, a tRNA analog, into the fish abdominal cavity results in puromycin incorporation into the muscle and elongation termination in nascent polypeptide chains. Thus, the extent of puromycin incorporation is proportional to the number of growing polypeptides, thereby allowing relative rates of translation to be measured. Briefly, fish were lightly anesthetized with 100 mg l−1 MS-222 until respiratory rate slowed slightly and voluntary swimming ceased. Puromycin (Life Technologies Corporation: A11138-03) was introduced into the fish via an intraperitoneal injection of 0.04 µmol g−1 fish body mass diluted in PBS for a total injection volume of 200 µl. Control fish were injected with 200 µl PBS. Full recovery from anesthesia occurred within 20–30 s in both cases. The fish were stimulated to swim vigorously for 2 min after recovery to encourage adequate puromycin perfusion into both the highly vascularized red muscle and the less vascularized white muscle. Muscle extraction was completed exactly 1.5 h after puromycin injection; muscle extraction and preparation were carried out as described above. Puromycin incorporation was detected via western blotting using a primary antibody (EMD Millipore Corp: MABE343, anti-puromycin) at a dilution of 1:1000 in 3% BSA and a secondary antibody (EMD Millipore Corp: AC111P, sheep-anti-mouse) at a 1:2000 dilution in 3% BSA.
ATP turnover rate
ATP turnover was determined by examining the rate of phosphocreatine (PCr) decrease over time during inhibition of ATP production pathways in red and white muscle. PCr serves as a reservoir of phosphoryl groups that can be transferred to ADP to form ATP:
 |
(2) |
where α is a partial proton, CK is creatine kinase and Cr is creatine. The primary function of the creatine kinase reaction is to prevent large fluctuations in ADP or ATP due to changes in ATP demand during contraction (Sahlin and Harris, 2011). The depletion rate of PCr is tightly coupled to ATP demand. Therefore, the depletion rate of PCr while inhibiting ATP production (oxidative phosphorylation and glycolysis) is equivalent to the ATP turnover rate in resting muscle (Jimenez et al., 2011, 2013).
For ATP turnover measurements, a total of 10 fish were used with white muscle extracted from 5 fish and red muscle extracted from an 5 additional fish. Muscle fiber bundles of about 2 cm in length were dissected by cutting parallel to the fiber long axis, after which tissues were immediately placed in saline (composition in mmol l−1: 133.5 NaCl, 2.4 KCl, 11.3 MgCl2, 0.47 CaCl2, 18.5 NaHCO3, pH 7.4) aerated with 99.5% O2–0.5% CO2. Muscle preparations were then quickly trimmed to a diameter of 1 mm to promote adequate oxygen diffusive flux to the muscle core. Muscle preparations were tied at resting length with surgical silk to glass tubing and allowed to stabilize in the aerated saline for 15 min.
To block muscle energy metabolism, muscle was placed in saline containing potassium cyanide (2.5 mmol l−1) and sodium iodoacetate (1.0 mmol l−1) to inhibit oxidative phosphorylation and glycolysis, respectively. Relative concentrations of PCr, ATP and inorganic P (Pi) were measured using 31P NMR. NMR spectra were acquired at 242 MHz on a Bruker AVANCE III 600 NMR spectrometer using a 45 deg excitation pulse and a 6 s relaxation delay. For white muscle, 112 scans were collected over a period of 2.5 min. For red muscle, 29 scans were collected over a period of 30 s because of the faster rate of PCr depletion in these muscles. A 10 Hz line broadening exponential multiplication was applied prior to Fourier transformation. The area under each peak was integrated using Topspin 3.6.0 (Bruker, Billerica, MA, USA) to yield relative concentrations. These values were converted to concentration by assuming an initial concentration of 7 mmol l−1 ATP in resting muscle.
Statistical analysis
All data were analyzed in JMP 14.2 (SAS Institute, Cary, NC, USA). A Dixon Q-test was used to detect outliers; outliers were removed from data analysis. Differences in body mass were analyzed via a Kruskal–Wallis test. Student's t-tests were used to compare red and white muscle groups. Linear regression was used to measure the rate of PCr decline for each red and white muscle replicate. ATP turnover in red and white muscle was compared via a Student's t-test by using the slope from the regression line of each sample. Differences were considered significant when P≤0.05.
RESULTS
Body mass
Across all experiments, body mass ranged from 26.6 to 100.2 g (wet mass). Kruskal–Wallis revealed a significant difference in body mass between experiments (P=0.03). This occurred because the fish were wild-caught at different times, and therefore we could not rigidly control animal size. However, within each type of experiment, fish of a similar size were analyzed. The exception to this was the TEM data, where one of the fish was larger than the others. However, even in this case, none of the TEM measurements were body mass dependent, based on the lack of a significant slope in regressions of each measurement against body mass.
Mitochondrial volume density (MVD)
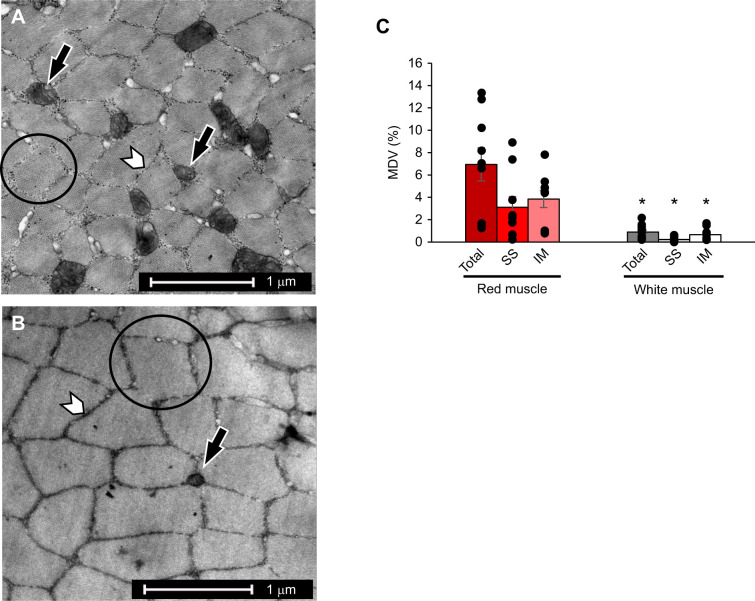
Aerobic red muscle and anaerobic white muscle samples were analyzed for total MVD, and these data were further divided into MVD fractions for the subsarcolemmal (SS) mitochondria, which lie at the periphery of the fiber, and intermyofibrillar (IM) mitochondria, which reside within the fiber core (Fig. 1A,B). Total MVD was 7-fold greater in the highly aerobic red muscle than in the largely anaerobic white muscle (Fig. 1C; Student's t-test, P<0.001), although as we did not measure mitochondrial function, cristae surface density or aerobic enzyme activity, we do not know the absolute difference in aerobic capacity. Mean total MVD was 6.9% in red muscle and 0.89% in white muscle. Likewise, SS and IM MVD fractions were greater in red muscle than in white muscle (Fig. 1C; Student's t-test, P=0.006 and P<0.001, respectively). MVD in red muscle was evenly split between the SS and IM populations, whereas in white muscle most of the mitochondria were in the IM population.
Fig. 1.
Mitochondrial volume density (MVD). Transmission electron micrographs of pinfish aerobic red (A) and anaerobic white (B) skeletal muscle fibers. Black arrows indicate examples of intermyofibrillar (IM) mitochondria, white arrowheads indicate the sarcoplasmic reticulum, and examples of myofibrils are outlined with a circle. (C) Total, subsarcolemmal (SS) and IM MVD in red (n=9) and white (n=11) muscle. Total, SS and IM MVD were significantly greater in red muscle than in white muscle (Student's t-test, *P<0.001, *P=0.006 and *P<0.001, respectively). Data are means±s.e.m.
Oxidative damage of proteins, lipids and DNA
The extent of oxidative damage to proteins, lipids and DNA was analyzed via dot blotting (Fig. 2A). There was significantly more protein carbonylation within red muscle than within white muscle (Fig. 2B; Student's t-test, P=0.008). Similarly, lipid peroxidation and DNA damage were significantly greater within red muscle than within white muscle (Fig. 2B; Student's t-test, both P<0.001).
Fig. 2.
Oxidative damage. (A) Images of dot blots with representative samples from red and white muscle for protein carbonylation (2,4-DNPH), lipid peroxidation (4-HNE) and DNA damage (8-OHdG) in red and white muscle. (B) Mean 2,4-DNPH, 4-HNE and 8-OHdG expression within red (n=8) and white (n=8) muscle. Protein carbonylation (Student's t-test, *P=0.008), lipid peroxidation (Student's t-test, *P<0.001) and DNA damage (Student's t-test, *P<0.001) were significantly higher in red muscle than in white muscle. ROS, reactive oxygen species. Data are means±s.e.m.
Antioxidant activity
To determine antioxidant activity in resting skeletal muscle, SOD and CAT activity were measured. Two forms of SOD were analyzed, Cu/Zn-SOD and Mn-SOD. Both Cu,Zn-SOD activity and Mn-SOD activity were significantly greater in red muscle (Fig. 3A,B; Student's t-test, both P<0.001). As we also measured cellular ultrastructure using TEM, Cu,Zn-SOD activity and Mn-SOD activity were normalized to the volume of cytoplasm and the volume of mitochondria, respectively, assuming all of the pellet was mitochondria for the mitochondrial SOD fraction. While Cu,Zn-SOD activity per gram of cytoplasm was significantly higher in red muscle (similar to that observed when normalized to grams of tissue), Mn-SOD activity per gram of mitochondria was significantly higher in white muscle (Student's t-test, both P<0.001; Fig. 3A,B insets). Thus, while total SOD activity was much greater in red muscle, the activity per gram of mitochondria was higher in white muscle. CAT activity in white muscle was largely below detection limits of the assay kit. Similar to SOD activity, CAT activity was significantly greater in red muscle than in white muscle (Student's t-test, P<0.001; Fig. 3C).
Fig. 3.

Antioxidant activity. (A) Cytosolic superoxide dismutase (SOD) activity was significantly greater in red muscle (n=8) than in white muscle (n=8; Student's t-test, *P<0.001). (B) Mitochondrial SOD activity was significantly greater in red muscle (n=8) than in white muscle (n=8, Student's t-test, *P<0.001). The insets show activity in per gram of cytoplasm (A) and per gram of mitochondria (B), calculated using TEM ultrastructural data assuming that 70% of tissue mass is intracellular, and that the centrifuged pellet was pure mitochondria. Cytosolic SOD activity in U g−1 cytoplasm was significantly higher in red muscle than in white muscle (*P<0.001), while mitochondrial SOD activity in U g−1 mitochondria was higher in white muscle (*P<0.001). (C) Catalase (CAT) activity in white muscle was below detection limits. CAT activity was significantly greater in red muscle (Student's t-test, *P<0.001). Data are means±s.e.m.
Autophagy–lysosomal system (APS)
Autophagy was assessed in red and white muscle via expression of LC3B-II (autophagosome membrane-bound protein) and TEM ultrastructural observations. As shown in Fig. 4A,B, LC3B-II expression was significantly greater within white muscle than within red muscle, suggesting greater autophagic activity in white muscle (Student's t-test, P<0.001). TEM revealed that the volume density of the SS and IM fractions of autophagosomes had the same pattern in both muscle types (Fig. 4C,D), so they were pooled into a single measure of total AVD. As shown in Fig. 4E, AVD was significantly greater in white muscle than in red muscle (Student's t-test, P=0.007), which is consistent with the findings for LC3B-II. Mean AVD was 0.10% in white muscle and 0.003% in red muscle.
Fig. 4.
Autophagic–lysosomal system (ALS). (A) Western blot image with representative samples of red (R) and white (W) muscle targeting LC3B-II. LC3B-I is cytosolic and is the precursor to LC3B-II. (B) Mean LC3B-II expression within red (n=8) and white (n=8) muscle. LC3B-II expression was significantly greater in white muscle (Student's t-test, *P<0.001). (C) Example transmission electron microscopy (TEM) image of an intermyofibrillar autophagosome, indicated by the arrowhead, in white muscle. The intermyofibrillar autophagosomes were typically round in shape. (D) Example TEM image of a subsarcolemmal autophagosome in white muscle, indicated by the arrowhead (arrowhead is within the autophagosome, pointing to the autophagosome membrane). The subsarcolemmal autophagosomes were typically elongated and in this example the autophagosome extends across the bottom of the image. (E) Mean autophagosome volume density (AVD) within red (n=9) and white (n=11) muscle. AVD was significantly greater in white than in red muscle (Student's t-test, *P=0.007). Data are means±s.e.m.
Ubiquitin–proteasome system (UPS)
The expression of ubiquitin and the E3 ubiquitin ligases MAFbx and MuRF1 was analyzed via dot and western blotting (Fig. 5A). Red muscle displayed significantly higher ubiquitin expression (Student's t-test, P<0.001) and 20S proteasome activity (Student's t-test, P=0.004) than white muscle (Fig. 5B,C), indicative of higher UPS activity. However, contrary to expectations, expression levels of the ubiquitin ligases MAFbx and MuRF1 were significantly greater in white muscle than in red muscle (Fig. 5B; Student's t-test, P=0.03 and P=0.005, respectively).
Fig. 5.
Ubiquitin–proteasome system (UPS). (A) Images of dot and western blots with representative samples for red and white muscle targeting ubiquitin and the E3 ubiquitin ligases MAFbx and MuRF1. (B) Mean expression of the UPS markers ubiquitin, MAFbx and MuRF1 within red (n=8) and white (n=8) muscle. Ubiquitin expression was significantly greater in red muscle (Student's t-test, *P<0.001) while MAFbx and MuRF1 expression was significantly greater in white muscle (Student's t-test, *P=0.03 and *P=0.005, respectively). (C) 20S proteasome activity of red (n=12) and white (n=12) muscle. 20S proteasome activity was significantly higher in red muscle (Student's t-test, *P=0.004). Data are means±s.e.m.
Relative rate of translation
The in vivo SUnSET method of quantifying relative translation rates involved puromycin incorporation into muscle tissue followed by western blotting (Fig. 6A). Control muscle with PBS injections displayed no puromycin expression. Significantly greater puromycin incorporation was found within red muscle than within white muscle, suggesting higher rates of protein turnover in red muscle, as expected for a highly aerobic tissue (Fig. 6B; Student's t-test, P<0.001).
Fig. 6.
Relative rate of translation. (A) Western blot images of puromycin incorporation within red (R) and white (W) muscle using the in vivo SUnSET technique. (B) Mean relative incorporation of puromycin in red (n=8) and white (n=8) muscle. Puromycin expression indicated significantly greater rates of translation in red muscle (Student's t-test, *P<0.001.) Data are means±s.e.m.
ATP turnover rate
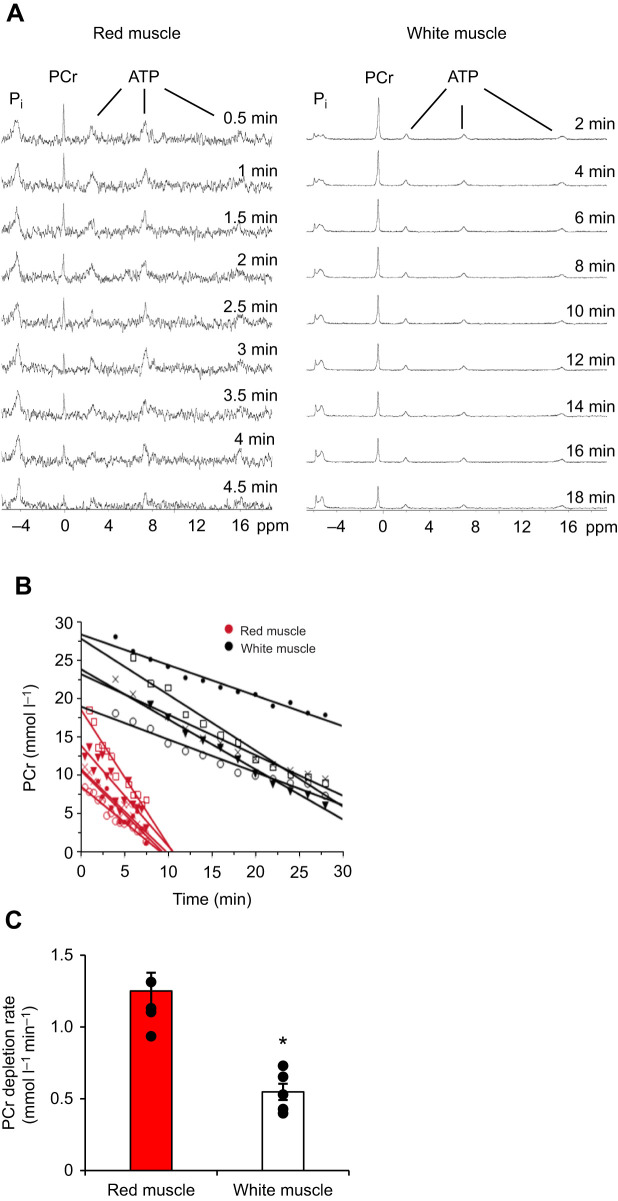
PCr depletion rates were measured in red and white muscle while oxidative phosphorylation and glycolysis were inhibited with potassium cyanide and sodium iodoacetate, respectively, to determine the rate of ATP turnover in resting muscle. Initial NMR spectra displayed a high PCr to ATP ratio and low Pi peaks (Fig. 7A). The fast depletion rate of PCr in red muscle required NMR spectra to be acquired every 30 s, which led to the lower signal to noise ratio in this tissue compared with that of white muscle, where data were acquired every 2 min. The total experiment time was 7 min for red muscle and 28 min for white muscle. As shown in Fig. 7B, red muscle had a 2.5-fold greater PCr depletion rate than white muscle (Fig. 7C; Student's t-test, P=0.005). The decrease in PCr concentration was accompanied by an increase in Pi concentration, a decrease in ATP concentration and minimal pH shift. PCr is in equilibrium with ATP as a result of creatine kinase activity in fish muscle. Thus, these results indicate that ATP turnover is 2.5 times greater in resting red muscle than in white muscle. This difference is smaller than the difference in MVD, suggesting that resting red muscle has a lower rate of ATP production per volume of mitochondria than white muscle.
Fig. 7.
ATP turnover rate. (A) Example 31P NMR spectra obtained while inhibiting ATP production in resting red and white muscle. Because of a fast phosphocreatine (PCr) depletion rate, red muscle NMR spectra were acquired every 30 s, leading to a lower signal to noise ratio, while spectra for white muscle were acquired every 2 min. (B) PCr depletion rates using 31P NMR to measure resting ATP consumption by inhibiting ATP production. Symbols indicate an individual muscle sample within each fiber type. (C) Mean PCr depletion rate for resting red (n=5) and white (n=5) muscle when ATP production was inhibited (absolute concentrations were calculated by assuming a concentration of 7 mmol l−1 ATP and relating the γ-ATP NMR peak to the peak for PCr). Linear regression was applied to each replicate within each muscle type to determine the PCr depletion rate. The significant difference in PCr depletion rate indicated that the resting ATP turnover rate was 2.5 times higher in red muscle than in white muscle (Student's t-test, *P=0.005). Data are means±s.e.m.
DISCUSSION
The current study investigated whether greater tissue aerobic capacity is associated with greater oxidative damage and higher rates of protein and ATP turnover. The major findings of this study were that, compared with white muscle, red muscle had (1) a higher aerobic capacity, demonstrated by an elevated MVD, (2) enhanced protein, lipid and DNA oxidative damage, (3) elevated activity of the antioxidant defense enzymes SOD and CAT, (4) greater expression of markers of the UPS protein degradation pathway, (5) higher relative rates of translation, and (6) a greater resting ATP turnover rate. Together, these results suggest that a higher aerobic capacity is associated with greater oxidative damage and increased cellular maintenance.
Contraction is powered by aerobic metabolism in red muscle and anaerobic PCr hydrolysis and glycolysis in white muscle, whereas both muscle types rely on aerobic metabolism for post-contraction recovery (Kinsey et al., 2007; Nyack et al., 2007). MVD reflects the functional differences of these muscle types; aerobic red muscle had a greater MVD than white muscle (6.9% compared with 0.9%, respectively). This is consistent with that found for red and white muscle in dolphinfish (Coryphaena hippurus; Pathi et al., 2012), and white muscle in bluefish (Pomatomus saltatrix), southern flounder (Paralichthys lethostigma) and black sea bass (Centropristis striata; Nyack et al., 2007; Burpee et al., 2010) although overall MVD in both fiber types is less than that found in some other marine fishes (Bone et al., 1986; Johnston, 2001). Additionally, resting aerobic ATP turnover rate was approximately 2.5 times greater in red muscle. The PCr depletion rate during inhibition of energy metabolism is a measure of the resting ATP turnover rate, and in white muscle of L. rhomboides is similar to that found in white muscle from several species of wild-caught marine fishes by Jimenez et al. (2013) and is also in agreement with other fish species including goldfish (Carassius auratus; van den Thillart et al., 1990) and tilapia (Oreochromis mossambicus; van Ginneken et al., 1999).
Although mitochondria are considered major sites for ROS production (Filho, 2007; St-Pierre et al., 2002), Zhang and Wong (2021) reviewed the available evidence and concluded that there is little direct data supporting mitochondria as the primary contributors to ROS production. Further, these authors showed that among a range of cell types, mitochondria were not the major producers of ROS in vitro. The current study indirectly looked at whether aerobic capacity, or mitochondrial density, is associated with ROS damage and ultimately protein turnover that may be influenced by ROS. Protein carbonylation, lipid peroxidation and DNA oxidative damage occurred in both muscle types, although all of these markers were found at a higher level in mitochondria-rich red muscle, which is consistent with the idea that elevated damage occurs as a consequence of increased ROS. These results are in agreement with those of Magherini et al. (2013), who demonstrated in a murine model that slow, oxidative muscle exhibits more protein carbonylation than fast, glycolytic muscle, and with Grim et al. (2015), who reported lipid peroxidation was higher in red than in white muscle of juvenile striped bass (Morone saxatilis). Protein carbonylation is of particular interest because it is irreversible and accumulates over time, requiring degradation of the damaged protein, and it was 40% higher in red muscle than white muscle in the present study. ROS are prone to attacking lipids with carbon–carbon double bonds, with a particular preference for polyunsaturated fatty acids and cholesterol, both of which are membrane components (Ayala et al., 2014), and we found that red muscle had 3-fold higher lipid peroxidation than white muscle. The current study also demonstrated a 3-fold increase in DNA damage in red muscle compared with white muscle. To the best of our knowledge, this is the first study to compare DNA damage in the form of 8-OHdG between aerobic and anaerobic skeletal muscle at rest. While most previous studies on DNA oxidation in skeletal muscle focused on the effect of exercise, diet or aging, both fast-glycolytic and slow-oxidative muscle have been shown to exhibit DNA damage under control conditions (dos Santos et al., 2018; Radák et al., 1999). Furthermore, Radak et al. (2007) has demonstrated that red fibers have significantly greater activity of oxoguanine DNA glycosylase (OGG1), an enzyme that removes oxidized guanines in mammals, than white fibers, and Mecocci et al. (1999) found there is a significant correlation between lipid peroxidation and DNA oxidation in the human vastus medialis muscle.
To limit oxidative damage, natural antioxidants in muscle including SOD, CAT and glutathione peroxidase (GPX) are produced to eradicate most ROS before excessive damage occurs. In most cases, antioxidant activity spikes following an oxidative event such as exercise and levels of antioxidants have been suggested to reflect exercise capacity in both mammals and fishes (Ji, 2008; Vélez-Alavez et al., 2015). SOD, CAT and GPX have been shown to be present in fish skeletal muscle (for a review on fish antioxidant defenses, see Martínez-Álvarez et al., 2005); however, in the present study, CAT activity was largely undetectable in white muscle, which appears to be common amongst marine teleosts (Otto and Moon, 1996; Vélez-Alavez et al., 2015; Vasylkiv et al., 2011). In addition to CAT, GPX also reduces H2O2 and is ubiquitous (Ighodaro and Akinloye, 2018). Both CAT and GPX have greater activity in slow, oxidative muscle than in fast, glycolytic muscle (Powers et al., 1994). It may be that the low levels of CAT in white muscle are offset by high levels of GPX. For example, Laughlin and colleagues (1990) demonstrated that rats exercised on a treadmill have higher GPX activity in both slow- and fast-twitch muscle than sedentary rats, while CAT activity does not change. The current study shows total SOD and CAT activity were significantly higher in red muscle. However, when normalized to mitochondrial mass, there was a higher activity of Mn-SOD in white muscle (Fig. 3B, inset). As SOD is the first line of defense in cells against ROS, catalyzing the conversion of O2·− into H2O2, this suggests that the simple mitochondria found in white muscle may produce more ROS than the more abundant and complex network of mitochondria found in red muscle. Consistent with our study, H2O2 production by mitochondria isolated from red and white muscle fibers of rainbow trout, Oncorhynchus mykiss, did not differ between fiber types when expressed relative to cytochrome oxidase (an indicator of metabolic potential), but when expressed per milligram of mitochondrial protein, expression was greater in white muscle (Leary et al., 2003). Similarly, murine studies using permeabilized fibers have shown that basal rates of H2O2 production are elevated in fast, glycolytic fibers expressed per milligram of tissue dry mass (Anderson and Neufer, 2006), and there is also greater H2O2 efflux in fast, glycolytic fibers when normalized to citrate synthase activity (a mitochondrial density marker; Picard et al., 2008). Nevertheless, the higher MVD in red muscle is associated with overall higher tissue antioxidant activity, but this tissue type still accrued more oxidative damage, suggesting total ROS production exceeds that scavenged by a greater margin in red than white muscle.
Proteins and lipids that are oxidized must be degraded/recycled or repaired, presumably incurring greater energetic costs. The present study indicates that red and white muscle fiber types have a different strategy with respect to turnover of cellular material, where the UPS is dominant in red muscle (based on higher poly-ubiquitin and 20S-proteasome activity) and the ALS is dominant in white muscle (based on higher LC3B-II and AVD). In vitro studies indicate that the ALS is accountable for >30% of protein degradation, while the UPS contributes only 3.5%, in white muscle myotubes of the rainbow trout, O. mykiss (Seiliez et al., 2014), and this pattern is similar to mammalian models (Fernando et al., 2019). Dominance of the UPS in red muscle has been documented in muscle lysates of the Arctic charr, Salvelinus alpinus (Cassidy et al., 2016), the rainbow trout, O. mykiss (Martin et al., 2002), and oxidative fibers of young and old mice (Fernando et al., 2019). Mofarrahi et al. (2013) illustrated that mRNA levels, protein expression of autophagy related genes as well as autophagic flux were greater in fast-glycolytic muscle than in slow-oxidative muscle of mice. Mizushima et al. (2004) found more autophagosomes in fast-twitch muscle both at basal levels and after starvation. Contrary to this, Lira et al. (2013) demonstrated that autophagic flux and LC3B-II expression was greater in slow-oxidative compared with fast-glycolytic muscle at basal levels in young mice. These tissue-specific differences have also been documented in aging mice, which is an area that needs further study as the results are not all in agreement (Triolo and Hood, 2021). Thus, our findings are consistent with most, but not all studies that have examined fiber-type variation in the ALS and UPS.
It is not entirely clear why protein degradation strategies vary by fiber type. Oxidized proteins can aggregate when ROS damage accumulates, resulting in protein aggregates that are often too large to be degraded by the UPS, leading to the possibility of cellular apoptosis (Grune et al., 2004). When in an oxidized environment, the 26S proteasome has been found to dissociate into 20S and 19S subunits (Aiken et al., 2011). The 20S subunit has been shown to degrade oxidized macromolecules after addition of H2O2 in vitro, independent of ATP (Aiken et al., 2011; Davies, 2001). In the current study, the higher 20S proteasome activity in red muscle may therefore be indicative of the more oxidized environment compared with that of the white muscle. Additionally, there is some evidence that ATP concentration (not the rate of production) controls autophagic flux and AVD (Plomp et al., 1987; Schellens et al., 1988). In mice, the resting ATP levels are higher in fast-twitch, anaerobic muscle than in slow-twitch, aerobic muscle (Kushmerick et al., 1992). While we did not measure absolute ATP concentration in the present study, a higher ATP in white muscle may lead to greater initiation of the ALS.
In addition to ubiquitin expression and proteasome activity, which were higher in red muscle, we also examined two ubiquitin E3 ligases, MAFbx and MuRF1, both of which function in skeletal and cardiac muscle (Bodine et al., 2001). Despite evidence of lower UPS activity in white muscle, MAFbx and MuRF1 expression was actually higher in this tissue, indicating that these ligases are involved in additional processes, or regulated differently in red and white muscle, or that there is increased ubiquitin-independent UPS activity in red muscle (as 20S proteasome activity was higher in red muscle). There are differences in target proteins for these ligases; MAFbx is thought to regulate protein synthesis by targeting MyoD (transcriptional factor regulating muscle differentiation) and eIF3f (eukaryotic transcription initiation factor 3 subunit f), and also label myosin heavy chain (MyHC) for degradation (Bodine and Baehr, 2014; Nemova et al., 2016). MuRF1 has been shown to target myofibrillar proteins including myosin light chain 1 and 2 (MyLC1 and MyLC2) and MyHC (Bodine and Baehr, 2014; Glass, 2010), although these proteins may not be the primary targets (Witt et al., 2005). In fact, proteins that are involved in glycolysis, including pyruvate dehydrogenase and its regulator PDK2, have both been shown to be downregulated in quadriceps (mostly fast, glycolytic muscle) of MuRF1 knockout mice (Hirner et al., 2008), which suggests a link between MuRF1 expression and anaerobic capacity. MuRF1 also had higher expression in type II fibers in mice after experiencing metabolic stress in the form of denervation (Moriscot et al., 2010) and hindleg immobilization (Okamoto et al., 2011). However, Okamoto et al. (2011) indicated that there was no difference in MuRF1 or MAFbx mRNA levels between fast and slow twitch muscle in the controls. Thus, the higher ubiquitin expression and proteasome activity are likely indicative of a larger role of the UPS in red muscle, and the fact that this is not associated with greater expression of the E3 ubiquitin ligases is not without precedent.
Maintaining protein homeostasis within muscle includes balancing protein degradation and protein synthesis. To the best of our knowledge, this is the first study to apply the in vivo SUnSET method to a teleost. This technique provides an effective alternative to measuring protein synthesis via radioactive tracers and stable isotopes (Goodman and Hornberger, 2013; Goodman et al., 2011). While this method has been criticized because it does not determine actual rates of protein synthesis (Marciano et al., 2018), the approach was appropriate for the present study because we focused on the incorporation of puromycin into nascent polypeptide chains as an indicator of the relative rates of translation in order to compare red and white muscle. An intraperitoneal injection of puromycin followed by anti-puromycin detection via western blot revealed relative puromycin incorporation approximately 2-fold greater in red muscle. This is consistent with prior studies in mammals that found that translation in type 2B fibers (fast-glycolytic) of the mouse hindleg is lower than that in type 2A fibers (fast-oxidative), which have a similar protein synthesis rate to type I fibers (slow-oxidative; Goodman et al., 2011). Our data are also consistent with those of McMillan and Houlihan (1989), who showed higher rates of protein synthesis in red muscle than white muscle of the rainbow trout, O. mykiss, via a flooding-dose technique using radioactivity of 3H-phenylalanine. Therefore, the greater resting ATP turnover rate in red muscle correlates with a higher rate of protein turnover. This suggests that at least some of the difference in resting ATP costs between muscle types arises from differential rates of protein synthesis and degradation. However, while oxidatively modified proteins are generally degraded and replaced, this does not always result in increased protein synthesis. In fact, elevated oxidative damage in red-blooded notothenioid fishes did not result in higher protein synthesis rates relative to icefishes that have low oxidative damage (Lewis et al., 2015).
The present study suggests that at least part of the reason for the higher ATP turnover rate in resting red muscle is that these fibers incur greater oxidative damage. However, other fiber type-specific factors almost certainly contribute to the difference in ATP turnover. For instance, we have previously shown that smaller fibers are more costly to maintain because of the increased surface area to volume ratio over which to maintain ionic gradients (Jimenez et al., 2011, 2013). Red fibers are considerably smaller than white fibers, which presumably leads to higher maintenance costs.
In summary, the current study provides evidence that skeletal muscle with a higher aerobic capacity accrues more oxidative damage and has an elevated rate of protein and ATP turnover, presumably leading to higher maintenance costs. These results also are consistent with previous studies indicating that protein degradation pathways are fundamentally different in red and white muscle.
Acknowledgements
We would like to thank Mark Gay for his guidance in microscopy techniques; the UNCW Richard Dillaman Microscopy Laboratory for allowing us use of the TEM; and Travis Ruffin for help collecting animals.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.M.N., S.T.K.; Methodology: J.M.N.; Formal analysis: J.M.N., E.T.P.; Investigation: J.M.N., E.T.P.; Writing - original draft: J.M.N.; Writing - review & editing: S.T.K.; Visualization: J.M.N.; Supervision: S.T.K.; Project administration: J.M.N.; Funding acquisition: S.T.K.
Funding
This work was supported by the National Institutes of Health [National Institute of Diabetes and Digestive and Kidney Diseases grant R15DK106688 to S.T.K.]. Deposited in PMC for release after 12 months.
References
- Aiken, C. T., Kaake, R. M., Wang, X. and Huang, L. (2011). Oxidative stress-mediated regulation of proteasome complexes. Mol. Cell. Proteomics 10, R110.006924. 10.1074/mcp.M110.006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, E. J. and Neufer, P. D. (2006). Type II skeletal myofibers possess unique properties that potentiate mitochondrial H2O2 generation. Am. J. Physiol. Cell Physiol. 290, C844-C851. 10.1152/ajpcell.00402.2005 [DOI] [PubMed] [Google Scholar]
- Ayala, A., Muñoz, M. F. and Argüelles, S. (2014). Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 1-31. 10.1155/2014/360438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banh, S., Wiens, L., Sotiri, E. and Treberg, J. R. (2016). Mitochondrial reactive oxygen species production by fish muscle mitochondria: Potential role in acute heat-induced oxidative stress. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 191, 99-107. 10.1016/j.cbpb.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Barreiro, E. and Hussain, S. N. A. (2010). Protein carbonylation in skeletal muscles: impact on function. Antioxid. Redox Signal. 12, 417-429. 10.1089/ars.2009.2808 [DOI] [PubMed] [Google Scholar]
- Bodine, S. C. and Baehr, L. M. (2014). Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 307, E469-E484. 10.1152/ajpendo.00204.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine, S. C., Latres, E., Baumhueter, S., Lai, V. K.-M., Nunez, L., Clarke, B. A., Poueymirou, W. T., Panaro, F. J., Na, E., Dharmarajan, K.et al. (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704-1708. 10.1126/science.1065874 [DOI] [PubMed] [Google Scholar]
- Bone, Q., Johnston, I. A., Pulsford, A. and Ryan, K. P. (1986). Contractile properties and ultrastructure of three types of muscle fibre in the dogfish myotome. J. Muscle Res. Cell Motil. 7, 47-56. 10.1007/BF01756201 [DOI] [PubMed] [Google Scholar]
- Brown, G. C. and Borutaite, V. (2012). There is no evidence that mitochondria are the main source of reactive oxygen species in mammalian cells. Mitochondrion 12, 1-4. 10.1016/j.mito.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Burpee, J. L., Bardsley, E. L., Dillaman, R. M., Watanabe, W. O. and Kinsey, S. T. (2010). Scaling with body mass of mitochondrial respiration from the white muscle of three phylogenetically, morphologically and behaviorally disparate teleost fishes. J. Comp. Physiol. B 180, 967-977. 10.1007/s00360-010-0474-x [DOI] [PubMed] [Google Scholar]
- Carpenè, E., Veggetti, A. and Mascarello, F. (1982). Histochemical fibre types in the lateral muscle of fishes in fresh, brackish and salt water. J. Fish Biol. 20, 379-396. 10.1111/j.1095-8649.1982.tb03932.x [DOI] [Google Scholar]
- Carrard, G., Bulteau, A.-L., Petropoulos, I. and Friguet, B. (2002). Impairment of proteasome structure and function in aging. Int. J. Biochem. Cell. B 34, 1461-1474. 10.1016/S1357-2725(02)00085-7 [DOI] [PubMed] [Google Scholar]
- Cassidy, A. A., Saulneir, R. J. and Lamarre, S. G. (2016). Adjustments of protein metabolism in fasting Arctic charr, Salvelinus alpinus. PLoS ONE 11, e0153364. 10.1371/journal.pone.0153364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, K. J. A. (2001). Degradation of oxidized proteins by the 20S proteasome. Biochimie 83, 301-310. 10.1016/S0300-9084(01)01250-0 [DOI] [PubMed] [Google Scholar]
- de Duve, C. and Wattiaux, R. (1966). Functions of lysosomes. Annu. Rev. Physiol. 28, 435-492. 10.1146/annurev.ph.28.030166.002251 [DOI] [PubMed] [Google Scholar]
- dos Santos, J. M., de Oliveira, D. S., Moreli, M. L. and Benite-Ribeiro, S. A. (2018). The role of mitochondrial DNA damage at skeletal muscle oxidative stress on the development of type 2 diabetes. Mol. Cell. Biochem. 449, 251-255. 10.1007/s11010-018-3361-5 [DOI] [PubMed] [Google Scholar]
- Eya, J. C., Yossa, R., Perera, D., Okubajo, O. and Gannam, A. (2017). Combined effects of diets and temperature on mitochondrial function, growth and nutrient efficiency in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 212, 1-11. 10.1016/j.cbpb.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Fedorova, M., Bollineni, R. C. and Hoffmann, R. (2014). Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom. Rev. 33, 79-97. 10.1002/mas.21381 [DOI] [PubMed] [Google Scholar]
- Fernando, R., Drescher, C., Deubel, S., Jung, T., Ost, M., Klaus, S., Grune, T. and Castro, J. P. (2019). Low proteasomal activity in fast skeletal muscle fibers in not associated with increased age-related oxidative damage. Exp. Gerontol. 117, 45-52. 10.1016/j.exger.2018.10.018 [DOI] [PubMed] [Google Scholar]
- Ferreira, L. F. and Laitano, O. (2016). Regulation of NADPH oxidases in skeletal muscle. Free Radic. Biol. Med. 98, 18-28. 10.1016/j.freeradbiomed.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filho, D. W. (2007). Reactive oxygen species, antioxidants and fish mitochondria. Front. Biosci. 12, 1229-1237. 10.2741/2141 [DOI] [PubMed] [Google Scholar]
- Fritz, K. S. and Petersen, D. R. (2011). Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem. Res. Toxicol. 24, 1411-1419. 10.1021/tx200169n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, D. J. (2010). Signaling pathways perturbing muscle mass. Curr. Opin. Clin. Nutr. Metab. Care 13, 225-229. 10.1097/MCO.0b013e32833862df [DOI] [PubMed] [Google Scholar]
- Goodman, C. A. and Hornberger, T. A. (2013). Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exerc. Sport Sci. Rev. 41, 107-115. 10.1097/JES.0b013e3182798a95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, C. A., Mabrey, D. M., Frey, J. W., Miu, M. H., Schmidt, E. K., Pierre, P. and Hornberger, T. A. (2011). Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 25, 1028-1039. 10.1096/fj.10-168799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim, J. M., Semones, M. C., Kuhn, D. E., Kriska, T., Keszler, A. and Crockett, E. L. (2015). Products of lipid peroxidation, but not membrane susceptibility to oxidative damage, are conserved in skeletal muscle following temperature acclimation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 308, R439-R448. 10.1152/ajpregu.00559.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati, P. and Bonaldo, P. (2012). Autophagy in skeletal muscle homeostasis and in muscular dystrophies. Cells 1, 325-345. 10.3390/cells1030325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune, T., Jung, T., Merker, K. and Davies, K. J. A. (2004). Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int. J. Biochem. Cell B 36, 2519-2530. 10.1016/j.biocel.2004.04.020 [DOI] [PubMed] [Google Scholar]
- Hirner, S., Krohne, C., Schuster, A., Hoffmann, S., Witt, S., Erber, R., Sticht, C., Gasch, A., Labeit, S. and Labeit, D. (2008). MuRF1-dependent regulation of systemic carbohydrate metabolism as revealed from transgenic mouse studies. J. Mol. Biol. 379, 666-677. 10.1016/j.jmb.2008.03.049 [DOI] [PubMed] [Google Scholar]
- Hoese, H. D. and Moore, R. H. (1998). Fishes of the Gulf of Mexico. Texas A&M University Press. [Google Scholar]
- Hsu, C.-Y., Chuanh, Y.-L. and Chan, Y.-P. (2014). Changes in cellular degradation activity in young and old worker honeybees (Apis mellifera). Exp. Gerontol. 50, 128-136. 10.1016/j.exger.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Ighodaro, O. M. and Akinloye, O. A. (2018). First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alexandria J. Med. 54, 287-293. 10.1016/j.ajme.2017.09.001 [DOI] [Google Scholar]
- Jackson, M. J. (2011). Control of reactive oxygen species production in contracting skeletal muscle. Antioxid. Redox Signal. 15, 2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, L. L. (2008). Modulation of skeletal muscle antioxidant defense by exercise: role of redox signaling. Free Radic. Biol. Med. 44, 142-152. 10.1016/j.freeradbiomed.2007.02.031 [DOI] [PubMed] [Google Scholar]
- Jimenez, A. G., Dasika, S. K., Locke, B. R. and Kinsey, S. T. (2011). An evaluation of muscle maintenance costs during fiber hypertrophy in the lobster Homarus americanus: are larger muscle fibers cheaper to maintain? J. Exp. Biol. 214, 3688-3697. 10.1242/jeb.060301 [DOI] [PubMed] [Google Scholar]
- Jimenez, A. G., Dillaman, R. M. and Kinsey, S. T. (2013). Large fiber size in skeletal muscle is metabolically advantageous. Nat. Commun. 4, 2150. 10.1038/ncomms3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, I. A. (2001). Muscle Development and Growth. San Diego, CA: Academic Press. [Google Scholar]
- Johnston, I. A. and Moon, T. W. (1981). Fine structure and metabolism of multiply innervated fast muscle fibres in teleost fish. Cell Tissue Res. 219, 93-109. 10.1007/BF00210021 [DOI] [PubMed] [Google Scholar]
- Johnston, I. A., Davison, W. and Goldspink, G. (1977). Energy metabolism of carp swimming muscles. J. Comp. Physiol. 114, 203-216. 10.1007/BF00688970 [DOI] [Google Scholar]
- Karnovsky, M. J. (1965). A formaldehyde-glutaraldehyde fixative of high osmolarity for use in. electron microscopy. J. Cell Biol. 27, 1A-149A.5857256 [Google Scholar]
- Kiessling, A., Ruohonen, K. and Bjornevik, M. (2006). Musclefibre growth and quality in fish. Arch. Tierz. Dummerstorf. 49, 137-146. [Google Scholar]
- Kinsey, S. T., Hardy, K. M. and Locke, B. R. (2007). The long and winding road: influences of intracellular metabolite diffusion on cellular organization and metabolism in skeletal muscle. J. Exp. Biol. 210, 3505-3512. 10.1242/jeb.000331 [DOI] [PubMed] [Google Scholar]
- Kushmerick, M. J., Moerland, T. S. and Wiseman, R. W. (1992). Mammalian skeletal muscle fibers distinguished by contents of phosphocreatine, ATP, and Pi. Proc. Natl. Acad. Sci. USA 89, 7521-7525. 10.1073/pnas.89.16.7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, C. A., Yoshimori, T. and Tooze, S. A. (2013). The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759-774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Laughlin, M. H., Simpson, T., Sexton, W. L., Brown, O. R., Smith, J. K. and Korthuis, R. J. (1990). Skeletal muscle oxidative capacity, antioxidant enzymes, and exercise training. J. Appl. Physiol. 68, 2337-2343. 10.1152/jappl.1990.68.6.2337 [DOI] [PubMed] [Google Scholar]
- Leary, S. C., Lyons, C. N., Rosenberger, A. G., Ballantyne, J. S., Stillman, J. and Moyes, C. D. (2003). Fiber-type differences in muscle mitochondrial profiles. Am. J. Physiol. Regul. Integr. Comp. Physiol. 285, R817-R826. 10.1152/ajpregu.00058.2003 [DOI] [PubMed] [Google Scholar]
- Lewis, J. M., Grove, T. J. and O'Brien, K. M. (2015). Energetic costs of protein synthesis do not differ between red- and white-blooded Antarctic notothenioid fishes. Biochem. Physiol. Part A Mol. Integr. Physiol. 187, 177-183. o. [DOI] [PubMed] [Google Scholar]
- Lira, V. A., Okutsu, M., Zhang, M., Greene, N. P., Laker, R. C., Breen, D. S., Hoehn, K. L. and Yan, Z. (2013). Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J. 27, 4184-4193. 10.1096/fj.13-228486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughna, P. T. and Goldspink, G. (1984). The effects of starvation upon protein turnover in red and white myotomal muscle of rainbow trout, Salmo gairdneri Richardson. J. Fish Biol. 25, 223-230. 10.1111/j.1095-8649.1984.tb04869.x [DOI] [Google Scholar]
- Lu, C.-Y. and Hsu, C.-Y. (2015). Ambient temperature reduction extends lifespan via activating cellular degradation activity in an annual fish (Nothobranchius rachovii). Age 37, 33. 10.1007/s11357-015-9775-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magherini, F., Gamberi, T., Pietrovito, L., Fiaschi, T., Bini, L., Esposito, F., Marini, M., Abrusso, P. M., Gulisano, M. and Modesti, A. (2013). Proteomic and carbonylation profile analysis of rat skeletal muscles following acute swimming exercise. PLoS ONE 8, e71839. 10.1371/journal.pone.0071839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano, R., Leprivier, G. and Rotblat, B. (2018). Puromycin labeling does not allow protein synthesis to be measured in energy-starved cells. Cell Death Dis. 9, 39. 10.1038/s41419-017-0056-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. A. M., Blaney, S., Bowman, A. S. and Houlihan, D. F. (2002). Ubiquitin-proteasome-dependent proteolysis in rainbow trout (Oncorhynchus mykiss): effect of food deprivation. Pflugers Arch. 445, 257-266. 10.1007/s00424-002-0916-8 [DOI] [PubMed] [Google Scholar]
- Martínez-Álvarez, R. M., Morales, A. E. and Sanz, A. (2005). Antioxidant defenses in fish: biotic and abiotic factors. Rev. Fish Biol. Fish. 15, 75-88. 10.1007/s11160-005-7846-4 [DOI] [Google Scholar]
- McMillan, D. N. and Houlihan, D. F. (1989). Short-term responses of protein synthesis to re-feeding in rainbow trout. Aquaculture 79, 37-46. 10.1016/0044-8486(89)90443-2 [DOI] [Google Scholar]
- Mecocci, P., Fanó, G., Fulle, S., MacGarvey, U., Shinobu, L., Polidori, M. C., Cherubini, A., Vecchiet, J., Senin, U. and Beal, M. F. (1999). Age-dependent increases in oxidative damage to DNA, lipids, and proteins in human skeletal muscle. Free Radic. Biol. Med. 26, 303-308. 10.1016/S0891-5849(98)00208-1 [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T. and Ohsumi, Y. (2004). In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101-1111. 10.1091/mbc.e03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofarrahi, M., Guo, Y., Haspel, J. A., Choi, A. M. K., Davis, E. C., Gouspillou, G., Hepple, R. T., Godin, R., Burelle, Y. and Hussain, S. N. A. (2013). Autophagic flux and oxidative capacity of skeletal muscles during acute starvation. Autophagy 9, 1604-1620. 10.4161/auto.25955 [DOI] [PubMed] [Google Scholar]
- Moriscot, A. S., Baptista, I. L., Bogomolovas, J., Witt, C., Hirner, S., Granzier, H. and Labeit, S. (2010). MuRF1 is a muscle fiber-type II associated factor and together with MuRF2 regulates type-II fiber trophicity and maintenance. J. Struct. Biol. 170, 344-353. 10.1016/j.jsb.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosse, P. R. L. and Hudson, R. C. L. (1977). The functional roles of different muscle fibre types identified in the myotomes of marine teleosts: a behavioural, anatomical and histochemical study. J. Fish Biol. 11, 417-430. 10.1111/j.1095-8649.1977.tb04136.x [DOI] [Google Scholar]
- Navon, A. and Ciechanover, A. (2009). The 26 S proteasome: from basic mechanisms to drug targeting. J. Biol. Chem. 284, 33713-33718. 10.1074/jbc.R109.018481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemova, N. N., Lysenko, L. A. and Kantserova, N. P. (2016). Degradation of skeletal muscle protein during growth and development of salmonid fish. Russ. J. Dev. Biol. 47, 161-172. 10.1134/S1062360416040068 [DOI] [PubMed] [Google Scholar]
- Nyack, A. C., Locke, B. R., Valencia, A., Dillaman, R. M. and Kinsey, S. T. (2007). Scaling of postcontractile phosphocreatine recovery in fish white muscle: effect of intracellular diffusion. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R2077-R2088. 10.1152/ajpregu.00467.2006 [DOI] [PubMed] [Google Scholar]
- Okado-Matsumoto, A. and Fridovich, I. (2001). Subcellular distribution of superoxide dismutases (SOD) in rat liver. J. Biol. Chem. 276, 38388-38393. 10.1074/jbc.M105395200 [DOI] [PubMed] [Google Scholar]
- Okamoto, T., Torii, S. and Machida, S. (2011). Differential gene expression of muscle-specific ubiquitin ligase MAFbx/Atrogin-1 and MuRF1 in response to immobilization-induced atrophy of slow-twitch and fast-twitch muscles. J. Physiol. Sci. 61, 537. 10.1007/s12576-011-0175-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orczewska, J. I., Hartleben, G. and O'Brien, K. M. (2010). The molecular basis of aerobic metabolic remodeling differs between oxidative muscle and liver of threespine sticklebacks in response to cold acclimation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 299, R352-R364. 10.1152/ajpregu.00189.2010 [DOI] [PubMed] [Google Scholar]
- Otto, D. M. E. and Moon, T. W. (1996). Endogenous antioxidant systems of two teleost fish, the rainbow trout and the black bullhead, and the effect of age. Fish Physiol. Biochem. 15, 349-358. 10.1007/BF02112362 [DOI] [PubMed] [Google Scholar]
- Passmore, L. A. and Barford, D. (2004). Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 379, 513-525. 10.1042/bj20040198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathi, B., Kinsey, S. T., Howdeshell, M. E., Priester, C., McNeill, R. S. and Locke, B. R. (2012). The formation and functional consequences of heterogeneous mitochondrial distributions in skeletal muscle. J. Exp. Biol. 215, 1871-1883. 10.1242/jeb.067207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard, M., Csukly, K., Robillard, M.-E., Godin, R., Ascah, A., Bourcier-Lucas, C. and Burelle, Y. (2008). Resistance to Ca2+-induced opening of the permeability transition pore differs in mitochondria from glycolytic and oxidative muscles. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R659-R668. 10.1152/ajpregu.90357.2008 [DOI] [PubMed] [Google Scholar]
- Picard, M., Hepple, R. T. and Burelle, Y. (2012). Mitochondrial functional specialization in glycolytic and oxidative muscle fibers: tailoring the organelle for optimal function. Am. J. Physiol. Cell Physiol. 302, C629-C641. 10.1152/ajpcell.00368.2011 [DOI] [PubMed] [Google Scholar]
- Plomp, P. J. A. M., Wolvetang, E. J., Groen, A. K., Meijer, A. J., Gordon, P. B. and Seglen, P. O. (1987). Energy dependence of autophagic protein degradation in isolated rat hepatocytes. Eur. J. Biochem. 164, 197-203. 10.1111/j.1432-1033.1987.tb11011.x [DOI] [PubMed] [Google Scholar]
- Powers, S. K., Criswell, D., Lawler, J., Ji, L. L., Martin, D., Herb, R. A. and Dudley, G. (1994). Influence of exercise and fiber type on antioxidant enzyme activity in rat skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 266, R375-R380. 10.1152/ajpregu.1994.266.2.R375 [DOI] [PubMed] [Google Scholar]
- Presnell, J. K. and Schreibman, M. P. (1997). Humason's Animal Tissue Techniques. Baltimore: The Johns Hopkins University Press. [Google Scholar]
- Radák, Z., Kaneko, T., Tahara, S., Nakamoto, H., Ohno, H., Sasvári, M., Nyakas, C. and Goto, S. (1999). The effect of exercise training on oxidative damage of lipids, proteins, and DNA in rat skeletal muscle: evidence for beneficial outcomes. Free Radic. Biol. Med. 27, 69-74. 10.1016/S0891-5849(99)00038-6 [DOI] [PubMed] [Google Scholar]
- Radak, Z., Kumagai, S., Nakamoto, H. and Goto, S. (2007). 8-Oxoguanosine and uracil repair of nuclear and mitochondrial DNA in red and white skeletal muscle of exercise-trained old rats. J. Appl. Physiol. 102, 1696-1701. 10.1152/japplphysiol.01051.2006 [DOI] [PubMed] [Google Scholar]
- Ramírez-Zarzosa, G., Gil, F., Vázquez, J. M., Arencibia, A., Latorre, R., López-Albors, O. and Moreno, F. (1998). The post-larval development of lateral musculature in gilthead sea bream Sparus aurata (L.) and sea bass Dicentrarchus labrax (L.). Anat. Histol. Embryol. 27, 21-29. 10.1111/j.1439-0264.1998.tb00151.x [DOI] [PubMed] [Google Scholar]
- Rani, V., Mishra, S., Yadav, T., Yadav, U. C. S. and Kohli, S. (2015). Hydrogen Peroxide Sensing and Signaling. In Free Radicals in Human Health and Disease (ed. Rani V. and Yadav U. C. S.), pp. 105-116. New York: Springer. [Google Scholar]
- Reid, M. B. (2001). Nitric oxide, reactive oxygen species, and skeletal muscle contraction. Med. Sci. Sports Exerc. 33, 371-376. 10.1097/00005768-200103000-00006 [DOI] [PubMed] [Google Scholar]
- Reynolds, E. S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17, 208-212. 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, C. E., Keshavarzian, A., Pasco, D. S., Frommel, T. O., Winship, D. H. and Holmes, E. W. (1999). Determination of protein carbonyl groups by immunoblotting. Anal. Biochem. 266, 48-57. 10.1006/abio.1998.2932 [DOI] [PubMed] [Google Scholar]
- Romanello, V. and Sandri, M. (2021). The connection between the dynamic remodeling of the mitochondrial network and the regulation of muscle mass. Cell. Mol. Life Sci. 78, 1305-1328. 10.1007/s00018-020-03662-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlin, K. and Harris, R. C. (2011). The creatine kinase reaction: a simple reaction with functional complexity. Amino Acids 40, 1363-1367. 10.1007/s00726-011-0856-8 [DOI] [PubMed] [Google Scholar]
- Sakellariou, G. K., Vasilaki, A., Palomero, J., Kayani, A., Zibrik, L., McArdle, A. and Jackson, M. J. (2013). Studies of mitochondrial and nonmitochondrial sources implicate nicotinamide adenine dinucleotide phosphate oxidase(s) in the increased skeletal muscle superoxide generation that occurs during contractile activity. Antioxid. Redox Signal. 18, 603-621. 10.1089/ars.2012.4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellens, J. P. M., Vreeling-Sindelárová, H., Plomp, P. J. A. M. and Meijer, A. J. (1988). Hepatic autophagy and intracellular ATP a morphometric study. Exp. Cell Res. 177, 103-108. 10.1016/0014-4827(88)90028-6 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval, R., Shvets, E., Fass, E., Shorer, H., Gil, L. and Elazar, Z. (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749-1760. 10.1038/sj.emboj.7601623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiliez, I., Dias, K. and Cleveland, B. M. (2014). Contribution of the autophagy-lysosomal and ubiquitin-proteasomal proteolytic systems to total proteolysis in rainbow trout (Oncorhynchus mykiss) myotubes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 307, R1330-R1337. 10.1152/ajpregu.00370.2014 [DOI] [PubMed] [Google Scholar]
- Shringarpure, R., Grune, T., Mehlhase, J. and Davies, K. J. A. (2003). Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J. Biol. Chem. 278, 311-318. 10.1074/jbc.M206279200 [DOI] [PubMed] [Google Scholar]
- Soukup, T., Zacharová, G. and Smerdu, V. (2002). Fibre type composition of soleus and extensor digitorum longus muscles in normal female inbred Lewis rats. Acta Histochem. 104, 399-405. 10.1078/0065-1281-00660 [DOI] [PubMed] [Google Scholar]
- Spurr, A. R. (1969). A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26, 31-43. 10.1016/S0022-5320(69)90033-1 [DOI] [PubMed] [Google Scholar]
- Staron, R. S., Kraemer, W. J., Hikida, R. S., Fry, A. C., Murray, J. D. and Campos, G. E. R. (1999). Fiber type composition of four hindlimb muscles of adult Fisher 344 rats. Histochem. Cell Biol. 111, 117-123. 10.1007/s004180050341 [DOI] [PubMed] [Google Scholar]
- St-Pierre, J., Buckingham, J. A., Roebuck, S. J. and Brand, M. D. (2002). Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 277, 44784-44790. 10.1074/jbc.M207217200 [DOI] [PubMed] [Google Scholar]
- Sturtz, L. A., Diekert, K., Jensen, L. T., Lill, R. and Culotta, V. C. (2001). A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane Space of mitochondria. J. Biol. Chem. 276, 38084-38089. 10.1074/jbc.M105296200 [DOI] [PubMed] [Google Scholar]
- Tacchi, L., Bickerdike, R., Secombes, C. J. and Martin, S. A. M. (2012). Muscle-specific RING finger (MuRF) cDNAs in Atlantic salmon (Salmo salar) and their role as regulators of muscle protein degradation. Mar. Biotechnol. 14, 35-45. 10.1007/s10126-011-9385-4 [DOI] [PubMed] [Google Scholar]
- Triolo, M. and Hood, D. A. (2021). Manifestations of age on autophagy, mitophagy and lysosomes in skeletal muscle. Cells 10, 1054. 10.3390/cells10051054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valavanidis, A., Vlachogianni, T. and Fiotakis, C. (2009). 8-hydroxy-2'-deoxyguanosine (8-OHdG): a critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 27, 120-139. 10.1080/10590500902885684 [DOI] [PubMed] [Google Scholar]
- van den Thillart, G., van Waarde, A., Muller, H. J., Erkelens, C. and Lugtenburg, J. (1990). Determination of high-energy phosphate compounds in fish muscle: 31P-NMR spectroscopy and enzymatic methods. Comp. Biochem. Physiol. B Comp. Biochem. 95, 789-795. 10.1016/0305-0491(90)90318-N [DOI] [Google Scholar]
- van Ginneken, V. J. T., van den Thillart, G. E. E. J. M., Muller, H. J., van Deursen, S., Onderwater, M., Visée, J., Hopmans, V., van Vliet, G. and Nicolay, K. (1999). Phosphorylation state of red and white muscle in tilapia during graded hypoxia: an in vivo31P-NMR study. Am. J. Physiol. Regul. Integr. Comp. Physiol. 277, R1501-R1512. 10.1152/ajpregu.1999.277.5.R1501 [DOI] [PubMed] [Google Scholar]
- Vasylkiv, O. Y., Kubrak, O. I., Storey, K. B. and Lushchak, V. I. (2011). Catalase activity as a potential vital biomarker of fish intoxication by the herbicide aminotriazole. Pestic. Biochem. Phys. 101, 1-5. 10.1016/j.pestbp.2011.05.005 [DOI] [Google Scholar]
- Vélez-Alavez, M., De Anda-Montañez, J., Galván-Magaña, F. and Zenteno-Savín, T. (2015). Comparative study of enzymatic antioxidants in muscle of elasmobranch and teleost fishes. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 187, 61-65. 10.1016/j.cbpa.2015.04.014 [DOI] [PubMed] [Google Scholar]
- Wilson, W. N., Baumgarner, B. L., Watanabe, W. O., Alam, M. S. and Kinsey, S. T. (2015). Effects of resveratrol on growth and skeletal muscle physiology of juvenile southern flounder. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 183, 27-35. 10.1016/j.cbpa.2014.12.014 [DOI] [PubMed] [Google Scholar]
- Witt, S. H., Granzier, H., Witt, C. C. and Labeit, S. (2005). MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. J. Mol. Biol. 350, 713-722. 10.1016/j.jmb.2005.05.021 [DOI] [PubMed] [Google Scholar]
- Wong, H.-S., Benoit, B. and Brand, M. D. (2019). Mitochondrial and cytosolic sources of hydrogen peroxide in resting C2C12 myoblasts. Free Radic. Biol. Med. 130, 140-150. 10.1016/j.freeradbiomed.2018.10.448 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. and Wong, H. S. (2021). Are mitochondria the main contributor of reactive oxygen species in cells? J. Exp. Biol. 224, jeb221606. 10.1242/jeb.221606 [DOI] [PubMed] [Google Scholar]