Summary
The engineering of C4 photosynthetic activity into the C3 plant rice has the potential to nearly double rice yields. To engineer a two‐cell photosynthetic system in rice, the rice bundle sheath (BS) must be rewired to enhance photosynthetic capacity. Here, we show that BS chloroplast biogenesis is enhanced when the transcriptional activator, Oryza sativa Cytokinin GATA transcription factor 1 (OsCGA1), is driven by a vascular specific promoter. Ectopic expression of OsCGA1 resulted in increased BS chloroplast planar area and increased expression of photosynthesis‐associated nuclear genes (PhANG), required for the biogenesis of photosynthetically active chloroplasts in BS cells of rice. A further refinement using a DNAse dead Cas9 (dCas9) activation module driven by the same cell‐type specific promoter, directed enhanced chloroplast development of the BS cells when gRNA sequences were delivered by the dCas9 module to the promoter of the endogenous OsCGA1 gene. Single gRNA expression was sufficient to mediate the transactivation of both the endogenous gene and a transgenic GUS reporter fused with OsCGA1 promoter. Our results illustrate the potential for tissue‐specific dCas9‐activation and the co‐regulation of genes needed for multistep engineering of C4 rice.
Keywords: C4 photosynthesis, dCas9‐mediated transcriptional activation, rice bundle sheath, chloroplast development
Introduction
Increasing demands placed on the global food supply indicate that a 50%–70% increase in agricultural output will be necessary by 2050 (Jaggard et al., 2010). Yet yields in rice, a primary staple consumed by over half of the world’s population, have remained relatively flat over the past decade, suggesting that yield gains driven by “green revolution” engineering have been maximized under our current production systems (Grassini et al., 2013). The installation of a C4 photosynthetic system into rice has the theoretical potential to double current rice yields (von Caemmerer et al., 2012; Hibberd et al., 2008) through improved photosynthetic efficiencies while reducing water and nitrogen demands. However, both fundamental discoveries and several technological hurdles must be overcome to achieve this goal (Sage and Zhu, 2011). As a central feature to most C4 systems, photosynthetic activities are partitioned between two cell types, the BS and the mesophyll (M) (Hibberd and Covshoff, 2010). The chloroplasts of the BS must decarboxylate malate and perform selective Calvin Cycle activities with reduced PSII activities, whereas the M cell plastids perform linear electron transport and reduce oxaloacetate into malate (Hatch, 1987). Unlike the BS plastids in a C3 plant, in a C4 context, the BS plastids serve an essential role in photosynthesis and therefore the number and or size of the BS plastids in rice must be increased to support the C4 cycle. Thus, in order to install a C4 system into rice, cell‐type specific promoters will be required to drive differential gene expression. It has been estimated that at least 20 genes will need to be differentially expressed (DE) between BS and M to install a minimal C4 system (Peterhansel, 2011). Another challenge is our limited understanding of the regulatory networks that drive both anatomical features and biochemical pathways underlying C4 photosynthesis. In lieu of these transcriptional regulators, single genes must be manipulated with the goal of targeting specific biochemical activities that may be insufficient to drive an operational C4 cycle.
Here, we have utilized a transcriptional activator that promotes chloroplast development through enhanced cytokinin responsiveness (Hudson et al., 2013; Naito et al., 2007; Reyes et al., 2004) to manipulate chloroplast architecture specifically in the BS of rice. This was achieved by driving OsCGA1 expression with the use of the glycine decarboxylase p‐subunit promoter (pFtGLDp), originally characterized in Flaveria trinervia (Engelmann et al., 2008 ). Ectopic expression of OsCGA1 resulted in increased accumulation of photosynthesis‐associated nuclear gene (PhANGs) transcripts including peripheral proteins for the plastid encoded RNA polymerase (PEP) complex, peripheral proteins of the plastid ribosome and light harvesting complexes in the BS. Histological characterizations and confocal imaging also revealed enhanced photosynthetic development in the BS cells of the transgenic plants. Importantly, the use of a two component dCas9/gRNA system to deliver a transcriptional activation module to the promoter of an endogenous rice gene opens the possibility of co‐regulating suites of genes in a cell‐type specific manner in rice and addresses a fundamental challenge in engineering C4 photosynthesis or similarly complex systems.
Results
Previous characterizations of Cytokinin GATA transcription factor 1 (CGA1) misexpression in O.sativa var. Nipponbare and Arabidopsis thaliana suggested that CGA1 functions cell autonomously to drive the expression of genes associated with chloroplast differentiation (Chiang et al., 2012; Hudson et al., 2011,2013). To investigate the phenotypic consequences of CGA1 overexpression in O.sativa var. Kitaake, a short‐statured and largely photoperiod insensitive rice variety, OsCGA1 was introduced into rice calli under the control of the maize ubiquitin 1 promoter (Christensen and Quail, 1996). Ectopic expression of OsCGA1 resulted in photosynthetic differentiation of callus tissue and partial shoot regeneration even when cytokinins were excluded from the growth media (Figure S1). Due to the pleiotropic growth defects associated with constitutive OsCGA1 expression on regeneration and growth, we opted for cell‐specific expression of OsCGA1 using the relatively weak FtGLDp promoter to enhance OsCGA1 expression in vascular tissues including the bundle sheath (Engelmann et al., 2008). In rice, pFtGLDp::GUS fusions conditioned a weak vascular profile of expression that was dependent on the absence of intronic sequences in the β‐glucuronidase reporter (Figure S2, Table S2). Interestingly, when intron sequences were inserted into the GUS reporter, it conditioned uniform leaf expression (Figure S2).
When the FtGLDP promoter was used to drive OsCGA1, calli were regenerated using standard tissue culture procedures and resulted in plants with general growth characteristics and phenotypes that were not easily distinguishable from the null segregant lines until after flowering when we noticed transgenic plants were slightly greener than null lines (Figure 1). Among 22 independent transgenic events, three single copy insertions conditioned a 4 ~ 13 fold increase in OsCGA1 expression as determined by RT‐PCR (Figure 1b). Siblings of three single copy transgenic events (#2, #12, #15) and one partial tandem duplication event (#4) were characterised at T1 and T2 generations (Table S2). Histological characterizations of transgenic lines revealed that chloroplast size in the BS was enhanced (Figure 1). Quantification of the plastid area using EM analysis indicated that the BS plastids of transgenics had increased up to 3.5 fold in size relative to the chloroplasts of nulls (Figure 2a). In two of the transgenic lines, the M chloroplast area was not significantly different in transgenics relative to nulls. However, in one line mesophyll cell size was reduced (# 15–19, Figure 2b). As this phenotype was observed in only one event it is not clear if ectopic expression of OsCGA1 may also mediate changes in M cell architecture (Figure 2b). Ultrastructural characterizations indicated that granal stacking and the size of starch granules were not significantly different from null lines. Thus, the increased size of the plastids did not obviously affect internal features of the plastids (Figure 1 and Figure S3). In addition to larger chloroplasts, mitochondria number and area were also increased in the BS of transgenic lines (Table S1), potentially indicative of enhanced photorespiratory activities in the BS plastids. Immunodetection of RUBISCO, RUBISCO activase, fructose 1,6‐bisphosphatase (FBPase) and the mitochondrial enzyme glycine decarboxylase (GLD) indicated these enzymes are present in the BS chloroplasts and mitochondria of transgenic and null lines suggesting that BS cells could support photosynthetic activities in both WT and transgenic lines (Figure S3). To determine if the increase in BS chloroplast area could enhance photosynthetic performance, leaf net CO2 assimilation rates (A) versus intercellular CO2 partial pressure (Ci) was measured in two independent pFtGLDp::OsCGA1 events and corresponding nulls under atmospheric O2 partial pressure of 18.4 kPa. Additionally, Rubisco activity, chlorophyll a + b content, chlorophyll a/b ratio, specific leaf mass (SLM) and total nitrogen were also measured. However, as shown in Figure S4, there were no significant differences between the transgenics and WT plants for any of these parameters. These results suggest that despite an increase in BS plastid area and possible enrichment of some photosynthetic enzymes, these enhancements were insufficient to impact global photosynthetic performance of the plants. This is perhaps not surprising given the limited access of CO2 to the BS chloroplasts that are embedded within leaf tissue and surrounded by multiple dense layers of photosynthetically active M cells. Nevertheless, these results suggest that manipulation of OsCGA1 could provide an avenue to enhance BS photosynthesis if a mechanism to deliver CO2 to the BS was also installed.
Figure 1.
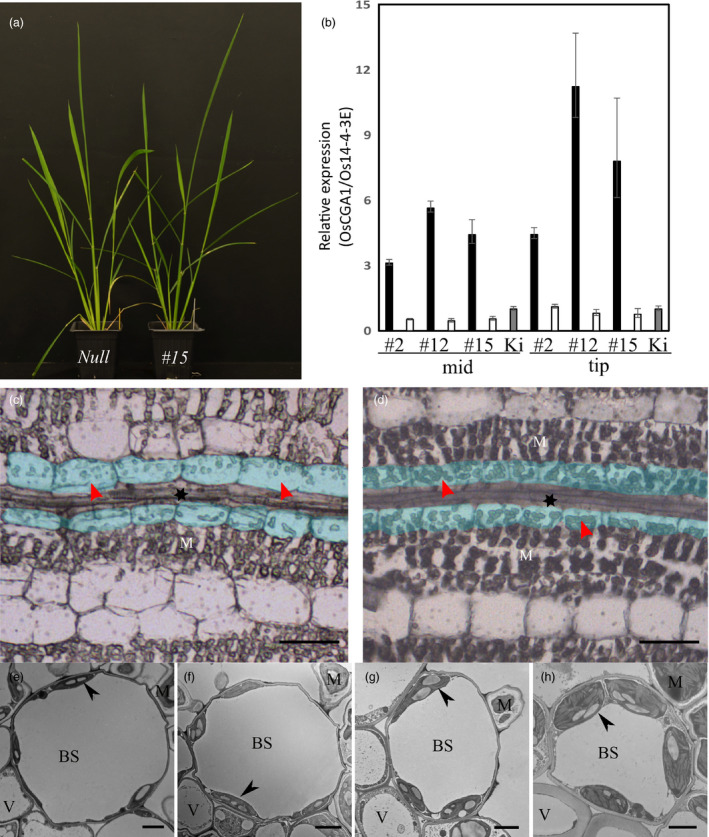
pFtGLDp::OsCGA1 plant appearance, transgene expression and BS chloroplast phenotypes. (a) pFtGLDp::OsCGA1 carrying plant (right) shows a pronounced green sheath compared to the null segregant (left). (b) Expression level of pFtGLDp::OsCGA1 in 3 independent transgenic events (#2, #12 and #15). Black bars and empty bars indicate segregating homozygotes and nulls, respectively. Kitaake WT is shown as a grey bar and 2 cm leaf segments from two individuals were pooled for each sample. The bars represent mean +SD and −SEM of three biological replicates. Mid, middle region of leaf; Tip, Tip region of leaf. (c and d) BS chloroplast phenotype of FtGLDp::OsCGA1 transgenic plant. Paradermal sections of leaf tissue were imaged using light microscopy prior to sampling for laser capture microdissection; (d) pFtGLDp::OsCGA1 #12 homozygote and (c) null segregant. The BS cells are shaded in light blue, M denotes mesophyll cells and * denotes vascular tissue. Arrowheads indicate the chloroplasts in BS. Bars, 5 μm. (e–h) Transmission electron micrographs of fourth leaves of (e) null segregant and transgenic lines (f) #2–13, (g) #12–15 and (H) #15–19. Arrows denote the chloroplasts. BS, bundle sheath; M, mesophyll; V, vascular tissue. Bars, 2 μm.
Figure 2.
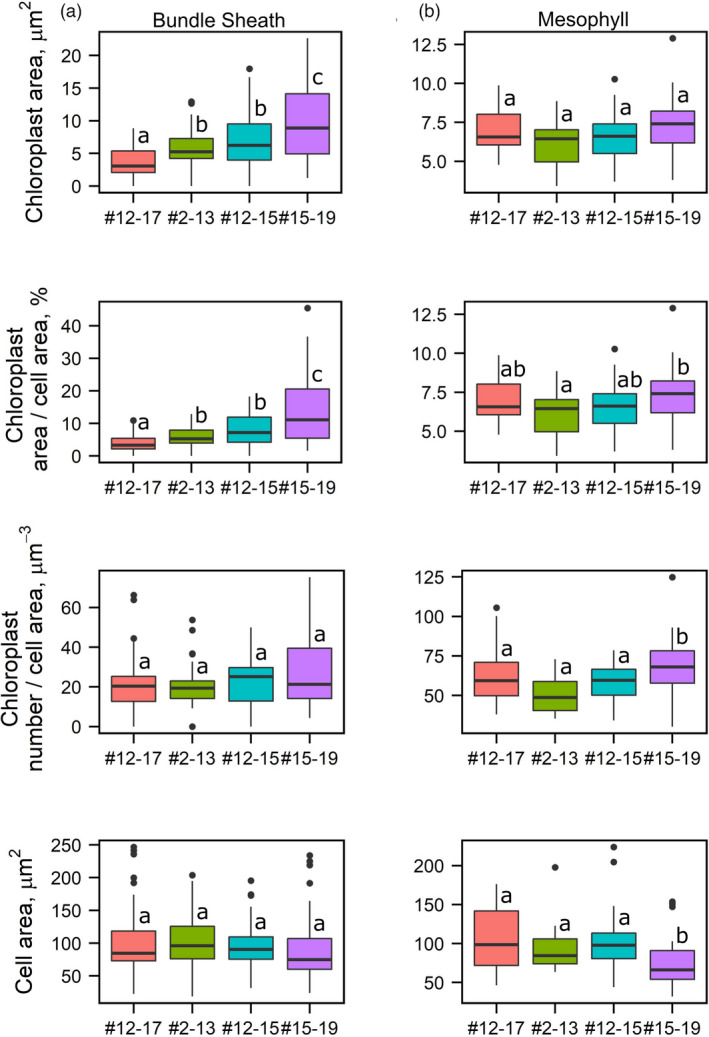
Quantification of chloroplast phenotypes in pFtGLDp::OsCGA1 transgenic plants. Quantification of chloroplast area and number in the bundle sheath (a) and mesophyll (b) cells from TEM images of WT (#12–17) and three independent transgenic events (#2–13, #12–15 and #15–19). Within each box, horizontal black lines denote median values. Top and bottom of boxes indicate the 25th and the 75th percentile of each group’s distribution of values. Vertical extending lines and dots denote adjacent values and observations outside the range, respectively. Values (a–d) with the same letters represent no significant difference (P > 0.05) when a Kruskal–Wallis one‐way analysis of variance was performed followed by a Dunn's post hoc test using an R script.
To further investigate the molecular consequences by OsCGA1 misexpression, we performed RNA‐seq analysis on four developmentally distinct leaf segments, base, −1 (a region covered by the outer leaf), +4 (middle region of emerged leaf) and tip from the emerging fourth leaves, similar to the leaf gradient scheme in (Li et al., 2010). As shown in Figure S5, these data suggest that given the low vascular activity of the FtGLDp promoter and the low BS to M cell ratio in leaf segments, we lacked the resolution to monitor global changes in gene expression using whole tissue segments.
To enhance the sensitivity of detecting DE genes, we performed Laser Capture Microdissection (LCM) on the mid‐region of leaves from T3 transgenic lines and their null segregants of strongest #12 event (Table S2). Cross‐sections of Steedman’s wax embedded leaf tissue were used to harvest bundle sheath strands (BSS) and Mesophyll (M) for RNA‐Seq analysis (Figure S6). An increase in BS chloroplast size was notable in transgenic lines under the LCM microscope (Figure 1c ~ d). LCM captured BS and M strips displayed the pronounced differential expression of vascular marker and mesophyll marker gene expression indicating little cross‐contamination (Figure 3b). As expected, endogenous OsCGA1 was expressed at higher levels in M cells relative to BS in both transgenic and null lines. However, CGA1 expression driven from the transgene was 6.2 times higher in BS than in M in transgenic lines, corroborating the plastid phenotypes associated with FtGLDp promoter activity in rice (Figure 3a). Differential expression analysis between transgenic and null lines led to the identification of 172 DE and 147 DE genes in BS and M strips, respectively, with an FDR cut off 0.05 (Figure 3c). Pageman analysis (Usadel et al., 2006) showed that photosystem II, electron transport chain, vitamin biosynthesis and plastid ribosomal machinery genes are enriched in transgenic BS transcriptome relative to the null counterpart (Figure S7). In contrast, immune response, protein degradation and vacuolar ATPase activity are markedly attenuated in the transgenic BS (Figure S7). Surprisingly, a total of 41.2% of DEG are proportioned to a broad range of photosynthesis‐associated genes (PhANGs), including nuclear‐encoded plastid ribosomal/translational machinery genes (21.5%), peripheral proteins for PEP complex (6.9%), chlorophyll biosynthesis, components of light harvesting complex and photosynthetic enzymes (FBPase and SSU) (Figure 3d). Interestingly, nucleus‐encoded PEP peripheral subunits, pTAC7, PAP6/FLN1, are up‐regulated together with the plastome‐located core units, rpoA, rpoB and rpoC2 (Figure 3e) in transgenic BS (He et al., 2018; Steiner et al., 2011). PEP complex activity is largely dependent on nucleus encoded PEP‐associated proteins (PAP) and is responsible for the photosynthetic gene expression in chloroplast development (Börner et al., 2015; Toyoshima et al., 2005). In contrast, nucleus encoded RNA polymerase (NEP), responsible for housekeeping gene expression in plastids (Demarsy et al., 2006; Hajdukiewicz et al., 1997), was not altered in OsCGA1 BS (data not shown). Other plastid transcription‐associated genes, including Os11g41910 (GTP binding protein) (Ye et al., 2017), Os11g23790 (homologous to AtPEP‐RELATED DEVELOPMENT ARRESTED 1) (Qiao et al., 2013), plastid post transcriptional machinery genes, nucleus encoded plastid translation machinery, electron transport chain components, light harvesting complexes, and chlorophyll biosynthesis pathway genes also accumulated to higher levels in the BS of transgenic plants (Figure 3e, Table S5). Collectively, these data suggest that ectopic expression of CGA1 is sufficient to mediate a transcriptional cascade leading to a global increase in photosynthetic machinery. Supporting this trend, we observed a significant enrichment in chloroplast biogenesis pathways genes with a total of 46.5%% of DEG with at least one predicted plastid localization signal (Sperschneider et al., 2017) (Table S5).
Figure 3.
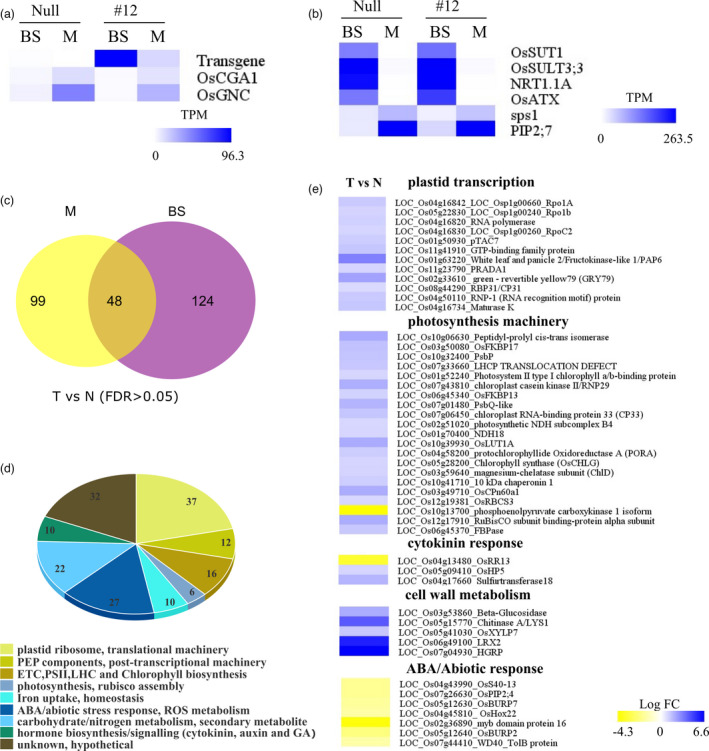
Expression of transgene and marker genes in LCM‐RNAseq and DE gene analysis in the BS of pFtGLDp::OsCGA1 plants. (a) Expression of endogenous OsCGA1, OsGNC and transgenic OsCGA1 in BS and M from LCM RNAseq. Transgenic OsCGA1 and endogenous OsCGA1 transcripts are discriminated by read mapping to unique regions in the 3’ end of transcripts (OsCGA1‐Nos terminator fusion transcripts and native OsCGA1 transcripts). Mean Transcripts Per Million (TPM) from three biological replicates of BS and M are represented as a color‐scale in the heat map. Data for the pFtGLDp::OsCGA1 #12 (#12) and the null are shown. (b) Expression patterns of vascular bundle and M markers in BS and M LCM RNAseq. Vascular bundle marker genes: OsSUT1 (LOC_Os03g07480), OsSULT3;3 (LOC_Os04g55800), NRT1.1A (LOC_Os08g05910), OsATX (LOC_Os01g67010); mesophyll marker genes: sps1 (LOC_Os01g69030), PIP2;7 (LOC_Os09g36930). (c) Summary of DE genes in BS strands and M strands between transgenics and nulls. There were 48 genes expressed in both BS and M but differentially expressed between transgenic and null plants. (d) Manually curated gene categorizations of DE genes from BS. Number of genes in each category are shown. (e) Heatmap of subsets of DE genes enriched in the gene clusters including plastid transcription, photosynthesis machinery, cytokinin response, cell wall metabolism and ABA/Abiotic responses. T vs. N, Transgenic versus Null.
In addition to photosynthesis‐associated genes, several cytokinin response and cell wall metabolism genes accumulated to higher levels in the transgenics. The exception was OsRR13, a potential inhibitor of cytokinin signal transduction based on its arabidopsis homolog ARR22 (Figure 3E) (Du et al., 2007; Wallmeroth et al., 2017,2019). This suggests that cytokinin response may be enhanced in BS cells ectopically expressing OsCGA1. Furthermore, a subset of ABA‐ and drought‐inducible genes are markedly decreased in BS of transgenics (Figure 3e, Table S5). As ABA is generally associated with the degradation of chloroplasts and increased senescence (Zhuang and Jiang, 2019), these data are consistent with an enhanced chloroplast biogenesis program in the BS of transgenics.
To exploit the FtGLDp promoter in driving multigene vascular specific expression, we employed the dCas9 activation strategy to up‐regulate the endogenous OsCGA1 in a tissue‐specific manner (Gilbert et al., 2013; Lowder et al., 2018). To demonstrate our proof‐of‐concept, we constructed a series of multigene cassettes to assess the transactivation capacity using a series of gRNAs directing dCas9‐activation modules to specific regions of the OsCGA1 promoter (Figure 4). To confer tissue specificity, we utilized the FtGLDp promoter to drive a dCas9 gene fused with VP64 and EDLL, two tandem activation domains (ADs) (Figure 4a). A 2.1 kb upstream sequence of the OsCGA1 gene was used to drive GUS reporter expression to validate tissue specific transactivation (Figure 4a). Multiple gRNAs were designed to target a region surrounding the TATA box of the OsCGA1 promoter and driven with the constitutive U6/U3 snRNApol promoter (Figure 4b). Prior to the generation of stable transgenics, we tested the positional effects of gRNAs on pOsCGA1::GUS gene activation using a constitutively expressed pZmUbi1::dCas9‐ADs construct in heterologous tobacco leaf assays (Figure S8). Among five gRNAs tested, gRNA2 and gRNA3 were the most effective in the activation of the pOsCGA1::GUS reporter (Figure S8) and resulted in mature plant phenotypes similar to pFtGLDp::OsCGA1. Although we introduced more constructs in rice, poor regeneration, low fertility and occasional loss of T‐DNA fragments were prevalent throughout the primary transgenic and subsequent generations (data not shown). Therefore, we limited our analysis to four stable constructs, dCas9‐ADs only, gRNA2, gRNA3 and multiple gRNA expression modules, in which we obtained more than three independent events (Figure 4). Due to the complexities associated with multigenic transgenes, we evaluated OsCGA1 transactivation and associated chloroplast phenotypes within single transgenic T1 siblings from low copy transgenic parents (Table S2). Activation levels of endogenous CGA1 varied depending on gRNA positions and transgenic events but showed an approximately 2 ~ 5 fold higher level compared to the expression of the dCas9‐ADs only transgenic (Figure 4g). Similar to transient assays, gRNA2 and gRNA3 were effective for endogenous OsCGA1 activation (Figure 4g). When multiple gRNAs were expressed with different U6/U3 promoters, we did not observe an additive effect on OsCGA1 expression, indicating a possible steric hindrance or competition between dCas9 proteins for access to the promoter region (Figure 4g). GUS staining of lines carrying pOsCGA1::GUS revealed strong staining along the veins (Figure 4f) that was much more distinct than the pattern conditioned by the pFtGLDp::GUS construct (Figure S9). Due to the large vacuole of the BS, GUS staining is only apparent in some BS cells when cytosolic portions are captured in 50 μm thick vibratome sections (Figure S9). Consistent with RT‐PCR analysis, gRNA2 and gRNA3 were most effective in driving tissue‐specific GUS reporter activation and also failed to display additive effects with gRNA multiplexing (Figure 4g and Figure S10). In summary, a dCas9 activation module was successfully used to drive transcript accumulation of the endogenous OsCGA1 and OsCGA driven‐GUS expression in a tissue‐specific manner. These results indicate that dCas9‐ADs act in a cell‐autonomous manner and can be utilized as shown here to drive tissue‐specific expression of multiple gene cassettes.
Figure 4.
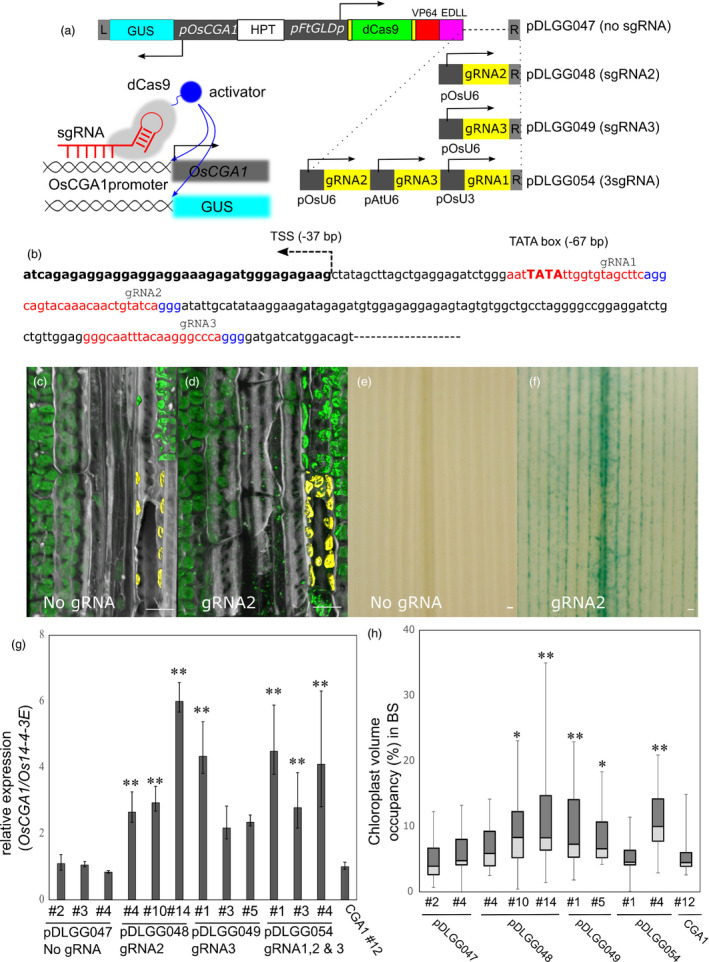
Tissue specific activation of OsCGA1 by dCas9‐AD constructs. (a) Schematic of dCas9‐mediated transcriptional activation. All dCas9 activation constructs have similar structures with the exception of gRNA sequences and number of gRNA expression modules to recruit dCas9‐ADs to the upstream region of both the endogenous OsCGA1 gene and OsCGA1promoter‐GUS reporters. The 2.1 kb upstream region of OsCGA1 was fused to the GUS gene as a proxy to monitor the tissue specificity of gene activation conferred by the FtGLDp promoter driven dCas9‐VP64‐EDLL. The pDLGG047 construct contains only the dCas9‐VP64‐EDLL cassette without a gRNA expression module and pDLGG048, pDLGG049 have a single gRNA module driven by the OsU6snRNApolymerase promoter. pDLGG054 contains three gRNAs expression modules driven by different snRNApolymerase promoters (detailed in Table S3). Left inset figure showing the schematic of dCas9‐mediated activation for both the endogenous OsCGA1 gene and the pOsCGA1::GUS reporters. HPT, hygromycin resistance cassette. Arrows indicate the direction of transcription. (b) gRNAs designed to the proximal region of the transcriptional start site of OsCGA1. gRNA binding regions and PAM sites are highlighted as red and blue, respectively. TSS, transcriptional start site. (c and d) 3D‐reconstructed confocal images of longitudinal leaf section in the BS of dCas9 activation lines, (c) dCas9‐ADs only transgenic (pDLGG047 #4) and (d) transgenic with gRNA2 (pDLGG048 #1). Chlorophyll autofluorescence and cell wall staining by calcofluor‐white are pseudo‐coloured as green and grey, respectively. Chloroplasts within a BS cell are rendered for volume quantification and highlighted as yellow pseudo colour. Scale bar, 10 μm. (e–f) GUS reporter activation by dCas9‐mediated transactivation, (e) dCas9‐ADs only transgenic (pDLGG047 #4) and (f) transgenic with gRNA2 (pDLGG48 #14). Scale bar, 10 μm. (g) Transcript accumulation difference of OsCGA1 by gRNA position. 2 cm long‐mid regions from the emerging (5th) leaf were harvested for RT‐qPCR. Three independent events from each construct are assessed for OsCGA1 expression. Each bar refers to the mean +SD and −SEM of indicated events (2 ~ 4 T1 siblings, three PCR replicates). The letters above the bars note significant differences (**P < 0.01 as analysed using one‐way ANOVA followed by a Tukey’s post hoc test) compared to the expression level of pDLGG047 (no gRNA) control lines. (h) Box plot of chloroplast volume occupancy in the BS of various OsCGA1‐dCas9 activation events. The letters above the bars denote significant differences (**P < 0.01, *P < 0.05 as analysed using one‐way ANOVA followed by a Tukey’s post hoc test) compared to the chloroplast volumes of pDLGG047 (no gRNA) control lines.
To assess dCas9‐activated BS chloroplast proliferation, we scored the chloroplast volume in BS of transgenic lines using a 3D‐rendering of chlorophyll autofluorescence using confocal laser scanning microscopy on thick longitudinal vibratome sections (Figure 4c ~ d). Chloroplast volume within the BS is highly variable and dependent on position within the vascular bundle. Nevertheless, there was a significant increase in chloroplast volume in the BS of OsCGA1 transactivation lines compared to the control, dCas9‐ADs only cassette (Figure 4h). Two insertion events in particular, pDLGG048 #14 (gRNA2) and pDLGG049 #1 (gRNA3), conditioned a nearly 2.2‐fold increase in average chloroplast volume occupancy relative to the dCas9 only (pDLGG047) event (Figure 4h). Despite this higher density of chloroplasts in the BS of OsCGA1 transactivated lines, we noted that higher variability in the BS chloroplast abundance across different events with many chloroplasts aggregated in the central vacuole and with abundant stromules suggestive of chloroplast degradation intermediates (Figure S10) (Gray et al., 2012; Izumi et al., 2017). We speculate that since these intermediate structures are rarely observed in wild‐type plants, they were revealed here by artificially enhancing plastid proliferation in the BS. These results suggest that an active chloroplast turnover process is operative in rice BS to maintain low plastid copy numbers (Zhuang and Jiang, 2019).
Discussion
The role of CGA1 in photosynthetic gene regulation
The CGA1/GNC gene regulators have long been known to integrate diverse developmental and environmental cues to promote photosynthetic differentiation in Arabidopsis (Behringer and Schwechheimer, 2015; Cortleven and Schmülling, 2015). DELLA and light‐mediated degradation of PIF proteins derepress CGA1/GNC1 transcription and promote PEP assembly and plastid gene expression (Yoo et al., 2019; Richter et al., 2010). In contrast, auxin and GA signalling act to inhibit CGA1/GNC transcription, fine‐tuning cell elongation and greening (Richter et al., 2013; Richter et al., 2010). The stunted dark green phenotype mediated by OsCGA1 overexpression in rice and Arabidopsis (Figure S1; Chiang et al., 2012; Hudson et al., 2013) is reminiscent of the GA signalling mutant, ga1 and auxin signalling mutant, arf2 in arabidopsis (Richter et al., 2010; Richter et al., 2013) suggesting that ectopic expression of CGA1 is sufficient to override endogenous GA and auxin signalling pathways that would otherwise restrict photosynthetic development of the BS. The biogenesis of photosynthetically active chloroplasts is linked to the assembly of plastid‐encoded RNA polymerase and the transcription of photosynthesis‐associated plastid‐encoded genes (PhAPG) in the plastid (Liere and Börner, 2006). Notably, the expression of genes encoding nucleus encoded plastid transcription associated factors and PEP‐associated proteins (PAPs) (Steiner et al., 2011) increased in the BS of pFtGLDp::OsCGA1. In addition, the increased accumulation of other PhANGs, including genes encoding post‐transcriptional/translational machinery, light harvesting complexes and chlorophyll biosynthetic components, indicated that OsCGA1 exerts its effect on chloroplast development by regulating nuclear and plastid gene expression. Similar to our observations, Hudson et al., 2013 has shown the primary effect of ectopic CGA1 expression is the increased expression of several nuclear‐encoded, chloroplast localized proteins involved in chlorophyll binding and photosynthesis. However, we did not detect the increased expression of genes involved in amino acid biosynthesis, starch biosynthesis and chloroplast division as detected by Hudson et al., 2013. This may be due to the differential promoter activities used in the two studies (i.e. a weak vascular‐specific promoter vs. the strong constitutive ZmUbi1 promoter).
Engineering orthogonal BS activities
To fine tune the expression of OsCGA1, we utilized the well‐characterized vascular‐specific promoter, FtGLDp, that was first identified in Flaveria trinervia and later characterized in Arabidopsis thaliana (Engelmann et al., 2008). In rice, we were surprised initially to see strong constitutive GUS staining of the pFtGLDp::Introns containing GUS and later realized that removal of introns in the GUS gene cassette that was inserted to enhance the reporter gene expression, conditioned the expected weak vascular‐specific pattern. In similar fashion, the use of an intron‐containing GUS reporter driven by the Zoyzia Japonica PEPCK promoter, converts a vascular‐specific to a constitutive leaf expression profile (data not shown; Nomura et al., 2005). Introns have often been introduced into reporter constructs to enhance gene expression in plants, especially when used in conjunction with weak promoters in an effort to mitigate transgene silencing and to discriminate reporter gene expression from bacterial expression during Agrobacterium‐mediated transformation (Callis et al., 1987; Tanaka et al., 1990). Though anecdotal, these findings suggest it is necessary to validate tissue‐ and cell‐specific profiles of gene expression across species using GUS reporters. Here, we confirmed the vascular‐specific activity of the FtGLDp promoter with LCM transcriptomics and tissue‐specific transactivation of OsCGA1.
Our successful dCas9‐mediated transactivation demonstrates the potential of regulating suites of genes in a tissue‐specific manner. In our studies, a single gRNA near the TSS was sufficient to increase expression of OsCGA1 and the pOsCGA1::GUS reporter simultaneously. By multiplexing gRNAs and expressing a dCas9‐activator (with a cell‐specific promoter) it should be feasible to drive multiple Kranz anatomy and/or C4 metabolic genes in a cell‐specific manner. Future studies to examine the placement of the gRNA relative to the TSS and multiplexing chimeric dCas9 and transcriptional repressors, such as SRDX or BRD domains could further extend the utility of the dCas9 system (Lowder et al., 2015). Similarly, the use of dCas12‐activator/repressor modules would enable the flexibility to simultaneously activate and repress gene expression in the same cell types (Bandyopadhyay et al., 2020). Although it was possible to manipulate chloroplast architecture in the BS, we also observed what is likely an increased activity of chloroplast degradation as well. Confocal imaging of BS cells expressing dCas9‐AD fusions revealed a proliferation of stromule projections from chloroplasts and chloroplast accumulation in central vacuoles. These features (increased stromules and vacuolar localization of plastids) can be induced by ABA‐mediated abiotic stress and UV light, respectively (Gray et al., 2012; Izumi et al., 2017) and are likely intermediate products of chloroplast degradation (Zhuang and Jiang, 2019). Consistent with this hypothesis, rice BS plastid integrity is more sensitive to drought than M plastids of rice (Yamane et al., 2003). As CK plays a protective role for the photosynthetic machinery by promoting antioxidant production under high light and dark‐induced senescence (Procházková et al., 2008; Zavaleta‐Mancera et al., 2007), it is likely that ABA‐ and CK‐mediated responses act antagonistically in the BS. In addition to ABA, protoxylem and procambium are rich in the phytohormones GA and auxin, respectively, which as discussed above may also inhibit the transcription of CGA1/GNC around vasculature (Scarpella et al., 2006; Yamazaki et al., 2018).
Engineering C4 activities in rice
In the context of engineering C4 activities into rice this study provides both promise and new challenges. A clear challenge in engineering C4 activities is the lack of BS‐specific promoters (Engelmann et al., 2008). To date only a single vascular‐specific promoter has been conclusively identified that can drive vascular‐specific expression, despite intensive investigations (Schuler et al., 2016). Here, we demonstrate that a single promoter can be used to drive expression from at least two genes. Furthermore, we demonstrate that a single gRNA is sufficient to drive ectopic gene expression, thus, in theory, dozens of genes could be co‐regulated using a single promoter driving expression of a dCas9‐AD by simply designing a tandem array of gRNA sequences targeted to the promoter regions of endogenous or transgenically delivered genes. A previous study suggested a minimum of 20 genes will need to be engineered in both BS and M cells to install a minimal C4 circuit in rice (Peterhansel, 2011). The dCas9 technology detailed here is one such enabling technology to realize this potential. Although the focus of this work was on the manipulation of the CGA1 network, work by Langdale and colleagues has suggested that BS plastid area could also be increased through the ectopic expression of GOLDEN2 and GOLDEN2‐like (Wang et al., 2017). Future studies to examine the potential for selectively increasing plastid numbers in the BS together with the BS‐specific expression of either CGA1 or GLK could result in additive effects that would increase both plastid size and number. However, this study also suggested the presence of a BS plastid degradation pathway that serves to limit the proliferation of BS chloroplasts in rice. Thus, to engineer a photosynthetic BS in rice, it may also be necessary to manipulate this autophagic response to maintain plastid integrity.
Material and Methods
Generation of constructs
All constructs were generated using the Golden Gate cloning system (Engler et al., 2014; Weber et al., 2011). Full‐length cDNA of OsCGA1 (Gene bank Accession No. AK099607) was obtained from NIAS DNA Bank (http://www.dna.affrc.go.jp/). Fragments were amplified by PCR and subcloned into pICH41308 using BbsI digestion and ligation, to domesticate (removal of internal BbsI and BsaI restriction sites). The Flaveria trinervia Glycine Decarboxylase p‐subunit promoter was received from Dr. Udo Gowik and was domesticated using PCR (Engelmann et al., 2008). For the dCas9‐AD module, dCas9 and two tandemly linked activation domains, VP64‐EDLL, were domesticated from pYPQ176 (Lowder et al., 2018). The gRNA expression modules from pOsU6snRNApol and pAtU6snRNApol were also domesticated from pYPQ141A and pYPQ141C. A 2126 bp upstream sequence of OsCGA1 was isolated from Kitaake gDNA and domesticated into the pL0M‐PU module. The gRNA sequences were designed using two web‐based applications (https://crispr.dbcls.jp/ and http://crispor.tefor.net/) and selected based on the proximity to the transcriptional start site of the OsCGA1 promoter. For the gRNA cloning, forward primer and complementary primer are annealed and cloned into the corresponding gRNA modules with appropriate positions of L1 modules using Esp3I and Eco31I. All constructs were generated via Golden Gate assembly using Thermo FastDigest restriction enzyme and T4 DNA Ligase (Thermo Fisher Scientific, Waltham,USA). All L0 module insert sequences, primer sequences for gRNA cloning and layout of assemblies used in this study are summarized in Table S4. All basal L0 modules and gRNA expression modules are cloned in the appropriate position of backbone and their insert sequence is provided between 4 bp overhangs in Table S4. Constructs received form ENSA projects were marked as EC*****. For L1 constructs and L2 constructs are assembled with the presented modules of lower order in Table S4.
Rice transgenic production
Agrobacterium‐mediated transformation of rice var. Kitaake was developed based on a previous protocol for Nipponbare (Ozawa, 2009) with major modifications in media composition and light conditions. Kitaake rice seeds were dehusked and sterilized with a 50% bleach solution for 30 min. Seeds were rinsed three times with sterile water and incubated in a 1% hydrogen peroxide solution for 1 h. Surface‐sterilized seeds were then placed on MSD (1x Murashige Skoog Basal salts (PhytoTechnology), 1x B5 Gamborg’s vitamins, 0.03% casamino acids, 0.11% L‐proline, 4% Maltose, 1 mg/mL 2,4‐D) with 0.3% gelrite to induce callus under continuous light at 28 °C for 2 weeks. Agro strain LBA4404 carrying the constructs was inoculated on AB liquid media (Wise et al., 2006) and resuspended in AAM media (Hiei et al., 1994) with 15 ųg/ml of acetosyringone. Calli were submerged in an agrobacterium suspension and blotted dry on sterile paper to remove excess Agrobacteria. Co‐cultivation was performed on three sheets of No. 2 filter papers (Advantec, JAPAN) wetted with MSD liquid supplemented with 0.01% L‐Cysteine, 0.5% glucose and 40 μg/mL of acetosyringone at 22 °C under dark for 3 days. Co‐cultivated tissues were washed with sterile water three times and 200 μg/mL timentin‐containing water. The washed calli were selected on the 0.3% gelrite MSD with hygromycin B (50 μg/mL) and timentin (100 μg/mL) for 2‐3 weeks. Newly arising nodal calli were transferred to MSR media (1× MS salts, 1× B5 Gamborg vitamin, 4% maltose, 0.2% casamino acid, 0.1 μg/mL NAA and 2 μg/mL Kinetin) with hygromycin B (50 μg/mL) and timentin (100 μg/mL) and 0.4% gelrite under continuous light at 28 °C. Regenerated explants with intact roots and shoots were transferred to soil under greater than 50% relative humidity to recover.
Plant growth
Plants were grown for seed propagation and phenotypic characterizations at the Donald Danforth Plant Science Center, St. Louis, MO under greenhouse growth conditions. Rice var. Kitaake and derived transgenic were grown in turface (Field & Fairway, PROFILE products LLC) in 175 mL square pots in flooded trays. Plants were given intermittent fertilizer with 200 ppm N. of 15–16–17 peat‐lite (ICL Special Fertilizers, OH) as needed. The greenhouse was maintained under daily photoperiod of 14 h, with radiation incident on the plant canopy composed by external sun radiation plus radiation emitted by supplemental HID lights mixed with Metal Halide and High Pressure Sodium lamps (the latter with relative intensity of 7: 3). In the time window from 10 a.m. to 2 p.m. standard time, the Photosynthetic Photon Flux Density (PPFD) incident on the plant canopy was in the range of 800–1000 µmol photons m−2 s−1. The air temperature was set at 28/25 °C during the day/night, respectively. Plants were given intermittently with fertilizer of 200 ppm N. of 15‐16‐17 peat‐lite (ICL Special Fertilizers, OH) as needed.
Plants used for leaf photosynthetic analysis were grown in a controlled environment growth chamber (Bigfoot series, BioChambers Inc., Winnipeg, MB, Canada) at the School of Biological Sciences at Washington State University, Pullman, WA (USA). The growth chamber was set with a daily photoperiod of 14 h with a maximum PPFD of 600 µmol photons m−2 s−1 incident on the plant canopy, air temperature of 26 °C and air relative humidity of ~70% (corresponding to air vapour pressure deficit of ~1.6 kPa); during the dark period air temperature was set at 22 °C. Plants were grown individually in 7.5‐L free drainage pots in a Sunshine Mix LC‐1 soil (Sun Gro Horticulture, Agawam, MA) mixed with turface (ratio of 3:1 in volume) under daily irrigation and fertilized twice a week to pot saturation.
For LCM analysis, plants were grown in 1:1 mixture of topsoil and sand for about 2 weeks in a growth room with a daily 12 h photoperiod, and PPFD of 300 μmol photons m−2 s−1 incident on the plant canopy. Air temperatures were set at 28 and 25 °C during the photoperiod and in the dark, respectively, and air relative humidity was 65% (corresponding to maximum air vapour pressure deficit of ~1.9 kPa).
RNA extraction and RT‐qPCR analysis
For the pFtGLDP::OsCGA1 transgene expression (Figure 1b), 2 cm of mid and tip regions of emerging fourth leaves were harvested into round‐bottom microfuge tubes and frozen in liquid N2. For dCas9‐mediated OsCGA1 activation, 2 cm sections of the mid region of the leaf were used (Figure 4g). Frozen tissues were ground by bead beating in a paint shaker with liquid N2. RNA was extracted using TriPure isolation reagent (Roche Diagnostics, Basel, Switzerland) and 1 μg RNA of individual samples were used for residual gDNA removal using RQ1 RNase‐Free DNase (Promega, Madison, USA). After ethanol precipitation, DNase‐treated RNA was used for cDNA synthesis using Improm‐II™ reverse transcriptase (Promega) and Oligo(dT)15 primer (promega) according to the manufacturer’s instructions. cDNA was used for the RT‐qPCR using a LightCycler®️ 480 Green I Master mix (Roche Diagnostics) and primer set (Table S3) for target and reference genes on LightCycler 480 II qPCR machine (Roche Diagnostics). Os14‐4‐3E (LOC_Os11g34450) were used for normalization in Figure 1b and Os14‐3‐3E was used solely for the normalization for Figure 4g. Normalized and relative expression were calculated based on the methods in (Taylor et al., 2019).
LCM sample preparation
The middle 1‐cm section of the third expanded leaf was sampled at 4 h after dawn and fixed in ice‐cold 100% acetone for 4 h on ice. The leaf tissue for LCM was then processed as described (Hua and Hibberd, 2019). In brief, leaf tissue was dehydrated through a series of ethanol gradients (v/v) of 70%, 85%, 95% and 2*100%, then kept in 100% ethanol overnight; the next day, leaf tissue was infiltrated with 25%, 50%, 75% and 2*100% Steedman’s wax in ethanol at 40 °C for 2 h each, then the leaf was infiltrated with 100% Steedman’s wax overnight, the next day, tissue was embedded in petri‐dish.
Laser capture microdissection and RNA extraction
Paradermal sections of 7 µm thickness were prepared with a Reichert‐Jung microtome and mounted on PEN membrane glass slides (Applied Biosystems) with DEPC‐treated water. Steedman’s wax was removed by incubating slides in 100% acetone for 1 min, then Laser capture microdissection was performed on ArcturusXT LCM System (Applied Biosystems) following the user manual. Bundle sheath strands and mesophyll cells were harvested on CapSure Macro LCM Caps (Applied Biosystems), RNA was extracted using PicoPure RNA Isolation Kit with on‐column DNase treatment according to manufacturer's instructions. RNA quality was examined using Bioanalyzer 2100 (Agilent, Santa Clara) in combination with RNA pico chip.
LCM Library preparation and sequencing
From 37 to100 ng bundle sheath strand (BSS) and M LCM isolated RNA from lines #12‐2‐4 and #12‐4‐4 were used for 3’ mRNA‐seq library preparation using a QuantSeq 3’ mRNA‐Seq Library Prep Kit (Lexogen, Vienna, Austria) according to the manufacturer's instructions. The RNA integrity number (RIN) values ranged from 5.6–6.7 as determined by analysis of electropherogram output from Bioanalyzer (Agilent). Sixteen PCR cycles were used to enrich libraries and libraries were sequenced using 75‐bp single end reads on an Illumina NextSeq 550 platform.
Confocal imaging and quantification of chloroplast volume
Corresponding fourth or fifth leaf segments were immersed and fixed for 30min in the freshly made 4% paraformaldehyde (80 mM sodium phosphate buffer, 0.1% Triton X‐100) with vacuum infiltration. After vacuum infiltrations, leaf tissue was placed in fixative for 1 ~ 2 h, rinsed and then stored in 0.1 M EDTA solution. Fixed tissues were stored in darkness at 4 °C and imaged within 6 months. Stored tissues were embedded in 7% low melting agarose and 50 μm sections generated with a vibratome. Both transverse and longitudinal sections were placed in a 5 μM calcofluor white solution for the cell wall imaging. Sections were observed with a Leica TCS SP8 confocal laser scanning microscope using a HC PL APO CS2 40×/1.10 WATER objective lens (Leica Microsystems, Manheim, Germany). Sections were imaged using a 405 nm UV laser for excitation. Calcofluor white and chlorophyll autofluorescence were obtained with the 460–480 nm and 680–700 nm band emission spectrum, respectively, with a 16 line average resolution. For chloroplast volume quantification, 90 nm thick optical section images were taken for each sample with fixed voxel size with pixel size 90 nm × 90 nm for the consistency. Optical stacks were trimmed and deconvoluted using Huygens Essential software (Scientific Volume imaging, Hilversum, Netherland). Chloroplast volumes were obtained by surface rendering on the IMARIS (Oxford instruments, UK) and chloroplasts within individual BS cells were manually picked based on the calcofluor staining. Longitudinally sliced BS cells were considered as partial cylinder and volume of each BS cells were roughly estimated by
measuring length (L), radius (R) and depth (D) of the cylinder.
Transmission electron microscopy
Leaf 4 was prepared for transmission electron microscopy (TEM), quantification of cellular features, and immunodetection of photosynthetic enzymes and glycine decarboxylase was previously described (Khoshravesh et al., 2017). Leaf sections from the middle of recent fully expanded leaves were fixed in 1% glutaraldehyde and 1% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 6.8) overnight at room temperature. Samples were subsequently dehydrated in ethanol: H2O with 10% increment increases of 10% to 100% ethanol for 1 h at each incremental increase. This was followed by two 1 h incubations in 100% ethanol before the tissue was infiltrated in London Resin White (LRW) using 1:3, 1:1, and 3:1 ratio of LRW to 100% ethanol (8 h each increment), followed by 2 × 100% LRW (8 h each). LRW infiltrated leaf samples were polymerized at 60 °C in an oxygen‐free environment for 12 h. Sections of 50–70 nm were collected for immunolocalization and TEM imaging. Images for TEM were captured on a Phillips 201 transmission electron microscope equipped with an Advantage HR camera system (Advanced Microscopy Techniques). For quantification of organelle numbers and area from EM images, Kruskal–Wallis one‐way analysis of variance was performed, followed by a Dunn's post hoc test in R.
Immunohistochemistry
Immunohistochemistry was performed as detailed in (Wang et al., 2017). In brief, 50–70 nm sections of LRW embedded tissue were rehydrated in 0.01 M phosphate saline buffer (PBS) pH 7.4, blocked for 20 min in 0.5% bovine serum albumin (BSA) in PBS, and then rinsed in PBS 3× for 15 min before incubating for 3 h in primary antibody at the following concentrations: 1:50 (anti‐glycine decarboxylase), 1:100 (anti‐RuBisCo and anti‐RuBisCo activase) and 1:400 (anti‐FBPase) in 0.1% BSA/PBS. Sections were then rinsed in PBS 3 × 15 min before incubation with secondary antibody (18 nm Colloidal Gold‐AffiniPure Goat Anti‐Rabbit IgG) for 1 h at a concentration of 1:20 (glycine decarboxylase) or 1:40 (photosynthetic enzymes) in 0.1% BSA/PBS. Samples were then rinsed in PBS three times for 15 min each, followed by ultrapure water (3 × 15 min) before being stained with 4% uranyl acetate for 10 min, and then lead citrate for 5 min (Reynolds, 1963).
RNA seq data analysis
Raw RNAseq reads were quality checked using FastQC (https://github.com/s‐andrews/FastQC). Adapter, quality trimming, and polyA tail removal was performed using TrimGalore (https://github.com/FelixKrueger/TrimGalore) with the following parameters (stringency 3, clip_R1 13, length 20, q 20). Gene annotation and transcript sequences (Osativa_323_v7.0) were obtained from Phytozome11. Transcript quantification and read counts were performed using Salmon (quasi‐mapping based, parameters: incompatPrio 0.0, noLengthCorrection, libType SF) (Patro et al., 2017). Read counts were imported with tximport R‐package (Soneson et al., 2015). Differential expression analysis was carried out with the edgeR package (FDR < 0.10) (Robinson et al., 2010).
Competing interests
The authors have no competing interests to declare.
Author contributions
D‐Y.L. conducted molecular genetics experiments and confocal imaging, L.H. performed LCM analysis, R.K. performed EM analysis and immunohistology, R.G. performed photosynthetic assays, I.K. conducted RNA seq analysis, A.C. analysed photosynthetic measurements, T.L.S. analysed EM and immunohistochemistry data, J.H. analysed LCM data, D‐Y.L and T.P.B conceptualized the project and wrote the manuscript. All authors contributed to editing the manuscript.
Materials & Correspondence
Please contact Dr. Thomas Brutnell (tom@viridisgenomics.com) for requests of materials and correspondence. Data is available through a CC BY license agreement. RNAseq data is accessible through NCBI accessions PRJNA736666 and PRJNA739378.
Supporting information
Figure S1. Overexpression analysis of OsCGA1 in Kitaake var. rice.
Figure S2. Flaveria trinervia Glycine Decarboxylase p‐subunit (FtGLDp) promoter activity and chloroplast proliferation in the BS of the pFtGLDp::OsCGA1 transgenic lines.
Figure S3. Accumulation of photosynthetic enzymes in WT and Transgenic lines.
Figure S4. Leaf photosynthetic and biochemical responses
Figure S5. Summary of leaf gradient RNA seq in two independent events of pFtGLDP::OsCGA1.
Figure S6. Microdissection images of BS strands and M strands and their RNA profiles from pFtGLDP::OsCGA1 transgenic and nulls.
Figure S7. Pageman analysis using DE genes in BSS LCM seq.
Figure S8. Transcriptional activation test on OsCGA1 promoter GUS reporter using dCas9‐mediated transactivation in heterologous system Nicotiana tabaccum.
Figure S9. Tissue specific expression of pOsCGA1::GUS reporter by dCas9 mediated activation in Kitaake transgenics.
Figure S10. Chloroplast morphologies in the bundle sheath cells of dCas9 activation lines.
Table S1. TEM and quantitative measurements of organelle phenotypes in pFtGLDp::OsCGA1 transgenic.
Table S2. Summary of transgenic lines used in this study.
Table S3. Primer and taqman probe sequences used in this study.
Table S4. Golden gate construct assembly and sequence information.
Table S5. DEGs in the BS LCM seq between pFtGLDp::OsCGA1 transgenic and their null (FDR<0.05).
Supplementary Material.
Acknowledgements
The authors would like to acknowledge Dr. Udo Gowik, to whom this manuscript is dedicated, for providing us FtGLDp promoter and many fruitful discussions on C4 biology and engineering efforts and Dr. Howard Berg for his invaluable advice on confocal imaging and tissue preparation. We would also like to thank Dr. Yiping Qi for providing vector backbones to enable this work. This work was supported by C4 Rice Project grants from the Bill & Melinda Gates Foundation to IRRI (Grant ID#51586) and the University of Oxford (OPP1129902), an NSF grant MCB‐1546882 to T.P.B. and a Korea NRF fellowship (2012R1A6A3A03040277) to D‐Y.L.
Lee, D.‐Y. , Hua, L. , Khoshravesh, R. , Giuliani, R. , Kumar, I. , Cousins, A. , Sage, T. L. , Hibberd, J. M. and Brutnell, T. P. (2021) Engineering chloroplast development in rice through cell‐specific control of endogenous genetic circuits. Plant Biotechnol. J., 10.1111/pbi.13660
References
- Bandyopadhyay, A. , Kancharla, N. , Javalkote, V.S. , Dasgupta, S. and Brutnell, T.P. (2020) CRISPR‐Cas12a (Cpf1): a versatile tool in the plant genome editing tool box for agricultural advancement. Front. Plant Sci. 11, 1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer, C. and Schwechheimer, C. (2015) B‐GATA transcription factors – insights into their structure, regulation, and role in plant development. Front. Plant Sci., 6, 1–12. 10.3389/fpls.2015.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner, T. , Aleynikova, A.Y. , Zubo, Y.O. and Kusnetsov, V.V. (2015) Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim. Biophys. Acta, 1847, 761–769. [DOI] [PubMed] [Google Scholar]
- von Caemmerer, S. , Quick, W. P. and Furbank, R.T. (2012) The Development of C4 Rice: Current Progress and Future Challenges. Science, 336(6089), 1671–1672. 10.1126/science.1220177 [DOI] [PubMed] [Google Scholar]
- Callis, J. , Fromm, M. and Walbot, V. (1987) Introns increase gene expression in cultured maize cells. Genes Dev. 1, 1183–1200. [DOI] [PubMed] [Google Scholar]
- Chiang, Y.‐H. , Zubo, Y.O. , Tapken, W. , Kim, H.J. , Lavanway, A.M. , Howard, L. , Pilon, M. et al. (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 160, 332–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, A.H. and Quail, P.H. (1996) Ubiquitin promoter‐based vectors for high‐level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218. [DOI] [PubMed] [Google Scholar]
- Cortleven, A. and Schmülling, T. (2015) Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 66, 4999–5013. [DOI] [PubMed] [Google Scholar]
- Demarsy, E. , Courtois, F. , Azevedo, J. , Buhot, L. and Lerbs‐Mache, S. (2006) Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol. 142, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L. , Jiao, F. , Chu, J. , Jin, G. , Chen, M. and Wu, P. (2007) The two‐component signal system in rice (Oryza sativa L.): a genome‐wide study of cytokinin signal perception and transduction. Genomics 89, 697–707. [DOI] [PubMed] [Google Scholar]
- Engelmann, S. , Wiludda, C. , Burscheidt, J. , Gowik, U. , Schlue, U. , Koczor, M. , Streubel, M. et al. (2008) The gene for the P‐subunit of glycine decarboxylase from the C4 species Flaveria trinervia: analysis of transcriptional control in transgenic Flaveria bidentis (C4) and Arabidopsis (C3). Plant Physiol. 146, 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler, C. , Youles, M. , Gruetzner, R. , Ehnert, T.‐M. , Werner, S. , Jones, J.D.G. , Patron, N.J. et al. (2014) A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Gilbert, L.A. , Larson, M.H. , Morsut, L. , Liu, Z. , Brar, G.A. , Torres, S.E. , Stern‐Ginossar, N. et al. (2013) CRISPR‐mediated modular RNA‐guided regulation of transcription in eukaryotes. Cell 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassini, P. , Eskridge, K.M. and Cassman, K.G. (2013) Distinguishing between yield advances and yield plateaus in historical crop production trends. Nat. Commun. 4, 2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J.C. , Hansen, M.R. , Shaw, D.J. , Graham, K. , Dale, R. , Smallman, P. , Natesan, S.K.A. et al. (2012) Plastid stromules are induced by stress treatments acting through abscisic acid. Plant J. 69, 387–398. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P.T. , Allison, L.A. and Maliga, P. (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16, 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, M.D. (1987) C4 photosynthesis: a unique elend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta (BBA) – Rev. Bioenerget., 895(2), 81–106. 10.1016/s0304-4173(87)80009-5 [DOI] [Google Scholar]
- He, L. , Zhang, S. , Qiu, Z. , Zhao, J. , Nie, W. , Lin, H. , Zhu, Z. , Zeng, D. et al. (2018) FRUCTOKINASE‐LIKE PROTEIN 1 interacts with TRXz to regulate chloroplast development in rice. J. Integr. Plant Biol. 60, 94–111. [DOI] [PubMed] [Google Scholar]
- Hibberd, J.M. and Covshoff, S. (2010) The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol. 61, 181–207. [DOI] [PubMed] [Google Scholar]
- Hibberd, J.M. , Sheehy, J.E. and Langdale, J.A. (2008) Using C4 photosynthesis to increase the yield of rice—rationale and feasibility. Curr. Opin. Plant Biol., 11(2), 228–231. 10.1016/j.pbi.2007.11.002 [DOI] [PubMed] [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hua, L. and Hibberd, J.M. (2019) An optimized protocol for isolation of high‐quality RNA through laser capture microdissection of leaf material. Plant Direct, 3, e00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, D. , Guevara, D. , Yaish, M.W. , Hannam, C. , Long, N. , Clarke, J.D. , Bi, Y.‐M. et al. (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd‐GOGAT) expression in Arabidopsis. PLoS One, 6, e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, D. , Guevara, D.R. , Hand, A.J. , Xu, Z. , Hao, L. , Chen, X. , Zhu, T. et al. (2013) Rice cytokinin GATA transcription Factor1 regulates chloroplast development and plant architecture. Plant Physiol. 162, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi, M. , Ishida, H. , Nakamura, S. and Hidema, J. (2017) Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell, 29, 377–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggard, K.W. , Qi, A. and Ober, E.S. (2010) Possible changes to arable crop yields by 2050. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 2835–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshravesh, R. , Lundsgaard‐Nielsen, V. , Sultmanis, S. and Sage, T.L. (2017) Light microscopy, transmission electron microscopy, and immunohistochemistry protocols for studying photorespiration. Methods Mol. Biol. 1653, 243–270. [DOI] [PubMed] [Google Scholar]
- Li, P. , Ponnala, L. , Gandotra, N. , Wang, L. , Si, Y. , Tausta, S.L. , Kebrom, T.H. et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Liere, K. and Börner, T. (2006) Transcription of plastid genes. Regulation of Transcription in Plants, Annual Plant Reviews. 29, pp. 184–224. Blackwell Publishing. [Google Scholar]
- Lowder, L.G. , Zhang, D. , Baltes, N.J. , Paul, J.W. 3rd , Tang, X. , Zheng, X. , Kebrom, T.H. et al. (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol., 169, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder, L.G. , Zhou, J. , Zhang, Y. , Malzahn, A. , Zhong, Z. , Hsieh, T.‐F. , Voytas, D.F. et al. (2018) Robust transcriptional activation in plants using multiplexed CRISPR‐Act2.0 and mTALE‐Act systems. Mol. Plant, 11, 245–256. [DOI] [PubMed] [Google Scholar]
- Naito, T. , Kiba, T. , Koizumi, N. , Yamashino, T. and Mizuno, T. (2007) Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 71, 1557–1560. [DOI] [PubMed] [Google Scholar]
- Nomura, M. , Higuchi, T. , Ishida, Y. , Ohta, S. , Komari, T. , Imaizumi, N. , Miyao‐Tokutomi, M. et al. (2005) Differential expression pattern of C4 bundle sheath expression genes in rice, a C3 plant. Plant Cell Physiol. 46, 754–761. [DOI] [PubMed] [Google Scholar]
- Ozawa, K. (2009) Establishment of a high efficiency Agrobacterium‐mediated transformation system of rice (Oryza sativa L.). Plant Sci. 176, 522–527. [DOI] [PubMed] [Google Scholar]
- Patro, R. , Duggal, G. , Love, M.I. , Irizarry, R.A. and Kingsford, C. (2017) Salmon provides fast and bias‐aware quantification of transcript expression. Nat. Methods, 14(4), 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhansel, C. (2011) Best practice procedures for the establishment of a C4 cycle in transgenic C3 plants. J. Exp. Bot. 62, 3011–3019. [DOI] [PubMed] [Google Scholar]
- Procházková, D. , Haisel, D. and Wilhelmová, N. (2008) Antioxidant protection during ageing and senescence in chloroplasts of tobacco with modulated life span. Cell Biochem. Funct. 26, 582–590. [DOI] [PubMed] [Google Scholar]
- Qiao, J. , Li, J. , Chu, W. and Luo, M. (2013) PRDA1, a novel chloroplast nucleoid protein, is required for early chloroplast development and is involved in the regulation of plastid gene expression in Arabidopsis. Plant Cell Physiol. 54, 2071–2084. [DOI] [PubMed] [Google Scholar]
- Reyes, J.C. , Muro‐Pastor, M.I. and Florencio, F.J. (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134, 1718–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, E.S. (1963) The use of lead citrate at high pH as an electron‐opaque stain in electron microscopy. J. Cell Biol. 17, 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, R. , Behringer, C. , Muller, I.K. and Schwechheimer, C. (2010) The GATA‐type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME‐INTERACTING FACTORS. Genes Dev. 24, 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, R. , Behringer, C. , Zourelidou, M. and Schwechheimer, C. (2013) Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA, 110, 13192–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D. , McCarthy, D.J. and Smyth, G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26(1), 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage, R.F. and Zhu, X.‐G. (2011) Exploiting the engine of C4 photosynthesis. J. Exp. Bot. 62, 2989–3000. [DOI] [PubMed] [Google Scholar]
- Scarpella, E. , Marcos, D. , Friml, J. and Berleth, T. (2006) Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M.L. , Mantegazza, O. and Weber, A.P.M. (2016) Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant J. 87, 51–65. [DOI] [PubMed] [Google Scholar]
- Soneson, C. , Love, M.I. and Robinson, M.D. (2015) Differential analyses for RNA‐seq: transcript‐level estimates improve gene‐level inferences. F1000Research, 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Catanzariti, A.‐M. , DeBoer, K. , Petre, B. , Gardiner, D.M. , Singh, K.B. , Dodds, P.N. et al. (2017) LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 7, 44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, S. , Schröter, Y. , Pfalz, J. and Pfannschmidt, T. (2011) Identification of essential subunits in the plastid‐encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 157, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A. , Mita, S. , Ohta, S. , Kyozuka, J. , Shimamoto, K. and Nakamura, K. (1990) Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and an efficient splicing of the intron. Nucleic Acids Res. 18, 6767–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S.C. , Nadeau, K. , Abbasi, M. , Lachance, C. , Nguyen, M. and Fenrich, J. (2019) The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 37, 761–774. [DOI] [PubMed] [Google Scholar]
- Toyoshima, Y. , Onda, Y. , Shiina, T. and Nakahira, Y. (2005) Plastid Transcription in Higher Plants. CRC Crit. Rev. Plant Sci., 24(1), 59–81. 10.1080/07352680590910438 [DOI] [Google Scholar]
- Usadel, B. , Nagel, A. , Steinhauser, D. , Gibon, Y. , Bläsing, O.E. , Redestig, H. , Sreenivasulu, N. et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics, 7, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallmeroth, N. , Anastasia, A.K. , Harter, K. , Berendzen, K.W. and Mira‐Rodado, V. (2017) Arabidopsis response regulator 22 inhibits cytokinin‐regulated gene transcription in vivo. Protoplasma, 254, 597–601. [DOI] [PubMed] [Google Scholar]
- Wallmeroth, N. , Jeschke, D. , Slane, D. , Nägele, J. , Veerabagu, M. , Mira‐Rodado, V. and Berendzen, K.W. (2019) ARR22 overexpression can suppress plant two‐component regulatory systems. PLoS One, 14, e0212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Khoshravesh, R. , Karki, S. , Tapia, R. , Balahadia, C.P. , Bandyopadhyay, A. , Paul Quick, W. et al. (2017) Re‐creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Curr. Biol. 27, 3278–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, E. , Engler, C. , Gruetzner, R. , Werner, S. and Marillonnet, S. (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS ONE, 6, e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, A.A. , Liu, Z. and Binns, A.N. (2006) Culture and maintenance of Agrobacterium strains. Methods Mol. Biol. 343, 3–13. [DOI] [PubMed] [Google Scholar]
- Yamane, K. , Hayakawa, K. , Kawasaki, M. , Taniguchi, M. and Miyake, H. (2003) Bundle sheath chloroplasts of rice are more sensitive to drought stress than mesophyll chloroplasts. J. Plant Physiol. 160, 1319–1327. [DOI] [PubMed] [Google Scholar]
- Yamazaki, K. , Kondo, Y. , Kojima, M. , Takebayashi, Y. , Sakakibara, H. and Fukuda, H. (2018) Suppression of DELLA signaling induces procambial cell formation in culture. Plant J. 94, 48–59. [DOI] [PubMed] [Google Scholar]
- Ye, L.‐S. , Zhang, Q. , Pan, H. , Huang, C. , Yang, Z.‐N. and Yu, Q.‐B. (2017) EMB2738, which encodes a putative plastid‐targeted GTP‐binding protein, is essential for embryogenesis and chloroplast development in higher plants. Physiol. Plant. 161, 414–430. [DOI] [PubMed] [Google Scholar]
- Yoo, C.Y. , Pasoreck, E.K. , Wang, H. , Cao, J. , Blaha, G.M. , Weigel, D. and Chen, M. (2019) Phytochrome activates the plastid‐encoded RNA polymerase for chloroplast biogenesis via nucleus‐to‐plastid signaling. Nat. Commun. 10, 2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaleta‐Mancera, H.A. , López‐Delgado, H. , Loza‐Tavera, H. , Mora‐Herrera, M. , Trevilla‐García, C. , Vargas‐Suárez, M. and Ougham, H. (2007) Cytokinin promotes catalase and ascorbate peroxidase activities and preserves the chloroplast integrity during dark‐senescence. J. Plant Physiol. 164, 1572–1582. [DOI] [PubMed] [Google Scholar]
- Zhuang, X. and Jiang, L. (2019) Chloroplast degradation: multiple routes into the vacuole. Front. Plant Sci. 10, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Overexpression analysis of OsCGA1 in Kitaake var. rice.
Figure S2. Flaveria trinervia Glycine Decarboxylase p‐subunit (FtGLDp) promoter activity and chloroplast proliferation in the BS of the pFtGLDp::OsCGA1 transgenic lines.
Figure S3. Accumulation of photosynthetic enzymes in WT and Transgenic lines.
Figure S4. Leaf photosynthetic and biochemical responses
Figure S5. Summary of leaf gradient RNA seq in two independent events of pFtGLDP::OsCGA1.
Figure S6. Microdissection images of BS strands and M strands and their RNA profiles from pFtGLDP::OsCGA1 transgenic and nulls.
Figure S7. Pageman analysis using DE genes in BSS LCM seq.
Figure S8. Transcriptional activation test on OsCGA1 promoter GUS reporter using dCas9‐mediated transactivation in heterologous system Nicotiana tabaccum.
Figure S9. Tissue specific expression of pOsCGA1::GUS reporter by dCas9 mediated activation in Kitaake transgenics.
Figure S10. Chloroplast morphologies in the bundle sheath cells of dCas9 activation lines.
Table S1. TEM and quantitative measurements of organelle phenotypes in pFtGLDp::OsCGA1 transgenic.
Table S2. Summary of transgenic lines used in this study.
Table S3. Primer and taqman probe sequences used in this study.
Table S4. Golden gate construct assembly and sequence information.
Table S5. DEGs in the BS LCM seq between pFtGLDp::OsCGA1 transgenic and their null (FDR<0.05).
Supplementary Material.


