Summary
The WUSCHEL‐related homeobox (WOX) transcription factors WOX11 and WOX12 regulate adventitious rooting and responses to stress. The underlying physiological and molecular regulatory mechanisms in salt stress tolerance remain largely unexplored. Here, we characterized the roles of PagWOX11/12a from 84K poplar (Populus alba × P. glandulosa) and the underlying regulatory mechanism in salt stress. PagWOX11/12a was strongly induced by salt stress in roots. Overexpression of PagWOX11/12a in poplar enhanced salt tolerance, as evident by the promotion of growth‐related biomass. In contrast, salt‐treated PagWOX11/12a dominant repression plants displayed reduced biomass growth. Under salt stress conditions, PagWOX11/12a‐overexpressed lines showed higher reactive oxygen species (ROS) scavenging capacity and lower accumulation of hydrogen peroxide (H2O2) than non‐transgenic 84K plants, whereas the suppressors displayed the opposite phenotype. In addition, PagWOX11/12a directly bound to the promoter region of PagCYP736A12 and regulated PagCYP736A12 expression. The activated PagCYP736A12 could enhance ROS scavenging, thus reducing H2O2 levels in roots under salt stress in PagWOX11/12a‐overexpressed poplars. The collective results support the important role of PagWOX11/12a in salt acclimation of poplar trees, indicating that PagWOX11/12a enhances salt tolerance through modulation of ROS scavenging by directly regulating PagCYP736A12 expression in poplar.
Keywords: cytochrome P450 monooxygenase gene, PagWOX11/12a, PagCYP736A12, root growth, ROS, salt stress
Introduction
Salt stress is a major environmental factor that adversely affects plant growth and production globally (Hasegawa, 2013). To cope with long‐term ionic stress, plants, especially perennials, have evolved a range of intrinsic mechanisms and adaptive strategies. Transcriptional regulation is important in controlling plant signalling pathways to regulate many biological processes in response to various stress conditions. These responses involve specific transcription factors (TFs), such as the WUSCHEL‐related homeobox (WOX), MYB, NAC and WRKY families (Ding et al., 2015; Fang et al., 2017; Liu et al., 2014a, 2014b; Mao et al., 2020; Zhao et al., 2009). Expression of some TFs is induced by salt stress, which activates downstream stress‐responsive genes that regulate the levels of reactive oxygen species (ROS) involved in plant developmental and physiological processes (Mittler et al., 2004; Suzuki et al., 2012). However, high concentrations of ROS can lead to cell injury and the oxidative destruction of cells (Dietz et al., 2016; Huang et al., 2016).
The WOX protein family of novel plant‐specific TFs plays crucial roles in meristem initiation, stem cell maintenance and organ formation (Der Graaff et al., 2009). The homeobox genes WOX11 and WOX12 are members of the WOX family. They are reportedly involved in root development. For example, rice OsWOX11 is a key regulator of crown root emergence, and the interaction between ERF3 and WOX11 is required to regulate RR2 expression in the cytokinin signalling pathway during crown root development (Zhao et al., 2009, 2015). Arabidopsis AtWOX11 and AtWOX12 have been reported to be involved in the first‐step cell fate transition for callus initiation during de novo root organogenesis (Liu et al., 2014c). Poplar WOX11 and WOX11/12 are essential for adventitious root development. Overexpression of WOX11 (PeWOX11a and PeWOX11b) or WOX11/12a gene increases the number of adventitious roots in transgenic cuttings (Liu et al., 2014a; Xu et al., 2015). A few studies have reported that WOX11 can regulate root growth in response to abiotic stress. Rice WOX11 expression is decreased by microcystin‐LR treatment, leading to significantly inhibited rice root growth (Chen et al., 2013). OsWOX11 is responsive to drought stress, and OsWOX12A and OsWOX12B are differentially regulated by drought, cold and high salt stress; overexpression of OsWOX11 can enhance drought tolerance by promoting root hair development and growth (Cheng et al., 2014, 2016). Furthermore, Jiang et al. (2017) reported that the transcriptional regulatory network of WOX11 is involved in the control of redox metabolic pathways in rice under drought stress. Recently, we isolated a homolog of Arabidopsis WOX11 and WOX12, PagWOX11/12a, from 84K poplar (Populus alba × P. glandulosa). PagWOX11/12a is a copy of the two PagWOX11/12 genes (PagWOX11/12a and PagWOX11/12b) in the 84K genome. PagWOX11/12a is strongly expressed under drought stress and can promote root elongation and biomass growth in transgenic poplars. In addition, PagERF35 was identified as an upstream regulator that directly activates the expression of PagWOX11/12a in response to drought stress (Wang et al., 2020a). However, whether WOX11 is involved in the response to salt stress remains unclear. If it is, the regulatory mechanisms and downstream target genes of WOX11 in salt stress need to be further investigated.
Recent studies have demonstrated that in addition to TFs, cytochrome P450 monooxygenase (CYP) genes are involved in diverse biological processes, including plant development and abiotic stress responses (Kurotani et al., 2015; Li and Wei, 2020; Ma et al., 2014). For instance, PtCYP714A3 from P. trichocarpa was observed to be markedly induced by salt stress, and transgenic rice plants displayed enhanced salt tolerance and reduced shoot growth (Wang et al., 2016). Panax ginseng PgCYP736A12 is up‐regulated by NaCl, H2O2 and salicylic acid, and overexpression of PgCYP736A12 in Arabidopsis was observed to slightly reduce plant height and enhance tolerance to phenylurea herbicide (Khanom et al., 2019). In addition, PgCYP736B, the homolog of PgCYP736A12, conferred enhanced salt tolerance by modulating H2O2 accumulation, carotenoid levels and abscisic acid biosynthesis in Arabidopsis (Balusamy et al., 2019). SoCYP85A1 from Spinacia oleracea contributes to drought tolerance by promoting root development and regulating ROS production and stress‐responsive gene expression (Duan et al., 2017). The wheat TaCYP81D5 gene is involved in the response to salt stress by regulating ROS scavenging during seedling and reproductive stages (Wang et al., 2020b). Finally, Arabidopsis WRKY33 regulates CYP94B1 expression and enhances suberin deposition in the endodermal cells of roots, which confers salt tolerance (Krishnamurthy et al., 2020). The foregoing highlights the need to clarify the regulation of CYP genes in the response to abiotic stress.
In the present study, we demonstrated that PagWOX11/12a is involved in the salt acclimation of poplars using both its overexpression and down‐regulation of transgenic poplars. In addition, we defined a molecular mechanism by which PagWOX11/12a enhances salt tolerance and ROS scavenging in roots by directly up‐regulating PagCYP736A12 expression. This study provides novel insights into the physiological roles of the WOX gene in response to salt stress.
Results
PagWOX11/12a responses to salt stress
PagWOX11/12a expression was significantly induced by salt stress and was transiently increased in the roots under salt stress, reaching a maximum 4.5‐fold increase 9 h after salt stress treatment (Figure 1a). β‐Glucuronidase (GUS) staining of ProPagWOX11/12a::GUS transgenic poplar plants revealed that PagWOX11/12a was mainly expressed in the root tips under control culture conditions. However, GUS staining was intense in whole roots treated with salt for 6 and 9 h (Figure 1b). The strong GUS activity was significantly increased by nearly 13‐fold in the roots at 9 h after salt stress (Figure 1c). The results indicate that PagWOX11/12a can be strongly induced by salt stress in poplar roots.
Figure 1.
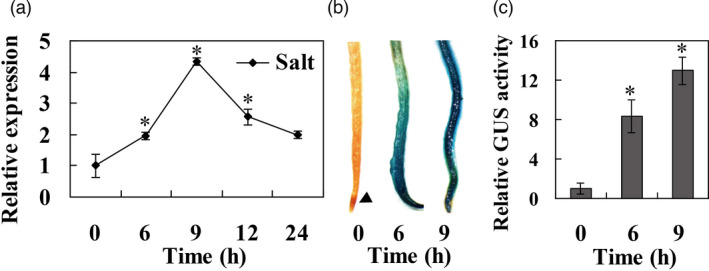
Expression profiles of PagWOX11/12a under salt stress. (a) RT‐qPCR analysis of PagWOX11/12a expression in total roots of non‐transgenic 84K poplar under salt stress. Four‐week‐old non‐transgenic 84K poplar plants were subjected to 200 mm NaCl for the indicated times. The root tissues were collected from 10 individuals and used for RT‐qPCR analysis. The bars represent means ± SD (n = 4). (b, c) Histological staining and GUS activity of the root tips of ProPagWOX11/12a::GUS transgenic poplar plants under salt stress. Four‐week‐old transgenic poplar plants were subjected to 200 mm NaCl for the indicated times. At least 20 roots were harvested from three biological replicates (more than 10 individuals each) for GUS staining and activity analysis. The values of GUS activity under control growth conditions were normalized to 1. The experiments were performed three times and the similar results were obtained. The bars represent means ± SD (n = 6) and the asterisks indicate significant differences with respect to the values for 0 h (*P < 0.05).
PagWOX11/12a positively regulates root growth under salt conditions
To investigate whether PagWOX11/12a regulates root growth under salt stress conditions, transgenic poplar plants overexpressing (OE) PagWOX11/12a (OE1 and OE3) and plants with dominant repression (DR) of PagWOX11/12a (DR7 and DR9) were chosen for subsequent studies (Wang et al., 2020a). Under different concentrations of NaCl treatment, the development of roots was retarded in each poplar genotype, and the growth was significantly different between transgenic and 84K plants, especially at high salt concentrations (Figure 2a). The main root (MR) number was markedly higher in OE lines but lower in DR lines than in 84K plants. In addition, MR development appeared completely suppressed in DR plants. Therefore, root biomass accumulation was reduced to a greater extent in 84K and DR plants than in OE lines under salt stress (Figure 2b, c). The relative fresh weight (FW) and dry weight (DW) in OE lines was significantly higher but were obviously lower in DR plants than in 84K plants treated with NaCl.
Figure 2.
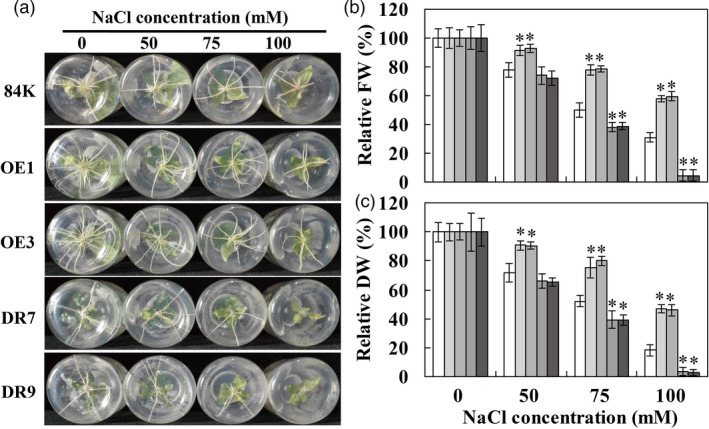
PagWOX11/12a enhances salt tolerance of root growth and biomass in poplar. (a) Root phenotypes of non‐transgenic 84K and transgenic (OE and DR) poplar plants. (b, c) Relative quantification of root biomass (fresh weight [FW] and dry weight [DW]) of non‐transgenic 84K and transgenic poplar plants. The cutting‐propagated non‐transgenic 84K and transgenic (OE and DR) poplar plants were cultivated on 1/2 MS solid medium supplemented with 0, 50, 75 and 100 mm NaCl for 20 days. The root biomass of individual plant was measured. The values for each plant under control growth conditions were normalized to 100%. The experiments were performed three times and the similar results were obtained. The bars represent means ± SD (n = 6) and the asterisks indicate significant differences between non‐transgenic 84K and transgenic plants (*P < 0.05).
PagWOX11/12a alleviates inhibition of biomass accumulation under salt conditions
To further investigate the function of PagWOX11/12a in response to salt stress in soil conditions, transgenic and 84K plants were cultured in soil under NaCl treatment. Under control growth conditions, although the transgenic lines and 84K plants grew well, the stem height was significantly higher in OE plants and obviously lower in DR plants than in 84K plants (Figure 3a, c). After salt stress treatment, the differences in plant height among different genotypes were more significant. Most leaves of 84K plants wilted and fell off, whereas those of OE plants remained turgid, except for the older leaves. In contrast, some DR plants died at this stage (Figure 3b, c). Although the growth rate of all genotypes was inhibited under salt stress conditions, the OE lines had significantly higher growth rates than the other plants (Figure 3d). In addition, the relative FW of shoots and roots was significantly higher in OE plants than in 84K plants under salt stress. In contrast, the shoot and root biomass of DR plants were markedly lower than the values of 84K plants (Figure 3e). Accordingly, the relative DW of shoots and roots showed the same trend (Figure 3f). These results suggest that the biomass of OE plants was less inhibited by salt stress. Consistent with the results obtained in half‐strength Murashige and Skoog (1/2 MS) solid medium, the biomass accumulation of 84K and DR plants was decreased to a greater extent than that in OE plants under salt conditions. The findings suggest that PagWOX11/12a overexpression can increase salt tolerance in transgenic poplar plants.
Figure 3.
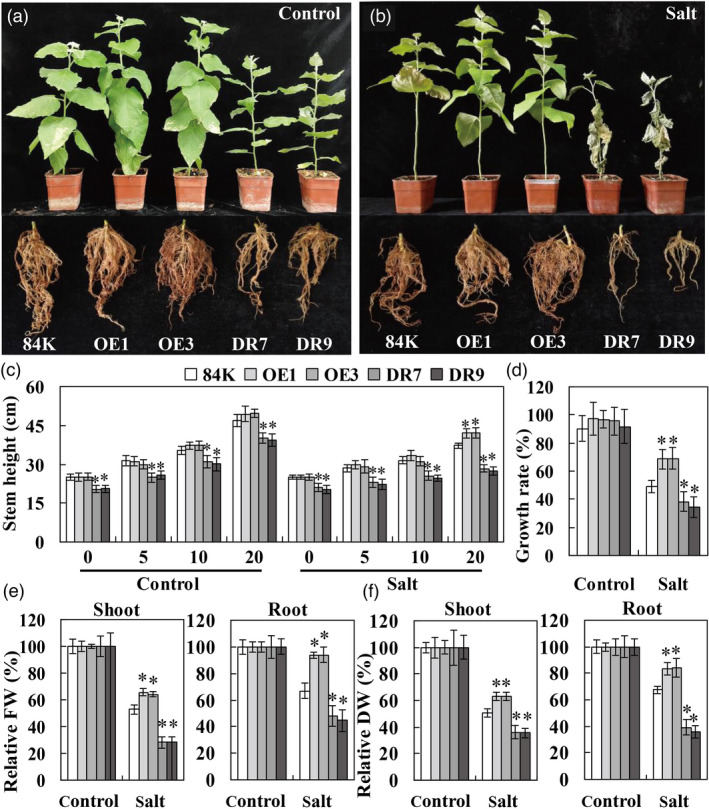
PagWOX11/12a enhances salt tolerance of shoot and root biomass in poplar. (a, b) Shoot and root phenotypes of non‐transgenic 84K and transgenic (OE and DR) plants. (c) Plant height of non‐transgenic 84K and transgenic plants. (d) Growth rate of non‐transgenic 84K and transgenic plants. (E, F) Relative quantification of shoot and root biomass (fresh weight [FW] and dry weight [DW]) of non‐transgenic 84K and transgenic plants. Four‐week‐old non‐transgenic 84K and transgenic plants cultivated in vitro were grown in soil for 20 days and irrigated subsequently with 0 (control) or 200 mm NaCl (salt treatment) every 2 days for 20 days. The plant height of individual plant was periodically measured. The growth rate, shoot and root biomass of individual plant were measured after 20 days of culture. The values for each plant under control growth conditions were normalized to 100%. The experiments were performed three times and the similar results were obtained. The bars represent means ± SD (n = 10) and the asterisks indicate significant differences between non‐transgenic 84K and transgenic plants (*P < 0.05).
PagWOX11/12a regulates genes involved in oxidation–reduction process under salt stress
To identify the target genes of PagWOX11/12a in response to salt stress, RNA‐sequencing (RNA‐seq) analysis was performed using the roots of transgenic OE, DR and 84K plants. Based on the Gene Ontology term enrichment analysis, we found 236 up‐regulated differentially up‐regulated genes (DEGs) of ‘OE vs 84K’ shared by control growth and salt stress conditions. Of these, 28 DEGs (11.86%) were mainly enriched in the oxidation–reduction process (Table S2). In contrast, 223 down‐regulated DEGs of ‘DR vs 84K’ were shared by control growth and salt stress conditions. Of these, 29 DEGs (13%) were involved in the oxidation–reduction process (Table S3). We further compared the two sets of DEGs (Figure S1) and found 17 DEGs in common, including two DEGs enriched in pathways related to the oxidation–reduction process (Table S4). Finally, we searched for cis‐acting elements in the 1‐kb region upstream of the transcription starting sites of the DEGs and found that only one DEG (PagCYP736A12) harboured a WOX‐binding motif (‘TTAATGC’) in its promoter region (Figure S2). It encodes a cytochrome P450 family protein with extensive homology to CYP736A12 from P. trichocarpa (XP_002310014.2). Phylogenetic analysis indicated that PagCYP736A12 exhibited a close phylogenetic relationship with PtCYP736A12 and PpCYP736A12 (Figure S3). The findings indicate that PagCYP736A12 is a downstream target gene of PagWOX11/12a in poplar.
PagWOX11/12a overexpression decreases oxidative damage
To investigate whether overexpression of PagWOX11/12a would decrease oxidative damage in the roots of transgenic poplar plants under salt stress, 2,7‐dichlorodihydrofluorescein diacetate (DCFH‐DA) staining was performed. The H2O2 levels in root tips of all poplars were not visibly different under control growth conditions. In contrast, the accumulation of H2O2 in root tips was obviously lower in transgenic OE plants, but was significantly higher in DR lines than in 84K plants under salt stress (Figure 4a). Quantification of H2O2 content yielded similar results (Figure 4b). In addition, there were no differences in malondialdehyde (MDA) content in roots between the transgenic lines and 84K plants under control growth conditions. However, the MDA content in OE lines was significantly lower than that in 84K plants after salt stress, whereas DR plants displayed higher MDA levels than 84K and OE plants (Figure 4c). These results suggest that transgenic plants overexpressing PagWOX11/12a accumulated less ROS under salt stress.
Figure 4.
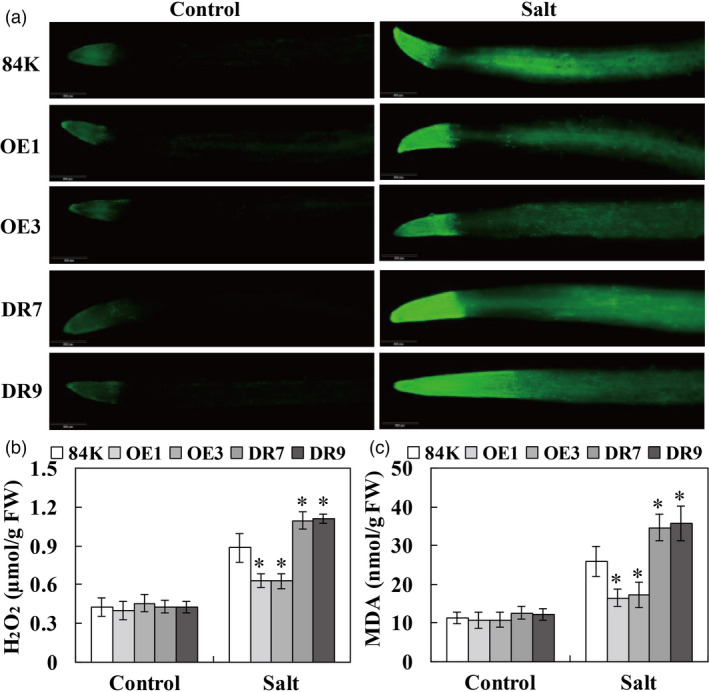
Overexpression of PagWOX11/12a decreased oxidative damage levels under salt stress. (a) Representative DCFH‐DA staining of H2O2 production in the root tips of non‐transgenic 84K and transgenic plants. (b, c) H2O2 and MDA content in the fresh roots of non‐transgenic 84K and transgenic plants. The cutting‐propagated non‐transgenic 84K and transgenic (OE and DR) poplar plants were cultivated and salinity treated with 0 and 75 mm NaCl as described in Figure 2. At least 20 roots were collected from three biological replicates (more than 10 individuals each) for DCFH‐DA staining and analysis of H2O2 and MDA contents. FW, fresh weight. The experiments were performed three times and the similar results were obtained. The bars represent means ± SD (n = 6) and the asterisks indicate significant differences between non‐transgenic 84K and transgenic plants (*P < 0.05).
PagWOX11/12a overexpression enhances ROS scavenging ability
Transgenic poplar plants overexpressing PagWOX11/12a showed less oxidative damage, perhaps due to their improved antioxidant capacity. To confirm this, the non‐enzymatic and enzymatic antioxidants in the ascorbate–glutathione (AsA–GSH) cycle and superoxide dismutase (SOD) activity between 84K and transgenic poplar plants were examined (Figure 5). No differences were apparent in the antioxidant compounds (AsA and GSH) and enzyme activity levels of ascorbate peroxidase (APX), catalase (CAT), glutathione reductase (GR) and SOD in the roots of the transgenic lines (OE and DR) and 84K plants under control growth conditions. However, the antioxidant compounds and enzyme activities were significantly higher in the OE lines, but lower in the DR lines compared with the values in 84K plants under salt stress. In addition, there were no significant differences in Na+ and K+ concentrations between 84K and transgenic plants under control and salt stress conditions (Figure S4). However, the Na+ content was increased and K+ content was decreased in both 84K and transgenic plants under salt stress. These results indicate that OE lines can increase their antioxidant capacity to decrease oxidative damage under salt stress, rather than maintain K+/Na+ homeostasis.
Figure 5.
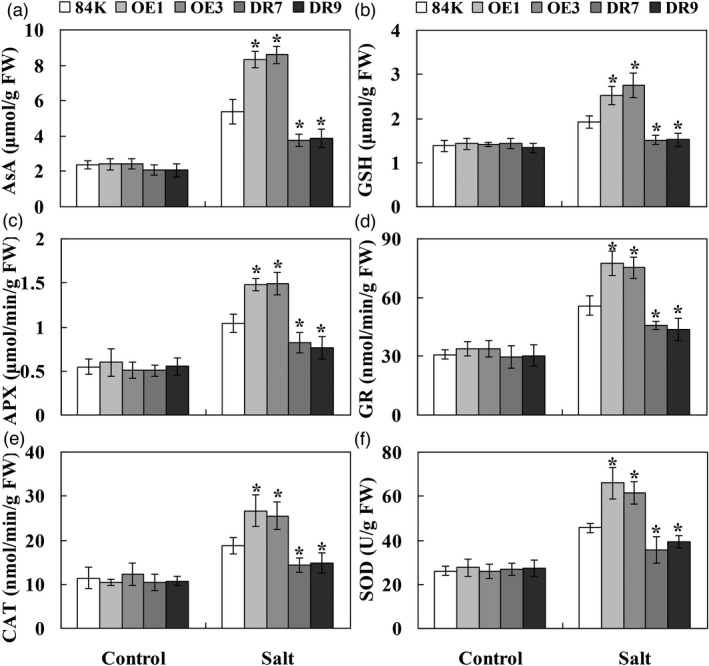
Overexpression of PagWOX11/12a enhanced ROS scavenging ability under salt stress. The cutting‐propagated non‐transgenic 84K and transgenic (OE and DR) poplar plants were cultivated and salinity treated with 0 and 75 mm NaCl as described in Figure 2. The fresh roots were collected and used for analysis of the non‐enzymatic [AsA (a) and GSH (b)] and enzymatic [APX (c), GR (d) and CAT (e)] antioxidants in the AsA–GSH cycle and the SOD activity (f). FW, fresh weight. The experiments were performed three times and the similar results were obtained. The bars represent means ± SD (n = 6) and the asterisks indicate significant differences between non‐transgenic 84K and transgenic plants (*P < 0.05).
PagWOX11/12a directly regulates PagCYP736A12 expression
To investigate whether PagWOX11/12a is involved in the salt stress response through direct regulation of PagCYP736A12 expression by binding to its promoter, yeast one‐hybrid (Y1H), chromatin immunoprecipitation (ChIP) and dual‐luciferase reporter assays were conducted. Yeast transformants harbouring the PagWOX11/12a and WOX11‐binding motif or PagCYP736A12 promoter fragment grew well on SD/−Leu/−Trp/−His medium containing 3‐amino‐1,2,4‐triazole (3‐AT) (Figure 6a). In addition, PagWOX11/12a‐green fluorescent protein (GFP) was used in the ChIP assay. The promoter fragment of PagCYP736A12 containing the WOX11‐binding motif was detected and was highly enriched in the ChIP sample compared with that in the mock. The enrichment was significantly higher under salt stress than under control growth conditions, indicating that salt stress enhanced this induction (Figure 6b). Luciferase luminescence was observed when 35S::PagWOX11/12a and Pro::LUC were co‐transformed into tobacco leaves, but no luminescence was evident when the tobacco leaves were co‐transformed with vector controls (62‐SK+0800‐LUC) and negative controls (62‐SK+Pro::LUC and 35S::PagWOX11/12a+0800‐LUC). The relative LUC/Renilla luciferase (REN) activity driven by the PagCYP736A12 promoter co‐transformed with the PagWOX11/12a effector was significantly higher than that of the vector controls and negative controls (Figure 6c).
Figure 6.
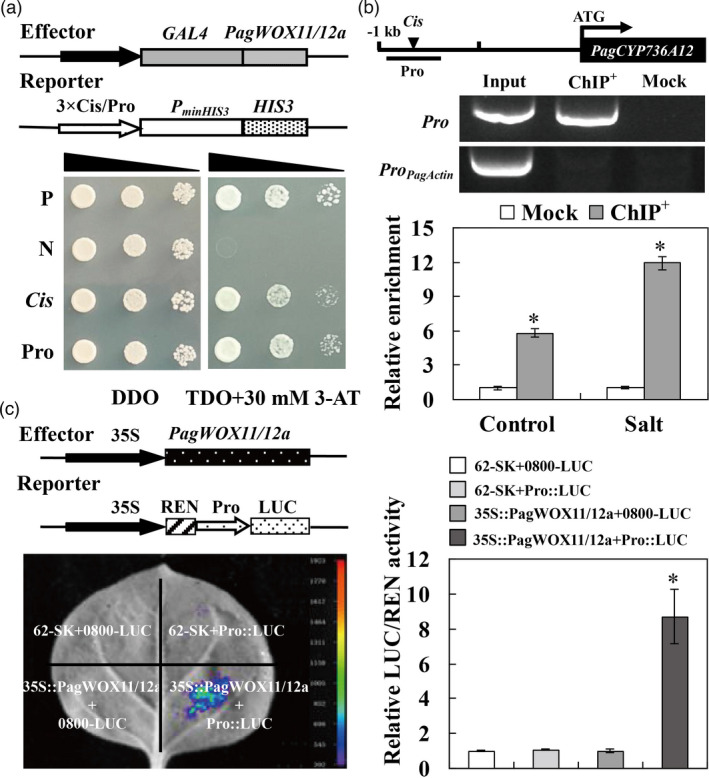
Identification of the targeted gene PagCYP736A12 of PagWOX11/12a. (a) Y1H analysis of the interaction between PagWOX11/12a and the PagCYP736A12 promoter. A diagram of the reporter and effector vectors is shown at the top. The reporter and effector constructs were co‐transformed into yeast Y187 cells, and the positive transformants were determined by spotting serial dilutions of yeast onto SD/−Leu/−Trp (DDO) and SD/−His/−Leu/−Trp (TDO) media supplemented with 3‐AT. P: positive control (p53HIS2 + pGAD‐53); N: negative control (p53HIS2 + pGAD‐PagWOX11/12a). (b) ChIP analysis of PagWOX11/12a binding to the PagCYP736A12 promoter. Simplified gene structure indicating the locations of the amplified promoter region (Pro) containing the ‘TTAATGC’ element. qPCR analysis of the abundance of the PagCYP736A12 promoter sequence. Transiently transgenic poplars were grown on 1/2 MS solid medium under normal conditions for 48 h or cultured on 1/2 MS medium for 36 h, and then subjected to 200 mm NaCl for 12 h. Input, chromatin preparation before immunoprecipitation; ChIP+, immunoprecipitated with anti‐GFP antibody; Mock, immunoprecipitated without anti‐GFP antibody. (c) Dual‐luciferase reporter analysis of PagWOX11/12a activating the PagCYP736A12 promoter in N. benthamiana leaves. Schematic of the reporter and effector constructs is shown at the top. The effector and reporter vectors were instantaneously co‐transformed into leaves of N. benthamiana and cultured in a greenhouse under control conditions for 48 h. The experiments were performed three times and the similar results were obtained. The bars represent means ± SD (n = 6) and the asterisks indicate significant differences (*P < 0.05).
In support of their close relationship, we found that the expression of both PagWOX11/12a and PagCYP736A12 was strongly induced by salt stress. The highest levels in OE and 84K roots were observed at 75 mm NaCl (Figure S5A). Notably, the two genes had similar expression patterns in OE and 84K roots under different salt concentrations (Figure S5A) and in 84K roots treated with 200 mm NaCl for 0, 6, 9, 12 and 24 h (Figure 1a and Figure S5B). Furthermore, PagCYP736A12 expression was strongly enhanced in OE lines, but was suppressed in DR lines under control and salt conditions (Figure S5C). Taken together, these results confirm that PagWOX11/12a directly binds to the promoter of PagCYP736A12, activating its transcription in response to salt stress.
PagCYP736A12 reduces H2O2 level and enhances salt tolerance
To determine whether PagCYP736A12 can enhance salt tolerance in poplar, transgenic poplars overexpressing PagCYP736A12 were generated, and the expression levels of PagCYP736A12 in transgenic lines were confirmed (Figure S6). The transgenic lines (OX1 and OX2) with high expression levels were selected for further study. DCFH‐DA staining results showed that the H2O2 levels of all poplars were not significantly different under control growth conditions. However, the accumulation of H2O2 in OX plant roots was obviously lower than that in 84K plants under salt stress (Figure 7a). Similarly, quantification of H2O2 and MDA content showed no significant differences between transgenic lines and 84K plants under control growth conditions. However, the content was significantly lower in OX lines than in 84K plants when exposed to salt stress (Figure 7b, c).
Figure 7.
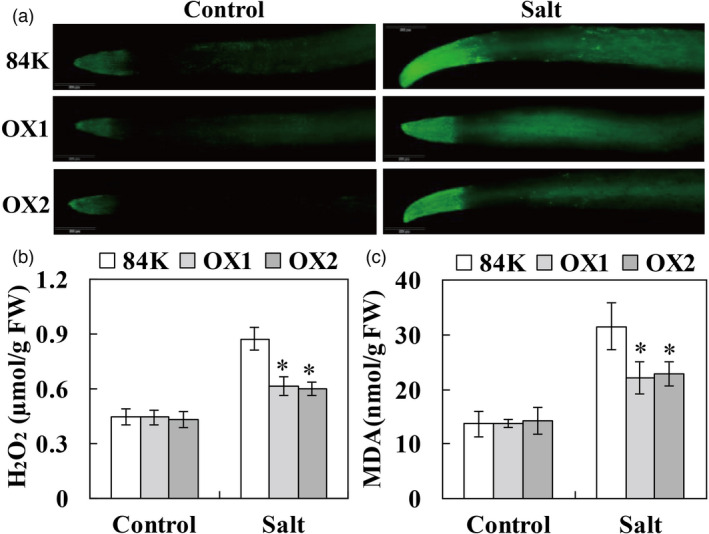
Overexpression of PagCYP736A12 decreased oxidative damage levels under salt stress. (a) Representative DCFH‐DA staining to detect H2O2 in the root tips of non‐transgenic 84K and transgenic plants. (b, c) H2O2 and MDA contents in the fresh roots of non‐transgenic 84K and transgenic plants. The cutting‐propagated non‐transgenic 84K and transgenic poplar lines overexpressing PagCYP736A12 (OX1 and OX2) were cultivated and salinity treated with 0 and 75 mm NaCl as described in Figure 2. At least 20 roots were collected from three biological replicates (more than 10 individuals each) for DCFH‐DA staining and analysis of H2O2 and MDA content. FW, fresh weight. The experiments were performed three times and the similar results were obtained. The bars represent means ± SD (n = 6) and the asterisks indicate significant differences between non‐transgenic 84K and transgenic plants (*P < 0.05).
OX and 84K plants showed the same phenotype in a greenhouse under control growth conditions. The stem height and root biomass were not significantly different between OX and 84K plants. After 10 days of salt stress, the older leaves of 84K plants wilted and fell off, whereas those of OX plants remained turgid (Figure 8a). Furthermore, the stem height and root biomass were significantly higher in OX lines than in 84K plants (Figure 8b, c). Taken together, these results showed that PagCYP736A12 overexpression, similar to the PagWOX11/12a OE plants, could enhance salt tolerance by decreasing ROS levels and thus accumulate more biomass than 84K plants under salt stress.
Figure 8.
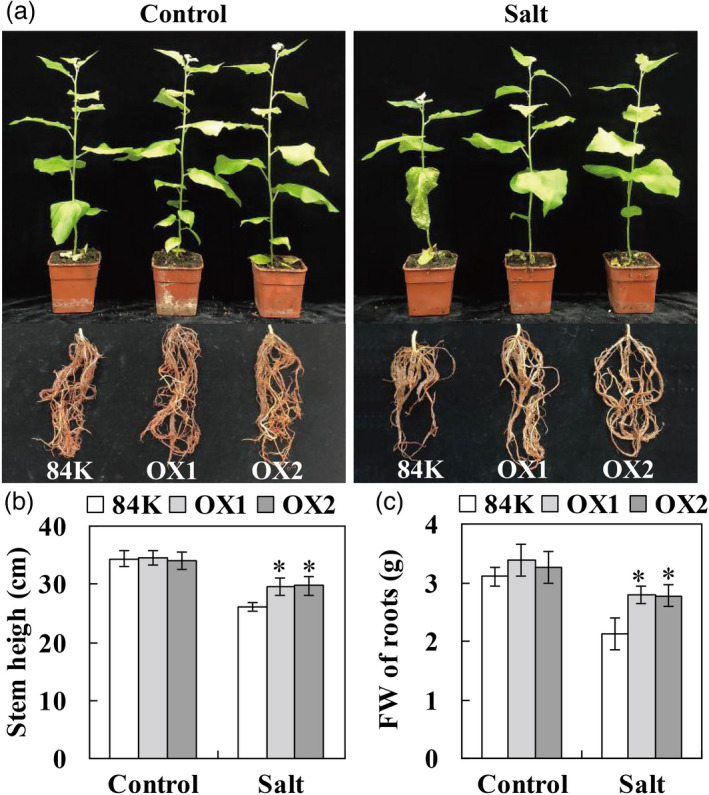
Enhanced salt tolerance in transgenic poplars overexpressing PagCYP736A12. (a) Shoot and root phenotypes of 84K and transgenic (OX) plants. (b, c) Plant height and root fresh weights (FW) of 84K and OX line plants. Four‐week‐old non‐transgenic 84K and transgenic (OX1 and OX2) poplar plants were cultivated and salinity (200 mm NaCl) for 10 days as described in Figure 3. The plant height and root FW of individual plant were measured. The experiments were performed three times and the similar results were obtained. The bars represent means ± SD (n = 10) and the asterisks indicate significant differences between non‐transgenic 84K and transgenic plants (*P < 0.05).
Discussion
WOX11 genes act as developmental regulators that mediate root formation and growth in plants (Liu et al., 2014c; Xu et al., 2015; Zhao et al., 2009, 2015). Recently, it was reported that the WOX11 and WOX12 genes are involved in salt and/or drought stress responses (Cheng et al., 2014, 2016; Wang et al., 2020a). In this study, we assessed the role and underlying mechanism of PagWOX11/12a involved in salt tolerance in poplar (Figure 9). PagWOX11/12a responds to salt stress and enhances poplar rooting in response to stress conditions. At the same time, PagWOX11/12a directly activates PagCYP736A12 expression, and the latter is involved in ROS scavenging to reduce ROS levels in roots under salt stress. Therefore, PagWOX11/12a plays a role in salt tolerance by regulating both the root system and physiological processes. Our study provides insight into the regulation of physiological processes by PagWOX11/12a under salt stress.
Figure 9.
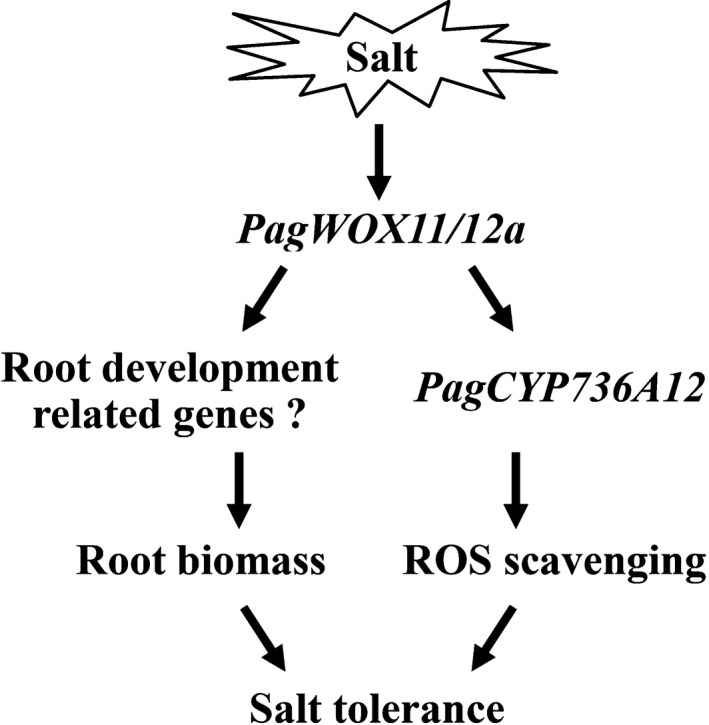
Proposed model of PagWOX11/12a gene involved in tolerance to salt stress. PagWOX11/12a is induced by salt stress then activates both the rooting‐related genes to promote root biomass and PagCYP736A12 to increase ROS scavenging capacity, which results in salt tolerance of poplar plants.
PagWOX11/12a is involved in tolerance to salt stress
Our previous study showed that PagWOX11/12a responds to drought stress and enhances drought tolerance in transgenic poplars (Wang et al., 2020a). In this study, PagWOX11/12a was strongly induced by salt stress (Figure 1), and transgenic poplars overexpressing PagWOX11/12a displayed promoted growth in biomass compared to 84K plants under salt stress. In contrast, the suppressed PagWOX11/12a plants exhibited inhibited root and shoot growth, and high sensitivity to salt stress, resulting in decreased stem and root growth (Figures 2 and 3). These results strongly suggest that salt‐induced PagWOX11/12a plays an important role in salt tolerance. It should be noted that the small seedlings were used in the NaCl (salt) treatment. Therefore, although statistically significant, the phenotypic differences were relatively small. Therefore, further studies are needed to determine the highest concentration of NaCl that the OE lines could be tolerant to and field trials are necessary to analyse the growth of transgenic poplars in saline regions. The results of these future studies will conclusively indicate the practical use of this gene.
Excess ROS, such as those produced by salt stress, lead to oxidative conditions that are harmful to cells (Kuge et al., 2010; Wang et al., 2008). Generally, H2O2 is a prominent ROS species and is a marker of ROS production (Bhattacharjee, 2012; de la Garma et al., 2015). Therefore, maintaining the steady‐state level of ROS by scavenging pathways is important to protect against oxidative damage caused by adverse environments (Jiang and Zhang, 2002; Miller et al., 2010; Mittler et al., 2004). In addition, MDA content is a valid indicator of cytomembrane oxidative damage in monitoring ROS levels (He et al., 2018; Shi et al., 2013). In this study, transgenic lines overexpressing PagWOX11/12a displayed lower H2O2 and MDA levels in roots than those in 84K plants under salt stress, whereas lines with PagWOX11/12a repressors exhibited severe ROS damage (Figure 4). These results suggest that the increased salt stress tolerance of the transgenic poplar plants overexpressing PagWOX11/12a was attributed to their low ROS levels.
Both SOD and the AsA–GSH cycle are crucial in scavenging ROS (Hernández et al., 2015; Shi et al., 2013). SOD is the first line of plant ROS defence. The enzyme catalyses the conversion of superoxide () to oxygen (O2) and H2O2 (Huang et al., 2016). H2O2 is then reduced to H2O by CAT and APX in the AsA–GSH cycle (Hernández et al., 2015). Transgenic poplar plants overexpressing PagWOX11/12a exhibited increased capacity to maintain ROS homeostasis by increasing both antioxidant compounds and antioxidant enzyme activity, but not enhanced capacity to control K+/Na+ under NaCl stress (Figure 5 and Figure S4). In contrast to the suppressed plants, low levels of antioxidant compounds and antioxidant enzyme activity were maintained under salt stress. These findings indicate that PagWOX11/12a may activate an antioxidant pathway to modulate ROS accumulation.
PagWOX11/12a regulates ROS scavenging by activating expression of PagCYP736A12
A recent transcriptomic analysis revealed considerable downstream genes of WOX11 in rice, which were demonstrated to be involved in drought stress response and redox homeostasis (Jiang et al., 2017). The findings indicated that WOXs may regulate genes involved in ROS. In this study, RNA‐seq analysis revealed that many DEGs were enriched in the oxidation–reduction process under salt stress (Table S2, S3). Seventeen DEGs were shared by PagWOX11/12a overexpressors and suppressors. The 17 DEGs included two involved in the oxidation–reduction process (Table S4). Interestingly, one of the DEGs, PagCYP736A12, contains the DNA sequence ‘TTAATGC’ in its promoter, which can be recognized by the WUSCHEL‐related homeobox TFs (Dai et al., 2007; Kamiya et al., 2003; Lohmann et al., 2001; Yadav et al., 2011; Zhao et al., 2009). Previous studies have shown that CYP genes confer tolerance to salt, drought and oxidative stress when overexpressed in transgenic plants (Balusamy et al., 2013, 2019; Duan et al., 2017; Mao et al., 2013; Wang et al., 2016; Wang et al., 2020b; Zhou et al., 2020), and that PgCYP736A12 is induced by salt and H2O2 stresses (Khanom et al., 2019). Therefore, we investigated whether PagWOX11/12a could directly regulate PagCYP736A12 using Y1H, ChIP and luciferase reporter assays. The results showed that PagWOX11/12a bound to the promoter and activated the expression of PagCYP736A12 (Figure 6). The similar expression patterns of PagWOX11/12a and PagCYP736A12 in both OE and 84K roots under salt stress support this mode of regulation (Figure S5A). In addition, we found that transgenic poplars overexpressing PagCYP736A12 showed increased salt tolerance (Figures 7 and 8), phenocopying the PagWOX11/12a OE plants. Moreover, overexpression of PagCYP736A12 in poplar led to lower H2O2 and MDA levels in roots, resulting in higher plant height and root biomass under salt stress (Figures 7 and 8). These results suggest that PagCYP736A12 confers salt tolerance by regulating H2O2 levels. Collectively, these results strongly support the hypothesis that PagWOX11/12a directly regulates PagCYP736A12 expression to improve salt tolerance by maintaining low ROS levels.
In summary, this work demonstrates that PagWOX11/12a enhances salt tolerance by regulating the expression of PagCYP736A12 as a direct downstream gene, which in turn maintains ROS homeostasis in roots. In addition, transgenic poplars with these two genes exhibited increased root biomass under salt stress. This study is the first to report the role of WOX11/12 gene in salt tolerance in woody plant species. Further research is needed to assess the performance of WOX11/12 transgenic poplars in saline regions.
Experimental procedures
Plant materials and stress treatments
Hybrid poplar ‘84K’ (Populus alba × P. glandulosa cv.) was used for gene cloning, expression analysis and genetic transformation. The generation of transgenic 84K poplars expressing the GUS gene under the control of the PagWOX11/12a promoter (PropagWOX11/12a::GUS), overexpressing (OE) and dominant repressors (DR) of PagWOX11/12a driven by the CaMV 35S promoter were described in our previous study (Wang et al., 2020a). PropagWOX11/12a::GUS transgenic polar plants were used for the spatial expression analysis of PagWOX11/12a. Two representative OE transgenic lines (OE1 and OE3) with high PagWOX11/12a expression levels and two DR transgenic lines (DR7 and DR9) with low expression levels were used for further functional analysis. Tissue cultures of transgenic and non‐transgenic 84K poplars were grown on 1/2 MS solid medium (pH 5.8−6.0) in a phytotron (25 °C, 16/8 h light/dark and 50 μm/m2/s light intensity). Seeds of Nicotiana benthamiana were surface‐sterilized and sown onto 1/2 MS solid medium. One‐week‐old seedlings were transplanted into pots filled with soil and perlite (3:1) in a growth chamber at 22 °C with a 16/8 h light/dark photoperiod and 70%–75% relative humidity. They were grown for 20 days and then used for luciferase reporter assays.
To analyse PagWOX11/12a gene expression under salt stress, 4‐week‐old non‐transgenic 84K and ProPagWOX11/12a::GUS poplar plants grown on 1/2 MS solid medium were subjected to a hydroponic solution of 200 mm NaCl for 0, 6, 9, 12 and 24 h for salt treatment or to fresh hydroponic solution as a control. Root tissues from 10 individuals were collected at each indicated time point, transferred to the staining solution and frozen immediately in liquid nitrogen for expression analyses. The treatment experiments were repeated three times.
RNA extraction and reverse transcription quantitative PCR (RT‐qPCR) analysis
The procedures for RNA isolation, quality examination, first‐strand cDNA synthesis and RT‐qPCR assays were identical to those used in our previous study (Wang et al., 2020a). In brief, total RNA was isolated from the samples using the RNeasy Plant Mini Kit (Qiagen, Dusseldorf, Germany) and was used to synthesize cDNA using the PrimeScriptTM RT reagent Kit (TaKaRa, Dalian, China). qPCR was performed using the SYBR Premix Ex TaqTM Kit (TaKaRa) with a Light Cycler 480 (Roche Applied Science, Penzberg, Germany). Relative gene expression was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001) with the reference gene PagActin as an internal standard. Three biological replicates with four technical replicates were performed for each sample. All primers used for RT‐qPCR are listed in Table S1.
GUS staining and activity analyses
To detect salt‐induced expression, 4‐week‐old ProPagWOX11/12a::GUS plants were subjected to salt treatment (200 mm NaCl) for 0, 6 and 9 h. At least 20 roots were harvested from three biological replicates (more than 10 individuals each) for histochemical staining and GUS activity assay. GUS staining was performed as previously described by Liu et al. (2014a) GUS activity was quantitatively measured using spectrophotometry (Jefferson et al., 1987). The experiments were repeated three times.
Genetic transformation of poplar
The full‐length PagCYP736A12 cDNA was cloned into the pMDC32 binary vector driven by the CaMV 35S promoter, introduced into Agrobacterium tumefaciens strain GV3101 (pSoup‐p19) according to Wang et al. (2020a) and then transformed into 84K poplar using the Agrobacterium‐mediated leaf disc method following the protocol described by Liu et al. (2014a) Putative transgenic plants were selected on1/2 MS medium supplemented with hygromycin (3 mg/L) and verified by RT‐qPCR. The details of the primers used are shown in Table S1.
Analysis of salt stress tolerance
To examine the rooting ability under salt stress, shoot segments of transgenic lines (OE and DR) and non‐transgenic 84K poplar plants (approximately 3 cm in length with 2–3 young leaves) were cut from sterilized plants and cultivated on 1/2 MS solid medium supplemented with 0, 50, 75 and 100 mm NaCl for 20 days. The phenotypes of the root systems were photographed and root biomass was quantified. To evaluate salt stress tolerance, 4‐week‐old non‐transgenic 84K and transgenic (OE and DR) plants grown on 1/2 MS solid medium were transplanted to plastic pots with a mixture of soil and perlite, grown in a greenhouse for 20 days and subsequently irrigated with 0 (control) or 200 mm NaCl (salt treatment) solution every 2 days for 20 days. Plant height was measured periodically after the beginning of the salt treatment. After 20 days of culture, the growth rate was calculated, the roots and aerial parts were collected, the FW was measured, and the DW was determined after incubation in an oven at 80 °C for 72 h. The experiments in vitro and soil conditions were performed three times, and at least 10 plants were used for each genotype per experiment.
Histochemical staining, physiological indices and ion analysis
To detect ROS levels, DCFH‐DA (Sigma‐Aldrich, St Louis) was used for H2O2 detection in the root tips of non‐transgenic 84K and transgenic poplar plants cultivated on 1/2 MS solid medium supplemented with 0 and 75 mm NaCl for 20 days. The confocal images were visualized by a fluorescence microscope (Olympus Bx51, Japan) as previously described (Shi et al., 2019). At least 20 roots were harvested from three biological replicates (more than 10 individuals each) for DCFH‐DA staining.
The H2O2 and MDA contents in the fresh roots were measured using customized kits (Beijing Solarbio Science & Technology Co., Ltd, Beijing, China) following the manufacturer’s instructions. The content of AsA and GSH, and the antioxidant enzyme activity levels of APX, GR, CAT and SOD were evaluated using the corresponding assay kits (Suzhou Keming Biotechnology Co. Ltd., Suzhou, China). The Na+ and K+ content was measured using atomic absorption spectroscopy (TAS990, Persee, Beijing, China). These experiments were performed three times.
RNA‐seq analysis
Transgenic OE (OE1 and OE3), DR (DR7 and DR9) and non‐transgenic 84K poplar plants were cut from sterilized plants and cultivated on 1/2 MS solid medium supplemented with 0 (control) and 75 mm NaCl (salt treatment) for 20 days. Root tissue samples were collected from 10 individuals for each of the three biological replicates and used for RNA‐seq analysis. Total RNA isolation, quality examination, library construction, sequencing, gene annotation and DEG analysis were performed by BioMarker Technologies. Gene function annotation, including Gene Ontology, eggNOG, Kyoto Encyclopedia of Genes and Genomes and Swiss‐Prot, was carried out using the 84K poplar genome database (https://doi.org/10.6084/m9.figshare.12369209) as previously described (Huang et al., 2021). DEGs were defined by a fold change ≥2 with a false discovery rate (P‐value) < 0.05. Finally, to verify candidate genes from the RNA‐seq analysis, RT‐qPCR was performed on roots from transgenic and 84K plants, prepared separately from the RNA‐seq sample. All primer sequences used are described in Table S1.
Y1H assay
The full‐length coding sequence (CDS) of PagWOX11/12a was inserted into the pGADT7‐Rec2 vector (Clontech) to generate the effector construct pGAD‐PagWOX11/12a. Three tandem copies of the WOX11‐binding motif ‘TTAATGC’ and the PagCYP736A12 promoter fragment upstream of the transcription starting sites (spanned from −1 to −1000) were PCR amplified and inserted into the pHIS2 vector (Clontech) to generate recombinant reporter constructs. The effector and reporter constructs were co‐transformed into yeast Y187 strain cells using a Matchmaker™ Yeast One‐Hybrid System (Clontech) and then placed on SD/−Leu/−Trp (DDO) and SD−Trp/−Leu/−His (TDO) media containing 30 mm 3‐AT. The Y1H assay was conducted as previously described (Wang et al., 2020a). The pGAD‐PagWOX11/12a and p53HIS2 were used as negative control. The pGAD‐Rec2‐53 and p53HIS2 were used as positive control. The experiments were performed in triplicates. All primers used are shown in Table S1.
ChIP assay
The procedures for vector construction, plant transient transformation and ChIP‐qPCR were identical to those used in previous studies (Ji et al., 2014; Li et al., 2014; Wang et al., 2020a). In brief, the CDS of PagWOX11/12a was inserted into the pBI121 vector containing the GFP reporter gene to generate a recombinant construct (35S::PagWOX11/12a‐GFP) and introduced into A. tumefaciens strain GV3101 for 84K poplar transient transformation. Transiently transgenic poplars were grown on 1/2 MS solid medium under normal conditions for 48 h or cultured on 1/2 MS medium for 36 h, and then subjected to 200 mm NaCl for 12 h. The roots of the transgenic plants were used for the ChIP assay. The sonicated chromatin was immunoprecipitated with an anti‐GFP antibody (ChIP+) or immunoprecipitated without an anti‐GFP antibody (Mock). qPCR was used to study the fold enrichment of the studied promoter fragments and was performed with the promoter fragment of the PagActin gene as an internal control. The PCR products were detected by 1.2% agarose gel electrophoresis. The experiments were performed three times. All primers used are shown in Table S1.
Dual‐luciferase reporter assay
The full‐length open reading frame of PagWOX11/12a was cloned into the pGreenII 62‐SK effector vector, and the promoter of PagCYP736A12 (1000 bp) used in the Y1H assay was cloned into the pGreenII 0800‐LUC reporter vector. The recombinant plasmids and negative control vectors were introduced into A. tumefaciens GV3101 (pSoup‐p19). The effector and reporter vectors were co‐transformed into N. benthamiana leaves, as previously described (Yao et al., 2020). D‐Luciferin (10 µm) was sprayed onto tobacco leaves and then photographed using an LB985 NightSHADE fluorescence imaging system (Berthold Technologies, Bad Wildbad, Germany). Dual‐luciferase (LUC) activity was determined using a GloMax 20/20 luminometer (Promega, Madison, WI) and a Dual‐Luciferase Assay Kit (Promega) according to the manufacturer’s instructions. The experiments were performed at least three times with six technical repeats. The primers used are listed in Table S1.
Statistical analysis
The experimental data were presented as the mean ± standard deviation (SD) and were subjected to inferential statistical analysis using Student’s t‐test. Significant differences between the two groups of data were evaluated for comparisons (*P < 0.05).
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Figure S1 Screening PagCYP736A12 gene from transcriptome data
Figure S2 Promoter sequence of the PagCYP736A12 gene
Figure S3 Sequence alignment and phylogenetic analysis of PagCYP736A12 and other plant CYP736A12 proteins
Figure S4 Na+ and K+ content in the roots of 84K and transgenic poplar plants
Figure S5 RT‐qPCR analysis of the expression of PagWOX11/12a and PagCYP736A12 in 84K and transgenic poplars
Figure S6 RT‐qPCR analysis of PagCYP736A12‐transformed poplars
Table S1 Primer sequences used in this study.
Table S2 Up‐regulated DEGs (n = 236) in comparisons between ‘OE vs 84K’ under control growth conditions and ‘OE vs 84K’ under salt conditions
Table S3 Down‐regulated DEGs (n = 223) in comparisons between ‘DR vs 84K’ under control growth conditions and ‘DR vs 84K’ under salt conditions.
Table S4 DEGs (n = 17) in both sets of data listed in Table S2 and S3.
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Non‐profit Research Institution of CAF (No. CAFYBB2018QB002) and National Natural Science Foundation of China (No. 31600539 and 31971620).
Wang, L.‐Q. , Wen, S.‐S. , Wang, R. , Wang, C. , Gao, B. and Lu, M.‐Z. (2021) PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant Biotechnol. J., 10.1111/pbi.13653
References
- Balusamy, S.R. , Kim, Y.J. , Rahimi, S. , Senthil, K.S. , Lee, O.R. , Lee, S. and Yang, D.C. (2013) Transcript pattern of Cytochrome P450, antioxidant and ginsenoside biosynthetic pathway genes under heavy metal stress in Panax ginseng Meyer. Bull. Environ. Contam. Toxicol. 90, 194–202. [DOI] [PubMed] [Google Scholar]
- Balusamy, S.R. , Rahimi, S. and Yang, D.C. (2019) Characterization of squalene‐induced PgCYP736B involved in salt tolerance by modulating key genes of abscisic acid biosynthesis. Int. J. Biol. Macromol. 121, 796–805. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S. (2012) The language of reactive oxygen species signaling in plants. J. Bot. 2012, 1–22. [Google Scholar]
- Chen, J. , Zhang, H.Q. , Hu, L.B. and Shi, Z.Q. (2013) Microcystin‐LR‐induced phytotoxicity in rice crown root is associated with the cross‐talk between auxin and nitric oxide. Chemosphere, 93, 283–293. [DOI] [PubMed] [Google Scholar]
- Cheng, S. , Huang, Y. , Zhu, N. and Zhao, Y. (2014) The rice WUSCHEL‐related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene, 549, 266–274. [DOI] [PubMed] [Google Scholar]
- Cheng, S. , Zhou, D.X. and Zhao, Y. (2016) WUSCHEL‐related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 11, e1130198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, M. , Hu, Y. , Zhao, Y. , Liu, H. and Zhou, D.X. (2007) A WUSCHEL‐LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 144, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der Graaff, E.V. , Laux, T. and Rensing, S.A. (2009) The WUS homeobox‐containing (WOX) protein family. Genome Biol. 10, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz, K.J. , Mittler, R. and Noctor, G. (2016) Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol. 171, 1535–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Z.J. , Yan, J.Y. , Li, C.X. , Li, G.X. , Wu, Y.R. and Zheng, S.J. (2015) Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84, 56–69. [DOI] [PubMed] [Google Scholar]
- Duan, F. , Ding, J. , Lee, D. , Lu, X. , Feng, Y. and Song, W. (2017) Overexpression of SoCYP85A1, a spinach cytochrome p450 gene in transgenic tobacco enhances root development and drought stress tolerance. Front. Plant Sci. 8, 1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Q. , Jiang, T. , Xu, L. , Liu, H. , Mao, H. , Wang, X. , Jiao, B.O. et al. (2017) A salt‐stress‐regulator from the Poplar R2R3 MYB family integrates the regulation of lateral root emergence and ABA signaling to mediate salt stress tolerance in Arabidopsis . Plant Physiol. Bioch. 114, 100–110. [DOI] [PubMed] [Google Scholar]
- de la Garma, J.G. , Fernandez‐Garcia, N. , Bardisi, E. , Pallol, B. , Rubio‐Asensio, J.S. , Bru, R. and Olmos, E. (2015) New insights into plant salt acclimation: the roles of vesicle trafficking and reactive oxygen species signalling in mitochondria and the endomembrane system. New Phytol. 205, 216–239. [DOI] [PubMed] [Google Scholar]
- Hasegawa, P.M. (2013) Sodium (Na+) homeostasis and salt tolerance of plants. Environ. Exp. Bot. 92, 19–31. [Google Scholar]
- He, F. , Wang, H.L. , Li, H.G. , Su, Y. , Li, S. , Yang, Y. , Feng, C.H. et al. (2018) PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA‐induced stomatal closure by ROS production in Populus . Plant Biotechnol. J. 16, 1514–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández, L.E. , Sobrino‐Plata, J. , Montero‐Palmero, M.B. , Carrasco‐Gil, S. , Flores‐Cáceres, M.L. , Ortega‐Villasante, C. and Escobar, C. (2015) Contribution of glutathione to the control of cellular redox homeostasis under toxic metal and metalloid stress. J. Exp. Bot. 66, 2901–2911. [DOI] [PubMed] [Google Scholar]
- Huang, S.B. , Van Aken, O. , Schwarzlander, M. , Belt, K. and Millar, A.H. (2016) The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol. 171, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Chen, S. , Peng, X. , Bae, E.K. , Dai, X. , Liu, G. , Qu, G. et al. (2021) An improved draft genome sequence of a hybrid tree Populus alba × Populus glandulosa . J. Forest Res. 32, 1663–1672. [Google Scholar]
- Jefferson, R.A. , Kavanagh, T.A. and Bevan, M.W. (1987) GUS fusions: beta‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, X. , Zheng, L. , Liu, Y. , Nie, X. , Liu, S. and Wang, Y. (2014) A transient transformation system for the functional characterization of genes involved in stress response. Plant Mol. Biol. Rep. 32, 732–739. [Google Scholar]
- Jiang, M. and Zhang, J. (2002) Water stress‐induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up‐regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 53, 2401–2410. [DOI] [PubMed] [Google Scholar]
- Jiang, W. , Zhou, S. , Zhang, Q. , Song, H. , Zhou, D.X. and Zhao, Y. (2017) Transcriptional regulatory network of WOX11 is involved in the control of crown root development, cytokinin signals, and redox in rice. J. Exp. Bot. 68, 2787–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, N. , Nagasaki, H. , Morikami, A. , Sato, Y. and Matsuoka, M. (2003) Isolation and characterization of a rice WUSCHEL‐type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 35, 429–441. [DOI] [PubMed] [Google Scholar]
- Khanom, S. , Jang, J. and Lee, O.R. (2019) Overexpression of ginseng cytochrome P450 CYP736A12 alters plant growth and confers phenylurea herbicide tolerance in Arabidopsis . J. Ginseng Res. 43, 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy, P. , Vishal, B. , Ho, W.J. , Lok, F.C.J. , Lee, F.S.M. and Kumar, P.P. (2020) Regulation of a cytochrome P450 gene CYP94B1 by WRKY33 transcription factor controls apoplastic barrier formation in roots to confer salt tolerance. Plant Physiol. 184, 2199–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge, S. , Tachibana, T. , Iwai, K. and Naganuma, A. (2010) Peroxiredoxin induced sensing and transduction of redox signal in response to oxidative stress and metabolism in yeast cells. J. Pharmacol. Sci. 112, 22p. [Google Scholar]
- Kurotani, K. , Hattori, T. and Takeda, S. (2015) Overexpression of a CYP94 family gene CYP94C2b increases internode length and plant height in rice. Plant Signal. Behav. 10, e1046667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Lin, Y.C. , Li, Q. , Shi, R. , Lin, C.Y. , Chen, H. , Chuang, L. et al. (2014) A robust chromatin immunoprecipitation protocol for studying transcription factor‐DNA interactions and histone modifications in wood‐forming tissue. Nat. Protoc. 9, 2180–2193. [DOI] [PubMed] [Google Scholar]
- Li, Y. and Wei, K. (2020) Comparative functional genomics analysis of cytochrome P450 gene superfamily in wheat and maize. BMC Plant Biol. 20, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Wang, L. , Zhang, J. , Li, J. , Zheng, H. , Chen, J. and Lu, M. (2014a) WUSCHEL‐related Homeobox genes in Populus tomentosa: diversified expression patterns and a functional similarity in adventitious root formation. BMC Genom. 15, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G. , Li, X. , Jin, S. , Liu, X. , Zhu, L. , Nie, Y. and Zhang, X. (2014b) Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS One, 9, e86895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Sheng, L. , Xu, Y. , Li, J. , Yang, Z. , Huang, H. and Xu, L. (2014c) WOX11 and 12 are involved in the first‐step cell fate transition during de novo root organogenesis in Arabidopsis . Plant Cell, 26, 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U. , Hong, R.L. , Hobe, M. , Busch, M.A. , Parcy, F. , Simon, R. and Weigel, D. (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis . Cell, 105, 793–803. [DOI] [PubMed] [Google Scholar]
- Ma, B. , Luo, Y. , Jia, L. , Qi, X. , Zeng, Q. , Xiang, Z. and He, N. (2014) Genome‐wide identification and expression analyses of cytochrome P450 genes in mulberry (Morus notabilis). J. Integr. Plant Biol. 56, 887–901. [DOI] [PubMed] [Google Scholar]
- Mao, C. , He, J. , Liu, L. , Deng, Q. , Yao, X. , Liu, C. , Qiao, Y. et al. (2020) OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnol. J. 18, 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, G. , Seebeck, T. , Schrenker, D. and Yu, O. (2013) CYP709B3, a cytochrome P450 monooxygenase gene involved in salt tolerance in Arabidopsis thaliana . BMC Plant Biol. 13, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, G. , Suzuki, N. , Ciftci‐Yilmaz, S. and Mittler, R. (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. [DOI] [PubMed] [Google Scholar]
- Mittler, R. , Vanderauwera, S. , Gollery, M. and Breusegem, F.V. (2004) Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Shi, H. , Ye, T. , Chen, F. , Cheng, Z. , Wang, Y. , Yang, P. , Zhang, Y. et al. (2013) Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: effect on arginine metabolism and ROS accumulation. J. Exp. Bot. 64, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, W.G. , Liu, W. , Yu, W. , Zhang, Y. , Ding, S. , Li, H. , Mrak, T. et al. (2019) Abscisic acid enhances lead translocation from the roots to the leaves and alleviates its toxicity in Populus × canescens . J. Hazard Mater., 362, 275–285. [DOI] [PubMed] [Google Scholar]
- Suzuki, N. , Koussevitzky, S. , Mittler, R. and Miller, G. (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Yang, Y. , Wang, H. , Ran, X. , Li, B. , Zhang, J. and Zhang, H. (2016) Ectopic expression of a cytochrome P450 monooxygenase gene PtCYP714A3 from Populus trichocarpa reduces shoot growth and improves tolerance to salt stress in transgenic rice. Plant Biotechnol. J. 14, 1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.Q. , Li, Z. , Wen, S.S. , Wang, J.N. , Zhao, S.T. and Lu, M.Z. (2020a) WUSCHEL‐related homeobox gene PagWOX11/12a responds to drought stress by enhancing root elongation and biomass growth in poplar. J. Exp. Bot. 71, 1503–1513. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Yuan, J. , Qin, L. , Shi, W. , Xia, G. and Liu, S. (2020b) TaCYP81D5, one member in a wheat cytochrome P450 gene cluster, confers salinity tolerance via reactive oxygen species scavenging. Plant Biotechnol. J. 18, 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Chen, S. , Zhou, X. , Shen, X. , Deng, L. , Zhu, H. , Shao, J. et al. (2008) Ionic homeostasis and reactive oxygen species control in leaves and xylem sap of two poplars subjected to NaCl stress. Tree Physiol. 28, 947–957. [DOI] [PubMed] [Google Scholar]
- Xu, M. , Xie, W. and Huang, M. (2015) Two WUSCHEL‐related HOMEOBOX genes, PeWOX11a and PeWOX11b, are involved in adventitious root formation of poplar. Physiol. Plantarum, 155, 446–456. [DOI] [PubMed] [Google Scholar]
- Yadav, R.K. , Perales, M. , Gruel, J. , Girke, T. , Jönsson, H. and Reddy, G.V. (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Gene Dev. 25, 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J. , Shen, Z. , Zhang, Y. , Wu, X. , Wang, J. , Sa, G. , Zhang, Y. et al. (2020) Populus euphratica WRKY1 binds the promoter of H +‐ATPase gene to enhance gene expression and salt tolerance. J. Exp. Bot. 71, 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Zhao, Y. , Cheng, S. , Song, Y. , Huang, Y. , Zhou, S. , Liu, X. and Zhou, D.X. (2015) The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell, 27, 2469–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Hu, Y. , Dai, M. , Huang, L. and Zhou, D.X. (2009) The WUSCHEL‐related homeobox gene WOX11 is required to activate shoot‐borne crown root development in rice. Plant Cell, 21, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Li, Z. , Xiao, G. , Zhai, M. , Pan, X. , Huang, R. and Zhang, H. (2020) CYP71D8L is a key regulator involved in growth and stress responses by mediating gibberellin homeostasis in rice. J. Exp. Bot. 71, 1160–1170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Screening PagCYP736A12 gene from transcriptome data
Figure S2 Promoter sequence of the PagCYP736A12 gene
Figure S3 Sequence alignment and phylogenetic analysis of PagCYP736A12 and other plant CYP736A12 proteins
Figure S4 Na+ and K+ content in the roots of 84K and transgenic poplar plants
Figure S5 RT‐qPCR analysis of the expression of PagWOX11/12a and PagCYP736A12 in 84K and transgenic poplars
Figure S6 RT‐qPCR analysis of PagCYP736A12‐transformed poplars
Table S1 Primer sequences used in this study.
Table S2 Up‐regulated DEGs (n = 236) in comparisons between ‘OE vs 84K’ under control growth conditions and ‘OE vs 84K’ under salt conditions
Table S3 Down‐regulated DEGs (n = 223) in comparisons between ‘DR vs 84K’ under control growth conditions and ‘DR vs 84K’ under salt conditions.
Table S4 DEGs (n = 17) in both sets of data listed in Table S2 and S3.


