Summary
Plants sense pathogen attacks using a variety of receptors at the cell surface. The LRR receptor‐like proteins (RLP) and receptor‐like kinases (RLK) are widely reported to participate in plant defence against bacterial and fungal pathogen invasion. However, the role of RLP and RLK in plant antiviral defence has rarely been reported. We employed a high‐throughput‐sequencing approach, transgenic rice plants and viral inoculation assays to investigate the role of OsRLP1 and OsSOBIR1 proteins in rice immunity against virus infection. The transcript of a rice LRR receptor‐like protein, OsRLP1, was markedly up‐regulated following infection by RBSDV, a devastating pathogen of rice and maize. Viral inoculation on various OsRLP1 mutants demonstrated that OsRLP1 modulates rice resistance against RBSDV infection. It was also shown that OsRLP1 is involved in the RBSDV‐induced defence response by positively regulating the activation of MAPKs and PTI‐related gene expression. OsRLP1 interacted with a receptor‐like kinase OsSOBIR1, which was shown to regulate the PTI response and rice antiviral defence. Our results offer a novel insight into how a virus‐induced receptor‐like protein and its adaptor kinase activate the PTI response and antiviral defence in rice.
Keywords: rice, receptor‐like protein, receptor‐like kinase OsSOBIR1, pattern‐triggered immunity, rice black‐streaked dwarf virus
Introduction
To protect themselves from damage, plants have various mechanisms that detect challenges from pathogens (fungi, bacteria and viruses). Plant innate immunity employs a two‐tiered system to activate defence responses, namely pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) and effector‐triggered immunity (ETI) (Boller and Felix, 2009; Dodds and Rathjen, 2010). PTI is activated through the perception and recognition of specific molecular patterns by plasma membrane (PM)‐localized pattern recognition receptors (PRRs). The danger signals include PAMPs, microbe pathogen‐associated molecular patterns (MAMPs) or damage‐associated molecular patterns (DAMPs) (Boller and Felix, 2009). PTI is generally considered to be a weak immune response that confers resistance to broad‐spectrum pathogens (Niks et al., 2015), while ETI is R gene‐mediated and typically confers race‐specific disease resistance (Wiesner‐Hanks and Nelson, 2016). Within a few minutes, PTI triggers a wide array of signalling cascade responses from cell to organism level, including Ca2+ influx, reactive oxygen species (ROS) production, the activation of mitogen‐activated protein kinases (MAPKs) and the expression of PTI‐related defence genes (Bigeard et al., 2015; Couto and Zipfel, 2016; Trapet et al., 2015; Wang et al., 2019).
Plants have hundreds of potential cell surface PRRs, including receptor‐like kinases (RLKs) and receptor‐like proteins (RLPs) (Macho and Zipfel, 2014; Shiu and Bleecker, 2003; Shiu et al., 2004). RLKs contain an extracellular ligand‐recognition domain (leucine‐rich repeats, LRR), a transmembrane (TM) domain and an intracellular Ser/Thr kinase domain, while receptor‐like proteins (RLPs) are similar but lack the cytoplasmic kinase domain (Fritz‐Laylin et al., 2005; Wang et al., 2008). Because RLPs lack a kinase domain, they need to interact with other proteins which contain a kinase domain such as SOBIR1, in order to activate downstream signalling (Liebrand et al., 2013; Liebrand et al., 2014). In Arabidopsis, 57 genes encoding LRR‐RLPs have been identified, while tomato has 176 genes that encode LRR‐RLPs (Fritz‐Laylin et al., 2005). Several reports have demonstrated that RLPs are as important as RLKs for plant innate immunity and development (Belkhadir et al., 2014; Stergiopoulos and de Wit, 2009). For example, the first characterized plant LRR‐RLP was tomato Cf‐9 protein that perceives the fungal effector Avr9 to resist the fungal pathogen Cladosporium fulvum (Stergiopoulos and de Wit, 2009). In Arabidopsis, a RECEPTOR OF ENIGMATIC MAMP OF XANTHOMONAS (ReMAX) was responsible for the recognition of eMAX in Xanthomonas (Jehle et al., 2013). In wheat (Triticum aestivum), TaRLP1.1 is involved in resistance to stripe rust caused by Puccinia striiformis f. sp. tritici (Jiang et al., 2013). In rice (Oryza sativa), 90 genes encoding LRR‐RLPs have been identified (Fritz‐Laylin et al., 2005) but while there is much information about the function of RLPs in Arabidopsis, their function in rice has rarely been studied. In addition, the involvement of RLPs in viral infection has received little attention.
Rice black‐streaked dwarf virus (RBSDV) is a member of the genus Fijivirus, family Reoviridae, and is transmitted by the small brown planthopper (Laodelphax striatellus Fallén; SBPH) (Wei and Li, 2016). RBSDV‐infected rice has growth abnormalities, such as darkening of leaves and dwarfism, there is a change of hormone homeostasis, and serious yield losses result (He et al., 2017; Xie et al., 2018; Zhang et al., 2019b). In this study, we found an LRR‐RLP gene, OsRLP1, that was significantly induced upon RBSDV infection. OsRLP1 activates MAPKs and PTI‐related gene expression. OsRLP1 interacts with an LRR‐RLK, OsSOBIR1, which participates in the regulation of rice PTI defence. In general, our findings provide novel insights into the roles of a receptor‐like protein associated with its adaptor kinase to mediate rice defence against a viral pathogen.
Results
A gene encoding an LRR receptor‐like protein was induced in rice by RBSDV infection
Our previous high‐throughput sequencing data had shown that many genes including an LRR‐RLP gene (OsRLP1, LOC_Os06g04830) were changed in response to RBSDV infection (He et al., 2017). The expression level of OsRLP1 was analysed in RBSDV‐infected plants by RT‐qPCR assay, and this confirmed that the OsRLP1 gene was significantly up‐regulated after RBSDV infection (Figure 1a). Sequence analysis showed that OsRLP1 encodes a protein of 980 amino acids, containing an N‐terminal signal peptide, multiple LRR domains and a transmembrane domain, but lacking a C‐terminal cytoplasmic kinase catalytic domain (Figure S1). An RT‐qPCR assay to investigate the expression pattern of the OsRLP1 gene in different tissues in NIP plants showed that it was most highly expressed in the rice stem (Figure S2). To observe the subcellular localization of OsRLP1, the full‐length cDNA sequence was fused to the N‐terminal end of enhanced green fluorescent protein (eGFP). First experiments were done in combination with a plasma membrane marker in rice protoplasts. Unfortunately, there was only a very weak expression of OsRLP1 in this system and it was not possible to observe the fluorescent signal. The subcellular localization of OsRLP1 was therefore further investigated using infiltrated leaves of Nicotiana benthamiana ( N. benthamiana). The results showed that OsRLP1 was localized to the cytoplasm and the plasma membrane, while the control 35S::eGFP construct was localized in both the nucleus and cytoplasm (Figure 1b).
Figure 1.
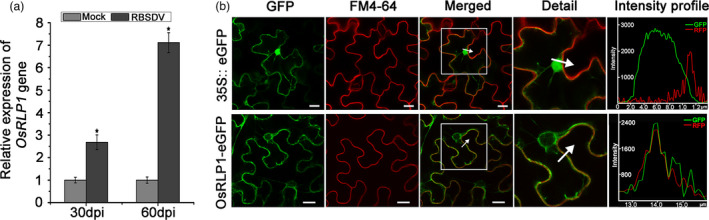
The expression pattern of the OsRLP1 gene. (a) RT‐qPCR analysis of OsRLP1 gene expression levels in RBSDV‐infected plants and controls at 30 and 60 dpi. Values are the means ± SD from 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (b) Subcellular localization of OsRLP1 protein. OsRLP1‐eGFP was transiently expressed in N. benthamiana leaves with 35S::eGFP construct as negative control. The infiltrated‐tobacco leaves were stained with plasma membrane dye FM4‐64 for 10 min. Excitation laser wavelengths of 514 nm and 563 nm were used for GFP and FM4‐64 signals, respectively. The white square in the merged image is magnified as a detailed picture. White arrows indicate the region of interest (ROI) and intensity profiles show the pixels grey value across the ROI in the eGFP and RFP channels. White bar represents 20 μm.
OsRLP1 positively modulates rice resistance to RBSDV infection
A reverse genetics assay was performed to investigate the role of OsRLP1 in RBSDV infection of rice. Having isolated and identified an OsRLP1 T‐DNA insertion mutant (osrlp1) (Figure S3a) (Jeon et al., 2000; Jeong et al., 2006), RT‐qPCR analysis then showed that the expression level of the OsRLP1 gene was significantly reduced in T‐DNA insertion mutant plants compared with the wild type (HY) (Figure S3b). In an inoculation assay, the T‐DNA mutant osrlp1 was obviously more susceptible than HY to RBSDV. The osrlp1 plants were more stunted (Figure 2a), and disease incidence was higher (90% v 70%; Figure 2b) than in HY control plants. An RT‐qPCR assay showed that the expression levels of RBSDV S6, S7 and S10 RNA were higher in the osrlp1 mutant than in control plants (Figure 2c), and there were corresponding differences in the accumulation of viral coat protein P10 (Figure 2d). These results suggest that OsRLP1 may be a positive regulator of rice immunity against viral infection.
Figure 2.
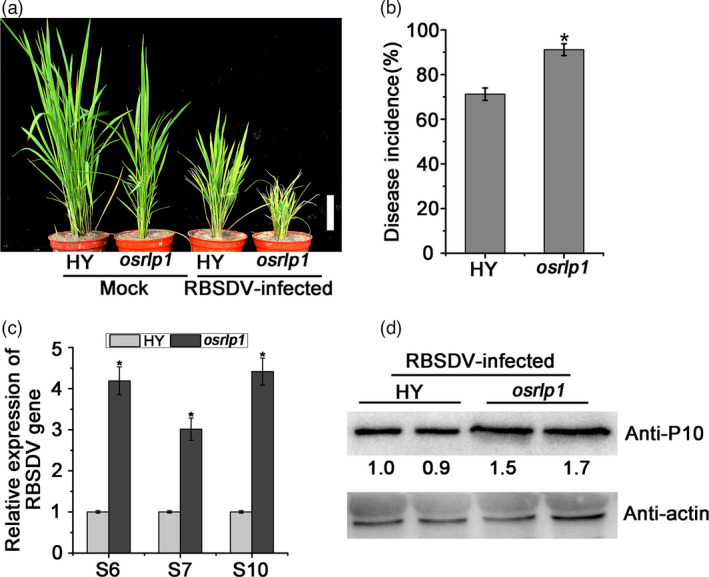
The effect of RBSDV infection on OsRLP1 T‐DNA mutant plants. (a) The phenotypes of RBSDV‐infected and mock‐inoculated plants photographed at 30 dpi. White bar represents 5 cm. (b) The viral incidence in HY (WT) and osrlp1 (OsRLP1 T‐DNA mutant) plants at 30 dpi. Disease incidence is the percentage of RBSDV‐infected plants based on the numbers of healthy and diseased plants as determined by RT‐PCR. (c) The relative expression levels of RBSDV S6, S7 and S10 genes in RBSDV‐infected HY and osrlp1 plants as detected by RT‐qPCR at 30 dpi. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD of 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (d) The expression level of viral P10 protein in RBSDV‐infected HY and osrlp1 plants as detected by Western blot. Actin and its corresponding antibody were used as a reference.
To further investigate the role of OsRLP1 in rice resistance to RBSDV infection, transgenic rice lines overexpressing OsRLP1 were constructed. The expression levels of OsRLP1 in transgenic plants were verified by RT‐qPCR and Western blot (Figure S4). Two homozygous T3 generation transgenic rice plants were selected for RBSDV testing. These transgenic plants were not obviously different in morphology to the wild‐type Nipponbare (NIP) but were less susceptible to RBSDV when inoculated using the insect vector. Symptoms were less severe on both OsRLP1‐18# and OsRLP1‐23# plants compared with the wild‐type control at 30 dpi (day post‐inoculation) (Figure 3a, left) and viral incidence was obviously lower in both transgenic lines (<40%) than in NIP plants (55%) (Figure 3b). RT‐qPCR and Western blot experiments also showed that the RBSDV RNA (S6, S7 and S10) and protein levels in both OsRLP1 transgenic lines were significantly less than in WT plants (Figure 3c,d). Thus, overexpressing OsRLP1 in rice enhanced resistance to RBSDV. Mutant plants with decreased expression of OsRLP1 were also generated via the CRISPR/Cas9 system in the NIP background. DNA sequencing showed that two independent mutants had been generated, osrlp1‐cas‐1# and osrlp1‐cas‐2#. The osrlp1‐cas‐1# mutant harboured a deletion of GC and osrlp1‐cas‐2# a deletion of A, both causing frameshift mutations and a premature stop codon (Figure S5a). When two homozygous mutants were inoculated with RBSDV, they had greatly reduced disease resistance (dwarfism symptom, viral incidence and viral accumulation) compared with the NIP controls (Figure 3a–d). Together, these results suggested that OsRLP1 indeed plays a critical role in modulating rice defence against RBSDV.
Figure 3.
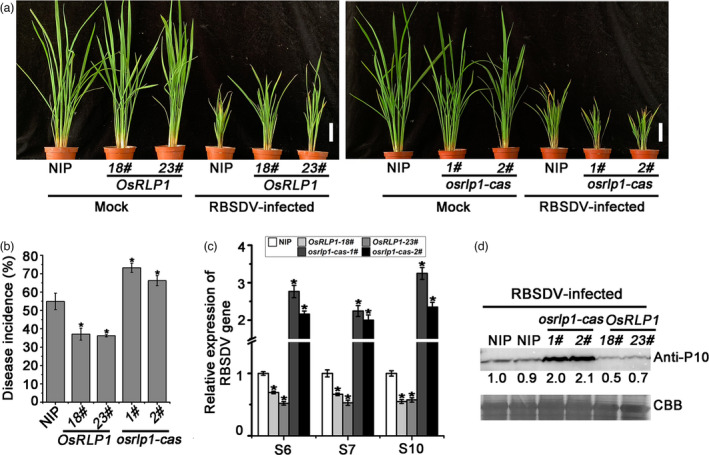
OsRLP1 positively regulates rice resistance to RBSDV. (a) The symptoms of RBSDV in WT (NIP), transgenic plants overexpressing OsRLP1 (OsRLP1) and mutant (osrlp1‐cas) plants. The phenotypes were photographed at 30 dpi. White bar represents 5 cm. (b) Disease incidence in NIP (control), OsRLP1 lines (18# and 23#) and osrlp1‐cas (1# and 2#) lines at 30 dpi. Disease incidence is the percentage of RBSDV‐infected plants based on the numbers of healthy and diseased plants as determined by RT‐PCR. (c) The relative expression levels of RBSDV S6, S7 and S10 genes in RBSDV‐infected NIP, OsRLP1 lines (18# and 23#) and osrlp1‐cas lines (1# and 2#) assessed by RT‐qPCR at 30 dpi. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD from 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (d) The accumulation of RBSDV P10 protein in RBSDV‐infected plants determined by Western blotting at 30 dpi. Total proteins were stained with Coomassie brilliant blue (CBB).
OsRLP1 is required for the activation of defence‐related genes in response to RBSDV infection
To further clarify the antiviral mechanism of OsRLP1, we used RNA high‐throughput sequencing to compare the expression patterns of other genes in the osrlp1‐cas1# mutant with those in WT plants in response to RBSDV infection. Ten‐day‐old (three to four‐leaf stage) rice seedlings were inoculated with either viruliferous or virus‐free SBPH. About 30 days after inoculation, symptoms of dwarfing and dark green leaves were displayed, while the mock‐inoculated plants were symptom‐free. Based on RT‐PCR assays, we collected leaves from mock‐inoculated and RBSDV‐infected plants of both osrlp1‐cas1# mutant and WT plants to analyse global transcriptomic changes at 30 dpi. Following viral inoculation of NIP plants, 3450 genes were differentially expressed (1840 repressed and 1610 induced) (Figure 4a,c) (Table S2). Compared to wild‐type (NIP) plants, osrlp1‐cas1# plants had 822 down‐regulated and 513 up‐regulated genes in response to RBSDV infection, (2‐fold‐change, P ≤ 0.05) (Figure 4a,c) (Table S2). In the absence of viral infection, the OsRLP1 mutant itself (osrlp1‐cas‐1#) has a significantly different transcriptome than wild‐type (NIP) plants (1456 down and 756 up) (Figure 4a,c) (Table S2). Comparative analysis showed that 62% (510/822) (red font) of OsRLP1‐dependent genes were down‐regulated in the osrlp1‐cas‐1#‐RB vs NIP‐RB comparison (Figure 4a, left). Interestingly, based on hierarchical cluster analysis, we found that these 510 down‐regulated genes were highly induced in RBSDV‐infected NIP plants compared with wild type (Figure 4a, right). Gene ontology (GO) analysis indicated that these down‐regulated genes were highly enriched in oxidation–reduction processes and plant defence, including response to jasmonic acid (JA) and peroxidase activity (Figure 4b). Further comparative analysis showed that 357 (red font) of OsRLP1‐dependent genes were up‐regulated in osrlp1‐cas‐1#‐RB vs NIP‐RB (Figure 4c, left). These 357 genes were mostly suppressed in RBSDV‐infected NIP plants compared with wild type (Figure 4c, right). GO analysis showed that these activated genes were highly enriched in the component of membrane category (Figure 4d).
Figure 4.
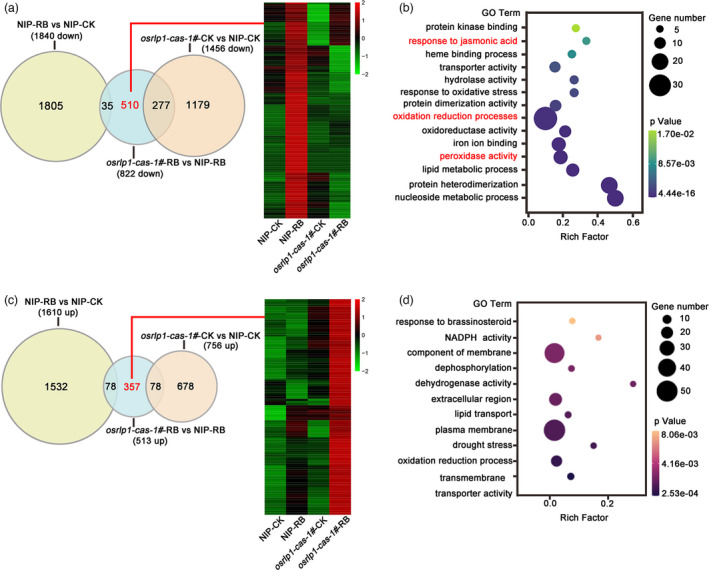
RNA‐seq analyses of NIP and osrlp1‐cas‐1# plants in response to RBSDV infection. (a), (c) Venn diagrams showing overlaps of down‐regulated (a) or up‐regulated (c) differentially expressed genes in NIP‐RB vs NIP‐CK, osrlp1‐cas‐1#‐RB vs NIP‐RB and osrlp1‐cas‐1#‐CK vs NIP‐CK samples at 30 dpi. The heat maps show hierarchical cluster analysis of the 510‐down or 357‐up (genes labelled in red) differentially expressed genes in NIP or osrlp1‐cas‐1# plants under mock or RBSDV inoculation. All differentially expressed genes were selected using cut‐off P‐value <0.05 and fold‐change >2 compared with controls. Data were collected from three biological replicates, each containing a pool of three plants. RB, RBSDV‐infected plants; CK, control plants. (b), (d) the gene ontology (GO) enrichment analyses of 510‐down or 357‐up (genes labelled in red) in (a) and (c). Oxidation–reduction processes and plant defence genes are labelled in red.
To further investigate how OsRLP1 is involved in rice immunity, we analysed the differentially expressed genes in the comparisons NIP‐RB vs NIP‐CK (1532 up‐regulated genes in Figure 4c) and osrlp1‐cas‐1#‐RB vs NIP‐RB (510 down‐regulated genes in Figure 4a). 295 genes (red font) from the two treatments were highly overlapping (Figure 5a, left and Table S3). Consistent with the findings above, GO analysis revealed that these overlapping genes were highly enriched in defence response, including response to JA, SA‐mediated systemic acquired resistance, peroxidase activity and chitin metabolism process (Figure 5b). These analyses demonstrate that OsRLP1 affects the plant–RBSDV interaction by regulating defence gene expression in rice. Notably, a significant number of defence genes, such as OsLOX, OsWRKY, OsCHIT, OsRLK and OsPRX, were activated in NIP in response to RBSDV infection based on hierarchical cluster analysis, but were not activated in osrlp1‐cas‐1# plants (Figure 5a, right and Table S3). The transcriptome data for these defence genes were further tested by RT‐qPCR, and the results were consistent with the RNA‐seq data (Figure 5c). Together, these results suggest that knockout of OsRLP1 affects the activation of defence genes in rice when challenged with RBSDV.
Figure 5.
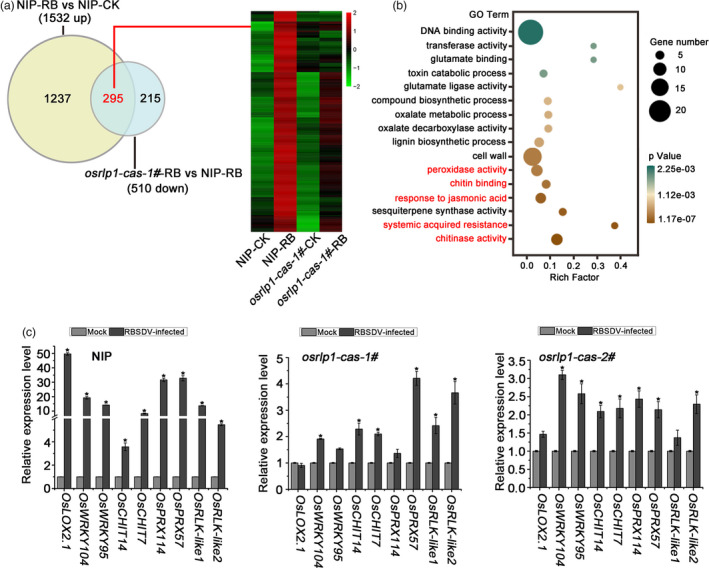
The interaction between NIP and osrlp1‐cas‐1# plants in response to RBSDV infection by transcriptome analyses. (a) Venn diagrams showing overlaps of 1532 up‐regulated genes in NIP‐RB vs NIP‐CK and 510 down‐regulated genes in osrlp1‐cas‐1#‐RB vs NIP‐RB. The heat maps show hierarchical cluster analysis of the 295 (genes labelled in red) differentially expressed genes in NIP or osrlp1‐cas‐1# plants under mock or RBSDV inoculation. All differentially expressed genes were selected using cut‐off P‐value <0.05 and fold‐change >2 compared with controls. Data were collected from three biological replicates, each containing a pool of three plants. RB, RBSDV‐infected plants; CK, control plants. (b) The gene ontology (GO) enrichment analyses of 295 (genes labelled in red) in (a). Plant defence and systemic acquired resistance are labelled in red. (c) The accuracy of the transcriptome data for these differentially expressed genes described in (a) was verified by RT‐qPCR in RBSDV‐infected NIP, osrlp1‐cas‐1# and osrlp1‐cas‐2# plants at 30 dpi. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD of 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test.
OsRLP1 is involved in PTI activation and antiviral defence
Since OsRLP1 modulates the expression of defence‐related genes, we next tested whether OsRLP1 participates in the PTI response. The expression levels of PTI‐related genes, such as OsKS4, OsPAL and OsWRKY70 (marker gene for the PTI response) (Bakshi and Oelmüller, 2014; Chen et al., 2014; Park et al., 2012), were notably induced in RBSDV‐infected NIP plants compared with healthy control plants (Figure 6a). However, the expression level of these PTI‐related genes did not obviously differ between RBSDV‐infected and mock‐inoculated osrlp1‐cas plants (Figure 6b). Another characteristic feature of PTI responses is phosphorylation of MAPKs. To determine whether OsRLP1 affects the expression of the MAPK response, we measured the expression levels of OsMPK3 and OsMPK6 in RBSDV‐infected and healthy plants. In NIP plants, RBSDV infection rapidly increased the expression level of OsMPK3 and OsMPK6 (Figure 6a), but the expression of OsMPK3 and OsMPK6 did not significantly change in osrlp1‐cas plants following viral infection (Figure 6b,c). Thus, OsRLP1 is required for virus‐induced expression of PTI‐related genes and activation of MAPK cascades.
Figure 6.
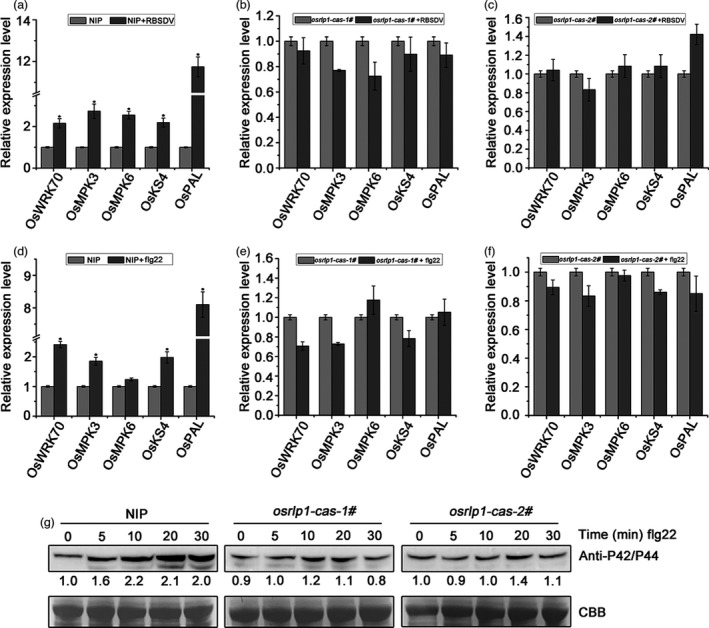
OsRLP1 modulates the expression of PTI‐related genes and MAPK cascades. (a) The relative expression levels of the PTI‐ and MAPK‐related genes in RBSDV‐infected NIP plants at 30 dpi assessed by RT‐qPCR. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD of 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (b), (c) The relative expression levels of the PTI‐ and MAPK‐related genes in RBSDV‐infected osrlp1‐cas‐1# and osrlp1‐cas‐2# plants at 30 dpi assessed by RT‐qPCR. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD from 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (d) The relative expression levels of the PTI‐ and MAPK‐related genes in response to flg22 in NIP plants at assessed by RT‐qPCR. NIP plants were infiltrated with 10 μm flg22, and leaves were collected 30 min post‐infiltration for transcript analysis. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD from 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (e), (f) the relative expression levels of the PTI‐ and MAPK‐related genes and MAPK in response to flg22 in osrlp1‐cas (1# and 2#) plants assessed by RT‐qPCR. osrlp1‐cas plants were infiltrated with 10 μm flg22, and leaves were collected 30 min post‐infiltration for transcript analysis. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD from 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (g) MAPK activation in NIP and osrlp1‐cas‐1# rice leaves after exogenous 10 μm flg22 treatment for 0, 5, 10, 20 or 30 min. Anti‐p42/44 antibody was used for Western blotting. At least two biological replicates were conducted for each experiment. Total proteins were stained with Coomassie brilliant blue (CBB) to show equal loading.
Previous studies have shown that flg22 acts as PAMPs and can induce PTI defence in rice (Meng et al., 2019). Therefore, in order to validate the involvement of OsRLP1 in PTI responses, we applied exogenous flg22 (a well‐known elicitor) to rice leaves. In response to flg22 application, NIP plants exhibited strongly increased expression of PTI‐related genes (such as OsWRKY70, OsMPK3, OsKS4 and OsPAL) compared with untreated control leaves (Figure 6d). However, the expression of these genes was slightly reduced in osrlp1‐cas mutant plants after flg22 application (Figure 6e and 6f). To check whether OsRLP1 is required for flg22‐induced MAPK activation, a Western blot assay was performed using anti‐phospho‐p44/42 (or MAPK3/6) antibodies (Hu et al., 2018) after treatment with 10 μm flg22 for between 5 and 30 min. In NIP plants, flg22 treatment rapidly induced MAPK activation, while the MAPK activation was reduced in osrlp1‐cas‐1# and osrlp1‐cas‐2# mutant plants (Figure 6g). These results suggest that OsRLP1 participates in the regulation of PTI responses in rice.
OsRLP1 physically associates with a receptor‐like kinase OsSOBIR1
Given that RLP lacks a cytoplasmic kinase domain, it needs additional components to transduce and activate cytoplasmic response outputs (Gust & Felix, 2014). Recently, an LRR‐RLK protein, SOBIR1 (SUPPRESSOR OF BIR1‐1), was found to interact with various RLPs involved in plant immunity in Arabidopsis and tomato (Gouveia et al., 2017; Gust & Felix, 2014; Liebrand et al., 2013) but there has been no report of SOBIR1 in rice. Based on protein homology, we found a rice LRR receptor‐like kinase (LOC_Os06g18000, which we named OsSOBIR1) that had 67% similarity to AtSOBIR1 (AT2G31880). As a kinase, AtSOBIR1 can trans‐autophosphorylate in plants (Aranka et al., 2019; Leslie et al., 2010). To investigate whether OsSOBIR1 is phosphorylated in vivo, we performed a phosphorylation assay. Because OsSOBIR1 contains a transmembrane (TM) domain and has strong hydrophobicity, we truncated the transmembrane domain and simply retained the complete kinase domain (OsSOBIR1‐T, 151‐510 aa of OsSOBIR1). The recombinant GST‐OsSOBIR1‐T was incubated in an in vitro kinase assay buffer, and to avoid protein degradation, we added DTT and a cocktail of proteinase inhibitors to the purified protein. The autophosphorylated OsSOBIR1 was detected in SDS‐PAGE using 32P labelled ATP, and it disappeared when 32P labelled ATP was removed (Figure 7a). The result confirms that OsSOBIR1 has kinase activity. To investigate the subcellular localization of OsSOBIR1, its full‐length cDNA sequence was fused to eGFP at the N‐terminal end and then expressed in rice protoplasts, using 35S:: eGFP as the negative control (Figure 7b). OsSOBIR1‐eGFP colocalized with the membrane‐localized marker in rice protoplasts, suggesting that OsSOBIR1 is a plasma membrane‐associated kinase. To test whether OsRLP1 interacts with OsSOBIR1, bimolecular fluorescence complementation (BiFC) assays were performed in N. benthamiana leaves. When OsRLP1‐nYFP and OsSOBIR1‐cYFP were co‐expressed, there were strong YFP fluorescence signals in the plasma membrane, but there were no signals in the combinations OsRLP1‐nYFP and cYFP or OsSOBIR1‐cYFP and nYFP (negative control) (Figure 7c). Furthermore, Co‐IP (Co‐immunoprecipitation) assays indicated that OsRLP1 interacts with OsSOBIR1 in rice protoplasts (Figure 7d). Luciferase complementation imaging (LCI) assays also showed that OsRLP1 protein interacts with OsSOBIR1 protein in N. benthamiana leaves (Figure 7e). Collectively, these results show that OsRLP1 associates with OsSOBIR1 in planta.
Figure 7.
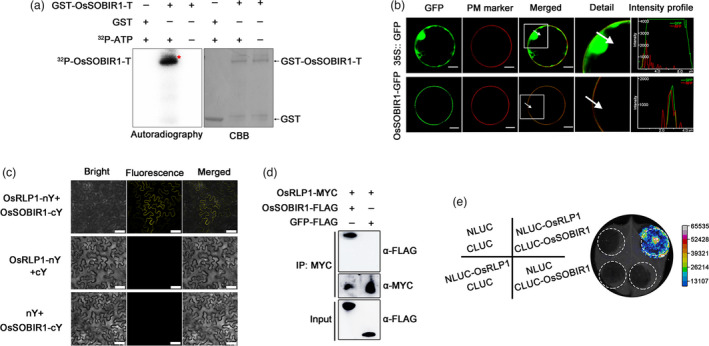
OsRLP1 interacts with OsSOBIR1 protein. (a) In vitro phosphorylation assay showing that OsSOBIR1 is a kinase. GST‐OsSOBIR1‐T is the region 151‐510 aa of OsSOBIR1 (which contains the integral kinase domain) fused to the GST tag. GST served as a negative control in the in vitro kinase assay. 32P‐ATP: [λ‐32P] ATP. The phosphorylated proteins were visualized by autoradiography. (b) Subcellular localization of OsSOBIR1 protein in rice protoplasts. OsSOBIR1‐eGFP was transiently expressed in rice protoplasts with the construct 35S:: eGFP as negative control. PM marker: Plasma Membrane marker. Merged images show co‐localization of OsSOBIR1‐eGFP and the plasma membrane. The white square in the merged image is magnified as a detailed picture. White arrows indicate the region of interest (ROI) and intensity profiles show the pixels grey value across the ROI in the eGFP and RFP channels. White bar represents 10 μm. *Significant difference at P < 0.05 frp, Fisher's LSD test. (c) BiFC assays showing the interaction between OsRLP1 and OsSOBIR1 in N. benthamiana cells. White bar represents 25 μm. (d) Co‐IP assays indicate that OsRLP1 interacts with OsSOBIR1 in rice protoplasts. Total proteins were extracted and immunoprecipitated by anti‐MYC magnetic beads. The coimmunoprecipitated proteins were probed with an anti‐FLAG antibody. (e) Luciferase complementation imaging (LCI) assays display OsRLP1 protein interacts with OsSOBIR1 protein in N. benthamiana leaves. NLUC and CLUC are negative controls.
OsSOBIR1 participates in PTI response and antiviral defence
To explore the role of the OsRLP1/OsSOBIR1 interaction in plant immunity, CRISPR/Cas9 system‐based OsSOBIR1 mutants were generated in the NIP background. Two homozygous mutants named ossobir1‐cas‐4# and ossobir1‐cas‐5# were identified and isolated (Figure S5b). The mutations consisted respectively of the insertion or deletion of a single A residue, leading to frameshifts and a premature stop codon. To test whether OsSOBIR1 was involved in the PTI response, we measured the expression of PTI‐related genes in mutant ossobir1‐cas and WT plants after flg22 stimulation. The expression levels of OsWRKY70, OsMPK3, OsKS4 and OsPAL were rapidly induced in NIP plants after flg22 stimulation, similar to the results with osrlp1‐cas mutant plants. However, in ossobir1‐cas‐4# and ossobir1‐cas‐5# plants the expression level of OsWRKY70 was obviously reduced compared with NIP plants while the levels of OsMPK3, OsKS4 and OsPAL were not obviously changed (Figure 8a–c). These results suggest that OsSOBIR1 may participate in PTI by activating the PTI response.
Figure 8.
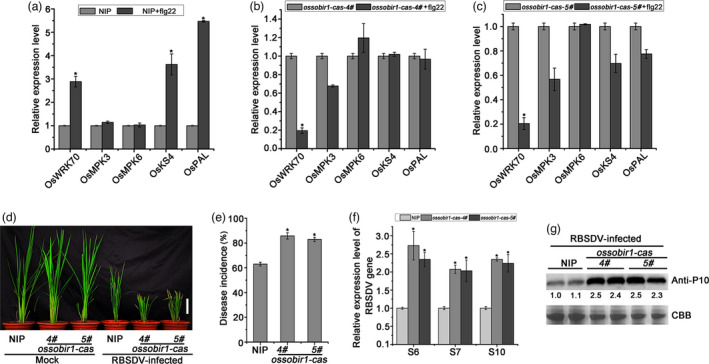
The effect of RBSDV infection on OsSOBIRI1 mutant plants. (a) The relative expression levels of the PTI‐ and MAPK‐related genes in response to flg22 in NIP plants as assessed by RT‐qPCR. NIP plants were infiltrated with 10 μm flg22, and leaves were collected 30 min post‐infiltration for transcript analysis. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD from 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (b), (c) The relative expression levels of the PTI‐ and MAPK‐ related genes and MAPK in response to flg22 in ossobir1‐cas (4# and 5#) plants assessed by RT‐qPCR. ossobir1‐cas plants were infiltrated with 10 μm flg22, and leaves were collected 30 min post‐infiltration for transcript analysis. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD of 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (d) The symptoms of RBSDV on infected WT (NIP) and ossobir1‐cas (4# and 5#) plants. The phenotypes were photographed at 30 dpi. White bar represents 5 cm. (e) Disease incidence in NIP (control) and ossobir1‐cas lines. Disease incidence is the percentage of RBSDV‐infected plants. (f) The relative expression levels of RBSDV S6, S7 and S10 genes in RBSDV‐infected NIP and ossobir1 mutant lines assessed by RT‐qPCR at 30 dpi. UBQ5 was used as the internal reference gene to normalize the relative expression. Values are the means ± SD from 3 biologically independent samples. *Significant difference at P < 0.05 from Fisher's LSD test. (g) The accumulation of RBSDV P10 protein in RBSDV‐infected NIP and ossobir1 mutant plants determined by Western blotting. Total proteins were stained with Coomassie brilliant blue (CBB) to show equal loading.
We next investigated the role of OsSOBIR1 in regulating rice defence against RBSDV infection. After viral inoculation, obvious dwarfing symptoms were observed in ossobir1‐cas‐4# and ossobir1‐cas‐5# mutants and these were more severe than in wild‐type plants (Figure 8d). RT‐PCR tests showed that viral incidence in the two mutant lines (83% and 82%) was significantly more than in WT plants (61%) (Figure 8e). The expression of RBSDV RNAs (S6, S7 and S10) in the two mutant lines was also significantly more than in control plants (Figure 8f), and there were corresponding differences in the accumulation of RBSDV P10 protein (Figure 8g). These results show that knockout of OsSOBIR1 in rice made plants more susceptible to RBSDV infection.
Discussion
Plant immunity and development both depend on transmembrane receptors to perceive extracellular signals. In recent years, several RLPs and RLKs have been identified as PRRs, which have critical functions in plant development and innate immunity (Böhm et al., 2014; Yu et al., 2017). In plant–virus interactions, plants may employ PTI to fight against virus infection as occurs with other pathogens (bacteria, fungi and oomycetes) (Niehl et al., 2016). For example, a well‐known receptor‐like kinase BAK1 acts as a regulator participating in PTI defence, and a bak1 mutant has decreased resistance to different RNA viruses in Arabidopsis (Kørner et al., 2013). However, little is known about the roles of receptor‐like proteins in response to viral infection. In the present study, we identified OsRLP1 as a positive regulator mediating rice defences against RBSDV. Upon RBSDV infection, the PTI‐related genes were significantly induced in NIP plants, but not in OsRLP1 mutant osrlp1‐cas plants (Figure 6a–c). These results indicated that OsRLP1 may take part in RBSDV‐induced PTI defence. Flg22 treatment showed that expression of PTI‐related genes and MAPK phosphorylation, which were significantly activated in wild‐type plants, did not obviously change in osrlp1‐cas plants (Figure 6d–f). The LRR receptor‐like proteins (RLP) play a prominent role in plant–pathogen interactions. Several LRR‐RLPs have been shown to be involved in responses triggered by fungi. AtRLP30 is important for Arabidopsis resistance to necrotrophic fungi (Zhang et al., 2013). In rice, a chitin elicitor binding protein (CEBiP, a lysine motif‐containing receptor‐like protein) recognizes chitin and induces chitin signalling to trigger plant immunity (Kaku et al., 2006). SERK3/BRI1‐ASSOCIATED KINASE 1 (BAK1) is an RLK that plays a key role in the activation of PTI‐triggered immunity by PAMPs or DAMPs in antiviral defence. These results indicated that different types of RLK or RLP mediate resistance to different types of pathogens. In this study, we showed that OsRLP1 participates in rice defence to RBSDV, and it will be interesting to clarify the roles of OsRLP1 in response to other types of pathogens in future work. Overexpression of OsRLP1 increased rice defence, while the OsRLP1 mutant was more susceptible to RBSDV infection, compared with wild‐type plants. Together, our results suggested that OsRLP1 is required for activation of the PTI response and antiviral defence.
RLPs are specific plasma membrane (PM)‐localized receptors and lack obvious cytoplasmic signalling domains (Wang et al., 2010). They therefore need to interact with adaptor kinases for signal transduction. It now appears that LRR‐RLPs specifically associate with adaptor kinases SOBIR1 to form functional equivalents of receptor kinases (Gust & Felix, 2014). In Arabidopsis, it seems that RLP30 depends on SOBIR1/EVR to mediate innate immunity, because RLP30‐mediated resistance against a necrotrophic fungal pathogen in sobir1 mutants was strongly impaired (Zhang et al., 2013). Furthermore, silencing of two tomato SOBIR1 homologues reduced resistance to two fungal pathogens (Cladosporium fulvum and Verticillium dahliae) (Liebrand et al., 2013). The function of SOBIR1 in rice has not previously been studied but we have now shown in this study that OsRLP1 physically associated with OsSOBIR1 in planta (Figure 7c–e). Phosphorylation and subcellular localization assays showed that OsSOBIR1 is a plasma membrane‐associated kinase (Figure 7a,b). The expression of PTI‐related genes was not significantly changed (except for OsWRKY70) in OsSOBIR1 mutants but was markedly induced in wild‐type plants after flg22 treatment (Figure 8a–c). Meanwhile, OsSOBIR1 mutant plants had more severe symptoms and greater virus content than wild‐type plants. We therefore infer that OsSOBIR1 plays important roles in the PTI response and antiviral defence. In Arabidopsis, the levels of Tobacco rattle virus (TRV) were mildly reduced in sobir1‐12 mutant plants, and a strong resistance phenotype against TRV was displayed in a double bir1‐1 sobir1‐1 mutant (Gao et al., 2009; Garnelo Gómez et al., 2019). These findings indicate that BAK1 and SOBIR1 act together to regulate the antiviral response, but their relationships during viral proliferation remain unclear. In rice, where OsRLP1 associates with its adaptor kinase OsSOBIR1, we suppose that this association forms a signalling receptor complex and then triggers PTI‐related responses, which ultimately activates rice immunity against RBSDV infection (Figure S6). Further investigation is needed to determine whether OsSOBIR1 phosphorylation plays a key role for its association with OsRLP1. It has been widely reported that viral PAMPs (VAMPs) can activate antiviral immunity in mammals, and the mechanisms by which viral effectors manipulate PTI defence have been well studied (Jensen and Thomsen, 2012; Yokota et al., 2010). Plants can specifically distinguish PAMP or DAMP to trigger their defence responses (Macho and Lozano‐Duran, 2019). In Arabidopsis, extracts of the green peach aphid (GPA; Myzus persicae) can trigger PTI responses. BAK1 was shown to be required for GPA elicitor‐induced PTI responses (Prince et al., 2014). Plant viruses depend on the mechanical injury caused by the insect vector to gain entry into plant cells and this process may induce endogenous DAMPs to activate the antiviral innate immune. It will be more challenging to discover what kind of viral molecule can act as a PAMP or DAMP. In this study, we did not find the RBSDV‐induced elicitor which was recognized by OsRLP1 to initiate the plant PTI response. In future, it will be pivotal to identify this elicitor and to explore how it is recognized by OsRLP1. In addition, successful viral infection is often accompanied by the accumulation of viral effectors, such as replicase, MPs and coat proteins (CPs) that suppress PTI and lead to disease (Teixeira et al., 2019). The Tomato yellow leaf curl virus (TYLCV) C4 protein targets plant RLK to inhibit cell‐to‐cell spread of RNAi (Rosas‐Diaz, et al., 2018). In addition, C4 also can manipulate RLK‐mediated signalling to defend against infection by geminiviruses (Garnelo Gómez, et al., 2019). The mechanism by which viral effectors limit OsRLP1‐mediated defence in RBSDV infection will be worth investigating in the future. In summary, our results provide the first evidence that a receptor‐like protein, associated with its adaptor kinase OsSOBIR1, mediates plant PTI defence against RBSDV infection in rice.
Experimental procedures
Plant materials
The rice (Oryza sativa subsp. japonica) varieties used in this study were Wuyujing No.3 and Nipponbare (NIP). We identified and obtained the T‐DNA insertion mutant osrlp1 in O. sativa L. subsp. japonica, cv. Hwayoung (HY) from the rice T‐DNA insertion sequence database. The OsRLP1 overexpression, OsRLP1 CRISPR/Cas9 mutant and OsSOBIR1 CRISPR/Cas9 mutant plants were created in a NIP background. RBSDV‐ infected plants were maintained in our laboratory. The plants were grown in the greenhouse at 28–30 °C with a 12 h/12 h light/dark cycle.
Isolation and characterization of T‐DNA insertion mutant plant
The identification of T‐DNA insertion sites in the mutant osrlp1 (PFG‐1C‐03744.L) was conducted according to http://signal.salk.edu/cgi‐bin/RiceGE. Right border primer LB1 was used to confirm integration of T‐DNA in osrlp1 and gene‐specific primers LP/RP to identify the corresponding wild‐type band (Yi and An, 2013). The primer sequences used in this study are listed in Table S1.
Total RNA extraction and RT‐qPCR assays
The total RNAs of mock and treated rice leaves were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA, Cat. no. 15596‐026) according to the manufacturer’s protocols. Subsequently, total RNA (1–2 μg) was used to synthesize cDNA using the fast quant RT kit (Tiangen, Beijing, China, Cat. no. KR106‐03). For RT‐qPCR assay, the ChamQTM SYBR qPCR Master Mix (Low ROX Premixed) and ABI7900HT Sequence Detection System (Applied Biosystems, Carlsbad, CA, USA) were used. The rice actin gene OsUBQ5 (AK061988) was used to normalize the statistic, and the results were analysed by the 2−ΔΔCt method (Sun et al., 2014). The experiments in this study were repeated at least three times with similar results. The RT‐qPCR primer sequences used in this study are listed in Table S1.
Western blot analysis
The RBSDV‐infected plant tissues to be tested were extracted with SDS lysis buffer (100 mm Tris‐HCl, pH = 6.8, 10% SDS) for Western blot analysis as previously described (Zhang et al., 2019a). Anti‐P10 polyclonal antibody (provided by Prof. Jianxiang Wu) at 1:3000 dilution was used for diagnosis of RBSDV infection. Anti‐actin antibody was used for diagnosis of the reference protein (Abbkine, A01050‐3). Anti‐Phospho‐p44/42 (or MAPK3/6) antibodies (Cell Signaling Technology, Danvers, MA, USA, Cat. no. 4370) at 1:2000 dilution were used to test seedlings treated with flg22. A chemiluminescent substrate (ECL; Pierce, Rockford, IL, USA, Cat. no. 32106) was used for the immunoblot signal. Total proteins were stained with Coomassie brilliant blue (CBB) to show equal loading. The experiments in this study were repeated at least three times with similar results.
Subcellular localization assay
Rice protoplasts were used in subcellular localization assays of OsSOBIR1. The rice protoplasts were isolated and transfected as described previously (Zhang et al., 2011). Firstly, rice seedlings were grown in 1/2 MS medium for 10 d and rice seedlings were cut into 0.5 mm strips. Then, the strips were soaked in 0.6 M mannitol for 10 min (in dark), subsequently, discarded the mannitol, added enzyme solution (1.5% cellulase RS (Yakult, Japan, Cat. no. 9033‐35‐6), 0.75% macerozyme R‐10 (Yakult, Japan, Cat. no. 180509‐01), 0.6 M mannitol, 10 mm CaCl2 and 0.1% BSA and 10 mm MES at pH = 5.7) and incubated for 5 h at 25 °C in the dark. After the enzymatic digestion, equal volume of W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl and 2 mm MES at pH = 5.7) was added. Using cell strainer (40 μm Nylon) to filter the tissues and centrifuging at 1000 g for 3 min to collect the pellets. The pellets were washed with 10 mL W5 solution (centrifugation at 1000 g for 3 min). Finally, using MMG solution (0.4 M mannitol,15 mm MgCl2 and 4 mm MES at pH = 5.7) to resuspend the pellets and check the protoplasts under microscope. To investigate the subcellular localization of OsSOBIR1 in rice protoplasts, the PM marker was transfected with 35S::OsRLP1‐eGFP, 35S::OsSOBIR1‐eGFP and 35S::eGFP (negative control) plasmid DNA, respectively, and then incubated in W5 solution with an equal volume of 40% (M/V) PEG solution for 12 h in dark at 25 °C. 5–10 μg of plasmid DNA was mixed with 100 μL protoplasts. After centrifugation at 1000 g for 3 min, transfected rice protoplasts were gently treated with 200 μL W5 solution, and then fluorescence signals were captured by confocal laser microscopy (Leica TCS SP8). Unfortunately, due to the weak expression of OsRLP1, we did not observe the fluorescent signal in rice protoplasts. Therefore, the assay to determine the subcellular localization of 35S::OsRLP1‐eGFP was performed using Agrobacterium strain GV3101 in N. benthamiana cells as described previously (Hecker et al., 2015). The 35S::eGFP vector was used as a negative control. To stain the plasma membrane, tobacco leaves were incubated with 5 μg/mL FM4‐64 for 10 min. Excitation laser wavelengths of 488 nm, 514 nm and 563 nm were used for GFP, FM4‐64 and RFP signals, respectively. Three biological repeats were done for all experiments.
Sequence analysis of OsRLP1
OsRLP1 and OsSOBIR1 protein transmembrane domain prediction was performed using TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM‐2.0/). Conserved domains of the OsRLP1‐encoded proteins were analysed using SMART (http://smart.embl‐heidelberg.de/).
Insect vectors and virus inoculation assay
RBSDV was transmitted by SBPH (small brown planthopper, Laodelphax striatellus). The RBSDV inoculation assay was done as described previously with some minor modifications (Zhang et al., 2019a). Briefly, RBSDV‐infected or virus‐free SBPH were transferred to ten‐days‐old (three to four‐leaf stage) rice seedlings (about three insects per seedling) and allowed to feed for 3–5 days. Then the insects were removed completely. The inoculated plants were grown in the greenhouse to obverse symptoms. Plants infected with RBSDV displayed stunted and darkened leaves at 30 dpi. RT‐PCR assays were used to test the plants and determine the number infected with RBSDV (viral incidence). The primers used to detect RBSDV are listed in Table S1. Samples of mock and RBSDV‐infected leaves were collected at 30 dpi, quick‐frozen in liquid nitrogen, and stored at −80 °C for use.
Vector construction and plant transformation
To generate transgenic plants, the full‐length ORF of the OsRLP1 gene was cloned into the pCV1300 vector which driven by the 35S promoter and with HA tag. The Escherichia coli strain used for cloning was E. coli DH5α. Then the recombinant plasmid was transferred into Agrobacterium tumefaciens strain GV3101 using electroporation and transformed into rice NIP background as described previously (Hiei et al., 1994). To generate osrlp1‐cas and ossobir1‐cas knockout mutants, the CRISPR/Cas9 plasmids were made as previously described (Lu et al., 2017). The constructs were introduced into rice NIP plants by Agrobacterium tumefaciens‐mediated transformation (BioRun, Wuhan, China). The CRISPR‐Cas9 knockout lines were confirmed by sequencing. To generate 35S::eGFP, 35S::OsRLP1‐eGFP and 35S::OsSOBIR1‐eGFP for investigating subcellular localization, the coding sequences of OsRLP1 and OsSOBIR1 were amplified and cloned into pCV‐GFP‐N1 vectors. To generate the constructs of OsRLP1‐MYC, OsSOBIR1‐FLAG and GFP‐MYC for Co‐IP assays in rice protoplast, the CDS of OsRLP1, OsSOBIR1 and GFP was amplified by PCR and cloned into pCV‐3×myc‐N1 or pCV‐GFP‐N1 vectors, respectively. For BiFC assays, the full‐length OsRLP1 and OsSOBIR1 genes were amplified by PCR and cloned into the N‐terminus of YFP or the C‐terminus of YFP, respectively, to generate constructs OsRLP1‐nYFP and OsSOBIR1‐cYFP as previously described (Sun et al., 2013). For luciferase complementation imaging assay (LCI), the CDS of OsRLP1 and OsSOBIR1 genes was cloned into the pCAMBIA1300‐CLUC or pCAMBIA1300‐NLUC vectors, respectively. For protein purification, the region 151‐510 aa of OsSOBIR1 was cloned into the pGEX6P1 vector to express GST‐tagged fusion protein GST‐OsSOBIR1‐T. All the primers used are listed in the Table S1.
BiFC assays
For BiFC assays, the different constructs combinations used were as follows: OsRLP1‐nY/Os‐SOBIR1‐cY, OsRLP1‐nY/cY and nY/Os‐SOBIR1‐cY. The recombinant binary expression vectors were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. After incubation in 6‐week‐old N. benthamiana leaves for 48 h, the YFP fluorescence was captured using confocal laser microscopy (Leica TCS SP10). An excitation laser wavelength of 514 nm was used for YFP signals. Three biological repeats were conducted for all experiments.
Protein interaction assay
For Co‐IP assays, the combinations of OsRLP1‐MYC/OsSOBIR1‐FLAG or GFP‐MYC/OsSOBIR1‐FLAG (as a negative control) were introduced into rice protoplasts for 12 h as previously described (Zhang et al., 2011). The Co‐IP assay was conducted as described previously (Zhang et al., 2020). Briefly, the native protein was extracted using IP buffer (50 mm Tris‐HCl, pH = 8.0, 1 mm MgCl2, 0.5 m sucrose, 10 mm EDTA with 10 mm DTT) (Thermo Scientific, Waltham, MA, USA, Cat. no. 87788) with the addition of 100 μm protease inhibitor cocktail (Roche, Basel, Switzerland, Cat. no. 04693132001) for 10 min at 4 °C and subsequently centrifuged at 12 000 g , 4 °C for 10 min (Wu et al., 2019). The supernatant was incubated with 10 μl Pierce™ anti‐c‐Myc magnetic beads (Thermo Scientific, Waltham, MA, USA, Cat. no. 88844) in 2 mL centrifuge tube for 1–2 h at 4 °C. Importantly, the beads were pre‐washed three times with 1 × PBS before using. The immunoprecipitates were then washed at least three times with 1 × PBS and then re‐suspended in 50 μL 2 × SDS sample buffer. Subsequently, the protein samples were boiled at 95–100 °C for 10 min and Western blots were performed.
LCI assay
To investigate the interaction between OsRLP1 and OsSOBIR1, an LCI assay was performed as previously described with minor modifications (Jin et al., 2016). Briefly, the combinations NLUC‐OsRLP1/CLUC‐OsSOBIR1, NLUC‐OsRLP1/CLUC, NLUC/CLUC‐OsSOBIR1 or NLUC/CLUC were transformed into Agrobacterium tumefaciens strain GV3101 using electroporation, and infiltrated into different areas of the same tobacco leaf with a final concentration of OD600 = 1.0 at 23 °C for 48 h. Then 0.2 mm luciferin (Perkin Elmer, EU, Cat. no. 122799) supplemented with 0.01% Triton X‐100 was infiltrated into the same areas of the N. benthamiana leaves in darkness for 3 min and the luciferase activity was determined using a low‐light cooled CCD imaging apparatus (NightOWL II LB983). Three biological repeats were conducted for all experiments.
Protein purification and in vitro kinase assay
The vector pGEX6P1‐OsSOBIR1‐T was transferred into Escherichia coli strain Rosetta (DE3) (TransGen Biotech, Beijing, China, Cat. no. CD801‐02) and the empty pGEX6P1 vector was used as the negative control. The bacteria were cultivated at 37 °C with shaking at 220 rpm. When the OD600 reached 0.4–0.6, the expression of GST‐OsSOBIR1‐T and GST proteins was induced by adding 0.8 mm IPTG to the culture medium and incubating at 28 °C for 5–7 h. The cultured cells were collected and disrupted at 4 °C. The recombinant proteins were purified using Glutathione–Sepharose (GE) (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. The in vitro kinase assay was performed as previously described with minor modifications (Wang et al., 2013). 0.4 μg of recombinant GST‐OsSOBIR1‐T was incubated in 50 μL reaction buffer which contained 25 mm Tris (pH 7.5), 6 mm MgCl2, 6 mm MnCl2, 1 mm DTT with 50 mm ATP and [λ32P] ATP at 25 °C for 30 min. The phosphorylated proteins were separated by SDS‐PAGE and then visualized by autoradiography. Total proteins were stained with Coomassie brilliant blue (CBB).
Agrobacterium tumefaciens transformation assays
The binary constructs were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation (constant pressure 2.2 V) and then grown in Luria–Bertani medium containing 50 μg/mL kanamycin, 25 μg/mL rifampicin and 10 μg/mL tetracycline at 28 °C for 48 h. The cultures were then collected by centrifuging at 5000 g for 2 min and re‐suspended in infiltration buffer (10 mm MgCl2, 10 mm MES (pH 5.6) and 0.2 mm acetosyringone) to a final concentration of OD600 = 1.0. The Agrobacterium cultures were kept at 28 °C for at least 2 h without shaking. For co‐infiltration, equal volumes of Agrobacterium suspensions including the designated constructs were infiltrated into N. benthamiana leaves. After mixing, equal quantities of the selected combinations were infiltrated into 6‐week‐old tobacco leaves. After infiltration, the plants were cultured at 23 °C for 48 h and then used for later experiments.
RNA library construction and sequencing
The leaf samples from mock and RBSDV‐infected rice plants were collected at 30 dpi, ground into fine powder and total RNA extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA, Cat. no. 15596‐026). Three leaves from different seedlings were collected as one biological replicate, and three biological replicates were used for each treatment. The methods of RNA library construction were described previously (He et al., 2017). The quantity and purity of RNA were assessed and the addition of adapters, size selection and RNA‐seq were all performed by Hangzhou Lianchuan (Hangzhou, China). RNA sequencing utilized the illumina HiSeqTM 2000 platform. Mapping of sequencing reads to the rice genome (The MSU Rice Genome Annotation Project Data base version 7.0) was done by Bowtie software. Blast2go program was used for data analysis including the gene ontology (GO) functional classes and KEGG pathways. A difference in gene expression was considered significant when the absolute value of log2 (fold‐change) ratio was ≥1 and P ≤ 0.05.
Detection of defence‐related genes expression
Defence‐related gene expression was detected as previously described with minor modification (Wang et al., 2019). Two‐week‐old leaves of rice seedlings were cut into 2 mm strips,; then, samples were placed in a 2‐mL microcentrifuge tube. The samples were infiltrated with 1 mL sterilized water in 2‐mL microcentrifuge tube overnight in darkness to recover from wounding stress. On the next day, sterilized water was replaced with 10 μm flg22 (MedChemExpress, Wilkinson way, Princeton, USA, Cat. no. 304642‐91‐9) or water as control. After flg22 treatment for 0 and 30 min, the samples were collected and quick‐frozen in liquid nitrogen and stored at −80 °C for RNA extraction and RT‐qPCR assays. Three leaves from different seedlings were collected as one biological replicate, and three biological replicates were used for each treatment. Two technical repeats were performed for RT‐qPCR in gene transcript analysis. The primer sequences for RT‐qPCR assays are listed in Table S1. After exogenous flg22 treatment for 0, 5, 10, 20 and 30 min, samples were also collected, quick‐frozen in liquid nitrogen and stored at −80 °C for Western blot assays.
Statistical analysis
Differences were analysed using one‐way or two‐way ANOVA with Fisher’s least significant difference (LSD) tests. Each experiment was repeated at least three times, and data were represented as the mean. A P‐value ≤0.05 was considered statistically significant. All analyses were performed using ORIGIN 8 software. For immunoblot quantification analysis, the intensities of bands were quantified with ImageJ. To generate a representative view hierarchical cluster analysis, RPKM expression values were normalized by z‐score transformation and presented in a boxplot. Gene Ontology (GO) enrichment analyses were performed with the compareCluster function of the R package and with the AGRIGO platform http://bioinfo.cau.edu.cn/agriGO/.
Conflict of interest
The authors declare no competing financial interests.
Author contributions
HZ, JC and ZS conceived the project and designed the experiments; HZ and CC carried out the experiments with assistance from LL, XT, ZW, YL, JL and FY; HZ and ZS wrote the manuscript.
Supporting information
Figure S1. Sequence and structure analysis of the OsRLP1 gene.
Figure S2. Results of RT‐qPCR showing expression of the OsRLP1 gene in different tissues (root, stem and leaf) of two‐old‐week NIP plants.
Figure S3. Identification of mutant plants with OsRLP1 T‐DNA insert.
Figure S4. Identification of OsRLP1 transgenic plants.
Figure S5. The mutations of osrlp1‐cas and ossobir1‐cas plants.
Figure S6. A model of OsRLP1‐mediated rice antiviral PTI response.
Table S1. The primers used in this study.
Table S2. The differentially expressed genes in rlp‐RB vs rlp‐CK vs Nip‐RB vs Nip‐CK.
Table S3. 295 highly overlapping genes in the comparisons NIP‐RB vs NIP‐CK (1532 up‐regulated genes) and osrlp1‐cas‐1#‐RB vs NIP‐RB (510 down‐regulated genes).
Acknowledgments
We thank Prof. Jianxiang Wu (Zhejiang University) for providing viral proteins antibody, Prof. Pengcheng Wang (Chinese Academy of Sciences Shanghai Center for Plant Stress Biology) for the assistance of kinase assay, Prof. Gynheung An for providing T‐DNA insertion mutant and Prof. Mike Adams for critically reading and improving the manuscript. This work was funded by China National Funds for Excellent Young Scientists (32022072), the Natural Science Foundation of China (32001888), Zhejiang Provincial Natural Science Foundation (LQ21C140005), Ningbo Natural Science Foundation (202003N4119), Ningbo Science and Technology Innovation 2025 Major Project (2019B10004). This work was sponsored by the K.C. Wong Magna Fund in Ningbo University.
Zhang, H. , Chen, C. , Li, L. , Tan, X. , Wei, Z. , Li, Y. , Li, J. , Yan, F. , Chen, J. and Sun, Z. (2021) A rice LRR receptor‐like protein associates with its adaptor kinase OsSOBIR1 to mediate plant immunity against viral infection. Plant Biotechnol. J., 10.1111/pbi.13663
Contributor Information
Jianping Chen, Email: jianpingchen@nbu.edu.cn.
Zongtao Sun, Email: sunzongtao@nbu.edu.cn.
References
- Aranka, M. , Jelle, P. , Silke, R. and Matthieu, H. (2019) Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol., 20, 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi, M. and Oelmüller, R. (2014) WRKY transcription factors. Plant Signal. Behav., 9, e27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir, Y. , Yang, L. , Hetzel, J. , Dangl, J. and Chory, J. (2014) The growth–defense pivot: crisis management in plants mediated by LRR‐RK surface receptors. Trends Biochem. Sci., 39, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard, J. , Colcombet, J. and Hirt, H. (2015) Signaling mechanisms in pattern‐triggered immunity (PTI). Mol. Plant, 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Böhm, H. , Albert, I. , Fan, L. , Reinhard, A. and Nürnberger, T. (2014) Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol., 20, 47–54. [DOI] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol., 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Zuo, S. , Schwessinger, B. , Chern, M. , Canlas, P.E. , Ruan, D. , Zhou, X. et al. (2014) An XA21‐associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant, 7, 874–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto, D. and Zipfel, C. (2016) Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol., 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Dodds, P. and Rathjen, J. (2010) Plant immunity: towards an integrated view of plant‐pathogen interactions. Nat. Rev. Genet., 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Fritz‐Laylin, L. , Krishnamurthy, N. , Tör, M. , Sjölander, K. and Jones, J. (2005) Phylogenomic analysis of the receptor‐like proteins of rice and Arabidopsis. Plant Physiol., 138, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M. , Wang, X. , Wang, D. , Xu, F. , Ding, X. , Zhang, Z. , Bi, D. et al. (2009) Regulation of cell death and innate immunity by two receptor‐like kinases in Arabidopsis . Cell Host Microbe, 6, 34–44. [DOI] [PubMed] [Google Scholar]
- Garnelo Gómez, B. , Zhang, D. , Rosas‐Díaz, T. , Wei, Y. , Macho, A. and Lozano‐Durán, R. (2019) The C4 Protein from Tomato yellow leaf curl virus can broadly interact with plant receptor‐like kinases. Viruses, 11, 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia, B. , Calil, I. , Machado, J. , Santos, A. and Fontes, E. (2017) Immune receptors and co‐receptors in antiviral innate immunity in plants. Front. Microbiol., 7, 2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust, A. and Felix, G. (2014) Receptor like proteins associate with SOBIR1‐type of adaptors to form bimolecular receptor kinases. Current Opinion in Plant Biology, 21, 104–111. [DOI] [PubMed] [Google Scholar]
- He, Y. , Zhang, H. , Sun, Z. , Li, J. , Hong, G. , Zhu, Q. , Zhou, X. et al. (2017) Jasmonic acid‐mediated defense suppresses brassinosteroid‐mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol., 214, 388–399. [DOI] [PubMed] [Google Scholar]
- Hecker, A. , Wallmeroth, N. , Peter, S. , Blatt, M. , Harter, K. and Grefen, C. (2015) Binary 2in1 vectors improve in planta (Co)localization and dynamic protein interaction studies. Plant Physiol., 168, 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J., 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hu, L. , Ye, M. , Kuai, P. , Ye, M. , Erb, M. and Lou, Y. (2018) OsLRR‐RLK1, an early responsive leucine‐rich repeat receptor‐like kinase, initiates rice defense responses against a chewing herbivore. New Phytol., 219, 1097–1111. [DOI] [PubMed] [Google Scholar]
- Jehle, A. , Lipschis, M. , Albert, M. , Fallahzadeh‐Mamaghani, V. , Fürst, U. , Mueller, K. and Felix, G. (2013) The receptor‐like protein ReMAX of Arabidopsis detects the microbe‐associated molecular pattern eMax from Xanthomonas. Plant Cell, 25, 2330–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S. and Thomsen, A. (2012) Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol., 86, 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J.‐S. , Lee, S. , Jung, K.‐H. , Jun, S.‐H. , Jeong, D.‐H. , Lee, J. , Kim, C. et al. (2000) T‐DNA insertional mutagenesis for functional genomics in rice. Plant J., 22, 561–570. [DOI] [PubMed] [Google Scholar]
- Jeong, D. , An, S. , Park, S. , Kang, H. , Park, G. , Kim, S. , Sim, J. et al. (2006) Generation of flanking sequence‐tag database for activation‐tagging lines in japonica rice. Plant J., 45, 123–132. [DOI] [PubMed] [Google Scholar]
- Jiang, Z. , Ge, S. , Xing, L. , Han, D. , Kang, Z. , Zhang, G. , Wang, X. et al. (2013) RLP1.1, a novel wheat receptor‐like protein gene, is involved in the defence response against Puccinia striiformis f. sp. tritici . J. Exp. Bot., 64, 3735–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L. , Qin, Q. , Wang, Y. , Pu, Y. , Liu, L. , Wen, X. , Ji, S. et al. (2016) Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog., 12, e1005847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. et al. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl Acad. Sci. USA, 103, 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kørner, C. , Klauser, D. , Niehl, A. , Domínguez‐Ferreras, A. , Chinchilla, D. , Boller, T. , Heinlein, M. et al. (2013) The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant Microbe Interact, 26, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Leslie, M. , Lewis, M. , Youn, J. , Daniels, M. and Liljegren, S. (2010) The EVERSHED receptor‐like kinase modulates floral organ shedding in Arabidopsis . Development, 137, 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T. , van den Berg, G. , Zhang, Z. , Smit, P. , Cordewener, J. , America, A. , Sklenar, J. et al. (2013) Receptor‐like kinase SOBIR1/EVR interacts with receptor‐like proteins in plant immunity against fungal infection. Proc. Natl Acad. Sci. USA, 110, 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T. , van den Burg, H. and Joosten, M. (2014) Two for all: receptor‐associated kinases SOBIR1 and BAK1. Trends Plant Sci., 19, 123–132. [DOI] [PubMed] [Google Scholar]
- Lu, Y. , Ye, X. , Guo, R. , Huang, J. , Wang, W. , Tang, J. , Tan, L. et al. (2017) Genome‐wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant, 10, 1242–1245. [DOI] [PubMed] [Google Scholar]
- Macho, A. and Lozano‐Duran, R. (2019) Molecular dialogues between viruses and receptor‐like kinases in plants. Mol. Plant Pathol., 20, 1191–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho, A. and Zipfel, C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell, 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Meng, Q. , Gupta, R. , Min, C.W. , Kim, J. , Kramer, K. , Wang, Y. , Park, S.‐R. et al. (2019) A proteomic insight into the MSP1 and flg22 induced signaling in Oryza sativa leaves. J. Proteomics, 196, 120–130. [DOI] [PubMed] [Google Scholar]
- Niehl, A. , Wyrsch, I. , Boller, T. and Heinlein, M. (2016) Double‐stranded RNAs induce a pattern‐triggered immune signaling pathway in plants. New Phytol., 211, 1018–1019. [DOI] [PubMed] [Google Scholar]
- Niks, R. , Qi, X. and Marcel, T. (2015) Quantitative resistance to biotrophic filamentous plant pathogens: concepts, misconceptions, and mechanisms. Annu. Rev. Phytopathol., 53, 445–470. [DOI] [PubMed] [Google Scholar]
- Park, C. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C. , Songkumarn, P. , Afzal, A. et al. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, D. , Drurey, C. , Zipfel, C. and Hogenhout, S. (2014) The leucine‐rich repeat receptor‐like kinase BRASSINOSTEROID INSENSITIVE1‐ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis . Plant Physiol., 164, 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas‐Diaz, T. , Zhang, D. , Fan, P. , Wang, L. , Ding, X. , Jiang, Y. , Jimenez‐Gongora, T. et al. (2018) A virus‐targeted plant receptor‐like kinase promotes cell‐to‐cell spread of RNAi. Proc. Natl Acad. Sci. USA, 115, 1388–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S. and Bleecker, A. (2003) Expansion of the receptor‐like kinase/Pelle gene family and receptor‐like proteins in Arabidopsis. Plant Physiol., 132, 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S. , Karlowski, W. , Pan, R. , Tzeng, Y. , Mayer, K. and Li, W. (2004) Comparative analysis of the receptor‐like kinase family in Arabidopsis and rice. Plant Cell, 16, 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos, I. and de Wit, P. (2009) Fungal effector proteins. Annu. Rev. Phytopathol., 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Sun, Z. , Yang, D. , Xie, L. , Sun, L. , Zhang, S. , Zhu, Q. , Li, J. , Wang, X. and Chen, J. (2013) Rice black‐streaked dwarf virus P10 induces membranous structures at the ER and elicits the unfolded protein response in Nicotiana benthamiana. Virology, 447, 131–139. [DOI] [PubMed] [Google Scholar]
- Sun, Z. , He, Y. , Li, J. , Wang, X. and Chen, J. (2014) Genome‐wide characterization of Rice black streaked dwarf virus‐responsive microRNAs in rice leaves and roots by small RNA and degradome sequencing. Plant Cell Physiol., 56, 688–699. [DOI] [PubMed] [Google Scholar]
- Teixeira, R. , Ferreira, M. , Raimundo, G. , Loriato, V. , Reis, P. and Fontes, E. (2019) Virus perception at the cell surface: revisiting the roles of receptor‐like kinases as viral pattern recognition receptors. Mol. Plant Pathol., 20, 1196–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapet, P. , Kulik, A. , Lamotte, O. , Jeandroz, S. , Bourque, S. , Nicolas‐Francès, V. , Rosnoblet, C. et al. (2015) NO signaling in plant immunity: a tale of messengers. Phytochemistry, 112, 72–79. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Ellendorff, U. , Kemp, B. , Mansfield, J.W. , Forsyth, A. , Mitchell, K. , Bastas, K. et al. (2008) A genome‐wide functional investigation into the roles of receptor‐like proteins in Arabidopsis . Plant Physiol., 147, 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Fiers, M. , Ellendorff, U. , Wang, Z. , de Wit, P.J.G.M. , Angenent, G.C. and Thomma, B.P.H.J. (2010) The diverse roles of extracellular leucine‐rich repeat‐containing receptor‐like proteins in plants. Crit. Rev. Plant Sci., 29, 285–299. [Google Scholar]
- Wang, J. , Liu, X. , Zhang, A. , Ren, Y. , Wu, F. , Wang, G. , Xu, Y. et al. (2019) A cyclic nucleotide‐gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res., 29, 820–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Xue, L. , Batelli, G. , Lee, S. , Hou, Y.‐j. , Van Oosten, M.j. , Zhang, H. et al. (2013) Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl Acad. Sci. USA, 27, 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. and Li, Y. (2016) Rice reoviruses in insect vectors. Annu. Rev. Phytopathol., 54, 99–120. [DOI] [PubMed] [Google Scholar]
- Wiesner‐Hanks, T. and Nelson, R. (2016) Multiple disease resistance in plants. Annu. Rev. Phytopathol., 54, 229–252. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Xu, S. , Zhao, P. , Zhang, X. , Yao, X. , Sun, Y. , Fang, R. et al. (2019) The Orthotospovirus nonstructural protein NSs suppresses plant MYC‐regulated jasmonate signaling leading to enhanced vector attraction and performance. PLoS Pathog., 15, e1007897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Li, L. , Zhang, H. , Wang, R. , Tan, X. , He, Y. , Hong, G. et al. (2018) Abscisic acid negatively modulates plant defence against rice black‐streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ., 41, 2504–2514. [DOI] [PubMed] [Google Scholar]
- Yi, J. and An, G. (2013) Utilization of T‐DNA tagging lines in rice. J. Plant Biol., 56, 85–90. [Google Scholar]
- Yokota, S. , Okabayashi, T. and Fujii, N. (2010) The battle between virus and host: modulation of Toll‐like receptor signaling pathways by virus infection. Mediators Inflamm., 2010, 184328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Feng, B. , He, P. and Shan, L. (2017) From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol., 55, 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Li, L. , He, Y. , Qin, Q. , Chen, C. , Wei, Z. , Tan, X. et al. (2020) Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc. Natl Acad. Sci. USA, 117, 9112–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Tan, X. , He, Y. , Xie, K. , Li, L. , Wang, R. , Hong, G. et al. (2019a) Rice black‐streaked dwarf virus P10 acts as either a synergistic or antagonistic determinant during superinfection with related or unrelated virus. Mol. Plant Pathol., 20, 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Tan, X. , Li, L. , He, Y. , Hong, G. , Li, J. , Lin, L. et al. (2019b) Suppression of auxin signalling promotes rice susceptibility to Rice black streaked dwarf virus infection. Mol. Plant Pathol., 20, 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Fraiture, M. , Kolb, D. , Löffelhardt, B. , Desaki, Y. , Boutrot, F.F.G. , Tör, M. et al. (2013) Arabidopsis RECEPTOR‐LIKE PROTEIN30 and receptor‐like kinase SUPPRESSOR OF BIR1‐1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell, 25, 4227–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Su, J. , Duan, S. , Ao, Y. , Dai, J. , Liu, J. , Wang, P. et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast‐related processes. Plant Methods, 7, 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence and structure analysis of the OsRLP1 gene.
Figure S2. Results of RT‐qPCR showing expression of the OsRLP1 gene in different tissues (root, stem and leaf) of two‐old‐week NIP plants.
Figure S3. Identification of mutant plants with OsRLP1 T‐DNA insert.
Figure S4. Identification of OsRLP1 transgenic plants.
Figure S5. The mutations of osrlp1‐cas and ossobir1‐cas plants.
Figure S6. A model of OsRLP1‐mediated rice antiviral PTI response.
Table S1. The primers used in this study.
Table S2. The differentially expressed genes in rlp‐RB vs rlp‐CK vs Nip‐RB vs Nip‐CK.
Table S3. 295 highly overlapping genes in the comparisons NIP‐RB vs NIP‐CK (1532 up‐regulated genes) and osrlp1‐cas‐1#‐RB vs NIP‐RB (510 down‐regulated genes).


