Abstract
Soybean is an important crop worldwide, but its production is severely affected by salt stress. Understanding the regulatory mechanism of salt response is crucial for improving the salt tolerance of soybean. Here, we reveal a role for nuclear factor Y subunit GmNFYA in salt tolerance of soybean likely through the regulation of histone acetylation. GmNFYA is induced by salt stress. Overexpression of GmNFYA significantly enhances salt tolerance in stable transgenic soybean plants by inducing salt‐responsive genes. Analysis in soybean plants with transgenic hairy roots also supports the conclusion. GmNFYA interacts with GmFVE, which functions with putative histone deacetylase GmHDA13 in a complex for transcriptional repression possibly by reducing H3K9 acetylation at target loci. Under salt stress, GmNFYA likely accumulates and competes with GmHDA13 for interaction with GmFVE, leading to the derepression and maintenance of histone acetylation for activation of salt‐responsive genes and finally conferring salt tolerance in soybean plants. In addition, a haplotype I GmNFYA promoter is identified with the highest self‐activated promoter activity and may be selected during future breeding for salt‐tolerant cultivars. Our study uncovers the epigenetic regulatory mechanism of GmNFYA in salt‐stress response, and all the factors/elements identified may be potential targets for genetic manipulation of salt tolerance in soybean and other crops.
Keywords: salt stress, NF‐Y, histone deacetylation, soybean, promoter variation
Introduction
As one of the most important crops worldwide, soybean (Glycine max) provides up to 69% of protein and 30% of oil to the human diet (Lam et al., 2010). The global soybean production must increase substantially to secure global food supplies for the rapid increase in world population. However, soybean yield is seriously threatened by unfavourable environment factors. Soil salinization is one of the major stresses of agriculture, which can greatly constrain plant growth and development, leading to a decrease in crop production. To cope with ion toxicity and osmotic stress caused by salt stress, plants have evolved different regulatory pathways for stress response and tolerance, for example hormone signalling pathways, the salt overly sensitive (SOS) pathway, chromatin modification and transcriptional regulation (Hanin et al., 2016; Kim et al., 2015; Yang and Guo, 2018; Zhu, 2016). The members of transcriptional regulators, such as bZIP, WRKY, MYB, NAC, PHD and HSF families conferring stress tolerance in soybean, have been characterized in our laboratory (Bian et al., 2020; Liao et al., 2008a; Liao et al., 2008b; Wang et al., 2015; Wei et al., 2017). We also demonstrated that miR172a, a long‐distance signal from root to shoot, improves salt tolerance mainly through cleaving its target genes to promote thiamine accumulation (Pan et al., 2016). Recently, GmSALT3/GmCHX1 and GmCDF1 have been identified to be involved in the regulation of salt tolerance by whole‐genome sequencing, quantitative trait locus (QTL) mapping and genome‐wide association studies (GWAS) (Guan et al., 2014; Qi et al., 2014; Zhang et al., 2019b).
Nuclear factor Y (NF‐Y) complex, also known as CCAAT‐binding factor CBF, is a conserved heterotrimeric transcription factor in eukaryotes, consisting of NF‐YA, NF‐YB and NF‐YC subunits (Mantovani, 1999). In contrast to yeast and human, where each NF‐Y subunit is encoded by a single gene, NF‐Y subunits have undergone an extensive expansion in plants. For example, soybean genome contains 21 NF‐YA genes, 32 NF‐YB genes and 15 NF‐YC genes (Quach et al., 2015). It has been demonstrated that NF‐Y proteins not only play a pivotal role in various development processes, but also act as key regulators of stress tolerance, especially drought stress. Arabidopsis NFYA5 and its soybean homologous gene GmNFYA3 function in drought stress tolerance by reducing water loss in leaves of the transgenic Arabidopsis plants (Li et al., 2008; Ni et al., 2013). NF‐YB and NF‐YC subgroups are also involved in drought tolerance regulation. The overexpression of drought‐inducible NF‐YB genes of Arabidopsis, maize and poplar significantly enhanced drought resistance and yield in transgenic plants (Han et al., 2013; Nelson et al., 2007). Bermudagrass Cdt‐NF‐YC1 gene confers tolerance to both drought and salinity in transgenic rice through regulating genes involved in both ABA‐dependent and ABA‐independent pathways (Chen et al., 2015). However, NF‐YA subunit gene seems to function independently in salinity and drought stresses, for example the overexpression of wheat TaNF‐YA10 enhanced drought tolerance but reduced salt tolerance in transgenic Arabidopsis plants (Ma et al., 2015). Whether NFYA gene plays any roles in salt‐stress tolerance has not been fully understood and should be further investigated.
In plants, chromatin regulation and gene expression changes in response to various environmental stresses, which are mediated by epigenetic regulatory mechanisms including DNA methylation, histone acetylation and histone methylation (Kim et al., 2015; Song et al., 2012). Among histone modifications, the acetylation level of histone is controlled by the action of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Generally, HATs are considered as transcriptional activators, whereas HDACs are associated with gene silencing (Xing and Poirier, 2012). HDACs in Arabidopsis are well characterized and categorized into three different families: the reduced potassium deficiency 3 (RPD3)‐like family, the plant‐specific HDAC (HD‐tuin) family and the silent information regulator 2 (SIR2)‐like (sirtuin) family (Hollender and Liu, 2008). Several studies indicated the involvement of HDACs in response to salt stress in plants. Both Arabidopsis HDA6 and HD2C mutants displayed hypersensitive phenotype to ABA and salt stress, indicating that HDA6 and HD2C function as positive regulators of stress responses (Chen et al., 2010; Luo et al., 2012). On the contrary, HDA9 was shown to negatively regulate salt and drought tolerance by modulating histone acetylation levels at a set of stress‐responsive genes (Zheng et al., 2016). A recent study demonstrated that HDA19 and HDA5/14/15/18 play opposite roles in regulating salt response in Arabidopsis (Ueda et al., 2017). AtSRT1, as a chromatin regulator, was reported to control both stress tolerance and primary metabolism (Liu et al., 2017).
MSI1‐like proteins are ubiquitous histone binding WD40‐repeat proteins in all eukaryotes and function in diverse chromatin‐associated complexes (Hennig et al., 2005). Arabidopsis MSI4/FVE and MSI5 are required to repress the FLOWERING LOCUS C (FLC) by forming histone deacetylase complexes with HDA6 and HDA5 (Jeon and Kim, 2011; Luo et al., 2015). Arabidopsis MSI1 is a subunit of Polycomb group protein complex, Chromatin assembly factor 1 and RPD3‐like HDAC complex. Transgenic plants with reduced levels of MSI1 exhibited pleiotropic phenotypes in development and abiotic stress response (Alexandre et al., 2009; Mehdi et al., 2016).
Previously, we discovered that a NF‐YA gene GmNFYA from soybean acts as a positive regulator of lipid biosynthesis (Lu et al., 2016). In this study, we find that GmNFYA is induced by salt stress and promotes salt tolerance in soybean plants possibly via histone modification. Competing with GmHDA13, GmNFYA interacts with GmFVE to derepress the expression of a large number of salt‐responsive genes. Moreover, three promoter haplotypes of GmNFYA were identified in cultivated soybeans. Haplotype I from salt tolerance cultivar Nannong 1138‐2 showed the highest promoter activity under NaCl treatment due to the stronger self‐activated transcription loop of GmNFYA. This haplotype may be selected during future breeding efforts. Our findings revealed the epigenetic regulatory mechanism of GmNFYA gene in salt response and should be useful for genetic improvement of salt tolerance in soybean and other crops.
Result
GmNFYA positively regulates salt‐stress tolerance in soybean
From our previous RNA‐seq analysis for salt‐responsive genes in wild soybeans and cultivated soybeans (Bian et al., 2020), GmNFYA was found to be strongly induced in roots by NaCl treatment. For the present concern, we suggest that GmNFYA may function in response to salinity stress. To examine the expression of GmNFYA, two‐week‐old seedlings of three wild soybeans (Y0532, Y20 and Y55) and one cultivar Kefeng1 (KF) were subject to salt stress (150 mM NaCl). Real‐time quantitative PCR (RT‐qPCR) showed that GmNFYA expression was highly induced in both leaves and roots under salinity condition (Figure 1a), which is consistent with the results from RNA‐seq analysis.
Figure 1.
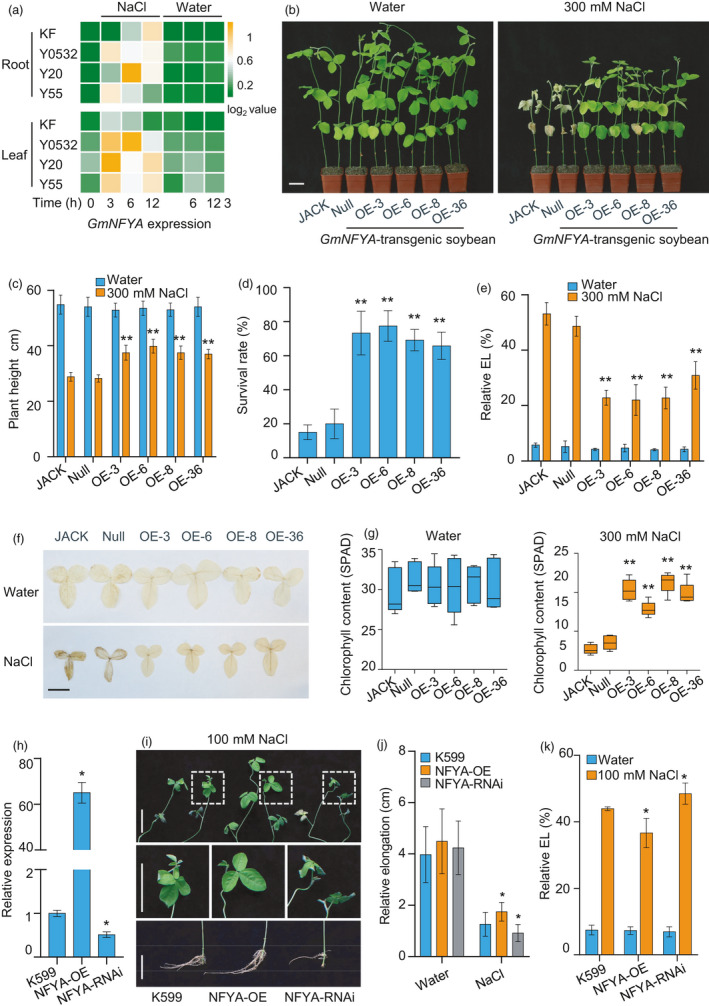
GmNFYA positively regulates soybean salt tolerance. (a) GmNFYA expression in response to salt stress in cultivar Kefeng1 (KF), and wild soybean Y0532, Y55 and Y20. (b) Overexpression of GmNFYA improves salt tolerance of stable transgenic soybean plants. Scale bar = 5 cm. (c) Plant heights of different transgenic lines after 300 mM NaCl treatment for 7 d. Bars indicate SD (n = 6). (d) Survival rates of control and GmNFYA‐overexpressing plants after NaCl treatment. Error bars indicate SD (n = 3). (e) Relative electrolyte leakage of trifoliolate leaves from different transgenic lines after NaCl treatment. Bars indicate SD (n = 4).(f) DAB staining of the first trifoliolate leaves from transgenic soybean plants after water or salt treatment. The brown colour indicates ROS level in each leaf. Bar = 5 cm. (G) Chlorophyll contents of the first trifoliate leaves from various soybean plants grown in water condition or under NaCl treatment. Bars indicate SD (n = 5). (h) Expression of GmNFYA in transgenic hairy roots. K599 indicates soybean plants with control hairy roots generated from A. rhizogenis strain K599. NFYA‐OE indicates soybean plants with transgenic hairy roots overexpressing GmNFYA. NFYA‐RNAi indicates soybean plants with transgenic hairy roots showing reduced expression of GmNFYA. Bars indicate SD (n = 3). (i) Phenotype of plants with GmNFYA‐transgenic hairy root after salt treatment. Scale bar = 5 cm. (j) Relative elongation of different transgenic hairy roots after NaCl treatment for 3 d. Bars indicate SD (n = 30). (k) Relative electrolyte leakage of trifoliolate leaves from different plants with transgenic hairy root. Bars indicate SD (n = 4). Asterisks indicate significant difference compared with the corresponding controls (*, P < 0,05; **, P < 0.01)
To determine the function of GmNFYA in salt response, stable transgenic soybean plants overexpressing the GmNFYA gene driven by the 35S promoter were generated in JACK background. We used JACK and Null plants (no T‐DNA insertion plants segregated from transgenic events) as control plants. The homozygous GmNFYA‐overexpression transgenic plants did not show significant phenotype change. However, they exhibited obvious dwarf phenotype at reproductive stage. Except this, seed size, pod number and other seed yield‐related traits in GmNFYA‐transgenic plants were not significantly affected (Figure S1).
Four individual transgenic lines were used to test salt tolerance phenotypes. Both transgenic and control plants grew well under normal condition. After salt treatment, transgenic plants had better performance than control plants, as revealed by the healthier leaves and the higher plants (Figure 1b, c). Under high‐salinity condition, the survival rates of GmNFYA‐transgenic plants were significantly higher than those of control plants (Figure 1d). Consistently, lower relative electrolyte leakage (EL) and ROS level were observed in the first trifoliolate leaves of GmNFYA‐overexpressing plants compared with those of control plants (Figure 1e, f). In addition, higher chlorophyll contents were detected in transgenic plants than those in control plants (Figure 1g). We also grew transgenic and control plants in a same pot to test whether GmNFYA‐transgenic plants still perform better than control plants after the salt treatment. As showed by the data of growing state, plant height, chlorophyll content and water content, transgenic plants grew much better than JACK, while null plants grew as bad as JACK (Figure S2). These results indicate that overexpression of GmNFYA confers salt tolerance in soybean.
To further investigate the function of GmNFYA in salt‐stress response, two constructs pBIN438‐GmNFYA and pZH01‐GmNFYA were made for generating GmNFYA‐overexpression (GmNFYA‐OE) or GmNFYA‐interference (GmNFYA‐RNAi) hairy roots using the Agrobaterium rhizogenis‐mediated hairy root transformation system (Figure 1h). Compared with control roots (hairy roots generated from A. rhizogenis strain K599), GmNFYA‐OE roots showed better performance and longer root length, whereas GmNFYA‐RNAi roots exhibited relatively shorter length under NaCl treatment (Figure 1i, j). Under salinity, the GmNFYA‐OE plants also had better leaves and lower EL than those of the control plants. However, the leaves of GmNFYA‐RNAi plants performed worse than those of control plants (Figure 1k). ROS levels in the control and transgenic plants were also examined, and low ROS levels were correlated with higher salt tolerance in plants with GmNFYA‐OE hairy roots (Figure S3). These results indicate that GmNFYA functions as a positive regulator of salt tolerance in soybean.
GmNFYA regulates the expression of genes in response to salinity stress
To better understand the molecular mechanism of GmNFYA in enhancing salt tolerance in soybean plants, we performed transcriptome analysis of roots and leaves in stable GmNFYA‐overexpressing plants and wild‐type plants under water and salinity conditions. The downstream salt‐responsive genes were roughly identified by comparing the differentially expressed genes (DEGs) in JACK (NaCl) vs JACK (Control) group with the DEGs in OE‐3 (Control) vs JACK (Control) group. Venn diagrams showed that approximately 50% of the DEGs in OE‐3 (Control) vs JACK (Control) group from roots were involved in response to salinity, and 57.8% of DEGs in leaves were involved in this process (Figure S4a and S3c). When a third comparison was introduced, OE‐3 (NaCl) vs OE‐3 (Control), 1065 common DEGs were identified. To understand the biological processes in which GmNFYA participates, gene ontology (GO) enrichment analysis was performed using the common DEGs in these comparisons. GO terms associated with oxidation–reduction, response to abiotic stress, defence response, ion transport, hormone signalling, cell wall metabolism, regulation of transcription and carbohydrate metabolic process were especially enriched (Figure 2b, c, Figure S4b and S3d). This was consistent with the conclusion that GmNFYA is involved in the regulation of plant response to salt stress in soybean.
Figure 2.
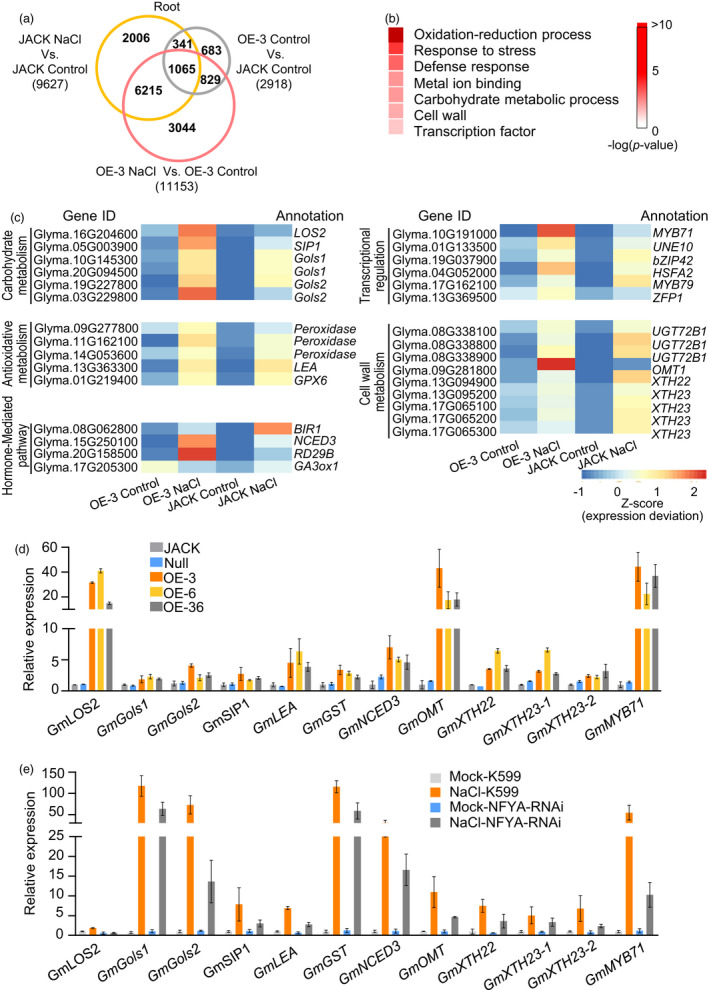
GmNFYA is involved in the regulation of the expression of salt‐responsive genes. (a) Venn diagram showing differentially expressed genes (DEGs) in root samples. Common DEGs were analysed between three ways of comparison: GmNFYA‐OE‐3 transgenic soybean (OE‐3_Control) versus wild‐type JACK (JACK_Control), NaCl‐treated JACK (JACK_NaCl) versus non‐NaCl‐treated JACK (JACK_Control) and NaCl‐treated transgenic soybean (OE‐3_NaCl) versus non‐NaCl‐treated transgenic soybean (OE‐3_Control). (b) GO terms that were statistically enriched in GmNFYA‐regulated genes involved in salt response, which were identified using the 1065 common DEGs in (a). (c) Heatmap of DEGs in the GO terms of carbohydrate metabolism, antioxidative metabolism, hormone signalling, cell wall metabolism and transcriptional regulation. (d) Expression levels of the salt‐responsive genes in stable GmNFYA‐overexpression transgenic soybean plants. Error bars indicate SD (n = 3). (e) Expression levels of the salt‐responsive genes in plants with GmNFYA‐RNAi transgenic hairy roots under water and salt‐stress condition. Error bars indicate SD (n = 3)
To confirm the results of transcriptome analysis, we extracted RNAs from three stable GmNFYA‐transgenic soybean lines and the control plants and examined gene expression by RT‐qPCR. Twelve DEGs were tested and found to be up‐regulated (Figure 2d), and the results were consistent with transcriptome analysis (Figure 2b, c). We also tested the effect of GmNFYA suppression on the expression of these genes and found that the gene transcript levels in the GmNFYA‐RNAi hairy roots were substantially lower than those in control roots after NaCl treatment (Figure 2e), indicating that GmNFYA is required for the response to salt stress in soybean.
GmNFYA interacts with GmFVE, a negative regulator of salt tolerance in soybean
The GmNFYA may function through interaction with some other factors in the control of salt tolerance. To obtain clues to the identities of potential interacting factors, soybean transgenic hairy roots harbouring 35S : GmNFYA‐GFP (NFYA‐GFP) were generated for immunoprecipitation‐mass spectrometry (IP‐MS) assay and GFP‐overexpressing hairy roots were used as control (Figure S5a). Only factors uniquely identified in NFYA‐GFP sample but not in control sample were taken into account. GmNFYA was found to co‐purify with GmFVE (Glyma.09G063100) (Figure 3a and Table S2). Yeast two‐hybrid assay was used to validate the IP‐MS results, and GmFVE was found to interact with GmNFYA in yeast cells (Figure 3b and Figure S5). To further support the interaction between GmNFYA and GmFVE, we conducted bimolecular fluorescence complementation (BiFC) and split‐luciferase complementation assay. These assays revealed that GmNFYA interacts with GmFVE in the nuclei of living plant cells (Figure 3c, d). In addition, co‐immunoprecipitation (Co‐IP) analysis showed that GmNFYA co‐precipitated with GmFVE (Figure 3e). Taken together, these results indicate that the GmNFYA physically interacts with the GmFVE.
Figure 3.
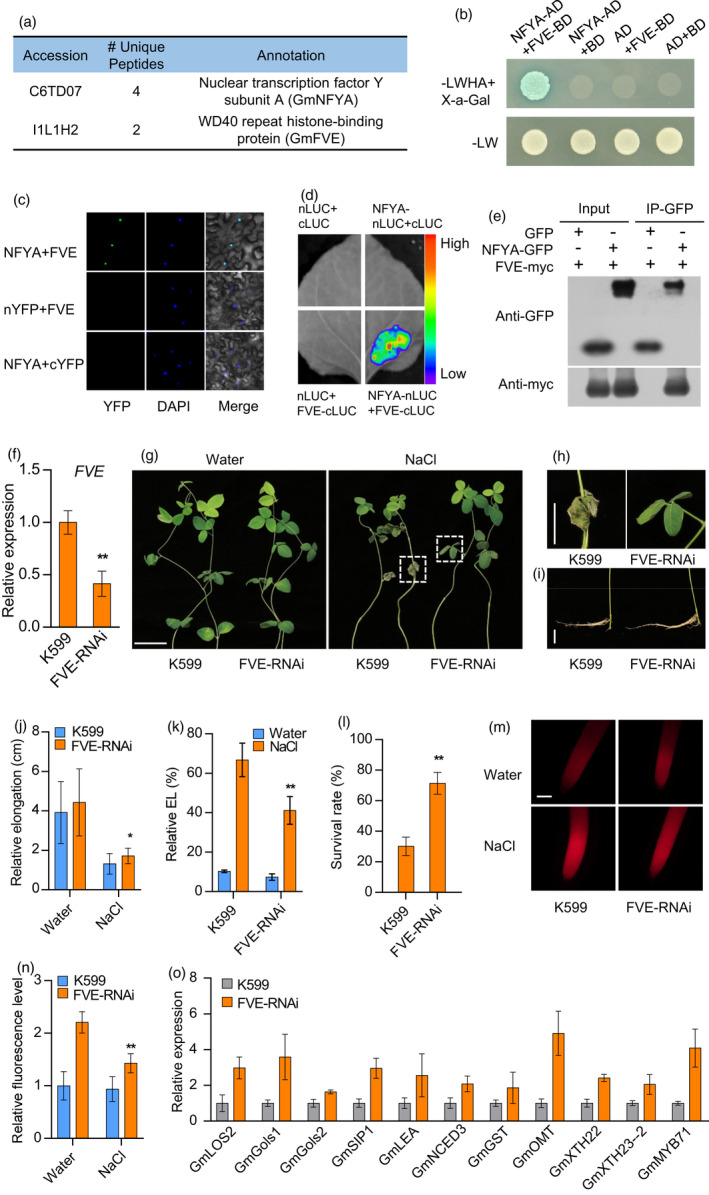
GmNFYA‐interacting protein GmFVE acts as a negative regulator of salt response. (a) Immunoprecipitation–mass spectrometry (IP‐MS) showing GmFVE is the interacting protein of GmNFYA. (b) GmNFYA interacts with GmFVE in a yeast two‐hybrid assay. ‐LWHA+X‐a‐Gal indicates Leu, Trp, His, Ade and X‐a‐Gal drop‐out plates. –LW indicates Leu and Trp drop‐out plates. (c) BiFC analysis of the interaction between GmNFYA and GmFVE in N. benthamiana leaf. (d) Split‐luciferase complementation assay of GmNFYA and GmFVE interaction in N. benthamiana leaf. (e) Co‐immunoprecipitation (Co‐IP) assay of GmNFYA and GmFVE interaction in vivo. (f) Expression of GmFVE in GmFVE‐RNAi transgenic hairy roots. FVE‐RNAi indicates transgenic hairy roots with reduced expression of GmFVE. Bars indicate SD (n = 3). (g) Phenotype of plants with GmFVE‐RNAi transgenic hairy roots after 100 mM NaCl treatment for 3 d. Scale bar = 5 cm. (h and i) The growth of leaves (h) and roots (i) from the plants with salt stress in (g). Bar = 2 cm. (j) Elongation of GmFVE‐RNAi transgenic hairy roots after 80 mM NaCl treatment for 3 d. Bars indicate SD (n = 30). (k) Relative electrolyte leakage (EL) of trifoliolate leaves from the plants with GmFVE‐RNAi hairy roots. Bars indicate SD (n = 4). (l) Survival rates of the plants with GmFVE‐RNAi transgenic hairy roots after 100 mm NaCl treatment for 3 d. Bars indicate SD (n = 3). (m and n) ROS staining (m) and ROS levels (n) of the transgenic and control roots after salt treatment for 6 h. Scale bar = 50 μm. Bars indicate SD (n = 6). (o) Expression of GmNFYA‐regulated genes in GmFVE‐RNAi transgenic hairy roots. Error bars indicate SD (n = 3). Asterisks indicate significant difference compared with the corresponding controls (*, P < 0,05; **, P < 0.01)
GmFVE is a homologous protein of Arabidopsis MSI4/FVE, which is a key epigenetic regulator of flowering time. However, the role of MSI4/FVE in stress response is still unknown. To investigate the function of GmFVE, we generated GmFVE‐OE hairy roots overexpressing GmFVE and GmFVE‐RNAi hairy roots with reduced GmFVE expression (Figure 3f and Figure S6). After salt stress with 100 mM NaCl, the plants with GmFVE‐RNAi hairy roots showed healthier leaves and greater root elongation than the control plants (Figure 3g, j), while the performance of the plants with GmFVE‐OE hairy roots was similar to the controls (Figure S6). Similarly, relative electrolyte leakage of leaves from plants with GmFVE‐RNAi hairy roots was lower than that in the control leaves (Figure 3k). Reduction in GmFVE also increased the survival rate in plants with GmFVE‐RNAi hairy roots under salt stress (Figure 3l). In addition, NaCl treatment increased ROS content in all roots. However, the GmFVE‐RNAi roots accumulated less ROS than the control roots (Figure 3m, n). To further investigate the role of GmFVE in salt response, we examined the expression of salt‐responsive genes regulated by GmNFYA in these GmFVE‐RNAi roots. Consistent with the salt‐tolerant phenotype, most of the downstream genes of GmNFYA were up‐regulated in GmFVE‐RNAi transgenic hairy roots (Figure 3o). These results demonstrate that GmFVE may function as a negative regulator of salt‐stress tolerance and affect a similar set of genes regulated by GmNFYA.
GmNFYA and GmFVE directly regulate the expression of salt‐responsive genes
Since the GmNFYA physically interacts with GmFVE and they play opposite roles in salt tolerance by regulating the similar set of salt‐responsive genes (Figure 1, 2, 3), we tested whether the two proteins are associated at the same region of gene promoters. Transgenic soybean hairy roots overexpressing 35S:GmNFYA‐GFP and 35S:GmFVE‐myc were generated and chromatin immunoprecipitation assay (ChIP) was performed. We found that GmNFYA mainly bound to the regions at P1 and P3 containing the CCAAT box in the promoters of GmLOS2 (LOW EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 2), GmXTH23‐2 (xyloglucan endotransglucosylase/hydrolase 23) and GmOMT (O‐methyltransferase 1). For the P2 regions without CCAAT box, there was no significant difference between transgenic and control samples (Figure 4b). Similarly, substantial enrichment of GmFVE to the P1 and P3 regions of GmLOS2, GmXTH23‐2 and GmOMT promoters was also detected in 35S:GmFVE‐myc hairy roots. Besides, GmFVE also bound to the P2 regions of GmXTH23‐2 and GmOMT (Figure 4c). These results indicate that GmNFYA and GmFVE may regulate their common target genes by directly binding to the gene promoters.
Figure 4.
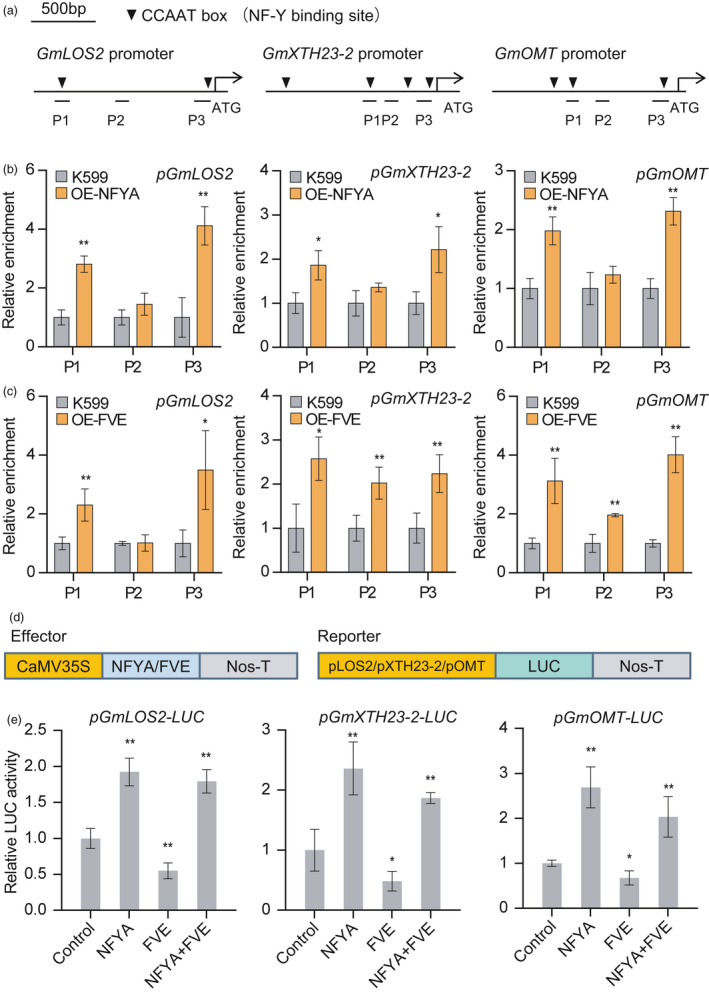
GmNFYA and GmFVE directly target to the promoters of salt‐responsive genes. (a) CCAAT box distribution in the promoters of GmLOS2, GmXTH23‐2 and GmOMT. P1 to P3 indicate fragments for ChIP–qPCR analysis. (b and c) ChIP analysis of binding of GmNFYA (b) and GmFVE (c) to the promoter regions of GmLOS2, GmXTH23‐2 and GmOMT. OE‐NFYA and OE‐FVE indicate transgenic hairy roots overexpressing GmNFYA‐GFP and GmFVE‐myc, respectively. K599 indicates the control hairy roots generated from A. rhizogenis K599. Error bars indicate SD (n = 3). (d) Schematic diagrams of the effector and reporter constructs used for transient expression assay in Arabidopsis protoplasts. (e) Regulation of GmNFYA and GmFVE on promoter activity of salt‐responsive genes. Error bars indicate SD (n = 3). Asterisks indicate significant difference compared with the corresponding controls (*, P < 0,05; **, P < 0.01)
To further determine whether GmNFYA and GmFVE could activate or repress the promoter activity of the target genes, we performed transient expression assay using a dual‐luciferase reporter assay in Arabidopsis mesophyll protoplast system (Figure 4d). The results showed that GmNFYA alone had a significant transcriptional activation effect on the downstream gene expression, whereas the promoter activities of these genes were largely inhibited by GmFVE. It is notable that this negative regulation of GmFVE on salt‐responsive genes was removed through co‐expression of GmNFYA (Figure 4e). All these results indicate that GmNFYA induces salt response through attenuating the repression activity of GmFVE.
GmHDA13 associates with GmFVE to negatively regulate salt tolerance in soybean
Arabidopsis MSI4/FVE acts as a component of HDACs complex, mainly forming transcriptional corepressor with RPD3‐like family HDACs (Jeon and Kim, 2011; Jung et al., 2013). Although RPD3‐like family HDACs have been demonstrated to function in response to salt stress, no MSI4/FVE‐HADCs complexes in this process have been reported so far. A recent study identified 28 HDAC genes in soybean genome, and 18 of these belong to RPD3‐like group (Yang et al., 2018). We preformed yeast two‐hybrid assay to screen the GmFVE‐interacting HDAC enzymes in this group. Of the HDACs tested, GmHDA13, GmHDA17 and GmHDA18 interacted strongly with GmFVE, while GmHDA5 and GmHDA9 interacted modestly with GmFVE in yeast cells (Figure 5a and Figure S7a). A phylogenetic analysis was performed to show the relationship of HADC proteins between Arabidopsis and soybean. GmHDA5 and GmHDA17 are grouped with AtHDA15; GmHDA9 is grouped with AtHDA19; GmHDA13 is grouped with AtHDA9; GmHDA18 is grouped with AtHDA2 (Figure S7b). Given that class I RPD3‐like HDAC, AtHDA9, plays a negative role in salt and drought responsiveness (Zheng et al., 2016), the function of its homolog, GmHDA13, may be conserved in soybean.
Figure 5.
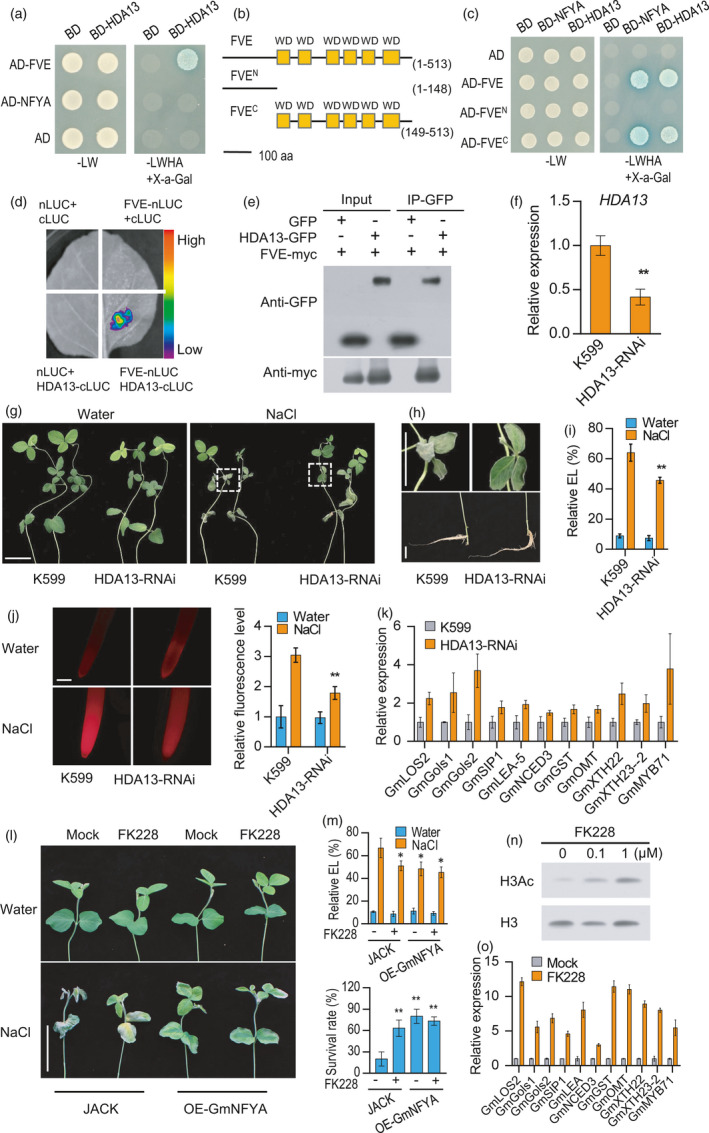
GmHDA13 associates with GmFVE to negatively regulate salt tolerance in soybean. (a) GmFVE interacts with GmHDA13 in a yeast two‐hybrid assay. (b) Different versions of GmFVE used for interaction analysis. Numbers indicate residue positions. WD, WD40 domain. aa, amino acids. (c) GmNFYA and GmHDA13 interact with the C terminal region of GmFVE in yeast cells. (d) Split‐luciferase complementation assay of GmFVE and GmHDA13 interaction in N. benthamiana leaf. (e) Co‐IP assay of the interaction between GmFVE and GmHDA13 in Arabidopsis protoplasts. (f) Expression of GmHDA13 in GmHDA13‐RNAi transgenic hairy roots. HDA13‐RNAi indicates transgenic hairy roots with reduced transcript level of GmHDA13. Bars indicate SD (n = 3). (g) Phenotype of the soybean plants with GmHDA13‐RNAi transgenic hairy roots after salt treatment. Scale bar = 5 cm. (h)The growth of leaf and root after salt treatment from plants in G. Bar = 2 cm. (i) Relative electrolyte leakage of trifoliolate leaves from the plants with GmHDA13‐RNAi transgenic hairy roots after 100 mM NaCl treatment for 3 d. Bars indicate SD (n = 4). (j) ROS levels of the GmHDA13‐RNAi transgenic and control roots after 80 mM NaCl treatment for 6h. Scale bar = 50 μm. (k) Expression of GmNFYA‐regulated genes in GmHDA13‐RNAi transgenic hairy root. Bars indicate SD (n = 3). (l) Effect of FK228 application on the performance of JACK and stable GmNFYA‐OE‐3 transgenic plants after 150 mM NaCl treatment for 3 days. Scale bar = 5 cm. (m) Relative electrolyte leakage and survival rates of JACK and stable GmNFYA‐transgenic plants with or without FK228 supply under salt‐stress condition. (n) Impact of FK228 on the histone acetylation level. The roots treated with different concentrations of FK228 for 16 h were harvested for Western blot assay. (o) Effect of FK228 application on the expression of salt‐responsive genes in roots of JACK. Asterisks indicate significant difference compared with the corresponding controls (*, P < 0,05; **, P < 0.01)
Interestingly, GmHDA13 only interacts with GmFVE, but not with GmNFYA in yeast two‐hybrid assay (Figure 5a). The assay using truncated GmFVE forms revealed that both GmNFYA and GmHDA13 interact with the C terminal region of GmFVE (Figure 5b, c). The interaction of GmFVE and GmHDA13 in planta was also verified by split‐luciferase complementation assay, consistent with results obtained in yeast cells (Figure 5d). We further conducted Co‐IP assay using Arabidopsis protoplasts to examine whether the interaction occurs in vivo. As a result, GmFVE protein was found to be immunoprecipitated with GmHDA13 (Figure 5e). These observations demonstrate that GmFVE associates with GmHDA13 to form protein complex.
We then investigated the effect of reduction in GmHDA13 level on salt tolerance using transgenic hairy root system. Compared with the control roots generated from wild‐type K599, GmHDA13 transcripts were decreased in GmHDA13‐RNAi hairy roots (Figure 5f). After NaCl treatment, the plants with GmHDA13‐RNAi roots performed better than control plants (Figure 5g, h), as revealed by reduced levels of relative electrolyte leakage in leaves and less ROS accumulation in root tips (Figure 5i, j). Because of the interaction of GmFVE and GmHDA13, we further determined whether GmHDA13 affects the similar set of downstream genes with GmFVE. We observed that the GmFVE‐regulated genes also showed higher expression in GmHDA13‐RNAi roots compared with control roots (Figure 5k). These results demonstrate that GmHDA13 also acts as a negative regulator of salt tolerance and may function in silencing salt‐responsive genes via associating with GmFVE.
We further employed FK228, an inhibitor of class I RPD3‐like HDACs (Ueda et al., 2017), to examine whether HDAC activity is related to salt response in soybean. The optimal concentration of this inhibitor that did not affect plant growth was selected and used for further phenotype analysis (Figure S8). Under salt stress, wild‐type plants treated with 0.1 μm FK228 were less sensitive to high‐salinity and exhibited lower relative electrolyte leakage of leaves and higher survival rate compared with the mock controls (Figure 5l, m). Unlike wild‐type plants, the application of FK228 could not further improve the performance of GmNFYA‐overexpression transgenic plants under NaCl treatment (Figure 5l, m). To confirm the inhibitory effect of FK228 on HDAC activity in soybean, histone acetylation level was evaluated after FK228 application. Notably, increased acetylation level of histone H3 at lysine 9 (H3K9) was observed with a dose effect after a 16‐htreatment of FK228 (Figure 5n). Histone acetylation is generally correlated with gene activation, and we, therefore, hypothesized that the increased histone acetylation caused by FK228 treatment may lead to up‐regulation of salt‐responsive genes. Similar to the observation in GmHDA13‐RNAi roots (Figure 5k), the expression of genes regulated by GmHDA13 was also up‐regulated in FK228‐treated plants (Figure 5o), indicating that FK228 conferred salt tolerance in soybean by up‐regulation of salt‐responsive genes via histone acetylation. All these results suggest that GmHDA13 may work as a negative regulator of salt tolerance through histone deacetylation.
GmNFYA antagonizes the action of GmFVE‐GmHDA13 complex and modulates histone acetylation
Since GmFVE interacts with both GmNFYA and GmHDA13, GmFVE may act as a bridge to make a connection between GmNFYA and GmHDA13. Unlike GmNFYA, whose expression is highly induced by NaCl treatment, the previous and our present study found that GmFVE and GmHDA13 transcripts were almost unchanged under salt stress (Yang et al., 2018; Table S3). It is possible that GmFVE and GmHDA13 form a co‐repression complex under normal condition, while GmNFYA is highly induced by salt stress and attenuates the activity of GmFVE and GmHDA13 in silencing salt‐responsive genes. To test this hypothesis, we performed yeast three‐hybrid assay. The coding region of the GmFVE was cloned into the pGADT7 vector, and the pBridge vector was used to express both GmNFYA and GmHDA13. As a result, the interaction between GmFVE and GmHDA13 when expressed alone was strong but substantially weakened upon co‐expression with GmNFYA (Figure 6a). To further corroborate this result, split‐luciferase complementation assay was performed. The assay revealed that GmFVE can physically interact with GmHDA13, while co‐expression of GmNFYA reduced this interaction (Figure 6b, c). These results most likely indicate that GmNFYA can disrupt the interaction between GmFVE and GmHDA13 to antagonize the action of GmFVE‐GmHDA13 complex.
Figure 6.
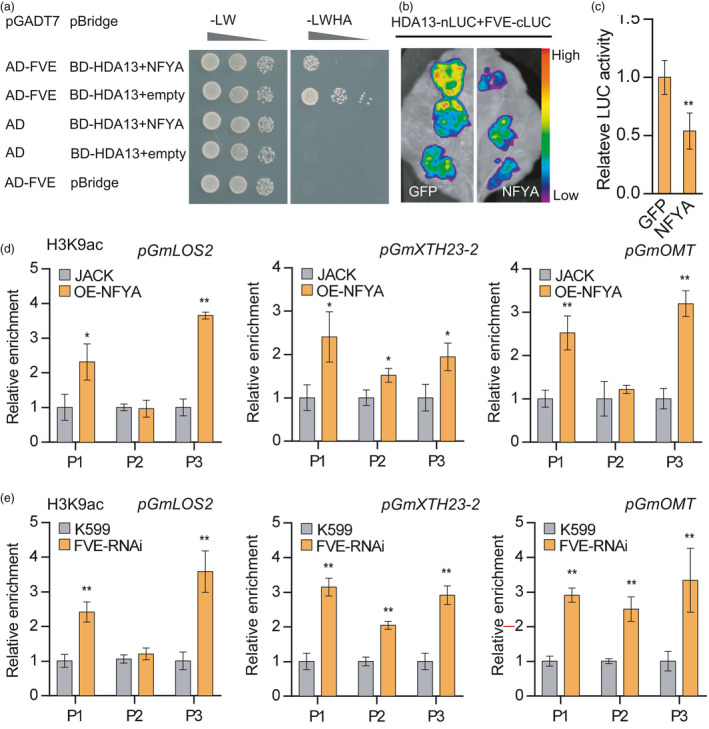
GmNFYA and GmFVE modulate histone H3 acetylation level at the chromatins of their target genes. (a) Yeast three‐hybrid assay analysing the GmFVE and GmHDA13 interaction in the presence or absence of co‐expressed GmNFYA. Yeasts cells were grown on SD‐Ade‐His‐Leu‐Trp medium to test the interaction and competition, respectively. (b) Split‐luciferase complementation assay showing the interaction of GmFVE and GmHDA13 is weakened by co‐expressed GmNFYA in N. benthamiana leaves. (c) Quantitative analysis of luminescence intensity in B. Bars indicate SD (n = 6). (d and e) ChIP analysis of H3K9ac levels at the promoter of GmLOS2, GmXTH23‐2 and GmOMT in stable GmNFYA‐overexpressing transgenic plants (d) and plants with GmFVE‐RNAi hairy roots (e). OE‐NFYA indicates stable GmNFYA‐overexpression transgenic soybean plants. FVE‐RNAi indicates plants with transgenic hairy roots showing reduced level of GmFVE. The indicated genomic regions (See Figure 4a) were analysed by ChIP‐qPCR. Error bars indicate SD (n = 3). Asterisks indicate significant difference compared with the corresponding controls (*, P < 0.05; **, P < 0.01)
We next investigated whether increased expression of salt‐responsive genes in GmNFYA‐OE and GmFVE‐RNAi plants is associated with increased histone acetylation by ChIP‐qPCR analysis with antibodies against H3K9ac. Compared with the controls, H3K9ac levels in chromatins at promoter P1 and P3 regions with CCAAT box of GmLOS2, GmXTH23‐2 and GmOMT were substantially increased in the plants with GmNFYA‐OE and GmFVE‐RNAi hairy roots, suggesting that GmNFYA and GmFVE regulate the expression of salt‐responsive genes via modulating H3K9ac modification (Figure 6d, e).
Correlation between GmNFYA promoter haplotypes and salt tolerance in cultivated soybean accessions
We compared the promoter sequences of GmNFYA from various soybean accessions to see whether there are any variations and whether these variations have any correlation with GmNFYA expression and salt tolerance. The 2.5 kb GmNFYA promoter sequences in 132 cultivated soybean accessions were sequenced and analysed. Three major haplotypes were identified through phylogenetic analysis, and haplotype I includes 86 accessions, haplotype II contains 18 accessions, and haplotype III contains 28 accessions (Figure 7a and Figure S9). We compared the GmNFYA expression in haplotype I accession Nannong 1138‐2 (NN) and haplotype II accession Kefeng 1 (KF) after salt treatment. NN (Hap 1) showed higher and faster induction of GmNFYA expression in both leaves and roots than KF (Hap 2) plants (Figure 7b).
Figure 7.
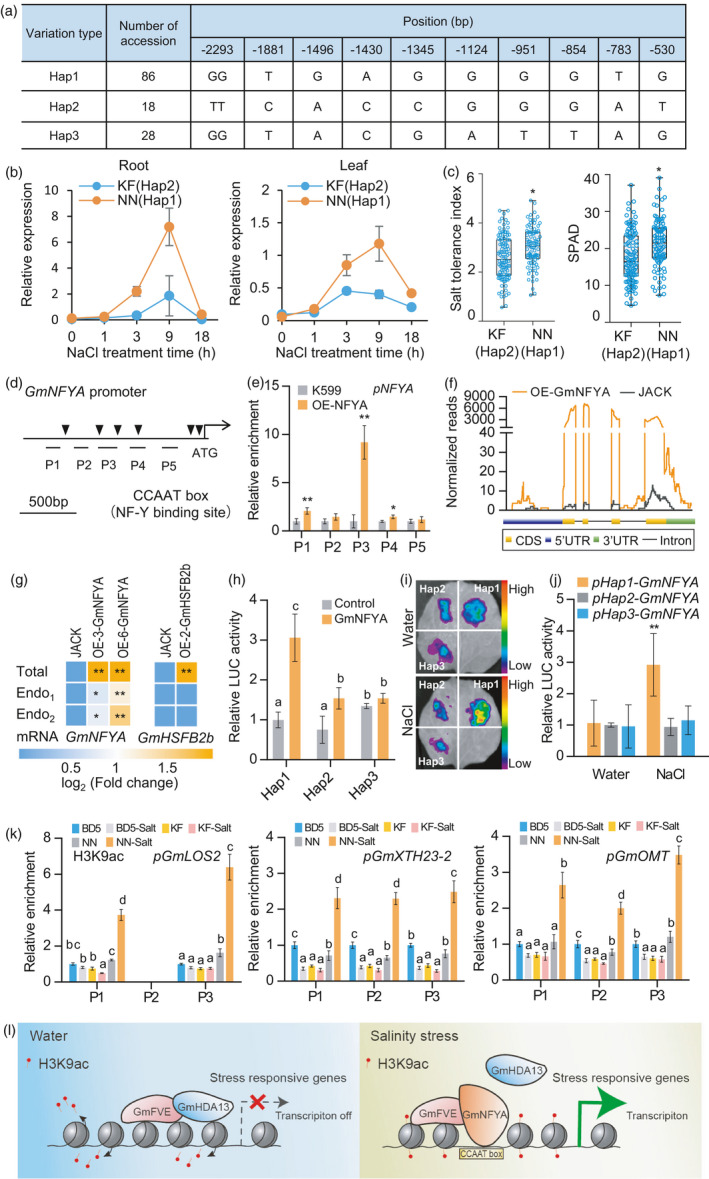
Correlation between GmNFYA expression and salt tolerance in cultivated soybeans. (a) Haplotype variation of cultivated soybeans in the GmNFYA promoter region. The start codon site is set at the ‘0’ position. (b) GmNFYA expression in response to salt stress in leaf and root of Kefeng1 (KF, Hap 2) and Nannong 1138‐2 (NN, Hap 1). Bars indicate SD (n = 3). (c) Salt tolerance index and chlorophyll content in the recombinant inbred lines (RILs) derived from a cross between NN (Hap 1) and KF (Hap2). The numbers of lines with Hap1 and Hap2 are 96 and 109, respectively. SPAD represents the chlorophyll content in the leaves. (d) Distribution of CCAAT box in the promoter region of GmNFYA. P1‐P5 are used in the following ChIP‐qPCR analysis. (e) ChIP‐qPCR assay showing that GmNFYA associates with its own promoters in GmNFYA‐GFP transgenic hairy root. Bars indicate SD (n = 3). (f) RNA‐seq analysis showing normalized reads of GmNFYA transcript in coding region (CDS) and untranslated region (UTR) in stable GmNFYA‐OE‐3 transgenic soybean and JACK roots. (g) Heatmaps showing the activation of GmNFYA on the endogeous GmNFYA gene examined by RT‐qPCR. Specific primers are designed at the coding region (Total) and 3’‐UTR (Endo1 and Endo2) of GmNFYA gene to distinguish total and endogenous GmNFYA expression. Stable GmHSFB2b transgenic plants are used as controls. (h) Activation ability of GmNFYA on the promoters of three haplotypes in Arabidopsis protoplast transient expression assay. Bars indicate SD (n = 3). (i) Promoter activity of GmNFYA haplotypes in the presence of GmNFYA by transient expression in N. benthamiana leaves. The LUC reporter gene fused with GmNFYA was driven by each haplotype and the luminescence intensity was determined after water or 300 mM NaCl treatment overnight. (j) Quantitative analysis of luminescence intensity in (i). Bars indicate SD (n = 4). (k) ChIP analysis of H3K9ac levels at the promoter of GmLOS2, GmXTH23‐2 and GmOMT in soybean plants belonging to three kinds of haplotypes, such as NN (Hap 1), KF (Hap 2) and BD5 (Hap 3). Bars indicate SD (n = 3). (l) Working model of GmNFYA in response to salinity stress. Under normal condition, GmFVE may interact with GmHDA13 to inhibit stress‐responsive genes through histone deacetylation. Under salt stress, GmNFYA accumulated and competitively interacted with GmFVE to remove GmHDA13 function for maintenance of histone acetylation and activation of stress‐responsive genes for salt tolerance. For (c), (e), (g) and (j), asterisks indicate significant difference compared with the corresponding controls (*, P < 0.05; **, P < 0.01). For (h) and (k), the lowercase ‘a’ to ‘d’ indicate significant differences (P < 0.05 or P < 0.01).
Since GmNFYA transcript can be induced to a higher level in NN than that in KF, the correlation between GmNFYA promoter haplotypes, namely Hap 1 and Hap 2, and salt tolerance was further studied in the recombinant inbred lines (RILs) derived from NN and KF. Totally, 96 lines with haplotype I and 109 lines with haplotype II were identified and subjected to 300 mM NaCl treatment for one week at the V1 stage. Salt tolerance index was used for evaluating the plant performance under high‐salinity condition as described previously (Bian et al., 2020). The results showed that salt tolerance index in NN and plants with haplotype I promoter was significantly higher than that in KF and plants with haplotype II promoter (Figure 7c). In addition, haplotype I lines also exhibited higher chlorophyll contents in leaves compared with haplotype II lines under salt stress (Figure 7c). These results indicate that promoter haplotype I likely contributes more to salt resistance than the haplotype II allele.
To investigate whether the GmNFYA expression difference was caused by the promoter variations, we further compared the promoter activity of different haplotypes using tobacco transient expression assay. The 2 kb promoter sequences of each haplotype promoter were fused to LUC reporter gene and transfected into tobacco leaves. Unexpectedly, there was no difference in promoter activity among the three haplotypes under water and salinity conditions (Figure S10). One possible explanation is that expression difference in GmNFYA mRNA in the presence of salt stress was regulated by additional regulators. Then, the promoter of GmNFYA was analysed, and several putative CCAAT box for potential nuclear factor Y binding upstream of the start codon were found (Figure 7d). We speculate that GmNFYA may target to its own promoter for self‐activation. To test this possibility, ChIP‐qPCR was performed to determine whether GmNFYA can directly bind to the regions of GmNFYA promoter. The result showed that GmNFYA selectively interacts with several regions especially at P3, revealing that GmNFYA directly binds to its own promoter (Figure 7e).
We further tested whether GmNFYA could activate the expression of the endogenous GmNFYA by binding to its own promoter. Based on our RNA‐seq data, we compared the normalized reads of GmNFYA transcript in the genomic region from 35S‐GmNFYA‐transgenic lines OE‐3 with that in control JACK and found that GmNFYA mRNA greatly increased not only in coding region but also in 5’‐untranslated region (5’‐UTR) and 3’‐UTR (Figure 7f). Specific primers were then designed to distinguish total (coding region) and endogenous (3’‐UTR) GmNFYA expression, respectively. When quantitative PCR using the above specific primers was performed, significant up‐regulation of both total and endogenous GmNFYA mRNA was observed in OE‐3 and OE‐6 plants (Figure 7g). However, as a control, endogenous GmHSFB2b expression could not be activated by GmHSFB2b in GmHSFB2b overexpression lines (Figure 7g; Bian et al., 2020). According to this observation, we further performed dual‐luciferase reporter assay in Arabidopsis protoplasts to investigate the effect of GmNFYA on the activity of three haplotype promoters. In the assay, the 35S : GmNFYA‐myc vector was constructed as the effector, and the LUC reporter gene was driven by each haplotype promoter. Our result showed that GmNFYA exhibited stronger transcriptional activation activity on the haplotype I promoter than those on the other two haplotypes (Figure 7h). Moreover, the promoter activity of GmNFYA in the presence of GmNFYA under high salinity was further compared in the tobacco leaf transient assay. Under salt condition, the promoter activity from haplotype I was higher than those from the other two haplotypes (Figure 7i, j). These results indicate that GmNFYA can activate the haplotype I promoter of GmNFYA gene with the highest promoter activity under salt stress for salt tolerance in soybean plants.
Three soybean accessions NN (Hap 1), KF (Hap 2) and BD5 (Hap 3, Figure S9), belonging to the three kinds of haplotypes, were tested for histone acetylation in the promoter regions of the three target genes. ChIP signals in promoter regions of GmLOS2, GmXTH23‐2 and GmOMT were enriched by H3K9ac antibody, except for the P2 region in GmLOS2. A 6‐h treatment of 150 mM NaCl was introduced to test whether H3K9ac levels of these downstream genes would change after salt stress. For BD5, H3K9ac levels in most regions were decreased after salt treatment. For KF, H3K9ac levels of target genes were not changed, except that H3K9ac level in the P1 region of GmLOS2 dropped after salt treatment. For NN, H3K9ac levels in the three target genes were all increased by salt treatment (Figure 7k). These results indicate that the histone in the target genes was acetylated differently among the three haplotypes, especially during salt stress.
Discussion
In this study, we discovered that a nuclear factor Y subunit GmNFYA from soybean functions in salt‐stress tolerance. GmNFYA can interact with GmFVE to reduce the association between GmFVE and GmHDA13 and hence weaken the histone deacetylation to maintain the H3K9 acetylation level for activation of salt‐responsive genes. A haplotype I promoter of the GmNFYA has been identified and can be activated by GmNFYA with high promoter activity under salt stress. This haplotype may be further selected during future breeding effort. Our study reveals a novel role for GmNFYA in salt resistance in addition to its function as an oil regulator in seeds (Lu et al., 2016).
NF‐YA, as one of the subunits of the NF‐Y complex, plays diverse functions in a wide range of processes in vegetative growth and reproductive development, as well as the response to environmental cues in plants (Li et al., 2013; Li et al., 2008; Mu et al., 2013). The present GmNFYA is mainly expressed in soybean seeds but less in other organs (Lu et al., 2016). However, its expression is highly induced by salt stress in both leaves and roots (Figure 1a), suggesting that GmNFYA may exert a role in stress response. Indeed, the overexpression of GmNFYA in both stable transgenic soybean plants and plants with transgenic hairy roots supports that GmNFYA has a positive role in response to salinity stress (Figure 1). Consistent with this fact, RNA‐seq analysis also showed that approximately half of the GmNFYA‐regulated genes are involved in salt response. GmNFYA activates genes involved in hormone‐mediated pathways in roots and hairy roots, such as the key enzyme gene in ABA biosynthesis, NCED3 (NINE CIS‐EPOXYCAROTENOID DIOXYGENASE 3) (Figure 2). The stress‐related hormones, such as ABA and strigolactones, can mediate root‐to‐shoot signal transduction and finally confer stress tolerance in aerial organs (Iuchi et al., 2001; Kuromori et al., 2018; Wei et al., 2017).
In animals, NF‐Y complexes have been shown to be essential both for gene activation and for repression of some of the NF‐Y targets associated with negative histone marks (Donati et al., 2008; Oldfield et al., 2014). Here, we demonstrated that GmNFYA is associated with promoters of its downstream genes and activates target gene expression by enhancing histone acetylation levels at their chromatins (Figure 4b and Figure 6d). Epigenetic control of NF‐Ys has also been described during flowering in Arabidopsis, where NF‐YCs directly activates the expression of SOC1, a major floral pathway integrator, by recruiting H3K27 demethylase REF6 (Hou et al., 2014). A recent study has shown that NF‐YCs function as transcriptional corepressors by interacting with HDA15 to modulate light‐controlled hypocotyl elongation (Tang et al., 2017). However, the partners of NF‐YA subunit and relevant epigenetic regulation mechanism in salt response remain unknown. In the present study, GmFVE, a MSI4/FVE homolog, was identified as the GmNFYA‐interacting protein and functioned as a negative regulator of salt tolerance (Figure 3). Consistently, increased expression and higher histone acetylation levels of salt‐responsive genes in the plants with transgenic GmFVE‐RNAi hairy roots are notified (Figure 3o and 6e). These results support that the GmNFYA may interact with GmFVE to inhibit GmFVE function in histone deacetylation.
MSI4/FVE is a component of HDAC corepressor complexes and mediates histone deacetylation and transcriptional silencing of the FLC gene and several cold‐responsive genes with HDA6 (Jeon and Kim, 2011; Jung et al., 2013). MSI1‐HDA19 complex has been reported to fine‐tune ABA signalling in Arabidopsis (Mehdi et al., 2016). Our data showed that GmFVE interacts with at least five HDACs: GmHDA5, GmHDA9, GmHDA13, GmHDA17 and GmHDA18 (Figure 5a and Figure S6). Among these HDACs, GmHDA13 and GmHDA17, which are most associated with GmFVE, are the homologs of HDA9 and HDA15 in Arabidopsis, respectively. Class I HDAC HDA9 is the chromatin proteins that negatively regulate salt‐stress tolerance, while class II HDA15 is involved in positive regulation of salt tolerance (Ueda et al., 2017; Zheng et al., 2016). Our study revealed a negative role of GmHDA13 in salt‐stress response using transgenic soybean hairy root system (Figure 5g, h), suggesting that GmHDA13 associates with GmFVE to co‐repress salt response in soybean. Furthermore, treatment with FK228, an inhibitor of class I HDAC (Ueda et al., 2017), also leads to activation of GmHDA13‐affected genes and improvement of plant performance in the presence of salt stress via inducing H3 hyperacetylation, indicating that GmHDA13 is a possible target enzyme to enhance salinity stress tolerance (Figure 5k–o). Since GmFVE binds to the promoters of several salt‐responsive genes, and reduction in GmFVE leads to increased H3K9 acetylation levels at these genes (Figures 3o and 6e), it is likely that GmFVE recruits GmHDA13 via protein interaction to repress gene expression by maintaining low histone acetylation levels at the chromatins of these target genes. In addition, MSI4/FVE has been demonstrated to interact with H3K4 demethylase, such as FLD, LDL1, LDL2 and LDL3, to repress the expression of FLC (Jung et al., 2013; Yu et al., 2016). It is intriguing to examine whether GmFVE also mediates salt response via regulation of histone methylation.
Identifying genetic variations from diverse germplasm resources is an important strategy for crop improvement. Several natural variations associated with important agronomic traits in crops were identified using various approaches, such as associative transcriptomic, whole‐genome resequencing and GWAS analysis. For example, in soybean, haplotype variation in the promoters of GmZF351, GmOLEO1 and GmHSFB2b influence the seed oil content and salt tolerance by driving different levels of gene expression (Bian et al., 2020; Li et al., 2017a; Zhang et al., 2019a). Natural variation in expression of the HECT E3 Ligase UPL3 is correlated with seed size and crop yields in Brassica napus (Miller et al., 2019). In rice, a natural allele of C2H2‐type TF Bsr‐d1, which causes a single nucleotide change in the promoter, contributes to broad‐spectrum blast resistance (Li et al., 2017b). In the present study, three haplotypes were identified in the GmNFYA promoter region (Figure 7a and Figure S9). Among them, haplotype I from salt‐tolerant soybean cultivar NN has the highest promoter activity after self‐activation by GmNFYA under salt stress (Figure 7i, j). Variation in promoter activity may lead to different levels of GmNFYA expression and influence salt tolerance in soybean accessions. Analysis of the contribution of GmNFYA promoter haplotypes to salt tolerance using the RIL population derived from the salt‐tolerant cultivar NN (with haplotype I) and salt sensitive cultivar KF (with haplotype II) supports the suggestion that haplotype I promoter correlates with strong self‐activated promoter activity, high gene expression and enhanced salt tolerance (Figure 7). It should be noted that, under normal or salt‐stress condition, the three promoter haplotypes did not show much difference in promoter activity (Figure S10). The inclusion of the GmNFYA in the assays drastically increased the haplotype 1 activity, suggesting the presence of a self‐activated transcription loop (Figure 7i, j). In addition, considering that GmNFYA allele with haplotype I promoter was still lacking in one third of cultivated soybean accessions examined, the introduction of the haplotype 1 allele into these cultivars in future breeding efforts may have potential benefits in the improvement of plant performance under salt stress (Figure 7a).
It should be mentioned that while we find a role for GmNFYA in salt tolerance in soybean, our previous study identified the protein as a regulator for lipid biosynthesis in soybean seeds (Lu et al., 2016). We propose that the GmNFYA may normally function in seeds since it shows very high expression in this organ. However, under stresses, this gene can be induced to exert tolerance in plants. This feature may represent a strategy for plants to use the gene efficiently at different developmental stages and/or environmental conditions.
Taken together, we propose that, under normal condition, GmFVE and GmHDA13 function in a histone deacetylation complex to maintain low transcript levels of salt‐responsive genes. Upon salt stress, GmNFYA accumulates and directly interacts with GmFVE to destabilize the GmFVE‐GmHDA13 complex, leading to histone acetylation and activation of salt‐responsive genes for salt tolerance (Figure 7l). A haplotype I promoter of GmNFYA is also identified with the highest self‐activation activity under salinity stress. All these factors/elements may be potential targets in genetic manipulation for salt tolerance in soybean and other crops.
Methods
Plant materials
Soybean plants were grown under long‐day condition (14h light/10h dark) at 25 °C in a greenhouse. Arabidopsis thaliana ecotype Columbia‐0 (Col‐0) and Nicotiana benthamiana were grown in a greenhouse at 22 °C with a photoperiod of 16 h light/8 h dark.
Plant transformation
The coding sequence of GmNFYA (Glyma02g303800) was cloned into pCambia1301 vector to generate overexpression construct. The resulting construct was transferred into soybean cultivar Jack (JACK) using the A. tumefaciens‐mediated cotyledon node transformation method (Bian et al., 2020). Positive transgenic soybean plants were selected with CP4 EPSPS test paper (You Long Biotech), and homozygous stable transgenic lines were used for further analysis.
Transcriptome analysis of transgenic soybean plants
The seedlings of JACK and GmNFYA‐overexpressing line OE‐3 at the V1 stage were treated with 150 mM NaCl for 0h or 6h. Roots and leaves were collected, and total RNA was extracted using TRIzol reagent (Invitrogen). RNA‐sequencing (RNA‐seq) was performed by Novogene (Beijing, China). There were three biological replicates for each sample. After adapter clipping and quality filter, clean data were remapped to the soybean genome sequence (Williams 82) using Hisat2 software and analysed with HTSeq (version). The edgeR software was used to analyse the differentially expressed genes (DEGs), which were defined as those having at least twofold change in expression compared with the corresponding controls (FDR<0.05). Gene ontology (GO) analysis was performed using agriGO v2.0 (Tian et al., 2017).
Production of transgenic hairy roots
Six‐day‐old soybean seedlings (Glycine max, Kefeng1) were infected with A. rhizogenes harbouring different recombinant construct through injection of the hypocotyls, as described previously (Wang et al., 2015). Transgenic hairy roots were generated from the infection sites 10‐12 d after infection and the original roots were removed. Then, the seedlings with transgenic hairy roots were transferred to water for 6 d for further analysis.
ChIP assay
ChIP assay was conducted as described previously (Bian et al., 2020). Briefly, about 3 g of 35S:GmNFYA‐GFP or 35S:GmFVE‐myc transgenic hairy roots were collected for fixation in 1% formaldehyde. Soluble chromatin was subjected to ChIP with anti‐GFP (Abcam) or anti‐myc (Abcam) antibodies. The protein/DNA complexes were collected on protein G‐conjugated magnetic beads (Invitrogen). Protein/DNA cross‐links were reversed, and DNA was purified and used for qPCR analysis.
Split‐luciferase complementation and BiFC assay
For the split‐luciferase complementation assay, the full‐length cDNA of GmNFYA GmFVE (Glyma.09G063100) and GmHDA13 (Glyma.12G086700) was amplified and cloned into pCAMBIA‐1300nLUC and pCAMBIA‐1300cLUC vectors, respectively, and co‐infiltrated into N. benthamiana leaves. LUC activities were detected 48 h post‐infiltration using Night‐SHADE LB 985 (Berthold). For the BiFC analysis, the GmNFYA and GmFVE coding sequences were cloned into pSPYCE(M) and pSPYNE(R)73. The resulting constructs were then introduced into A. tumefaciens strain EHA105 for transient expression in N. benthamiana leaves. YFP fluorescence was analysed with a confocal laser‐scanning microscope (Carl Zeiss; LSM710).
Yeast two‐hybrid assay
The coding regions of GmNFYA, GmFVE and GmHDA13 were amplified and cloned into pGBKT7 and pGADT7 vectors, respectively. The yeast strain AH109 was transfected with different constructs, and the transfected cells were grown on SD/‐Ade/‐His/‐Leu/‐Trp/X‐α‐Gal medium for interaction test.
Yeast three‐hybrid assay
The pBridge vector was used to express both GmHDA13 and GmNFYA. The pGADT7 vector was used to express GmFVE. The recombinant constructs were co‐transfected into yeast AH109 cells. Yeast transformation and growth assay were performed as described in the Yeast Protocols Handbook (Clontech).
Co‐immunoprecipitation assay
The cDNA of GmNFYA, GmFVE and GmHDA13 was cloned into pCAMBIA‐2300 vector to generate 35S:GmNFYA‐myc, 35S:GmFVE‐GFP and 35S:HDA13‐myc constructs. Pairwise constructs were transfected into Arabidopsis protoplasts as described previously (Wei et al., 2017). After incubation at 22 °C for 16 h in darkness, transformed protoplasts were harvested, and total proteins were extracted in Rippa buffer (Sigma). The protein extracts were purified with anti‐myc/anti‐GFP antibody (Sigma/Chromotek) and the immunoprecipitated proteins were separated by SDS–PAGE gel and detected by anti‐myc (Abmart) or anti‐GFP (Abmart) antibodies.
Immunoprecipitation‐mass spectrometry (IP‐MS) assay
For immunoprecipitation followed by mass spectrometry, soybean transgenic hairy roots harbouring 35S:GmNFYA‐GFP (NFYA‐GFP) were generated and GFP‐overexpressing hairy roots were used as control. Three gram of transgenic hairy roots were harvested and total proteins were extracted in Rippa buffer (Sigma). Anti‐GFP Trap‐A beads (Chromotek) was used for IP. After IP, the proteins were separated by 10% SDSS‐PAGE. The stained protein band was cut and in‐gel digestion was performed by Novogene (Beijing, China). Mass spectrometry measurements were performed on a Q ExactiveTM series mass spectrometer (Thermo Fisher). The resulting spectra were searched against UNIPROT protein database, using soybean as a target organism. Gene ontology (GO) and InterPro (IPR) functional analysis were conducted using the interproscan programme against the non‐redundant protein database (including Pfam, PRINTS, ProDom, SMART, ProSite, PANTHER), and the databases of COG (Clusters of Orthologous Groups) and KEGG (Kyoto Encyclopedia of Genes and Genomes) were used to analyse the protein family and pathway. Proteins uniquely identified in NFYA‐GFP sample but not in control sample were taken into account.
Transcriptional activation assay
The 3 Kb promoter regions of GmNFYA or GmNFYA‐regulated genes were cloned into pCambia2300 vectors containing LUC sequence to generate reporter vectors. Transient expression assay was performed in N. benthamiana leaves or in Arabidopsis mesophyll protoplasts as described previously (Li et al., 2017).
FK228 treatment
HDACs inhibitor FK228 was dissolved in dimethylsulfoxide (DMSO) and DMSO treatment served as the control. Soybean plants were treated with FK228 for 16 h and subsequently transferred to 150 mM NaCl for 5 days to observe phenotype and calculate survival rate.
Measurement of physiological parameters and ROS examination
Relative electrolyte leakage assay was performed as described previously (Wei et al., 2017). DAB (3, 3′‐Diaminobenzidine) staining was used for analysis of reactive oxygen species (ROS) as described before (Bian et al., 2020). ROS staining was performed as described before (Wang et al,l., 2015).
Phylogenetic analysis
For the analysis of GmNFYA promoter, 2.5 kb promoter sequences from 132 cultivated soybean accessions were sequenced and analysed. For the analysis of HDA proteins, protein sequence of 18 soybean HDACs and 13 Arabidopsis HDACs were analysed. The alignment was performed with ClustalX 2.0 software. Phylogenetic trees were generated by MEGA 5 based on the neighbour‐joining method with PHYLIP (Tamura et al., 2011). The number of bootstrap replications was 1000.
Accession numbers
Sequence data from this article can be obtained from the Plant Genome Database (www.plantgdb.org) under the following accession numbers: GmNFYA (Glyma.02G303800), GmFVE (Glyma.09G063100) and GmHDA13 (Glyma.12G086700).
Author Contributions
LL and WW performed most of the work and initiated the draft. J‐JT helped to conduct phenotypic analysis of RILs. LX identified candidate genes. X‐HB and YH participated in soybean cultivation. TC and C‐CY were involved in data analysis. J‐SZ, S‐YC and W‐KZ conceived of the study, obtained funding and revised the final version of the manuscript. All authors read and approved the final article.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1. Phenotype of stable GmNFYA‐transgenic soybean plants.
Figure S2. Performance of GmNFYA‐transgenic and control plants under salt treatment when grown in a same pot.
Figure S3. ROS levels in GmNFYA‐transgenic hairy roots.
Figure S4. GmNFYA regulates a set of genes involved in response to salt stress in leaf.
Figure S5. Identification of the GmNFYA‐interacting proteins by Immunoprecipitation‐mass spectrometry.
Figure S6. Knockdown of GmFVE enhances salt tolerance of the plants with FVE‐RNAi transgenic hairy root.
Figure S7. Interactions of GmFVE and RPD3‐like histone deacetylases (HDACs) and the cluster analysis of HDACs.
Figure S8. Application of HDAC inhibitor FK228 increases salt tolerance of Kefeng1 seedlings.
Figure S9. Phylogenetic tree of GmNFYA promoter sequences from 132 cultivated soybeans.
Figure S10. There is no significant difference in promoter activity among three GmNFYA haplotypes under water and salt conditions.
Table S1. List of primers used in this study.
Table S2. GmNFYA‐interacting proteins identified by IP‐MS.
Table S3. Transcriptome data of GmNFYA‐transgenic soybean plant.
Acknowledgements
This work is supported by the CAS project (ZDRW‐ZS‐2019‐2), NSFC project (32090063, U1906203 and 31971896) and the State Key Lab of Plant Genomics, IGDB, CAS.
Lu, L. , Wei, W. , Tao, J.‐J. , Lu, X. , Bian, X.‐H. , Hu, Y. , Cheng, T. , Yin, C.‐C. , Zhang, W.‐K. , Chen, S.‐Y. and Zhang, J.‐S. (2021) Nuclear factor Y subunit GmNFYA competes with GmHDA13 for interaction with GmFVE to positively regulate salt tolerance in soybean. Plant Biotechnol. J., 10.1111/pbi.13668
Contributor Information
Wan‐Ke Zhang, Email: wkzhang@genetics.ac.cn.
Shou‐Yi Chen, Email: sychen@genetics.ac.cn.
Jin‐Song Zhang, Email: jszhang@genetics.ac.cn.
Data Availability Statement
The clean reads of RNA‐seq in this paper have been deposited in the GenBank database (GEO accession number: GSE173640).
References
- Alexandre, C. , Moller‐Steinbach, Y. , Schonrock, N. , Gruissem, W. and Hennig, L. (2009) Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol. Plant, 2, 675–687. [DOI] [PubMed] [Google Scholar]
- Bian, X.H. , Li, W. , Niu, C.F. , Wei, W. , Hu, Y. , Han, J.Q. , Lu, X. et al . (2020) A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 225, 268–283. [DOI] [PubMed] [Google Scholar]
- Chen, L.T. , Luo, M. , Wang, Y. and Wu, K. (2010) Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. , Zhao, Y. , Zhuo, C. , Lu, S. and Guo, Z. (2015) Overexpression of a NF‐YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 13, 482–491. [DOI] [PubMed] [Google Scholar]
- Donati, G. , Gatta, R. , Dolfini, D. , Fossati, A. , Ceribelli, M. and Mantovani, R. (2008) An NF‐Y‐dependent switch of positive and negative histone methyl marks on CCAAT promoters. PLoS One, 3, e2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, R. , Qu, Y. , Guo, Y. , Yu, L. , Liu, Y. , Jiang, J. , Chen, J. et al . (2014) Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80, 937–950. [DOI] [PubMed] [Google Scholar]
- Han, X. , Tang, S. , An, Y. , Zheng, D. , Xia, X. and Yin, W. (2013) Overexpression of the poplar NF‐YB7 transcription factor confers drought tolerance and improves water‐use efficiency in Arabidopsis. J. Exp. Bot. 64, 4589–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin, M. , Ebel, C. , Ngom, M. , Laplaze, L. and Masmoudi, K. (2016) New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 7, 1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig, L. , Bouveret, R. and Gruissem, W. (2005) MSI1‐like proteins: An escort service for chromatin assembly and remodeling complexes. Trends Cell. Biol. 15, 295–302. [DOI] [PubMed] [Google Scholar]
- Hollender, C. and Liu, Z. (2008) Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 050, 875–885. [DOI] [PubMed] [Google Scholar]
- Hou, X. , Zhou, J. , Liu, C. , Liu, L. , Shen, L. and Yu, H. (2014) Nuclear factor Y‐mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis . Nat. Commun. 5, 4601. [DOI] [PubMed] [Google Scholar]
- Iuchi, S. , Kobayashi, M. , Taji, T. , Naramoto, M. , Seki, M. , Kato, T. , Tabata, S. et al. (2001) Regulation of drought tolerance by gene manipulation of 9‐cis‐epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis . Plant J. 27, 325–333. [DOI] [PubMed] [Google Scholar]
- Jeon, J. and Kim, J. (2011) FVE, an Arabidopsis homologue of the retinoblastoma‐associated protein that regulates flowering time and cold response, binds to chromatin as a large multiprotein complex. Mol. Cells, 32, 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J.H. , Park, J.H. , Lee, S. , To, T.K. , Kim, J.M. , Seki, M. and Park, C.M. (2013) The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short‐term cold stress in Arabidopsis . Plant Cell, 25, 4378–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.M. , Sasaki, T. , Ueda, M. , Sako, K. and Seki, M. (2015) Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori, T. , Seo, M. and Shinozaki, K. (2018) ABA Transport and Plant Water Stress Responses. Trends Plant Sci. 23, 513–522. [DOI] [PubMed] [Google Scholar]
- Lam, H.M. , Xu, X. , Liu, X. , Chen, W. , Yang, G. , Wong, F.L. , Li, M.W. et al. (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. [DOI] [PubMed] [Google Scholar]
- Li, Q.T. , Lu, X. , Song, Q.X. , Chen, H.W. , Wei, W. , Tao, J.J. , Bian, X.H. et al. (2017a) Selection for a zinc finger protein contributes to seed oil increase during soybean domestication. Plant Physiol. 173, 2208–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.X. , Oono, Y. , Zhu, J. , He, X.J. , Wu, J.M. , Iida, K. , Lu, X.Y. et al. (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell, 20, 2238–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Zhu, Z. , Chern, M. , Yin, J. , Yang, C. , Ran, L. , Cheng, M. et al. (2017b) A natural allele of a transcription factor in rice confers broad‐spectrum blast resistance. Cell, 170, 114–126. [DOI] [PubMed] [Google Scholar]
- Li, Y.J. , Fang, Y. , Fu, Y.R. , Huang, J.G. , Wu, C.A. and Zheng, C.C. (2013) NFYA1 is involved in regulation of postgermination growth arrest under salt stress in Arabidopsis . PLoS One, 8, e61289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. Zou, H.‐F. , Wang, H.‐W. , Zhang, W.‐K. , Ma, B. , Zhang, J.‐S. and Chen, S.‐Y. (2008a) Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 18, 1047. [DOI] [PubMed] [Google Scholar]
- Liao, Y. , Zou, H.F. , Wei, W. , Hao, Y.J. , Tian, A.G. , Huang, J. , Liu, Y.F. et al. (2008b) Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis . Planta, 228, 225–240. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Wei, W. , Zhu, W. , Su, L. , Xiong, Z. , Zhou, M. , Zheng, Y. et al. (2017) Histone deacetylase AtSRT1 links metabolic flux and stress response in Arabidopsis . Mol. Plant, 10, 1510–1522. [DOI] [PubMed] [Google Scholar]
- Lu, X. , Li, Q.T. , Xiong, Q. , Li, W. , Bi, Y.D. , Lai, Y.C. , Liu, X.L. et al. (2016) The transcriptomic signature of developing soybean seeds reveals the genetic basis of seed trait adaptation during domestication. Plant J, 86, 530–544. [DOI] [PubMed] [Google Scholar]
- Luo, M. , Tai, R. , Yu, C.W. , Yang, S. , Chen, C.Y. , Lin, W.D. , Schmidt, W. et al. (2015) Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis . Plant J. 82, 925–936. [DOI] [PubMed] [Google Scholar]
- Luo, M. , Wang, Y.‐Y. , Liu, X. , Yang, S. , Lu, Q. , Cui, Y. and Wu, K. (2012) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis . J. Exp. Bot. 63, 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Li, C. and Wang, M. (2015) Wheat NF‐YA10 functions independently in salinity and drought stress. Bioengineered, 6, 245–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani, R. (1999) The molecular biology of the CCAAT‐binding factor NF‐Y. Gene, 239, 15. [DOI] [PubMed] [Google Scholar]
- Mehdi, S. , Derkacheva, M. , Ramström, M. , Kralemann, L. , Bergquist, J. and Hennig, L. (2016) The WD40 domain protein MSI1 functions in a HDAC complex to fine‐tune ABA signaling. Plant Cell, 28, 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C. , Wells, R. , McKenzie, N. , Trick, M. , Ball, J. , Fatihi, A. , Dubreucq, B. et al. (2019) Variation in Expression of the HECT E3 Ligase UPL3 Modulates LEC2 Levels, Seed Size, and Crop Yields in Brassica napus . Plant Cell, 31, 2370–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, J. , Tan, H. , Hong, S. , Liang, Y. and Zuo, J. (2013) Arabidopsis transcription factor genes NF‐YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol. Plant, 6, 188–201. [DOI] [PubMed] [Google Scholar]
- Nelson, D.E. , Repetti, P.P. , Adams, T.R. , Creelman, R.A. , Wu, J. , Warner, D.C. , Anstrom, D.C. et al. (2007) Plant nuclear factor Y (NF‐Y) B subunits confer drought tolerance and lead to improved corn yields on water‐limited acres. Proc. Natl. Acad. Sci. USA, 104, 16450–16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, Z. , Hu, Z. , Jiang, Q. and Zhang, H. (2013) GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82, 113–129. [DOI] [PubMed] [Google Scholar]
- Oldfield, A.J. , Yang, P. , Conway, A.E. , Cinghu, S. , Freudenberg, J.M. , Yellaboina, S. and Jothi, R. (2014) Histone‐fold domain protein NF‐Y promotes chromatin accessibility for cell type‐specific master transcription factors. Mol. Cell, 55, 708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, W.J. , Tao, J.J. , Cheng, T. , Bian, X.H. , Wei, W. , Zhang, W.K. , Ma, B. et al. (2016) Soybean miR172a improves salt tolerance and can function as a long‐distance signal. Mol. Plant, 9, 1337–1340. [DOI] [PubMed] [Google Scholar]
- Qi, X. , Li, M.W. , Xie, M. , Liu, X. , Ni, M. , Shao, G. , Song, C. et al. (2014) Identification of a novel salt tolerance gene in wild soybean by whole‐genome sequencing. Nat. Commun. 5, 4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach, T.N. , Nguyen, H.T.M. , Valliyodan, B. , Joshi, T. , Xu, D. and Nguyen, H.T. (2015) Genome‐wide expression analysis of soybean NF‐Y genes reveals potential function in development and drought response. Mol. Genet. Genomics, 290, 1095–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. , Ji, D. , Li, S. , Wang, P. , Li, Q. and Xiang, F. (2012) The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS One, 7, e41274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y. , Liu, X. , Liu, X. , Li, Y. , Wu, K. and Hou, X. (2017) Arabidopsis NF‐YCs mediate the light‐controlled hypocotyl elongation via modulating histone acetylation. Mol. Plant, 10, 260–273. [DOI] [PubMed] [Google Scholar]
- Tian, T. , Liu, Y. , Yan, H. , You, Q. , Yi, X. , Du, Z. , Xu, W. et al. (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, M. , Matsui, A. , Tanaka, M. , Nakamura, T. , Abe, T. , Sako, K. , Sasaki, T. et al. (2017) The distinct roles of class I and II RPD3‐like histone deacetylases in salinity stress response. Plant Physiol. 175, 1760–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Chen, H.W. , Li, Q.T. , Wei, W. , Li, W. , Zhang, W.K. , Ma, B. et al. (2015) GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. [DOI] [PubMed] [Google Scholar]
- Wei, W. , Tao, J.J. , Chen, H.W. , Li, Q.T. , Zhang, W.K. , Ma, B. , Lin, Q. et al. (2017) A histone code reader and a transcriptional activator interact to regulate genes for salt tolerance. Plant Physiol. 175, 1304–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, S. and Poirier, Y. (2012) The protein acetylome and the regulation of metabolism. Trends Plant Sci., 17, 423–430. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Shen, W. , Chen, H. , Chu, L. , Xu, Y. , Zhou, X. , Liu, C. et al. (2018) Characterization and subcellular localization of histone deacetylases and their roles in response to abiotic stresses in soybean. BMC Plant Biol., 18, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. and Guo, Y. (2018) Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. [DOI] [PubMed] [Google Scholar]
- Yu, C.W. , Chang, K.Y. and Wu, K.Q. (2016) Genome‐wide analysis of gene regulatory networks of the FVE‐HDA6‐FLD complex in Arabidopsis . Front. Plant Sci. 7, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Zhang, H. , Hu, Z. , Chu, S. , Yu, K. , Lv, L. , Yang, Y. et al. (2019a) Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet. 15, e1008267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Liao, X. , Cui, Y. , Ma, W. , Zhang, X. , Du, H. , Ma, Y. et al. (2019b) A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean. PLoS Genet. 15, e1007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. , Ding, Y. , Sun, X. , Xie, S. , Wang, D. , Liu, X. , Su, L. et al. (2016) Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis . J. Exp. Bot. 67, 1703–1713. [DOI] [PubMed] [Google Scholar]
- Zhu, J.K. (2016) Abiotic stress signaling and responses in plants. Cell, 167, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phenotype of stable GmNFYA‐transgenic soybean plants.
Figure S2. Performance of GmNFYA‐transgenic and control plants under salt treatment when grown in a same pot.
Figure S3. ROS levels in GmNFYA‐transgenic hairy roots.
Figure S4. GmNFYA regulates a set of genes involved in response to salt stress in leaf.
Figure S5. Identification of the GmNFYA‐interacting proteins by Immunoprecipitation‐mass spectrometry.
Figure S6. Knockdown of GmFVE enhances salt tolerance of the plants with FVE‐RNAi transgenic hairy root.
Figure S7. Interactions of GmFVE and RPD3‐like histone deacetylases (HDACs) and the cluster analysis of HDACs.
Figure S8. Application of HDAC inhibitor FK228 increases salt tolerance of Kefeng1 seedlings.
Figure S9. Phylogenetic tree of GmNFYA promoter sequences from 132 cultivated soybeans.
Figure S10. There is no significant difference in promoter activity among three GmNFYA haplotypes under water and salt conditions.
Table S1. List of primers used in this study.
Table S2. GmNFYA‐interacting proteins identified by IP‐MS.
Table S3. Transcriptome data of GmNFYA‐transgenic soybean plant.
Data Availability Statement
The clean reads of RNA‐seq in this paper have been deposited in the GenBank database (GEO accession number: GSE173640).


