Abstract
Sclerotinia sclerotiorum and Botrytis cinerea are typical necrotrophic pathogens that can attack more than 700 and 3000 plant species, respectively, and cause huge economic losses across numerous crops. In particular, the absence of resistant cultivars makes the stem rot because of S. sclerotiorum the major threat of rapeseed (Brassica napus) worldwide along with Botrytis. Previously, we identified an effector‐like protein (SsSSVP1) from S. sclerotiorum and a homologue of SsSSVP1 on B. cinerea genome and found that SsSSVP1 could interact with BnQCR8 of rapeseed, a subunit of the cytochrome b‐c1 complex. In this study, we found that BnQCR8 has eight homologous copies in rapeseed cultivar Westar and reduced the copy number of BnQCR8 using CRISPR/Cas9 to improve rapeseed resistance against S. sclerotiorum. Mutants with one or more copies of BnQCR8 edited showed strong resistance against S. sclerotiorum and B. cinerea. BnQCR8‐edited mutants did not show significant difference from Westar in terms of respiration and agronomic traits tested, including the plant shape, flowering time, silique size, seed number, thousand seed weight and seed oil content. These traits make it possible to use these mutants directly for commercial production. Our study highlights a common gene for breeding of rapeseed to unravel the key hindrance of rapeseed production caused by S. sclerotiorum and B. cinerea. In contrast to previously established methodologies, our findings provide a novel strategy to develop crops with high resistance against multiple pathogens by editing only a single gene that encodes the common target of pathogen effectors.
Keywords: Brassica napus, cytochrome b‐c1 complex, CRISPR/Cas9, white mould, grey mould, disease resistance
Introduction
There is a large group of necrotrophic pathogens that can circumvent the resistance of numerous plants. Such pathogens usually have many hosts and encounter little if any resistance from plants in the field. They destroy tremendous amounts of economically important crops and, as a result, constantly threaten food safety and security as well as ecological security (Dean et al., 2012). Sclerotinia sclerotiorum and Botrytis cinerea are two such cosmopolitan pathogens. S. sclerotiorum can attack more than 700 species of plants, most of which are dicotyledonous, whereas Botrytis cinerea, which is in the same Sclerotiniaceae family as is S. sclerotiorum, has more than 3000 host species, including both dicotyledonous plants and monocotyledonous plants (https://nt.ars‐grin.gov/fungaldatabases/). Importantly, almost all hosts of S. sclerotiorum can be attacked by B. cinerea. Diseases caused by S. sclerotiorum are called white mould or stem rot sometimes, whereas those caused by B. cinerea are called grey moulds. Both cause widespread and extensive economic losses across numerous crop species, including oil crops, bean, fruit and vegetable crop species, and in postharvest products (Amselem et al., 2011; Bolton et al., 2006; van Kan, 2006; Williamson et al., 2007). To date, no cultivars or germplasm resistant to both the two fungal species has been found in any host crop and both have developed strong resistance against commercial fungicides (Matthias, 2014; Petrasch et al., 2019; Zhou et al., 2014). Hence, here is an urgent need worldwide to establish an environmental‐friendly approach to control crop diseases caused by S. sclerotiorum and B. cinerea.
Rapeseed (Brassica napus) is an allotetraploid plant species that provides major edible plant oil worldwide (Chalhoub et al., 2014). In China, the cultivated area of rapeseed is about 6.7 million ha (Hu et al., 2017). Because no resistant cultivars are available and lack of efficient control measures (Derbyshire and Denton‐Giles, 2016; Hu et al., 2017; Taylor et al., 2018), stem rot caused by S. sclerotiorum is the major disease of rapeseed causing losses of 8.4‐billion‐yuan RMB loss each year in China alone. Meanwhile, like stem rot, grey mould caused by B. cinerea also heavily damages rapeseed simultaneously, inducing similar symptom on rapeseed and even rots seeds in the field. Endowing rapeseed with strong resistance against S. sclerotiorum and B. cinerea is the most effective way to improve rapeseed production efficiency and recover economic losses.
CRISPR/Cas 9 serves as an unprecedented genome‐editing tool for improving crop traits to solve food security problems for the growing human population (Armin and David, 2017; Chen et al., 2019; Gao, 2021; Hickey et al., 2019; Kwon et al., 2020; Rodríguez‐Leal et al., 2017; Zaidi et al., 2019). Previously, MLO, a gene involved in the susceptibility to powdery mildew within barley and wheat, was edited by using TALEN‐based gene editing and CRISPR/Cas9 techniques, and the resulting MLO mutants revealed strong resistance against powdery mildew and could be used for breeding durable and broad‐spectrum resistance (Wang et al., 2014). Genome editing of the promotor of three SWEETs, which encode the binding sites of bacterial transcription activator‐like effectors (TALes), provides broad‐spectrum resistance to rice bacterial blight (Li et al., 2012; Oliva et al., 2019; Xu et al., 2019), and similar results were also observed in citrus plants (Peng et al., 2017). However, when genes involved in susceptibility were edited, these disease‐resistant mutants could not be directly applied in the field because their agronomic traits were affected to varying degrees (Oliva et al., 2019; Xu et al., 2019).
QCR8 is a conserved subunit of the cytochrome b‐c1 complex in the respiratory chain of plants, and the silencing of QCR8 in Nicotiana benthamiana with VIGS results in abnormal development and cell necrosis (Lyu et al., 2016). A small secreted virulence‐related protein (SsSSVP1) in S. sclerotiorum that can interact with QCR8 has been identified. SsSSVP1 ‘hijacks’ QCR8 and changes its location from mitochondria to the plant cytoplasm. It not only disables the biological function of QCR8 but also leads to cell death that may be beneficial to the growth of S. sclerotiorum (Lyu et al., 2016).
There are eight BnQCR8 copies in B. napus cultivar Westar. The expression level of BnQCR8 copies is various, and the expression of BnQCR8.C09 and BnQCR8.A10 is higher than other copies (http://cbi.hzau.edu.cn/bnapus/) in eight cultivars tested. The expression of BnQCR8.C09 and BnQCR8.A10 in Westar is significantly up‐regulated by 2.8‐ and 2.7‐fold under S. sclerotiorum infection, respectively (Girard et al., 2017), and is down‐regulated by 1.1‐ and 1.3‐fold in the resistant cultivar compared with those in the susceptible cultivar (Wu et al., 2016), indicating that the two copies of QCR8 might play an important role in responding to the infection of S. sclerotiorum. In this study, we used the CRISPR/Cas9 genome‐editing tool to introduce mutations on some copies of BnQCR8 and found that the copy number reduced genome‐editing lines showed strong resistance against both S. sclerotiorum and B. cinerea but did not affect their major agronomic traits.
Results
Design of sgRNAs to knock out QCR8 genes in B. napus
QCR8 has eight homologous proteins in B. napus according to BLAST searches of the NtQCR8 and AtQCR8 proteins in the B. napus cultivar Westar genome (http://www.cbi.hzau.edu.cn/bnapus/), and the genes encoding for these proteins include BnQCR8.A10 (BnaA10G0270400WE), BnQCR8.C09 (BnaC09G0564700WE), BnQCR8.A03 (BnaA03G0028100WE), BnQCR8.A05 (BnaA05G0386800WE), BnQCR8.A01 (BnaA01G0290500WE), BnQCR8.C01 (BnaC01G0399200WE), BnQCR8.C05 (BnaC05G0478600WE) and BnQCR8.C02 (BnaC02G0015500WE). The identity among all the homologous copies ranged from 79.66% to 100%, and the identity of amino acid sequence was 100% in BnQCR8.A10/C09, BnQCR8.A01/C01 and BnQCR8.A05/C05. BnQCR8.A05/C05 was near identical to BnQCR8.A01/C01 (98.61% identity), BnQCR8.A03 was highly identical to BnQCR8.A10/C09 (97.22% identity), and the identity shared among BnQCR8.C02, BnQCR8.A03 and BnQCR8.A05/C05 was 83.05% (Figure S1c). Phylogenetically, BnQCR8.A10 and BnQCR8.C09 formed a clade, and these two genes together with BnQCR8.A03 further formed a branch. All BnQCR8 homologous copies showed a close relationship with AtQCR8 but were distantly related to other QCR8 sequences examined (Figure 1a). All other copies of BnQCR8, except BnQCR8.C02, could interact with SsSSVP1 (Figure 1b).
Figure 1.
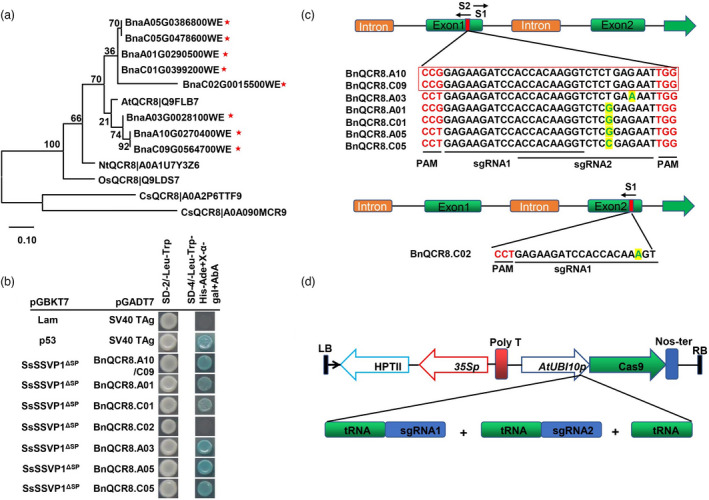
Strategy of reducing the copy number of BnQCR8 in cultivar Westar of rapeseed. (a) The phylogenetic tree of eight homologous copies of BnQCR8 and QCR8 from other plants selected. The phylogenetic tree was made with the maximum‐likelihood algorithm. Branch lengths are proportional to the average probability of change for characters on that branch. The red pentagram showed the BnQCR8s in rapeseed. (b) The interaction between SsSSVP1ΔSP and BnQCR8. The pGADT7‐SV40 tag co‐transferred with pGBKT7‐Lam pGBKT7‐p53 were used as negative and positive controls, respectively. Photos were taken 2 dpi. (c) The structure of BnQCR8 and the sgRNAs in cultivar Westar of rapeseed. The sgRNA1 and sgRNA2 sites are in the exon shown with red vertical line and the direction shown with the black arrow and the different base is shown in green. The PAM is in red. The sequences in the red box are the sgRNA1 and sgRNA2 in the vector. (d) The schematic of the T‐DNA region of the pRGEB35‐Cas9‐BnQCR8 vector. The two sgRNAs were driven by AtUBI10
To generate mutations in BnQCR8.A10 and BnQCR8.C09 by the CRISPR/Cas9 system, sgRNA1 (S1) and sgRNA2 (S2) were designed by using the CRISPR‐P program (Lei et al., 2014); they were located in the exon of BnQCR8 (Figure 1c and Figure S1a) and were linked with tRNA and formed a tandemly arrayed tRNA‐gRNA architecture based on the CRISPR/Cas9 multiplex editing system (Figure 1d) (Ding et al., 2018; Xie et al., 2015). The vector formed was named pRGEB35‐Cas9‐BnQCR8. The two sgRNAs and Cas9 were driven by AtUBI10P (Figure 1d). sgRNA1 could target all of the BnQCR8 copies, and sgRNA2 could target both BnQCR.A10 and BnQCR.C09 at least, with only 1‐bp mismatch with the other 5 copies.
Identification of CRISPR/Cas9 induced mutations in BnQCR8
Vector pRGEB35‐Cas9‐BnQCR8 was transformed into the B. napus cultivar Westar by Agrobacterium‐mediated genetic transformation and tissue culture methods. Finally, twenty‐three positive T0 individual transgenic lines (Figure S2) were obtained and confirmed by PCR using HPTII gene‐specific primers.
To examine the editing efficiency, we examined the sequences of the targeted genes using Hi‐TOM (Liu et al., 2019) and Sanger DNA sequencing. In all 23 HPTII‐positive T0 individual transgenic lines, fourteen lines were edited, and the editing efficiency was 60.9%. The editing of BnQCR8 produced three types of mutation, including single‐base insertion, single‐base deletion and multi‐base deletion, and most loci of mutation were within the 3rd–4th bp region upstream of the PAM (Figure 2a‐e). The editing events mainly occurred in BnQCR8.A10 and BnQCR8.C09 (Figure 2c), but the other homologous copies were also edited to different extents (Figure 2a‐d). Among the T0 mutants, BnQCR8.A01/C01 was edited only in BnQCR8‐CR43 (Figure 2a). Single‐base insertions and single‐base deletions constituted the main types of mutations, accounting for 41.5% and 18.3%, respectively (Figure 2f).
Figure 2.
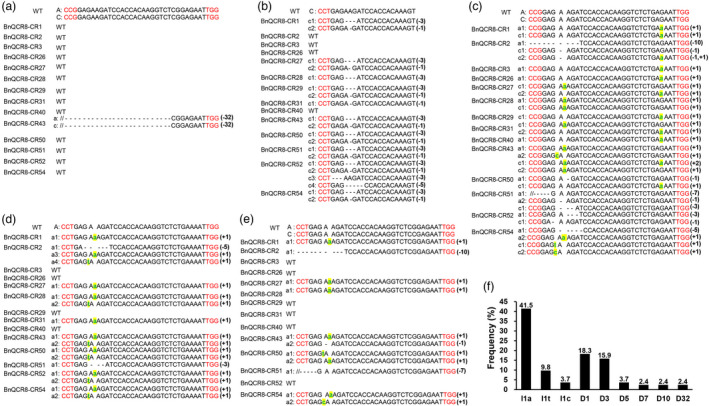
Genotype of BnQCR8‐edited T0 mutants of rapeseed cultivar Westar. (a‐e) Fourteen BnQCR8‐edited T0 plants showing mutation sites at eight homologous copies of BnQCR8 (BnQCR8.A01/C01, BnQCR8.C02, BnQCR8.A10/C09, BnQCR8.A03 and BnQCR8.A05/C05). Insertions and deletions are indicated in highlight green font and “‐” hyphens, respectively, and the red fonts show the PAM sites. On the left of each copy, A and C are the WT allele of the BnQCR8 copies, and a# and c# show the mutant allele numbers. On the right, the number in brackets on the right is the base numbers of insertion and deletion. (f) Mutation types and frequency at the sgRNA target sites in 14 T0 mutants. The X‐axis: I# and D# are the numbers of base pairs inserted and deleted at the sgRNA target sites
To obtain stable and different genotypes of the mutation lines, seven individual T0 transgenic lines of BnQCR8‐edited mutants were used to produce T1 and T2 progeny lines by self‐pollination. All the T1 progeny were analyzed by sequencing, and the results revealed that the allelic mutations of the T0 lines were transmitted to the T1 generation at an average transmission rate of 68.21% in BnQCR8.A10/C09 (Table S1). Moreover, the incidence of foreign DNA introduction in the T1 lines was also detected by PCR assays. Of the 58 T1 lines, 11 plants presented no HPTII gene (Table S1 and Figure S3). We further obtained different T2 mutants from the T1 generation by self‐pollination. Five different genotypes in which at least one BnQCR8 homologous copy had been edited, each genotype mutant contained four or more plants. The mutants of these genotypes were named WCR1, WCR2, WCR3, WCR4 and WCR5, and their mutation sites and parents are presented in Figure 3f and Table S4, respectively. The five mutants of these genotypes were used for resistance assays and major agronomic trait assays.
Figure 3.
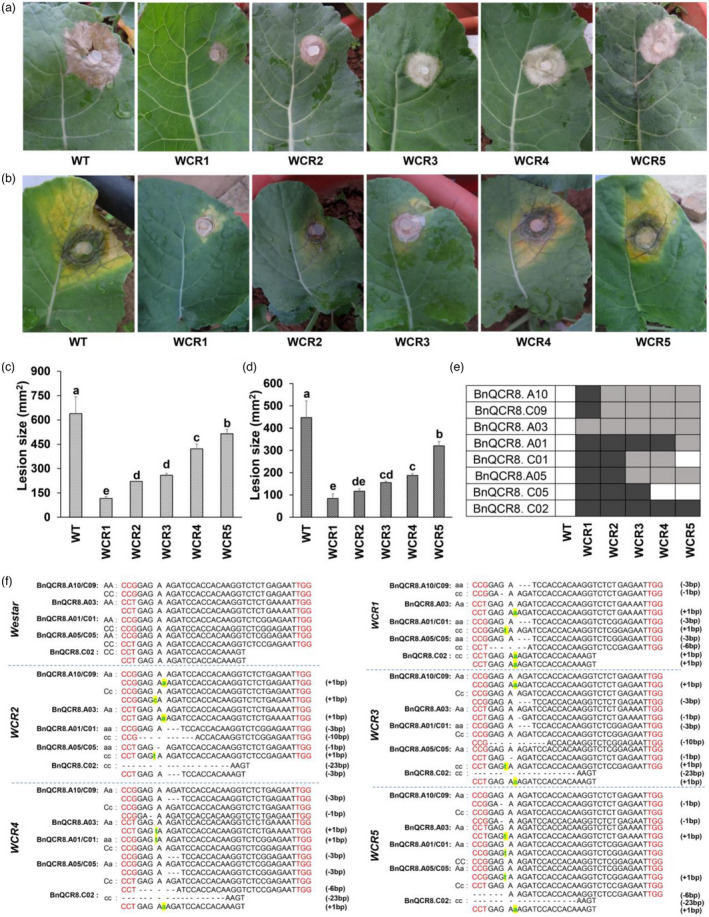
Resistance of the five genotypes of BnQCR8‐edited T2 mutants to S. sclerotiorum and B. cinerea in Westar of rapeseed. (a and b) The lesion caused by S. sclerotiorum and by B. cinerea, respectively. Plants were grown under field conditions for 56 days and 33 days, and leaves were inoculated with activating hyphal agar discs (Ф = 5.0 mm) of S. sclerotiorum strain 1980 or B. cinerea strain B05.10, respectively. Inoculated leaves were kept moist within a transparent plastic bag for additional 96 h for S. sclerotiorum and 120 h for B. cinerea under the same conditions, with a temperature about 20 °C in the day and 10 °C at night. (c and d) The average size of lesions induced by S. sclerotiorum (c) and B. cinerea (d). For each genotype mutant, four lesions were measured. (e) The genotype of BnQCR8 mutants of Westar. White squares indicate nonmutated copy of BnQCR8. French grey squares indicate heterozygous copy, and dark grey squares indicate homozygous copy of BnQCR8. The detail of genotype of mutant WCR1, WCR2, WCR3, WCR4 and WCR5 is presented in Figure 1e. The experiment was repeated two times independently and with comparable results. The values are presented as the means ± s.d for n = 4 replicates. Data were analysed by one‐way ANOVA, followed by DMRT. Different lowercase letters on top of each column indicate significant differences at P < 0.05. (f) The five genotypes of BnQCR8‐edited T2 mutant, namely, WCR1, WCR2, WCR3, WCR4 and WCR5. Insertions and deletions are indicated in highlighted green font and “‐” hyphens, respectively, the red fonts show the PAM sites. A and C show the WT allele; a and c show the mutant allele; the number in brackets on the right is the base numbers of insertion and deletion
BnQCR8‐edited mutants displayed strong resistance to S. sclerotiorum
To analyse the resistance of the mutants against S. sclerotiorum, WCR1, WCR2, WCR3, WCR4 and WCR5 T2 mutants were used for a resistance assay. Young leaves of plants growing in the field for 56 days were each inoculated with a hyphal agar disc of strain 1980 and the inoculated plants kept on growing under the same conditions for 96 h. The lesions formed on all mutants were significantly smaller than those formed on the wild‐type (WT) cultivar Westar. The lesions on WCR1 were the smallest among the mutants, followed by WCR2, WCR3, WCR4 and WCR5. The average lesion size on the four WCR1 plants was 116.5 ± 9.89 mm2, and that on the WT plants was 638.8 ± 103.71 mm (Figure 3a, c). The resistance to S. sclerotiorum of each mutant was also determined in a growth chamber. Young leaves of plants growing in the chamber for 40 days were inoculated, and the results were consistent with those under field conditions (Figure S4a, c). These results demonstrated that all the mutants have high resistance to S. sclerotiorum, and WCR1 with only one copy of BnQCR8.A03 showed the strongest resistance. WCR2, which had one copy of BnQCR8.A03 and one copy of BnQCR8.C09, also showed strong resistance; the other three mutants of these genotypes had more copies of QCR8, and their resistance to S. sclerotiorum was weaker than that of WCR1 and WCR2. By combining the results of sequence mutation analysis, we found that mutants had increased resistance when more copies of BnQCR8 were edited.
BnQCR8‐edited mutants displayed strong resistance against B. cinerea
We found gene BcSSVP1 (XP_024549683.1), a homologue of SsSSVP1, in the genome of B. cinerea and BcSSVP1 could also interact with BnQCR8 (Figure 4a). Hence, we presumed that the mutants obtained might also show strong resistance against B. cinerea. To probe whether QCR8‐edited Westar mutants were resistant to B. cinerea, we inoculated the hyphal discs of strain B05.10 of B. cinerea onto the young leaves of 49‐day‐old and 33‐day‐old mutants in vivo in a growth chamber and in the field, respectively. We found that mutants also showed very strong resistance against B. cinerea (Figure 3b, d; Figure S4b, d). Furthermore, the resistance of the mutants to B. cinerea was consistent with that against S. sclerotiorum (Figure 3a; Figure S4a).
Figure 4.
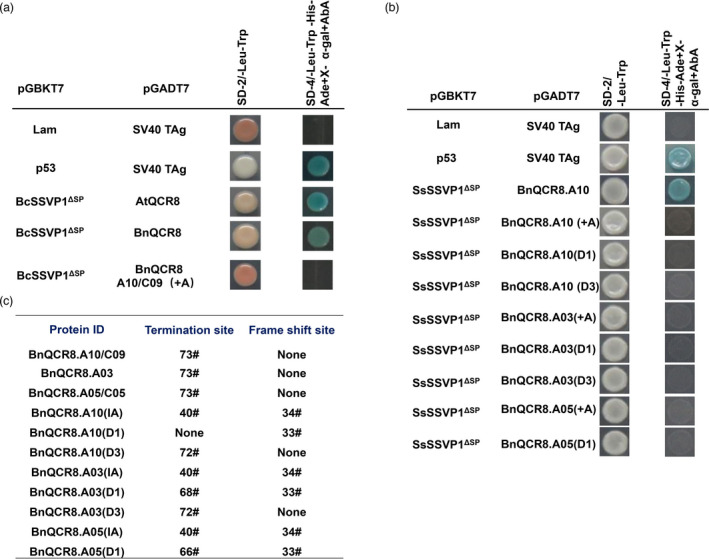
Interactions between BnQCR8 and its mutated BnQCR8 and BcSSVP1 of B. cinerea or SsSSVP1 of S. sclerotiorum. (a) Yeast two‐hybrid (Y2H) assays showed the interaction between BcSSVP1ΔSP and BnQCR8 (BnQCR8.A10) or AtQCR8. pGADT7‐SV40 TAg co‐transferred with pGBKT7‐Lam or pGBKT7‐p53 was used as negative and positive controls. SD–Ade/–His/–Leu/–Trp contains125 ng mL−1 aureobasidin A (ABA) and 40 μg mL−1 X‐α‐Gal was used. Photos were taken 2 dpi. (b) Interaction of edited BnQCR8 with SsSSVP1ΔSP by Yeast two‐hybrid (Y2H) assays. “+A” indicate the insertion of adenine. D1 and D3 indicate one and three bases deletions, respectively. (c) The termination and frame shift amino acid site of the edited copies of BnQCR8, and ‘none’ indicates no events occurred in the edited copies of BnQCR8
BnQCR8‐edited mutations did not affect major agronomic traits and respiration
To assay the effects of editing BnQCR8 on rapeseed, we firstly compared the phenotypes of the T1 mutants of Westar (BnQCR8‐CR2, BnQCR8‐CR27, BnQCR8‐CR28, BnQCR8‐CR43, BnQCR8‐CR51, BnQCR8‐CR52 and BnQCR8‐CR54) with Westar (WT). We found no significant changes in phenotypes among T1 mutant and the WT seedlings (Figure 5a). We then further assayed the effects of the edited BnQCR8 on the yield of B. napus. We collected the seeds of the T1 mutants separately and found that there was no significant difference in the yield or thousand seed weight between the mutants and the WT (Figure 5b, c). The seed yield per plant of the T1 mutants was approximately 23.5–24.5 g, with the average 23.9 g, and the average yield of the WT was 23.7 g; the thousand seed weight for the T1 mutants was approximately 3.73–3.75 g, and that for the WT was 3.72 g.
Figure 5.
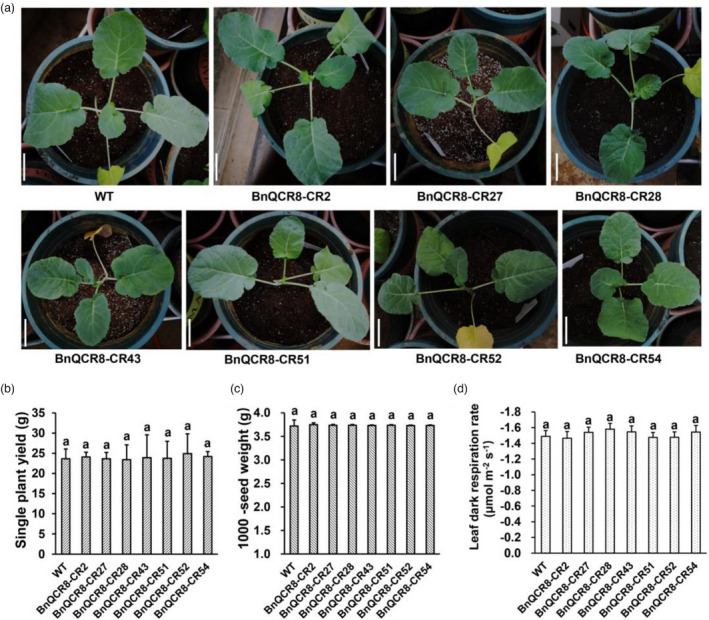
Partial phenotype and main agronomic traits of T1 mutants. (a) 40‐day‐old seedlings of BnQCR8‐edited T1 mutants. Bar is 5.45 cm. (b) The seed yields of the BnQCR8‐edited T1 mutants. The values are presented as means ± s.d for n = 15 replicates. (c) The thousand seed weight of the BnQCR8‐edited T1 mutants and the WT. (d) The leaf dark respiratory rate of the T1 mutants by using Li‐Cor 6800 portable photosynthesis system. The values are presented as means ± s.d for n = 8 replicates. All data were analysed by one‐way ANOVA followed by DMRT. Different lowercase letters on top of each column indicate significant differences at P < 0.05. WT represents the wild type of Westar
The major agronomic traits of the five genotypes of T2 mutants, WCR1, WCR2, WCR3, WCR4 and WCR5, were further examined. There was no significant phenotypic difference between the WT and T2 mutants at any growth stage of rapeseed; their phenotypes were the same at the seedling, bolting, flowering and maturation stages, and they had similar plant shapes at the flowering stage (Figure 6a). The silique length between the T2 mutants and the WT was not significantly different, and each was approximately 60 mm (Figure 6b).
Figure 6.
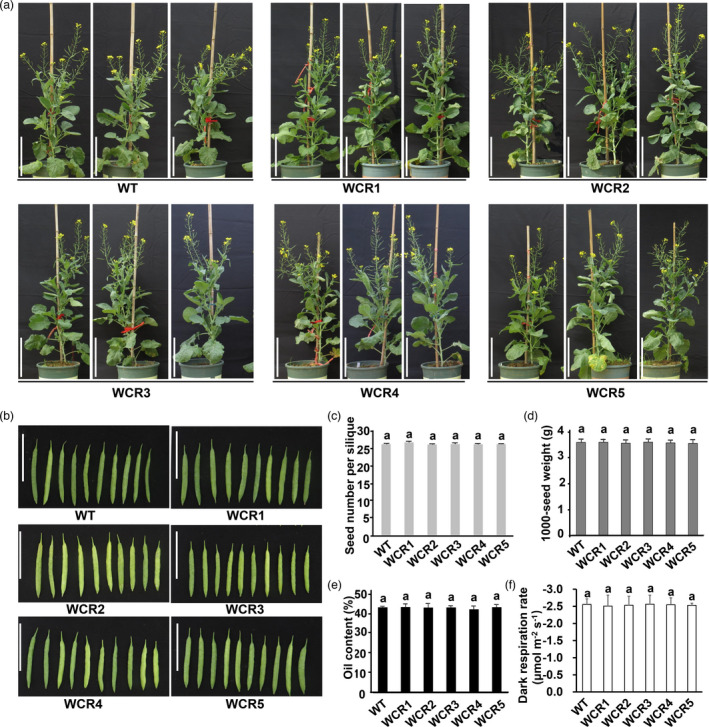
Major agronomic traits of the five genotype T2 mutants of QCR8‐edited rapeseed cultivar Westar. (a) Mutants at the flowering stage, each genotype had four or more plants, but only three typical mutant plants are presented. Bar is 27.5 cm. (b) The silique shape and length of mutants. Siliques of only one plant of each genotype were presented. Bar is 5 cm. (c) Seed numbers per silique of the mutants. Three individual plants of each mutant were sampled, and ten siliques of each plant were counted. The values are presented as the means ± s.d for n = 3 replicates. (d) The thousand seed weight of mutants. Seven plants of WT and WCR3, six plants of WCR2 and four plants of WCR1, WCR4 and WCR5 were used for thousand seed weight assessment. The values are presented as the means ± s.d for n ≥ 4 replicates. (e) The oil content of mutants. The oil content of seeds collected from each plant of the mutants were determined using Near‐infrared spectrometer (NIRSystems 3750), and the average oil content of four plants of each mutant was calculated. (f) The leaf dark respiratory rate of the T2 mutants by using Li‐Cor 6800 portable photosynthesis system. Seven plants of WT and WCR3, six plants of WCR2 and four plants of WCR1, WCR4 and WCR5, were used for thousand seed weight assessment. The values are presented as the means ± s.d for n ≥ 4 replicates. All data were analysed by one‐way ANOVA followed by DMRT. Different lowercase letters on top of each column indicate significant differences at P < 0.05. The genotype of mutant WCR1, WCR2, WCR3, WCR4 and WCR5 is shown in Figure 1e; WT represents the wild type of Westar
The seed number per silique of both the T2 mutants and the WT was approximately 26 (Figure 6c), and the average thousand seed weight of the mutants was approximately 3.53–3.58 g, and that of the WT was 3.58 g (Figure 6d). The average oil contents of the seeds of the WCR1, WCR2, WCR3, WCR4 and WCR5 T2 mutants were 43.2 ± 1.44%, 42.9 ± 2.16%, 43.1 ± 1.11%, 42.1 ± 1.65% and 43.1 ± 1.50%, respectively, whereas the oil content of the seeds of Westar was 43.1 ± 0.53%. Statistically, there was no significant difference in the seed oil content between the T2 mutants and the WT (Figure 6e). In addition, there was no significant difference in the content of oleic acid, linoleic acid, linolenic acid and glucosinolate among the seeds of the mutants or in comparison with the WT (Figure S6a‐d). Overall, the major agronomic traits were not altered as a result of editing of one or more copies of BnQCR8.
The leaf respiratory rate of T1 and T2 mutants under dark condition was determined, and the results showed that there was no significant difference in the leaf dark respiratory rate between T1 mutants and the WT growing in the chamber under 22 °C at the seedling stage. The leaf dark respiratory rate of the mutants was approximately −1.51 μmol m−2 s−1 to −1.55 μmol m−2 s−1, and for the WT it was −1.53 μmol m−2 s−1 (Figure 5d). The respiratory rates between T2 mutants and the WT growing under natural conditions at the flowering stage did not show any significant difference either. The leaf dark respiratory rate of the T2 mutants was approximately −2.51 μmol m−2 s−1 to −2.56 μmol m−2 s−1, and it was −2.56 μmol m−2 s−1 under 15 °C for the WT (Figure 6f).
Mutations affected the interaction between BnQCR8 and SsSSVP1
We further found that a single‐base insertion or deletion and a short‐segment deletion produced frameshift mutations in BnQCR8 (Figure S5). To confirm whether edited BnQCR8 could affect the interaction with SsSSVP1ΔSP, the cDNA sequences of some edited BnQCR8 were cloned and inserted into vector pGADT7. Y2H assay showed that edited BnQCR8 could not interact with SsSSVP1ΔSP (Figure 4b), maybe resulting from an amino acid deletion or a frameshift mutation (Figure 4c; Figure S5).
Off‐target mutation analysis of T0 BnQCR8 mutants
The results of the predicted off‐targets in mutations showed that the off‐scores were low, and the related information of potential off‐target sites was listed in Table S3. Except for off1‐1 and off1‐2 within S1, the off‐scores of other off‐targets were less than 37%. We chose off1‐1, off1‐2, off1‐3, off1‐4 and off1‐5 of S1 as well as off2‐1, off2‐2, off2‐3 and off2‐4 of S2 to determine the off‐target mutation in 14 T0 mutants by Sanger sequencing and the results showed no mutations in any of the mutants. Therefore, the pRGEB35/Cas9 system can highly specifically generate targeted mutations in the B. napus genome.
Discussion and conclusion
In this study, we successfully edited some copies of BnQCR8 in rapeseed by using CRISPR/Cas9 and found that mutants showed strong resistance against both S. sclerotiorum and B. cinerea, two major pathogens infecting numerous economically important crop species and for which no resistant commercial cultivars are available. Compared with those of the original cultivar Westar, the agronomic traits of the BnQCR8‐edited Westar mutants, such as the plant shape, flowering time, thousand seed weight, yield, oil content and oil components, were not significantly different. The BnQCR8 mutants mainly afforded rapeseed with disease resistance and no significant changes in other agronomic traits, giving them potential to be directly deployed into the field. Thus, our finding provides a common gene for rapeseed resistance‐breeding against stem rot, a disease that has severely limited rapeseed production efficiency for decades (Derbyshire and Denton‐Giles, 2016; Hu et al., 2017; Taylor et al., 2018).
Although we currently do not clearly know the mechanism through which the BnQCR8‐edited mutant is resistant to S. sclerotiorum and B. cinerea, we found that the putative proteins encoded by edited BnQCR8 could not interact with SsSSVP1 and the lesion size on WCR1, which has only one copy of BnQCR8, was significantly smaller than that on WCR2 and the other mutants with two or more copies of BnQCR8 (Figure 3a). These results suggest that the copy number of BnQCR8 may be involved in resistance and that relatively fewer copies of BnQCR8 led to stronger resistance. We also reduced the copy number of BnQCR8 in another commercial rapeseed cultivar, Zhongshuang 6, and found that the mutants similarly showed strong resistance against S. sclerotiorum and B. cinerea (data not shown). Additional copies of gene are usually functionally redundant. Recent discoveries found that mutation on additional copies could not affect gene function (El‐Brolosy et al., 2019; Ma et al., 2019; Rossi et al., 2015), and in this study, mutation on some copies of BnQCR8 could not affect the respiratory rate of rapeseed (Figure 5d; Figure 6f). This study suggests that one copy of BnQCR8 may be sufficient for rapeseed.
Both S. sclerotiorum and B. cinerea are worldwide spread destructive necrotrophic pathogens. Importantly, B. cinerea ranks No. 2 among top 10 plant fungal pathogens (Dean et al., 2012). How to breeding resistant cultivars of other economic‐important crops against grey mould and white mould is critical for food safety and security. QCR8 is a highly conserved gene in plants, and its function is indispensable. In this study, we also found that SsSSVP1 could interact with QCR8 in many host plant species, such as B. rapa, Capsicum annuum, Glycine max, Helianthus annuus and Solanum lycopersicum (Figure S7). Therefore, QCR8 is a potential target of S. sclerotiorum and B. cinerea in these plant species, and QCR8 is most likely to be an ideal gene for genome editing to generate resistant cultivars of these crop species that are the hosts of S. sclerotiorum and/or B. cinerea. For plants with multiple copies of QCR8 in their genome, mutants resistant to S. sclerotiorum and B. cinerea might be acquired through the genome‐editing methods used in this study. For plants that have only one copy of QCR8, it is possible to obtain highly resistant mutants by using CRISPR/Cas9 gene‐editing technology, which is capable of replacing only the amino acid residues of QCR8 required for SsSSVP1 binding and not affecting its biochemical function. Recent developments in prime editing and base editing could be used to test this possibility (Yan et al., 2021; Zhu et al., 2020).
Editing susceptibility genes conferring crops strong resistance against plant diseases has been exhibited. Editing wheat MLO gene and the TALE‐binding elements of rice SWEET genes afforded wheat and rice high resistance against wheat powdery mildew and rice bacterial blight, respectively (Li et al., 2012; Oliva et al., 2019; Wang et al., 2014; Xu et al., 2019). Controlling transcription and translation of key immune regulator genes via uORF enhanced plant disease resistance (Xu et al., 2017). Effectors are a large group of pathogenicity factors among biotrophic, hemibiotrophic and necrotrophic pathogens, and most pathogens can transfer multiple effectors to plants, either directly into cells or through the apoplast, to interfere with plant functions (Cui et al., 2015). Some effectors target essential plant proteins that function on mitochondria or chloroplasts to weaken plant resistance (Cui et al., 2015; Kretschmer et al., 2019; Lyu et al., 2016; Tang et al., 2020). In this study, we found that edition of the effector target gene could improve plant resistance yet not change major agronomic traits. Hence, our study provides a common strategy to develop gene‐edited crop plants with high resistance based on effector biology, which is different from previously established methods.
The reality is that crops in the field face a variety of pathogens, and in most cases, plant resistance involves only specific diseases, that is, plants can be resistant to one disease while susceptible to other diseases. How to generate crop varieties with strong resistance to multiple important diseases is an urgent problem needing resolution. If pathogens have similar effectors that can target the same plant protein, editing the gene coding for the target could provide multi‐resistance. Hence, the discovery of common targets or susceptibility genes for pathogens in crop plants could now be a very important focus. In fact, pathogens sharing the same plant targets have been frequently reported (Ali and Yun, 2020; Park et al., 2019; Ray et al., 2019; Shen et al., 2020). Our study provides an approach to develop highly resistant plants against multiple pathogens by editing only a single gene.
Briefly, we provided a gene for rapeseed resistance‐breeding against S. sclerotiorum and B. cinerea, and this gene can also be potentially used for resistance breeding of other crops which are severely attacked by these fungi. We also exhibit a novel strategy to confer strong resistance in plants against multiple pathogens based on editing only one gene which encodes common target of pathogen effectors. This strategy is very important for crop production because crops must deal with different multiple pathogens in the field.
Materials and methods
Plant materials and fungal pathogens
The rapeseed cultivar Westar was used as the transformation recipient in the present research. T0 plants were generated by Wuhan Towin Biotechnology Co., Ltd., and T1 and T2 plants were generated by self‐pollination. The T0 and T1 Westar mutants were grown in medium‐sized pots (diameter 27.5 cm, height 31 cm) containing garden soil (Zhenjiang Peilei Substrate Technology Development) in the growth chamber with a temperature of 22 °C, with a photoperiod of 16‐h light and 8‐h dark and a light intensity of approximately 10 000 lux. These mutants were used for self‐pollination and for the phenotypic, thousand seed weight, and yield assessments. Some Westar T2 mutants were grown in small pots (diameter 12 cm, height 12 cm) containing garden soil (Zhenjiang Peilei Substrate Technology Development) in a growth chamber with a temperature of 22 °C and a photoperiod of 16 h light and 8 h dark for resistance assessment. Westar T2 mutants were grown in the same medium‐sized pots, but containing field soil, were used for phenotypic, silique, thousand seed weight, yield and seed oil content assessments. Westar T2 mutants grown in large pots (diameter 51 cm, height 35 cm) containing field soil on September 20 under field conditions were used for resistance assessment. All plants were managed as needed, with imidacloprid (Bayer, China) to control insects according to the manufacturer’s instructions. Fungal pathogens, S. sclerotiorum strain 1980 and Botrytis cinerea strain B05.10, were grown on potato dextrose agar (PDA) medium under 20 °C and maintained in PDA slants under 4 °C.
gRNA design and CRISPR/Cas9 vector construction
In this study, the web‐based tool CRISPR‐P (http://cbi.hzau.edu.cn/cgi‐bin/CRISPR) was used to design the sequence‐specific sgRNAs of BnQCR8 and to predict off‐targets. Two target sites within the genes were designed. A pRGEB35 vector, which was provided by Professor Kabin Xie (Huazhong Agricultural University), was used for rapeseed genome editing. This vector contains a hygromycin B phosphotransferase (HPTII) selection marker. The two sgRNAs were connected by tRNA and formed a tandemly arrayed tRNA‐gRNA architecture and driven by AtUBI10 (Figure 1d). The oligos used for constructing the vector are listed in Table S2.
Agrobacterium‐mediated rapeseed transformation
The pRGEB35 vector construct was transformed into hypocotyls of Westar rapeseed plants by Wuhan Towin Biotechnology Co., Ltd., using the Agrobacterium tumefaciens‐mediated transformation technique according to the method previously described by Zhou et al., (2002).
Identification of the mutant transgenic plants
Rapeseed genomic DNA was extracted by using the hexadecyltrimethylammonium bromide method, as described by Porebski et al., (1997). To detect the presence of the foreign T‐DNA of the vector, HPTII genes were amplified with specific primers (Table S2). Similarly, to detect mutagenesis, the CRISPR target sites were amplified by PCR with specific primers (Table S2). The PCR products were identified via agarose gel electrophoresis and purified by a gel extraction kit (OMEGA kit, D2500‐02). The purified fragments were sequenced by Illumina sequencing, and the mutagenesis and genotypes of the mutants were analysed via the Hi‐TOM website (http://www.hi‐tom.net/hi‐tom/) as described by Liu et al. (2016). The mutants of the genotype were further identified by Sanger and TA clone sequencing, and the heterozygous mutations were decoded by using the degenerate sequence decoding method (Ma et al., 2015a).
Resistance, yield and seed oil content assays
The seeds of mutants were harvested individually at maturity after growing for 240 days and then dried under natural conditions to assay the effects of edited BnQCR8 on Westar plants. Fifteen plants of each T1 mutant and the WT were weighed individually for the yield assay. Ten of the fifteen plants were used for a T1 thousand seed weight assay, in which one thousand seeds of each plant were weighed (three replications). T2 mutants of five different Westar genotypes (WCR1, WCR2, WCR3, WCR4 and WCR5) and T1 mutants of three different genotypes (Table S4) were selected to analyse the resistance, phenotype, thousand seed weight, yield and seed oil content. In the T2 mutants, ten siliques selected randomly from a single plant of each mutant and WT were used for silique phenotypic analysis and seed number analysis, and three plants of each mutant and WT (serving as three replicates) were selected for the experiment. The following plants were used for the thousand seed weight and yield assessments according to the same methods described earlier: seven WT and WCR3 plants, six WCR2 plants and four plants each for WCR1, WCR4 and WCR5. In addition, 2 g of seed from each plant was used for oil content assays using the near‐infrared spectrometry (NIRSystems 3750).
For resistance assays in the growth chamber, T2 mutants of each genotype of Westar were grown in a growth chamber at a temperature of 22 °C, a 16:8‐h light/dark photoperiod, and a light intensity of approximately 10 000 lux for 35 days. Young leaves of the same stage were inoculated in vivo with hyphal discs (Ф = 5.0 mm) of S. sclerotiorum strain 1980 by using a method described by Yang et al., (2018) with minor modifications. The inoculated leaves were kept moist using transparent plastic bags, and the plants were further grown under the same conditions for an additional 36 h. After imaging and measuring the size of the lesions, the inoculated leaves were removed with sterilized scissors, and the plants were further grown in the same chamber for another eight days. The new grown leaves were inoculated with hyphal discs (Ф = 5.0 mm) of B. cinerea strain B05.10 using the same methodology. Plants were then grown under the same conditions for an additional 36 h and the lesion sizes were measured.
For resistance assays under field conditions, the mutants of each genotype were grown in field soil and under field conditions within large pots for 28 days and leaves were inoculated with hyphal discs (Ф = 5.0 mm) of B. cinerea strain B05.10. The inoculated leaves were covered by a transparent plastic bag to keep moist, and lesion sizes were measured at 120 h after post inoculation. Subsequently, the inoculated leaves were removed with sterilized scissors. The plants were then grown for an additional 28 days, the newly grown leaves were inoculated with hyphal agar discs (Ф = 5.0 mm) of S. sclerotiorum strain 1980 in the same way and the plants were grown under the same conditions for an additional 96 h. The lesion sizes were then measured, and the lesions were imaged. The day and night temperatures were approximately 20 °C/10 °C during the inoculation periods in the field. There were four individual mutant plants of each genotype, and the field experiments were repeated twice.
Dark respiratory rate assay
The T1 mutant and WT were grown in pots (diameter 12 cm, height 12 cm) containing garden soil, in the same growth chamber mentioned before, for 20 days. The young leaves of the T1 plants (seedling stage) and 166‐day‐old T2 mutants (WCR1, WCR2, WCR3, WCR4 and WCR5) (flowering stage) were used for dark respiratory assay (in the dark) using the Li‐Cor 6800 portable gas‐exchange system (Li‐Cor, Lincoln, NE, USA) on a 2‐cm2 area of a leaf that had been adapted to dark for 30 min. The flow setpoint was set to 150 μmol s−1, the CO2 injector was set to 400 μmol mol−1 and the saturated vapor pressure was set to 1.5 kPa. The dark respiratory rate was recorded after the instrument reading stabilization. The temperature was 22 °C for T1 mutants, and 15 °C for T2 mutants. There were eight individual mutant plants of each mutant used for measurement.
Yeast two‐hybrid system and western blotting
Yeast two‐hybrid (Y2H) assays were performed to determine the effects of edited BnQCR8 and QCR8 of other plant species on the interaction with SsSSVP1ΔSP. The cDNA of QCR8 and edited BnQCR8 were cloned and inserted into a pGADT7 vector as ‘prey’, and SsSSVP1ΔSP and BcSSVP1ΔSP were cloned and inserted into pGBKT7 as ‘bait’. These vectors were subsequently co‐transformed into a Y2H Gold strain (Clontech, Palo Alto, CA) by using PEG‐mediated transformation according to the manufacturer’s instructions. The transformed yeast cells were subsequently assayed, as described previously by Lyu et al., (2016).
Off‐target mutation analysis
The potential off‐target sites in T0 BnQCR8 mutants were predicted by the CRISPR‐P analysis system (http://cbi.hzau.edu.cn/cgi‐bin/CRISPR). The sequences covering each off‐target site were amplified with specific primers, and the purified products were cloned into pMD19‐T vectors, and then three clones were selected randomly for Sanger sequencing. These results were compared with those of the WT sequences.
Bioinformatics analysis
The reference sequences of gene and protein of BnQCR8 and its homologous proteins were obtained from B. napus cultivar Westar genome (http://cbi.hzau.edu.cn/bnapus/) and UniProt database (https://www.uniprot.org/). The protein sequences were aligned by using online tool ESPRIPT 3.0 (http://espript.ibcp.fr/ESPript/cgi‐bin/ESPript.cgi). The phylogenetic tree was constructed with MEGA 6.0 by using the maximum‐likelihood method.
Statistical Analysis
The data concerning plant traits and the resistance to S. sclerotiorum and B. cinerea strains were subjected to one‐way analysis of variance in SPSS (SPSS Institute, version 19.0) followed by Duncan’s new multiple range test (DMRT). Treatment means for each index followed by different letters (as shown in the figures) are significantly different at P < 0.05.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Author contributions
Z.X. designed the research, performed all experiments, analysed the data and wrote the manuscript. J.D. designed and supervised the research, analysed the data and wrote the manuscript. C.J., F.Y., X.J., L.B. and L.Y. contributed to design the research, analysed the data and wrote the manuscript. B.X., F.Y., T.Q., L.W., Z.H., L.X., Z.Y. and L.C. contributed to perform experiments.
Supporting information
Figure S1. Sequence alignment of the homologous copies of BnQCR8 in cultivar Westar of rapeseed. (a) The CDS sequence alignment of BnQCR8.A10/C09 and their homologous copies. The target sequences are underlined and highlighted in yellow, respectively, and the PAM is highlighted in red. * shows the same base. (b) Protein sequence alignment of BnQCR8.A10/C09 and their homologous copies. * shows the same amino acid. “.” shows the number of the different amino acid. (c) The amino acid identity among eight homologous copies of BnQCR8 in cultivar Westar.
Figure S2. Identification of T0 positive BnQCR8‐edited mutants of rapeseed cultivar Westar. (a) Detection of T0 mutations by PCR amplification using the specific primers of the HPT II gene. Westar (WT) and ddH2O were as negative controls. The DNA contained HPT II gene was used as a positive control provided by Wuhan Towin Biotechnology Co. LTD. (b) Sequencing chromatogram of the S1 target site in three T0 lines to detect the mutated sites (Red box) in BnQCR8.A10.
Figure S3. Detection of T1 positive transgenic lines of BnQCR8‐edited rapeseed cultivar Westar by PCR amplification assay using the specific primers of the HPT II gene. Westar (‐) was used as a negative control. DNA sample of T0 positive rapeseed with the HPT II gene was used as a positive control (+). BnQCR8‐CR2 (2), BnQCR8‐CR27 (27), BnQCR8‐CR28 (28), BnQCR8‐CR43 (43), BnQCR8‐CR51 (51), BnQCR8‐CR52 (52) and BnQCR8‐CR54 (54) were derived from T0 mutants by self‐pollinating.
Figure S4. Resistance of five genotypes of BnQCR8‐edited T2 mutants to S. sclerotiorum and B. cinerea in rapeseed cultivar Westar tested in the growth chamber. (a and b) Lesions caused by S. sclerotiorum and by B. cinerea, respectively. The plants were grown in a growth chamber for 40 days and 49 days, and leaves were inoculated activating hyphal agar discs (Ф = 5.0 mm) of S. sclerotiorum strain 1980 or B. cinerea strain B05.10, respectively. Inoculated leaves were kept moist with a transparent plastic bag, and the inoculated plants were grown for an additional 36 h for S. sclerotiorum and B. cinerea at the same conditions, and the temperature was about 22 °C with a 16:8‐h light/dark photoperiod and a light intensity of approximately 10 000 lux. (c and d) The average size of lesions induced by S. sclerotiorum (c) and B. cinerea (d). For each genotype mutant, four lesions were measured. The values are presented as means ± s.d for n = 4 replicates. Data were analysed by one‐way ANOVA followed by DMRT. Different lowercase letters on top of each column indicate significant differences at P < 0.05.
Figure S5. Putative proteins or peptides of edited BnQCR8 copies of rapeseed cultivar Westar and their alignments. Insertions and deletions are indicated in “I” and “D”, and the red number shows the number of base and “‐” means the missing amino acid. Insertions and deletions are indicated in “I” and “D”, and the red number shows the number of base and “‐” means missing amino acid.
Figure S6. Content of partial unsaturated fatty acids and glucosinolate content in the oil of each genotype BnQCR8‐edited mutant of rapeseed cultivar Westar. (a‐c) The content of oleic acid, linoleic acid and linolenic acid, respectively. (d) The content of glucosinolate in oil. WCR1, WCR2, WCR3, WCR4 and WCR5 are different genotype mutants of Westar (WT). The values (a‐d) are presented as the means ± s.d for n ≥ 4 replicates. Data were analysed by one‐way ANOVA followed by DMRT. Different lowercase letters on top of each column indicate significant differences at P < 0.05. The oil composition was determined by using near‐infrared spectrometer (NIRSystems 3750), see Figure 4 for detail.
Figure S7. S. sclerotiorum effector SsSSVP1 interacts with QCR8 of other selected plants tested with Yeast two‐hybrid (Y2H). Bn, Sl, Br, Ca, Ha and Gm represent B. napus, Solanum lycopersicum, B. rapa, Capsicum annuum, Helianthus annuus and Glycine max, respectively. The pGADT7‐SV40 TAg co‐transferred with pGBKT7‐Lam and pGBKT7‐p53 were used as negative and positive controls for protein‐protein interaction, respectively. SD–Ade/–His/–Leu/–Trp contained 125 ng mL−1 aureobasidin A (ABA) and 40 μg mL−1 X‐α‐Gal was used.
Table S1. Molecular and genetic analysis of CRISPR/Cas9‐induced mutations in BnQCR8.9/10 of rapeseed cultivar Westar.
Table S2. Primers used in this study.
Table S3. Detection of potential off‐target effect of BnQCR editing at each sgRNA target site on rapeseed cultivar Westar
Table S4. The parent lines of BnQCR8‐edited T2 mutants of rapeseed cultivar Westar.
Acknowledgments
The study was financially supported by China Agriculture Research System of MOF and MARA. We thank Professor Kabin Xie (Huazhong Agricultural University) for providing the pRGEB35 vectors and for his generous suggestions.
Zhang, X. , Cheng, J. , Lin, Y. , Fu, Y. , Xie, J. , Li, B. , Bian, X. , Feng, Y. , Liang, W. , Tang, Q. , Zhang, H. , Liu, X. , Zhang, Y. , Liu, C. and Jiang, D. (2021) Editing homologous copies of an essential gene affords crop resistance against two cosmopolitan necrotrophic pathogens. Plant Biotechnol. J., 10.1111/pbi.13667
References
- Ali, A. and Yun, D.J. (2020) Arabidopsis HOS15 is a multifunctional protein that negatively regulate ABA‐signaling and drought stress. Plant Biotechnol. Rep. 14, 163–167. [Google Scholar]
- Amselem, J. , Cuomo, C.A. , van Kan, J.A. , Viaud, M. , Benito, E.P. , Couloux, A. , Coutinho, P.M. et al. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genet. 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armin, S. and David, E. (2017) Plant breeding genome editors take on crops genome editing technologies may help to enhance global food security. Science, 355, 1122–1123.28302807 [Google Scholar]
- Bolton, M.D. , Thomma, B.P. and Nelson, B.D. (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I.A. , Tang, H. , Wang, X. , Chiquet, J. et al. (2014) Plant genetics. Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Wang, Y. , Zhang, R. , Zhang, H. and Gao, C. (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. et al. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire, M.C. and Denton‐Giles, M. (2016) The control of sclerotinia stem rot on oilseed rape (Brassica napus): current practices and future opportunities. Plant Pathol. 65, 859–887. [Google Scholar]
- Ding, D. , Chen, K. , Chen, Y. , Li, H. and Xie, K. (2018) Engineering introns to express RNA guides for Cas9‐ and Cpf1‐mediated multiplex genome editing. Mol. Plant, 11, 542–552. [DOI] [PubMed] [Google Scholar]
- El‐Brolosy, M.A. , Kontarakis, Z. , Rossi, A. , Kuenne, C. , Günther, S. , Fukuda, N. , Kikhi, K. et al. (2019) Genetic compensation triggered by mutant mRNA degradation. Nature, 568, 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, I.J. , Tong, C. , Becker, M.G. , Mao, X. , Huang, J. , de Kievit, T. , Fernando, W. et al. (2017) RNA sequencing of Brassica napus reveals cellular redox control of Sclerotinia infection. J. Exp. Bot. 68, 5079–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey, L.T. , Hafeez, N. , Jackson, S.A. , Leal‐Bertioli, S.C.M. , Tester, M. , Gao, C. et al. (2019) Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. [DOI] [PubMed] [Google Scholar]
- Hu, Q. , Hua, W. , Yin, Y. , Zhang, X. and Liu, L. (2017) Rapeseed research and production in China. Crop J. 5, 127–135. [Google Scholar]
- van Kan, J.A.L. (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11(5), 247–253. [DOI] [PubMed] [Google Scholar]
- Kretschmer, M. , Damoo, D. , Djamei, A. and Kronstad, J. (2019) Chloroplasts and plant immunity: Where are the fungal effectors? Pathogens, 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C.T. , Heo, J. , Lemmon, Z.H. , Capua, Y. , Hutton, S.F. , Van Eck, J. , Park, S.J. et al. (2020) Rapid customization of Solanaceae fruit crops for urban agriculture. Nat. Biotechnol. 28, 182–188. [DOI] [PubMed] [Google Scholar]
- Lei, Y. , Lu, L. , Liu, H.Y. , Li, S. , Xing, F. and Chen, L.L. (2014) CRISPR‐P: a web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Mol. Plant, 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2012) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nat. Biotechnol. 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Wang, C. , Jiao, X. , Zhang, H. , Song, L. , Li, Y. , Gao, C. et al. (2019) Hi‐TOM: a platform for high‐throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China life Sci. 62, 1–7. [DOI] [PubMed] [Google Scholar]
- Lyu, X. , Shen, C. , Fu, Y. , Xie, J. , Jiang, D. , Li, G. and Cheng, J. (2016) A small secreted virulence‐related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 12, e1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Chen, L. , Zhu, Q. , Chen, Y. and Liu, Y. (2015a) Rapid decoding of sequence‐specific nuclease‐induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol. Plant, 8, 1285–1287. [DOI] [PubMed] [Google Scholar]
- Ma, Z. , Zhu, P. , Shi, H. , Guo, L. , Zhang, Q. , Chen, Y. , Chen, S. et al. (2019) PTC‐bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature, 568, 259–263. [DOI] [PubMed] [Google Scholar]
- Matthias, H. (2014) The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J. Chem. Biol. 7, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva, R. , Ji, C. , Atienza‐Grande, G. , Huguet‐Tapia, J.C. , Perez‐Quintero, A. , Li, T. , Eom, J.S. et al. (2019) Broad‐spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.J. , Baek, D. , Cha, J.Y. , Liao, X. , Kang, S.H. , McClung, C.R. , Lee, S.Y. et al. (2019) HOS15 Interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA . Plant Cell, 31, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, A. , Chen, S. , Lei, T. , Xu, L. , He, Y. , Wu, L. , Yao, L. et al. (2017) Engineering canker‐resistant plants through CRISPR/Cas9‐targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasch, S. , Knapp, S.J. , van Kan, J. and Blanco‐Ulate, B. (2019) Grey mould of strawberry, a devastating disease caused by the ubiquitous necrotrophic fungal pathogen Botrytis cinerea . Mol. Plant Pathol. 20, 877–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski, S. , Bailey, L.G. and Baum, B.R. (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. [Google Scholar]
- Ray, S.K. , Macoy, D.M. , Kim, W.Y. , Lee, S.Y. and Kim, M.G. (2019) Role of RIN4 in regulating PAMP‐triggered immunity and effector‐triggered immunity: Current status and future perspectives. Mol. Cells, 42, 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Leal, D. , Lemmon, Z.H. , Man, J. , Bartlett, M.E. and Lippman, Z.B. (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell, 171, 470–480. [DOI] [PubMed] [Google Scholar]
- Rossi, A. , Kontarakis, Z. , Gerri, C. , Nolte, H. , Hölper, S. , Krüger, M. and Stainier, D.Y. (2015) Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature, 524, 230–233. [DOI] [PubMed] [Google Scholar]
- Shen, M. , Lim, C.J. , Park, J. , Kim, J.E. , Baek, D. , Nam, J. , Lee, S.Y. et al. (2020) HOS15 is a transcriptional corepressor of NPR1‐mediated gene activation of plant immunity. Proc. Natl. Acad. Sci. USA, 117, 30805–30815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, L. , Yang, G. , Ma, M. , Liu, X. , Li, B. , Xie, J. , Fu, Y. et al.(2020) An effector of a necrotrophic fungal pathogen targets the calcium‐sensing receptor in chloroplasts to inhibit host resistance. Mol Plant Pathol. 21, 686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A. , Rana, K. , Handy, C. and Clarkson, J.P. (2018) Resistance to Sclerotinia sclerotiorum in wild Brassica species and the importance of Sclerotinia subarctica as a Brassica pathogen. Plant Pathol. 67, 433–444. [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Williamson, B. , Tudzynski, B. , Tudzynski, P. and van Kan, J.A. (2007) Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Zhao, Q. , Yang, Q. , Liu, H. , Li, Q. , Yi, X. , Cheng, Y. et al. (2016) Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus . Sci. Rep. 6, 19007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Minkenberg, B. and Yang, Y. (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proc. Natl. Acad. Sci. USA, 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. , Yuan, M. , Ai, C. , Liu, L. , Zhuang, E. , Karapetyan, S. , Wang, S. et al. (2017) uORF‐mediated translation allows engineered plant disease resistance without fitness costs. Nature, 545, 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Xu, X. , Gong, Q. , Li, Z. , Li, Y. , Wang, S. , Yang, Y. et al. (2019) Engineering broad‐spectrum bacterial blight resistance by simultaneously disrupting variable TALE‐binding elements of multiple susceptibility genes in rice. Mol Plant, 12, 1434–1446. [DOI] [PubMed] [Google Scholar]
- Yan, D. , Ren, B. , Liu, L. , Yan, F. , Li, S. , Wang, G. , Sun, W. et al. (2021) High‐fficiency and multiplex adenine base editing in plants using new TadA variants. Mol. Plant, 14, 722–731. [DOI] [PubMed] [Google Scholar]
- Yang, G. , Tang, L. , Gong, Y. , Xie, J. , Fu, Y. , Jiang, D. , Li, G. et al. (2018) A cerato‐platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum . New Phytol. 217, 739–755. [DOI] [PubMed] [Google Scholar]
- Zaidi, S.S. , Vanderschuren, H. , Qaim, M. , Mahfouz, M.M. , Kohli, A. , Mansoor, S. and Tester, M. (2019) New plant breeding technologies for food security. Science, 363, 1390–1391. [DOI] [PubMed] [Google Scholar]
- Zhou, F. , Zhang, X. , Li, J. and Zhu, F. (2014) Dimethachlon resistance in Sclerotinia sclerotiorum in China. Plant. Dis. 98, 1221–1226. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Wang, H. , Gilmer, S. , Whitwill, S. , Keller, W. and Fowke, L.C. (2002) Control of petal and pollen development by the plant cyclin‐dependent kinase inhibitor ICK1 in transgenic Brassica plants. Planta, 215, 248–257. [DOI] [PubMed] [Google Scholar]
- Zhu, H. , Li, C. and Gao, C. (2020) Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence alignment of the homologous copies of BnQCR8 in cultivar Westar of rapeseed. (a) The CDS sequence alignment of BnQCR8.A10/C09 and their homologous copies. The target sequences are underlined and highlighted in yellow, respectively, and the PAM is highlighted in red. * shows the same base. (b) Protein sequence alignment of BnQCR8.A10/C09 and their homologous copies. * shows the same amino acid. “.” shows the number of the different amino acid. (c) The amino acid identity among eight homologous copies of BnQCR8 in cultivar Westar.
Figure S2. Identification of T0 positive BnQCR8‐edited mutants of rapeseed cultivar Westar. (a) Detection of T0 mutations by PCR amplification using the specific primers of the HPT II gene. Westar (WT) and ddH2O were as negative controls. The DNA contained HPT II gene was used as a positive control provided by Wuhan Towin Biotechnology Co. LTD. (b) Sequencing chromatogram of the S1 target site in three T0 lines to detect the mutated sites (Red box) in BnQCR8.A10.
Figure S3. Detection of T1 positive transgenic lines of BnQCR8‐edited rapeseed cultivar Westar by PCR amplification assay using the specific primers of the HPT II gene. Westar (‐) was used as a negative control. DNA sample of T0 positive rapeseed with the HPT II gene was used as a positive control (+). BnQCR8‐CR2 (2), BnQCR8‐CR27 (27), BnQCR8‐CR28 (28), BnQCR8‐CR43 (43), BnQCR8‐CR51 (51), BnQCR8‐CR52 (52) and BnQCR8‐CR54 (54) were derived from T0 mutants by self‐pollinating.
Figure S4. Resistance of five genotypes of BnQCR8‐edited T2 mutants to S. sclerotiorum and B. cinerea in rapeseed cultivar Westar tested in the growth chamber. (a and b) Lesions caused by S. sclerotiorum and by B. cinerea, respectively. The plants were grown in a growth chamber for 40 days and 49 days, and leaves were inoculated activating hyphal agar discs (Ф = 5.0 mm) of S. sclerotiorum strain 1980 or B. cinerea strain B05.10, respectively. Inoculated leaves were kept moist with a transparent plastic bag, and the inoculated plants were grown for an additional 36 h for S. sclerotiorum and B. cinerea at the same conditions, and the temperature was about 22 °C with a 16:8‐h light/dark photoperiod and a light intensity of approximately 10 000 lux. (c and d) The average size of lesions induced by S. sclerotiorum (c) and B. cinerea (d). For each genotype mutant, four lesions were measured. The values are presented as means ± s.d for n = 4 replicates. Data were analysed by one‐way ANOVA followed by DMRT. Different lowercase letters on top of each column indicate significant differences at P < 0.05.
Figure S5. Putative proteins or peptides of edited BnQCR8 copies of rapeseed cultivar Westar and their alignments. Insertions and deletions are indicated in “I” and “D”, and the red number shows the number of base and “‐” means the missing amino acid. Insertions and deletions are indicated in “I” and “D”, and the red number shows the number of base and “‐” means missing amino acid.
Figure S6. Content of partial unsaturated fatty acids and glucosinolate content in the oil of each genotype BnQCR8‐edited mutant of rapeseed cultivar Westar. (a‐c) The content of oleic acid, linoleic acid and linolenic acid, respectively. (d) The content of glucosinolate in oil. WCR1, WCR2, WCR3, WCR4 and WCR5 are different genotype mutants of Westar (WT). The values (a‐d) are presented as the means ± s.d for n ≥ 4 replicates. Data were analysed by one‐way ANOVA followed by DMRT. Different lowercase letters on top of each column indicate significant differences at P < 0.05. The oil composition was determined by using near‐infrared spectrometer (NIRSystems 3750), see Figure 4 for detail.
Figure S7. S. sclerotiorum effector SsSSVP1 interacts with QCR8 of other selected plants tested with Yeast two‐hybrid (Y2H). Bn, Sl, Br, Ca, Ha and Gm represent B. napus, Solanum lycopersicum, B. rapa, Capsicum annuum, Helianthus annuus and Glycine max, respectively. The pGADT7‐SV40 TAg co‐transferred with pGBKT7‐Lam and pGBKT7‐p53 were used as negative and positive controls for protein‐protein interaction, respectively. SD–Ade/–His/–Leu/–Trp contained 125 ng mL−1 aureobasidin A (ABA) and 40 μg mL−1 X‐α‐Gal was used.
Table S1. Molecular and genetic analysis of CRISPR/Cas9‐induced mutations in BnQCR8.9/10 of rapeseed cultivar Westar.
Table S2. Primers used in this study.
Table S3. Detection of potential off‐target effect of BnQCR editing at each sgRNA target site on rapeseed cultivar Westar
Table S4. The parent lines of BnQCR8‐edited T2 mutants of rapeseed cultivar Westar.


