Abstract
Canine distemper of domestic dogs is caused by canine distemper virus (CDV), a member of the morbilliviruses. It has been a highly contagious disease of great veterinary importance for centuries, but for the last several decades it has been controlled satisfactorily by modified live vaccines. In the 1990s, however, it was described that CDV strains genetically different from vaccine strains may have caused the disease in vaccinated dogs. The highest antigenic variation is found in the H protein. Therefore, in the present study, hemagglutinin (H) genes obtained from current vaccines and field isolates and amplified directly from clinical specimens were genetically analyzed by restriction fragment length polymorphism assay and sequencing. Phylogenetic analysis of the H-gene amino acid sequences revealed that at least two CDV genotypes are circulating among dogs in Japan; one is a genotype to which almost all Japanese CDV isolates belong and the other has not been previously described. Both are separate and independent from the other lineages or genotypes of vaccine strains, as well as European and U.S. CDV isolates. The results suggest that CDV has also evolved in Japan, and further studies will be needed for an evaluation and possible improvement of the efficacies of current CDV vaccines.
Canine distemper virus (CDV), a member of the genus Morbillivirus in the Paramyxoviridae family, is closely related to human measles virus and bovine rinderpest virus. The disease caused by CDV has been known for centuries and throughout the world still remains one of the important contagious diseases of dogs as well as other carnivores, including Old World large felids (3, 34). When dogs that possess no protective immunity are infected with CDV, acute to subacute systemic diseases with high mortality rates develop. Targets of infection for CDV are mainly mucous membranes and lymphoid tissues throughout the body; thus, the disease is typically characterized by manifestations of pyrexia, anorexia, nasal discharge, conjunctivitis, and diarrhea. Skin pustules, hyperkeratosis, and central nervous system signs are also seen in some animals. The disease has been kept under control since introduction of the modified live CDV vaccines several decades ago (for reviews, see references 1, 2, and 13).
CDV has an envelope composed of a membrane protein termed M and two glycoproteins termed H (hemagglutinin, the attachment protein of CDV) and F (fusion protein), and the last two proteins are major target antigens for the host immune system. The highest antigenic variation has been found in the H protein (5), and this protein is the most appropriate protein to be monitored for detection of genetic changes of the virus, as has been pointed out previously (15, 26). Accordingly, it has been reported from European countries (6, 14, 15), the United States (18), and Japan (20) that it is apparent that there is pronounced genetic diversity in the H gene of recent field CDV isolates (referred to here as “new CDV”) compared to the genetic diversity of currently available modified live CDV vaccine strains, for example, Onderstepoort and Convac. These vaccine strains were produced in the 1950s and 1960s (hence, they are referred to as “old CDV”). Antigenic differences between new CDVs and old CDVs have been found in neutralization tests (12, 19), whereas this may not necessarily invalidate the current vaccines for protecting dogs against new CDV infections (14, 15).
An additional notable aspect of new CDVs is the presence of geographically distinct lineages revealed by phylogenetic analysis of the H gene (6, 7, 15, 18, 20), as was also found for other morbilliviruses (8, 28, 31, 33). These lineages or genotypes are not grouped exclusively according to the host species of the viruses (6, 7, 18), although there seem to be host species-specific lineages (15). In Japan, one genotype has been recognized in new CDVs since the 1980s, and it was found to differ from those of the European and U.S. new CDVs (20, 25).
We have routinely detected CDV RNA in clinical specimens by the reverse transcriptase PCR (RT-PCR) assay instead of by a cell culture method because of the fastidious nature of CDV in vitro. When CDV was detected in clinical specimens by the RT-PCR, a restriction fragment length polymorphism (RFLP) assay was also applied to the RT-PCR product for tentative genotyping of the H gene. Here we describe the genotype profiles of CDVs recently prevalent among dogs in Japan. A strain of a new CDV genotype was detected in the sample taken from a puppy clinically diagnosed with canine distemper. Phylogenetic analysis of the H-gene amino acid sequence showed that it is genotypically distinct from any previously known genotypes.
MATERIALS AND METHODS
Viruses and clinical specimens.
Seven CDV strains were used as references; six (strains Onderstepoort, Fromm, DFE-HC, Lederle VR-128, Rockborn, and FXNO) are vaccine strains presently distributed in Japan. One strain, the attenuated Snyder Hill strain in a commercial vaccine product, could not be applied for the experiment because the amplification of the H gene from the vaccine was unsuccessful. However, a laboratory wild-type Snyder Hill strain was used as an alternative. These reference CDVs are considered to be old CDVs on the basis of the dates of their isolation. Reconstituted vaccine and cell culture fluids were applied for the RT-PCR.
Strain KDK-1, a field isolate obtained from an infected animal in 1991, has been passaged in a Vero cell culture. Culture fluid from the sixth passage was used for H-gene sequencing and RFLP experiments. The Vero cells were cultured in Eagle’s minimal essential medium supplemented with 10% tryptose phosphate broth and 8% fetal bovine serum. Four additional CDV isolates obtained in 1994, namely, C710D, C714D, C717D, and C720D, were also passaged in the Vero cell culture 10 times and were then used.
A total of 101 clinical specimens, 11 oral and 90 rectal swab specimens, were taken from dogs manifesting respiratory or enteric disease signs. These samples were submitted by animal hospitals in various parts of Japan during the years 1997 and 1998, and of these, three oral and four rectal swab specimens were found to be CDV RNA positive by the RT-PCR assay. The profiles of these field CDV isolates and CDV RNA-positive clinical specimens are summarized in Table 1.
TABLE 1.
CDV isolates and CDV-positive clinical specimens
| Strain source and strain | Date and place of collection (Japan) | Vaccination historya | Age and clinical signs of patient | Type by RFLP analysis with:
|
|
|---|---|---|---|---|---|
| NdeI | FbaI | ||||
| Field isolates | |||||
| KDK-1 | 1991, Ibaraki | − | Puppy; canine distemper? (no specification) | KDK-1 | KDK-1 |
| C710D | 1994, Mie | Unknown | 2 mo; respiratory and nervous signs, bloody diarrhea | KDK-1 | KDK-1 |
| C714D | 1994, Hokkaido | − | 6 mo; lameness of right hind leg, nasal discharge | KDK-1 | KDK-1 |
| C717D | 1994, Hokkaido | + | 10 mo; diarrhea, convulsion | KDK-1 | KDK-1 |
| C720D | 1994, Hokkaido | + | 7 mo; nervous signs | KDK-1 | KDK-1 |
| Rectal swab | |||||
| 97-050b | 1997, Okayama | − | 2 mo; diarrhea, coughing, eye discharge, hard pad | NPc | NP |
| 97-063b | 1997, Tochigi | Unknown | 4 mo; canine distemper? (no specification) | NP | NP |
| 98-001d | 1998, Hokkaido | Unknown | Puppy; diarrhea | KDK-1 | KDK-1 |
| 98-002e | 1998, Chiba | − | 2 mo; diarrhea, hypersalivation, nervous signs | 98-002 | KDK-1 |
| Oral swab | |||||
| 98/001e | 1998, Chiba | − | 2 mo; diarrhea, hypersalivation, nervous signs | 98-002 | KDK-1 |
| 98/003 | 1998, Chiba | − | 2 mo; nasal discharge, vomiting, tonsillitis | KDK-1 | KDK-1 |
| 98/005 | 1998, Chiba | − | 2 mo; vomiting, respiratory signs | KDK-1 | KDK-1 |
Combined modified live CDV vaccine.
Samples 97-050 and 97-063 were positive for coronavirus.
NP, not performed.
Sample 98-001 was positive for canine parvovirus and reovirus.
Taken from the same patient.
Primer selection.
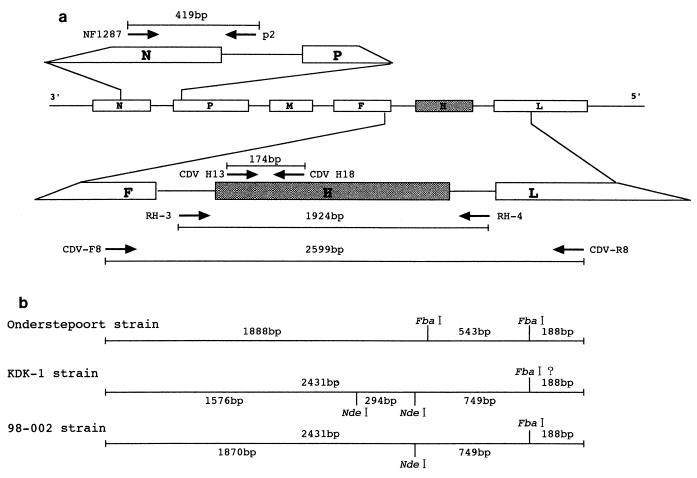
Four sets of oligonucleotide primers were used in the present study according to the sequence information for the Onderstepoort strain (Fig. 1; Table 2). For routine CDV detection, a set of primers (CDV H13 and CDV H18) inside of the H gene (20) was applied. To obtain a PCR product for RFLP analysis and sequencing of the H gene, two sets of primers were used: one was designed from the sequence of the untranslated regions between the F and H genes (primer RH-3) and between the H- and large (L)-protein genes (primer RH-4) (18), and another was from the F-gene region (primer CDV-F8) and the L-gene region (primer CDV-R8). For detection and sequencing of the nucleocapsid (N) gene, a previously described set of primers (primers NF1287 and p2) (35) was applied.
FIG. 1.
Outline of the RT-PCR and scheme for RFLP analysis of the CDV H gene. (a) Genomic organization of the CDV genome, the set of primers, and the lengths of the products of the RT-PCRs. The various CDV genes are represented by boxes. The genes for nucleocapsid protein (N), the phosphoprotein (P), the matrix protein (M), the fusion protein (F), the hemagglutinin protein (H), and the large protein (L) are indicated. (b) Scheme for the restriction enzyme patterns of CDV H genes digested with FbaI and NdeI. Strains Onderstepoort and KDK-1 were used as representative strains of old and new CDVs, respectively. For strain KDK-1, the third restriction site, which was obtained by digestion with NdeI and which generated the smallest fragment (approximately 100 bp), cannot be specified because no sequence information is available for either end of a PCR product for primers CDV-F8 and CDV-R8.
TABLE 2.
Oligonucleotide primers used in the RT-PCR assay
| Primer | Sequence (5′ to 3′) | Positiona | Orientation | Reference or source |
|---|---|---|---|---|
| NF1287 | GTGTCAGAAATAGCATCCAAG | 1287–1307 | Sense | 35 |
| p2 | GTGGGATCCAGACTGGTCTTGAATAT | 1705–1680 | Antisence | 35 |
| CDV H13 | CAAGACAAGGTGGGTGCCTT | 7091–7110 | Sense | 20 |
| CDV H18 | CTTGGTGAAATCGAACTCCA | 7265–7246 | Antisence | 20 |
| RH-3 | AGGGCTCAGGTACTCCAGC | 7059–7077 | Sense | 18 |
| RH-4 | AATGCTAGAGATGGTTTAATT | 8995–8975 | Antisence | 18 |
| CDV-F8 | GTTGTTGCTGATTTACTGTT | 6800–6819 | Sense | Present study |
| CDV-R8 | CCCCGTCTGTTATTTTGCTA | 9399–9380 | Antisence | Present study |
Numerical position on the genome of CDV Onderstepoort.
Reverse transcription and PCR amplification.
Total RNA was obtained from 250 μl of clarified infected cell culture fluid or swab extract by using Isogen-LS (Nippon Gene, Toyama, Japan) according to the manufacturer’s instructions and was dissolved in 20 μl of TE (Tris-EDTA) buffer (pH 8.0). Complementary DNA was synthesized by using a random 9-mer primer (Takara, Tokyo, Japan). After 1 μl of the random primer (50 pmol/μl) was added to 9 μl of the RNA solution, the mixture was incubated at 70°C for 10 min and cooled on ice. To this mixture 4 μl of 5× reaction buffer (250 mM Tris-HCl [pH 8.3], 500 mM KCl, 20 mM dithiothreitol, 50 mM MgCl2; Takara) 1 μl of 133 U RNase inhibitor (Takara) per μl, and 4 μl of a deoxynucleoside triphosphate mixture (with a 2.5 mM concentration of each dNTP; dNTP Mixture; Takara) were added, the mixture was incubated at 25°C for 5 min, and then 1 μl of 37 U of avian myeloblastosis virus RT XL (Takara) per μl was finally added. A total of 20 μl of the reaction mixture was then incubated at 25°C for 10 min, 42°C for 50 min, and 70°C for 10 min.
For the routine detection of CDV in clinical specimens, PCR was performed as follows. A total of 50 μl of a reaction mixture was made by mixing 0.5 μl of the cDNA, 0.25 μl of Taq polymerase (5 U/μl; Takara), 5 μl of 10× PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl; Takara), 3 μl of 25 mM MgCl2, 4 μl of 2.5 mM dNTP Mixture, 36.25 μl of distilled water, and 0.5 μl each of primers CDV H13 and CDV H18 (50 pmol/μl). The mixture was placed in a Trio-Thermoblock (Biometra, Tampa, Fla.). The temperature cycling protocol consisted of 30 cycles of 30 s for each denaturation at 94°C, primer annealing at 55°C, and extension at 72°C. Exactly the same protocol was used for amplification of part of the N gene by using 0.5 μl each of primers NF1287 and p2 (50 pmol/μl).
To obtain a PCR product for sequencing of the H gene of strain KDK-1, 0.5 μl each of primers RH-3 and RH-4 (50 pmol/μl) was added to a total of 49 μl of the reaction mixture mentioned above. Amplification was conducted by a temperature cycling protocol consisting of 25 cycles of 30 s of denaturation at 94°C, 1 min of primer annealing at 55°C, and 1 min of extension at 72°C, followed by 10 min of the final extension phase at 72°C.
To obtain PCR products for the RFLP analysis and sequencing of the H gene from rectal swab sample 98-002, 0.5 μl each of primers CDV-F8 and CDV-R8 (50 pmol/μl) was added to a total of 49 μl of the reaction mixture consisting of 0.5 μl of the cDNA, 0.25 μl of Ex Taq polymerase (5 U/μl; Takara), and 5 μl of 10× PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 20 mM MgCl2; Takara), 4 μl of 2.5 mM dNTP Mixture, and 39.75 μl of distilled H2O. Amplification was conducted by a temperature cycling protocol consisting of 35 cycles of 1 min of denaturation at 94°C, 2 min of primer annealing at 50°C, and 2 min of extension at 72°C, followed by 2 min of the final extension phase at 72°C.
Sequencing.
The PCR products prepared from strain KDK-1 and rectal swab sample 98-002 were purified with the Wizard PCR Preps DNA Purification System (Promega, Madison, Wis.). Then, TA cloning was performed with the Regular pT7Blue(R)T-vector Kit (Novagen, Madison, Wis.), and ligation mixtures were transformed into Epicurian Coli XL2-Blue MRF′ competent cells (Stratagene, La Jolla, Calif.). For each sample, 10 plasmids containing the PCR product were purified with the Wizard Minipreps DNA Purification System (Promega), sequenced with Cy5-labelled m13 primers, and further sequenced with Cy5-labelled primers designed from the sequences that were obtained by using the AutoRead Sequencing Kit (Amersham Pharmacia Biotech, Tokyo, Japan).
RFLP analysis of PCR-amplified products.
The PCR product obtained with primers CDV-F8 and CDV-R8 was concentrated by using the Wizard PCR Preps DNA Purification System, and the resultant concentrate was digested with restriction enzymes FbaI and NdeI (Takara), respectively. As shown in Fig. 1, the enzymes were adopted because of a probability that they distinguish between old and new CDVs on the basis of information on their sequences from the nucleotide sequence database. An 8-μl aliquot was digested with 1.2 and 1.0 U of FbaI and NdeI, respectively, at 37°C for 2 h under the manufacturer’s recommended conditions. The resulting restriction fragments were resolved by 2% agarose gel electrophoresis, and the bands were visualized after staining with ethidium bromide.
Phylogenetic analysis.
Nucleic acid sequences were translated into amino acid sequences, and the latter were aligned with known CDV H-gene amino acid sequences by using Genetyx-Mac, version 8.5 (Software Development Co., Tokyo, Japan). The alignments were input into Clustal W, version 1.74 (32), and PHYLIP, version 3.5 (11), modules to produce phylogenetic trees. Tree construction was done without outgroup specification but with bootstrap analysis (Clustal W; 1,000 trials with seed 111, including gap positions, without multiple-substitution correction).
The term genotype has been used occasionally in the grouping of CDVs (6, 15), but a formal analysis has not yet been published. In the present study genotypes were defined by the phylogenetic properties of the H-gene amino acid sequence; that is, strains in the same clade showing more than 95% amino acid homology are considered to belong to the same genotype.
Nucleotide sequence accession numbers.
The references for and the nucleotide sequence accession numbers in the GenBank database of the H-gene sequences of the indicated strains used in this study are as follows: Snyder Hill (15); Vaccine/Onderstepoort (4, 9), AF014953 and D00758; Vaccine/Convac (22), Z35493; Chinese leopard/A92-27/4 (18), Z54156; Black leopard/A92-6 (18), Z54166; Dog/US89 (6), Z47762; Leopard/US91 (6), Z47763; Raccoon/US89 (6), Z47765; Javelina/US89 (6), Z47765; Dog/DK91, B+C (6), Z47761; Mink/DK86 (6), Z47759; Dog/GR88 (6), Z47760; Dog/404 (15), Z77671; Dog/2544 (15), Z77672; Dog/4513 (15), 77673; Siberian seal/PDV-2 (23), X84998; Ferret/1493/Han89 (23), X84999; Dog/5804/Han90 (23), X85000; Dog/Ueno (20), D85753; Dog/Hamamatsu (20), D85754; Dog/Yanaka (20), D85755; and Racoon dog/Tanu96, AB016776. The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession nos. AB025270 and AB025271 for the strain from sample 98-002 and strain KDK-1, respectively.
RESULTS
RFLP analysis of H gene from reference CDV strains and CDV RNA-positive clinical specimens.
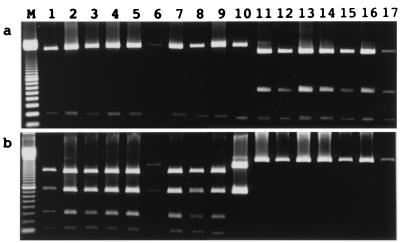
At first, the reference CDV strains and KDK-1 strain were compared. As shown in Fig. 2a, all seven reference CDV strains were digested with FbaI, resulting in three fragments of approximately 1,800, 600, and 200 bp, but only two fragments of approximately 2,400 and 200 bp were obtained for strain KDK-1. When NdeI was applied (Fig. 2b), KDK-1 was digested into four fragments of approximately 1,500, 300, and 750, and 100 bp, but the reference CDV strains were not digested at all. These results indicated that strain KDK-1, which was isolated in 1991, was genetically different from the reference old CDV strains.
FIG. 2.
Restriction enzyme patterns of CDV H genes digested with FbaI (a) and NdeI (b). Lane M, 100-bp ladder size marker; lanes 1 to 5, field isolates KDK-1, C710D, C714D, C717D, and C720D, respectively; lanes 6 to 10, clinical samples 98/001, 98/003, 98/005, 98-001, and 98-002, respectively; lanes 11 to 17, reference strains Onderstepoort, Fromm, DFE-HC, Lederle VR-128, Rockborn, and FXNO and wild-type strain Snyder Hill, respectively.
Of a total of seven clinical specimens which were found to be CDV RNA positive by RT-PCR (Table 1), three oral and two rectal swab samples were used for RFLP analysis of the H gene region. A PCR product appropriate for analysis was not obtained from two rectal swab samples (samples 97-050 and 97-063) because of an interfering background of nonspecific PCR products. Consequently, four CDV isolates and five clinical samples were analyzed (Table 1 and Fig. 2). CDV C710D, C714D, C717D, and C720D, oral swab samples 98/002 and 98/003, and rectal swab sample 98-001 showed RFLP profiles similar to those of strain KDK-1. The RFLP profiles of oral swab sample 98/001 and rectal swab sample 98-002, which were taken from the same patient (Table 1), were similar to that of strain KDK-1 when the samples were digested with FbaI but different from that of KDK-1 when they were digested with NdeI. By RFLP analysis two fragments of approximately 1,900 and 750 bp were observed (Fig. 1 and 2b).
Amino acid sequence analysis of H genes of KDK-1 and rectal sample 98-002. (i) KDK-1.
As a representative of new Japanese CDV isolates, the H gene of strain KDK-1 was sequenced and the predicted amino acid sequence was analyzed. The H gene was composed of a fragment of 1,824 bp and a single open reading frame capable of encoding 607 amino acids. The amino acid homologies to the published amino acid sequences of vaccine strains Onderstepoort and Convac and the wild-type Snyder Hill strain (15) were 89.6, 90.3, and 90.9%, respectively. On the other hand, the highest degrees of homology (98.5 to 98.7%) were found to the sequences of Japanese new CDV strains Ueno, Yanaka, and Hamamatsu (20), but the degrees of homology to new CDV strains reported from other geographical regions and Siberian seal morbillivirus PDV-2 were rather lower (93.1 to 95.9%). A total of eight potential glycosylation sites were recognized at positions 149 to 151, 309 to 311, 391 to 393, 422 to 424, 456 to 458, 584 to 586, 587 to 589, and 603 to 605. The glycosylation site at positions 19 to 21, which is shared by all CDV strains reported so far, was absent from strain KDK-1 but the rest of the sites were shared with those of the Japanese CDV strains, which possess nine sites (20).
(ii) 98-002.
Since the RFLP analysis indicated that oral swab sample 98/001 and rectal swab sample 98-002 were most likely identical, only sample 98-002 was used for sequencing. The nucleotide length of the H-gene fragment of 98-002 was also 1,824 bp, and a single open reading frame capable of encoding 607 amino acids was identified. The degrees of amino acid homology to the old CDV strains Onderstepoort and Convac and to wild-type strain Snyder Hill were again the lowest (89.2 to 90.1%). However, the degrees of homology not only to the other Japanese CDV strains including KDK-1 (93.2 to 93.4%) but also to new CDV strains from other geographical regions (92.6 to 93.9%) were lower than those found in the analysis of strain KDK-1. Different from the other Japanese CDV strains (20), a total of eight potential glycosylation sites were recognized in the H-gene amino acid sequence at positions 19 to 21, 149 to 151, 309 to 311, 391 to 393, 422 to 424, 456 to 458, 587 to 589, and 603 to 605; and amino acid changes were found to be randomly distributed throughout the gene. The glycosylation site at positions 584 to 586, which is shared by all Japanese CDV strains including KDK-1 but not by CDVs from the other regions including old CDVs, was absent from specimen 98-002.
In both strain KDK-1 and specimen 98-002, cysteine residues were completely conserved at 12 positions (positions 139, 154, 188, 283, 296, 377, 382, 390, 490, 566, 575, and 602), and these are identical to those of all CDV strains in the database.
Phylogenetic analysis of amino acid sequence of H genes.
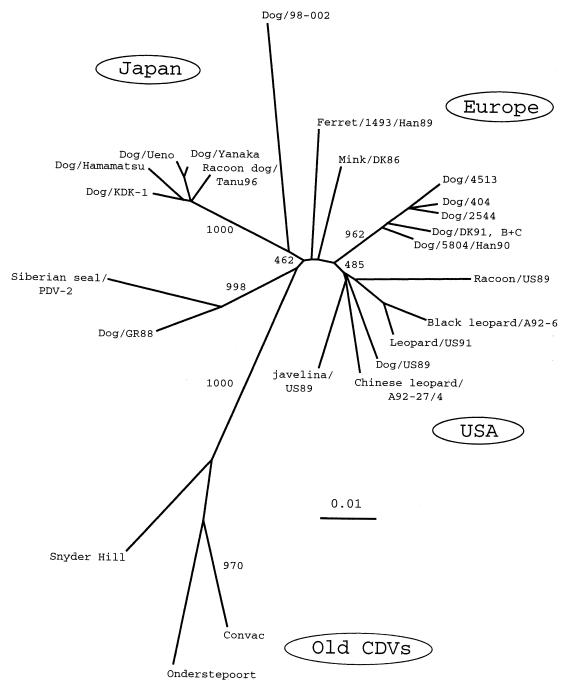
Figure 3 shows the inferred phylogenetic relationships between old and new CDV strains including that in specimen 98-002 on the basis of the alignments of the amino acids of the H gene. Old CDV strains, such as Onderstepoort, Convac, and Snyder Hill, form a distinct clade, while the new CDV strains grouped in several separate clades independent from old CDV strains. Strain KDK-1 joined a tight cluster of the Japanese new CDV isolates, which formed a distinct clade. The U.S. isolates appear to be in one clade, and there are several lineages among the European isolates, as described previously (6, 15, 18). The phylogenetic tree with high bootstrap values indicates that the CDV in specimen 98-002 branched out from the root of the Japanese clade, and it does not correspond to any known CDV clades.
FIG. 3.
Phylogenetic analysis of the amino acid sequences of the coding regions of CDV H proteins. The tree was constructed by using Clustal W, version 1.74 (32), and PHYLIP, version 3.5 (11), modules. Tree topology was based on the neighbor-joining method. The bootstrap values indicate the number of times that each branching was found in 1,000 bootstrap analyses. Branch lengths indicate phylogenetic distances calculated from distance martices of deduced amino acid sequences. Sequences of CDV strain KDK-1 (accession no. AB02527) and the strain from sample 98-002 (accession no. AB025270) were generated in this study. Other sequences were extracted from the database for the following strains (nucleotide sequence accession numbers are given in parentheses): vaccine strains, Onderstepoort (AF014953, D00758) and Convac (Z35493); U.S. isolates, Chinese leopard/A92-27/4 (Z54156), Black leopard/A92-6 (Z54166), dog/US89 (Z47762), Leopard/US91 (Z47763), Raccoon/US89 (Z47765), Javelina/US89 (Z47765); European isolates, Dog/DK91,B+C (Z47761), Mink/DK86 (Z47759), Dog/404 (Z77671), Dog/2544 (Z77672), Dog/4513 (77673), Ferret/1493/Han89 (X84999), Dog/5804/Han90 (X85000); Asian isolate, Siberian seal/PDV-2 (X84998); Greenland isolate, Dog/GR88 (Z47760); isolates of Japanese origin, Dog/Ueno (D85753), Dog/Hamamatsu (D85754), Dog/Yanaka (D85755), and Racoon dog/Tanu96 (AB016776). The data for the Snyder Hill strain were from reference 15.
Detection and sequence of N-protein gene from rectal sample 98-002.
A fragment of the expected size (approximately 420 bp) was amplified, and its nucleotide sequence was obtained (data not shown). The amplified region consisted of 419 bp, and the homology of a 372-bp length, which did not include the primer-binding regions, to the known sequence of Onderstepoort strain was 92.2%.
DISCUSSION
Comparative studies of CDV strains have been hampered for a long time because only a few cultivable strains have been available for in vitro experiments. However, in the 1990s several papers which, in particular, described the genetic properties of CDV strains that originated from domestic canids as well as wild animals appeared (6, 7, 12, 14, 15, 17–20, 23). In such studies viruses were generally isolated by passage in a cell culture system, which may result in changes to the amino acids of the gene during cultivation. However, in the present study and in other studies (7, 17, 25), molecular examinations were carried out directly with clinical specimens, and a new CDV genotype, represented by the CDV strain in specimen 98-002, was unexpectedly found. Indeed, we attempted to isolate viruses from swab extracts but did not succeed (24). It seems, therefore, that this direct method is more appropriate for elucidation of the epidemiology and molecular virology of fastidious CDV strains, especially when fresh and sufficient material for virus isolation is not available. On the other hand, primer selection may be a critical point for the experiment.
The current Japanese CDV isolates are in a close phylogenetic cluster (Fig. 3) and are clearly separated from old CDV strains as well as new CDV strains of European and U.S. origin, as described previously (20). Our CDV strain, strain KDK-1, showed the closest relationship to this clade, showing more than 98.5% amino acid homology, indicating that it belongs to the same genotype. The H genes amplified from the other CDV isolates and all clinical samples except samples 98/001 and 98-002 were also found to belong to the same genotype as KDK-1 by RFLP analysis. These results confirm the findings of a recent Japanese study (25) that most CDVs circulating among dogs since the 1980s in Japan belong to the same genotype as strain KDK-1 used in the present study. However, our findings of the distinct properties of the H gene from a new specimen (specimen 98-002) revealed in the present study suggest that there is another lineage or genotype among CDVs in the field.
It may be questioned whether CDV really existed in our swab extracts, because no virus was recovered from the extracts, nor could it be visualized in the extracts by electron microscopy. When referring to the computerized genetic databases for homology to the sequence in sample 98-002 obtained with the FASTA program, the first 20 best-matching sequences obtained were found to be of CDV origin, strongly suggesting that the virus in sample 98-002 is also of CDV origin. Furthermore, we succeeded in amplifying a 419-bp product from rectal swab extract 98-002, and this product was shown to represent part of the N-protein gene. This indicates that it is highly likely that actual CDV was present in the sample.
The puppy from which sample 98-002 was recovered died after showing clinical signs clearly indicative of canine distemper. Although no CDV (or other viral agents) could be recovered from sample 98-002, for a definite diagnosis, the evidence is convincing that the puppy was infected and killed by a CDV strain with such H-gene properties. This raises two questions; one is about the origin of this new CDV genotype. When considering the close genetic relationship and high rate of detection of the new Japanese CDVs (Fig. 3), the strain in sample 98-002 is novel enough that one can guess that it has not been an endemic strain. If this virus originated outside of Japan, it is unlikely that it was introduced into Japan by this puppy, given the young age of the dog (Table 1). However, it can be speculated that the CDV strain in sample 98-002 was introduced by other dogs imported into Japan and that the puppy was infected in the pet store. As possible evidence of this, the H gene of the strain in sample 98-002 possesses the same glycosylation sites as CDV strains of foreign origin, and glycosylation sites different from those of the other new Japanese CDV strains.
Another question is the efficacy of the present vaccines against infection and illness caused by new CDVs, as has been noted by others (6, 15, 20). The genotype described in this report is defined by the properties of the H gene, which is a key protein for both CDV itself and its animal hosts (1, 13). CDV uses this protein for attachment to receptors on the cell as the first step of infection, whereas an adequate host immune response against the H protein is essential for the prevention of CDV infection. Our preliminary in vitro experiments showed one-way cross-neutralization between strain Onderstepoort and new CDV strain KDK-1. (16). This however, contradicts previous studies that found very low cross-reactivities between Onderstepoort and Japanese new CDVs (12) and no significant antigenic difference between old CDVs and new CDVs (5, 15). On the other hand, no information is available about vaccine performance with new CDVs in vivo. In our laboratory, we tried to examine this by using a vaccine composed of old CDV strain Fromm (Fig. 2) and KDK-1 as a challenge virus. The vaccinated dogs were not infected with the challenge strain, whereas unvaccinated control dogs did become infected but did not conclusively manifest the typical clinical signs of distemper, making it impossible to fully confirm the efficacy of the vaccine (16). Therefore, a priority should be to develop a dependable in vivo challenge system with virulent new CDVs, although it is well-known that experimentally reproducible clinical canine distemper is hard to achieve (10, 18).
Cultivation of CDV in vitro from samples 98/001 and 98-002 was unsuccessful, so it is unlikely that we will be able to succeed in producing a conventional modified live vaccine from the tiny volume remaining from the swab extracts. Theoretically, however, at least cDNA and the cloned H gene of the CDV strain in sample 98-002 are sufficient to produce new types of CDV vaccines, as described recently, such as a recombinant subunit vaccine (10), an experimental DNA vaccine (29), and experimental as well as commercial vaccines made with recombinant vectors (21, 27, 30). These vaccines are expected to elicit a cellular immunity essential for prophylaxis of canine distemper.
In conclusion, a new H-protein gene was discovered in the clinical sample derived from a puppy manifesting typical signs of canine distemper. Genotyping by RFLP analysis and amino acid sequencing of the H gene obtained from both the recent isolates and isolates in clinical specimens revealed that at least two genotypes of CDV are circulating among dogs in Japan. It is not useful to resume a discussion of the issue of the efficacies of presently available commercial modified live CDV vaccines at the present stage because even the relationship between the H-gene genotypes and the antigenicities of CDV isolates in the field is not fully understood. If a modification were required, it should be designed on a global scale as the world’s transportation network has progressed enough to disseminate pathogens rapidly.
ACKNOWLEDGMENT
We thank Frank Roerink for critically reviewing the manuscript and a valuable discussion of phylogenetic analysis.
REFERENCES
- 1.Appel M. Canine distemper virus. In: Appel M J, editor. Virus infections of carnivores. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1987. pp. 133–159. [Google Scholar]
- 2.Appel M J G, Summers B A. Pathogenicity of morbilliviruses for terrestrial carnivores. Vet Microbiol. 1995;44:187–191. doi: 10.1016/0378-1135(95)00011-x. [DOI] [PubMed] [Google Scholar]
- 3.Appel M J G, Yates R A, Foley G L, Bernstein J J, Santinelli S, Spelman L H, Miller L D, Arp L H, Anderson M, Barr M, Pearce-Kelling S, Summers B A. Canine distemper epizootic in lions, tigers, and leopards in North America. J Vet Diagn Invest. 1994;6:277–288. doi: 10.1177/104063879400600301. [DOI] [PubMed] [Google Scholar]
- 4.Barrett T, Clarke D K, Evans S A, Rima B K. The nucleotide sequence of the gene encoding the F protein of canine distemper virus: a comparison of the deduced amino acid sequences with other paramyxoviruses. Virus Res. 1987;8:373–386. doi: 10.1016/0168-1702(87)90009-8. [DOI] [PubMed] [Google Scholar]
- 5.Blixenkrone-Møller M, Svansson V, Appel M, Krogsrud J, Have P, Orvell C. Antigenic relationship between field isolates of morbilliviruses from different carnivores. Arch Virol. 1992;123:279–294. doi: 10.1007/BF01317264. [DOI] [PubMed] [Google Scholar]
- 6.Bolt G, Jensen T D, Gottschalck E, Arctander P, Appel M J G, Buckland R, Blixenkrone-Møller M. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J Gen Virol. 1997;78:367–372. doi: 10.1099/0022-1317-78-2-367. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter M A, Appel M J G, Roelke Parker M E, Munson L, Hofer H, East M, O’Brien S J. Genetic characterization of canine distemper virus in Serengeti carnivores. Vet Immunol Immunopathol. 1998;65:259–266. doi: 10.1016/s0165-2427(98)00159-7. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain R W, Wamwayi H, Hockley E, Subbarao S M, Goatley L, Knowles N J, Barrett T. Evidence for different lineages of rinderpest virus reflecting their geographic isolation. J Gen Virol. 1993;72:443–447. doi: 10.1099/0022-1317-74-12-2775. [DOI] [PubMed] [Google Scholar]
- 9.Curran M D, Clarke D K, Rima B K. The nucleotide sequence of the gene encoding the attachment protein H of canine distemper virus. J Gen Virol. 1991;72:443–447. doi: 10.1099/0022-1317-72-2-443. [DOI] [PubMed] [Google Scholar]
- 10.De Vries P, UytdeHaag F C G M, Osterhaus A D M E. Canine distemper virus (CDV) immunestimulating complexes (ISCOMs) but not measles virus ISCOMs protect dogs against CDV infection. J Gen Virol. 1988;69:2071–2083. doi: 10.1099/0022-1317-69-8-2071. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. PHYLIP: phylogeny inference package, version 3.5. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 12.Gemma T, Iwatsuki K, Shin Y-S, Yoshida E, Kai C, Mikami T. Serological analysis of canine distemper virus using an immunocapture ELISA. J Vet Med Sci. 1996;58:791–794. doi: 10.1292/jvms.58.791. [DOI] [PubMed] [Google Scholar]
- 13.Greene C E, Appel M J. Canine distemper. In: Greene C E, editor. Infectious diseases of the dog and cat. 2nd ed. Philadelphia, Pa: The W. B. Saunders Co.; 1998. pp. 9–22. [Google Scholar]
- 14.Haas L, Harder T, Liermann H, Martens W, Greiser-Wilke I, Maack D, von Messling V, Liess B. Zur Situation der Hundestaupe in Deutschland. Kleintierpraxis. 1997;42:613–620. [Google Scholar]
- 15.Haas L, Martens W, Greiser-Wilke I, Mamaev L, Butina T, Maack D, Barrett T. Analysis of the haemagglutinin gene of current wild-type canine distemper virus isolates from Germany. Virus Res. 1997;48:165–171. doi: 10.1016/s0168-1702(97)01449-4. [DOI] [PubMed] [Google Scholar]
- 16.Hagiwara, S., Y. Sanada, K. Toriyabe, and T. Sagawa. 1994. Unpublished data.
- 17.Harder T C, Kenter M, Appel M J G, Roelke-Parker M E, Barret T, Osterhaus A D M E. Phylogenetic evidence of canine distemper virus in Serengeti’s lions. Vaccine. 1995;13:521–523. doi: 10.1016/0264-410x(95)00024-u. [DOI] [PubMed] [Google Scholar]
- 18.Harder T C, Kenter M, Vos H, Siebelink K, Huisman W, van Amerongen G, Örvell C, Barrett T, Appel M J G, Osterhaus A D M E. Canine distemper virus from diseased large felids: biological properties and phylogenetic relationships. J Gen Virol. 1996;77:397–405. doi: 10.1099/0022-1317-77-3-397. [DOI] [PubMed] [Google Scholar]
- 19.Harder T C, Klusmeyer K, Frey H-R, Örvell C, Liess B. Intertypic differentiation and detection of intratypic variants among canine and phocid distemper morbillivirus isolates by kinetic neutralization using a novel immunoplaque assay. J Virol Methods. 1993;41:77–92. doi: 10.1016/0166-0934(93)90164-m. [DOI] [PubMed] [Google Scholar]
- 20.Iwatsuki K, Miyashita N, Yoshida E, Gemma T, Shin Y-S, Mori T, Hirayama N, Kai C, Mikami T. Molecular and phylogenetic analyses of the haemagglutinin (H) proteins of field isolates of canine distemper virus from naturally infected dogs. J Gen Virol. 1997;78:373–380. doi: 10.1099/0022-1317-78-2-373. [DOI] [PubMed] [Google Scholar]
- 21.Jones L, Tenorio E, Gorham J, Yilma T. Protective vaccination of ferrets against canine distemper with recombinant pox virus vaccines expressing the H or F genes of rinderpest virus. Am J Vet Res. 1997;58:590–593. [PubMed] [Google Scholar]
- 22.Kövamees J, Blixenkrone-Møller M, Norrby E. The nucleotide and predicted amino acid sequence of the attachment protein of canine distemper virus. Virus Res. 1991;19:223–234. doi: 10.1016/0168-1702(91)90048-z. [DOI] [PubMed] [Google Scholar]
- 23.Mamaev L V, Denikina N N, Belikov S I, Volchkov V E, Visser I K G, Fleming M, Kai C, Harder T C, Liess B, Osterhaus A D M E, Barrett T. Characterisation of morbilliviruses isolated from Lake Baikal seals (Phoca sibirica) Vet Microbiol. 1995;44:251–259. doi: 10.1016/0378-1135(95)00018-6. [DOI] [PubMed] [Google Scholar]
- 24.Mochizuki, M., and M. Hashimoto. 1998. Unpublished data.
- 25.Ohashi K, Iwatsuki K, Nakamura K, Mikami T, Kai C. Molecular identification of a recent type of canine distemper virus in Japan by restriction fragment length polymorphism. J Vet Med Sci. 1998;60:1209–1212. doi: 10.1292/jvms.60.1209. [DOI] [PubMed] [Google Scholar]
- 26.Örvell C, Blixenkrone-Møller M, Svansson V, Have P. Immunological relationships between phocid and canine distemper virus studied with monoclonal antibodies. J Gen Virol. 1990;71:2085–2092. doi: 10.1099/0022-1317-71-9-2085. [DOI] [PubMed] [Google Scholar]
- 27.Pardo M C, Bauman J E, Mackowiak M. Protection of dogs against canine distemper by vaccination with a canarypox virus recombinant expressing canine distemper virus fusion and hemagglutinin glycoproteins. Am J Vet Res. 1997;58:833–836. [PubMed] [Google Scholar]
- 28.Rima B K, Earle J A P, Yeo R P, Herlihy L, Baczko K, ter Meulen V, Carabana J, Caballero M, Celma M L, Fernandes-Munoz R. Temporal and geographical distribution of measles virus genotypes. J Gen Virol. 1995;76:1173–1180. doi: 10.1099/0022-1317-76-5-1173. [DOI] [PubMed] [Google Scholar]
- 29.Sixt N, Cardoso A, Vallier A, Fayolle J, Buckland R, Wild T F. Canine distemper virus DNA vaccination induces humoral and cellular immunity and protects against a lethal intracerebral challenge. J Virol. 1998;72:8472–8476. doi: 10.1128/jvi.72.11.8472-8476.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephensen C B, Welter J, Thaker S R, Taylor J, Tartaglia J, Paoletti E. Canine distemper virus (CDV) infection of ferrets as a model for testing morbillivirus vaccine strategies: NYVAC- and ALVAC-based CDV recombinants protect against symptomatic infection. J Virol. 1997;71:1506–1513. doi: 10.1128/jvi.71.2.1506-1513.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor M J, Godfrey E, Baczko K, ter Meulen V, Wild T F, Rima B K. Identification of different lineages of measles virus. J Gen Virol. 1991;72:83–88. doi: 10.1099/0022-1317-72-1-83. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wamwayi H M, Fleming M, Barrett T. Characterization of African isolates of rinderpest virus. Vet Microbiol. 1995;44:151–163. doi: 10.1016/0378-1135(95)00008-x. [DOI] [PubMed] [Google Scholar]
- 34.Wood S L, Thomson G W, Haines D M. Canine distemper virus-like infection in a captive African lioness. Can Vet J. 1995;36:34–35. [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida E, Iwatsuki K, Miyashita N, Gemma T, Kai C, Mikami T. Molecular analysis of the nucleocapsid protein of recent isolates of canine distemper virus in Japan. Vet Microbiol. 1998;59:237–244. doi: 10.1016/s0378-1135(97)00194-6. [DOI] [PubMed] [Google Scholar]