I read with interest the recent publication by Bayarri-Olmos et al. (1) and would like to express some concerns regarding the study.
The SARS-CoV-2 receptor-binding domain (RBD) variant investigated by Bayarri-Olmos et al. (1), Y453F, is not the “cluster 5” variant (2). We initially described the “cluster 5” variant (3, 4), which has five spike amino acid changes (Fig. 1; strain: hCoV-19/Denmark/DCGC-3024/2020; GISAID number: EPI_ISL_616802). The mislabeling and out-of-context comparison of the Y453F properties with that of the multispike mutation variant “cluster 5” are incorrect and misleading. This kind of reporting is not only careless, but also irresponsible in light of the ongoing disputes surrounding the mink culling in Denmark. Furthermore, the “cluster 5” preliminary report clearly states that a microneutralization assay with an antinucleocapsid ELISA read-out was used (3), not a “bona fide plaque reduction neutralization assay.” This further casts into doubt the thoroughness by which the authors studied the cited report and their understanding of SARS-CoV-2 virus neutralization assays.
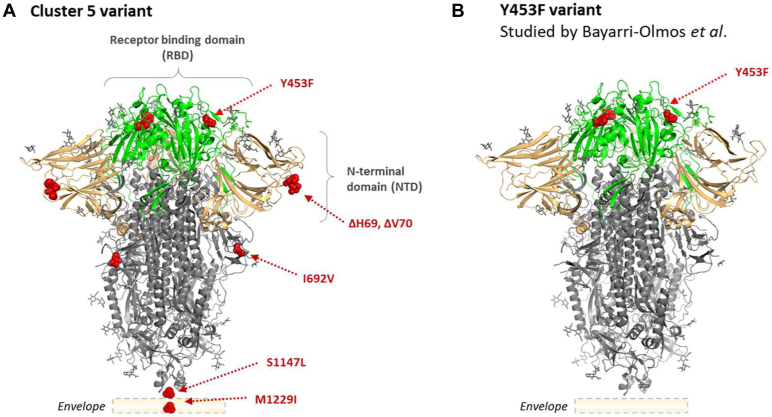
Figure 1.
The crystal structure of a closed prefusion SARS-CoV-2 spike trimer [PDB:6ZGE] indicating the amino acid changes oftwo different variants.A, the SARS-CoV-2 cluster 5 variant described in a preliminary report by Statens Serum Institut (3, 4); and B, the SARS-CoV-2 Y453F receptor-binding domain (RBD) variant studied by Bayarri-Olmos et al. (1). Red spheres indicate the positions of amino acid changes. The RBD is presented in green, the N-terminal domain (NTD) in beige, and the S2 domain in gray. The regions encompassing the S1147L and M1229I substitutions are not within the crystal structure; however, their relative positions are indicated.
The Y453F variant has some degree of neutralization resistance (5). Due to the heterogeneity of antibody responses, mutations will differentially affect convalescent plasma. It is therefore relevant to investigate the effect of the Y453F variant on an individual level and not only grouped. Lastly, the authors expressed surprise upon the observed higher affinity of the 453F RBD for ACE2 compared with the wild-type. However, this finding is not novel. Starr et al. (6) already demonstrated enhanced binding of the Y453F RBD to human ACE2 in September 2020.
Conflict of interest
The author declares no conflicts of interests with the contents of this article.
Edited by Craig Cameron
References
- 1.Bayarri-Olmos R., Rosbjerg A., Johnsen L.B., Helgstrand C., Bak-Thomsen T., Garred P., Skjoedt M.O. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J. Biol. Chem. 2021;296:100536. doi: 10.1016/j.jbc.2021.100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen H.D., Fonager J., Lomholt F.K., Dalby T., Benedetti G., Kristensen B., Urth T.R., Rasmussen M., Lassaunière R., Rasmussen T.B., Strandbygaard B., Lohse L., Chaine M., Møller K.L., Berthelsen A.-S.N. Preliminary report of an outbreak of SARS-CoV-2 in mink and mink farmers associated with community spread, Denmark, June to November 2020. Eurosurveillance. 2021;26:2100009. doi: 10.2807/1560-7917.ES.2021.26.5.210009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lassaunière R., Fonager J., Rasmussen M., Frische A., Polacek Strandh C., Rasmussen T.B., Bøtner A., Fomsgaard A. Statens Serum Insititut; Denmark: 2020. SARS-CoV-2 Spike Mutations Arising in Danish Mink and Their Spread to Humans. Statens Serum Institut Webpage. [Google Scholar]
- 4.Lassauniere R., Fonager J., Rasmussen M., Frische A., Polacek C., Rasmussen T.B., Lohse L., Belsham G.J., Underwood A., Winckelmann A.A., Bollerup S., Bukh J., Weis N., Sækmose S.G., Aagaard B. In vitro characterization of fitness and convalescent antibody neutralization of SARS-CoV-2 Cluster 5 variant emerging in mink at Danish farms. Front. Microbiol. 2021;12:698944. doi: 10.3389/fmicb.2021.698944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deepmutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]