Summary
Highly transmissible SARS-CoV-2 variants identified in India and designated B.1.617, Kappa (B.1.617.1), Delta (B.1.617.2), B.1.618, and B.1.36.29 contain spike mutations L452R, T478K, E484K, E484Q, and N440K located within the spike receptor-binding domain and thus could contribute to increased transmissibility and potentially allow re-infection or cause resistance to vaccine-elicited antibody. To address these issues, we used lentiviruses pseudotyped by variant spikes to measure their neutralization by convalescent sera, vaccine-elicited and Regeneron therapeutic antibodies, and ACE2 affinity. Convalescent sera and vaccine-elicited antibodies neutralized viruses with Delta spike with 2- to 5-fold decrease in titer in different donors. Regeneron antibody cocktail neutralized virus with the Delta spike with a 2.6-fold decrease in titer. Neutralization resistance to serum antibodies and monoclonal antibodies was mediated by L452R mutation. These relatively modest decreases in antibody neutralization titer for viruses with variant spike proteins suggest that current vaccines will remain protective against the family of Delta variants.
Subject areas: Biological sciences, Immune response, Virology
Graphical abstract
Highlights
-
•
Vaccine-elicited antibodies neutralize Delta spike with 4- to 5-fold decrease in titer
-
•
Delta variant is resistant to REGN10933 monoclonal antibody
-
•
Neutralization resistance is mediated by L452R
Biological sciences; Immune response; Virology
Introduction
Despite efforts to contain the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) the virus has continued to spread throughout the world's population, generating novel variants with mutations selected for immunoevasion and increased transmissibility. The variants contain point mutations and deletions in the spike protein driven by selective pressure for increased affinity for its receptor, ACE2, and escape from neutralizing antibody. They have been defined as Variants of Concern or Variants of Interest. The increased transmissibility of the variants is, at least in part, the result of mutations in the spike protein that allow for increased affinity of the spike protein for ACE2 and/or resistance to antibody neutralization. The N501Y mutation in B.1.1.7 (Andrew Rambaut et al., 2020; Volz et al., 2021) results in increased affinity for ACE2 (Gu et al., 2020; Liu et al., 2021a; Starr et al., 2020), whereas the E484K mutation in the B.1.351 (Tegally et al., 2020) and P.1 (Faria et al., 2021) spike proteins provides partial resistance to neutralizing antibodies in recovered individuals and antibodies elicited by vaccination (Garcia-Beltran et al., 2021; Wang et al., 2021; Wibmer et al., 2021; Wu et al., 2021; Xie et al., 2021).
Recent months have seen a dramatic increase in the rate of spread of highly transmissible SARS-CoV-2 variants Delta (B.1.617.2) accompanied by increased frequency. In addition, a highly transmissible family of Delta variants was identified in India, and the variants were designated B.1.617, Kappa (B.1.617.1), B.1.618, and B.1.36.29.
Here, we measured the neutralizing titer of convalescent sera and vaccine-elicited and Regeneron therapeutic antibodies against virus pseudotyped with B.1.617 and B.1.618 spikes. Although we used lentivirus pseudotyped virus, titers determined with this approach reflect the live virus neutralization test (Noval et al., 2021). Convalescent sera and vaccine-elicited antibodies neutralized viruses with B.1.617 and B.1.618 spike with modest decrease in titer. This suggests that current vaccines will remain protective against B.1.617 and B.1.618.
Results
Generation of SARS-CoV-2 variant spike protein-pseudotyped lentiviruses
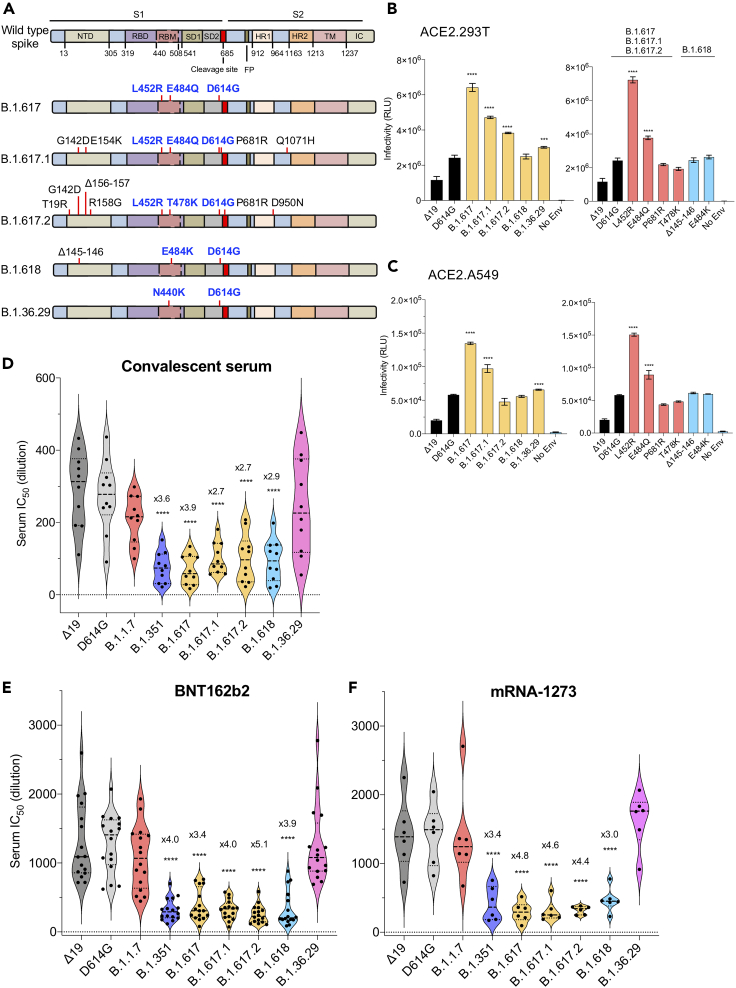
The variant spike proteins contain amino acid changes in the receptor-binding domain (RBD) and N-terminal domain (NTD) that could influence viral transmissibility and resistance to antibody neutralization. The B.1.617 and Kappa (B.1.617.1) spike proteins have L452R and E484Q mutations in the RBD in addition to D614G mutation near the proteolytic processing site (Figures 1A and S1A). The Delta (B.1.617.2) spike protein has L452R and T478K mutations in the RBD in addition to D614G and P618R mutations near the proteolytic processing site (Figures 1A and S1A). The B.1.618 spike has E484K in the RBD in addition to D614G and the N-terminal deletion Δ145-146 (Figures 1A and S1A). Residues 452 and 484 are in the RBD and thus could play a role in immunoevasion and/or resistance to antibody neutralization. Δ145-146 lies in the NTD, which is known to be a site of monoclonal antibody binding. The Delta (B.1.617.2) spike has both L452R and T478K mutations in the RBD, whereas the B.1.36.29 variant has a novel N440K mutation (Figures 1A and S1A). Which of these mutations contribute to increased affinity for ACE2 and antibody neutralization resistance that might account for the increased transmissibility of the variants is not well understood.
Figure 1.
Neutralization of spike protein variants by convalescent sera and antibodies elicited by BNT162b2 and mRNA-1273 vaccine
(A) The domain structure of the SARS-CoV-2 spikes of B.1.617, Kappa (B.1.617.1), Delta (B.1.617.2), B.1.618, and B.1.36.29 is diagrammed. NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor-binding motif; SD1 subdomain 1; SD2, subdomain 2; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane region; IC, intracellular domain.
(B) Infectivity of virus pseudotyped by B.1.617, Kappa (B.1.617.1), Delta (B.1.617.2), B.1.618, B.1.36.29 variant spikes and no envelope (no Env) pseudotyped lentivirus in ACE2.293T cells. Expression vectors for the variant spike proteins were generated and used to produce pseudotyped viruses. Viruses were normalized for RT activity and applied to target cells. Infectivity of viruses pseudotyped with the B.1.617, B.1.618, and B.1.36.29 spike protein (left) or individual mutations (right) was tested on ACE2.293T cells. The experiment was done three times with similar results. (∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001).
(C) Infectivity of virus pseudotyped by B.1.617, Kappa (B.1.617.1), Delta (B.1.617.2), B.1.618, B.1.36.29 variant spikes and no envelope (no Env) pseudotyped lentivirus in ACE2.A549 cells. Infectivity of viruses pseudotyped with the B.1.617, B.1.618, and B.1.36.29 spike protein (left) or individual mutations (right) was tested on ACE2.A549 cells. The experiment was done three times with similar results. (∗∗∗∗p ≤ 0.0001).
(D) Neutralization of viruses pseudotyped by Δ19 (dark), D614G (gray), B.1.1.7 (red), B.1.351 (blue), B.1.617 (yellow), B.1.618 (light blue), and B.1.36.29 (pink) spikes by convalescent serum samples from 10 donors was tested. Each dot represents the IC50 for a single donor. The experiment was done three times with similar results. (∗∗∗∗p ≤ 0.0001).
(E) Neutralizing titers of serum samples from BNT162b2 vaccinated individuals (n = 16) was measured. IC50 of neutralization of virus from individual donors is shown. Significance was based on two-sided testing (∗∗∗∗p ≤ 0.0001). The experiment was done three times with similar results.
(F) Neutralizing titers of serum samples from mRNA-1273 vaccinated donors (n = 6) was measured. IC50 of neutralization of virus from individual donors are shown. Significance was based on two-sided testing. (∗∗∗∗p ≤ 0.0001). The experiment was done three times with similar results.
To address these questions, we generated lentiviral pseudotypes containing a genome encoding GFP and luciferase reporters with the B.1.617, Kappa (B.1.617.1), Delta (B.1.617.2), B.1.618, and B.1.36.29 spike proteins and those with the individual point mutations and used these to quantify neutralizing antibody titers. Titers determined with this approach are highly consistent with those obtained with the live virus plaque reduction neutralization test (Noval et al., 2021). Immunoblot analysis of transfected pseudotype virus producer cells and virus-containing supernatants showed that the variant spike proteins were expressed well, were proteolytically processed to a similar extent, and were incorporated into lentiviral virions at a level similar to that of the parental D614G spike protein (Figures S1B and S1C). Analysis of the infectivity of each virus, normalized for particle number, on ACE2.293T cells showed that the Kappa (B.1.617.1) spike protein had a >2-fold increase in infectivity, whereas B.1.618 and B.1.36.29 were similar to the D614G spike (Figure 1B left). Analysis of the individual mutations showed that the increased infectivity of the B.1.617 spikes was attributed to L452R, which itself caused a 3.5-fold increase in infectivity and in combination with E484Q caused a 2-fold increase. The other point mutations had no significant effect on infectivity (Δ145-146, E484K, T478K, P681R) (Figure 1B right). Analysis of the same panel of pseudotyped viruses on ACE2.A549 cells showed a similar pattern of relative infectivity of each spike protein with an overall decrease of 50-fold in infectivity on these cells (Figure 1C).
Neutralization of the Delta and related variants by convalescent sera and vaccine-elicited antibody
To determine the sensitivity of the B.1.617 and B.1.618 pseudotyped viruses to antibody neutralization, we tested the serum specimens from convalescent patients who had been infected prior to the emergence of the variants for neutralization of the panel of pseudotyped viruses. The results showed that viruses with the B.1.617 and B.1.618 spikes were 2- to 4-fold resistant to neutralization by convalescent sera compared with wild type, a finding that was similar to the 3.6-fold resistance of the B.1.351 variant (Figure 1D). The resistance of B.1.617 was caused by the L452R and E484Q mutations, and B.1.618 resistance was caused by the E484K mutation, as is the case for B.1.351 (Figure S2A). Δ145-146 and P681R had no significant effect on neutralization resistance. B.1.36.29 was neutralized well by the convalescent sera.
To determine the resistance of the variants to neutralization by vaccine-elicited antibodies, we tested sera from individuals vaccinated with Pfizer BNT162b2 and Moderna mRNA-1273 vaccines for neutralization of the variant spike pseudotyped viruses. The results showed a similar pattern of resistance to neutralization as for the convalescent sera except that overall antibody titers were about 5-fold higher (Figures 1E and S2B). Virus with the B.1.617 spike was about 3- to 5-fold more resistant to neutralization and B.1.618 was about 4-fold resistant to antibodies in BNT162b2-elicited sera (Figure 1E). Similarly, Moderna mRNA-1273-elicited sera neutralized virus with B.1.617 and B.1.618 spikes with a 3- to 5-fold decrease in titer (Figure 1F). The resistance was attributed to L452R and E484Q in B.1.617 and E484K in B.1.618 (Figure S2C).
Variant pseudotype neutralization by REGN10933 and REGN10987 monoclonal antibodies
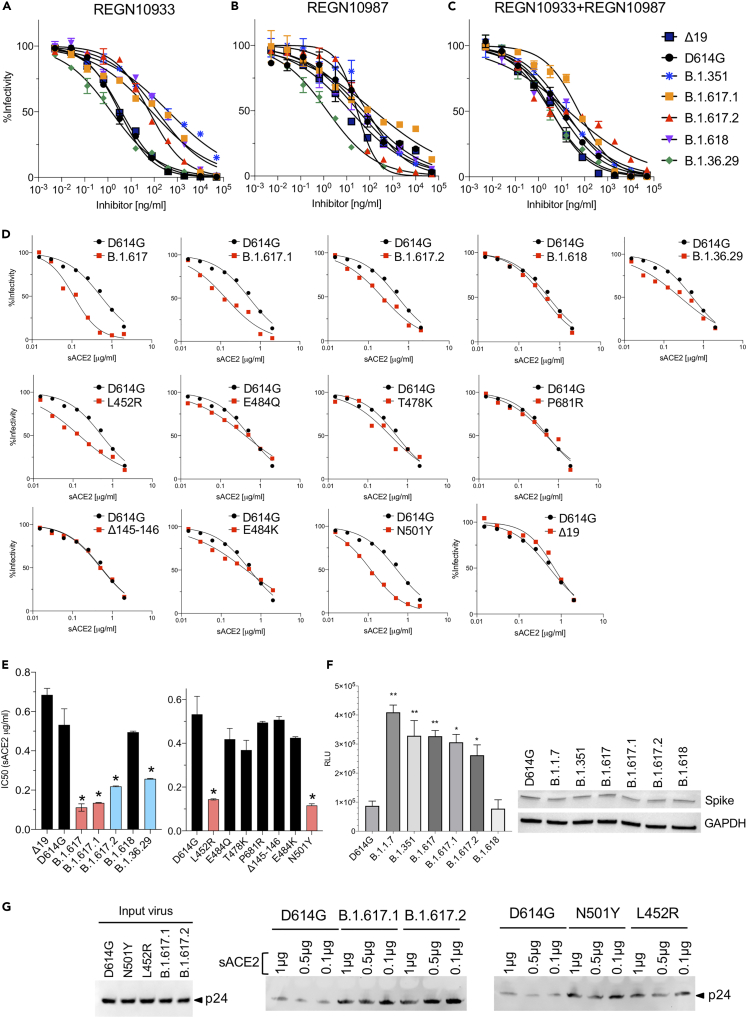
Monoclonal antibody therapy for COVID-19 has been shown to reduce disease symptoms and hospitalization (Weinreich et al., 2021). However, the treatment is subject to becoming less effective for patients infected with a variant in which the epitopes engaged by the antibodies are mutated. To address this question, we tested the ability of monoclonal antibodies REGN10933 and REGN10987 that constitute the Regeneron REGN-COV2 therapy to neutralize viruses pseudotyped by the variant spikes. The results showed that the neutralizing titer of REGN10933 for Kappa (B.1.617.1) and Delta (B.1.617.2) viruses was decreased by about 20-fold, similar to what was found for virus with the E484K variant spike (Figures 2A, S2D and Table 1), and was decreased on virus with the B.1.618 spike protein 43-fold (Figure 2A, Table 1). The Δ145-146 and P681R mutations had no effect on neutralizing titer. REGN10987, which is less subject to the effect of variant mutations, neutralized viruses with the Kappa (B.1.617.1) spike or with the L452R mutation with an approximate 3-fold decrease in titer (Figures 2B, S2E and Table 1). The REGN10933 and REGN10987 cocktail neutralized virus with the Kappa (B.1.617.1) and Delta (B.1.617.2) spikes with a 2- to 7-fold decreased titer, whereas the neutralizing titer of virus with the B.1.618 spike was unchanged (Figures 2C, S2F and Table 1). To test for possible synergy, we tested cocktails containing different ratios of the two monoclonal antibodies. We found no apparent synergy and optimal neutralization at a ratio of 1:1 (Figure S2G).
Figure 2.
Neutralization of spike protein variants by monoclonal antibodies REGN10933 and REGN10987 and soluble ACE2
(A–C) Neutralization of B.1.617 and B.1.618 spike protein variants by REGN10933 and REGN10987. Neutralization of D614G, B.1.351, Kappa (B.1.617.1), Delta (B.1.617.2), B.1.618, B.1.36.29 pseudotyped viruses by REGN10933 (A), REGN10987 (B), and 1:1 mixture of REGN10933 and REGN10987 (C) was measured. The experiment was done twice with similar results.
(D) Neutralization of B.1.617, B.1.618, and B.1.36.29 spike proteins and individual mutated spike variants by soluble ACE2 (sACE2). Viruses pseudotyped with variant spike proteins were incubated with a serially diluted recombinant sACE2 and then applied to ACE2.293T cells. Each plot represents the percent infectivity of D614G (black) and B.1.617 and B.1.618 (red) pseudotyped virus. The experiment was done twice with similar results.
(E) The histogram shows the calculated IC50 for each curve from (D). (∗p ≤ 0.05).
(F) Soluble ACE2 protein binds to variant spikes with high avidity. Variant spikes were transiently expressed in HEK293T cells for 1 day. sACE2-Nluc protein, 2.5 μg, was then incubated with equal variant spikes-expressing cells for 30 min. Cells were washed twice with PBS and followed by measurement of luciferase activity. Relative luciferase unit 105 counts from non-transfected cells were subtracted in each group. Cell lysates of variant spike-expressing cells were probed with anti-spike and anti-GAPDH antibodies. (∗p ≤ 0.05, ∗∗p ≤ 0.01).
(G) Soluble ACE2 proteins bind with high affinity to SARS-CoV-2 pseudotyped virions. Nickel agarose beads were coated with different amounts (1, 0.5, and 0.1 μg) of soluble ACE2 proteins (sACE2). D614G, N501Y, L452R, Kappa (B.1.617.1), and Delta (B.1.617.2) SARS-CoV-2 pseudotyped virions were incubated with the beads. After 1 h of incubation, the bound virions were analyzed on an immunoblot probed with antibody p24 antibody. Input virions were analyzed on an immunoblot probed with anti-p24 antibody. The experiment was done twice with similar results.
Table 1.
Neutralization of viruses by REGN10933, REGN10987, and REGN10933 + REGN10987 antibodies
| IC50 (ng/mL) |
|||||||
|---|---|---|---|---|---|---|---|
| Δ19 | D614G | B.1.351 | B.1.617.1 | B.1.617.2 | B.1.618 | B.1.36.29 | |
| REGN10933 | 3.9 | 3.2 | 424.6 | 78.8 | 62.1 | 139.8 | 1.4 |
| REGN10987 | 13.9 | 50.3 | 73.5 | 151.7 | 28.2 | 31.4 | 1.4 |
| REGN10933 + REGN10987 | 3.7 | 8.1 | 14.6 | 55.3 | 20.1 | 11.8 | 3.4 |
B.1.617, B.1.618 have increased affinity for ACE2
To determine whether the increased transmissibility of the variants could be caused by increased affinity for ACE2, we measured the relative ACE2 affinities of virion avidity using an assay we have previously reported, in which pseudotyped viruses were incubated with soluble ACE2, and then tested for infectivity on ACE2.293T cells. The results showed a 6-fold increase in the avidity of B.1.617 spikes for ACE2 (Figures 2D, 2E and 2F). The spike and sACE2-Nluc binding assay showed around 3- to 4-fold increased binding of B.1.617 spikes and ACE2 (Figure 2G). Variants with L452R showed a 3.7-fold increase in ACE2 affinity. T478K, E484Q, E484K caused smaller 1.3- to 1.4-fold increases in affinity. The findings suggest that the L452R mutation in the spike RBD increases virus binding to ACE2, likely contributing to the increased transmissibility of the variants.
Discussion
We show here that the serum neutralizing antibody titers elicited by BNT162b2 and mRNA-1273 vaccination against the B.1.617 and B.1.618 variants showed only a modest 2- to 5-fold decrease. Average serum neutralizing antibody titers were approximately 1:500, which is higher than that of individuals who have recovered from infection with the earlier unmutated virus. According to the model of Khoury et al., 50% protection is achieved with a neutralizing antibody titer that is 20% of the mean convalescent serum titer (Khoury et al., 2021). In our study, 20% of the mean titer from convalescent serum was approximately 60, and thus the model predicts that the neutralizing antibody titers achieved by vaccination will provide a high degree of protection against infection with the two variants.
The neutralization resistance to the vaccine-elicited antibody was mapped to the L452R, E484Q, and E484K mutations. The L452R mutation was found to result in increased affinity of the spike protein for ACE2, which is likely to have been the selection for the mutation in the virus. Amino acid 452 does not directly contact ACE2 but is located in proximity to a negatively charged patch in ACE2 (E35, E37, and D38) (Motozono et al., 2021). The introduction of a positive charge by the L452R mutation may increase electrostatic complementarity resulting in increased affinity for ACE2. The L452R mutation is present in the California B.1.427/B.1.429 variant and was found to cause a 2-fold increase in virus shedding by infected individuals, increased infectivity in cell culture, and a 4- to 6.7-fold and 2-fold decrease in neutralizing titer by the antibodies of convalescent and vaccinated donors, respectively (Deng et al., 2021).
The degree of resistance of B.1.617, Kappa (B.1.617.1), Delta (B.1.617.2), and B.1.618 we found was similar to that of B.1.351, P.1, and the New York B.1.526 (E484K) variants. The E484K mutation in the B.1.351, B.1.526, and P.1 spike proteins has been shown to cause partial resistance to neutralization (Garcia-Beltran et al., 2021; Tada et al., 2021; Wang et al., 2021; Wibmer et al., 2021; Wu et al., 2021; Xie et al., 2021). Mutation of the same position, E484Q, in B.1.617 and Kappa (B.1.617.1) caused a 2- to 4-fold decrease of neutralization by serum, demonstrating the importance of this residue as an epitope for antibody recognition. Our findings are similar to those of Wall et al. who found that both Delta (B.1.617.2) and B.1.351 were partially resistant to neutralization (Wall et al., 2021); some previous reports found that Delta (B.1.617.2) was highly sensitive to neutralization by mRNA vaccine-elicited serum antibody (Choi et al., 2021; Liu et al., 2021b; Planas et al., 2021). The reason for this difference is not clear, but it could be caused by differences in the study population or the time point post infection at which sera were collected (Choi et al., 2021; Liu et al., 2021b; Planas et al., 2021).
The B.1.617 and B.1.618 variants were partially resistant to REGN10933, one of the two monoclonal antibodies constituting the REGN-COV2 therapy, and virus with the B.1.617 spike was also partially resistant to the antibody cocktail. Viruses with the B.1.617 and B.1.618 spikes were partially resistant to neutralization, with an average 3.1-fold and 4.2-fold decrease in IC50 for convalescent sera and antibodies elicited by Pfizer and Moderna mRNA vaccines, respectively.
The analyses in this study were limited to the mRNA-based vaccines; it will be of interest to determine whether vector-based and protein-based vaccines induce a similar resistance profile. Although novel variants may emerge that are more resistant to vaccine-elicited antibody and more difficult to protect against, this has not yet occurred. The mRNA vaccines induce a robust T cell response to spike protein epitopes, which is more resistant to mutational alterations found in the variants (Gallagher et al., 2021; Sahin et al., 2020), and thus provide a second level of vaccine protection against current and future variants that may emerge. The findings reported here highlight the importance of widespread adoption of vaccination, which will protect from disease, will decrease virus spread, and slow the emergence of novel variants.
Limitations of the study
The number of donor sera analyzed in this study was sufficient to provide statistically significant data, but analysis of large number would have achieved higher confidence level and accuracy. The study number of sera analyzed was restricted by the number of subjects enrolled in the clinical studies at the NYULH Vaccine Center. Neutralizing antibody titers were determined at a single time point post vaccination. It will be of interest to test later time points to gauge the durability of antibody titers. The donors analyzed were recruited from a single medical center. As a result, donor sampling could introduce a bias that could account for the differences in results obtained in other studies that analyzed donors at different geographic locations and with different ethnicities, ages, or medical histories.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-His | Invitrogen | Cat# 37-2900, RRID:AB_2533309 |
| anti-GAPDH | Life Technologies | Cat# AM4300, RRID:AB_437392 |
| anti-p24 antibody (AG3.0) | AIDS respository | Cat# 4121 |
| Anti-spike antibody | GeneTex | Cat# GTX632604, RRID:AB_2864418 |
| goat anti-mouse HRP-conjugated second antibody | Sigma | Cat# A4416, RRID:AB_258167 |
| Bacterial and virus strains | ||
| VSV-G pseudotyped reporter virus | This paper | N/A |
| SARS-CoV-2 Δ19 S pseudotyped reporter virus | This paper | N/A |
| SARS-CoV-2 Δ19 S (B.1.617, B.1.617.1, B.1.617.2, B.1.618 and B.1.36.29) pseudotyped reporter virus | This paper | N/A |
| Biological samples | ||
| convalescent sera | NYU Vaccine Center with written consent under I.R.B. approval | IRB 20-00595 and IRB 18-02037 |
| Chemicals, peptides, and recombinant proteins | ||
| Soluble ACE2 | This paper | N/A |
| Critical commercial assays | ||
| Nano-Glo® Luciferase Assay System | Promega | Cat# N1120 |
| Nickel-nitrilotriacetic acid-agarose beads | QIAGEN | Cat# D-40724 |
| Experimental models: Cell lines | ||
| 293T | ATCC | N/A |
| ACE2.293T | This paper | N/A |
| ACE2.A549 | This paper | N/A |
| Software and algorithms | ||
| MacVector ver 17.0.0 (27) | MacVector Inc | N/A |
| GraphPad Prism 8 Software | GraphPad Prism Software, Inc. | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nathaniel R. Landau (nathaniel.landau@med.nyu.edu).
Materials availability
All unique DNA constructs, proteins and pseudotyped virus generated in this study are available from the Lead Contact upon request.
Experimental model and subject details
Plasmids
pLenti.GFP.NLuc based on pLenti.CMV.GFP.puro containing a GFP/nanoluciferase cassette separated by a picornavirus P2A self-processing amino acid motif cloned into the BamH-I and Sal-I sites (Addgene plasmid #17448, provided by Eric Campeau and Paul Kaufman). pcCOV2.Δ19S is based on pCDNA6 in which the CMV promoter drives transcription of a synthetic, codon-optimized SARS-CoV-2 spike gene based on Wuhan-Hu-1/2019 with a termination codon at position 1255 that deletes the carboxy-terminal 19 amino acids. B.1.617, Kappa (B.1.617.1), Delta (B.1.617.2), B.1.618 and B.1.36.29 spike mutations were introduced into pcCOV2.Δ19.D614G.S by overlap extension PCR and confirmed by DNA nucleotide sequencing. Plasmids used in the production of lentiviral pseudotypes (pMDL and HIV-1 Rev expression vector pRSV.Rev) have been previously described (Tada et al., 2021). sACE2-Nluc was introduced into pcDNA6 by overlap extension PCR and confirmed by DNA nucleotide sequencing.
Human Sera
Convalescent serum samples were taken from patients who had positive COVID-19 PCR tests between 32 to 57 days post-symptom onset. Sera from BNT162b2 or Moderna-vaccinated individuals were collected on day 28 and 35, respectively, 7 days post-second immunization, at the NYU Vaccine Center with written consent under IRB approved protocols (IRB 18-02035 and IRB 18-02037). Donor age and gender were not reported.
Cells
293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S) at 37°C in 5% CO2. ACE2.293T and ACE2.A549 are clonal cell-lines that stably express a transfected human ACE2. The cells were maintained in DMEM/1 μg/ml puromycin/10% FBS/1% P/S.
Method details
SARS-CoV-2 spike lentiviral pseudotypes
Lentiviral pseudotypes with variant SARS-CoV-2 spikes were produced by cotransfecting 293T cells with pMDL, pLenti.GFP-NLuc, pcCoV2.S (including variant spikes) and pRSV.Rev at a mass ratio of 4:3:4:1 by calcium phosphate coprecipitation. Virus lacking an envelope glycoprotein (No Env) was produced by using pcDNA6 in place of pcCoV2.S. The viruses were concentrated by ultracentrifugation and normalized for reverse transcriptase (RT) activity as previously described (Tada et al., 2021).
SARS-CoV-2 pseudotypes infectivity assay
ACE.293T or ACE2.A549 cells were plated in a 96 well tissue culture dish at 1 X 104 cells/well. Cells were infected with pseudotyped virus normalized for RT activity at MOI=0.2. After 2 days, the culture medium was removed, Nano-Glo luciferase substrate (Promega) was added and plate was read in an Envision 2103 microplate luminometer (PerkinElmer). To determine neutralizing antibody titers, sera or monoclonal antibodies were serially diluted 2-fold and then incubated with pseudotyped virus (approximately 2.5 X 107 cps) for 30 minutes at room temperature and then added to target cells. After 2 days, medium was removed, same amount of fresh medium and Nano-Glo luciferase substrate (Promega) was added and transferred to a microtiter plate which was read in an Envision 2103 microplate luminometer (PerkinElmer).
Soluble ACE2 neutralization assay
Serially diluted recombinant sACE2 protein prepared from transfected CHO cells was incubated with pseudotyped virus for 30 minutes at room temperature and added to 1 X 104 ACE2.293T cells. After 2 days, luciferase activity was measured using Nano-Glo luciferase substrate (Nanolight) in an Envision 2103 microplate luminometer (PerkinElmer).
Virion-sACE2 pull-down assay
Different amounts (1, 0.5 and 0.1 μg) of sACE2 was incubated with 30 μl of nickel-nitrilotriacetic acid-agarose beads (QIAGEN). After 1 hour of incubation, beads were washed with PBS, then added 30 μl of virus. Post 1 hour of incubation, the beads were washed, and bound protein was eluted with Laemmli loading buffer. The proteins were analyzed on an immunoblot probed with anti-p24 mAb (AG3.0) and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG secondary antibody (Sigma-Aldrich).
Spike-sACE2-Nluc binding assay
HEK293T cells were transfected with variants spike expression vectors by lipofectamine 2000 (Invitrogen). 2.5 μg purified sACE2-Nuc protein prepared from transfected CHO cells was incubated with variant spikes-expressing cells for 30 minutes. Free sACE2-Nuc protein was washed away with PBS twice and luciferase substrates were added to the cells. Luciferase activity was measured using Nano-Glo luciferase substrate (Nanolight) in an Envision 2103 microplate luminometer (PerkinElmer).
Immunoblot analysis
Spike proteins were analyzed on immunoblots probed with transfected cells that were lysed in buffer containing 50 mM HEPES, 150 mM KCl, 2 mM EDTA, 0.5% NP-40, and protease inhibitor cocktail. Protein (40 μg) was separated by SDS-PAGE. The proteins were transferred to polyvinylidene difluoride membranes and probed with anti-spike mAb (1A9) (GeneTex), anti-p24 mAb (AG3.0) and anti-GAPDH mAb (Life Technologies) followed by goat anti-mouse HRP-conjugated secondary antibody (Sigma). The proteins were visualized by luminescent substrate (Millipore) on iBright CL1000.
Quantification and statistical analysis
All experiments were in technical duplicates or triplicates and the data were analyzed using GraphPad Prism 8. Statistical significance was determined by the two-tailed, unpaired t-test. Significance was based on two-sided testing and attributed to p < 0.05. Confidence intervals are shown as the mean ± SD or SEM. (∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001, ∗∗∗∗P ≤ 0.0001). The PDB file of D614G SARS-CoV-2 spike protein (PDB: 7BNM) (Benton et al., 2021) was downloaded from the Protein Data Bank. 3D view of protein was obtained using PyMOL.
Acknowledgments
The work was funded by grants from the NIH to N.R.L. (DA046100, AI122390 and AI120898) and to M.J.M. (UM1AI148574). T.T. was supported by the Vilcek/Goldfarb Fellowship Endowment Fund.
Author contributions
T.T. and N.R.L. designed the experiments. H.Z., T.T., and B.M.D. carried out the experiments and analyzed data. T.T., H.Z., and N.R.L. wrote the manuscript. M.I.S. and M.J.M. provided key reagents and useful insights. All authors provided critical comments on the manuscript.
Declaration of interests
The authors declare no competing interests, except M.J.M. who received research grants from Lilly, Pfizer, and Sanofi and serves on advisory boards for Pfizer, Merck and Meissa Vaccines.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103341.
Supplemental information
Data and code availability
Additional information and data reported in this paper is available from the lead contact upon request.
References
- Andrew Rambaut N.L., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563; 2020. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations. [Google Scholar]
- Benton D.J., Wrobel A.G., Roustan C., Borg A., Xu P., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2022586118. e2022586118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A., Koch M., Wu K., Dixon G., Oestreicher J., Legault H., Stewart-Jones G.B.E., Colpitts T., Pajon R., Bennett H., et al. Serum neutralizing activity of mRNA-1273 against SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1128/JVI.01313-21. 2021.2006.2028.449914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S., et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184:3426–3437.e8. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Claro I.M., Candido D., Moyses Franco L.A., Andrade P.S., Coletti T.M., Silva C.A.M., Sales F.C., Manuli E.R., Aguiar R.S., et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological. 2021 https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586. [Google Scholar]
- Gallagher K.M.E., Leick M.B., Larson R.C., Berger T.R., Katsis K., Yam J.Y., Brini G., Grauwet K., Collection M.C.-., Team P., et al. SARS -CoV-2 T-cell immunity to variants of concern following vaccination. bioRxiv. 2021 2021.2005.2003.442455. [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Liu H., Zhang Q., Wei P., Chen Z., Aviszus K., Yang J., Downing W., Jiang C., Liang B., Reynoso L., et al. The basis of a more contagious 501Y.V1 variant of SARS-CoV-2. Cell Res. 2021;31:720–722. doi: 10.1038/s41422-021-00496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu Y., Xia H., Zou J., Weaver S.C., Swanson K.A., Cai H., Cutler M., Cooper D., Muik A., et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;596:273–275. doi: 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., Ngare I., Kimura I., Uriu K., Kosugi Y., et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136.e11. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noval M.G., Kaczmarek M.E., Koide A., Rodriguez-Rodriguez B.A., Louie P., Tada T., Hattori T., Panchenko T., Romero L.A., Teng K.W., et al. Antibody isotype diversity against SARS-CoV-2 is associated with differential serum neutralization capacities. Scientific Rep. 2021;11:5538. doi: 10.1038/s41598-021-84913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of infectious SARS-CoV-2 variant B.1.617.2 to monoclonal antibodies and sera from convalescent and vaccinated individuals. bioRxiv. 2021 2021.2005.2026.445838. [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310 e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Dcosta B.M., Samanovic M.I., Herati R.S., Cornelius A., Zhou H., Vaill A., Kazmierski W., Mulligan M.J., Landau N.R. Convalescent-phase sera and vaccine-elicited antibodies largely maintain neutralizing titer against global SARS-CoV-2 variant spikes. mBio. 2021;12:e0069621. doi: 10.1128/mBio.00696-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 2020.2012.2021.20248640. [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv. 2021 2020.2012.2030.20249034. [Google Scholar]
- Wall E.C., Wu M., Harvey R., Kelly G., Warchal S., Sawyer C., Daniels R., Hobson P., Hatipoglu E., Ngai Y., et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. 2021;397:2331–2333. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional information and data reported in this paper is available from the lead contact upon request.