Abstract
MYC oncoprotein promotes cell proliferation and serves as the key driver in many human cancers; therefore, considerable effort has been expended to develop reliable pharmacological methods to suppress its expression or function. Despite impressive advances, MYC-targeting drugs have not reached the clinic. Recent advances suggest that within a limited expression range unique to each tumor, MYC oncoprotein can have a paradoxical, pro-apoptotic function. Here we introduce a counterintuitive idea that modestly and transiently elevating MYC levels could aid chemotherapy-induced apoptosis and thus benefit the patients as much, if not more than MYC inhibition.
Keywords: oncogene addiction, apoptosis, chemotherapy, MYC
Introduction
MYC has long been considered a major cancer driver, on par with activated K-Ras. For that reason, therapeutic targeting of this oncogene has become an article of faith in precision medicine. The overall rationale for targeting initiating oncogenes is based on the phenomenon known as “oncogene addiction” where tumor cells become dependent on one protein or signaling pathway in a way their normal counterparts never are [1]. Thus, the former can be specifically killed while leaving the latter mostly unharmed, which is not the case for cytotoxic therapies like chemotherapy. The concept of addiction to MYC has been amply validated in genetically engineered mouse models [2], and several MYC-targeting compounds are now in clinical development or in clinical trials. They range from broad blockers of MYC transcription such as Brd4 inhibitors (BRD4i) [3, 4] and G-quadruplex-promoting compounds [5] to very specific blockers of MYC protein function such as 10074-G5 [6], MYCi361 [7], and Omomyc [8]. However, all these compounds are yet to produce tangible successes in the clinic, and in fact some of them such as MYC-targeting siRNA [9] resulted in terminated clinical trials.
One could argue that this is because the MYC locus frequently finds ways to bypass transcriptional inhibition, while specific and potent inhibitors of MYC itself are proving to be elusive. Interestingly, even though BRD4i are ineffective against most MYC-overexpressing cancers, they show acceptable potency against bona fide Brd4-driven cancers such as midline carcinoma [10], arguing that Brd4i are in fact good drugs, just not necessarily for MYC-driven cancers. The alternative explanation – unproven, but hard to rule out - is that genetically complex human cancers are far less dependent on MYC than commonly thought.
The second concern is that in both preclinical and clinical settings, Brd4i, Omomyc, and their brethren are sometimes tested as monotherapies, setting aside their interactions with existing standards of care in such scenarios. Thus, in parallel to inhibiting MYC expression or function as an anti-growth strategy, a complementary approach would be to exploit unique vulnerabilities associated with high MYC expression as an anti-survival strategy. One such vulnerability is the propensity of MYC to engage cell death pathways. In this review, we highlight this paradoxical pro-apoptotic function of MYC and the often-overlooked fact that MYC-driven tumors live (and sometimes die) by a Goldilocks Principle, according to which the levels of this lethal oncoprotein have to be just right: not too low, but not too high either. The corollary of this balancing act is that transiently elevating MYC levels aids chemotherapy-induced apoptosis and thus could directly benefit patients and potentially overcome chemotherapy resistance.
The many facets of MYC
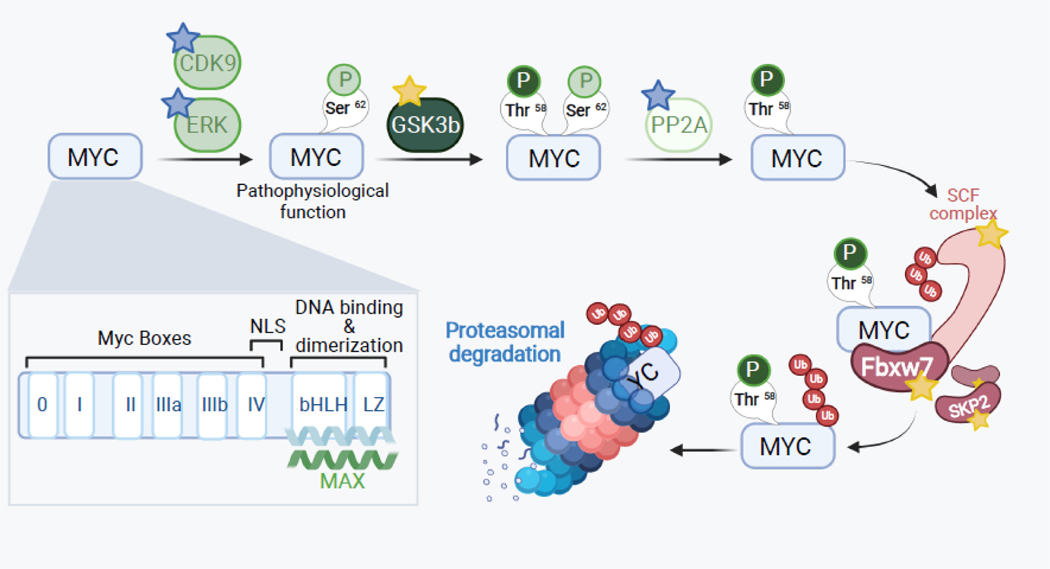
The MYC gene is deregulated in over half of all cancers, making it the most frequently altered oncogene [11]. Gene amplification of MYC and its paralogs MYCN and MYCL alone was observed in almost 30% of all The Cancer Genome Atlas (TCGA) samples. MYC deregulation also has been found to occur by various other means including point mutations, activation of upstream signaling pathways resulting in elevated transcription and protein stabilization, and more recently – by enhancer hijacking within core regulatory circuitries, which are especially important for MYCN activation [12, 13]. Breakthroughs in deciphering the mechanisms of MYC-driven tumorigenesis came from the characterization of MYC protein domains and identification of MYC target genes and gene networks. MYC (formerly referred to as c-Myc) is an oncoprotein and a nuclear transcription factor that belongs to a family that also includes N-MYC and L-MYC. It has distinct structural modules and several key phosphorylation sites, which control its function and well as turnover via proteasomal degradation (see Figure 1 for details) (highlighted in [14, 15]).
Figure 1. MYC protein structure and regulation.
MYC protein is comprised of 5 major domains: MBI and MBII (MYC Box I and II) in the N-terminus, and the NLS (nuclear localization signal), bHLH and LZ domains (basic helix-loop-helix and leucine zipper) in the C-terminus. This C-terminal domain also allows MYC to hetero-dimerize with its binding partner Max and associate with E-box DNA sequences (CACGTG), which is essential for its transcriptional and transforming activity. Another box (MB0) has been recently identified and shown to positively control transcription elongation. There are also multiple protein-protein interactions, which either enhance (blue stars) or counteract (yellow stars) MYC function. Thus, inhibitors of CDK9 and Erk, activators of PP2A, and the synthetic peptide Omomyc (which disrupts MYC:MAX binding) bring about either reduced MYC levels or impaired MYC function. Conversely, inhibitors of GSK3 and E3 ligases (Fbxw7, Skp2, etc.) stabilize MYC protein levels and boost its function.
In addition to its well-established role as a transcriptional activator, MYC has been shown to function as a transcriptional repressor, which in most cases involves complex formation with another nuclear protein Miz-1 [16]. Its opposite effects on gene expression notwithstanding, MYC has been shown to bind the majority of actively transcribed human genes and regulate both protein-coding and non-coding RNA genes, suggesting that it acts as a global transcriptional regulator (reviewed in [17, 18]). A large proportion of MYC-regulated genes aid tumor growth and upkeep. This set includes genes that control cell cycle and growth, metabolism, protein synthesis, cell migration, angiogenesis, and chromosomal instability. All these pathways contribute to MYC-mediated transformation of normal cells into cancerous ones (reviewed in [19]). More recently, the role of MYC in creating the immunosuppressive microenvironment (for example, through its effects on CD47 and PDL1 expression) has come to the fore [20]. Thus, it comes as no surprise that MYC is one of the few oncogenes that could single-handedly drive rapid neoplastic growth. Consequently, in many systems MYC inactivation led to regression of established tumors [21–24]. Numerous subsequent studies have confirmed that MYC plays an important role not only in tumor initiation, but also in tumor maintenance in multiple organ systems (reviewed in [25]). For this reason, considerable efforts have been expended to learn how to inhibit MYC activity.
Therapeutic targeting of MYC: opportunities and challenges
Transcription factors are notoriously challenging to target pharmacologically due to their lack of hydrophobic pockets and large interaction surface areas, which are at odds with the standard binding models of small drug molecules [26]. Only very recently some advances have been achieved with direct MYC inhibitors [7]. As an alternative to disrupting MYC with small molecules, the MYC dominant-negative peptide Omomyc has been developed to inhibit its function [27–29]. This 92-amino acid polypeptide homodimerizes and binds to both MYC and Max, thus preventing MYC:Max heterodimerization. While Omomyc has displayed anti-tumor activity in experimental models of non-small cell lung cancer, its efficacy in the patient setting is currently being determined (ClinicalTrials.gov Identifier: NCT04808362) [8].
Thus, complementary efforts were expanded to inhibit MYC expression and function at the mRNA and protein levels and to target signaling pathways that activate MYC. There are approaches to inhibit MYC function at just about every level of its regulation in the cell. Numerous upstream signaling pathways deregulate MYC activity, such as Notch, WNT, PI3K, and MAPK pathways [30]. Many small molecule inhibitors exist for each of these pathways and are currently undergoing pre-clinical and clinical trials.
One recently emerged strategy to target MYC function is the inhibition of BET bromodomain family members. BET inhibition was first shown to down-regulate the MYC transcriptional program [31]. Following the discovery that the BET transcriptional regulator BRD4 can bind to the MYC promoter and regulate MYC expression, investigators began to explore the therapeutic use of BRD4 inhibitors such as JQ1 to treat MYC-driven cancers [32–34]. There are currently several BRD4 inhibitors in clinical trials as monotherapies for a variety of cancer types. Despite the promising pre-clinical studies, the clinical trial results have thus far been mixed. Positive anti-tumor effects have been observed, but so have detrimental side effects at below-efficacy doses (reviewed in [35]). Additionally, pre-clinical studies have identified resistance mechanisms to BET inhibition, such as increased expression of anti-apoptotic or autophagy proteins, that could likewise end up leading to BET inhibitor resistance in patient tumors [36]. It is also notable that BET inhibition affects key MYC-independent oncogenic pathways (see for instance [37]), making interpretation of results less straightforward.
In addition to BRD4, another targetable protein that affects the function of MYC is Aurora kinase A. This enzyme was first identified to enhance N-MYC stability in neuroblastoma, and since then has been tested pre-clinically as a druggable target for MYC-driven cancers [38–40]. These findings led to clinical trials of Aurora kinase inhibitors as monotherapy in solid and hematologic tumors; these inhibitors were unable to produce durable responses in solid tumors but were somewhat more effective at treating the hematologic malignancies (reviewed in [41]). As the mixed responses in clinical trials would suggest, BET and Aurora kinase inhibition may only be suitable for certain subsets of MYC-driven cancers.
A complementary approach to MYC destabilization has been to boost the activity of the PP2A phosphatase using compounds collectively known as SMAPs (small-molecule activators of PP2A) [42].
Together with mTOR inhibition, this approach has shown efficacy in pre-clinical models of highly aggressive pancreatic ductal adenocarcinoma [43], but SMAPs are yet to be tested in human patients.
Finally, there is considerable evidence that some therapeutic benefits could be reaped from inactivating MYC targets. One salient example is ornithine decarboxylase (ODC). In the 2005 proof-of-principle paper, deleting the ODC gene or inhibiting the enzyme with difluoromethyl-ornithine (DFMO) delayed lymphoma development in the Eμ-MYC mouse model [44]. However, in the realm of experimental oncology, this approach appears to be re-directed towards N-MYC-driven tumors, such as neuroblastoma [45]. In summary, while multiple independent strategies are concurrently being pursued to inhibit MYC either directly or indirectly, each has its own limitations, which make it difficult to ascertain whether any of them would ever emerge as blockbuster drugs to successfully treat MYC-driven malignancies.
MYC-driven apoptosis: a potential vulnerability
Given that the efforts to inhibit pro-oncogenic activities of MYC are yet to come to fruition clinically, unorthodox approaches might be in order. One such approach would be to exploit the long-known function of MYC to promote programmed cell death, or apoptosis, during normal development. Indeed, in a recent study MYC was found to be highly expressed in young tissues that were exquisitely primed to undergo apoptosis, and loss of a MYC allele resulted in a reduced response to apoptotic stimuli [46]. Rather paradoxically, oncogenic MYC also retains the conflicting functions of driving proliferation and cell death. The anti-survival effects of MYC first reported 30 years ago are achieved by a variety of mechanisms (comprehensively reviewed in [47]) in both immortalized and cancerous cells lines as well as in vivo tumor models.
In fact MYC-dependent apoptosis is a hallmark of many cancers such as Burkitt lymphoma, in which tumor cells are highly proliferative but at the same time also display high levels of apoptosis [48]. In the realm of solid tumors such as lung, liver, and ovarian cancers, where MYC is usually activated via copy number gains [49], activation of MYC was reported to correlate with higher apoptotic indices and/or focal amplification of anti-apoptotic genes [50–52]. Admittedly, correlation does not prove causation, but there is also a large body of evidence demonstrating direct pro-apoptotic effects of MYC, as discussed below.
Early studies revealed that stimuli such as low serum, cytokine withdrawal, or T-cell activation induce apoptosis, and that this cell death response was dependent on MYC [53–55]. Not long after it was discovered that MYC triggers apoptosis via ARF upregulation and ensuing activation of the tumor suppressor p53; this p53-driven apoptotic response was found to require the activity of the DNA damage response protein ATM [56, 57]. Subsequently, it was noted that MYC-driven murine transgenic lymphomas and other MYC-driven tumors inactivate this pro-apoptotic pathway by disabling Arf or p53 [58]. The inactivation of MYC-driven apoptosis is thought to be crucial for tumor initiation and progression, because upon loss of the ARF-MDM2-p53 pathway in mouse models of MYC-driven cancers, there was a robust acceleration of tumorigenesis [59, 60]. Two mutant forms of MYC commonly found in Burkitt lymphoma (P57S and T58A) are unable to induce apoptosis owing to their failure to upregulate BIM and inhibit Bcl-2 function, further demonstrating how critical it is for cancer to evade MYC-driven apoptosis during tumorigenesis [61]. Of note, adjacent serine and proline residues are recurrently mutated in other histotypes as well, including endometrial salivary and gland cancers ([62]; in the COSMIC database T58 is annotated as T73, owing to the existence of the longer CTG codon-initiated MYC Isoform 2 with the Uniprot Identifier P01106–2).
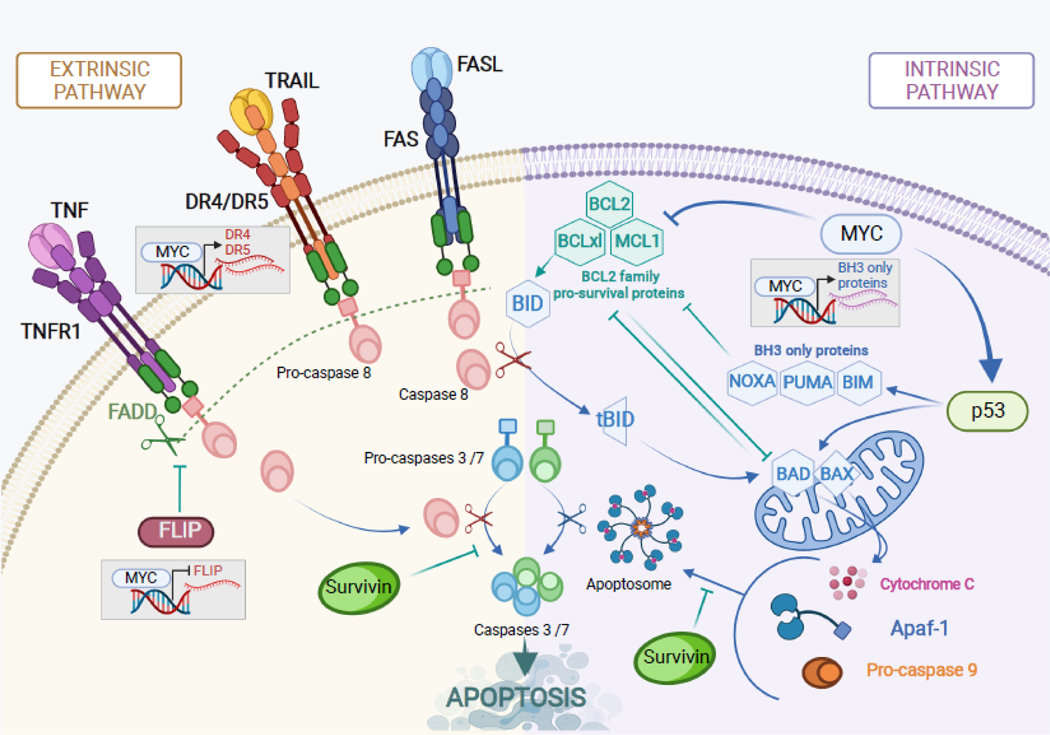
While it is well established that p53 is the main effector of MYC-mediated apoptosis, p53-independent mechanisms have also been identified (reviewed in [47]). It has been often proposed that MYC exerts its pro-apoptotic function through direct modulation of gene transcription. Later it was found that numerous intrinsic and extrinsic apoptotic genes (Figure 2) are in fact direct transcriptional targets of MYC and are bound by MYC at their promoters. In Eμ-MYC murine lymphomas MYC activity led to the suppression of the anti-apoptotic proteins BCL-2 and BCL-XL [63, 64], and in melanoma MYC-driven transcription of NOXA, a pro-apoptotic protein in the Bcl-2 family of proteins, was observed [65]. One of the major pro-apoptotic proteins, Bax, is also a direct transcriptional target of MYC [66], although non-transcriptional regulation of Bax activity by MYC has been described as well [67]. Additionally, MYC has been shown to bind to and activate the promoters of the pro-apoptotic proteins BIM and BID and in doing so contribute to the priming of mitochondria to respond to apoptotic stimuli. This was observed in multiple models including transformed rodent fibroblasts [67, 68] and B-cell neoplasms [69, 70], but also pancreatic [71], and breast, ovarian, and colon [66, 72] carcinomas. MYC was also found to bind the promoter of mtCLIC, a pro-apoptotic mitochondrial chloride ion channel; when the expression of this gene was suppressed, so was MYC-driven apoptosis, suggesting that it is yet another player in MYC-mediated cell death [73].
Figure 2. Regulation of apoptosis by MYC.
MYC can promote apoptosis through both intrinsic (mitochondrial) and extrinsic (death receptor-mediated) mechanisms by regulating many key components of these pathways. Common anti-cancer drugs (e.g., chemotherapeutics) often trigger both apoptotic pathways, thereby increasing their reliance on MYC. Additionally, the extrinsic pathway can be specifically engaged by death receptor agonists or DR4-activating antibodies (e.g., mapatumumab).
In addition to amplifying intrinsic, or mitochondrial cell death pathway, MYC has also been found to modulate extrinsic, or death-receptor mediated cell death (Figure 2). Early studies demonstrated that MYC could sensitize cells to signaling though the death receptor CD95/Fas [74]. MYC expression was also found to greatly sensitize cells to apoptosis triggered by the extrinsic ligand TRAIL [75]. Just as with intrinsic apoptosis, many extrinsic apoptosis genes are direct transcriptional targets of MYC. This includes genes like death receptor-4 (DR4) and the extrinsic ligand FasL which are both positively regulated by MYC, as well as CFLAR/FLIP, the negative regulator of Caspase 8, whose transcriptional expression is repressed by MYC [76–78]. The TRAIL receptor DR5 was found to be upregulated at the cell surface upon MYC activation [79, 80]. But do these well-established axes represent valid therapeutic targets in MYC-driven tumors? Some preliminary answers are beginning to emerge from studies exploiting the concept of synthetic lethality: perturbations that uniquely affect cells with a certain genetic background, in this case MYC dysregulation.
Lessons from synthetic lethality and correlative studies
Genome-wide screens have proved an invaluable tool for global assessment of MYC-synthetic lethality genes. Using shRNA, siRNA, or CRISPR pooled libraries screens, numerous genes and gene networks have emerged as synthetically lethal with high MYC expression. Some of these gene networks include components of RNA polymerase complexes, transcription initiation complexes, DNA repair and cell cycle checkpoint proteins, metabolic enzymes, and notably – components of the apoptotic pathways (reviewed in [81, 82]). For example, CDK1 knockdown was identified from RNAi screens to be synthetically lethal with MYC overexpression, and inhibiting CDK1 in MYC-driven lymphoma and neuroblastoma models lead to apoptosis and decreased tumor growth owing to dysregulation of a direct CDK1 target BIRC5 (a.k.a. survivin), one of the Inhibitor of Apoptosis (IAP) gene family members and a caspase 3/7 inhibitor (Figure 2) [83]. Of note, survivin is now considered a good drug target, with multiple compounds in clinical development [84].
Admittedly, many other MYC synthetic lethal genes emerging from genome-wide screens have no known direct connection to cell death pathways and are involved instead in a range of other functions ranging from cell cycle regulation (checkpoint kinase 1) to metabolism (glutaminase) [85]. However, there is little overlap between targets identified in independent screens, questioning the broad application of specific synthetic lethal targets across different tumor types. In contrast, targeted synthetic lethality approaches aimed at apoptotic pathways might be more enlightening. For example, it has long been appreciated that MYC sensitizes cells to apoptosis through the extrinsic death receptor Fas, and subsequently it was found that the expression of high MYC lead to cells being sensitized to the extrinsic ligand TRAIL via MYC-dependent upregulation of death receptor-5 (DR5) [79]. Another recent study similarly identified that while high MYC expression drives brain metastases of breast cancer, it also generates a synthetically lethal interaction with TRAIL [86]. Through these studies, the field has been able to identify unique vulnerabilities of MYC-driven cancers, many of which are now being pursued in the pre-clinical or clinical settings ([87] and references therein).
However, for the majority of tumor types, new investigational drugs targeting specific genetic lesions are tested in combination with standards of care [88]. Thus, the complex effects of MYC on pro-survival and pro-apoptotic pathways might be particularly relevant in the context of chemotherapy, where MYC-directed and conventional therapies would need to function together. There is considerable evidence that they might. Almost 20 years ago, it was demonstrated that in human colorectal cancers, MYC amplification (combined with wild type p53 expression) increases susceptibility to 5-fluorouracil in vivo [89]. Subsequently, sensitizing effects of MYC were observed in fibroblasts [90] and in a non-lymphoid hematologic malignancy with dismal outcomes: multiple myeloma [91]. More recently, the presence of high MYC was shown to sensitize multiple cancer types to anti-mitotic chemotherapy agents through upregulation of pro-apoptotic BH3 proteins and suppression of BCL-XL [92]. Similarly, in breast cancer the anti-tumor activity of Bcl-2 inhibitors combined with AMPK activators was found to be dependent on high MYC expression [93]. In yet another study using an in vivo model of MYC-driven small cell lung cancer, tumors were sensitized to Aurora kinase inhibitors which synergized with chemotherapy to induce apoptosis [94]. Conversely, numerous studies on transformed fibroblasts, B-cell lymphoma, adrenocortical cancer and other cancer types have demonstrated that the loss of MYC expression confers resistance to chemotherapeutic drugs like doxorubicin, etoposide, and paclitaxel [68, 95–97] as well as the proteasome inhibitor bortezomib [98]. In fairness, there are several cell lines where inactivation of MYC was reported to render cells more susceptible to chemotherapy, e.g., M14 melanoma [99, 100] and MCF-7 breast carcinoma [101]. This complexity indicates that the role of MYC in chemotherapy is either narrowly histotype-specific or more likely of bi-phasic nature, where for the tumor to withstand the onslaught of anticancer drugs MYC levels have to be just right: not too low, but not too high either. This latter Goldilocks scenario has broad translational implications.
Boosting MYC-dependent therapeutic apoptosis
The intrinsic ability of MYC to drive apoptosis is clearly insufficient to offset high cell proliferation rates found in most MYC-driven tumors. However, by employing strategies to enhance MYC-mediated apoptosis it should be possible to tip the scale in favor of cancer cell death and ensuing tumor regression. Sophisticated in vivo studies with finely controlled MYC alleles have suggested that there are thresholds for MYC activity: a modest increase in the level of MYC led to oncogenesis, but yet higher levels were required to trigger apoptosis [92, 102]. This would indicate that in the context of cancer, there could be a certain threshold for MYC that when surpassed, could activate cell death.
Conceptually, there are precedents for the counterintuitive overexpression of oncogenes as a therapeutic strategy. As early as in 1998, it was demonstrated that overexpression of adenovirus 5 E1A oncogene suppresses tumor growth in vivo via increased apoptosis [103]. This concept has been advanced to Phase I clinical studies using intratumoral E1A cancer gene therapy [104, 105]. Although these formulations are currently not in clinical development, recent studies demonstrated the validity of this approach in genetically engineered mouse models, where transgenically expressed E1A blocked chemical skin carcinogenesis [106]. However, up until recently, it remained to be determined how exactly the pro-apoptotic function of MYC could be re-engaged in various cancers.
Several laboratories have reported that strengthening the CD19-PI3K-AKT axis is a reliable method to boost MYC protein stability in B-lymphoid cells [107–111]. This finding is consistent with the propensity of glycogen synthase kinase 3 beta (GSK-3β), which is inhibited by Akt, to phosphorylate MYC at Thr-58, marking MYC for recognition by the E3 ubiquitin ligase Fbxw7 and subsequent degradation [112–115] [reviewed in [116] (Figure 1). Given that activation of the PI3K pathway is one of the most frequent alteration in human cancers [11, 117], and that it is a known suppressor of MYC-induced apoptosis [118, 119]it would need to be manipulated only transiently, for example with short-lived GSK-3 inhibitors.
Further work demonstrated that transient (90–120 min) stabilization of MYC protein using the GSK-3β inhibitor CHIR99021 [120] (see Table 1) is sufficient to sensitize therapy-resistant B-cell lymphomas to chemotherapy and direct engagers of the extrinsic apoptotic pathway such as TRAIL or DR4 agonistic antibodies [121] (Figure 2). Several lines of evidence suggest that the bulk of these sensitizing effects was MYC-dependent. First, inhibition of GSK-3 had no discernable effect on sensitivity to chemotherapy in Epstein-Barr virus-transformed B-lymphoid P493–6 cells with tet-repressible MYC alleles [122] when they were maintained in the MYCOFF state (with the caveat that these cells were also non-proliferating and potentially refractory to genotoxic stresses.) Second, in Burkitt lymphoma cell lines the effects of GSK3i were cancelled following JQ1–mediated MYC downregulation. Lastly, no chemosensitizing effects were observed in lymphoma cells bearing MYC Thr-58 mutations, where MYC protein levels are no longer regulated by GSK-3β nor transiently increased by GSK3i. Of note, while this mutation is relatively frequent in Burkitt’s lymphoma, it is not common in human cancers in general. For example, COSMIC Cancer Mutation Census estimates its frequency to be 2.3×10−3 and recurrence - 144 (https://cancer.sanger.ac.uk/cmc/gene/myc).
Table 1.
MYC-stabilizing compounds in clinical development
| Inhibitor | Mechanism of action | Indication | Stage of development | References |
|---|---|---|---|---|
|
| ||||
| CHIR99021 | GSK-3 inhibitor | various xenograft models | Preclinical | Harrington et al., 2019; O’Flaherty et al., 2019 |
| tideglusib | GSK-3 inhibitor | Alzheimer’s disease, myotonic dystrophy | Phase II completed | Lovestone et al., 2015 |
| LY2090314 | GSK-3 inhibitor | acute leukemia, metastatic solid cancers | Phase II trials | Rizzieri et al., 2016; Gray et al., 2015 |
| 9-ING-41 | GSK3-inhibitor | pediatric and adult cancers | Ongoing Phase I/II | Ugolkov et al., 2018 |
| LiCl | GSK-3 inhibitor | bipolar disorders | FDA approved | O’Brien and Klein, 2009 |
| Compound A | Skp2 inhibitor | hematologic malignancies | Preclinical | Chen et al., 2008 |
| C1/C2 | Skp2 inhibitor | soft tissue sarcoma | Preclinical | Wu et al., 2012; Li et al., 2020 |
| C25 | Skp2 inhibitor | T-ALL, other cancers | Preclinical | Chan et al., 2013; Rodriguez et al., 2020 |
| Dioscin | Degradation of SKP2 | colorectal carcinoma | Preclinical | Zhou et al., 2020 |
Beyond B-cell malignancies, GSK-3 inhibition has been shown to induce apoptosis via MYC-dependent mechanisms in certain cancer types, such as KRAS-mutant pancreatic adenocarcinoma, neuroblastoma, and glioma even as a monotherapy [123–125]. Other groups demonstrated that GSK-3 inhibition can increase the sensitivity of melanoma cells to the extrinsic ligand TRAIL and to inhibitors of mutant BRAF [126, 127]. Although in these studies the authors attributed the sensitization to the activation of the Wnt pathway, the caveat is that MYC itself is a well-known transcriptional target of that pathway [128], making it difficult to invoke MYC-independent effects. Additionally, the role of GSK-3 in cancer is complicated by contradictory findings that this protein promotes apoptosis in some cell lines while inhibiting it in others. Yet recent studies by several groups provide rationale and supporting evidence for the therapeutic targeting of GSK-3 in cancer ([129], also [130] and references therein, ).
Whatever pharmacological approach one might take, timing will be of essence. Certainly, long-term stabilization of MYC must be avoided, not only because of its pro-growth properties, but also because of well documented immunosuppressive effects of MYC, for example via controlling PD-L1 expressing (first reported in [131]; reviewed in [132]). In several murine models, inhibition of MYC allowed recruitment of immune effector cells, which contributed to anti-tumor effects (see for instance [7]). Curiously, in yet other mouse models, including the above-referenced AMPK study, pharmacological reactivation of MYC increased, not suppressed susceptibility to anti-PD-1 immunotherapy [93]. One possible explanation for this is that high levels of MYC have been documented to increase genomic instability [133], which at least in principle could lead to the increased expression of neoantigens. But regardless of the exact effects of MYC on the immune system, adaptive immune responses unravel slowly, within days, and are unlikely to be affected by two hour-long elevation of MYC levels, as achieved with GSK-3i [121]. In fact, one can envision a bi-phasic form of therapy where long-term MYC inhibition (to sustain anti-tumor responses) is combined with metronomic burst of short-term MYC induction scheduled to coincide with cycles of chemotherapy.
Repeated, “metronomic” administration of MYC agonists might also avoid another commonly encountered problem: that of clonal selection. First, short-term nature of MYC induction removes continuous pressure needed to select for MYC-resistant clones. Second, as drug selection often favors quiescent or dormant cells, which are inevitably characterized by low MYC expression, MYC agonists could reverse or at least ameliorate this unfavorable trend.
Concluding remarks
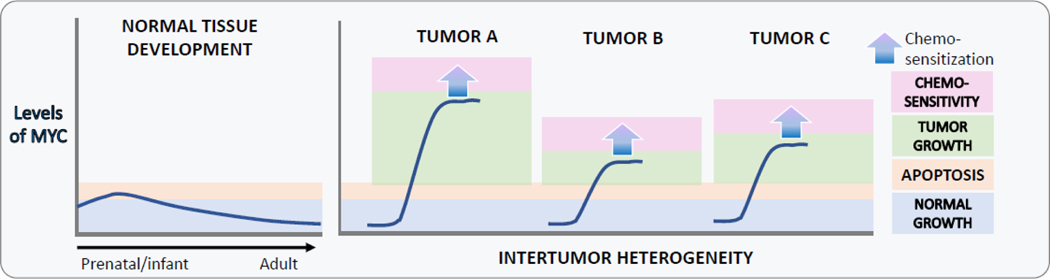
Work by many laboratories have shown that the ability of MYC to engage cell death pathways is the core feature of this oncoprotein. The pro-apoptotic activity of MYC could be leveraged for improved treatment outcomes even in chemoresistant tumors, if one considers that tumor cells require enough MYC to initiate tumorigenesis and sustain growth, but cannot have too much MYC, which would induce cell death. What constitutes “too much MYC” is likely going to be different among individual tumors, depending at least in part on the genetic make-up of each particular neoplasm (TP53 status, BCL2 expression, etc.) One can envision a simple scenario where tumors A, B, and C have already achieved their own maximum tolerated levels of MYC, and further increases, however small, would push them over the edge when combined with chemotherapy (Figure 3).
Figure 3. The role of MYC in development and cancer.
During normal development, MYC levels start out high and decline in aging tissues, with a concomitant decline in both proliferation and apoptosis (left). During the development of cancer, MYC levels are increased and so is neoplastic growth, which compensates for the increase in apoptosis and results in tumorigenesis (right). Both proliferative and apoptotic thresholds are not absolute and vary in individual neoplasms, resulting in inter-tumor heterogeneity. Here tumors A, B, and C have already achieved their own maximum tolerated levels of MYC, and further increases, however small, would push them over the edge when combined with chemotherapy.
Indeed, a recent study has demonstrated that transiently increasing MYC levels immediately prior to chemotherapy through GSK-3β inhibition with CHIR99021 [120] improves apoptotic response in p53-mutated, highly chemoresistant lymphomas [121]; similar data exist for solid cancers such as lung adenocarcinoma as well [129]. Besides CHIR99021, there is a handful of GSK-3 inhibitors currently in clinical trials [134], such as tideglusib which has undergone trials for Alzheimer’s disease [135], and LY2090314 which is in phase II trials for the treatment of acute leukemia [136] and has been tested in phase I clinical trials in many other cancer types [137]. The potential to re-purpose the FDA approved GSK-3β inhibitor lithium chloride [138] makes this adjuvant therapy strategy particularly viable as transitioning this psychiatric drug to cancer therapy would be relatively unchallenging. Of note, long-term usage of lithium chloride is not correlated with an increase in cancer incidence and appears to be a safe adjuvant [139]. Interestingly, inhibition of GSK-3α was recently shown to sensitize drug-resistant leukemias to asparaginase by a mechanism at least partly dependent of WNT signaling, which is the upstream regulator of MYC [140] and relies on it for some of its key functions in several organ systems [141].
In addition to GSK-3β, there are many other regulators of MYC protein stability that could be considered as therapeutic targets. Besides FBXw7, MYC is regulated by a host of E3 ubiquitin ligases including but not limited to Skp2 [reviewed by [116]. Thus, therapeutic inhibition of Skp2 [142–147] and other degradation complexes could potentially be another way to boost MYC expression and re-engage apoptosis. Furthermore, it was recently discovered that CDK9, which was already known to be required for transcription of the MYC gene, positively regulates MYC protein stability and prevents its degradation [148]. Although CDK9 agonists are yet to be developed, it is clear that there are numerous avenues to manipulating MYC levels for therapeutic benefit.
Finally, given that much of MYC pro-apoptotic effects are realized through the extrinsic apoptotic pathway, one could also revisit the use of death receptor agonists as anti-cancer agents. For example, TRAIL and its analogs had undergone extensive Phase I/II clinical trials where they well tolerated but displayed minimal efficacy against tumors [149]. Similar fate befell death receptor agonist antibodies such as mapatumumab [150]; however, the addition of a GSK-3β inhibitor could well bring these compounds back into clinical relevance. More broadly speaking, re-designing MYC synthetic lethality screens to incorporate standards of care appears to be a highly promising approach.
In fairness, many questions pertaining to MYC stabilization therapies remain unanswered (see Outstanding Questions Box). Thus, the goal of this review is not to advocate for the immediate use of GSK3i or similar molecules in the clinic, but rather to promote further research on the subject in preclinical models utilizing both chemotherapeutic drugs and death receptor agonists.
Outstanding Questions
Are pro-apoptotic effects of MYC limited to certain tumor types, such as hematologic malignancies, or are they a hallmark of most, if not all, human cancers?
What other small molecules, beyond GSK3i, could be used to hyperactivate MYC?
How would tumors with stabilized MYC respond to radiation therapy?
What effects would MYC stabilization therapy have on anti-tumor immunity and more specifically - on the efficacy of immune checkpoint inhibitors?
Could tumors become resistant to MYC-and-chemotherapy-driven apoptosis?
Could inhibitors and activators of MYC be combined into single therapeutic regimens to alternatily target cell proliferation and cell survival?
Highlights
As genomic and transciptomic profiling of human cancers is becoming a routine diagonstic and prognostic tool, the MYC oncogene has emerged as a pervasive force in human cancers, whose gain-of-function alterations are apparent in the plurality of tumor types and specimens
Thus, inhibitors of the MYC oncoprotein are under active development and show promise in model systems, but are yet to prove their utility in clinical settings
On the other hand, a considerable body of evidence indicates that expressing too much MYC can be counterproductive for neoplastic growth, as it drives tumor cell apoptosis and correlates with favorable responses to chemotherapy
Indeed, some very recent studies demonstrate that as antagonists of MYC degradation pathways (such as small molecule inhibitors of the GSK3 kinase) transiently elevate MYC levels, they confer chemosensity within that narrow window.
Acknowledgements
We thank all members of our laboratories for helpful discussions and our many Penn colleagues, in particular Michael Hogarty, Peter Klein, and Xiaolu Yang, for invaluable advice. Our own research was supported by the V Foundation for Cancer Research and the NIH grants T32 GM007229 & F31 CA217004 (CTH), P30 CA010815, R01 CA057341 & R01 CA051497 (CVD), and R01CA196299 & R21 CA183445 (ATT).
Declaration of Interests
ATT and ES are inventors on the US Patent US10751356B2 “Compositions and methods for transient up-regulation of MYC in B-cell lymphomas for enhancing P53 independent apoptotic responses to chemotherapy”. This intellectual property is held by CHOP and to date has not been licensed or otherwise commercialized. ATT receives funding from Pfizer’s ASPIRE Program for research unrelated to the topic of this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weinstein IB and Joe AK (2006) Mechanisms of disease: Oncogene addiction--a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol 3 (8), 448–57. [DOI] [PubMed] [Google Scholar]
- 2.Felsher DW (2003) Cancer revoked: oncogenes as therapeutic targets. Nat Rev Cancer 3 (5), 375–380. [DOI] [PubMed] [Google Scholar]
- 3.Piha-Paul SA et al. (2020) Phase 1 Study of Molibresib (GSK525762), a Bromodomain and Extra-Terminal Domain Protein Inhibitor, in NUT Carcinoma and Other Solid Tumors. JNCI Cancer Spectr 4 (2), pkz093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amorim S et al. (2016) Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol 3 (4), e196–204. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez V and Hurley LH (2010) The c-MYC NHE III(1): function and regulation. Annu Rev Pharmacol Toxicol 50, 111–29. [DOI] [PubMed] [Google Scholar]
- 6.Yin X et al. (2003) Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 22 (40), 6151–9. [DOI] [PubMed] [Google Scholar]
- 7.Han H et al. (2019) Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell 36 (5), 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaulieu ME et al. (2019) Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy. Sci Transl Med 11 (484), eaar5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tolcher AW et al. (2015) Safety and activity of DCR-MYC, a first-in-class Dicer-substrate small interfering RNA (DsiRNA) targeting MYC, in a phase I study in patients with advanced solid tumors. J Clin Oncol 33 (15_suppl), 11006. [Google Scholar]
- 10.Grayson AR et al. (2014) MYC, a downstream target of BRD-NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene 33 (13), 1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaub FX et al. (2018) Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst 6 (3), 282–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durbin AD et al. (2018) Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet 50 (9), 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sin-Chan P et al. (2019) A C19MC-LIN28A-MYCN Oncogenic Circuit Driven by Hijacked Super-enhancers Is a Distinct Therapeutic Vulnerability in ETMRs: A Lethal Brain Tumor. Cancer Cell 36 (1), 51–67 e7. [DOI] [PubMed] [Google Scholar]
- 14.Baluapuri A et al. (2020) Target gene-independent functions of MYC oncoproteins. Nat Rev Mol Cell Biol 21 (5), 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalkat M et al. (2018) MYC protein interactome profiling reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol Cell 72 (5), 836–848. [DOI] [PubMed] [Google Scholar]
- 16.Wanzel M et al. (2003) Transcriptional repression by Myc. Trends Cell Biol. 13 (3), 146–150. [DOI] [PubMed] [Google Scholar]
- 17.Patel JH et al. (2004) Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer 4 (7), 562–568. [DOI] [PubMed] [Google Scholar]
- 18.Psathas JN and Thomas-Tikhonenko A. (2014) MYC and the art of microRNA maintenance. Cold Spring Harb Perspect Med 4 (8), a014175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang CV (2012) MYC on the path to cancer. Cell 149 (1), 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casey SC et al. (2018) The MYC oncogene is a global regulator of the immune response. Blood 131 (18), 2007–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain M et al. (2002) Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297 (5578), 102–104. [DOI] [PubMed] [Google Scholar]
- 22.Choi PS et al. (2011) Lymphomas that recur after MYC suppression continue to exhibit oncogene addiction. Proc Natl Acad Sci U S A 108 (42), 17432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flores I et al. (2004) Defining the temporal requirements for Myc in the progression and maintenance of skin neoplasia. Oncogene 23 (35), 5923–30. [DOI] [PubMed] [Google Scholar]
- 24.Soucek L et al. (2013) Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev 27 (5), 504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabay M et al. (2014) MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 4 (6), a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang CV et al. (2017) Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer 17 (8), 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savino M et al. (2011) The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS One 6 (7), e22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soucek L et al. (2002) Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Research 62 (12), 3507–3510. [PubMed] [Google Scholar]
- 29.Soucek L et al. (2008) Modelling Myc inhibition as a cancer therapy. Nature 455 (7213), 679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eilers M and Eisenman RN (2008) Myc’s broad reach. Genes Dev. 22 (20), 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuber J et al. (2011) RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478 (7370), 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delmore JE et al. (2011) BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146 (6), 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filippakopoulos P et al. (2010) Selective inhibition of BET bromodomains. Nature 468 (7327), 1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muhar M et al. (2018) SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science 360 (6390), 800–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alqahtani A et al. (2019) Bromodomain and extra-terminal motif inhibitors: a review of preclinical and clinical advances in cancer therapy. Future Sci OA 5 (3), FSO372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conery AR et al. (2016) Preclinical anticancer efficacy of BET bromodomain inhibitors is determined by the apoptotic response. Cancer Res 76 (6), 1313–9. [DOI] [PubMed] [Google Scholar]
- 37.Gollavilli PN et al. (2018) EWS/ETS-Driven Ewing Sarcoma Requires BET Bromodomain Proteins. Cancer Res 78 (16), 4760–4773. [DOI] [PubMed] [Google Scholar]
- 38.Dauch D et al. (2016) A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat Med 22 (7), 744–53. [DOI] [PubMed] [Google Scholar]
- 39.Gustafson WC et al. (2014) Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 26 (3), 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otto T et al. (2009) Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell 15 (1), 67–78. [DOI] [PubMed] [Google Scholar]
- 41.Bavetsias V and Linardopoulos S. (2015) Aurora kinase inhibitors: current status and outlook. Front Oncol 5, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farrington CC et al. (2020) Protein phosphatase 2A activation as a therapeutic strategy for managing MYC-driven cancers. J Biol Chem 295 (3), 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen-Petersen BL et al. (2019) Activation of PP2A and inhibition of mTOR synergistically reduce MYC signaling and decrease tumor growth in pancreatic ductal adenocarcinoma. Cancer Res 79 (1), 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nilsson JA et al. (2005) Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell 7 (5), 433–44. [DOI] [PubMed] [Google Scholar]
- 45.Bachmann AS and Geerts D. (2018) Polyamine synthesis as a target of MYC oncogenes. J Biol Chem 293 (48), 18757–18769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarosiek KA et al. (2017) Developmental regulation of mitochondrial apoptosis by c-Myc governs age- and tissue-specific sensitivity to cancer therapeutics. Cancer Cell 31 (1), 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMahon SB (2014) MYC and the control of apoptosis. Cold Spring Harb Perspect Med 4 (7), a014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruf IK et al. (2001) EBV regulates c-MYC, apoptosis, and tumorigenicity in Burkitt’s lymphoma. Curr Top Microbiol Immunol 258, 153–60. [DOI] [PubMed] [Google Scholar]
- 49.Vita M and Henriksson M. (2006) The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol 16, 318–330. [DOI] [PubMed] [Google Scholar]
- 50.Kim YH et al. (2006) Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene 25 (1), 130–138. [DOI] [PubMed] [Google Scholar]
- 51.Ladu S et al. (2008) E2F1 Inhibits c-Myc-driven apoptosis via PIK3CA/Akt/mTOR and COX-2 in a mouse model of human liver cancer. Gastroenterology 135 (4), 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iba T et al. (2004) Expression of the c-myc gene as a predictor of chemotherapy response and a prognostic factor in patients with ovarian cancer. Cancer Science 95 (5), 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Askew DS et al. (1991) Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 6 (10), 1915–1922. [PubMed] [Google Scholar]
- 54.Evan GI et al. (1992) Induction of apoptosis in fibroblasts by c-myc protein. Cell 69, 119–128. [DOI] [PubMed] [Google Scholar]
- 55.Shi Y et al. (1992) Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science 257, 212–214. [DOI] [PubMed] [Google Scholar]
- 56.Zindy F et al. (1998) Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 12 (15), 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pusapati RV et al. (2006) ATM promotes apoptosis and suppresses tumorigenesis in response to Myc. Proc Natl Acad Sci U S A 103 (5), 1446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eischen CM et al. (1999) Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 13 (20), 2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacobs JJ et al. (1999) Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 13 (20), 2678–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmitt CA and Lowe SW (1999) Apoptosis and therapy. J Pathol 187 (1), 127–37. [DOI] [PubMed] [Google Scholar]
- 61.Hemann MT et al. (2005) Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436 (7052), 807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forbes SA et al. (2015) COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43, D805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eischen CM et al. (2001) Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene 20 (48), 6983–93. [DOI] [PubMed] [Google Scholar]
- 64.Eischen CM et al. (2001) Apoptosis triggered by Myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol Cell Biol 21 (15), 5063–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nikiforov MA et al. (2007) Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc Natl Acad Sci USA 104 (49), 19488–19493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitchell KO et al. (2000) Bax is a transcriptional target and mediator of c-myc-induced apoptosis. Cancer Res 60 (22), 6318–6325. [PubMed] [Google Scholar]
- 67.Soucie EL et al. (2001) Myc potentiates apoptosis by stimulating Bax activity at the mitochondria. Mol Cell Biol 21 (14), 4725–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Albihn A et al. (2006) c-Myc-dependent etoposide-induced apoptosis involves activation of Bax and caspases, and PKCdelta signaling. J Cell Biochem 98 (6), 1597–614. [DOI] [PubMed] [Google Scholar]
- 69.Egle A et al. (2004) Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA 101 (16), 6164–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eischen CM et al. (2001) Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol Cell Biol 21 (22), 7653–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dansen TB et al. (2006) Specific requirement for Bax, not Bak, in Myc-induced apoptosis and tumor suppression in vivo. J Biol Chem 281 (16), 10890–5. [DOI] [PubMed] [Google Scholar]
- 72.Nieminen AI et al. (2007) c-Myc primed mitochondria determine cellular sensitivity to TRAIL-induced apoptosis. EMBOJ 26 (4), 1055–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shiio Y et al. (2006) Quantitative proteomic analysis of myc-induced apoptosis: a direct role for Myc induction of the mitochondrial chloride ion channel, mtCLIC/CLIC4. J Biol Chem 281 (5), 2750–6. [DOI] [PubMed] [Google Scholar]
- 74.Hueber AO et al. (1997) Requirement for the CD95 receptor-ligand pathway in c-Myc-induced apoptosis. Science 278 (5341), 1305–1309. [DOI] [PubMed] [Google Scholar]
- 75.Ricci MS et al. (2007) Reduction of TRAIL-induced Mcl-1 and cIAP2 by c-Myc or sorafenib sensitizes resistant human cancer cells to TRAIL-induced death. Cancer Cell 12 (1), 66–80. [DOI] [PubMed] [Google Scholar]
- 76.Kasibhatla S et al. (2000) A ‘non-canonical’ DNA-binding element mediates the response of the Fas-ligand promoter to c-Myc. Curr Biol 10 (19), 1205–8. [DOI] [PubMed] [Google Scholar]
- 77.Ricci S et al. (2004) Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol Cell Biol 24 (19), 8541–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sussman RT et al. (2007) Chemotherapy-resistant side-population of colon cancer cells has a higher sensitivity to TRAIL than the non-SP, a higher expression of c-Myc and TRAIL-receptor DR4. Cancer Biol Ther 6 (9), 1490–5. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y et al. (2004) Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell 5 (5), 501–12. [DOI] [PubMed] [Google Scholar]
- 80.Chen W et al. (2021) Large remodeling of the Myc-induced cell surface proteome in B cells and prostate cells creates new opportunities for immunotherapy. Proc Natl Acad Sci U S A 118 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cermelli S et al. (2014) Synthetic lethal screens as a means to understand and treat MYC-driven cancers. Cold Spring Harb Perspect Med 4 (3), a014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stine ZE et al. (2015) MYC, Metabolism, and Cancer. Cancer Discov 5 (10), 1024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goga A et al. (2007) Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med 13 (7), 820–7. [DOI] [PubMed] [Google Scholar]
- 84.Li F et al. (2019) Cancer therapeutics using survivin BIRC5 as a target: what can we do after over two decades of study? J Exp Clin Cancer Res 38 (1), 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen H et al. (2018) Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduction and Targeted Therapy 3 (1), 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee HY et al. (2019) c-MYC drives breast cancer metastasis to the brain, but promotes synthetic lethality with TRAIL. Mol Cancer Res 17 (2), 544–554. [DOI] [PubMed] [Google Scholar]
- 87.Thng DKH et al. (2021) Capitalizing on synthetic lethality of MYC to treat cancer in the digital age. Trends Pharmacol Sci 42 (3), 166–182. [DOI] [PubMed] [Google Scholar]
- 88.Donoghue MTA et al. (2020) Discovery through clinical sequencing in oncology. Nature Cancer 1 (8), 774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arango D et al. (2001) c-myc/p53 interaction determines sensitivity of human colon carcinoma cells to 5-fluorouracil in vitro and in vivo. Cancer Res 61 (12), 4910–4915. [PubMed] [Google Scholar]
- 90.Maclean KH et al. (2003) c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol Cell Biol 23 (20), 7256–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nawrocki ST et al. (2008) Myc regulates aggresome formation, the induction of Noxa, and apoptosis in response to the combination of bortezomib and SAHA. Blood 112 (7), 2917–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Topham C et al. (2015) MYC is a major determinant of mitotic cell fate. Cancer Cell 28 (1), 129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Haikala HM et al. (2019) Pharmacological reactivation of MYC-dependent apoptosis induces susceptibility to anti-PD-1 immunotherapy. Nat Commun 10 (1), 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mollaoglu G et al. (2017) MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to Aurora kinase inhibition. Cancer Cell 31 (2), 270–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Adachi S et al. (2001) c-Myc is necessary for DNA damage-induced apoptosis in the G(2) phase of the cell cycle. Mol Cell Biol 21 (15), 4929–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cerquetti L et al. (2015) c-MYC modulation induces responsiveness to paclitaxel in adrenocortical cancer cell lines. Int J Oncol 46 (5), 2231–40. [DOI] [PubMed] [Google Scholar]
- 97.Grassilli E et al. (2004) Loss of MYC confers resistance to doxorubicin-induced apoptosis by preventing the activation of multiple serine protease- and caspase-mediated pathways. J Biol Chem 279 (20), 21318–26. [DOI] [PubMed] [Google Scholar]
- 98.Sotillo E et al. (2011) Myc overexpression brings out unexpected antiapoptotic effects of miR-34a. Oncogene 30 (22), 2587–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Biroccio A et al. (2001) c-Myc down-regulation increases susceptibility to cisplatin through reactive oxygen species-mediated apoptosis in M14 human melanoma cells. Mol Pharmacol 60 (1), 174–182. [DOI] [PubMed] [Google Scholar]
- 100.Bucci B et al. (2005) Myc down-regulation sensitizes melanoma cells to radiotherapy by Inhibiting MLH1 and MSH2 mismatch repair roteins. Clin Cancer Res 11 (7), 2756–2767. [DOI] [PubMed] [Google Scholar]
- 101.Bidwell GL 3rd and Raucher D. (2006) Enhancing the antiproliferative effect of topoisomerase II inhibitors using a polypeptide inhibitor of c-Myc. Biochem Pharmacol 71 (3), 248–56. [DOI] [PubMed] [Google Scholar]
- 102.Murphy DJ et al. (2008) Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell 14 (6), 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Deng J et al. (1998) Adenovirus 5 E1A-mediated tumor suppression associated with E1A-mediated apoptosis in vivo. Oncogene 17 (17), 2167–75. [DOI] [PubMed] [Google Scholar]
- 104.Yoo GH et al. (2001) Phase I trial of intratumoral liposome E1A gene therapy in patients with recurrent breast and head and neck cancer. Clin Cancer Res 7 (5), 1237–45. [PubMed] [Google Scholar]
- 105.Hortobagyi GN et al. (2001) Cationic liposome-mediated E1A gene transfer to human breast and ovarian cancer cells and its biologic effects: a phase I clinical trial. J Clin Oncol 19 (14), 3422–3433. [DOI] [PubMed] [Google Scholar]
- 106.Cimas FJ et al. (2017) E1a is an exogenous in vivo tumour suppressor. Cancer Letters 399, 74–81. [DOI] [PubMed] [Google Scholar]
- 107.Chung EY et al. (2012) CD19 is a major B cell receptor-independent activator of MYC-driven B-lymphomagenesis. J Clin Invest 122 (6), 2257–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Psathas JN et al. (2013) The Myc-miR-17–92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: a novel lymphomagenic feed-forward loop. Blood 122 (26), 4220–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sander S et al. (2012) Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 22 (2), 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ren Y et al. (2018) PLK1 stabilizes a MYC-dependent kinase network in aggressive B cell lymphomas. J Clin Invest 128 (12), 5517–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Varano G et al. (2017) The B-cell receptor controls fitness of MYC-driven lymphoma cells via GSK3beta inhibition. Nature 546 (7657), 302–306. [DOI] [PubMed] [Google Scholar]
- 112.Welcker M et al. (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA 101 (24), 9085–9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yada M et al. (2004) Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO Journal 23 (10), 2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.King B et al. (2013) The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153 (7), 1552–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gregory MA et al. (2003) Phosphorylation by glycogen synthase kinase-3 controls c-Myc proteolysis and subnuclear localization. J Biol Chem 278 (51), 51606–51612. [DOI] [PubMed] [Google Scholar]
- 116.Farrell AS and Sears RC (2014) MYC degradation. Cold Spring Harb Perspect Med 4 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sanchez-Vega F et al. (2018) Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 173 (2), 321–337 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kennedy SG et al. (1997) The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev 11 (6), 701–13. [DOI] [PubMed] [Google Scholar]
- 119.Kauffmann-Zeh A et al. (1997) Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature 385 (6616), 544–548. [DOI] [PubMed] [Google Scholar]
- 120.Ring DB et al. (2003) Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 52 (3), 588–95. [DOI] [PubMed] [Google Scholar]
- 121.Harrington CT et al. (2019) Transient stabilization, rather than inhibition, of MYC amplifies extrinsic apoptosis and therapeutic responses in refractory B-cell lymphoma. Leukemia 33, 2429–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pajic A et al. (2000) Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int J Cancer 87 (6), 787–793. [DOI] [PubMed] [Google Scholar]
- 123.Duffy DJ et al. (2014) GSK3 inhibitors regulate MYCN mRNA levels and reduce neuroblastoma cell viability through multiple mechanisms, including p53 and Wnt signaling. Mol Cancer Ther 13 (2), 454–67. [DOI] [PubMed] [Google Scholar]
- 124.Kazi A et al. (2018) GSK3 suppression upregulates beta-catenin and c-Myc to abrogate KRas-dependent tumors. Nat Commun 9 (1), 5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kotliarova S et al. (2008) Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-kappaB, and glucose regulation. Cancer Res 68 (16), 6643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Biechele TL et al. (2012) Wnt/β-catenin signaling and AXIN1 regulate apoptosis triggered by inhibition of the mutant kinase BRAFV600E in human melanoma. Sci Signal 5 (206), ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zimmerman ZF et al. (2013) Activation of Wnt/β-catenin signaling increases apoptosis in melanoma cells treated with trail. PLoS One 8 (7), e69593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.He TC et al. (1998) Identification of c-MYC as a target of the APC pathway. Science 281 (5382), 1509–1512. [DOI] [PubMed] [Google Scholar]
- 129.O’Flaherty L et al. (2019) Tumor growth suppression using a combination of taxol-based therapy and GSK3 inhibition in non-small cell lung cancer. PLoS One 14 (4), e0214610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Walz A et al. (2017) Molecular pathways: revisiting glycogen synthase kinase-3β as a target for the treatment of cancer. Clin Cancer Res 23 (8), 1891–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Casey SC et al. (2016) MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352 (6282), 227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Truica MI et al. (2021) Turning Up the Heat on MYC: Progress in Small-Molecule Inhibitors. Cancer Res 81 (2), 248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kuzyk A and Mai S. (2014) c-MYC-induced genomic instability. Cold Spring Harb Perspect Med 4 (4), a014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ugolkov AV et al. (2018) 9-ING-41, a small-molecule glycogen synthase kinase-3 inhibitor, is active in neuroblastoma. Anticancer Drugs 29 (8), 717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lovestone S et al. (2015) A phase II trial of tideglusib in Alzheimer’s disease. J Alzheimers Dis 45 (1), 75–88. [DOI] [PubMed] [Google Scholar]
- 136.Rizzieri DA et al. (2016) An open-label phase 2 study of glycogen synthase kinase-3 inhibitor LY2090314 in patients with acute leukemia. Leuk Lymphoma 57 (8), 1800–6. [DOI] [PubMed] [Google Scholar]
- 137.Gray JE et al. (2015) A first-in-human phase I dose-escalation, pharmacokinetic, and pharmacodynamic evaluation of intravenous LY2090314, a glycogen synthase kinase 3 inhibitor, administered in combination with pemetrexed and carboplatin. Invest New Drugs 33 (6), 1187–1196. [DOI] [PubMed] [Google Scholar]
- 138.O’Brien WT and Klein PS (2009) Validating GSK3 as an in vivo target of lithium action. Biochem Soc Trans 37 (Pt 5), 1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cohen Y et al. (1998) Cancer morbidity in psychiatric patients: influence of lithium carbonate treatment. Med Oncol 15 (1), 32–6. [DOI] [PubMed] [Google Scholar]
- 140.Hinze L et al. (2019) Synthetic lethality of Wnt pathway activation and asparaginase in drug-resistant acute leukemias. Cancer Cell 35 (4), 664–676 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sansom OJ et al. (2007) Myc deletion rescues Apc deficiency in the small intestine. Nature 446 (7136), 676–679. [DOI] [PubMed] [Google Scholar]
- 142.Rodriguez S et al. (2020) Therapeutic targeting of the E3 ubiquitin ligase SKP2 in T-ALL. Leukemia 34 (5), 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chan CH et al. (2013) Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell 154 (3), 556–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu L et al. (2012) Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol 19 (12), 1515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen Q et al. (2008) Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood 111 (9), 4690–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li GZ et al. (2020) Rb and p53-Deficient Myxofibrosarcoma and Undifferentiated Pleomorphic Sarcoma Require Skp2 for Survival. Cancer Res 80 (12), 2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhou L et al. (2020) Cdh1-mediated Skp2 degradation by dioscin reprogrammes aerobic glycolysis and inhibits colorectal cancer cells growth. EBioMedicine 51, 102570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Blake DR et al. (2019) Application of a MYC degradation screen identifies sensitivity to CDK9 inhibitors in KRAS-mutant pancreatic cancer. Sci Signal 12 (590), eaav7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lemke J et al. (2014) Getting TRAIL back on track for cancer therapy. Cell Death Differ 21 (9), 1350–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maddipatla S et al. (2007) Augmented antitumor activity against B-cell lymphoma by a combination of monoclonal antibodies targeting TRAIL-R1 and CD20. Clin Cancer Res 13 (15 Pt 1), 4556–64. [DOI] [PubMed] [Google Scholar]