Abstract
Better detection of minority human immunodeficiency virus type 1 (HIV-1) populations containing gene mutations may improve the usefulness of antiretroviral resistance testing for clinical management. Molecular cloning of HIV-1 PCR products which might improve minority detection can be slow and difficult, and commercially available recombinant virus assays test drug susceptibility of virus pools. We describe novel plasmids and simple methods for rapid cloning of HIV-1 PCR products from patient specimens and their application to generate infectious recombinant virus clones for virus phenotyping and genotyping. Eight plasmids with differing deletions of sequences encoding HIV-1 protease, reverse transcriptase, or Gag p7/p1 and Gag p1/p6 cleavage sites were constructed for cloning HIV-1 PCR products. A simple HIV-1 sequence-specific uracil deglycosylase-mediated cloning method with the vectors and primers designed here was more rapid than standard ligase-mediated cloning. Pooled and molecularly cloned infectious recombinant viruses were generated with these vectors. Replicative viral fitness and drug susceptibility phenotypes of cloned infectious viruses containing patient specimen-derived sequences were measured. Clonal resistance genotyping analyses were also performed from virus isolates, plasma HIV-1 RNA, and infected cell DNA. Sequencing of a limited number of molecular clones detected minorities of resistant virus not identified in the pooled population PCR product sequence and linkage of minority mutations.
Human immunodeficiency virus type 1 (HIV-1) gene mutations conferring resistance to reverse transcriptase (RT) and protease (PR) inhibitors may lead to virologic failure of the antiretroviral drugs being used to treat HIV-1 infection. Such mutations, called resistance mutations, can also confer cross-resistance to subsequent, alternative drugs of the same class. One specific drug may select different resistant mutants with varying degrees of cross-resistance to separate antiretroviral drugs in different patients (3, 28, 30, 31). Recent data suggest that resistance mutations need not be selected early by each drug in a triple combination regimen during rebound of plasma HIV-1 RNA (4, 9). Resistance testing may be useful in the clinical management of antiretroviral failure, as an adjunct to clinical, immunologic, and viral load responses, by identifying drugs to which the patient’s virus population is resistant and those to which the virus may still be susceptible (12). Mutations should be interpreted in the context of other mutations; for example, lamivudine-selected RT M184V may suppress phenotypic effects of zidovudine resistance mutations to varying degrees in different clinical isolates (20). New recombinant virus assays speed phenotypic drug susceptibility testing (10, 11, 15) and may have an advantage over genotyping because they can directly assess mutational interactions as well as crossresistance.
All current recombinant virus assays generating virus with both PR and RT from a patient-derived PCR product test a pool of virus variants or strains. Therefore, a variant present as a small minority of the HIV-1 population may not be routinely detected. This may limit diagnostic resistance testing, as previously used antiretroviral drugs may have selected for specific resistance mutations. Because these drugs and their specific selection pressures are no longer present, virus strains containing these mutations may be reduced to only a small percentage of the total viral population. Some recombinant virus cloning vectors (10, 29) use standard molecular cloning methods and could potentially better detect minority strains. However, these standard cloning methods are relatively technically demanding. Only one of these previously described vectors can be used to clone both PR and RT reading frames (10). The presence of both HIV-1 long terminal repeats (LTRs) in a recombinant clone risks causing deletions in patient-derived sequences during the plasmid propagation in Escherichia coli necessary for any clonal analysis (23).
A series of novel plasmids and simple methods for rapid cloning of HIV-1 PCR products from patient specimens are described in this report and applied to generate infectious recombinant virus clones and pools. Clonal analyses using these versatile vectors and new methods were better able to identify minority strains of resistant virus than a parallel analysis of the viral pool. These vectors and methods will facilitate development of clinically applicable clonal analytic approaches to antiretroviral drug resistance testing. Some advantages over previously described recombinant virus cloning vectors for research purposes are presented.
MATERIALS AND METHODS
Patient specimens.
Specimens from HIV-1-infected patients included plasma, uncultured peripheral blood mononuclear cells (PBMCs), semen mononuclear cells, and HIV-1 isolates cocultured from PBMCs (13).
Oligonucleotide primers.
Oligonucleotides (Gibco-BRL, Gaithersburg, Md., or Integrated DNA Technologies, Coralville, Calif.) (Table 1) used as reverse transcription and amplification primers were purified by desalting. Primers used for HIV-1 sequence-specific uracil deglycosylase (UDG) cloning (see below) included dUMP residues incorporated into the 5′ end of each primer instead of TMP at positions where T occurs in the HIV-1 sequence and were purified from polyacrylamide gels. All primers were designed with attention to sequence conservation (12a).
TABLE 1.
Oligonucleotide primers
| Vector | Primer | PCR | Sequence | Fragment size (bp) | Temp (°C)a | Cotransfection DNAb |
|---|---|---|---|---|---|---|
| pJM11ΔGPR | Apa1988 | 5′-ACATAGCCAIAAATTGCAGGGCCCCTAG | 878 | 65 | p83-10 | |
| ApaI | ||||||
| Sse2835 | 5′-CTGATTTTTTCTGTTTTAACCCTGCAGGATG | |||||
| SseB387I | ||||||
| pJM13ΔGPRT | 5CAI1964Bc | 5′-GUTUCAAUTGUGGUAAAGAAGGUCACATAGCTAAAAATTGCAGGG | 2,232 | 55 | p83-10 | |
| ApaI | ||||||
| 3CAI4155LIGc | 5′-ACCCAUGCUAGGUAGACCUTTTCCTTTTTAATTAACTGCTC | |||||
| PacI | ||||||
| pJM14ΔRT | 2574U29 | 5′-CCAGTAAAATTAAAGCCCGGGATGGATGG | 1,581 | 55 | p83-10 | |
| XmaI | ||||||
| 4120L35 | 5′-CCATGCCAGGTAGACTTTTTCCTTTTTAATTAACT | |||||
| PacI | ||||||
| pJM20ΔGPRT | 3CAV1982c | Vector | 5′-TCUTUACCACAAUUGAAACACUTAACAGTCTTTCTTTGGTTCCTA | 3,381 | 59 | pJM31ΔGPRT |
| 5V4142c | Vector | 5′-CUACCUAGCAUGGGTACCAGCACACAAAGGAATTGGAGGAAATGA | ||||
| 3R4226d | RT, first round | 5′-TGGGCCTTATCTATTCCATCTAAAAATAGT | 2,268 | 55 | ||
| pJM31ΔGPRT | 1607U25 | First round | 5′-TAGTAAGAATGTATAGCCCTACCAG | 2,939 | 55 | pJM20GPRT or PCR product |
| 4522L24 | RT, first round | 5′-CTGCTAATTTTAAGAGGAAGTATG | ||||
| 1811U24 | Second round (nested) | 5′-CTACACTAGAAGAAATGATGACAG | 2,549 | 62 | ||
| 4335L25 | Second round (nested) | 5′-GACATTTATCACAGCTGGCTACTAT | ||||
| pJM2ΔPRe | V5CA2611c | Vector | 5′-UAAACAAUGGCCAUTGACUGAAGAAA | >7,343 | 65 | None |
| V3CA2260-30Bc | Vector | 5′-GAUCUGAGGGAAGCUAAAGGAUACAGUTCC | ||||
| I5CA2238PCRc | Second round (nested) | 5′-GUAUCCUTUAGCUTCCCUCAGAUCACUCTTTG | 418 | 65 | None | |
| I3CA2626PCRc | Second round (nested) | 5′-CAAUGGCCAUTGUTUAACUTTTGGGCCATC | ||||
| pJM5ΔPRT | V5CA4147cf | Vector | 5′-AGCAUGGGUACCAGCACAUAAAGGAATTGG | >6,355 | 62 | None |
| I3CA4163cg | Second round (nested) | 5′-GUGCUGGUACCCAUGCUAGAUAGAC | 1,950 | 65 |
Optimal annealing temperature for PCR empirically determined.
Plasmid or PCR product used to cotransfect MT-2 cells in order to produce infectious virus in vitro (after deletion reconstructed in vectors other than pJM31ΔGPRT). p83-10 was from J. Gibbs, D. Regier, and R. Desrosiers (6).
HIV-1 sequence-specific UDG cloning primers.
First-round PCR primer used was Apa1988, described for pJM11ΔGPR. Second-round (nested) PCR primers for UDG cloning were 5CAI1964B and 5CAI4155LIG, described for pJM13ΔGPRT.
RT and first-round PCR with primers 3R4226, described for pJM20ΔGPRT, and Apa1988, described for pJM11ΔGPR.
In combination with V3CA2260-30B, described for pJM2ΔPR.
In combination with I5CA2238PCR, described for pJM2ΔPR.
RT PCR and PCR.
One milliliter of plasma was centrifuged (21,000 × g for 60 min) prior to HIV-1 RNA purification (viral RNA preparation kit; Qiagen, Valencia, Calif.). RNA was reverse transcribed with Superscript II RT (Gibco-BRL) at 42°C for 60 min with a specific primer (3R4226 described for pJM20GPRT or 4522L24 described for pJM31GPRT [Table 1]).
PCRs used XL recombinant Tth DNA polymerase (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.) with a hot start and three temperature steps in each thermal cycle. The magnesium concentration was optimized empirically, and the annealing temperature was optimized by using Oligo (version 5.0; National Biosciences, Inc., Plymouth, Maine) for each primer pair; 30 to 35 cycles and 0.2 mM (each) deoxynucleoside triphosphate were used unless otherwise stated. The cDNA from 3R4226 was amplified in a first round at a 54°C annealing temperature with Apa1988 and 3R4226, and a nested reaction was amplified with 5CAI 1964B and 5CAI4155LIG at a 55°C annealing temperature. cDNA from 4522L24 was amplified in a first round at a 55°C annealing temperature with 1607U25 and 4522L24, and a nested reaction was amplified with 1811U24 and 4335L25 at a 62°C annealing temperature.
HIV-1 cultures.
Wild-type (WT) HIV-1 was produced by electroporation (as described below) of pNL4-3 (1) or coelectroporation of two half HIV-1 genome plasmids, 5′-half HIV-1 genome p83-2 and 3′-half HIV-1 genome p83-10 (6), into MT-2 cells (7, 8). MT-2 cells were maintained in RPMI 1640 (Cellgro) supplemented with 10% fetal calf serum (FCS) (Sigma, St. Louis, Mo.), 2 mM l-glutamine (Cellgro), 10 mM HEPES buffer (Cellgro), and 50 μg of penicillin and streptomycin (Cellgro) per ml. Supernatant fluids of all cultures were replenished with media and tested by HIV-1 p24 antigen enzyme-linked immunosorbent assay (Alliance; NEN Life Science Products, Boston, Mass.) every 3 or 4 days. Cell-free supernatant fluids (filtered through Millex HV, 0.45-μm pore size; Millipore, Bedford, Mass.) from HIV-1-electroporated MT-2 cells which were p24 antigen positive were used to infect phytohemagglutinin (PHA)-stimulated PBMCs (2). PBMC cultures were refed with PHA-stimulated PBMCs every 7 days. Virus stocks were made from PBMC cultures and titrated on PBMCs (14). Infected PBMC DNA was prepared from frozen infected PBMC pellets from day 7 or 10 of culture (Puregene; Gentra Systems Inc., Minneapolis, Minn.).
Cloning vectors.
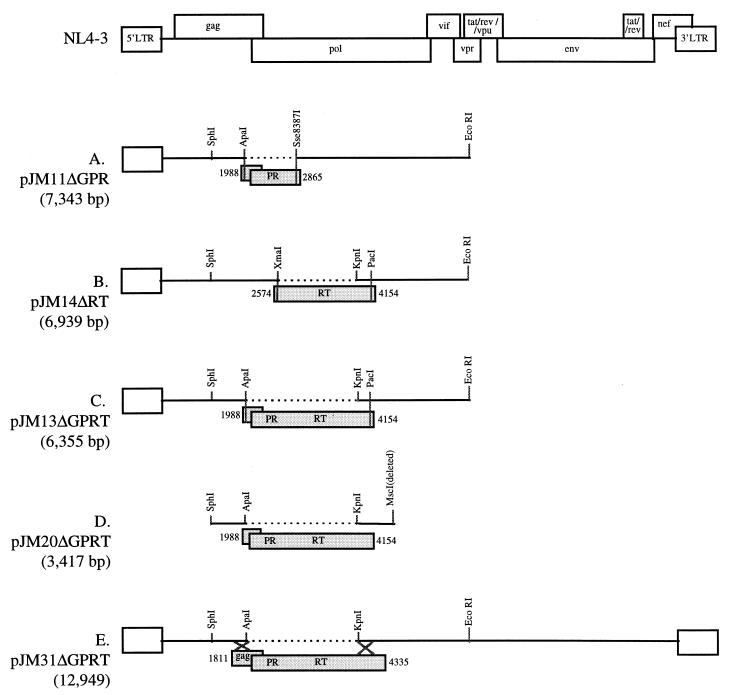
Molecular cloning was used to construct several plasmids with specific HIV-1 deletions (details are available on request). All of the plasmids with deletions can be used for standard ligase-mediated cloning, and some can also be used for HIV-1 sequence-specific UDG cloning. Ligase-mediated cloning could be used to restore gag PR PCR products into the 5′-half HIV-1 genome plasmid pJM11ΔGPR with a deletion of gag PR (gag PR-deleted) (Fig. 1A), gag PR-RT (GPRT) PCR products into the gag PRT-deleted 5′-half genome plasmid pJM13ΔGPRT (Fig. 1C) or a smaller gag PRT-deleted plasmid pJM20ΔGPRT (Fig. 1D), RT PCR products into the RT-deleted 5′-half genome plasmid pJM14ΔRT (Fig. 1B), PRT PCR products into the PRT-deleted 5′-half genome plasmid pJM5ΔPRT (data not shown). HIV-1 sequence-specific UDG cloning (see below) could also be used to clone gag PRT PCR products into pJM13ΔGPRT or pJM20ΔGPRT (Fig. 1C and D), PR PCR products into pJM2ΔPR (data not shown), and PR PCR products into pJM5ΔPRT (data not shown). A gag PRT-deleted plasmid, pJM31ΔGPRT (Fig. 1E), was also constructed for the generation of either pooled or molecularly cloned infectious recombinant viruses.
FIG. 1.
HIV-1 sequences in cloning vectors for antiretroviral resistance testing. (A) pJM11ΔGPR is used to clone HIV-1 sequences of Gag p7/p1 and Gag p1/p6 cleavage sites and PR (GPR). pJM11ΔGPR and the HIV-1 PCR product amplified with insert PCR primers Apa1988 and Sse2835 (in gray) are digested with restriction enzymes ApaI and Sse8387I and subsequently are ligated with T4 DNA ligase. Recombinant clones are cotransfected with p83-10 (6) into MT-2 cells to produce infectious virus. A polylinker (not shown) including the only XbaI and BamHI restriction sites in the plasmid substitutes for the deleted HIV-1 sequence in each of the vectors depicted in Fig. 1. Plasmid sequences are not shown for this or any other vector. (B) pJM14ΔRT is used to clone HIV-1 RT. pJM14ΔRT and the HIV-1 DNA PCR product amplified with insert PCR primers 2574U29 and 4120L35 (in gray) are digested with restriction enzymes XmaI and PacI and subsequently are ligated with T4 DNA ligase. Recombinant clones are cotransfected with p83-10 (6) into MT-2 cells to produce infectious virus. (C) pJM13ΔGPRT is used to clone HIV-1 sequences encoding Gag p7/p1 and Gag p1/p6 cleavage sites, PR, and RT (GPRT). pJM13ΔGPR and pJM20ΔGPRT are used to clone an HIV-1 PCR product amplified with the same insert PCR primers, 5CAI1964B and 3CAI4155LIG (in gray). pJM13ΔGPRT and the PCR product are digested with ApaI and PacI and subsequently are ligated with T4 DNA ligase. Recombinant clones are cotransfected with p83-10 (6) into MT-2 cells to produce infectious virus. (D) pJM20ΔGPRT is used to clone HIV-1 sequences encoding Gag p7/p1 and Gag p1/p6 cleavage sites, PR, and RT (GPRT). pJM20ΔGPRT is used for HIV-1 sequence-specific UDG cloning. It is digested with BamHI and then amplified with vector PCR primers 3CAV1982 and 5V4142 (see Fig. 2). Vector and insert (in gray) PCR products are digested with UDG and directly used to transform E. coli. Recombinant clones are cotransfected with pJM31ΔGPRT into MT-2 cells to produce infectious virus. (E) pJM31ΔGPRT is used to generate either pooled or molecularly cloned infectious HIV-1 with patient specimen-derived sequences of Gag p7/p1 and Gag p1/p6 cleavage sites, PR, and RT (GPRT). pJM31ΔGPRT digested with BamHI and the HIV-1 insert PCR product amplified with primers 1811U24 and 4522L24 (in gray) is directly cotransfected into MT-2 cells to produce infectious virus as a pool of different variants. A reconstructed pJM20GPRT plasmid is cotransfected with pJM31ΔGPRT into MT-2 cells to produce a cloned infectious virus.
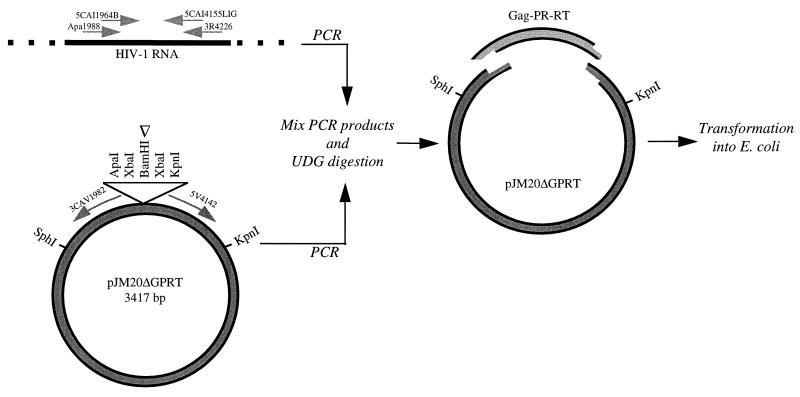
HIV-1 sequence-specific UDG cloning.
Only one target amplicon defined by a set of insert PCR primers can be cloned into a specific vector PCR product amplified by a specific primer pair designed for inverse PCR of that plasmid: PR into pJM2ΔPR, insert primers I5CA2238PCR and I3CA2626PCR and vector primers V5CA2611 and V2CA2260-30B; PRT into pJM5ΔPRT, insert primers I5CA2238PCR and I3CA4163 and vector primers V5CA4147 and V3CA2260-30B; gag PRT into pJM13ΔGPRT and pJM20ΔGPRT, insert primers 5CAI1964B and 3CAI4155Lig and vector primers 3CAV1982 and 5V4142 (Table 1). An example is depicted in Fig. 2, including the orientation of the vector PCR product primers for pJM20ΔGPRT. The pJM20ΔGPRT plasmid was inverse PCR amplified for 10 cycles; 20 cycles were used for inverse PCR of pJM2ΔPR and for pJM5ΔPRT. Inverse PCR of vector plasmid was performed with 25 to 100 ng of plasmid DNA linearized by restriction digestion. After mixing of the insert, patient-derived PCR product and the vector PCR product amplified from the respective plasmid with the dUMP-containing primers, UDG digestion (Gibco-BRL or Epicenter Technologies) was performed for 30 min at 37°C (1 U of UDG in annealing buffer containing 20 mM Tris-HCl [pH 8.4], 50 mM KCl, and 1.5 mM MgCl2). This generates overlapping, complementary, single-stranded ends; UDG digestion is depicted for pJM20ΔGPRT and the appropriate PCR products amplified from primers 5CAI1964B and 3CAI4155Lig (Table 1) in Fig. 2. Annealing of the two PCR products also occurs during this time. E. coli cells are then directly transformed by the annealed mixture. A transformation is performed with a vector PCR product without an insert PCR product to control in each cloning for uncut deletion vector plasmid DNA which may yield transformants without inserts.
FIG. 2.
HIV-1 sequence-specific UDG cloning. In this schematic representation of HIV-1 sequence-specific UDG cloning, pJM20ΔGPRT is inverse PCR amplified and the vector PCR product is directly mixed and annealed with the Gag p7/p1 and Gag p1/p6 cleavage site, PR, and RT (GPRT) insert PCR amplicon obtained from HIV-1 RNA. After the UDG digestion, the 5′ ends of both PCR products become long sticky ends, up to the last dUMP incorporated in the primer, and stably anneal. The mix is then electroporated into E. coli, where gaps are repaired. The white arrowhead indicates the restriction site used to linearize pJM20ΔGPRT before inverse vector PCR.
Recombinant virus production.
Infectious HIV-1 molecular clones were generated by cotransfection of 5′-half genome plasmids (pJM11GPR, pJM14RT, or pJM13GPRT with deletions reconstructed by cloning of gag PR, RT, or gag PRT deletion PCR products, respectively) with equimolar amounts of p83-10 (6). p83-10 contains the complementary 3′-half HIV-1NL4-3 genome (including partial vpr and complete tat, rev, vpu, env, and nef genes, as well as the 3′ LTR). Five micrograms of each half genome clone was digested with EcoRI, ethanol precipitated together, and resuspended in 20 μl of water before electroporation into MT-2 cells. p83-2 and p83-10 cotransfection was a positive control in each experiment (6). Clones of recombinant virus were also generated by cotransfecting a reconstructed pJM20GPRT plasmid with pJM31ΔGPRT (Fig. 1D and E). Homologous recombination in MT-2 cells between the overlapping ends of reconstructed pJM20GPRT and pJM31ΔGPRT regenerates a complete HIV-1 genome. Five micrograms of each plasmid was used; pJM20GPRT was double digested with SphI and EcoRI, and pJM31ΔGPRT was linearized with BamHI. Pools of recombinant virus were also generated by coelectroporation of pJM31ΔGPRT (5 μg) with PCR products (2 μg). PCR products were purified (QIAquick; Qiagen) before coelectroporation.
MT-2 cells were subcultured on the day before transfection at a density of 0.2 × 106 cells/ml in RPMI 1640 with 10% FCS, 2 mM l-glutamine, 10 mM HEPES buffer, and 50 μg of penicillin and streptomycin per ml. MT-2 cells were pelleted and resuspended in electroporation media (RPMI 1640 supplemented with 10 mM dextrose and 0.1 mM dithiothreitol in the absence of FCS) at room temperature at a concentration of 1 × 107 cells/ml. A 0.5-ml portion (5 × 106 cells), along with the resuspended DNA, was used in each electroporation with the Bio-Rad gene pulser in 0.4-cm-diameter electrode cuvettes. Cells were electroporated at 960 μF at a constant voltage of 250 V. After 15 min, 10 ml of fresh growth medium (R10) and 0.5 × 106 fresh cells were added and incubated at 37°C in 5% CO2, with the p24 antigen tested every 3 to 4 days to monitor viral production.
RESULTS
Generation of molecular clones of infectious HIV-1.
HIV-1 PCR products were cloned into the new cloning vectors constructed here. WT HIV-1 was amplified from HIV-1NL4-3-infected PBMC lysates with insert primers designed for a particular vector (Table 1). A PCR product including gag p7/p1 and gag p1/p6 cleavage sites and all PR coding sequences was cloned into pJM11ΔGPR (Fig. 1A). An amplicon with the 3′ end of gag, PR, and RT was cloned into both pJM13ΔGPRT and pJM20ΔGPRT (Fig. 1C and D). An RT amplicon was cloned into pJM14ΔRT (Fig. 1B). In each case, these plasmids had the sequences of the HIV-1 reading frame(s) deleted from the cloning vector restored with a PCR product. MT-2 cell cultures were transfected with each reconstructed WT HIV-1 plasmid clone and the appropriate complementary plasmid needed to test whether virus could be produced from the recombinant plasmid (Table 1). Each transfected culture became HIV-1 p24 antigen positive after 4 to 10 days. The viruses produced from these different plasmids were each derived from a molecularly cloned PCR product rather than a pool of PCR products. These molecularly cloned viruses can be used for genotyping, drug susceptibility testing, and assessment of viral fitness.
The vector pJM31ΔGPRT (Fig. 1E), used for generating a molecularly cloned virus by cotransfection with a reconstructed pJM20GPRT, was also used to produce a pool of infectious variants directly from PCR products without intervening cloning. Pooled virus was recovered as early as 4 days after MT-2 cell cotransfection of pJM31ΔGPRT with a 2,939-bp PCR product amplified from HIV-1NL4-3-infected cell DNA (primers 1607U25 and 4522L24 [Table 1]); vector and amplicon ends overlap between 380 and 386 bp. Identical results were seen with a 2,549-bp PCR product (primers 1811U24 and 4335L25 [Table 1]) with vector and amplicon ends that overlap between 176 and 199 bp.
Characterization of fitness and drug susceptibility phenotypes of infectious mutant virus clones.
One of the plasmids, pJM11ΔGPR, was used for site-directed mutagenesis to construct specific mutant virus clones for comparison to a WT clone. Protease mutants D30N, L63P, L90M, D30N/L63P, and L90M/L63P and the WT were each constructed by recombinant PCR from NL4-3-infected cell lysates by methods previously described (2, 24, 25, 34) and were cloned into pJM11ΔGPR by ligase-mediated methods. Clonal variants with WT PR and mutations in PR codons 30, 63, and 90 replicated in MT-2 and PBMC cultures, supporting the infectivity of the virions made from a gag PR-restored pJM11ΔGPR. The replicative fitness of the mutant clones in the absence of drug was repeatedly tested in several different assays relative to the WT virus, including experiments measuring kinetics of p24 antigen production in culture and direct competitive mixed cultures (16). Differences in a phenotype of these PR mutants, relative replicative fitness compared to that of the WT, were noted. The D30N mutant was the least replicative (16).
The drug susceptibility phenotypes of the WT and D30N and D30N/L63P cloned mutant viruses were also tested in a PBMC-based assay (14) (Table 2). There were no significant differences in susceptibility to nelfinavir between the molecularly cloned D30N and D30N/L63P mutant viruses. This indicates the utility of these recombinant virus cloning vector systems for generating cloned viruses for drug susceptibility testing. As expected based on previous findings that L63P is a common polymorphism in virus from drug-naïve patients, the data also showed that L63P did not significantly modify D30N-mediated nelfinavir resistance.
TABLE 2.
Drug susceptibility phenotypes of molecularly cloned recombinant viruses
| Virus clone | Nelfinavir IC (fold resistance) ata:
|
|
|---|---|---|
| 50% | 90% | |
| WT | 76.7 | 146.0 |
| D30N | 339.7 (4.4) | 1148.0 (7.9) |
| D30N/L63P | 549.6 (7.2) | 1190.0 (8.1) |
IC, inhibitory concentration expressed in nanomolar concentration. The ICs were determined in PBMC cultures by using supernatant fluid p24 antigen as the end point. Fold resistance indicates the IC of HIV variants relative to the WT (NL4-3).
HIV-1 sequence-specific UDG cloning.
Some of these new vectors were designed to simplify and facilitate molecular cloning of HIV-1 PCR products for clinical diagnostic laboratories by use of the HIV-1 sequence-specific UDG cloning methodology, although clinical specimens were also cloned into pJM11ΔGPR, pJM13ΔGPRT, and pJM14ΔRT by standard ligase-mediated methods. The HIV-1 sequence-specific UDG cloning method does not use enzymatic ligation of restriction-digested DNA, which makes it more rapid and simpler than cloning by standard methods. It differs from a commercially available method for UDG cloning (CloneAmp system; Gibco-BRL) by using UDG digestion to generate single-stranded ends of specific HIV-1 sequences common to both the plasmid vector and the corresponding insert PCR product (Fig. 2). PCR products were cloned by this method from clinical isolate-infected PBMC DNA, patient PBMC proviral DNA, and patient plasma HIV-1 RNA into pJM2ΔPR, pJM5ΔPRT, or pJM20ΔGPRT (and its derivatives pJM21ΔGPRT and pJM22ΔGPRT) with primers specific for each vector (Table 1). To clone PCR products into pJM13ΔGPRT, it is an option to use either standard ligase-mediated methods or the HIV-1 sequence-specific UDG cloning methodology. Although the 5′-half HIV-1 genome vectors (pJM2ΔPR, pJM5ΔPRT, and pJM13ΔGPRT) can be used for HIV-1-specific UDG cloning, the large vector inverse PCR product requires 20 cycles for adequate amplification yield with a proofreading PCR polymerase. Amplifications requiring fewer cycles would further minimize potential for misincorporation during vector PCR (i.e., throughout the HIV-1 sequences in the vector). Therefore, the smaller pJM20ΔGPRT and its derivatives were developed to UDG clone HIV-1 gag PRT PCR fragments. Only 10 cycles of proofreading PCR were adequate for producing enough vector PCR product from these 3.4-kb plasmids to allow cloning.
Insert PCR product amplification yield was adequate for cloning in all HIV-1 sequence-specific UDG cloning attempts. However, a low yield of insert PCR product and storage of the vector PCR product at 4°C for >2 weeks did decrease cloning efficiency. HIV-1 sequence-specific UDG cloning into pJM20ΔGPRT yielded 4 × 104 colonies per μg of vector PCR product DNA (using an approximate 2:1 molar excess of vector to insert PCR products), with about 70% of the transformants containing the correct insert. Correctly reconstructed cloning junctions were sequenced from each of 30 clinical specimens cloned by using HIV-1 sequence-specific UDG cloning. Standard ligase-mediated cloning into pJM11ΔGPR yielded 5 × 105 colonies of pJM11 DNA per μg, with >95% correct recombinants.
Application of clonal analyses to clinical resistance genotyping.
Direct comparisons with the bulk PCR product sequence indicated that clonal analysis with these cloning vectors improved the detection of small minorities of resistant virus strains. Analyses of up to only 15 clones derived from a clinical isolate of virus from patient PBMCs (13) detected minorities of resistant virus not identified in bulk PCR product sequencing of the same amplicon. Minority strains of resistant mutant virus were found in clones, but not in bulk PCR product sequences, from 6 of 16 patient virus isolates (17). In addition, physical linkage of minority resistance mutations was determined from the sequences of molecular clones. Multiple linked PR and RT mutations were identified in 2 of 15 clones of an insert PCR product cloned into pJM5ΔPRT; no mutations were identified in the bulk PCR product sequence of the same amplicon (Table 3). Amplicons were also cloned from plasma HIV-1 RNA from 30 patients who had an early rebound of plasma HIV-1 RNA levels during investigational antiretroviral therapy (4). Nested RNA PCR was performed with primer set 5CAI1964B and 5CAI4155LIG for cloning into the pJM20ΔGPRT series (Table 1). Insert PCR product amplification and HIV-1 sequence-specific UDG cloning were successful in every attempt. Sequencing of about 10 clones of a PCR product derived from each patient’s plasma HIV-1 RNA identified a minority resistant mutant in 18 of the 30 (60%) patients which was not found in the bulk sequencing analysis of the same PCR product. In some cases, cloning identified additional mutations not evident in the bulk PCR product sequence, and in other cases, the only evidence for any resistance mutation was in clonal analyses. This supports both the feasibility of and justification for clonal analysis with this system.
TABLE 3.
Clonal analysis detects an HIV-1 minority subpopulation with multiple physically linked resistance mutations
| HIV-1 wild type or clone | Amino acid of strain at the indicated position of:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PR
|
RT
|
||||||||||||
| 10 | 36 | 46 | 48 | 54 | 63 | 82 | 41 | 67 | 69 | 184 | 210 | 215 | |
| NL4-3 | Leu | Met | Met | Gly | Ile | Leu | Val | Met | Asp | Thr | Met | Leu | Thr |
| 578 bulk | —a | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT1 | — | — | — | — | — | — | — | ||||||
| 578PRT2 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT3 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT8 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT9 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT10 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT11 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT12 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT13 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT14 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 578PRT16 | — | — | — | — | — | — | — | Leu | — | — | — | — | — |
| 578PRT4 | — | — | — | — | NDb | — | — | — | — | — | — | — | — |
| 578PRT6 | — | — | — | — | — | Pro | — | — | — | — | — | — | — |
| 578PRT5 | — | — | — | — | — | Pro | — | — | — | — | — | — | — |
| 578PRT7 | Ile | Ile | Ile | Val | Thr | Gln | Ala | Leu | Asn | Asp | Val | Trp | Tyr |
| 578PRT15 | Ile | Ile | Ile | Val | Thr | Gln | Ala | Leu | Asn | Asp | Val | Trp | Tyr |
—, no change from amino acid of WT (NL4-3).
ND, not determined.
The vectors were also applied to cloning PCR products from patient cell DNA. The vector pJM2ΔPR was used to UDG clone PR amplified from uncultured semen cell DNA of two patients treated with PR inhibitor-containing combinations. Indeed, the PCR product yield from these specimens was suboptimal for bulk PCR product sequencing but adequate for cloning. Clonal sequence analysis revealed at least a minority of PR mutants among 9 or 10 semen clones sequenced from each patient (19). A blood cell specimen from the same time point was also amplified and cloned into pJM2ΔPR for one patient. In this case, clonal analysis helped discern compartmental differences in resistant virus populations based, in part, on detection of minority strains. The L90M resistance mutation in the PR was found in two of nine blood cell DNA-derived clones, but it was not identified among any of the nine semen cell DNA-derived clones. Seven of the nine semen cell DNA-derived clones had a PR V77I resistance mutation which was not found in a single blood cell DNA-derived clone (19).
Pools of infectious recombinant virus were also made from patient specimens of plasma HIV-1 RNA. Three plasma HIV-1 RNAs with viral loads from 4,000 to 26,000 copies/ml (as measured for clinical monitoring in another laboratory by the standard Roche Amplicor assay) were amplified by nested PCR (1607U25 and 4522L24 for the first round and 1811U24 and 4335L25 for the second round [Table 1]). The nested second-round PCR product (2 μg) was cotransfected into MT-2 cells with pJM31ΔGPRT to generate a pool of infectious recombinant virus from each of these plasma HIV-1 RNAs within 4 days of transfection. These supernatant fluids were used to successfully infect PHA-stimulated PBMCs from uninfected donors. The first-round PCR products used to generate pooled infectious recombinant viruses can also be reamplified in a second-round nested reaction with different primers (5CAI1964B and 3CAI4155LIG [Table 1]) to generate molecular clones in pJM13ΔGPRT or pJM20ΔGPRT.
DISCUSSION
This set of versatile recombinant virus vectors for molecular cloning of HIV-1 PCR products has several advantages for the development of improved drug resistance testing of clinical specimens and for basic research into antiretroviral resistance. Either infectious recombinant virus clones or pools can be produced from these vectors. Their application to studies of pathogenesis was demonstrated in laboratory studies of replicative fitness and drug susceptibility of site-directed mutant virus clones and detailed molecular characterization of cloned clinical specimens. The rationale for development of clonal analyses for identification of drug resistance mutations in clinical specimens from patients with virologic failure, as indicated by confirmed rebound of plasma HIV-1 RNA levels, was demonstrated by improved detection of minority strains and genetic linkage of resistance mutations. The feasibility of using sequence-specific UDG cloning with these vectors was also demonstrated.
The major advantages for development of improved clinical resistance testing involve simplified cloning to facilitate better detection of minorities of resistant virus and linkage of multiple resistance mutations. The data presented here with the recombinant virus vectors designed for HIV-1 sequence-specific UDG cloning (pJM2ΔPR, pJM5ΔPRT, pJM13ΔGPRT, and the pJM20ΔGPRT series) indicate that detection of minorities of resistant mutants is improved by sequencing as few as 15 clones of PCR products compared to sequencing bulk PCR products from clinical HIV-1 isolates or plasma HIV-1 RNA. Clonal analysis is the only method by which genetic linkage of different minority resistance mutations can be determined. One example (Table 3) illustrates the clinical relevance of determining linkage: a mutant virus strain with several mutations conferring broad PR inhibitor and RT inhibitor resistance in the same genome would not be expected to be inhibited by a combination of these agents, whereas a viral population with a mixture of different genomes, each with less than a full complement of mutations, might be inhibited to some extent. Even if minority mutant strains were detected as mixed bases in the bulk PCR product sequence, it would be impossible to differentiate several different mutant strains each with a subset of the mutations from one single minority mutant strain containing all the identified mutations. The differentiation of these two possibilities by a clonal analysis might be useful for choosing a salvage regimen for a patient with multiple resistance mutations or, alternatively, for determining that few treatment options remain. (Note that Table 3 depicts an unusual phenomenon. Detailed drug treatment history was not available for the patient from whom this specimen was obtained, but withdrawal of all drugs for several weeks was reported for other instances of such minority, linked mutants we have studied.)
The first RT-deleted recombinant virus vectors (15), as well as more updated versions with a deletion of PRT (11) or of Gag p1/p7 and Gag p1/p6 cleavage sites as well as PR and RT (22), do not allow molecular cloning of PCR products. A pool of infectious recombinant virus is generated by homologous recombination after cotransfection of vector plasmid DNA and patient virus-derived PCR product DNA into T-cell lines (11, 15). Thus, genotyping, phenotyping, or assessments of replicative fitness of amplicons from genetically heterogeneous clinical specimens are limited to characterization of a pool of recombinant virus. An RT-deleted, HIV-1LAI background plasmid cloning vector (29) and a PRT-deleted HIV-1 background plasmid cloning vector (10) have also been used for clinical diagnostic purposes. The former plasmid uses standard cloning methods and does not allow cloning of gag PR. The latter also uses standard cloning methods and has been reported as being used for phenotypic drug susceptibility testing of pools of molecular clones of recombinant virus including PR and RT sequences. Other plasmids have been used for molecularly cloning different HIV-1 PCR products for research purposes (18, 21, 32); none of these allows cloning of all the sequences of relevance for current combinations of PR inhibitors and RT inhibitors (e.g., Gag cleavage sites, PR, and RT). The standard ligase-mediated cloning methods are more complex and time-consuming than the method presented here.
The HIV-1 sequence-specific UDG cloning method can speed cloning. PCR product purification is not needed, and neither amplicons nor the vector requires restriction enzyme digestion, phosphatase treatment, or ligation, each of which are needed for the standard ligase-mediated methods. The HIV-1 sequence-specific UDG cloning into these vectors is also simple enough for those without molecular biology expertise. The use of these HIV-specific vectors eliminates the need for subcloning from a general-purpose UDG cloning plasmid vector (pAMP, CloneAmp system; Gibco-BRL) for functional studies of phenotypes. Although the cloning efficiency observed with these vectors was about 10-fold less than that of standard cloning or UDG cloning into pAMP, it yielded more than enough transformants for these purposes and virtually all transformants contained the desired inserts. Preliminary data also suggest that primers can be optimized to improve cloning efficiency. Every cloning junction sequenced to date has been correct. The pJM20ΔGPRT plasmid is preferred among those described here for UDG cloning, because of its small size. This permits the vector PCR product to be generated with only 10 cycles of high-fidelity conditions and a proofreading polymerase to minimize, and probably effectively preclude, unintended in vitro misincorporation within vector HIV-1 sequences. In other studies, PCR-introduced mutations were seen with a proofreading polymerase only in the later of 35 cycles of amplification (data not shown).
The amplification primers developed here also add versatility to this system, because the same first-round PCR product can be used with one of two nested second-round PCR primer pairs to generate either a pool of recombinant virus or clones of recombinant virus (pJM31ΔGPRT and pJM20ΔGPRT primer pairs [Table 1]). This may facilitate laboratory testing algorithms for speeding diagnostic testing involving clonal analyses. For example, a pool of recombinant virus can be rapidly generated by cotransfection of PCR products with pJM31ΔGPRT. If a screening phenotype of the pool (or bulk genotype of the PCR products) suggests minimal or no resistance or if other indications suggest that minority mutants be sought, then another aliquot of the same first-round PCR products can be amplified with the alternate second-round primers to permit molecular cloning into pJM20ΔGPRT. This would allow specimens without easily detectable dominant resistance to be triaged for a clonal analysis.
Some aspects of the genetic background of these deletion plasmids are advantages for research purposes. Unlike the HxB2 genome (26), on which most other recombinant virus vector systems are based, HIV-1NL4-3 encodes functional forms of some accessory genes of HIV-1 (e.g., vif, vpr, and nef). This will facilitate laboratory research of gag-pol variants selected in vivo in the background of these gene products. The availability of the NL4-3-based and the hybrid HxB2–NL4-3-based vectors constructed here also may help to further define poorly understood differences in viral fitness and drug susceptibility of the same PR mutants in NL4-3 versus HxB2 genetic backgrounds (27). Another advantage is the flexibility for studying clinical specimen-derived clones of the reading frames selected by current PR and RT inhibitors either separately or together as a unit in the same genetic background. This is relevant for further research into interactions between PR inhibitor and RT inhibitor-selected mutations (33). None of the previously described vectors allow cloning of PR and RT either separately or together as a unit into the same genetic background (10, 11, 15, 18, 21, 29, 32). The vectors described here allow cloning of either PR, RT, or PRT as a unit from a patient specimen into an isogenic HIV-1 background for comparative study. They also allow study of gag PR versus PR, gag PR versus gag PRT, or gag PRT versus PRT. The Gag p7/p1 and Gag p1/p6 cleavage sites are selected during PR inhibitor therapy and may compensate for deleterious effects on fitness of PR active-site mutations (5, 35).
Molecular cloning into the plasmids in which the deletion includes sequences encoding Gag p7/p1 and Gag p1/p6 cleavage sites (pJM11ΔGPR, pJM13ΔGPRT, and the pJM20ΔGPRT series) also ensures that the sequences of these cleavage sites derived from the clinical specimen are retained in the molecularly cloned recombinant virus, in contrast to some other systems where these sequences may be lost if they are outcompeted by other virus variants in the recombinant virus pool (11, 15). The reconstructed 5′-half genome plasmids (pJM11GPR, pJM13GPRT, and pJM14RT) also have potential for cotransfection with any 3′-half genome plasmid, allowing for the study of a patient-derived gag-pol sequence in a virus with HIV-1 envelope tropism other than the CXCR-4 tropism of HxB2 and NL4-3. Also, cloning into subgenomic constructs (pJM11ΔGPR, pJM13ΔGPRT, pJM20ΔGPRT, pJM2ΔPR, or pJM5ΔPRT) avoids potential deletions due to recombination between HIV-1 LTRs during plasmid growth in E. coli (23), to which complete genome vectors are liable (10, 18, 21, 29, 32). Mutant E. coli strains and reduced temperatures of growth can minimize this potential, as was done here for the 2-LTR plasmid pJM31ΔGPRT (which is not used for molecular cloning of patient-derived specimens); this slows plasmid DNA preparation needed for clonal analyses, however.
The molecular tools described here can help to reconstruct recombinant virus containing Gag p7/p1 and Gag p1/p6, PR, and/or RT sequences amplified from clinical or laboratory specimens. Thus, they simplify and speed assays that determine gag PRT genotype as well as phenotypic drug sensitivity and replicative capacity from a given gag PRT genotype for laboratory research and may help in the development of improved resistance testing for clinical management.
ACKNOWLEDGMENTS
N. Kartsonis helped with the construction and use of pPRdel. Helpful discussions with J. Kaplan are greatly appreciated. The following were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3 from Malcolm Martin, p83-2 and p83-10 from R. Desrosiers, D. Regier, and J. Gibbs, and MT-2 cells from D. Richman.
This work was supported by grants from NIH (AI-29193 and the Virology Advanced Technology Laboratory subcontract for AI 27659). J.M.-P. was supported by a postdoctoral fellowship from the Spanish Ministry of Education.
REFERENCES
- 1.Adachi A, Gendelman H E, Koening S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caliendo A M, Savara A, An D, DeVore K, Kaplan J C, D’Aquila R T. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J Virol. 1996;70:2146–2153. doi: 10.1128/jvi.70.4.2146-2153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelsi L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Pasquale M P, Murphy R, Kuritzkes D, Martinez-Picado J, Sommadossi J P, Gulik R, Smeaton L, DeGruttola V, Caliendo A, Sutton L, Savara A V, D’Aquila R T. Resistance during early virological rebound on amprenavir plus zidovudine plus lamivudine triple therapy or amprenavir monotherapy in ACTG protocol 347. Antivir Ther. 1998;3(Suppl. 1):50–51. . (Abstract 71.) [Google Scholar]
- 5.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. Second locus involved in human immunodeficiency virus type resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Hum Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 7.Haertle T, Carrera C J, Wasson D B, Sowers L C, Richman D D, Carson D A. Metabolism and anti-human immunodeficiency virus-1 activity of 2-halo-2′,3′-dideoxyadenosine derivatives. J Biol Chem. 1988;263:5870–5875. [PubMed] [Google Scholar]
- 8.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 9.Havlir D V, Petropoulos C J, Hellmann N S, Whitcomb J M, Richman D D the ACTG 343. Evolution of drug resistance associated with loss of viral suppression in patients treated with indinavir, lamivudine and zidovudine. Antivir Ther. 1998;3(Suppl. 1):52–53. . (Abstract 74.) [Google Scholar]
- 10.Hecht F M, Grant R M, Petropoulos C J, Dillon B, Chesney M A, Tian H, Hellmann N S, Brandrapalli N I, Digilio L, Branson B, Kahn J O. Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N Engl J Med. 1998;339:307–311. doi: 10.1056/NEJM199807303390504. [DOI] [PubMed] [Google Scholar]
- 11.Hertogs K, de Bethune M P, Miller V, Ivens T, Schel P, Van Cauwenberge A, Van Den Eynde C, Van Gerwen V, Azijn H, Van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirsch M S, Conway B, D’Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. International AIDS Society—USA panel. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 12a.HIV sequence database. 21 May 1999, revision date. [Online.] Theoretical Biology and Biophysics Division, Los Alamos National Laboratory, Los Alamos, N.Mex. http://HIV-web.lanl.gov. [15 July 1999, last date accessed.]
- 13.Jackson J B, Coombs R W, Sannerud K, Rhame F S, Balfour H H. Rapid and sensitive viral culture method for human immunodeficiency virus type 1. J Clin Microbiol. 1988;26:1416–1418. doi: 10.1128/jcm.26.7.1416-1418.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J M, Lane J, Black R J, Reichelderfer P S, D’Aquila R T, Crumpaker C S The RV-43 Study Group; The AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellam P, Larder B A. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1994;38:23–30. doi: 10.1128/aac.38.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Picado J, Savara A, Sutton L, D’Aquila R T. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Picado J, Sutton L, Helfant A H, Savara A, D’Aquila R T. Novel clonal analyses of resistance to HIV-1 RT and protease inhibitors. Antivir Ther. 1997;2(Suppl. 5):47. . (Abstract 45.) [Google Scholar]
- 18.Maschera B, Furfine E, Blair E D. Analysis of resistance to human immunodeficiency virus type 1 protease inhibitors by using matched bacterial expression and proviral infection vectors. J Virol. 1995;69:5431–5436. doi: 10.1128/jvi.69.9.5431-5436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer K H, Boswell S, Goldstein R, Lo W, Xu C, Tucker L, De Pasquale M P, D’Aquila R T, Anderson D J. HIV persistence in semen after adding indinavir to combination antiretroviral therapy. Clin Infect Dis. 1999;28:1252–1259. doi: 10.1086/514775. [DOI] [PubMed] [Google Scholar]
- 20.Miller V, Sturmer M, Staszewski S, Groschel B, Hertogs K, de Bethune M P, Pauwels R, Harrigan P R, Bloor S, Kemp S D, Larder B A. The M184V mutation in HIV-1 reverse transcriptase (RT) conferring lamivudine resistance does not result in broad cross-resistance to nucleoside analogue RT inhibitors. AIDS. 1998;12:705–712. doi: 10.1097/00002030-199807000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Patick A K, Rose R, Greytok J, Bechtold C M, Hermsmeier M A, Chen P T, Barrish J C, Zahler R, Colonno R J, Lin P F. Characterization of a human immunodeficiency virus type 1 variant with reduced sensitivity to an aminodiol protease inhibitor. J Virol. 1995;69:2148–2152. doi: 10.1128/jvi.69.4.2148-2152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauwels R, Hertogs K, Kemp S, Bloor S, Van Acker K, Hansen J, De Beukeleer W, Roelant C, Larder B, Stoffels P. Comprehensive HIV drug resistance monitoring using rapid, high-throughput phenotypic and genotypic assays with correlative data analysis. Antivir Ther. 1998;3(Suppl. 1):35–36. . (Abstract 51.) [Google Scholar]
- 23.Peden K W. Instability of HIV sequences in high copy number plasmids. J Acquir Immune Defic Syndr. 1992;5:313–315. [PubMed] [Google Scholar]
- 24.Rashtchian A, Buchman D M, Schuster D M, Beringer M S. Uracil DNA glycosylase-mediated cloning of PCR-amplified DNA: application to genomic and cDNA cloning. Anal Biochem. 1992;206:91–97. doi: 10.1016/s0003-2697(05)80015-6. [DOI] [PubMed] [Google Scholar]
- 25.Rashtchian A, Thorton C G, Heidecker G. A novel method for site-directed mutagenesis using PCR and uracil DNA glycosylase. PCR Methods Appl. 1992;2:124. doi: 10.1101/gr.2.2.124. [DOI] [PubMed] [Google Scholar]
- 26.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Jr, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 27.Rose R E, Gong Y-F, Greytok J A, Bechtold C M, Terry B J, Robinson B S, Alam M, Colonno R J, Lin P F. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc Natl Acad Sci USA. 1996;93:1648–1653. doi: 10.1073/pnas.93.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shafer R W, Kozal M J, Winters M A, Iversen A K, Katzenstein D A, Ragni M V, Meyer W A, III, Gupta P, Rasheed S, Coombs R, Katzman M, Fiscus S, Merigan T C. Combination therapy with zidovudine and didanosine selects for drug-resistant human immunodeficiency virus type 1 strains with unique patterns of pol gene mutations. J Infect Dis. 1994;169:722–729. doi: 10.1093/infdis/169.4.722. [DOI] [PubMed] [Google Scholar]
- 29.Shi C, Mellors J W. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1997;41:2781–2785. doi: 10.1128/aac.41.12.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirasaka T, Kavlick M F, Ueno T, Gao W Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirasaka T, Yarchoan R, O’Brien M C, Husson R N, Anderson B D, Kojima E, Shimada T, Broder S, Mitsuya H. Changes in drug sensitivity of human immunodeficiency virus type 1 during therapy with azidothymidine, dideoxycytidine, and dideoxyinosine: an in vitro comparative study. Proc Natl Acad Sci USA. 1993;90:562–566. doi: 10.1073/pnas.90.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winslow D L, Anton E D, Horlick R A, Zagursky R J, Tritch R J, Scarnati H, Ackerman K, Bacheler L T. Construction of infectious molecular clones of HIV-1 containing defined mutations in the protease gene. Biochem Biophys Res Commun. 1994;205:1651–1657. doi: 10.1006/bbrc.1994.2857. [DOI] [PubMed] [Google Scholar]
- 33.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D, Caliendo A M, Eron J J, DeVore K M, Kaplan J C, Hirsch M S, D’Aquila R T. Resistance to 2′,3′-dideoxycytidine conferred by a mutation in codon 65 of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1994;38:282–287. doi: 10.1128/aac.38.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]