Abstract
The potential of artificial intelligence (AI) applied to clinical data from electronic health records (EHRs) to improve early detection for pancreatic and other cancers remains underexplored. The Kenner Family Research Fund, in collaboration with the Cancer Biomarker Research Group at the National Cancer Institute, organized the workshop entitled: “Early Detection of Pancreatic Cancer: Opportunities and Challenges in Utilizing Electronic Health Records (EHR)” in March 2021. The workshop included a select group of panelists with expertise in pancreatic cancer, EHR data mining, and AI-based modeling. This review article reflects the findings from the workshop and assesses the feasibility of AI-based data extraction and modeling applied to EHRs. It highlights the increasing role of data sharing networks and common data models in improving the secondary use of EHR data. Current efforts using EHR data for AI-based modeling to enhance early detection of pancreatic cancer show promise. Specific challenges (biology, limited data, standards, compatibility, legal, quality, AI chasm, incentives) are identified, with mitigation strategies summarized and next steps identified.
Key Words: artificial intelligence, electronic health records, machine learning, natural language processing, pancreatic cancer, early detection
Progress remains painstakingly slow in early detection of pancreatic cancer, and rigorous prevention and identification strategies are lacking. The most prevalent form of pancreatic cancer is pancreatic ductal adenocarcinoma (PDAC), which involves the malignant transformation of the exocrine duct cells. The cancer is more common with age and slightly more common in men than women. Both racial and socioeconomic disparities have been identified in the diagnosis, treatment, and prognosis of this cancer.1 Genetic/familial factors contribute to 10% to 20% of those diagnosed. Although only 3.2% of all new cancers in the United States are PDAC, the disease contributes to a substantial portion of cancer deaths. Recent statistics indicate that PDAC has the third highest number of cancer deaths after lung/bronchial and colorectal cancers. Importantly, data suggest that an estimated 60,430 new cases will be diagnosed in 2021, and more than 48,000 individuals will die of the disease within the year.1
The onset of PDAC is often asymptomatic, and many late-stage signs are nonspecific. Reported symptoms include stomach and/or backache, changes in bowels, jaundice, new onset of diabetes, and emotional variability.2 Such symptoms could be due to multiple etiologies, where a busy general practitioner may not have the time or experience to connect the dots leading to further evaluation and a timely pancreatic cancer diagnosis. Although the progression of the disease is thought to silently take place over multiple years, most patients are diagnosed at a late stage.3 As a result, early-stage PDAC is frequently under-identified. This is a missed opportunity for timely, effective treatment. When a malignant lesion is detected early and at a resectable stage, potentially curative treatment options are available to the patient. In other words, early detection saves lives, but current risk assessment and screening tools are failing to capture this opportunity.2
Early detection is critical to maximizing the number of people who survive PDAC.4,5 A singular opportunity arises from identifying and monitoring high-risk individuals. Inherent in this is a stage shift, where more individuals are diagnosed at a local stage versus a distant stage. On the other hand, intensive surveillance may raise concerns about overtesting, particularly for older patients who tend to have additional comorbidities, making testing itself a potential risk. These concerns highlight the need for better discernment of who should or should not be monitored for the first signs of PDAC. Such triaging could contribute to meaningful and cost-effective screening or early detection programs.6–8
To enhance early detection of pancreatic cancer, researchers are increasingly turning to advanced computational approaches for risk assessment and stratification. Recently, artificial intelligence (AI) has emerged as a state-of-the-art tool for early detection of cancer and other diseases.9–12 Artificial intelligence algorithms have been applied to detect imaging abnormalities, extract relevant information from EHR files, identify patients who need intensive care unit care, and prevent medication errors. In cancer research, AI-based algorithms have been instrumental in facilitating biomarker discovery, improving diagnostic imaging workflows, and accelerating drug development.13
As noted by Thomas J. Fuchs, DSC, Dean of Artificial Intelligence and Human Health at Icahn School of Medicine at Mount Sinai, New York, NY, “Without a doubt, the impact of artificial intelligence will eclipse that of the Industrial Revolution, personal computing, and the internet combined. In medicine, it's just getting started.”14 Could this technology be applied to risk assessment and stratification for early detection of pancreatic cancer? Could AI mitigate the continuous diagnostic challenges and speed the tempo of early detection?
ELECTRONIC HEALTH RECORDS
Although AI and its subfield, machine learning (ML), are increasingly applied to imaging data to improve early detection for pancreatic and other cancers, clinical data from electronic health records (EHRs) remain underexplored. Only a handful of research studies have used ML to build predictive models with EHR data in this field.11,15,16 These studies have demonstrated that by leveraging AI/ML and EHRs, subpopulations at high risk for PDAC can be identified 1 to 2 years before diagnosis. Such efforts also highlight specific challenges and opportunities for improving the secondary use of EHR data with AI and innovative data science solutions. Combined with natural language processing (NLP), another subfield of AI, ML algorithms can greatly facilitate processing and analyzing information from EHRs. The growth of health data sharing networks and AI techniques has provided new opportunities to improve early detection of cancers.15 This availability of longitudinal data for a large swath of patient populations is particularly promising for cancers with relatively low incidence rates such as PDAC.
Discussion at the recent AI and Early Detection of Pancreatic Cancer 2020 Summit organized by Kenner Family Research Fund and the American Pancreatic Association indicated that a singular opportunity might exist in data found in EHRs to improve risk stratification for this cancer.13 As a follow-up to the Summit, Kenner Family Research Fund, in collaboration with the Cancer Biomarker Research Group at the National Cancer Institute (NCI), Bethesda, Md, presented the “Early Detection of Pancreatic Cancer: Opportunities and Challenges in Utilizing Electronic Health Records (EHR)” in March 2021 (workshop: https://www.kennerfamilyresearchfund.org/early-detection-of-pancreatic-cancer-electronic-health-records/ and workshop videos: https://www.kennerfamilyresearchfund.org/videos-opportunities-and-challenges-in-utilizing-electronic-health-records/). The workshop included a select group of panelists with expertise in PDAC, EHR data mining, and AI-based modeling. Experts shared their experiences using different common data models (CDMs) and data sharing networks for AI-based data extraction and predictive risk modeling. Also discussed were strategies used by these networks to mitigate challenges inherent to the use of EHR data, including protection of patient privacy and confidentiality, data transfer agreements, consideration of racial/ethnicity diversity, ongoing updates in institutional EHR systems, and funding. A partial list of networks is presented in Table 1.
TABLE 1.
Health Data Networks and CDMs
| Network/Funders | CDM | Reference and/or URL |
|---|---|---|
| TriNetX | TriNetX |
17 https://trinetx.com |
| OHDSI/Reagan-Udal Foundation, FDA, PhARMA, FNIH | OMOP | https://www.ohdsi.org |
| Cleveland Clinic | UMLS-based | 18 |
| i2b2/NIH NCBC | i2b2/ACT |
19,20 https://www.i2b2.org |
| MSK Cancer Center | MSK-specific | 21–23 |
| PCORI | PCORNet | https://pcornet.org |
| REP | REP-specific | 24 |
| Cancer Research Network Consortium/NCI | VDW |
25 http://www.hcsrn.org/crn/en/ |
| VSD/CDC | CDC-specific | https://www.cdc.gov/vaccinesafety/ensuringsafety/monitoring/vsd/index.html |
| Sentinel Data Partners/ FDA | Sentinel | https://www.sentinelinitiative.org |
ACT indicates the Accrual to Clinical Trials project; CDC, the Centers for Disease Control and Prevention; FNIH, the Foundation for the National Institutes of Health; NCBC, the National Centers for Biomedical Computing; NIH, the National Institutes of Health; OHDSI, the Occupational Health Data Sciences and Informatics group; PhARMA, the Pharmaceutical Research and Manufacturers of America; PCORNet, the National Patient-Centered Clinical Research Network; URL, Uniform Resource Locator; VDW, the Virtual Data Warehouse; VSD, the Vaccine Safety Datalink project.
There is a growing interest in applying AI-based risk stratification models to pancreatic cancer. Limor Appelbaum from Beth Israel Deaconess Medical Center, Boston, Mass, and colleagues from Martin Rinard's laboratory at Massachusetts Institute of Technology's Computer Science and Artificial Intelligence Laboratory, Cambridge, Mass, developed and validated a prediction model that can identify individuals at high risk for PDAC up to 1 year before diagnosis through diagnostic codes extracted from EHRs.15 More recently, they were able to improve the model's performance using an independent, multicenter data set, and additional laboratory test features.26 Instead of using predefined feature sets for model development, they used multiple indicators to mitigate the potential risk of overfitting. The risk stratification models based on concatenated laboratory test and diagnostic feature sets outperformed diagnostic-based and laboratory test–based models for early prediction of PDAC development.
Appelbaum and her colleagues15,17 are now deploying PDAC risk models in a federated manner using TriNetX, Cambridge, Mass, a global federated network. According to Matvey Palchuk, Vice President of Informatics at TriNetX, this growing network enables access to patient clinical data (eg, demographics, encounters, diagnoses, procedures, medications, laboratory test results, vital signs, etc) from more than a hundred health care organizations across the globe, of which 70 are in the United States. Palchuk believes in using “just the right amount” of federation and aggregation to ensure privacy protection while enabling creative problem solving. Merging data sets across institutions and countries not only increases power but also reduces the possibility of bias. In federated learning, the models are trained separately on the different data sets and then merged. The data can be stored locally with a federated layer applied on top to ensure site-agnostic training, validation, and deployment.
Studies are made possible owing to increased availability of EHR-derived clinical data on millions of patients accumulated by health data sharing networks and development of CDMs focused on using clinical patient data for research. Examples of such CDMs are the Informatics for Integrating Biology and the Bedside (i2b2),27 The Observational Medical Outcomes Partnership (OMOP),28 National Patient-Centered Clinical Research Network (PCORnet),29 All of Us Research,30 and others. TriNetX is also such a CDM, albeit based on public-private partnerships instead of being grant-funded.
Clinical data from EHRs are part of real-world data, defined as any data collected at the point of care and outside of the context of a clinical trial, and can also include sources such as disease registries, claims databases, and wearables. Recently, secondary use of EHR data has emerged as a critical component of clinical research studies, as research involving EHR is frequently exempt from institutional review board (IRB) review. This has resulted in a patchwork of health data sharing networks, most of which use different CDMs.31 Each network was designed to enable data sharing among participating organizations but not between networks. Sometimes mappings are available between CDMs (eg, between i2b2 and OMOP), a characteristic that may simplify the harmonization process for CDMs developed and used by various networks. Latest work by the National Coronavirus Disease of 2019 (COVID-19) Cohort Collaborative32,33 led to development and deployment of mappings from i2b2/ACT, PCORnet, and TriNetX to OMOP as part of their aggregated data enclave to study patients with COVID-19.
In addition, multiple efforts have been underway for over a decade to bridge the gap between health and clinical research data standards, but the process is still in its early stages.31 The US Food and Drug Administration (FDA) led a project, funded by the PCOR Trust Fund, aiming to harmonize CDMs with the Biomedical Research Integrated Domain Group (BRIDG) domain information model.34 The BRIDG model can guide transfer of EHR data generated by clinical trials into electronic data capture systems and links biomedicine and health care concepts in the areas of clinical imaging and pathology. The FDA also supports adoption of the Fast Healthcare Interoperability Resources, as the data standard for exchanging EHRs.35,36
Michael Rosenthal from Dana-Farber Cancer Center, Boston, Mass, introduced a conceptual framework based on risk factors as indirect and direct signs of pancreatic disease, on a continuum over time (Fig. 1). These factors could be amenable to advanced computational techniques such as AI/ML and integrated into a cumulative assessment of risk or a screening program. However, successful application of AI/ML techniques to EHRs requires CDMs to enable effective data extraction and analysis across populations and sites. Currently, EHR-based studies face many challenges starting with ambiguous definitions, custom Current Procedural Terminology codes, accurately excluding negative diagnoses, and navigating third-party systems that load legacy data into EHR without updating. In other work, many health systems have gone through different versions of EPIC, contributing to a Tower of Babel situation, according to Rosenthal.
FIGURE 1.
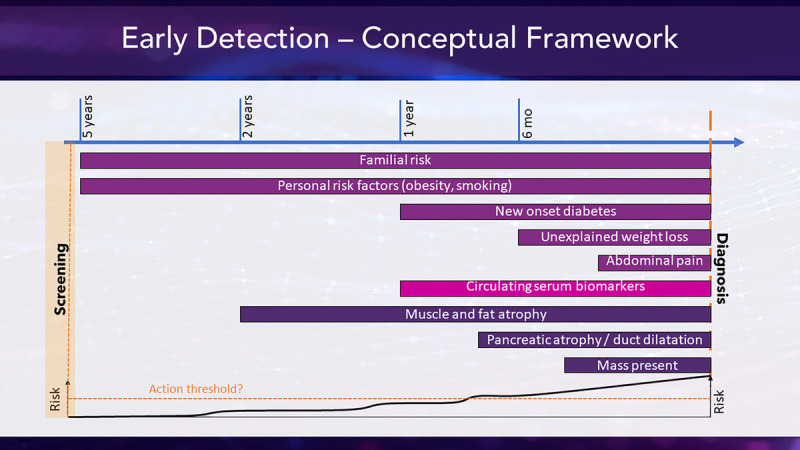
Early detection conceptual framework. Courtesy of Michael Rosenthal.
Rosenthal shared his experience using the OMOP CDMs for multicenter research.37 This model has been adopted by the Occupational Health Data Sciences and Informatics group, which developed a suite of open-source tools to support CDM implementation and use. Current data models can improve accessibility of data, uniformity of data fields, and data encoding consistency. The main advantage of using OMOP is the ability to include multiple data sources from collaborators, who can adapt the same CDM. This approach is also compatible with federated learning, in which all Patient Health Information stays local, and just the models move around, addressing privacy and confidentiality concerns. In addition, new sites can be added with very minimal central work. A disadvantage of the OMOP model, according to Rosenthal, includes a lack of integrated high-level conceptual mappings as in the Unified Medical Language System (UMLS).
Alex Milinovich from the Cleveland Clinic Health System, Cleveland, Ohio, discussed his approach to EHR data extraction based on the UMLS unique concept identifiers.38 With 9 million patients, Cleveland Clinic has one of the largest EHR systems in the world. It hosts 17,000 tables in the database and 5 billion laboratory results. The system supports 260,000 users, with 68,000 users entering data currently. To enable secondary use of these data, Milinovich and his team applied a 4-step mapping process to normalize health data from multiple sources into a UMLS-based CDM. The UMLS system combines more than 100 different medical vocabularies and thousands of term concepts.38 The process uses classic data mining and NLP techniques to extract data from the EHR data and map it to UMLS concept identifiers. As a result, all data collected over the years is stored in a research-ready repository. Given the barriers frequently associated with data ascertainment, having repositories like this is a significant step toward meeting research goals.
Aside from data extraction and normalization, secondary use of EHRs requires de-identification and review by the IRB. Like many other health systems, the Cleveland Clinic strongly prefers keeping the patient data private while providing researchers with summary statistics for building prediction models. Milinovich's team pulls the data and completes the analysis, which obviates the need for de-identification. However, many projects involve clinicians' notes, and de-identifying notes is complicated and time-consuming. In addition, it is challenging to obtain an IRB approval for such collaborative multisite studies, given the waiver of consent that research like this often requires.39
Another challenge involves validation of the diagnoses that are annotated in the record in EHR systems. For example, electronically identifying patients with new-onset diabetes is not always straightforward given the variable diagnostic approaches and definition of diabetes used by clinicians. Some institutions may have 50 codes for measuring blood glucose levels. Some patients are fasting, and some are not. Some are in the emergency department, some are inpatient, some are at home using a Blood Sugar Fingerprint Scanner. Shawn Murphy from Harvard Medical School, Boston, Mass, illustrated how his team tackled this problem. They developed the i2b2 system, which allows investigators at Mass General Brigham, Boston, Mass, to manage project-related clinical research data.19,27 To improve accuracy and enable validation, Murphy and his team applied NLP to match data from multiple sources, including doctors' notes and billing data.
The i2b2 system enables fast access to aggregate numbers for diverse patient cohorts across hospital systems without needing an IRB approval. However, detailed data only stay where the patients receive services, and therefore, IRB can approve these requests. The Mass General Brigham system contains records for more than 6.7 million patients, including 3.5 billion searchable facts: diagnoses, medications, genomics, procedures, and so forth. Access to the system is authorized by a person's faculty status, and collaborators are authorized by faculty to use this tool at Mass General Brigham. The i2b2 software is broadly distributed along with demonstration data and allows many organizations to extend its modular framework.20 There are multiple i2b2 implementations across the United States and internationally as well.
In cancer research, there is an additional challenge in knowledge extraction from EHRs involving the “oncology phenotype gap.” Electronic health record data alone cannot provide sufficient information to explain the variation and treatment outcomes or guide precision prevention strategies for cancer patients. Peter Stetson from Memorial Sloan Kettering (MSK), New York, NY, talked about the MSK Cancer Center approach to closing this gap via augmentation of EHRs with multimodal phenotyping of the whole human. The MSK platform is based on information exchange “among patients foremost and then on strategic relationships with specific partners.”21
Memorial Sloan Kettering collects patient-reported data for clinical and research purposes, including MSK Engage, an electronic questionnaire tool enabling the collection and visualization of patient-generated health data. Another platform, Precision Insight Support Engine (PRECISE), was designed to match patients to investigational therapies based on molecular and clinical criteria.22 The MSK toolset also includes the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) system, which stores information on genetic alterations implicated in cancer predisposition syndromes.23 These platforms are used to extract a key set of core cancer data elements that come from OMOP, Nasser (a tumor registry data standard), and the Minimal Common Oncology Data Elements often referred to as mCODE. Some challenges remain, such as extensions of OMOP for oncology or further automation of data extraction with NLP.
Jason Block from the Harvard Medical School shared his experience of leveraging PCORnet for epidemiologic studies focused on obesity and chronic disease. This extensive, distributed network was funded by the Patient-Centered Outcomes Institute (PCORI) to serve as the infrastructure to support comparative effectiveness research.40 PCORnet uses a CDM, which has similarities to OMOP and the FDA Sentinel CDM used for health plan claims data. The FDA has been working with PCORI to advance their postmarketing surveillance research enterprise called the Sentinel program. PCORnet includes diagnoses, laboratory results, demographics, procedures, and medications for a relatively diverse population with robust longitudinal data. According to Block, PCORnet shares many of the challenges common to other networks and aims to address them with advanced computing techniques. For example, NLP is increasingly being used to bring in unstructured data.
Many risk modeling projects involve linking back to the full text of the medical records. For that reason, full de-identification of the data for research purposes is not always feasible. Jennifer St. Sauver at Mayo Clinic, Rochester, Minn, talked about her approach to this challenge. She is co-principal investigator of the National Institutes of Health–funded Rochester Epidemiology Project (REP), which aggregates information from medical care available to residents of southeast Minnesota and southwest Wisconsin.41 The REP initiative involves a dynamic platform for EHR and AI research, which includes a comprehensive medical record linkage system, going back for nearly half a century. The project intends to help researchers optimally define and identify specific study populations and follow them over time. Various NLP algorithms have been developed to facilitate information extraction for poorly coded conditions in the medical record. For example, one project aims to develop an algorithm to identify persons with delirium.42 The data are structured in models like those of other large groups such PCORnet and the Cleveland Clinic, so there is some degree of interoperability.
DISCUSSION
The final portion of the workshop was dedicated to various discussions, and a potential roadmap was summarized by Suresh Chari, The University of Texas MD Anderson Cancer Center, Houston, Tex, and Sudhir Srivastava, Cancer Biomarkers Research Group, Division of Cancer Prevention, NCI, Bethesda, Md. Dr Srivastava remarked, “Electronic health records are a goldmine with many clinical as well as biological parameters that can help” predict progress in pancreatic cancer or any other cancer type in the future, improve data accuracy, and promote clinical trial efficiency.
Current efforts of AI in the pancreatic cancer space are primarily used in imaging with some limited application in tissue and liquid biomarker assays. The use of AI in the analysis of patient characteristics (ie, EMR analysis, social media, pharmacy, and insurance claim records, lifestyle) is in the development stage.13 Most current studies use classical techniques of AI rather than more advanced tools that may eventually yield better performance and improved insights. Although current AI technologies may not be able to demonstrate a causal relationship, they may effectively flag predictors of high risk, prompting further investigation and earlier detection.
The AI bias challenge is particularly relevant to pancreatic cancer owing to its heterogeneity and relatively low prevalence in the general population. Federated learning and CDMs are expected to help overcome the data scarcity and mitigate this bias, which in turn may motivate AI developers to focus on early detection of cancer instead of the next Twitter optimization algorithm.
To expand efforts in this area, close collaboration across disciplines and between AI experts and pancreatic cancer researchers is critical. There is a need for a mechanism that links people in clinical pancreatic research with the AI community to bring modern techniques into the analytic processes of very established groups and to initiate interactions that smooth the way for efficient and effective collaborations across institutions. In addition, a central source for information on data sources and AI efforts in PDAC is essential.
FUTURE DIRECTIONS
Research efforts, including studies presented at the 2021 workshop, demonstrate that longitudinal patient records offer a singular opportunity for improving risk assessment and stratification for pancreatic cancer in presymptomatic people. Some longitudinal information can be obtained from observational cohort studies such as the New Onset Diabetes study sponsored by the NCI and the National Institute of Diabetes and Digestive and Kidney Diseases. However, EHRs and other records obtained in routine health care represent a significant resource, which can transform predictive risk modeling for pancreatic cancer.43,44
Advancements in AI and data science continue to accelerate both data extraction and risk modeling based on longitudinal patient data. Because of heterogeneity and size, EHR data are particularly amenable to cutting-edge AI techniques such as ML/DL and NLP. However, there are several key challenges to using these techniques on EHR data to improve risk assessment or stratification of pancreatic cancer. These challenges include sample size requirements, need for CDMs, and an acceptable level of data quality, among others. Furthermore, clinical relevance and generalizability of AI models, multisite collaborations or partnerships, and incentives for prevention research are critical to developing trustworthy and unbiased AI systems (Table 2).
TABLE 2.
Challenges, Potential Mitigation Strategies, and Next Steps
| Challenge | Mitigation Strategy | Next Steps |
|---|---|---|
| Complex PDAC biology | Tracking multiple potential predictors over time Observational, natural history studies |
• Develop better understanding of interrelationship between PDAC and other diseases of the pancreas • Apply AI-based modeling to improve early detection of PDAC and diseases with similar etiology or symptoms |
| Limited availability of longitudinal data for AI-based risk modeling | Secondary use of EHR and other relevant patient data collected and harmonized by clinical data sharing networks | • Facilitate retrospective cohort discovery by leveraging centralized and federated EHR data repositories compiled by data sharing networks • Incentivize multinetwork partnerships to extract data from EHRs relevant to PDAC risk modeling • Extend AI-based risk modeling to diseases with similar signs and symptoms |
| Gap between health data standards and clinical research data standards | Fast Healthcare Interoperability Resource and related efforts | • Incentivize common adoption of standards by clinical and research communities • Incentivize interoperability of EHR and electronic data capture systems • Exploring EHRs as an alternative to eCRFs • Encourage the use of IT products certified by the ONC Health IT Certification program |
| Incompatible CDMs used by health care systems | Data sharing networks, which normalize EHRs using network specific CDMs | • Extend CDM mappings relevant to PDAC • Incentivize data sharing networks to develop more interoperable CDMs • Facilitate multinetwork partnerships focused on specific use cases such as AI-based risk modeling for PDAC |
| Legal, intellectual property, privacy related, data sharing barriers across organizations and networks | Develop privacy-protecting data sharing approaches like federated learning and other private-sector innovations | • Validate and leverage federated learning and other private-sector innovations in data sharing for risk modeling of PDAC • Expand privacy-preserving methods for linking multi-modal patient records • Incentivize data sharing networks to extract data from EHRs relevant to PDAC and other diseases of the pancreas • Encourage health systems to engage in patient care optimization and building a learning health system focused on disease prevention and population health management |
| Insufficient data quality and reliability | Approaches for validation of data reliability and quality developed by data sharing networks | • Develop shared criteria for evaluating data reliability and quality • Develop shared criteria for validation of the approaches • Encourage adoption of best practices for data extraction by clinical centers participating in PDAC research • Developing methods for normalizing unstructured data, such as clinician's notes • Incentivizing use of standardize software for diagnostic codes • Expanding standardized vocabularies for oncology and PDAC |
| AI chasm: poor clinical utility and generalizability of AI systems | Transparency of validation, benchmarking competitions and possibly other certification process (ie, FDA, NCCN, USPSTF) | • Incentivize interdisciplinary partnerships and AI validation and benchmarking efforts • Establish predefined validity specific to use cases and the endpoints of interest • Incentivize external validation of AI systems on external multi-institutional data sets • Expand the development of multimodal AI-based risk models by combining EHRs with imaging, omics, and other data • Private-public partnerships for software benchmarking competitions |
| Lack of incentives for efforts focused on precision prevention of PDAC and public health management | Multidisease partnerships and collaborations to build a virtual network of researchers and clinicians | • Leverage existing networks to promote academic-industry partnerships • Facilitate multi-network partnerships focused on specific AI use cases relevant to PDAC and related conditions or diseases |
eCRFs indicates electronic case report forms; IT, information technology; NCCN, the National Comprehensive Cancer Network; ONC, the Office of the National Coordinator for Health Information Technology; USPSTF, the US Preventive Services Task Force.
Research has implemented multiple creative solutions to the challenges in applying AI to EHRs. To take these promising developments further, appropriate measures need to be put in place to ensure successful use of promising strategies to improve risk assessment and stratification for PDAC. Lastly, a concerted effort similar to the effort initiated by the Alliance of Pancreatic Cancer Consortia toward AI for prediagnostic imaging of pancreatic cancer45 is necessary to properly address these challenges, use mitigating strategies, and initiate next steps.
CONCLUSIONS
Applying AI techniques to population data can benefit both patients and society at large. However, no single entity can achieve success in this area alone. It will require a diverse group of stakeholders and experts to make this possible. The potential benefits of AI for early detection stand a better chance of success if all partners and stakeholders are aligned and take part in developing the plan. Government and industry collaboration in these endeavors should be encouraged. Likewise, patient advocacy groups play an essential role as they serve as the hub between patients, government agencies, private sectors, and academia. Time is of the essence with this disease. The frustration level of patients, their families, and medical practitioners linked to the absence of an earlier detection protocol cannot be underestimated.
I am reminded that our work in science is meant not just to enrich the scientific literature & culture—but to serve the improvement of human lives—even when that is hard and takes time to come to fruition. One is humbled by the challenge.
Chris Sander, April 9, 2021
ACKNOWLEDGMENTS
Presenters for the “Early Detection of Pancreatic Cancer: Opportunities and Challenges in Utilizing Electronic Health Records (EHR)” were David S. Klimstra, MD, of Memorial Sloan Kettering Cancer Center (MSKCC); Sudhir Srivastava, PhD, MPH, MS, of the NCI Cancer Biomarkers Research Group, Division of Cancer Prevention; Alex Milinovich, BS, of Lerner Research Institute at Cleveland Clinic; Shawn Murphy, MD, PhD, of Mass General Brigham; Michael Rosenthal, MD, PhD, of Dana-Farber Cancer Institute; Jason Perry Block, MD, MPH, of Harvard Medical School; Jennifer St. Sauver, MPH, PhD, Mayo Clinic; Pete Stetson, MD, MA, of Memorial Sloan Kettering Cancer Center; Limor Appelbaum, MD, Beth Israel Deaconess Medical Center; Matvey Palchuk, MD, MS, FAMIA, TriNetX, LLC; and Suresh Chari, MD, of The University of Texas MD Anderson Cancer Center. Workshop was moderated by William Hoos, Canopy Cancer Collective. Workshop Planning Committee Members were Sudhir Srivastava, PhD, MPH, MS; Natalie Abrams, PhD, and Matthew Young, PhD (National Cancer Institute); Vay Liang Go, MD; Suresh Chari, MD; David Klimstra, MD; Bruce Field; and William Hoos, MBA (Kenner Family Research Fund scientific board members); Laura Rothschild, MBA; Ann Goldberg, BA; and Barbara Kenner, PhD (Kenner Family Research Fund board members). Editing assistance was provided by Tara Coffin, PhD.
Footnotes
This article was prepared for the 2021 workshop entitled: “Early Detection of Pancreas Cancer: Opportunities and Challenges in Utilizing Electronic Health Records” through the support of Kenner Family Research Fund.
The authors declare no conflict of interest. S.T.C., B.F.F., V.L.W.G., W.A.H., and D.S.K. are Kenner Family Research Fund scientific advisors. A.E.G., B.J.K., and L.J.R. are Kenner Family Research Fund board members.
Contributor Information
Natalie D. Abrams, Email: natalie.abrams@nih.gov.
Suresh T. Chari, Email: STChari@mdanderson.org.
Bruce F. Field, Email: bfield@minn.net.
Ann E. Goldberg, Email: anngoldberg21@gmail.com.
William A. Hoos, Email: williamhoos@gmail.com.
David S. Klimstra, Email: klimstrd@mskcc.org.
Laura J. Rothschild, Email: ljrchild@gmail.com.
Sudhir Srivastava, Email: srivasts@mail.nih.gov.
Matthew R. Young, Email: matthew.young@nih.gov.
Vay Liang W. Go, Email: VLWGo@mednet.ucla.edu.
REFERENCES
- 1.Cancer Stats Facts: Pancreatic Cancer. Available at: https://seer.cancer.gov/statfacts/html/pancreas.html. Accessed June 28, 2021.
- 2.Chari ST Kelly K Hollingsworth MA, et al. Early detection of sporadic pancreatic cancer: summative review. Pancreas. 2015;44:693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singhi AD Koay EJ Chari ST, et al. Early detection of pancreatic cancer: opportunities and challenges. Gastroenterology. 2019;156:2024–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira SP Oldfield L Ney A, et al. Early detection of pancreatic cancer. Lancet Gastroenterol Hepatol. 2020;5:698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn EH. Cancer interception. Cancer Prev Res (Phila). 2011;4:787–792. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz NRM Matrisian LM Shrader EE, et al. Potential cost-effectiveness of risk-based pancreatic cancer screening in patients with new-onset diabetes. J Natl Compr Canc Netw. 2021;1–9. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 7.Serrano J Rinaudo JA Srivastava S, et al. The national institutes of health's approach to address research gaps in pancreatitis, diabetes and early detection of pancreatic cancer. Curr Opin Gastroenterol. 2021;37:480–485. [DOI] [PubMed] [Google Scholar]
- 8.Sharma A Kandlakunta H Nagpal SJS, et al. Model to determine risk of pancreatic cancer in patients with new-onset diabetes. Gastroenterology. 2018;155:730–739.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estiri H Strasser ZH Klann JG, et al. Predicting COVID-19 mortality with electronic medical records. NPJ Digit Med. 2021;4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang N Khan A Polubriaginof F, et al. Medical records-based chronic kidney disease phenotype for clinical care and “big data” observational and genetic studies. NPJ Digit Med. 2021;4:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malhotra A Rachet B Bonaventure A, et al. Can we screen for pancreatic cancer? Identifying a sub-population of patients at high risk of subsequent diagnosis using machine learning techniques applied to primary care data. PloS One. 2021;16:e0251876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharpless NE, Kerlavage AR. The potential of AI in cancer care and research. Biochim Biophys Acta Rev Cancer. 1876;2021:188573. [DOI] [PubMed] [Google Scholar]
- 13.Kenner B Chari ST Kelsen D, et al. Artificial intelligence and early detection of pancreatic cancer: 2020 summative review. Pancreas. 2021;50:251–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs TJ. The AI revolution. Mount Sinai Science & Medicine. Spring/Summer 2021;4–7. Available at: https://physicians.mountsinai.org/news/the-ai-revolution-discover-how-artificial-intelligence-is-moving-to-the-forefront-of-innovative-patient-care-at-mount-sinai. Accessed July 3, 2021.
- 15.Appelbaum L Cambronero JP Stevens JP, et al. Development and validation of a pancreatic cancer risk model for the general population using electronic health records: an observational study. Eur J Cancer. 2021;143:19–30. [DOI] [PubMed] [Google Scholar]
- 16.Placido D Yuan B Hu JX, et al. Pancreatic cancer risk predicted from disease trajectories using deep learning. bioRxiv. June 28, 2021. Available at: 10.1101/2021.06.27.449937. Accessed July 1, 2021. [DOI] [Google Scholar]
- 17.Topaloglu U, Palchuk MB. Using a federated network of real-world data to optimize clinical trials operations. JCO Clin Cancer Inform. 2018;2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reimer AP, Milinovich A. Using UMLS for electronic health data standardization and database design. J Am Med Inform Assoc. 2020;27:1520–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy SN Mendis ME Berkowitz DA, et al. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc. 2006;2006:1040. [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy SN Weber G Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc. 2010;17:124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao JJ Eubank MH Schram AM, et al. Real-world outcomes of an automated physician support system for genome-driven oncology. JCO Precis Oncol. 2019;3:PO.19.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vickers AJ, Chen LY, Stetson PD. Interfaces for collecting data from patients: 10 golden rules. J Am Med Inform Assoc. 2020;27:498–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng DT Prasad M Chekaluk Y, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics. 2017;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rocca WA Grossardt BR Brue SM, et al. Data resource profile: Expansion of the Rochester Epidemiology Project medical records-linkage system (E-REP). Int J Epidemiol. 2018;47:368–368j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner EH Greene SM Hart G, et al. Building a research consortium of large health systems: the Cancer Research Network. J Natl Cancer Inst Monogr. 2005;35:3–11. [DOI] [PubMed] [Google Scholar]
- 26.Appelbaum L Berg A Cambronero JP, et al. Development of a pancreatic cancer prediction model using a multinational medical records database. J Clin Oncol. 2021;39(3 suppl):394.abstract. [Google Scholar]
- 27.Murphy SN Mendis M Hackett K, et al. Architecture of the open-source clinical research chart from informatics for integrating biology and the bedside. AMIA Annu Symp Proc. 2007;2007:548–552. [PMC free article] [PubMed] [Google Scholar]
- 28.Overhage JM Ryan PB Reich CG, et al. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Califf RM. The patient-centered outcomes research network: a national infrastructure for comparative effectiveness research. N C Med J. 2014;75:204–210. [DOI] [PubMed] [Google Scholar]
- 30.All of Us Research Program Investigators; Denny JC Rutter JL Goldstein DB, et al. The “all of us” research program. N Engl J Med. 2019;381:668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weeks J, Pardee R. Learning to share health care data: a brief timeline of influential common data models and distributed health data networks in U.S. health care research. EGEMS (Wash DC). 2019;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National COVID Cohort Collaborative. Available at: https://covid.cd2h.org. Accessed July 6, 2021.
- 33.Haendel MA Chute CG Bennett TD, et al. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2021;28:427–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becnel LB Hastak S Ver Hoef W, et al. BRIDG: a domain information model for translational and clinical protocol-driven research. J Am Med Inform Assoc. 2017;24:882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehne M Luijten S Vom Felde Genannt Imbusch P, et al. The use of FHIR in digital health—a review of the scientific literature. Stud Health Technol Inform. 2019;267:52–58. [DOI] [PubMed] [Google Scholar]
- 36.Braunstein ML. Health care in the age of interoperability part 6: the future of FHIR. IEEE Pulse. 2019;10:25–27. [DOI] [PubMed] [Google Scholar]
- 37.Hripcsak G Duke JD Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 38.Milinovich A, Kattan MW. Extracting and utilizing electronic health data from Epic for research. Ann Transl Med. 2018;6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taksler GB Dalton JE Perzynski AT, et al. Opportunities, pitfalls, and alternatives in adapting electronic health records for health services research. Med Decis Making. 2021;41:133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Block JP Bailey LC Gillman MW, et al. Early antibiotic exposure and weight outcomes in young children. Pediatrics. 2018;142:e20180290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.St Sauver JL Grossardt BR Yawn BP, et al. Use of a medical records linkage system to enumerate a dynamic population over time: the Rochester epidemiology project. Am J Epidemiol. 2011;173:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu S Lopes GS Pagali SR, et al. Ascertainment of delirium status using natural language processing from electronic health records. J Gerontol A Biol Sci Med Sci. 2020 Oct 30. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Use of Electronic Health Record Data in Clinical Investigations: Guidance for Industry. Rockville, MD: U.S. Department of Health and Human Services, Food and Drug Administration; 2018. [Google Scholar]
- 44.Collaborative learning without sharing data. Nat Mach Intell. 2021;3:459. [Google Scholar]
- 45.Young MR Abrams N Ghosh S, et al. Prediagnostic image data, artificial intelligence, and pancreatic cancer: a tell-tale sign to early detection. Pancreas. 2020;49:882–886. [DOI] [PubMed] [Google Scholar]


