Abstract
Background:
Hypertrophic scar is a fibroproliferative disorder caused by skin injury. The incidence of hypertrophic scar following trauma or burns is 40 to 70 percent or 70 percent, respectively. It has been shown that transforming growth factor (TGF) β1/Smad signaling plays a crucial role in hypertrophic scar, and that USP15 can regulate the activity of TGFβ1/Smad signaling to affect the progression of the disease. However, the underlying mechanism of USP15 in hypertrophic scar remains unclear. The authors hypothesized that USP15 was up-regulated and enhanced the proliferation, migration, invasion, and collagen deposition of hypertrophic scar–derived fibroblasts by deubiquitinating TGF-β receptor I (TβRI) in vitro.
Methods:
Fibroblasts were isolated from human hypertrophic scars in vitro. The knockdown and overexpression of USP15 in hypertrophic scar–derived fibroblasts were performed using lentivirus infection. The effect of USP15 on hypertrophic scar–derived fibroblast proliferation, migration, and invasion, and the expression of TβRI, Smad2, Smad3, α-SMA, COL1, and COL3, were detected by Cell Counting Kit-8, scratch, invasion, quantitative real-time polymerase chain reaction, and Western blot assays. The interaction between USP15 and TβRI was detected by co-immunoprecipitation and ubiquitination assays.
Results:
The authors demonstrated that USP15 knockdown significantly inhibited the proliferation, migration, and invasion of hypertrophic scar–derived fibroblasts in vitro and down-regulated the expression of TβRI, Smad2, Smad3, α-SMA, COL1, and COL3; in addition, USP15 overexpression showed the opposite trends (p < 0.05). Co-immunoprecipitation and ubiquitination assays revealed that USP15 interacted with TβRI and deubiquitinated TβRI.
Conclusion:
USP15 enhances the proliferation, migration, invasion, and collagen deposition of hypertrophic scar–derived fibroblasts by deubiquitinating TβRI in vitro.
Hypertrophic scar is a fibroproliferative disorder that results from injury of the skin.1 The overall incidence of hypertrophic scar caused by skin trauma is 40 to 70 percent, whereas the incidence of burn scars is up to 70 percent.2 It manifests as abnormal fibroblast proliferation and excessive collagen deposition.3 Hypertrophic scar is often accompanied by pain, pruritus, contracture, and deformities, which affect the patient’s aesthetics and cause dysfunction, and have impacts on the physical and mental health of the affected individuals.4 To date, the main therapies for hypertrophic scar are surgical resection, cryotherapy, compression therapy, intralesional corticosteroid injection, and laser therapy, among others.5 None of these methods are satisfactory therapeutic options for hypertrophic scar. The recurrence rate of a single surgical resection is 45 to 100 percent; cryotherapy is not effective for patients with hypertrophic scar longer than 12 months; compression therapy requires the continuous use of pressure suits for 6 months, and the daily duration should not be less than 23 hours.6 Therefore, there is an urgent need to elucidate the mechanism of hypertrophic scar to guide clinical treatment.
Substantial evidence has revealed that transforming growth factor (TGF) β1/Smad signaling plays a crucial role in hypertrophic scar.7 The increased activity of TGFβ1/Smad stimulates the proliferation of fibroblasts and leads to overproduction and excess deposition of collagen by fibroblasts, resulting in hypertrophic scar formation.8 Inversely, weakening the activity of TGFβ1/Smad signaling has been reported to inhibit hypertrophic scar formation.9 TGFβ1/Smad signaling is mediated by a pair of transmembrane serine-threonine kinase receptors known as TGF-β receptor I (TβRI) and TβRII. In response to TGFβ1 binding, TβRII autophosphorylation occurs first and then phosphorylates TβRI, leading to the activation of TβRI. Subsequently, it phosphorylates and activates intracellular receptor-regulated Smad2 and Smad3 (R-Smads); activated R-Smads then interact with the co-Smad (Smad4) and enter the nucleus to regulate gene transcription.10 Expression of these genes, including α-smooth muscle actin (α-SMA) collagen I (COL1), and collagen III (COL3), is closely associated with the myofibroblast phenotype and progression of fibrosis.11
The ubiquitin-proteasome pathway regulates intracellular protein degradation and affects signal transduction. Deubiquitination is the reverse process of ubiquitination mediated by deubiquitinating enzymes and plays an essential role in regulating the signal pathway.12,13 USP15 is an important member of the deubiquitinating enzyme family.14 Accumulating evidence has revealed that USP15 exhibits abnormal expression in various diseases and can regulate TGFβ1/Smad signaling. For example, USP15 is overexpressed in glioblastoma, breast cancer, ovarian cancer. Eichhorn et al. have reported that USP15 is highly expressed in glioma and deubiquitinates and stabilizes TβRI, leading to an enhanced TGFβ1/Smad signaling.15 Similarly, Inui et al. revealed that USP15 could directly bind to R-Smads and deubiquitinate TβRI, thereby promoting its stability and up-regulating the activity of TGFβ1/Smad signaling.16 Galant et al. indicated that overexpression of USP15 in systemic sclerosis fibroblasts increases response to TGFβ1, leading to fibrosis.17
However, the role of USP15 in hypertrophic scar remains unclear. Liu et al. have indicated that TGF-β up-regulates the translation of USP15 through the PI3K/AKT pathway in HEK293 cells.18 We hypothesized that USP15 deubiquitinated TGF-β receptor I to enhances TGF-β/Smad signaling, which in turn up-regulated USP15, leading to enhancement of the proliferation, migration, invasion, and collagen deposition of hypertrophic scar–derived fibroblasts in vitro. This positive feedback loop possibly persists in hypertrophic scar–derived fibroblasts.
PATIENTS AND METHODS
Specimen Collection
This study was approved by the Medical Ethics Committee of the First Affiliated Hospital of Nanchang University, and written informed consent was obtained from each patient. Hypertrophic scar and matched normal skin tissues of six patients (three female and three male patients; age range, 5 to 41 years) were collected from the Department of Plastic Surgery in our hospital. Specimens were divided into three parts. One part was used for hematoxylin and eosin staining and Masson staining, another part was used for RNA and protein extraction, and the third part was used for isolating fibroblasts.
Hematoxylin and Eosin and Masson Staining
Tissues were fixed, embedded, sliced into 4-µm-thick sections, and stained with the hematoxylin and eosin staining kit (catalogue no. AR1180; Boster Biological Technology, Wuhan, People’s Republic of China) and Masson staining kit (catalogue no. G1340; Solaibio, Beijing, People’s Republic of China), respectively, according to the manufacturer’s instructions. The histology feature of hypertrophic scar was characterized by increased number of fibroblasts, disordered collagen fibers, and excessive collagen deposition.19 Images were captured using an Olympus microscope (Olympus Corp., Tokyo, Japan). To count the number of fibroblasts, collagen fiber arrangement, and collagen content, three random fields were evaluated by two pathologists with blinding. Relative collagen levels were quantified by comparing optical density values using Image-Pro Plus (Media Cybernetics, Rockville, Md.).
Cell Culture and Identification
Hypertrophic scar–derived fibroblasts were isolated according to a previously described protocol.20 When the hypertrophic scar–derived fibroblasts reached 90 percent confluence, the fibroblasts were subcultured. Hypertrophic scar–derived fibroblasts from the third to the fifth passages were used for subsequent experiments.
For immunofluorescence staining, fibroblasts growing on slides were fixed in 4% paraformaldehyde for 15 minutes and permeabilized with 0.05% Triton X-100 for 15 minutes at room temperature. Fibroblasts were incubated for 1 hour at room temperature by means of 3% goat serum to block nonspecific binding. After the cells were washed in phosphate-buffered saline, they were treated with primary antibodies specific for vimentin (1:200, ab92547; Abcam, Cambridge, Mass.) at 4°C overnight. The Alexa Fluor 488–conjugated secondary antibodies (1:200, ab150117; Abcam) were used to detect the primary antibodies. Then, 4′,6-diamidino-2-phenylindole was used for nuclear counterstaining. Finally, images were obtained by a laser scanning confocal microscope (Olympus). The cytoplasm of fibroblasts stained green was regarded as positive. To calculate the percentage of positive-stained cells, three random fields were evaluated by two operators with blinding.
Lentivirus Transfection and Grouping of Fibroblasts
The USP15 knockdown and USP15-overexpressing lentiviruses were purchased from Genechem Company (Shanghai, People’s Republic of China). The human USP15 small hairpin shRNA-specific target sequences used were as follows:
Small hairpin USP15-1: 5′-CCCATTGATAACTCTGGACTT-3′
Small hairpin USP15-2: 5′-AAATACCAGATGGGAGATCAA-3′
Small hairpin USP15-3: 5′-TAGTCGATAGTCGCTGGTTCA-3′
Cells were seeded in six-well plates and cultured overnight, and the appropriate lentivirus (multiplicity of infection = 20) was added into the wells. Twelve hours later, the medium containing the lentivirus was discarded and replaced with fresh medium. After 72 hours of transfection, lentivirus-infected hypertrophic scar–derived fibroblasts were subjected to puromycin for 1 week to select stably infected cells. Finally, total RNA or proteins were extracted to assess the knockdown and overexpression efficiency. The fibroblasts were then divided into blank control group (blank group), negative control group (small hairpin negative control group), lentivirus-USP15 knockdown group (small hairpin USP15 group), lentivirus–empty vector group (vector group), and lentivirus-USP15 overexpression group (ubiquitin-specific protease 15 group).
Real-Time Quantitative Polymerase Chain Reaction
Total RNA of hypertrophic scar and matched normal skin tissues or fibroblasts was extracted using the TRIzol reagent (Invitrogen, Waltham, Mass.).The cDNA was synthesized using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, People’s Republic of China), and quantitative polymerase chain reaction was performed using the real-time polymerase chain reaction system (Bio-Rad, Hercules, Calif.). The reaction conditions were as follows: predenaturation at 95°C for 10 minutes, followed by 40 cycles of polymerase chain reaction denaturation at 95°C for 10 seconds, annealing at 58°C for 30 seconds, and extension at 72°C for 30 seconds. Human glyceraldehyde-3-phosphate dehydrogenase was used as an internal control and fold changes were calculated using the relative quantification (2-ΔΔCt) method. The primer sequences are listed in Table 1.
Table 1.
Real-Time Polymerase Chain Reaction Primers
| Gene | Sequence |
|---|---|
| USP15 | |
| Forward | 5′-ACGCTGCTCAAAACCTCG-3′ |
| Reverse | 5′-ACATACCCTGTTCAACCACCT-3′ |
| TβRI | |
| Forward | 5′-ACAAAAAGGTACATGGCCCC-3′ |
| Reverse | 5′-TCCACCAATGGAACATCGTC-3′ |
| Smad2 | |
| Forward | 5′-ATCTCCTACTACTCTTTCCCCTGTT-3′ |
| Reverse | 5′-TTTCTACCGTGGCATTTCG-3′ |
| Smad3 | |
| Forward | 5′-GAGGAGAAATGGTGCGAGAA-3′ |
| Reverse | 5′-CAGGCGGCAGTAGATGACA-3′ |
| α-SMA | |
| Forward | 5′-GCGATCTCACCGACTACCTG-3′ |
| Reverse | 5′-GCCGACTCCATACCGATGAA-3′ |
| COL-1 | |
| Forward | 5′-AGACATCCCACCAATCACCT-3′ |
| Reverse | 5′-CGTCATCGCACAACACCTT-3′ |
| COL-3 | |
| Forward | 5′-TGGCATCAAAGGACATCG-3′ |
| Reverse | 5′-CATAATACGGGGCAAAACC-3′ |
| GAPDH | |
| Forward | 5′-TGACTTCAACAGCGACACCCA-3′ |
| Reverse | 5′-CACCCTGTTGCTGTAGCCAAA-3′ |
Western Blot Analysis
The proteins were extracted from hypertrophic scar and matched normal skin tissues or fibroblasts for Western blot analysis using the RIPA lysis buffer (Invitrogen) containing protease inhibitors. Protein levels were quantified using a bicinchoninic acid protein assay kit. Protein samples were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred onto a polyvinylidene fluoride membrane. The membranes were blocked with 5% nonfat milk for 2 hours at room temperature and the following primary antibodies: USP15 (ab71713; Abcam), TβR1 (ab31013; Abcam), Smad2 (no. 5339; Cell Signaling Technology, Inc., Danvers, Mass.), Smad3 (no. 5923; Cell Signaling Technology), α-SMA (Proteintech, Rosemont, Ill.), COL1 (ab138492; Abcam), and COL3 (ab7778; Abcam) incubated overnight at 4°C. The membranes were then incubated with horseradish peroxidase–conjugated secondary antibodies for 2 hours at room temperature. An enhanced chemiluminescence substrate (Thermo Fisher) was added to the membranes and scanned using the ChemiDoc XRS Imaging System (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. Relative protein levels were quantified by comparing the gray values by using Image-Pro Plus.
Cell Counting Kit-8 Assay
The Cell Counting Kit-8 assay was used to measure cell proliferation. The different group cells were seeded in 96-well plates at a density of 5 × 103 cells/ml and cultured for 0-, 24-, 48-, and 72-hour time points. Ten microliters of the Cell Counting Kit-8 reagent was added to each well and incubated for 2 hours. The absorbance at 450 nm was measured using a microplate reader.
Scratch Assays
The scratch wound assay was used to evaluate the migration of different group cells. The cells were seeded in six-well plates at a density of 2 × 105 cells/ml and cultured until they reached approximately 100 percent confluence. A scratch wound was generated on the cell surface using a sterile 200-μl pipette; phosphate-buffered saline was used to remove the floating cells and then change to serum-free Dulbecco’s Modified Eagle Medium. Digital images of each scratch distance at 0-, 12-, 24-, and 36-hour time points were captured using the Leica microscope (Leica, Wetzlar, Germany).
Matrigel Invasion Assays
Matrigel (BD Biosciences, San Diego, Calif.) was diluted with serum-free Dulbecco’s Modified Eagle Medium and added to the Transwell upper chamber, then incubated at 37°C overnight until Matrigel solidified. The cell density was adjusted to 2 × 105 cells/ml in serum-free medium, and 100 μl of cell suspension was added to the Transwell upper chamber with preplated Matrigel, and 700 μl of 10% fetal bovine serum Dulbecco’s Modified Eagle Medium was added to the lower chambers. After incubation for 48 hours, the invasion cells were fixed with 4% paraformaldehyde, stained with 0.05% crystal violet, and counted in three random fields under microscopy.
Co-Immunoprecipitation and Ubiquitination Assays
Cells were extracted with the immunoprecipitation lysis buffer (Thermo Fisher Scientific, Waltham, Mass.) containing protease inhibitors. Protein A/G agarose beads were added to the whole-cell lysates to remove nonspecific proteins before immunoprecipitation. Equal amounts of cell lysates were incubated with specific primary antibody overnight at 4°C under rotation conditions. The next day, protein A/G agarose beads were added and incubated on a rotator for 2 hours at 4°C. The samples were then centrifuged, the supernatant was discarded, and the beads were washed four times with immunoprecipitation lysis/wash buffer. The immunoprecipitated complex was analyzed by Western blot.
For the ubiquitination assay, stably infected hypertrophic scar–derived fibroblasts were co-transfected with TβRI and HA-ubiquitin plasmid, and cells were treated with proteasome inhibitor MG-132 (10 µM) for 6 hours. Then, proteins were extracted and co-immunoprecipitation assay was performed to detect the ubiquitination of TβRI.
Statistical Analysis
Data are presented as mean ± SEM of three independent experiments. The results were analyzed using IBM SPSS Version 21.0 (IBM Corp., Armonk, N.Y.). The t test or one-way analysis of variance was used to determine significant differences. The Bonferroni correction was used to correct for multiple comparisons. A value of p < 0.05 was considered statistically significant.
RESULTS
The Number of Fibroblasts and Collagen Content Were Increased in Hypertrophic Scar
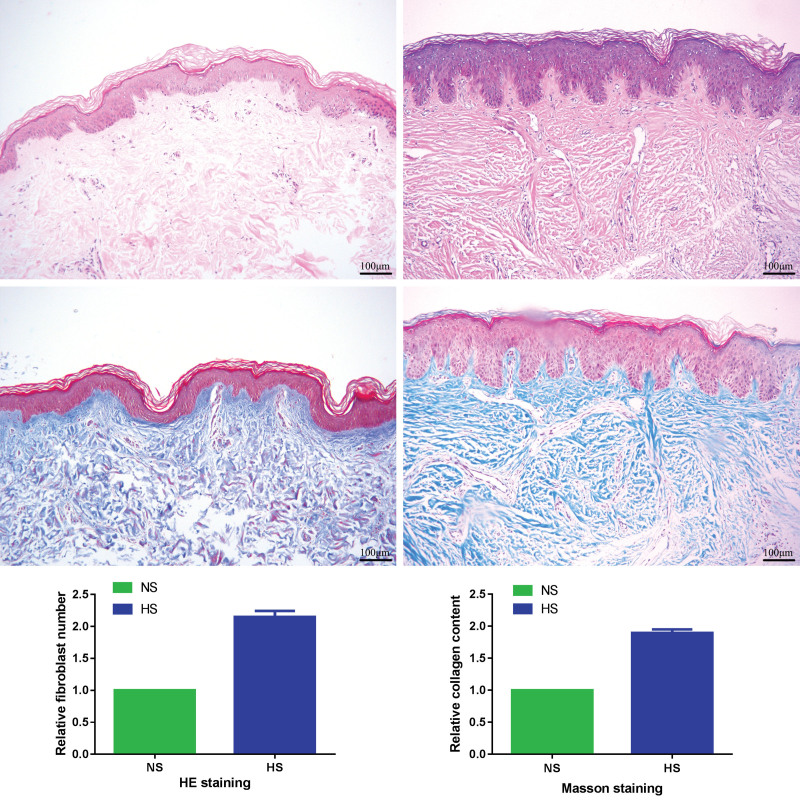
Hematoxylin and eosin staining showed that the disordered collagen fibers in the dermis layer and the number of fibroblasts were increased in hypertrophic scar tissues compared with the matched normal skin tissues (2.15 ± 0.09-fold; p < 0.05). Masson staining showed that collagen fibers were distributed in a disorderly fashion and the collagen content was increased in hypertrophic scar tissues compared with the matched normal skin tissues (1.89 ± 0.06-fold; p < 0.05) (Fig. 1).
Fig. 1.
Histologic features of hypertrophic scars and normal skin tissues. (Above, left) Hematoxylin and eosin staining for normal skin tissues (original magnification, × 100). (Above, right) Hematoxylin and eosin staining for hypertrophic scar tissues (original magnification, × 100). (Center, left) Masson staining for normal skin tissues (original magnification, × 100). (Center, right) Masson staining for hypertrophic scar tissues (original magnification, × 100). (Below, left) Relative fibroblast number of hematoxylin and eosin staining. (Below, right) Relative collagen content of Masson staining. HS, hypertrophic scar; NS, normal skin; HE, hematoxylin and eosin. Scale bar = 100 μm.
USP15 Is Up-Regulated and TGFβ/Smad Signaling Is Overactivated in Hypertrophic Scar
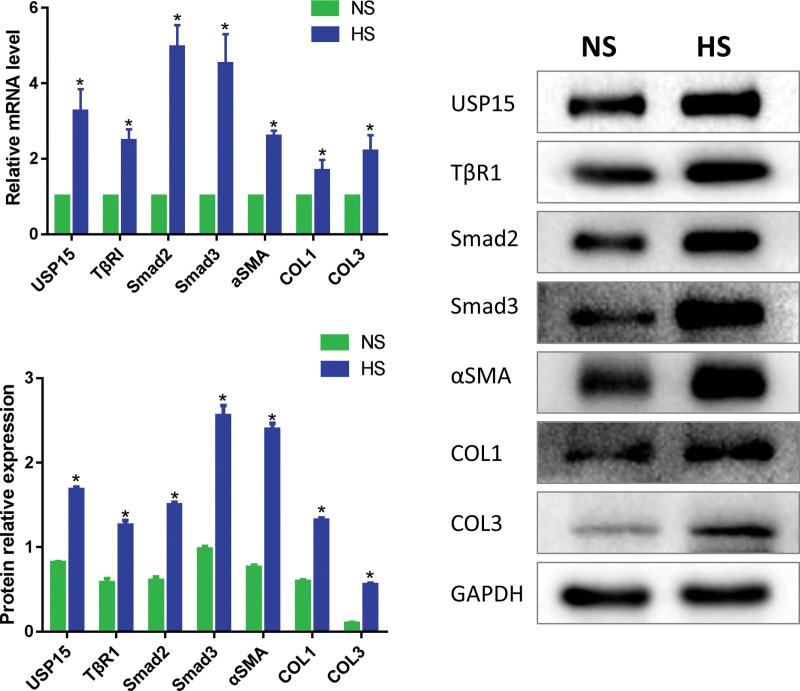
The mRNA and protein expression levels of USP15 in hypertrophic scar compared with the matched normal skin tissues were detected by quantitative real-time polymerase chain reaction and Western blot, respectively. Quantitative real-time polymerase chain reaction showed that the mRNA expression level of USP15 was significantly up-regulated in hypertrophic scar compared with the matched normal skin tissues (3.26 ± 0.34-fold; p < 0.05). Western blot showed that the protein expression level of USP15 was significantly up-regulated in hypertrophic scar compared with the matched normal skin tissues (1.68 ± 0.03-fold versus 0.82 ± 0.01-fold; p < 0.05). These results indicated that USP15 was up-regulated in hypertrophic scar. In addition, quantitative real-time polymerase chain reaction and Western blot results showed that compared with the matched normal skin tissues, TGFβ1/Smad signaling–related molecules TβRI, Smad2, Smad3, and fibrosis markers α-SMA, COL1, and COL3 were highly expressed in hypertrophic scar tissues (1.26 ± 0.06-fold versus 0.58 ± 0.05-fold, 1.50 ± 0.03-fold versus 0.61 ± 0.04-fold, 2.56 ± 0.12-fold versus 0.98 ± 0.03-fold, 2.40 ± 0.07-fold versus 0.76 ± 0.03-fold, 1.32 ± 0.03-fold versus 0.59 ± 0.02-fold, and 0.55 ± 0.02-fold versus 0.10 ± 0.01-fold, respectively; p < 0.05) (Fig. 2).
Fig. 2.
USP15 is significantly up-regulated and the TGFβ/Smad signaling pathway is over-activated in hypertrophic scar tissues. (Above, left) Real-time polymerase chain reaction was used to measure the mRNA expression level of USP15, TβRI, Smad2, Smad3, and fibrosis markers α-SMA, COL1, and COL3 in hypertrophic scar tissues and matched normal skin tissues (*p < 0.05). (Below, left) Western blot was used to measure the protein expression level of USP15, TβRI, Smad2, Smad3 and α-SMA, COL1, and COL3 in hypertrophic scar tissues and matched normal skin tissues. (Right) Densitometry analysis of Western blot results (*p < 0.05). HS, hypertrophic scar; NS, normal skin.
Hypertrophic Scar–Derived Fibroblasts Were Successfully Isolated from Hypertrophic Scar In Vitro
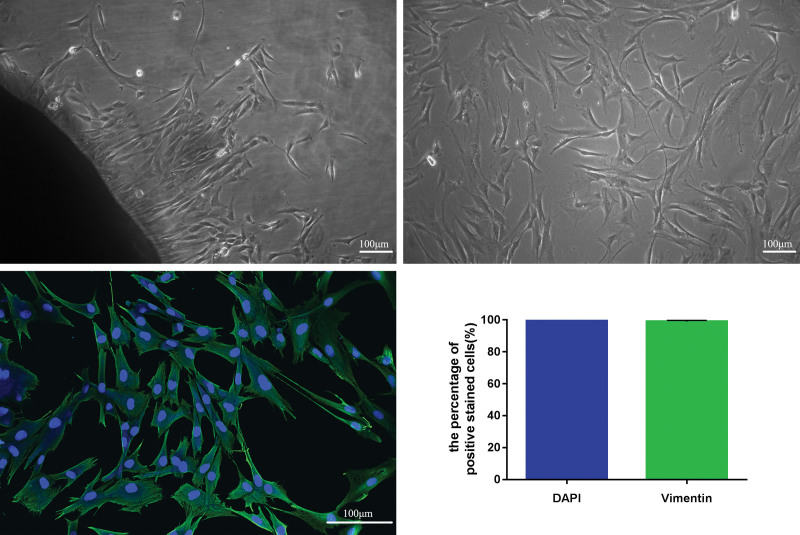
Tissue pieces combined with enzymatic digestion methods were used for primary cell culture of hypertrophic scar–derived fibroblasts. After 1 week, few fibroblasts emerged from the hypertrophic scar tissue edges. Hypertrophic scar–derived fibroblasts exhibit long spindle-shaped, triangular, or irregular morphologies. After the cells were grown to 90 to 100 percent confluence, the fibroblasts were subcultured. Hypertrophic scar–derived fibroblasts from the third to the fifth passages were used for subsequent experiments.
Consistent with previous research, the vimentin was a fibroblast-specific marker.21 Immunofluorescence staining showed that the nuclei of fibroblasts were stained blue and the cytoplasm of fibroblasts was stained green because a vimentin-specific marker was regarded as positively stained cell. The proportion of positively stained cells was nearly 100 percent (Fig. 3). It was confirmed that the cells cultured in vitro were fibroblasts.
Fig. 3.
Optical and fluorescence micrographs of cell morphologies of hypertrophic scar–derived fibroblasts. (Above, left) The primary fibroblasts were cultured derived from hypertrophic scar tissues (original magnification, × 100). (Above, right) Third-passage fibroblasts derived from hypertrophic scar tissues were observed under inverted microscopy (original magnification, × 100). (Below, left) Cell morphologies of hypertrophic scar–derived fibroblasts were detected by immunofluorescence staining assay (original magnification, × 200). (Below, right) The percentage of positively stained cells. DAPI, 4′,6-diamidino-2-phenylindole. Scale bar = 100 μm (above, right and left, and below, left).
USP15 Promotes the Proliferation, Migration, and Invasion of Hypertrophic Scar–Derived Fibroblasts
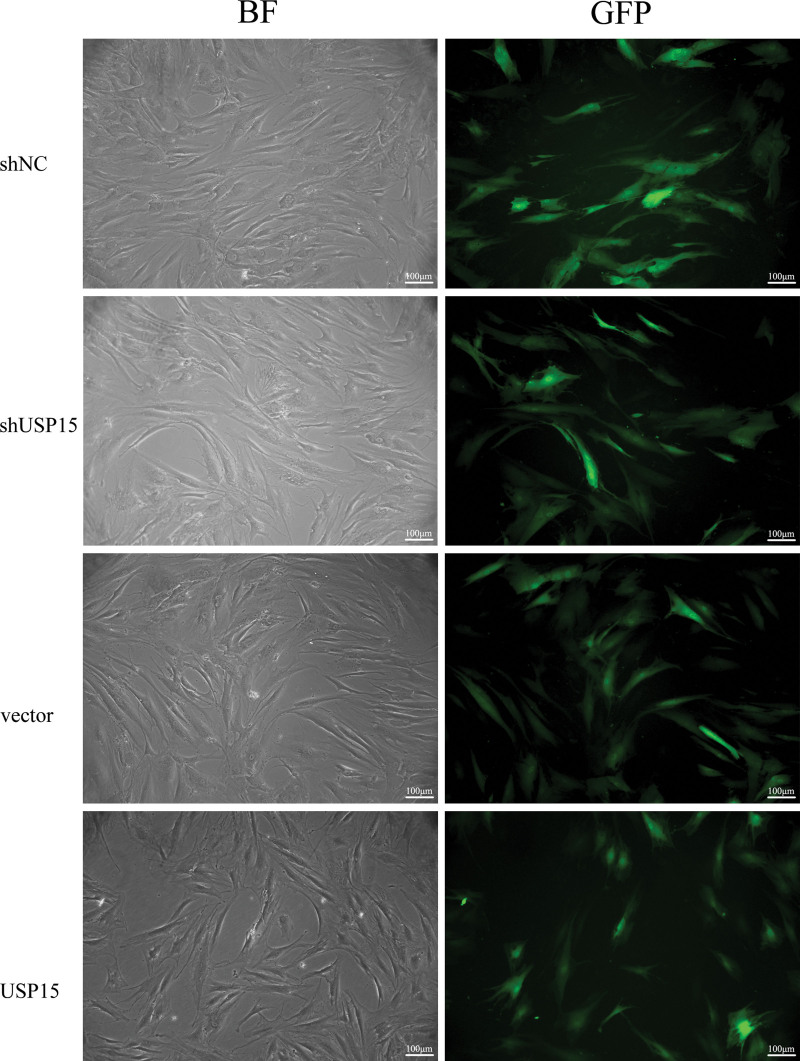
USP15 knockdown and overexpressing lentiviruses with enhanced green fluorescent protein were successfully transfected into hypertrophic scar–derived fibroblasts (Fig. 4). Transfection efficiency was confirmed by quantitative real-time polymerase chain reaction and Western blot. As expected, the mRNA and protein expression level of USP15 was significantly down-regulated in the small hairpin USP15 group compared with the blank group and small hairpin negative control group (0.12 ± 0.02-fold versus 0.64 ± 0.02-fold and 0.62 ± 0.02-fold; p < 0.05). On the contrary, the mRNA and protein expression level of USP15 was significantly up-regulated in the USP15 group compared with the blank group and vector group (0.95 ± 0.01-fold versus 0.21 ± 0.01-fold and 0.22 ± 0.01-fold; p < 0.05). [See Figure, Supplemental Digital Content 1, which shows to establish stable USP15 knockdown and overexpression in hypertrophic scar fibroblasts. (Above, left) The real-time polymerase chain reaction was used to measure the expression level of USP15 in blank, small hairpin negative control, and small hairpin USP15 groups (*p < 0.05). (Above, right) The real-time polymerase chain reaction was used to measure the expression level of USP15 in blank, vector, and USP15 groups (*p < 0.05). (Center, left) Western blot was used to measure the protein expression level of USP15 in blank, small hairpin negative control, and small hairpin USP15 groups (*p < 0.05). (Center, right) Western blot was used to measure the protein expression level of USP15 blank, vector, and USP15 groups (*p < 0.05). (Below) Densitometry analysis of Western blot results (*p < 0.05). shNC, lentivirus USP15 negative control group; shUSP15, lentivirus USP15 knockdown group; vector, lentivirus empty vector group; USP15, lentivirus USP15 overexpression group, http://links.lww.com/PRS/E644.] The stably infected cells were used for subsequent experiments.
Fig. 4.
USP15 knockdown and overexpression lentiviruses with enhanced green fluorescent protein were successfully transfected into hypertrophic scar–derived fibroblasts. (Above, left) Small hairpin negative control group in bright field. (Above, right) Small hairpin negative control in fluorescent light. (Second row, left) Small hairpin USP15 group in bright field. (Second row, right) Small hairpin USP15 group in fluorescent light. (Third row, left) Vector in bright field. (Third row, left) Vector in fluorescent light. (Below, left) USP15 in bright field. (Below, right) USP15 in fluorescent light. shNC, lentivirus USP15 negative control group; shUSP15, lentivirus USP15 knockdown group; vector, lentivirus empty vector group; USP15, lentivirus USP15 overexpression group. BF, bright field; GFP, green fluorescent protein. Scale bar = 100 μm.
We further verified whether USP15 could affect the biological behavior of hypertrophic scar–derived fibroblasts. Cell Counting Kit-8 assays showed that USP15 knockdown significantly inhibited the proliferation of hypertrophic scar–derived fibroblasts (62.06 ± 10.46 percent; p < 0.05), whereas USP15 overexpression significantly promoted the proliferation of hypertrophic scar–derived fibroblasts (147.97 ± 10.49 percent; p < 0.05.) [See Figure, Supplemental Digital Content 2, which shows that USP15 knockdown and overexpression affect the proliferation and migration of hypertrophic scar fibroblasts. (Above, left) The proliferation of hypertrophic scar fibroblasts in blank, small hairpin negative control, and small hairpin USP15 groups was detected by the Cell Counting Kit-8 assay (*p < 0.05). (Above, right) The proliferation of hypertrophic scar fibroblasts in blank, vector, and USP15 group was detected by Cell Counting Kit-8 assay (*p < 0.05). (Center, left) The migration of hypertrophic scar fibroblasts in blank, short hairpin negative control, and short hairpin USP15 groups was detected by scratch assays (*p < 0.05). (Center, right) The migration of hypertrophic scar fibroblasts in blank, vector, and USP15 groups was detected by scratch assays (*p < 0.05), http://links.lww.com/PRS/E645.] Scratch assays showed that USP15 knockdown significantly inhibited the migration of hypertrophic scar–derived fibroblasts (79.25 ± 0.71 percent; p < 0.05), whereas USP15 overexpression significantly promoted the migration of hypertrophic scar–derived fibroblasts (163.84 ± 0.21 percent; p < 0.05) (see Figure, Supplemental Digital Content 2, http://links.lww.com/PRS/E645). Matrigel invasion assays showed that USP15 knockdown significantly inhibited the invasion of hypertrophic scar–derived fibroblasts (0.37 ± 0.03-fold; p < 0.05), whereas USP15 overexpression significantly promoted the invasion of hypertrophic scar–derived fibroblasts (2.17 ± 0.11-fold; p < 0.05). [See Figure, Supplemental Digital Content 3, which shows that USP15 knockdown and overexpression affect the invasion of hypertrophic scar fibroblasts. (Left) The invasion of hypertrophic scar fibroblasts in blank, short hairpin negative control, and short hairpin USP15 groups was detected by Matrigel invasion assays (*p < 0.05). (Right) The invasion of hypertrophic scar fibroblasts in blank, vector, and USP15 groups was detected by Matrigel invasion assays (*p < 0.05), http://links.lww.com/PRS/E646.] These results suggest that USP15 promotes the proliferation, migration, and invasion of hypertrophic scar–derived fibroblasts.
USP15 Increases the Expression of TβRI, Smad2, Smad3, α-SMA, COL1, and COL3
We further verified whether USP15 could affect TGFβ1/Smad signaling. Quantitative real-time polymerase chain reaction and Western blot analysis indicated that USP15 knockdown significantly reduced the mRNA and protein expression levels of TβRI, Smad2, Smad3, α-SMA, COL1, and COL3 (0.51 ± 0.01-fold versus 1.68 ± 0.06-fold, 0.58 ± 0.01-fold versus 1.30 ± 0.03-fold, 0.25 ± 0.03-fold versus 1.23 ± 0.04-fold, 0.44 ± 0.06-fold versus 0.88 ± 0.06-fold, 0.83 ± 0.09-fold versus 1.61 ± 0.07-fold, and 0.42 ± 0.03-fold versus 1.03 ± 0.03-fold; p < 0.05). Inversely, USP15 overexpression significantly increased the mRNA and protein expression levels of TβRI, Smad2, Smad3, α-SMA, COL1, and COL3 (1.11 ± 0.01-fold versus 0.53 ± 0.01-fold, 0.95 ± 0.01-fold versus 0.56 ± 0.01-fold, 1.29 ± 0.03-fold versus 0.73 ± 0.03-fold, 0.89 ± 0.01-fold versus 0.42 ± 0.02-fold, 0.74 ± 0.02-fold versus 0.36 ± 0.01-fold, and 0.64 ± 0.04-fold versus 0.33 ± 0.04-fold; p < 0.05). [See Figure, Supplemental Digital Content 4, which shows that USP15 knockdown and overexpression affect the expression of the TGFβ1/Smad signaling–related molecules TβRI, Smad2, Smad3, and the fibrosis markers α-SMA, COL1, and COL3. (Above, left) The mRNA expression of TβRI, Smad2, Smad3, and fibrosis markers α-SMA, COL1, and COL3 in blank, small hairpin negative control, and small hairpin USP15 groups was detected by real-time polymerase chain reaction (*p < 0.05). (Above, right) The mRNA expression of TβRI, Smad2, and Smad3 and the fibrosis markers α-SMA, COL1, and COL3 in blank, vector, and USP15 group was detected by real-time polymerase chain reaction (*p < 0.05). (Below, left) The protein expression of TβRI, Smad2, a Smad3 and the fibrosis markers α-SMA, COL1, and COL3 in blank, small hairpin negative control, and small hairpin USP15 groups was detected by Western blot (*p < 0.05). (Below, right) The protein expression of TβRI, Smad2, and Smad3 and the fibrosis markers α-SMA, COL1, and COL3 in blank, vector, and USP15 groups was detected by Western blot (*p < 0.05), http://links.lww.com/PRS/E647.) Considered together, these results demonstrated that USP15 could enhance TGFβ1/Smad signaling.
USP15 Interacted and Deubiquitinated TβRI in Hypertrophic Scar Fibroblasts
Previous research suggests that TβRI is regulated by ubiquitination.22 We further explored whether the ubiquitin-proteasome pathway degraded TβRI in hypertrophic scar–derived fibroblasts. Western blot analysis indicated that treatment with the proteasome inhibitor MG-132 for the indicated time resulted in significant accumulation of endogenous TβRI protein in hypertrophic scar–derived fibroblasts. In addition, the co-immunoprecipitation results showed that endogenous TβRI and ubiquitin were directly bound in hypertrophic scar–derived fibroblasts. These results demonstrated that the ubiquitin-proteasome pathway degraded TβRI in hypertrophic scar–derived fibroblasts.
Furthermore, we further assessed the role of USP15 in regulating the degradation of TβRI protein. Co-immunoprecipitation assay identified endogenous USP15 and TβRI bound in hypertrophic scar–derived fibroblasts. USP15 is an important member of the deubiquitinating enzyme family; we further verified whether USP15 could deubiquitinate TβRI in hypertrophic scar–derived fibroblasts. The ubiquitination assay showed that USP15 knockdown significantly increased the TβRI ubiquitination levels in hypertrophic scar–derived fibroblasts, whereas USP15 overexpression significantly reduced the TβRI ubiquitination levels in hypertrophic scar–derived fibroblasts. [See Figure, Supplemental Digital Content 5, which shows that USP15 interacted with and deubiquitinated TβRI in hypertrophic scar fibroblasts. (Above, left) Hypertrophic scar–derived fibroblasts were treated with MG132 (15 μM) for the indicated times, and the protein expression of TβRI levels was detected by Western blotting. (Above, right) Hypertrophic scar–derived fibroblasts cell lysates were subjected to immunoprecipitation with immunoglobulin G or antiubiquitin antibody and then analyzed by immunoblotting with TβRI antibody. Input cell lysates were immunoblotted with TβRI antibody. (Center) Cell lysates from hypertrophic scar–derived fibroblasts were immunoprecipitated with anti-USP15 or anti-TβRI antibody, followed by immunoblotting with anti-TβRI or anti-USP15 antibody, respectively. Immunoglobulin G was used as a negative control. (Below) Hypertrophic scar–derived fibroblasts were treated with proteasome inhibitor MG132 (10 μM) for 6 hours. Then, cell lysates were subjected to immunoprecipitation with TβRI antibody and the ubiquitination of TβRI was detected by immunoblotting, http://links.lww.com/PRS/E648.] The above-mentioned evidence indicated that USP15 interacted with TβRI and deubiquitinated TβRI to enhance TGFβ1/Smad signaling. (See Figure, Supplemental Digital Content 6, which shows a schematic depiction of USP15 enhancing TGFβ1/Smad signaling by deubiquitinating TβRI in hypertrophic scar, http://links.lww.com/PRS/E649.)
DISCUSSION
Studies on fibrotic disease over the past decades have demonstrated that TGFβ1/Smad signaling plays a pivotal role in hypertrophic scar.23 Accumulating studies have reported that the activity of TGFβ1/Smad signaling is abnormally increased in hypertrophic scar.24–26 The hyperactive TGFβ1/Smad signaling leads to fibroblast proliferation, induces transformation of fibroblasts to myofibroblasts, increases collagen synthesis, inhibits matrix degradation, and eventually leads to hypertrophic scar formation.27–30
Numerous studies have demonstrated that the ubiquitin-proteasome pathway regulates intracellular protein degradation and affects TGFβ1/Smad signal transduction.31,32 Deubiquitination is the reverse process of ubiquitination, which is mediated by deubiquitinating enzymes and plays an essential role in regulating the activity of TGFβ1/Smad signaling.33,34 USP15 is an important member of the deubiquitinating enzyme family. Previous research reported that USP15 has abnormal expression in various tumors or fibrotic diseases and can regulate the activity of TGFβ1/Smad signaling. However, the role of USP15 in hypertrophic scar remains unclear.
In the present study, we first found that USP15 was up-regulated in hypertrophic scar compared with the matched normal skin tissues detected by quantitative real-time polymerase chain reaction and Western blot. Because the fibroblasts are the primary effector cells of hypertrophic scar, we further verified whether USP15 could affect the biological behavior of hypertrophic scar–derived fibroblasts. The Cell Counting Kit-8, scratch, and invasion assays showed that USP15 knockdown significantly inhibited the proliferation, migration, and invasion of the hypertrophic scar–derived fibroblasts, and USP15 overexpression showed the opposite trends. Furthermore, we verified whether USP15 could affect TGFβ1/Smad signaling. Quantitative real-time polymerase chain reaction and Western blot analysis indicated that USP15 knockdown significantly reduced the mRNA and protein expression levels of TβRI, Smad2, and Smad3 and the fibrosis markers α-SMA, COL1, and COL3; USP15 overexpression showed the opposite trends. These results demonstrated that USP15 could enhance TGFβ1/Smad signaling. The expression level of TβRI protein reflects the activity of TGFβ1/Smad signaling. We confirmed that the ubiquitin-proteasome pathway degraded TβRI in hypertrophic scar–derived fibroblasts. Furthermore, we assessed the role of USP15 in regulating the degradation of the TβRI protein. Co-immunoprecipitation and ubiquitination assays showed that USP15 knockdown significantly increased the TβRI ubiquitination levels in hypertrophic scar–derived fibroblasts, whereas USP15 overexpression significantly reduced the TβRI ubiquitination levels in hypertrophic scar–derived fibroblasts. These data demonstrated that USP15 increased the activity of TGFβ1/Smad signaling by deubiquitinating TβRI in hypertrophic scar–derived fibroblasts. Thus, USP15 may be a novel target for the treatment of hypertrophic scar.
As the deubiquitinating enzymes are druggable proteins, two companies, Progenra (Malvern, Pa.) and Hybrigenics (Gard, France), have developed inhibitors of deubiquitinating enzymes (P5091 and HBX 41108, respectively) with the hope of anticancer treatment.35 Foreseeable, developing pharmaceutical agents that target USP15 for the treatment of hypertrophic scar will be commercially feasible.36 Before clinical application, safety tests should be conducted to evaluate the effects of target USP15 on cardiovascular, respiratory, and nervous system functions and the toxicity of drugs, such as general toxicity, genetic toxicity, carcinogenic toxicity, and immunotoxicity.37
The limitations of the present study are as follows. First, we elucidated the role of USP15 in hypertrophic scar–derived fibroblasts in vitro only. However, whether USP15 plays the same role in vivo is unclear. In future experiments, we aim to develop an animal model of hypertrophic scar and to further clarify the feasibility of USP15 as a new target for hypertrophic scar treatment. Second, we discovered that USP15 was up-regulated in hypertrophic scar but did not explore the factors responsible for the up-regulation of USP15. Several studies have reported that TGF-β may up-regulate USP15.15,16 Liu et al. have indicated that TGF-β enhances the translation of USP15 through the PI3K/AKT pathway in HEK293 cells.18 However, the underlying mechanism of the up-regulation of USP15 in hypertrophic scar requires further exploration.
CONCLUSION
In conclusion, USP15 enhances the proliferation, migration, and collagen deposition of hypertrophic scar–derived fibroblasts by deubiquitinating TβRI in vitro.
ACKNOWLEDGMENTS
This study was supported by grants of National Natural Science Foundation of China (81860340), the Project of Education Department of Jiangxi Province (no. 60119), and the Special Fund for Graduate Innovation Project of Jiangxi Province (YC2019-B019).
Supplementary Material
Footnotes
Related digital media are available in the full-text version of the article on www.PRSJournal.com.
Disclosure: The authors have no financial interest to declare in relation to the content of this article.
REFERENCES
- 1.Padmanabhan J, Maan ZN, Kwon SH, Kosaraju R, Bonham CA, Gurtner GC. In vivo models for the study of fibrosis. Adv Wound Care (New Rochelle) 2019;8:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rotatori RM, Starr B, Peake M, et al. Prevalence and risk factors for hypertrophic scarring of split thickness autograft donor sites in a pediatric burn population. Burns 2019;45:1066–1074. [DOI] [PubMed] [Google Scholar]
- 3.Lingzhi Z, Meirong L, Xiaobing F. Biological approaches for hypertrophic scars. Int Wound J. 2020;17:405–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ault P, Plaza A, Paratz J. Scar massage for hypertrophic burns scarring: A systematic review. Burns 2018;44:24–38. [DOI] [PubMed] [Google Scholar]
- 5.Lee HJ, Jang YJ. Recent understandings of biology, prophylaxis and treatment strategies for hypertrophic scars and keloids. Int J Mol Sci. 2018;19:E711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coentro JQ, Pugliese E, Hanley G, Raghunath M, Zeugolis DI. Current and upcoming therapies to modulate skin scarring and fibrosis. Adv Drug Deliv Rev. 2019;146:37–59. [DOI] [PubMed] [Google Scholar]
- 7.Beanes SR, Dang C, Soo C, Ting K. Skin repair and scar formation: The central role of TGF-beta. Expert Rev Mol Med. 2003;5:1–22. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Chu J, Wen CJ, et al. Functional characterization of TRAP1-like protein involved in modulating fibrotic processes mediated by TGF-β/Smad signaling in hypertrophic scar fibroblasts. Exp Cell Res. 2015;332:202–211. [DOI] [PubMed] [Google Scholar]
- 9.Walraven M, Gouverneur M, Middelkoop E, Beelen RH, Ulrich MM. Altered TGF-β signaling in fetal fibroblasts: What is known about the underlying mechanisms? Wound Repair Regen. 2014;22:3–13. [DOI] [PubMed] [Google Scholar]
- 10.Massagué J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. [DOI] [PubMed] [Google Scholar]
- 11.Lim MJ, Ahn J, Yi JY, et al. Induction of galectin-1 by TGF-β1 accelerates fibrosis through enhancing nuclear retention of Smad2. Exp Cell Res. 2014;326:125–135. [DOI] [PubMed] [Google Scholar]
- 12.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. [DOI] [PubMed] [Google Scholar]
- 13.Kim SY, Baek KH. TGF-β signaling pathway mediated by deubiquitinating enzymes. Cell Mol Life Sci. 2019;76:653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou C, Chang Y, Korinek M, et al. The regulations of deubiquitinase USP15 and its pathophysiological mechanisms in diseases. Int J Mol Sci. 2017;18:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichhorn PJA, Rodón L, Gonzàlez-Juncà A, et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat Med. 2012;18:429–435. [DOI] [PubMed] [Google Scholar]
- 16.Inui M, Manfrin A, Mamidi A, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13:1368–1375. [DOI] [PubMed] [Google Scholar]
- 17.Galant C, Marchandise J, Stoenoiu MS, et al. Overexpression of ubiquitin-specific peptidase 15 in systemic sclerosis fibroblasts increases response to transforming growth factor β. Rheumatology (Oxford) 2019;58:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu WT, Huang KY, Lu MC, et al. TGF-β upregulates the translation of USP15 via the PI3K/AKT pathway to promote p53 stability. Oncogene 2017;36:2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuo J, Chen Z, Zhong X, Lan W, Kuang Y, Huang D. FBP1 is highly expressed in human hypertrophic scars and increases fibroblast proliferation, apoptosis, and collagen expression. Connect Tissue Res. 2018;59:120–128. [DOI] [PubMed] [Google Scholar]
- 20.Tu L, Huang Q, Fu S, et al. Aberrantly expressed long noncoding RNAs in hypertrophic scar fibroblasts in vitro: A microarray study. Int J Mol Med. 2018;41:1917–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nejaddehbashi F, Bayati V, Mashali L, et al. Isolating human dermal fibroblasts using serial explant culture. Stem Cell Investig. 2019;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Zhang X, Xie F, et al. The regulation of TGF-β/SMAD signaling by protein deubiquitination. Protein Cell 2014;5:503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song J, Li X, Li J. Emerging evidence for the roles of peptide in hypertrophic scar. Life Sci. 2020;241:117174. [DOI] [PubMed] [Google Scholar]
- 24.Xiao K, Luo X, Wang X, Gao Z. MicroRNA-185 regulates transforming growth factor-β1 and collagen-1 in hypertrophic scar fibroblasts. Mol Med Rep. 2017;15:1489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan C, Dong Y, Xie Y, et al. Shikonin reduces TGF-β1-induced collagen production and contraction in hypertrophic scar-derived human skin fibroblasts. Int J Mol Med. 2015;36:985–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhai XX, Tang ZM, Ding JC, Lu XL. Expression of TGF-β1/mTOR signaling pathway in pathological scar fibroblasts. Mol Med Rep. 2017;15:3467–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun GF, Li HC, Zhan YP, et al. SnoN residue (1-366) attenuates hypertrophic scars through resistance to transforming growth factor-β1-induced degradation. Lab Invest. 2019;99:1861–1873. [DOI] [PubMed] [Google Scholar]
- 28.Guo B, Hui Q, Xu Z, Chang P, Tao K. miR-495 inhibits the growth of fibroblasts in hypertrophic scars. Aging (Albany NY) 2019;11:2898–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinz B, Lagares D. Evasion of apoptosis by myofibroblasts: A hallmark of fibrotic diseases. Nat Rev Rheumatol. 2020;16:11–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rippa AL, Kalabusheva EP, Vorotelyak EA. Regeneration of dermis: Scarring and cells involved. Cells 2019;8:E607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamura T, Oshima Y, Hikita A. Regulation of TGF-β family signalling by ubiquitination and deubiquitination. J Biochem. 2013;154:481–489. [DOI] [PubMed] [Google Scholar]
- 32.Hanzl A, Winter GE. Targeted protein degradation: Current and future challenges. Curr Opin Chem Biol. 2020;56:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggarwal K, Massagué J. Ubiquitin removal in the TGF-β pathway. Nat Cell Biol. 2012;14:656–657. [DOI] [PubMed] [Google Scholar]
- 34.Nan L, Jacko AM, Tan J, et al. Ubiquitin carboxyl-terminal hydrolase-L5 promotes TGFβ-1 signaling by de-ubiquitinating and stabilizing Smad2/Smad3 in pulmonary fibrosis. Sci Rep. 2016;6:33116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colland F, Formstecher E, Jacq X, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286–2295. [DOI] [PubMed] [Google Scholar]
- 36.Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 2010;143:686–693. [DOI] [PubMed] [Google Scholar]
- 37.Clark M, Steger-Hartmann T. A big data approach to the concordance of the toxicity of pharmaceuticals in animals and humans. Regul Toxicol Pharmacol. 2018;96:94–105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.