Supplemental Digital Content is available in the text.
Keywords: autophagy; cell cycle; heart defects, congenital; hypoplastic left heart syndrome; induced pluripotent stem cells; unfolded protein response; whole exome sequencing
Abstract
Background:
Complex molecular programs in specific cell lineages govern human heart development. Hypoplastic left heart syndrome (HLHS) is the most common and severe manifestation within the spectrum of left ventricular outflow tract obstruction defects occurring in association with ventricular hypoplasia. The pathogenesis of HLHS is unknown, but hemodynamic disturbances are assumed to play a prominent role.
Methods:
To identify perturbations in gene programs controlling ventricular muscle lineage development in HLHS, we performed whole-exome sequencing of 87 HLHS parent–offspring trios, nuclear transcriptomics of cardiomyocytes from ventricles of 4 patients with HLHS and 15 controls at different stages of heart development, single cell RNA sequencing, and 3D modeling in induced pluripotent stem cells from 3 patients with HLHS and 3 controls.
Results:
Gene set enrichment and protein network analyses of damaging de novo mutations and dysregulated genes from ventricles of patients with HLHS suggested alterations in specific gene programs and cellular processes critical during fetal ventricular cardiogenesis, including cell cycle and cardiomyocyte maturation. Single-cell and 3D modeling with induced pluripotent stem cells demonstrated intrinsic defects in the cell cycle/unfolded protein response/autophagy hub resulting in disrupted differentiation of early cardiac progenitor lineages leading to defective cardiomyocyte subtype differentiation/maturation in HLHS. Premature cell cycle exit of ventricular cardiomyocytes from patients with HLHS prevented normal tissue responses to developmental signals for growth, leading to multinucleation/polyploidy, accumulation of DNA damage, and exacerbated apoptosis, all potential drivers of left ventricular hypoplasia in absence of hemodynamic cues.
Conclusions:
Our results highlight that despite genetic heterogeneity in HLHS, many mutations converge on sequential cellular processes primarily driving cardiac myogenesis, suggesting novel therapeutic approaches.
Clinical Perspective.
What Is New?
Exome sequencing and in vivo cardiomyocyte transcriptome analyses suggest intrinsic disturbances of specific gene programs contributing to the pathogenesis of hypoplastic left heart syndrome (HLHS) during fetal ventricular cardiogenesis and argue against a solely hemodynamic origin.
Patient induced pluripotent stem cell models of specific HLHS anatomic subsets indicate a primary disease onset occurring during initial stages of cardiogenesis in cardiac progenitors and translating later into disrupted cardiomyocyte subtype lineage specification and maturation.
Electromechanical preconditioning of ventricular HLHS induced pluripotent stem cell–derived cardiomyocytes in 3D heart patches revealed that impaired maturation limits their ability to respond to growth cues, resulting in premature cell cycle exit and increased apoptosis, contributing to left ventricular hypoplasia.
What Are the Clinical Implications?
Our results indicate that a shared, cell-autonomous mechanism exists for discrete pathologic subsets of HLHS, extending the classical “flow-volume hypoplasia” model.
The existence of intrinsic cardiomyocyte defects in distinct HLHS subsets offers an explanation for the unpredictable results of in utero aortic valvuloplasty as treatment to enable left ventricular growth and suggests a path toward rational therapeutic catheter-based interventions.
Knowledge of these specific molecular defects may inform strategies, either by stratification or adjunctive therapies, permitting biventricular repair in less serious forms of left ventricular hypoplasia.
Hypoplastic left heart syndrome (HLHS) is a severe form of congenital heart disease (CHD) characterized by underdevelopment of left-sided cardiac structures, including left ventricle (LV) hypoplasia, hypoplastic ascending aorta, intact interventricular septum, and aortic or mitral valve atresia or stenosis, or both, in the setting of concordant ventriculoarterial connections.1–3 Although a genetic etiology is supported by an increased recurrence risk and familial clustering, the largely sporadic occurrence suggests a complex genetic model.4,5
Historically, ventricular and aortic hypoplasia in HLHS has been attributed to reduced growth arising as consequence of restricted blood flow caused by maldeveloped mitral or aortic valve.6,7 Recently, an anatomic study of HLHS hearts indicated that ventricular and valvular morphology are poorly correlated and 3 phenotypic LV variants (slit-like, thickened, and miniature) are present, extending earlier suggestions that a pathogenic mechanism based on reduced blood flow alone may be insufficient.8,9 In the Ohia mouse, specific mutations drive LV hypoplasia by perturbing cardiomyocyte proliferation and differentiation.10 Ohia mutants display aspects of HLHS, although some can have double-outlet right ventricle that does not uniformly resemble human HLHS. Transcriptional and structural alterations found in the LV of the HLHS Ohia mice are present, though less severe, in the right ventricle (RV).10,11 This suggests that intrinsic defects in myogenic programs of both ventricles may manifest differently, depending on combinations of the cardiac progenitor (CP) population affected and physiologic milieu.
During cardiogenesis, 2 main CP lineages provide cardiomyocytes to the developing heart with distinct temporal and spatial contributions. The first heart field (FHF) CPs are fated to differentiate early, forming the primitive heart tube, the LV, and portion of the atria; the second heart field cells show delayed differentiation into cardiomyocytes and represent initially a reservoir of multipotent CPs.12,13 Later, second heart field CPs give rise to the RV, the proximal outflow tract, and part of the atria.14
Work using cardiomyocytes derived from human induced pluripotent stem cells (iPSCs) has begun to elucidate molecular mechanisms of CHD in patient-specific in vitro models of cardiogenesis.15,16 Studies in iPSC-derived cardiomyocytes (iPSC-CMs) from patients with HLHS carrying variants in the Notch signaling pathway demonstrated impaired differentiation17–19; other studies implicated transcriptional repression of NKX2-5, HAND1, and NOTCH1 or activation of atrial gene programs.20,21 Lacking is a unifying picture of how the profound genetic heterogeneity of HLHS converges in common perturbations of sequential cellular processes driving heart morphogenesis, how these processes are altered, and how such alterations contribute to the disease.
We combined whole-exome sequencing (WES) of parent–offspring trios, transcriptome profiling of cardiomyocytes from ventricular biopsies, iPSC-derived CP/cardiomyocyte models of 2D/3D cardiogenesis, and single-cell gene expression analysis to decode the cellular and molecular principles of HLHS phenotypes. Our results show that initial aberrations in the cell cycle/unfolded protein response (UPR)/autophagy hub lead to disrupted CP lineage commitment. Consequently, impaired maturation of ventricular cardiomyocytes limits their ability to respond to growth cues, resulting in premature cell cycle exit and increased apoptosis under biomechanical stress in 3D heart structures. Together, these studies provide evidence that HLHS pathogenesis is not exclusively of hemodynamic origin and reveal novel potential nodes for rational design of therapeutic interventions.
Methods
Data Availability
All data needed to evaluate the conclusions of this study are present in the article or the Data Supplement. The raw omics data are deposited at publicly available databases (bulk and single-cell RNA sequencing, #GSE135411; nuclear RNA sequencing, #PRJNA353755; proteomics, #PXD014812).
Patients and Controls
All patients with HLHS harbored hypoplastic LV and ascending aorta, mitral or aortic valve atresia or stenosis, and intact interventricular septum. Two additional patients with LV hypoplasia and a ventricular septal defect not meeting strict HLHS criteria were included in the WES analysis but omitted from further studies.
Blood for WES was collected from 87 probands with sporadic HLHS and their respective parents (clinical information summarized in Table IA in the Data Supplement). Human ventricle samples from controls and patients with HLHS were obtained from aborted fetuses or during Norwood Stage I palliation. All donors or their legal representatives provided informed consent. The study was performed according to the Helsinki Declaration and approved by local ethics committees at the respective institutions (KaBi-DHM [cardiovascular biobank, German Heart Center Munich]: 5943/13, 247/16S; Institute of Human Genetics: 5360/13; Human Subjects Management: 15–00696).
Statistics
Statistical analyses were performed using R and GraphPad Prism. Analysis of variance (post hoc Tukey honestly significant difference) or the Student t test were used for normally distributed data and Kruskal-Wallis (post hoc Dunn test) or Mann-Whitney U tests for non-normally distributed data, as appropriate. Categorical data were analyzed using χ2 tests. P values and P-adjusted <0.05 were considered statistically significant.
Results
Damaging De Novo Mutations in Patients With HLHS Are Linked to Cardiac Development, Chromatin Organization, and Cell Cycle Phases
WES from 87 patients with sporadic HLHS and their parents recovered 94 nonsynonymous de novo mutations (DNMs), of which 12 were loss-of-function and 76 predicted damaging missense variants (Table IB in the Data Supplement). A total of 1 to 5 nonsynonymous DNMs were found in 56 patients (Figure IA and IB in the Data Supplement). Among the 51 patients with damaging DNMs (D-DNMs), 34 carried mutations in genes or HGNC gene families that harbored multiple hits (Figure 1A). D-DNMs in MYRF, MACF1, and LRP6 were found twice, with 2 individuals carrying variants in 2 of the 3 genes (Figure 1A). MYRF was recently associated with CHD22 and HLHS.23 LRP6 and MACF1 are linked to Wnt signaling.24,25 Gene families with multihits were likewise related to pivotal signaling pathways implicated in cardiac development (Hedgehog, FGF, and Notch) or to the histone modification H3K4me-H3K27me pathway that has been associated with CHD.26,27
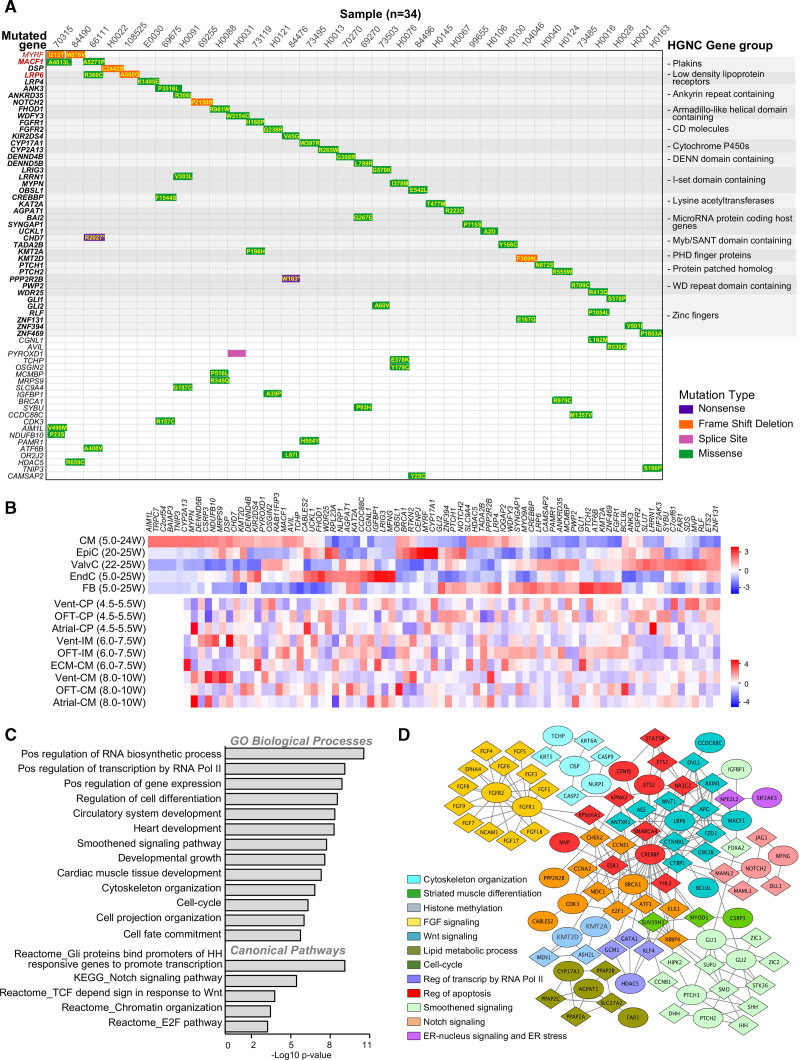
Figure 1.
Characterization of damaging de novo mutations in patients with hypoplastic left heart syndrome. A, Waterfall plot of de novo multihit genes (red) or gene family (bold) identified in the hypoplastic left heart syndrome (HLHS) cohort. “Multihit” refers to the occurrence/presence of variants in the same gene or family of genes among different individuals. Additional de novo genes in each subject are in plain black. B, Cell type–specific expression of HLHS damaging de novo mutation (D-DNM) genes based on single-cell RNA sequencing from normal fetal hearts between 4.5 and 25 weeks (W) of gestation. Data from Ciu et al.29 and Sahara et al.28 were used to generate the upper and lower heatmap, respectively. C and D, Bar chart of Gene Ontology (GO) enrichment analysis (C) and protein–protein network analysis (D) of D-DNM genes. In D, each Netbox module is coded by a different color, with mutated genes illustrated as circles and linker genes as diamonds. CM indicates cardiomyocytes; CP, cardiac progenitor; ECM, extracellular matrix; EndC, endothelial cells; EpiC, epicardial cells; ER, endoplasmic reticulum; FB, fibroblasts; HGNC, HUGO Gene Nomenclature Committee; IM, intermediates; OFT, outflow tract; ValvC, valvular cells; and Vent, ventricular.
Analyzing single-cell transcriptomics from human embryonic (4.5 to 25 weeks postconception) cardiac cells,28,29 we found 96% of the 84 D-DNM genes expressed in different cell types, significantly more than random gene sets (76.7%±8.8%; P<10e-15). Interestingly, high expression was observed in at least 1 stage of chamber or outflow tract development (Figure 1B and Figure II in the Data Supplement), suggesting an intrinsic cardiomyocyte lineage dysfunction as a possible determinant of HLHS. Gene set enrichment analysis (GSEA) of D-DNM genes revealed a significant enrichment in gene categories related to cardiac and embryonic development and growth, cell fate commitment and differentiation, as well as cell cycle and G1/S phase transition (E2F pathway; Figure 1C and Table IC in the Data Supplement). Network analysis identified 12 interconnected protein–protein interaction modules (0.63 modularity; 11.84 scaled modularity), significantly different from random networks (0.36±0.023 modularity [mean±SD; n=100]; P<10e-05), comprising 32 of the 84 D-DNM genes (Figure 1D). Genes in these modules act in biological processes critical for CP specification and cardiomyocyte maturation, including cell cycle and apoptosis; response to endoplasmic reticulum (ER) stress; signaling by Smoothened, Wnt, FGF, and Notch; transcriptional regulation; and histone methylation.
We interrogated 459 independent HLHS parents–offspring trios within the CHD cohort of the Pediatric Cardiac Genomics Consortium27 for de novo variants. The occurrence of nonsynonymous DNMs and the distribution of variant classes were comparable, with 3 genes related to known syndromic CHD forms (CHD7, KMT2D, MYRF) being common D-DNM multihits (Figure IA through IC in the Data Supplement). GSEA and network interactome analyses of the combined cohort yielded similar results to our cohort alone (Figure ID and IE in the Data Supplement). Compared with 1789 control trios comprising parents and unaffected siblings of probands with autism,30 most GSEA-retrieved pathways showed substantial burden enrichment in D-DNMs among cases (Figure ID in the Data Supplement). Randomization of DNMs between HLHS cases and controls confirmed the nonstochastic nature of our results (Table ID in the Data Supplement).
Dysregulated Genes in Infant HLHS Cardiomyocytes Belong to Dynamic Transcriptional Networks During Cardiogenesis
To verify dynamic expression of HLHS D-DNM genes during cardiomyocyte development and identify temporal and spatial processes of cardiogenesis critical to HLHS pathogenesis, we performed nuclear RNA sequencing (RNA-Seq) of RV cardiomyocytes from healthy hearts at 3 developmental stages—fetal (16 to 23 weeks postconception), infancy (10 to 40 weeks), and adulthood (21 to 50 years)—and from infants with HLHS (5 to 11 days; Figure 2A and Table IIA in the Data Supplement). The choice of RV tissue was based on the fact that LV is inaccessible from living patients with HLHS and that both ventricles may share altered transcriptional signatures, as found in Ohia mutants.
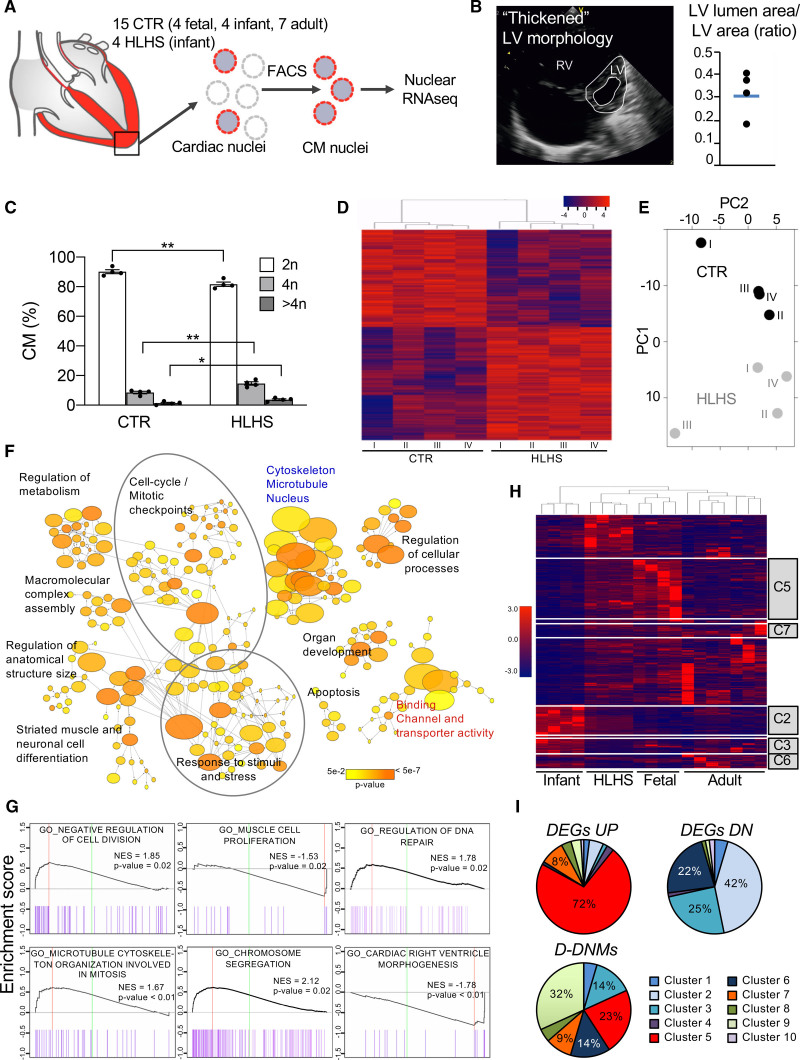
Figure 2.
Gene expression analysis of cardiomyocyte nuclei from hypoplastic left heart syndrome and control hearts. A, Workflow of cardiomyocyte nuclei isolation for RNA sequencing (RNA-Seq). B, Representative echocardiogram of patients with hypoplastic left heart syndrome (HLHS) with a distinct left ventricular (LV) phenotype. Dot plot shows the ratio of LV lumen area/LV area for the 4 patients with HLHS and the average value (blue line). C, Ploidy level of cardiomyocyte nuclei in HLHS and control (CTR) hearts. Data are mean±SEM. *P<0.05, **P<0.01 (t test). D, Heatmap depicting normalized RNA-Seq expression values of differentially expressed genes (DEGs; 1.5-fold expression, P≤0.05). Gene regulations are reported as a color code and hierarchical clustering result as a dendrogram. E, Principal component analysis performed on rlog-normalized (DESeq2) counts for all nuclear RNA-Seq samples. F, Network visualization of the enriched Gene Ontology (GO) of HLHS DEGs using the Cytoscape plugins BinGO. Nodes represent enriched GO terms, node size corresponds to the gene number, and color intensity to the P value. Edges represent GO relation of biological process (black), molecular function (red), and cellular component (blue). G, Representative enrichment plots from gene set enrichment analysis. Normalized enrichment score (NES) and P value are specified. H, Heatmap illustrating the expression of HLHS DEGs during fetal, infant, and adult stages of normal cardiac development. The dendrogram shows clustering of the HLHS infant samples with control fetal samples. Genes belonging to developmentally regulated gene clusters from Figure IIIC in the Data Supplement are highlighted. I, Pie charts showing the percentage of HLHS upregulated (DEGs UP), downregulated (DEGs DN), and damaging de novo affected (D-DNM) genes belonging to the developmentally regulated gene clusters from Figure IIIC in the Data Supplement. CM indicates cardiomyocytes; FACS, fluorescence-activated cell sorting; and RV, right ventricle.
We first compared RV tissues from 4 healthy infants and 4 infants with HLHS, who presented thickened morphologic features of the hypoplastic LV8,9 (Figure 2B). An equal proportion of cardiomyocyte nuclei was found in both groups (Figure IIIA in the Data Supplement). HLHS cardiomyocytes showed a substantial increase in tetraploid nuclei at the expense of diploid (Figure 2C). Because polyploidization accompanies terminal cardiomyocyte differentiation and permanent cell cycle withdrawal,31 this suggests premature cell cycle arrest in HLHS. RNA-Seq analysis revealed 2286 differentially expressed genes (DEGs) in patients with HLHS compared with controls (Figure 2D and Table IIB in the Data Supplement). Comparative principal component analysis separated patients with HLHS and controls (Figure 2E). Enrichment map of the Gene Ontology categories recovered from DEGs demonstrated high interconnection of functionally related gene sets associated with cell cycle and mitotic checkpoints, response to stimuli and stress, organ development, cell differentiation and apoptosis, cytoskeleton/microtubule/nucleus, and regulation of metabolism (Figure 2F and Table IIC in the Data Supplement). Most pathways overlapped with the modules recovered from the HLHS D-DNM genes. In HLHS, GSEA revealed reduced expression of genes important for cardiac ventricle morphogenesis and increased expression of genes controlling DNA repair (Figure 2G and Table IID in the Data Supplement). Moreover, negative regulators of cell division were upregulated, whereas genes associated with muscle cell proliferation were down, supporting the notion of premature cell cycle exit.
To assess whether DEGs and D-DNM genes in HLHS are dynamically regulated during development, we next established a global expression atlas of fetal, infant, and adult healthy RV genes. Hierarchical clustering demonstrated distinct expression profiles among developmental stages (Figure IIIB in the Data Supplement). Investigation of DEGs identified 10 transcriptome clusters dynamically changing during cardiac development (Figure IIIC in the Data Supplement). DEGs in infants with HLHS resembled the fetal rather than the infant stage (Figure 2H). A total of 60% of HLHS downregulated DEGs belonged to dynamic transcriptome clusters, being 56% distributed in gene clusters 1, 2, 3, and 6. These contained genes upregulated from the embryonic to newborn phases and mainly involved in cellular transport and metabolic processes (Figure 2I and Figure IIIC and Table IIE in the Data Supplement). A total of 21% of the upregulated transcripts fitted to clusters 5 and 7, representing genes regulating morphogenesis and cell cycle and whose expression levels decreased in infant stage (Figure 2I and Figure IIIC and Table IIE in the Data Supplement). A total of 22 of the 84 D-DNM genes in our HLHS cohort were found in most clusters.
Together, these findings indicate that HLHS may result from alterations in specific gene programs critical during fetal ventricular cardiogenesis and suggest a role of cell cycle and cardiomyocyte maturation as potential disease drivers.
Specific Gene Networks Are Dynamically Altered During Differentiation of HLHS iPSCs Toward CPs and Cardiomyocytes
To better determine to which extent transcriptional alterations of native HLHS cardiomyocytes underlie disease pathogenesis rather than physiologic differences at the time of RV sampling, we generated iPSCs from 3 patients with HLHS and 3 healthy controls (Figures IV and V in the Data Supplement) and mechanistically analyzed developmental processes during early in vitro cardiogenesis. Selection of patients with HLHS was based on echocardiographic LV morphology (thickened phenotype) and presence of D-DNMs in multihit genes or gene families (Figure VI in the Data Supplement).
Control and HLHS iPSC lines were directed to early CPs32 and cardiomyocytes33 using stepwise differentiation protocols (Figure 3A). We performed RNA-Seq at various time points and explored functional characteristics of HLHS DEGs by GSEA (Figure 3B and Table IIIA in the Data Supplement). Dynamic alterations in several gene categories were common in both CP and cardiomyocyte differentiation protocols, including heart and aorta development, cell cycle, and chromatin modification. Unique aberrations in autophagy terms were present when directing HLHS iPSCs toward early CPs, whereas apoptosis-associated pathways appeared solely affected in later CPs (day 6) and cardiomyocytes (day 8 and day 14; Figure 3B and Table IIIB in the Data Supplement).
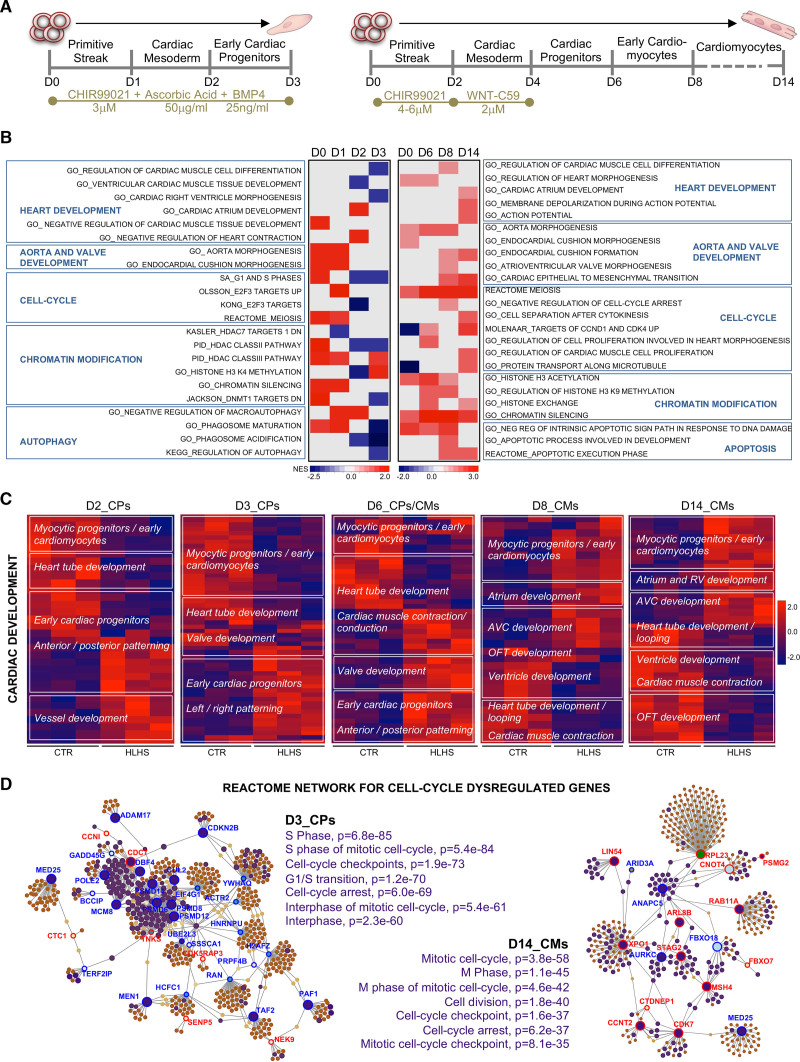
Figure 3.
Gene expression analysis during induced pluripotent stem cell–based cardiogenesis reveals networks of dysregulated genes in hypoplastic left heart syndrome. A, Directed induced pluripotent stem cell cardiac differentiation protocols used in the study. B, Heatmap of normalized enrichment scores for selected gene set enrichment analysis terms. Red and blue denote terms with positive and negative normalized enrichment scores, respectively. C, Heatmaps showing gene expression of differentially expressed genes (1.5-fold expression, P≤0.05) involved in cardiac development at the indicated days of differentiation. Values are row-scaled to show their relative expression. Blue and red are low and high levels, respectively. D, Network analyst–generated protein–protein interactome of differentially expressed genes involved in cell cycle at day 3 and day 14. Upregulated (red) and downregulated (blue) genes are shown. In purple are highlighted the genes belonging to the enriched Gene Ontology categories specified on the side of the plots. Protein–protein interactions are indicated as solid gray lines between genes. AVC indicates atrioventricular canal; CM, cardiomyocytes; CP, cardiac progenitor; CTR, control; HLHS, hypoplastic left heart syndrome; OFT, outflow tract; and RV, right ventricle.
In CPs at day 2 and day 3, detailed analysis of DEGs involved in cardiac development revealed that genes expressed in committed myocytic precursors and important for heart tube formation (ID2, TPM2, XIRP1, SRF, ETV1) were downregulated in HLHS, whereas genes involved in anterior and posterior patterning (HOXB9) and in vessel and valve development (VEGFB, TGFB2, GATA5) were upregulated (Figure 3C). Transcripts typical of early CPs were decreased at day 2, but augmented at day 3, suggesting incomplete or delayed CP lineage specification. At day 8 and day 14, HLHS cardiomyocytes showed upregulation of genes distinctive of myocytic progenitors and early immature cardiomyocytes and altered expression of transcripts important for outflow tract and atrioventricular canal (MEIS1, ISL1, TGFB2, JUN) as well as heart chamber development (NR2F1, WNT2, ETV2, RXRA; Figure 3C and Table IIIC in the Data Supplement), supporting dysregulated lineage-specific cardiomyocyte differentiation.
In CPs at day 3, 74 of the 890 DEGs related to cell cycle Gene Ontology categories (Figure VIIA and Table IIID and IIIE in the Data Supplement). Thirty-four of them generated a functional interactome network encompassing cell cycle interphase pathways as the top enriched, with most leading genes being downregulated (Figure 3D and Table IIIE in the Data Supplement). In cardiomyocytes at day 14, cell cycle DEGs (42/754) generated 18 functional interaction nodes; top enriched terms within the interactome and regulation of the leading genes pointed to alteration in M phase, with active separation of chromatids (STAG2, XPO1) but defective progression through mitosis (ANAPC5) and cytokinesis (AURKC; Figure 3D and Table IIID in the Data Supplement). Cell cycle defects in HLHS cardiomyocytes were confirmed by proteomic analysis (Expanded Results, Figure VIII, and Table IIIF–IIIJ in the Data Supplement).
Together, the specific transcriptional alterations detected during early CP specification and cardiomyocyte differentiation of HLHS iPSCs suggest the primary onset of the disease occurs at the initial stages of cardiogenesis when cardiomyocyte lineage decisions arise within CP populations.
Defects in UPR–Induced Autophagy Lead to Delayed and Disrupted CP Lineage Specification
To investigate the role of cell cycle disturbances in HLHS pathogenesis, we analyzed cell cycle patterns during early CP formation and compared them with the emergence of CP lineages marked by ISL1, NKX2-5, and TBX5. Whereas control lines demonstrated G1 lengthening starting between day 1 and day 2, patient lines had a prolonged G1 phase with a 24-hour delay (Figure 4A). Concurrently, at day 1, activation of ISL1 and NKX2-5 transcripts was dramatically reduced in HLHS (Figure VIIB in the Data Supplement) and correlated with significantly lower proportions of cells expressing ISL1 and NKX2-5 proteins at day 2 (Figure 4B), supportive of a retarded CP specification. Four patterns of ISL1 and NKX2-5 expression were detected at day 3: ISL1low/NKX2-5low, representing early committed CPs; ISL1high/NKX2-5high, denoting fully committed early CPs; ISL1high/NKX2-5low, typical of second heart field progenitors; and ISL1low/NKX2-5high, distinctive of FHF cells (Figure 4C). TBX5, a specific FHF marker, was mainly found in ISL1low/NKX2-5high cells (Figure 4C), confirming FHF identity. The relative distribution of CP subgroups was altered in HLHS settings (Figure 4D). All HLHS lines failed to upregulate TBX5 during CP specification (Figure VIIB in the Data Supplement) and few of the ISL1low/NKX2-5high FHF cells expressed TBX5 (Figure 4E), indicating common defective transcriptional programs within the FHF lineage. To assess the contribution of apoptosis and proliferation to the observed CP phenotypes, we analyzed caspase-3 activation and EdU incorporation. Apoptosis was barely detectable in both HLHS and controls (Figure VIIC in the Data Supplement), arguing against any cell selection of HLHS CPs. Consistent with the alteration in cell cycle patterns, global changes in cell proliferation rates between controls and HLHS were only significantly different at day 2 (Figure VIID in the Data Supplement). However, when analyzed separately at day 3, ISL1low/NKX2-5high FHF progenitors demonstrated higher proliferation in all HLHS lines (Figure 4F), consistent with the reported role of TBX5 as negative regulator of cell proliferation during early cardiac development.34 Collectively, these data indicate common defects in CP lineage commitment and imbalance of both progenitor fields during initial steps of HLHS cardiogenesis.
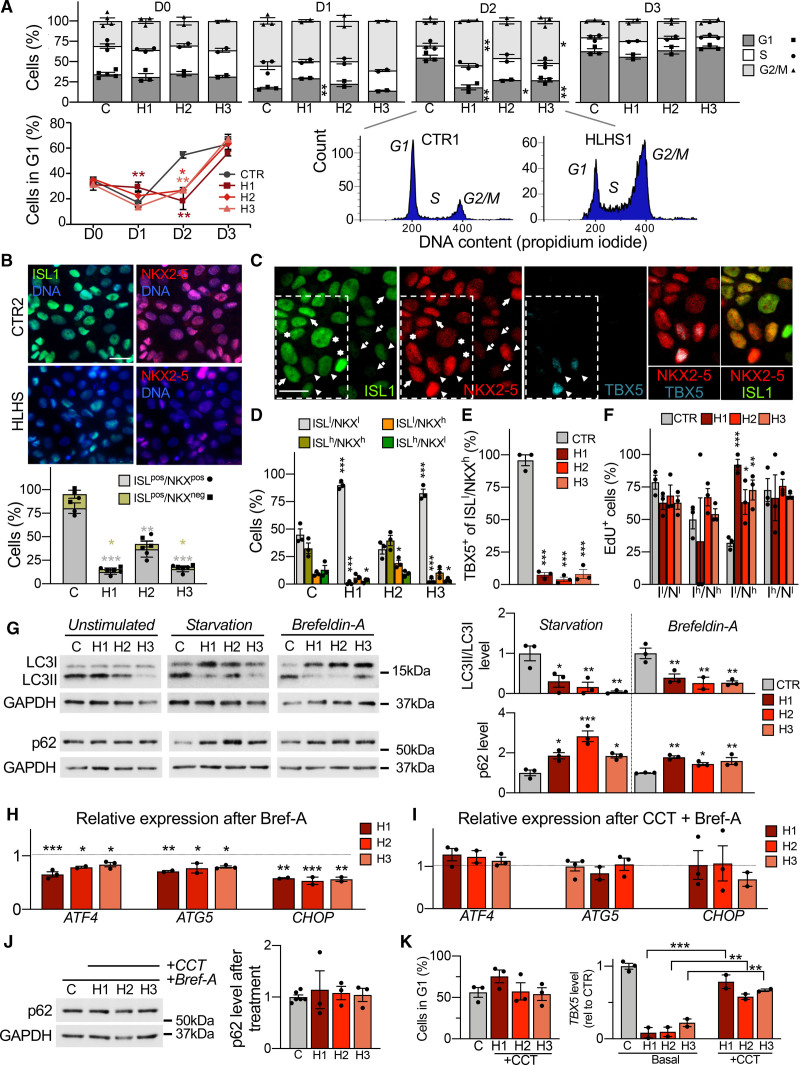
Figure 4.
Defects in unfolded protein response and autophagy delay and disrupt cardiac progenitor lineage specification. To better discriminate possible phenotypic variations among diseased lines, functional results from each hypoplastic left heart syndrome (HLHS) line are shown separately. Data from control lines have been combined. A, Propidium iodide staining analysis of HLHS (H) and control cells (C) during cardiac progenitor (CP) differentiation. Data are mean±SEM; 2 to 4 differentiations per line, N≥20 000 cells per sample at each time point. *P<0.05, **P<0.01 compared with controls (CTR; 2-way analysis of variance [ANOVA]). B, Immunofluorescence analysis of ISL1 and NKX2-5 in HLHS and control CPs at day 2. Scale bar, 25 µm. Data are mean±SEM; 431 (CTR), 463 (HLHS1), 442 (HLHS2), and 396 (HLHS3) cells from 3 differentiations per line. *P<0.05, **P<0.01, ***P<0.001 compared with CTR (1-way ANOVA). C, Representative immunofluorescence of ISL1, NKX2-5, and TBX5 in control CPs (CTR3) at day 3. Four ISL1/NKX2-5 expression patterns are highlighted: ISL1low/NKX2-5low (dotted arrows), ISL1high/NKX2-5high (arrows), ISL1high/NKX2-5low (asterisks), and ISL1low/NKX2-5high (arrowheads). Scale bar, 20 µm. D, Distribution of cells with ISL1/NKX2-5 expression patterns from (C) in HLHS and control CPs at day 3. Data are mean±SEM; 369 (CTR), 347 (HLHS1), 322 (HLHS2), 357 (HLHS3) cells from 3 differentiations per line. *P<0.05, ***P<0.001 compared with CTR (1-way ANOVA). E, Percentage of ISL1low/NKX2-5high cells expressing TBX5 in HLHS and control CPs at day 3. Data are mean±SEM; 114 (CTR), 70 (HLHS1), 182 (HLHS2), and 123 (HLHS3) cells from 3 differentiations per line. ***P<0.001 compared with CTR (1-way ANOVA). F, Quantification of EdU+ cells in HLHS and control CP subpopulations at day 3. Data are mean±SEM; 369 (CTR), 347 (HLHS1), 322 (HLHS2), 357 (HLHS3) cells from 3 differentiations per line. *P<0.05, **P<0.005, ***P<0.001 compared with CTR (1-way ANOVA). G, Western blot of LC3 and p62 in HLHS and control CPs at day 3 with and without starvation or brefeldin-A. For detection of LC3, all 3 conditions were carried out in presence of chloroquine. Data are mean±SEM; 2 to 3 differentiations per line. *P<0.05, **P<0.01, ***P<0.001 compared with CTR (1-way ANOVA). H and I, Expression analysis of ATF4 and its downstream targets ATG5 and CHOP in HLHS and control CPs at day 3 after treatment with brefeldin-A alone (H) or in combination with the PERK activator CCT020312 (I). Shown are expression levels relative to controls. Data are mean±SEM; 2 to 4 differentiations per line. *P<0.05, **P<0.01, ***P<0.001 compared with CTR (1-way ANOVA). J, Western blot of p62 in HLHS and control CPs at day 3 after treatment with brefeldin-A and CCT020312. Data are mean±SEM; 3 to 6 differentiations per line. K, Propidium iodide staining–based quantification of cells in G1 phase in HLHS and control CPs at day 2 (left) and TBX5 expression by qRT-PCR at day 3 (right) after 6-hour treatment of HLHS cells with CCT020312 at day 1.5. Data are mean±SEM; 2 to 3 differentiations per line, N≥20 000 cells per sample in left panel. **P<0.01, ***P<0.001 compared with own basal (1-way ANOVA).
Autophagy and cell cycle are coordinated and reciprocally regulated.35 Because autophagy was altered in HLHS CPs from day 1 on (Figure 3B), we measured autophagic flux in cells at day 3 by analyzing the levels of LC3II, a marker of autophagosomes, and p62, a substrate for autophagic degradation. Under basal conditions, LC3II and p62 proteins were normal in all HLHS lines (Figure 4G and Figure VIIE in the Data Supplement). However, after activation of autophagy (starvation or brefeldin-A), defective autophagosome formation and p62 degradation were evident in all HLHS lines (Figure 4G). This was confirmed by Cyto-ID staining of autophagic vacuoles (Figure VIIF in the Data Supplement). Brefeldin-A triggers autophagy through ER stress and activation of the UPR,36 a pathway also challenged by starvation. UPR plays an important role in cell fate acquisition of embryonic stem cells and progenitors.37–39 Given the recovery of HLHS D-DNMs in ER-stress/UPR genes (Figure 1D and Table IB in the Data Supplement), we asked whether the defective autophagy in HLHS CPs is caused by impaired UPR. On ER stress, UPR is transduced by derepression of 3 ER membrane proteins—IRE1α, PERK, and ATF6—that work alone or in concert to restore normal cellular function.40 Neither an increased splicing of XBP1 mRNA, occurring downstream of IRE1α stimulation, nor activation of ATF6 were altered in HLHS CPs at day 3 on brefeldin-A treatment (Figure VIIG in the Data Supplement). Instead, we measured a specific common defect of all HLHS lines in activating the PERK pathway, as indicated by decreased PERK-mediated phosphorylation of eIF2α (Figure VIIH in the Data Supplement) and reduced increase of ATF4 and its downstream targets including CHOP (Figure 4H and Figure VIIG in the Data Supplement). A 6-hour treatment of HLHS cells with the selective PERK activator CCT020312 normalized ATF4 and CHOP levels and rescued the defective activation of autophagy (Figure 4I and 4J). Early application of CCT020312 at day 1.5 of CP differentiation was sufficient to revert the HLHS phenotype, as indicated by normalization of the number of cells in G1 at day 2 and the TBX5 level at day 3 (Figure 4K). Conversely, HLHS-like disturbances in G1-phase lengthening and TBX5 upregulation could be induced in control cells by inhibiting autophagy at day 1.5 using chloroquine, with no influence on ISL1 expression (Figure VIII in the Data Supplement).
Taken together, these results suggest that defects in UPR and autophagy activation in the early phase of CP specification contribute to delayed and disrupted CP lineage commitment in HLHS.
Single-Cell RNA-Seq Defines Impaired Cardiomyocyte Lineage Segregation and Maturation in HLHS
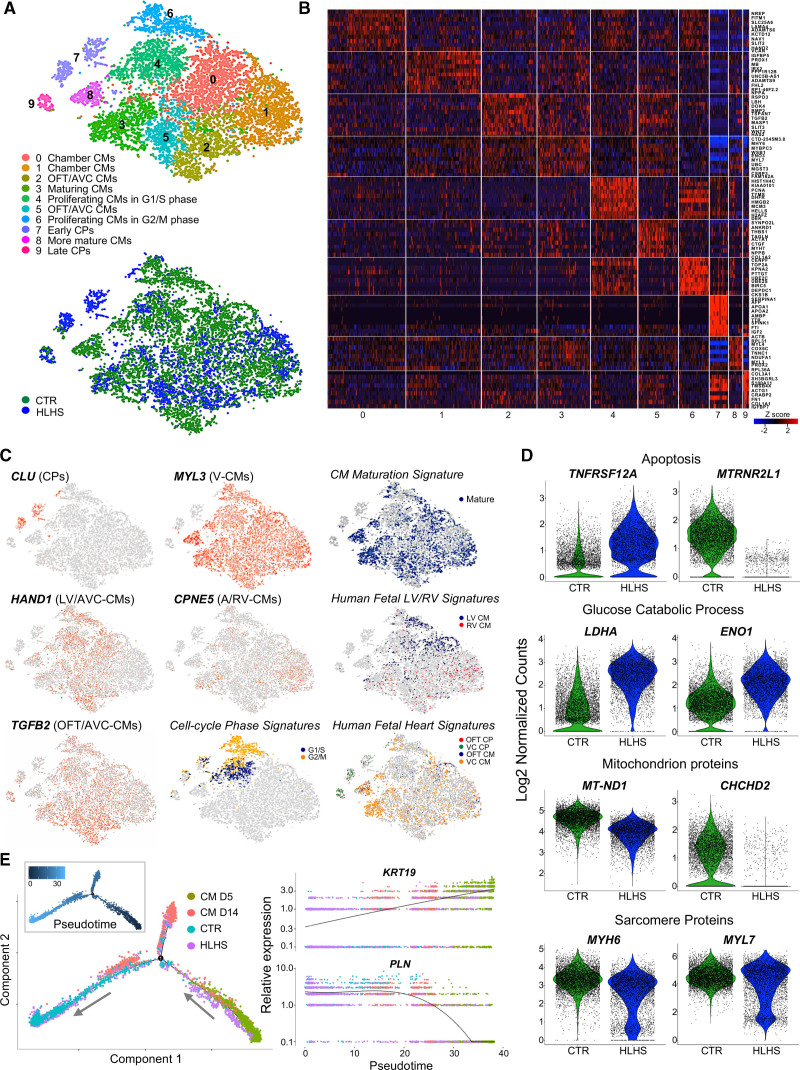
To investigate the consequences of altered CP specification on cardiomyocyte subtype formation, we performed single-cell RNA-Seq of early cardiomyocytes at day 14. Transcriptomes of 6431 control and 4439 HLHS cells were recovered. Unsupervised clustering analysis identified 10 distinct subpopulations (clusters 0–9; Figure 5A and Table IIIK in the Data Supplement). We assigned identities to each population by cross-referencing the most highly and uniquely expressed genes in each cluster with known cardiac subtype markers from human and mouse single-cell studies (Figure 5B and Expanded Results in the Data Supplement).28,29,41–43 We captured transcriptome characteristics of primary (outflow tract and atrioventricular canal) and chamber myocardium, early and late CPs, maturing cardiomyocytes, and proliferating cardiomyocytes in G1/S and G2/M phases (Figure 5A–5C and Figure IXA in the Data Supplement). HLHS cells contributed almost exclusively to CP clusters (cluster 7: controls, 40; HLHS, 450; cluster 9: controls, 35; HLHS, 117) and were strongly underrepresented in the cluster containing terminally differentiated cardiomyocytes (cluster 8: controls, 318; HLHS, 37), together suggestive of an intrinsic differentiation delay. Moreover, we noticed a significant reduced proportion of HLHS cells in the proliferating cell clusters (cluster 4: controls, 922; HLHS, 284; cluster 6: controls, 675; HLHS, 103), corroborating a premature cell cycle exit in HLHS cardiomyocytes. Coexpression of LV-enriched genes was observed mainly in cardiomyocytes of cluster 0, which was scarce in HLHS cells (cluster 0: controls, 1446; HLHS, 497). This correlated with an overall downregulation of LV transcripts in HLHS (TBX5, HAND1, SLIT2; Figure IXB in the Data Supplement), hinting to a potentially reduced LV commitment. Activated UPR program was detected exclusively in CPs (Figure X in the Data Supplement). Differential expression analysis within each cluster revealed increased levels of proapoptotic genes (TNFRSF12A, FAM162A) and reduced expression of antiapoptotic regulators (MTRNR2L1) in HLHS cardiomyocytes (Figure 5D and Figure IXC in the Data Supplement), confirming bulk RNA-Seq results. Moreover, HLHS cardiomyocytes presented an overall upregulation of genes involved in glucose catabolism, downregulation of mitochondrial transcripts, and lower expression of sarcomeric genes; exceptions were MYL7 and TNNI1, isoforms highly expressed in immature cardiomyocytes (Figure 5D and Figure IXC and IXD in the Data Supplement), collectively suggesting reduced cellular differentiation and maturation. To verify this, we used recently published single-cell profiles of healthy iPSC-CMs at different stages of differentiation.44 We ordered cardiomyocytes captured at day 5 and day 14 together with our cells in pseudotime. The resulting differentiation trajectory began with cardiomyocytes at day 5 and bifurcated into 2 lineages where cardiomyocytes at day 14 were allocated (Figure 5E). A large proportion of HLHS cardiomyocytes, although at day 14, clustered with day 5 cardiomyocytes and expressed high level of KRT19, a gene specific of early immature cardiomyocytes (Figure 5E).
Figure 5.
Single-cell RNA sequencing of induced pluripotent stem cell–derived cardiomyocytes reveals defects in cardiac lineage segregation and maturation in hypoplastic left heart syndrome. A, t-Distributed stochastic neighbor embedding plot of all hypoplastic left heart syndrome (HLHS) and control (CTR) cell populations captured at day 14 colored by cluster identity (top) and genotype (bottom). Data are from 2 control (CTR2 and CTR3) and 2 HLHS (HLHS2 and HLHS3) lines. B, Heatmap showing Z score scaled average expression levels of the top 10 differentially expressed genes for each cellular cluster. C, Expression of selected genes marking subpopulations on t-distributed stochastic neighbor embedding plot. Single gene panels: red and gray indicate high and low expression, respectively. Signature panels: color key indicates cells matching with the gene signatures tested (see Expanded Methods in the Data Supplement). D, Violin plots of selected differentially expressed genes between CTR and HLHS cells. All genes represented have P<0.05. E, Branching analysis of HLHS and CTR cardiomyocytes at day 14 together with cardiomyocytes at day 5 and day 1444 colored by genotype and estimated pseudotime along the inferred cell trajectory (inset). Pseudotime dynamics of early (KRT19) and mature (PLN) cardiomyocyte genes in dependence on inferred cell identity. A, atria; VC, ventricular chamber. AVC indicates atrioventricular canal; CM, cardiomyocyte; CP, cardiac progenitor; LV, left ventricle; OFT, outflow tract; and RV, right ventricle.
Together, these results indicate that both early cardiomyocyte subtype lineage specification and cardiomyocyte differentiation and maturation are disrupted in HLHS. Moreover, they confirm, at a single-cell level, impaired UPR in early diseased CPs and premature cell cycle withdrawal of cardiomyocytes.
3D Heart Patches Unravel Dysregulated Nodes for Cardiomyocyte Contractility, Maturation, and Survival in HLHS
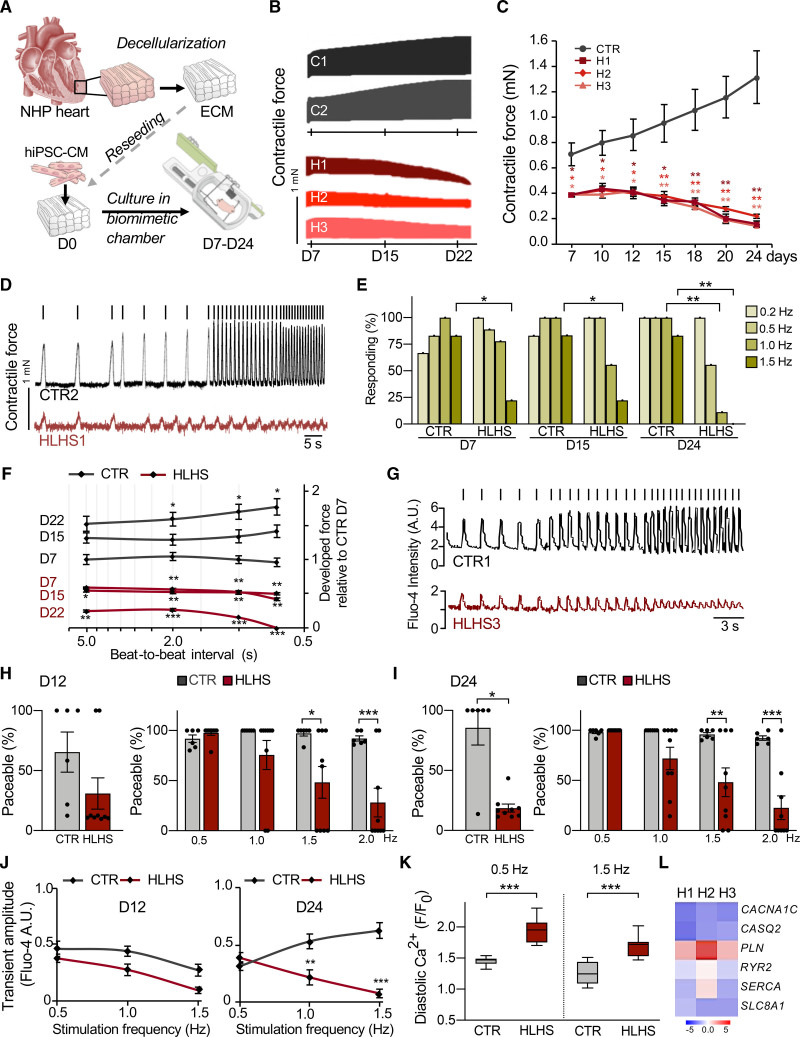
We analyzed HLHS phenotypes in a multicellular 3D context by repopulating decellularized scaffolds from nonhuman primate LV heart slices with early day 14 iPSC-CMs. Standardized cell seeding was achieved using bioprinting and constructs underwent electromechanical conditioning (1 Hz pacing, 1 mN diastolic preload) in customized biomimetic chambers45 for 24 days (Figure 6A). Contractile force of HLHS patches was significantly reduced compared with controls and did not increase, but rather decreased, over time (Figure 6B and 6C). Whereas progressive electromechanical maturation was evident in controls, HLHS tissues failed to respond to high stimulation frequencies and to develop a positive force–frequency relationship (Figure 6D–6F). Concurrent Ca2+ imaging of single cardiomyocytes within the patches demonstrated a gradual decline in the number of electrically responsive HLHS cells (Figure 6G–6I) and intrinsic Ca2+ handling defects, with failed increase of systolic and abnormal elevation of diastolic Ca2+ at high stimulation rates (Figure 6J and 6K). Expression profile of HLHS cardiomyocytes isolated from 3D patches at day 12 confirmed inherent molecular changes of pivotal genes involved in electromechanical coupling and Ca2+ homeostasis (Figure 6L). Live evaluation of cell viability revealed a progressive increase of dead cells in HLHS tissues (Figure XIA in the Data Supplement), suggesting apoptosis as an additional mechanism underlying the functional deterioration. Immunohistochemistry demonstrated a gradual increase of apoptotic cardiomyocytes (clCasp3+/TUNEL+) in diseased patches (Figure 7A). Not all TUNEL+ cardiomyocytes expressed activated caspase-3, although the opposite was true; the percentage of clCasp3–/TUNEL+ cells was also significantly elevated in HLHS (Figure 7A), indicating apoptosis-independent DNA damage. To better assess this, we performed coimmunofluorescence analysis of activated caspase-3 and activated p53 (phP53), a tumor suppressor that induces either cell cycle arrest to facilitate DNA repair or apoptosis.46 We detected clCasp3+/phP53+ and clCasp3–/phP53+ cardiomyocytes, with a significant increase of both in HLHS tissues (Figure 7B). The percentage of clCasp3–/phP53+ cells matched well with the proportion of clCasp3–/TUNEL+ cells, suggesting that DNA damage might be the first event leading to apoptosis of HLHS cardiomyocytes.
Figure 6.
Three-dimensional culture of induced pluripotent stem cell–derived cardiomyocytes under electromechanical stress reveals hypoplastic left heart syndrome–related functional abnormalities. A, Schematic of the experimental setup for 3D culture of human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CM) within decellularized heart patches kept in biomimetic chambers providing mechanical load and electric stimulation while allowing continuous monitoring of force development. All measurements were done in patches generated from 2 control and 3 hypoplastic left heart syndrome (HLHS) lines. Unless otherwise illustrated, results from different control and HLHS lines have been pooled. B and C, Representative plots (B) and statistical analysis (C) of contractile force in HLHS and control patches over 24 days of culture. In (C), data are mean±SEM of serial measurements at the indicated days; n=8 (CTR), n=3 (HLHS1 and HLHS2), n=6 (HLHS3) patches. *P<0.05, **P<0.01 compared with CTR (2-way repeated-measures analysis of variance). D, Representative traces of contraction force at increasing stimulation frequencies in 1 control and 1 HLHS line from an experiment aimed at assessing the force–frequency relationship (FFR) and the paceability at different stimulation frequencies. Stimulation pulses are indicated as vertical bars above the respective tracing. E, Percentage of patches responding to stimulation at indicated pacing frequencies; n=6 (CTR) and n=9 (HLHS). *P<0.05, **P<0.01 compared with CTR at the same day and frequency (Fisher’s exact test). F, FFR, depicted as the developed force (normalized to the mean force developed by CTR patches at day 7) as a function of the beat-to-beat interval (depicted on a logarithmic scale). Serial FFR values obtained from HLHS (n=11) and control (n=8) patches at indicated time points are shown. Data are mean±SEM. *P<0.05, **P<0.01, ***P<0.001 compared with CTR at day 7 at the same beat-to-beat interval (mixed effects model). G, Representative images of Fluo-4-based intracellular calcium transients from single control and HLHS cardiomyocytes within the patch at increasing pacing rates. Vertical bars over the tracings represent the stimulation pulses. H and I, The left graph shows the overall percentage of paceable cardiomyocytes within control and HLHS patches at day 12 (H) and day 24 (I) of 3D culture. The right graph shows, only considering cells that were paceable with at least 1 of the applied pacing rates, the percentage of cells responding at the indicated frequencies at day 12 (H) and day 24 (I). Data are mean±SEM; n=6 (CTR) and n=9 (HLHS) patches; left panels: n=183 and 156 (CTR), n=466 and 70 (HLHS) cells in (H) and (I), respectively; right panels: N≥67 and 90 (CTR), N≥21 and 17 (HLHS) cells for each frequency in (H) and (I), respectively. *P<0.05, **P<0.01, ***P<0.001 compared with CTR (Mann-Whitney test for left panels and 2-way analysis of variance for right panels). J, Amplitude of the systolic calcium transients plotted as a function of the stimulation frequency for cardiomyocytes within control (n=6) and HLHS (n=9) patches at day 12 and day 24 of 3D culture; N≥65 and 81 (CTR), N≥3 and 35 (HLHS) cells for each frequency at day 12 and day 24, respectively. Data are mean±SEM. **P<0.01, ***P<0.001 compared with CTR (2-way analysis of variance). K, Diastolic calcium level (expressed as ratio of the diastolic Fluo-4 intensity at the indicated pacing frequency [F] and the basal Fluo-4 intensity at the beginning of the experiment [F0]) of single cardiomyocytes in day 24 control (n=6) and HLHS (n=9) patches at 0.5 Hz and 1.5 Hz pacing rates; N=91 and 83 (CTR), N=70 and 44 (HLHS) cells at 0.5 Hz and 1.5 Hz, respectively. Data are mean±SEM. ***P<0.001 (Mann-Whitney test). L, Expression level of key genes for electromechanical coupling (black) and Ca2+ homeostasis (red) in cardiomyocytes isolated from HLHS and control 3D patches at day 12. Data are log2 mean fold changes relative to controls; n=3 patches per line. ECM indicates extracellular matrix; and NHP, nonhuman primate.
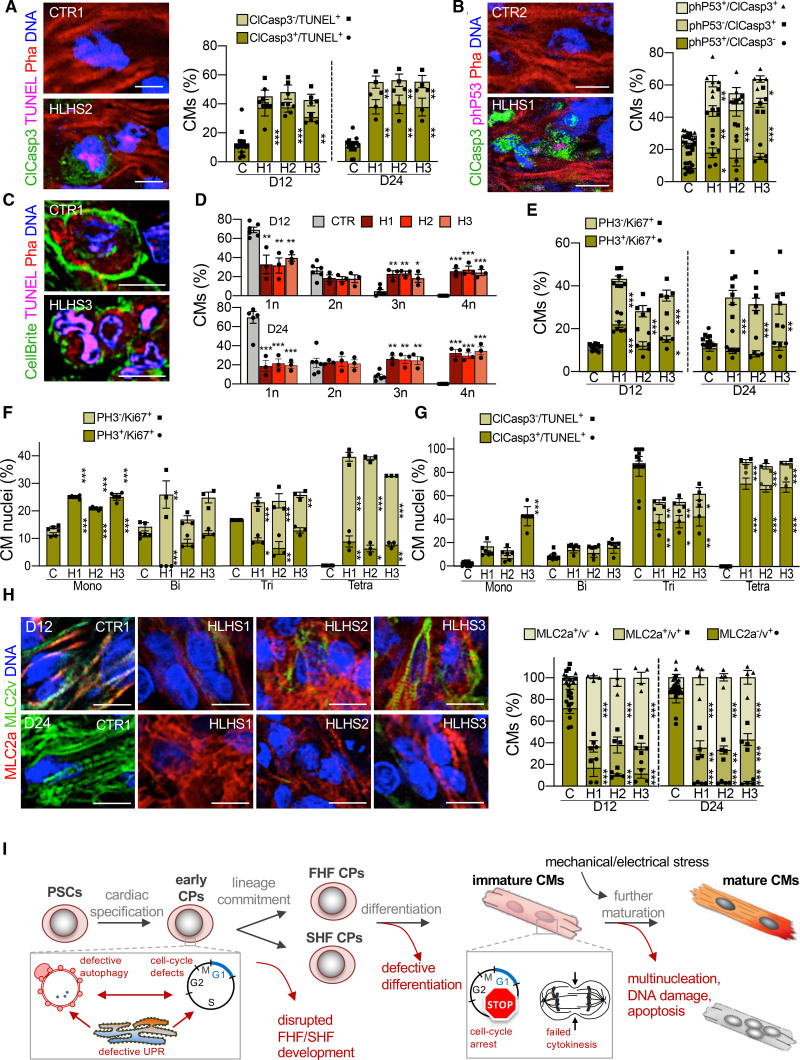
Figure 7.
Aberrant apoptosis, multinucleation, and maturation of hypoplastic left heart syndrome cardiomyocytes in 3D biomimetic culture. All measurements were done in patches generated from 2 control and 3 hypoplastic left heart syndrome (HLHS) lines. Results from the 2 different controls have been pooled. A, Representative fluorescence images of day 24 control and HLHS patches after immunostaining for activated caspase 3 (ClCasp3) in conjunction with TUNEL labeling. Phalloidin (Pha) marks F-actin and distinguishes cardiomyocytes. Scale bar, 10 µm. Bar graph shows the statistical evaluation of day 12 and day 24. Data are mean±SEM; n=8 (control [CTR], N=226 cells) and n=4 (each HLHS line, N≥200 cells per line) patches at day 12; n=7 (CTR, N=534) and n=3 (each HLHS line, N≥434 cells per line) patches at day 24. **P<0.01, ***P<0.001 compared with CTR (1-way analysis of variance [ANOVA]). B, Representative immunostaining for activated caspase 3 (ClCasp3) and phosphorylated P53 (phP53) in day 24 control and HLHS patches. Scale bar, 10 µm. Bar graph illustrates the percentages of cardiomyocytes (identified by Pha) expressing one or both markers. Data are mean±SEM, n=10 (CTR, N=652 cells), n=6 (HLHS1, N=572 cells), n=5 (HLHS2, N=470 cells), and n=5 (HLHS3, N=314 cells) patches. *P<0.05, **P<0.01, ***P<0.001 compared with CTR (1-way ANOVA). C, Representative fluorescence images of cardiomyocytes (identified by Pha) after plasmamembrane (CellBrite), TUNEL, and DNA labeling within control and HLHS patches at day 24. Scale bar, 10 µm. D, Bar graphs showing the percentage of cardiomyocytes with 1 (1n), 2 (2n), 3 (3n), and 4 (4n) nuclei in the different cell lines at day 12 and day 24. Data are mean±SEM; n=6 (CTR, N=340 cells) and n=3 (each HLHS line, N≥182 cells per line) patches at day 12; n=6 (CTR, N=341) and n=3 (each HLHS line, N≥147 cells per line) patches at day 24. *P<0.05, **P<0.01, ***P<0.001 compared with CTR (1-way ANOVA). E, Bar graphs showing the percentage of cardiomyocytes positive for phosphorylated histone 3 (PH3) and Ki67 at day 12 and day 24. Data are mean±SEM; n=7 (CTR, N=528 cells), n=7 (HLHS1, N=698 cells), n=5 (HLHS2, N=463), and n=5 (HLHS3, N=668) patches at day 12; n=7 (CTR, N=673 cells), n=7 (HLHS1, N=682 cells), n=5 (HLHS2, N=486), and n=5 (HLHS3, N=485) patches at day 24. *P<0.05, **P<0.01, ***P<0.001 compared with CTR (1-way ANOVA). F, Bar graph showing the percentage of nuclei positive for PH3 and Ki67 in mono-, bi-, tri-, and tetranucleated cardiomyocytes at day 24. Data are mean±SEM; n=3 patches per line; N=331 (CTR), N=258 (HLHS1), N=228 (HLHS2), and N=240 (HLHS3) cells. *P<0.05, **P<0.01, ***P<0.001 compared with CTR (1-way ANOVA). G, Bar graph showing the percentage of nuclei positive for ClCasp3 and phP53 in mono-, bi-, tri-, and tetranucleated cardiomyocytes at day 24. Data are mean±SEM; n=7 (CTR) and n=3 (each HLHS line) patches; N=401 (CTR), N=194 (HLHS1), N=175 (HLHS2), and N=237 (HLHS3) cells. *P<0.05, **P<0.01, ***P<0.001 compared with CTR (1-way ANOVA). H, Left, representative immunostains for MLC2a and MLC2v in control and HLHS patches. Scale bar, 10 µm. Right, bar graph shows statistical evaluation. Data are mean±SEM, n=8 (CTR, N=951 cells) and n=4 (each HLHS line, N≥428 cells per line) patches at day 12; n=8 (CTR, N=949) and n=4 (each HLHS line, N≥442 cells per line) patches at day 24. **P<0.01, ***P<0.001 compared with CTR (1-way ANOVA). I, Scheme depicting the identified steps during cardiac development at which HLHS-related abnormalities interfere with normal development and contribute to the complex CHD phenotype. CM indicates cardiomyocyte; CP, cardiac progenitor; FHF, first heart field; PSC, pluripotent stem cell; SHF, second heart field; and UPR, unfolded protein response.
An interesting finding emerging from histologic analyses was abnormal multinucleation of cardiomyocytes in HLHS patches, already evident at day 12 (Figure 7C). Polyploidy and binucleation are characteristic features of mammalian cardiomyocytes that develop shortly after birth when most differentiated cardiomyocytes exit cell cycle.31 In humans, polyploidy is often increased in pathologic conditions; however, binucleation occurs in only 25% of cardiomyocytes, with no evidence of tri- or tetranucleation.47 In control patches, most cardiomyocytes carried 1 (∼70%) or 2 (∼25%) nuclei, and few cells presented 3 or more; conversely, in HLHS tissues, ∼50% of cardiomyocytes were tri- or tetranucleated (Figure 7C and 7D), indicating an intrinsic failure to complete cytokinesis and premature cell cycle withdrawal. Markers of cell cycle activity (Ki67) and mitosis (PH3 [phospho-histone H3]) demonstrated an overall increase in the number of active cardiomyocytes in HLHS patches, with the proportion of cells in mitosis being similar (Figure 7E). However, detailed analysis of Ki67 and PH3 expression revealed that, in mononucleated cardiomyocytes, most nuclei were in M phase (Ki67+/PH3+) and their percentage was higher in HLHS (Figure 7F). The greater the degree of multinucleation in HLHS, the higher was the proportion of Ki67+/PH3– versus Ki67+/PH3+ nuclei (Figure 7F), suggesting that polyploidization was also occurring in multinucleated diseased cardiomyocytes. To assess whether and how the abortive cell cycle mode of HLHS cardiomyocytes related to apoptosis, we evaluated the distribution of TUNEL+ nuclei and clCasp3+/TUNEL+ cells (Figure 7G). We found a striking correlation between the number of TUNEL+ nuclei and the level of multinucleation, with the most damaged nuclei being in the tri- or tetranucleated cardiomyocytes. The latter also showed the highest percentage of activated caspase-3 (Figure 7G), together indicating chromosomal instability acquired during aberrant cell cycle as possible apoptotic trigger in HLHS cardiomyocytes.
We hypothesized that the observed HLHS cardiomyocyte phenotype might be a specific reaction to strong cues for cell division and growth and maturation, which are provided by the electromechanical preconditioning in the 3D tissue environment. Because our transcriptome profiling of early HLHS cardiomyocytes developed in 2D monolayer pointed to defects in differentiation and maturation, we performed coimmunofluorescence analysis for MLC2a (expressed in all immature cardiomyocytes and becoming atrial-specific after terminal chamber differentiation) and MLC2v (expressed in maturing ventricular cardiomyocytes). In HLHS patches, we measured a striking increase of MLC2a+ cells with reduction of MLC2v+ cardiomyocytes at day 12 and day 24 (Figure 7H); moreover, sarcomere organization was impaired (Figure XIB in the Data Supplement), indicative of reduced maturation. Similar results were obtained in HLHS cardiomyocytes (day 30 and day 60) in 2D monolayers (Figure XIC–XIE in the Data Supplement). Expression analysis of terminally differentiated atrial, ventricular, and cardiomyocyte maturation markers confirmed that the abnormal distribution of HLHS MLC2a+/MLC2v+ cells was a result of defective ventricular differentiation and maturation rather than an atrial lineage switch (Figure XIF in the Data Supplement). This was corroborated by measuring intracellular Ca2+ cycling of 2D cardiomyocytes (Figure XIIA–XIIC in the Data Supplement). No aberrant multinucleation or apoptosis was visible in 2D (Figure XIID–XIIF in the Data Supplement).
Together, these results indicate that impaired maturation and inability to respond to cues for developmental growth by normal progression through the cell cycle leads to increased apoptosis of ventricular cardiomyocytes, contributing to HLHS pathogenesis.
Discussion
Our work provides a multidisciplinary framework for studying human heart development and its disruption in CHD. WES of HLHS parent–offspring trios and transcriptomics of patient RV cardiomyocytes identified consistent perturbations in gene expression programs and associated features of abnormal cardiac development. Human iPSC lines derived from patients with HLHS facilitated dynamic evaluation of transcriptional and cellular phenotypes during progression of cardiogenesis in single cells and 3D functional modeling of ventricular chamber development. Together, our data indicate that initial perturbations of the cell cycle/UPR/autophagy hub result in disrupted differentiation of early CP lineages and disproportionate allocation of ventricular cardiomyocyte subtypes in HLHS. Moreover, impaired maturation and premature cell cycle exit of ventricular cardiomyocytes reduce their ability to respond to cues for tissue growth, resulting in increased apoptosis and ventricular hypoplasia (Figure 7I).
Identification of Dysregulated Transcriptional Nodes in HLHS
Pathway analysis of D-DNMs in HLHS cases showed significant enrichment in heart development and its related signaling pathways. Comprehensive investigation of single cell expression profile and dynamically regulated gene programs in cardiomyocytes indicated an occurrence of D-DNMs in gene modules typical of the embryonic stage encompassing cell cycle. Transcriptomes of infant HLHS ventricular cardiomyocytes closely resembled a fetal stage, demonstrating that embryonic gene programs persist in HLHS cardiomyocytes after birth. We identified dysregulated transcriptional nodes common to both CP and cardiomyocyte states or unique of one state. Moreover, single-cell RNA-Seq of HLHS cardiomyocytes recognized distinct regulatory defects in specific cellular subsets indicating that early cardiomyocyte subtype lineage specification, maturation, and cell cycle are disrupted in the disease.
Previous studies in HLHS iPSCs showed reduced cardiac differentiation and structural cardiomyocyte maturation in presence of dysfunctional Notch signaling17–19 and suggested defective commitment to the ventricular lineage,21 corroborating our results. Moreover, abnormalities in endocardial cells were shown to influence cardiomyocyte proliferation and maturation in HLHS.48
Evidence of cell cycle arrest and impaired maturation was reported in cardiomyocytes of the hypoplastic LV of HLHS fetuses49 and Ohia mutants.10 Loss of replication potential during fetal growth likely affects cardiac chamber development and function. Our 3D functional studies revealed, both on the tissue and single cardiomyocyte level, abnormalities of excitation–contraction coupling reminiscent of failing human myocardium. Besides the valvular perturbations, chamber-specific differences in proliferation rates at early stages of development may explain why loss of cardiomyocyte proliferation in HLHS affects LV more than RV. By analyzing cell cycle gene signature of single cardiomyocytes from RV and LV regions of human embryonic hearts from 5 to 25 weeks of gestation,29 we found LV cardiomyocytes proliferate more than their RV counterparts at 5 weeks (58% LV versus 48% RV), but these differences are extinguished later in development (45% and 55% LV versus 43% and 53% RV at 6 and 7 weeks, respectively). Concordantly, similar analysis using our single-cell RNA-Seq data of day 14 control iPSC-CMs with LV and RV transcriptome revealed a ∼3-fold increased proliferation of LV compared with RV cells (34.5% LV versus 11.8% RV; P<0.05).
Linking Clinical Phenotype to Mechanism
HLHS is a spectrum of disease that includes LV and aortic hypoplasia with aortic and mitral valve malformations ranging from stenosis to complete atresia. Historically, the “flow-volume hypoplasia” hypothesis has been supported by experiments in multiple model systems. However, recent observations in patients8,9 argue that hemodynamic conditions alone are insufficient to explain variable LV morphology. We focused on the “thickened” LV morphology with a clearly visible LV lumen8,9 and were able to identify a common mechanism (Figure 7I) for the initial arrested LV development in this specific anatomic subtype. Intrauterine fetal valvuloplasty in patients with HLHS is being studied as a treatment option to enable LV growth. The existence of intrinsic cardiomyocyte defects such as loss of replication and increased apoptosis in distinct subsets of patients with HLHS suggest a way to potentially improve the unpredictable results of this aortic valvuloplasty approach.50 Furthermore, knowledge of these specific defects allows development of rational strategies, either by stratification or adjunctive therapies, potentially permitting biventricular repair in those with less serious forms of LV hypoplasia.
Study Limitations
The small number of our HLHS parent–offspring trios limited identification of new disease candidate genes. However, the combination of WES data from our and the Pediatric Cardiac Genomics Consortium HLHS cohorts identified disturbed genetic pathways with substantial burden enrichment in D-DNMs, which we functionally validated.
Our analysis of HLHS heart tissue was limited to RV and may not recover all critical changes driving HLHS pathology. Moreover, differences in RV versus LV physiology, age, medications, and the intrinsic variability among different iPSC lines are potential confounders. However, the recovery of consistent molecular and cellular alterations in native RV cardiomyocytes and in vitro iPSC models reinforces their causal relation to HLHS pathogenesis.
Taken together, our results suggest a shared mechanism for subsets of HLHS and highlight that reduced LV growth in HLHS is likely not solely the consequence of disrupted valve formation and impaired blood flow; instead, intrinsic defects of ventricular cardiomyocyte lineage development are primary contributors to the disease pathogenesis. More broadly, our work illustrates that, despite the extensive genetic heterogeneity underlying CHD, studying cardiac developmental processes in patients with CHD using converging multidimensional technologies can provide deep mechanistic insight into these complex diseases to suggest novel therapeutic approaches.
Acknowledgments
The authors thank the patients with hypoplastic left heart syndrome, their family members, and the healthy volunteers for participation; Birgit Campbell and Christina Scherb for technical assistance in cell culture; Gabi Lederer (Cytogenetic Department, Technical University of Munich) for karyotyping; members of the KaBi-DHM Biobank (cardiovascular biobank, German Heart Center Munich) for patient sample collection; Alma Kuechler (Institute of Human Genetics, University Hospital Essen) for providing a hypoplastic left heart syndrome parent-offspring trio; and Rabea Hinkel (Deutsches Primatenzentrum Göttingen) for providing nonhuman primate heart samples.
Sources of Funding
This work was supported by grants from the European Research Council (788381 to Dr Moretti and 261053 to Dr Laugwitz); the German Research Foundation (KR3770/7-3, KR3770/11-1, and KR3770/14-1 to Dr Krane); Transregio Research Unit (152 to Dr Moretti and Dr Laugwitz and 267 to Dr Moretti, Dr Laugwitz, and Dr Kupatt); and the German Center for Cardiovascular Research (DZHK_B 19 SE to Dr Krane and FKZ 81Z0600601 to Dr Moretti and Dr Laugwitz). Dr Santamaria was funded by Fondazione Umberto Veronesi.
Disclosures
None.
Supplemental Materials
Expanded Methods
Expanded Results
Data Supplement Figures I–XII
Data Supplement Tables I–VI
Supplemental Appendix (contains Tables IV–VI)
Supplementary Material
Nonstandard Abbreviations and Acronyms
- CHD
- congenital heart disease
- CP
- cardiac progenitor
- D-DNM
- damaging de novo mutation
- DEG
- differentially expressed gene
- DNM
- de novo mutation
- ER
- endoplasmic reticulum
- FHF
- first heart field
- GSEA
- gene set enrichment analysis
- HLHS
- hypoplastic left heart syndrome
- iPSC-CM
- induced pluripotent stem cell–derived cardiomyocyte
- iPSC
- induced pluripotent stem cell
- LV
- left ventricle
- PH3
- phospho-histone H3
- RNA-Seq
- RNA sequencing
- RV
- right ventricle
- UPR
- unfolded protein response
- WES
- whole-exome sequencing
M. Krane, M. Dreßen, G. Santamaria, I. My, C.M. Schneider, T. Dorn, and S. Laue contributed equally.
The Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.121.056198.
For Sources of Funding and Disclosures, see page 1426.
Contributor Information
Markus Krane, Email: krane@dhm.mhn.de.
Martina Dreßen, Email: dressen@dhm.mhn.de.
Gianluca Santamaria, Email: santamariagianluca@tum.de.
Ilaria My, Email: ilariamy@libero.it.
Christine M. Schneider, Email: pedro.j.schneider@gmail.com.
Tatjana Dorn, Email: tatjana.dorn@mytum.de.
Svenja Laue, Email: svenja.laue@gmx.net.
Elisa Mastantuono, Email: mastantuonoelisa@gmail.com.
Riccardo Berutti, Email: riccardo.berutti@tum.de.
Hilansi Rawat, Email: hilansirawat@tum.de.
Ralf Gilsbach, Email: gilsbach@vrc.uni-frankfurt.de.
Pedro Schneider, Email: pedro.j.schneider@gmail.com.
Harald Lahm, Email: lahm@dhm.mhn.de.
Sascha Schwarz, Email: sascha.schwarz@tum.de.
Stefanie A. Doppler, Email: steffidoppler@gmail.com.
Sharon Paige, Email: spaige@stanford.edu.
Nazan Puluca, Email: nazan.puluca@web.de.
Sophia Doll, Email: doll@omicera.com.
Irina Neb, Email: neb@dhm.mhn.de.
Thomas Brade, Email: thomas.meitinger@mri.tum.de.
Zhong Zhang, Email: dor.zhangzhong@gmail.com.
Claudia Abou-Ajram, Email: abou@dhm.mhn.de.
Bernd Northoff, Email: bernd.northoff@med.uni-muenchen.de.
Lesca M. Holdt, Email: lesca.holdt@med.uni-muenchen.de.
Stefanie Sudhop, Email: stefanie.sudhop@hm.edu.
Makoto Sahara, Email: makoto.sahara@ki.se.
Alexander Goedel, Email: Alexander.Goedel@gmx.de.
Andreas Dendorfer, Email: andreas.dendorfer@med.uni-muenchen.de.
Fleur V.Y. Tjong, Email: f.v.tjong@amsterdamumc.nl.
Maria E. Rijlaarsdam, Email: mariarijlaarsdam@gmail.com.
Julie Cleuziou, Email: cleuziou@dhm.mhn.de.
Nora Lang, Email: langn@dhm.mhn.de.
Christian Kupatt, Email: Christian.kupatt@tum.de.
Connie Bezzina, Email: c.r.bezzina@amc.uva.nl.
Rüdiger Lange, Email: lange@dhm.mhn.de.
Neil E. Bowles, Email: neil.bowles@oehs.utah.edu.
Matthias Mann, Email: mmann@biochem.mpg.de.
Bruce D. Gelb, Email: bruce.gelb@mssm.edu.
Lia Crotti, Email: liacrotti@yahoo.it.
Lutz Hein, Email: lutz.hein@pharmakol.uni-freiburg.de.
Thomas Meitinger, Email: thomas.meitinger@mri.tum.de.
Sean Wu, Email: smwu@stanford.edu.
Daniel Sinnecker, Email: dsinneck@yahoo.de.
References
- 1.Lev M. Pathologic anatomy and interrelationship of hypoplasia of the aortic tract complexes. Lab Invest. 1952;1:61–70 [PubMed] [Google Scholar]
- 2.Noonan JA, Nadas AS. The hypoplastic left heart syndrome: an analysis of 101 cases. Pediatr Clin North Am. 1958;5:1029–1056. doi: 10.1016/s0031-3955(16)30727-1 [DOI] [PubMed] [Google Scholar]
- 3.Anderson RH, Spicer DE, Crucean A. Clarification of the definition of hypoplastic left heart syndrome. Nat Rev Cardiol. 2021;18:147–148. doi: 10.1038/s41569-020-00500-5 [DOI] [PubMed] [Google Scholar]
- 4.Hinton RB, Jr, Martin LJ, Tabangin ME, Mazwi ML, Cripe LH, Benson DW. Hypoplastic left heart syndrome is heritable. J Am Coll Cardiol. 2007;50:1590–1595. doi: 10.1016/j.jacc.2007.07.021 [DOI] [PubMed] [Google Scholar]
- 5.Øyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M. Recurrence of congenital heart defects in families. Circulation. 2009;120:295–301. doi: 10.1161/CIRCULATIONAHA.109.857987 [DOI] [PubMed] [Google Scholar]
- 6.Hickey EJ, Caldarone CA, McCrindle BW. Left ventricular hypoplasia: a spectrum of disease involving the left ventricular outflow tract, aortic valve, and aorta. J Am Coll Cardiol. 2012;59(1 Suppl)S43–S54. doi: 10.1016/j.jacc.2011.04.046 [DOI] [PubMed] [Google Scholar]
- 7.Harh JY, Paul MH, Gallen WJ, Friedberg DZ, Kaplan S. Experimental production of hypoplastic left heart syndrome in the chick embryo. Am J Cardiol. 1973;31:51–56. doi: 10.1016/0002-9149(73)90810-2 [DOI] [PubMed] [Google Scholar]
- 8.Crucean A, Alqahtani A, Barron DJ, Brawn WJ, Richardson RV, O’Sullivan J, Anderson RH, Henderson DJ, Chaudhry B. Re-evaluation of hypoplastic left heart syndrome from a developmental and morphological perspective. Orphanet J Rare Dis. 2017;12:138. doi: 10.1186/s13023-017-0683-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossfeld P, Nie S, Lin L, Wang L, Anderson RH. Hypoplastic left heart syndrome: a new paradigm for an old disease? J Cardiovasc Dev Dis. 2019;6:E10. doi: 10.3390/jcdd6010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Yagi H, Saeed S, Bais AS, Gabriel GC, Chen Z, Peterson KA, Li Y, Schwartz MC, Reynolds WT, et al. The complex genetics of hypoplastic left heart syndrome. Nat Genet. 2017;49:1152–1159. doi: 10.1038/ng.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xinxiu Xu KJ, Bais AS, Zhu W, Yagi H, Feinstein TN, Nguyen P, Criscione J, Liu X, Beutner G, Karunakaran KB, et al. IPSC modeling shows uncompensated mitochondrial mediated oxidative stress underlies early heart failure in hypoplastic left heart syndrome. BioRxiv. Preprint posted online May 10, 2021. doi: 10.1101/2021.05.09.443165 [DOI] [Google Scholar]
- 12.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029 [DOI] [PubMed] [Google Scholar]
- 13.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002 [DOI] [PubMed] [Google Scholar]
- 14.Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135:193–205. doi: 10.1242/dev.001883 [DOI] [PubMed] [Google Scholar]
- 15.Kodo K, Ong SG, Jahanbani F, Termglinchan V, Hirono K, InanlooRahatloo K, Ebert AD, Shukla P, Abilez OJ, Churko JM, et al. iPSC-derived cardiomyocytes reveal abnormal TGF-β signalling in left ventricular non-compaction cardiomyopathy. Nat Cell Biol. 2016;18:1031–1042. doi: 10.1038/ncb3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, et al. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016;167:1734, e22–1749. doi: 10.1016/j.cell.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theis JL, Hrstka SC, Evans JM, O’Byrne MM, de Andrade M, O’Leary PW, Nelson TJ, Olson TM. Compound heterozygous NOTCH1 mutations underlie impaired cardiogenesis in a patient with hypoplastic left heart syndrome. Hum Genet. 2015;134:1003–1011. doi: 10.1007/s00439-015-1582-1 [DOI] [PubMed] [Google Scholar]
- 18.Yang C, Xu Y, Yu M, Lee D, Alharti S, Hellen N, Ahmad Shaik N, Banaganapalli B, Sheikh Ali Mohamoud H, Elango R, et al. Induced pluripotent stem cell modelling of HLHS underlines the contribution of dysfunctional NOTCH signalling to impaired cardiogenesis. Hum Mol Genet. 2017;26:3031–3045. doi: 10.1093/hmg/ddx140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hrstka SC, Li X, Nelson TJ; Wanek Program Genetics Pipeline Group. NOTCH1-dependent nitric oxide signaling deficiency in hypoplastic left heart syndrome revealed through patient-specific phenotypes detected in bioengineered cardiogenesis. Stem Cells. 2017;35:1106–1119. doi: 10.1002/stem.2582 [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi J, Yoshida M, Tarui S, Hirata M, Nagai Y, Kasahara S, Naruse K, Ito H, Sano S, Oh H. Directed differentiation of patient-specific induced pluripotent stem cells identifies the transcriptional repression and epigenetic modification of NKX2-5, HAND1, and NOTCH1 in hypoplastic left heart syndrome. PLoS One. 2014;9:e102796. doi: 10.1371/journal.pone.0102796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Habibollah S, Tilgner K, Collin J, Barta T, Al-Aama JY, Tesarov L, Hussain R, Trafford AW, Kirkwood G, et al. An induced pluripotent stem cell model of hypoplastic left heart syndrome (HLHS) reveals multiple expression and functional differences in HLHS-derived cardiac myocytes. Stem Cells Transl Med. 2014;3:416–423. doi: 10.5966/sctm.2013-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinz H, Pyle LC, Li D, Izumi K, Skraban C, Tarpinian J, Braddock SR, Telegrafi A, Monaghan KG, Zackai E, et al. De novo variants in Myelin regulatory factor (MYRF) as candidates of a new syndrome of cardiac and urogenital anomalies. Am J Med Genet A. 2018;176:969–972. doi: 10.1002/ajmg.a.38620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi H, Yu L, Zhou X, Wynn J, Zhao H, Guo Y, Zhu N, Kitaygorodsky A, Hernan R, Aspelund G, et al. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS Genet. 2018;14:e1007822. doi: 10.1371/journal.pgen.1007822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song L, Li Y, Wang K, Zhou CJ. Cardiac neural crest and outflow tract defects in Lrp6 mutant mice. Dev Dyn. 2010;239:200–210. doi: 10.1002/dvdy.22079 [DOI] [PubMed] [Google Scholar]
- 25.Wu X, Shen QT, Oristian DS, Lu CP, Zheng Q, Wang HW, Fuchs E. Skin stem cells orchestrate directional migration by regulating microtubule-ACF7 connections through GSK3β. Cell. 2011;144:341–352. doi: 10.1016/j.cell.2010.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahara M, Santoro F, Sohlmér J, Zhou C, Witman N, Leung CY, Mononen M, Bylund K, Gruber P, Chien KR. Population and single-cell analysis of human cardiogenesis reveals unique LGR5 ventricular progenitors in embryonic outflow tract. Dev Cell. 2019;48:475–490.e7. doi: 10.1016/j.devcel.2019.01.005 [DOI] [PubMed] [Google Scholar]
- 29.Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, Hu Y, Dong J, Li Q, Wu X, et al. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 2019;26:1934, e5–1950. doi: 10.1016/j.celrep.2019.01.079 [DOI] [PubMed] [Google Scholar]
- 30.Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–588. doi: 10.1038/ng.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- 32.Cao N, Liang H, Huang J, Wang J, Chen Y, Chen Z, Yang HT. Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell Res. 2013;23:1119–1132. doi: 10.1038/cr.2013.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatcher CJ, Kim MS, Mah CS, Goldstein MM, Wong B, Mikawa T, Basson CT. TBX5 transcription factor regulates cell proliferation during cardiogenesis. Dev Biol. 2001;230:177–188. doi: 10.1006/dbio.2000.0134 [DOI] [PubMed] [Google Scholar]
- 35.Mathiassen SG, De Zio D, Cecconi F. Autophagy and the cell cycle: a complex landscape. Front Oncol. 2017;7:51. doi: 10.3389/fonc.2017.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H, Tsang KS, Wang Y, Chan JC, Xu G, Gao WQ. Unfolded protein response is required for the definitive endodermal specification of mouse embryonic stem cells via Smad2 and β-catenin signaling. J Biol Chem. 2014;289:26290–26301. doi: 10.1074/jbc.M114.572560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laguesse S, Creppe C, Nedialkova DD, Prévot PP, Borgs L, Huysseune S, Franco B, Duysens G, Krusy N, Lee G, et al. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev Cell. 2015;35:553–567. doi: 10.1016/j.devcel.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 39.Mao C, Tai WC, Bai Y, Poizat C, Lee AS. In vivo regulation of Grp78/BiP transcription in the embryonic heart: role of the endoplasmic reticulum stress response element and GATA-4. J Biol Chem. 2006;281:8877–8887. doi: 10.1074/jbc.M505784200 [DOI] [PubMed] [Google Scholar]
- 40.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270 [DOI] [PubMed] [Google Scholar]
- 41.Li G, Tian L, Goodyer W, Kort EJ, Buikema JW, Xu A, Wu JC, Jovinge S, Wu SM. Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Development. 2019;146doi: 10.1242/dev.173476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia G, Preussner J, Chen X, Guenther S, Yuan X, Yekelchyk M, Kuenne C, Looso M, Zhou Y, Teichmann S, et al. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat Commun. 2018;9:4877. doi: 10.1038/s41467-018-07307-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asp M, Giacomello S, Larsson L, Wu C, Fürth D, Qian X, Wärdell E, Custodio J, Reimegård J, Salmén F, et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell. 2019;179:1647, e19–1660. doi: 10.1016/j.cell.2019.11.025 [DOI] [PubMed] [Google Scholar]
- 44.Guo H, Tian L, Zhang JZ, Kitani T, Paik DT, Lee WH, Wu JC. Single-cell RNA sequencing of human embryonic stem cell differentiation delineates adverse effects of nicotine on embryonic development. Stem Cell Reports. 2019;12:772–786. doi: 10.1016/j.stemcr.2019.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer C, Milting H, Fein E, Reiser E, Lu K, Seidel T, Schinner C, Schwarzmayr T, Schramm R, Tomasi R, et al. Long-term functional and structural preservation of precision-cut human myocardium under continuous electromechanical stimulation in vitro. Nat Commun. 2019;10:117. doi: 10.1038/s41467-018-08003-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015 [DOI] [PubMed] [Google Scholar]
- 47.Foglia MJ, Poss KD. Building and re-building the heart by cardiomyocyte proliferation. Development. 2016;143:729–740. doi: 10.1242/dev.132910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao Y, Tian L, Martin M, Paige SL, Galdos FX, Li J, Klein A, Zhang H, Ma N, Wei Y, et al. Intrinsic endocardial defects contribute to hypoplastic left heart syndrome. Cell Stem Cell. 2020;27:574, e8–589. doi: 10.1016/j.stem.2020.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaber N, Gagliardi M, Patel P, Kinnear C, Zhang C, Chitayat D, Shannon P, Jaeggi E, Tabori U, Keller G, et al. Fetal reprogramming and senescence in hypoplastic left heart syndrome and in human pluripotent stem cells during cardiac differentiation. Am J Pathol. 2013;183:720–734. doi: 10.1016/j.ajpath.2013.05.022 [DOI] [PubMed] [Google Scholar]
- 50.Freud LR, McElhinney DB, Marshall AC, Marx GR, Friedman KG, del Nido PJ, Emani SM, Lafranchi T, Silva V, Wilkins-Haug LE, et al. Fetal aortic valvuloplasty for evolving hypoplastic left heart syndrome: postnatal outcomes of the first 100 patients. Circulation. 2014;130:638–645. doi: 10.1161/CIRCULATIONAHA.114.009032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gramlich M, Pane LS, Zhou Q, Chen Z, Murgia M, Schötterl S, Goedel A, Metzger K, Brade T, Parrotta E, et al. Antisense-mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol Med. 2015;7:562–576. doi: 10.15252/emmm.201505047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moretti A, Fonteyne L, Giesert F, Hoppmann P, Meier AB, Bozoglu T, Baehr A, Schneider CM, Sinnecker D, Klett K, et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med. 2020;26:207–214. doi: 10.1038/s41591-019-0738-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679 [DOI] [PubMed] [Google Scholar]
- 54.Burridge PW, Holmström A, Wu JC. Chemically defined culture and cardiomyocyte differentiation of human pluripotent stem cells. Curr Protoc Hum Genet. 2015;87:21.3.1–21.3.15. doi: 10.1002/0471142905.hg2103s87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorn T, Kornherr J, Parrotta EI, Zawada D, Ayetey H, Santamaria G, Iop L, Mastantuono E, Sinnecker D, Goedel A, et al. Interplay of cell-cell contacts and RhoA/MRTF-A signaling regulates cardiomyocyte identity. EMBO J. 2018;37:e98133. doi: 10.15252/embj.201798133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilsbach R, Schwaderer M, Preissl S, Grüning BA, Kranzhöfer D, Schneider P, Nührenberg TG, Mulero-Navarro S, Weichenhan D, Braun C, et al. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat Commun. 2018;9:391. doi: 10.1038/s41467-017-02762-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takata A, Ionita-Laza I, Gogos JA, Xu B, Karayiorgou M. De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron. 2016;89:940–947. doi: 10.1016/j.neuron.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 2010;5:e8918. doi: 10.1371/journal.pone.0008918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375 [DOI] [PubMed] [Google Scholar]
- 60.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–W311. doi: 10.1093/nar/gkp427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Churko JM, Garg P, Treutlein B, Venkatasubramanian M, Wu H, Lee J, Wessells QN, Chen SY, Chen WY, Chetal K, et al. Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat Commun. 2018;9:4906. doi: 10.1038/s41467-018-07333-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, et al. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016;39:480–490. doi: 10.1016/j.devcel.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- 68.Scheltema RA, Hauschild JP, Lange O, Hornburg D, Denisov E, Damoc E, Kuehn A, Makarov A, Mann M. The Q Exactive HF, a benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol Cell Proteomics. 2014;13:3698–3708. doi: 10.1074/mcp.M114.043489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelstrup CD, Jersie-Christensen RR, Batth TS, Arrey TN, Kuehn A, Kellmann M, Olsen JV. Rapid and deep proteomes by faster sequencing on a benchtop quadrupole ultra-high-field Orbitrap mass spectrometer. J Proteome Res. 2014;13:6187–6195. doi: 10.1021/pr500985w [DOI] [PubMed] [Google Scholar]
- 70.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 71.Amps K, Andrews PW, Anyfantis G, Armstrong L, Avery S, Baharvand H, Baker J, Baker D, Munoz MB, Beil S, et al. ; International Stem Cell Initiative. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol. 2011;29:1132–1144. doi: 10.1038/nbt.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taapken SM, Nisler BS, Newton MA, Sampsell-Barron TL, Leonhard KA, McIntire EM, Montgomery KD. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835 [DOI] [PubMed] [Google Scholar]
- 73.Chan LL, Shen D, Wilkinson AR, Patton W, Lai N, Chan E, Kuksin D, Lin B, Qiu J. A novel image-based cytometry method for autophagy detection in living cells. Autophagy. 2012;8:1371–1382. doi: 10.4161/auto.21028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chu CT, Plowey ED, Dagda RK, Hickey RW, Cherra SJ, III, Clark RS. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 2009;453:217–249. doi: 10.1016/S0076-6879(08)04011-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions of this study are present in the article or the Data Supplement. The raw omics data are deposited at publicly available databases (bulk and single-cell RNA sequencing, #GSE135411; nuclear RNA sequencing, #PRJNA353755; proteomics, #PXD014812).