Abstract
Steroid receptors mediate responses to lipophilic hormones in a tissue- and ligand-specific manner. To identify nonreceptor proteins that confer specificity or regulate steroid signaling, we screened a human cDNA library in a steroid-responsive yeast strain. One of the identified cDNAs, isolated in the screen as ligand effect modulator 6, showed no homology to yeast or Caenorhabditis elegans proteins but high similarity to the recently described mouse coactivator PGC-1 and was accordingly termed hPGC-1. The hPGC-1 DNA encodes a nuclear protein that is expressed in a tissue-specific manner and carries novel motifs for transcriptional regulators. The expression of hPGC-1 in mammalian cells enhanced potently the transcriptional response to several steroids in a receptor-specific manner. hPGC-1-mediated enhancement required the receptor hormone-binding domain and was dependent on agonist ligands. Functional analysis of hPGC-1 revealed two domains that interact with steroid receptors in a hormone-dependent manner, a potent transcriptional activation function, and a putative dimerization domain. Our findings suggest a regulatory function for hPGC-1 as a tissue-specific coactivator for a subset of nuclear receptors.
Steroid hormones play important roles in development, growth, glucose and mineral homeostasis, stress responses, sexual differentiation, and reproduction. The effects of steroids are mediated by intracellular receptors that, together with receptors for thyroid hormones, retinoids, vitamin D, and other small lipophilic molecules, belong to the superfamily of nuclear receptors (2, 43). Binding of hormone to these receptors triggers a conformational change that leads to the release of associated proteins, such as molecular chaperones or corepressors, recruitment of new proteins, such as coactivators, binding to specific DNA sites termed hormone response elements (HREs), and regulation of transcription from promoters in the vicinity of the HREs (reviewed in references 2, 47, 61, and 75).
The major determinant of the ability of a cell to respond to a specific steroid hormone is the presence of the cognate receptor. In addition, nonreceptor proteins contribute to the cellular response by enabling or regulating distinct steps in the hormone response pathway. For example, membrane proteins that regulate the transport of hormone across the plasma membrane modify the availability of hormone to the intracellular receptors (32, 63); chaperones such as Hsp90 and p23 interact with steroid receptors in the absence of hormone and support a receptor conformation competent for hormone binding (reviewed in reference 61); DNA-binding proteins such as HMG-1 enhance the ability of steroid receptors to bind DNA (3, 57, 80); and the chromatin-remodeling SWI-SNF complex enables steroid receptors to regulate transcription (6, 14, 49, 78). Finally, several nuclear proteins are recruited by the receptors in a hormone-dependent manner and are thought to mediate their transcriptional regulatory activity (reviewed in references 13, 47, and 75). Among them, corepressors such as SMRT and NcoR bind to receptors in the absence of hormone or the presence of an antagonist ligand, connect the receptors to histone deacetylases, and promote the silencing of neighboring promoters (9, 21, 23, 35, 51). In contrast, coactivators (e.g., SRC-1 [also called NcoA1], TIF2 [the human homologue of mouse GRIP1, also known as NcoA2], pCIP [also known as AIB1, ACTR, and RAC3], CREB-binding protein [CBP], p300, pCAF, and others) are recruited by hormone-activated receptors and enhance transcription (13, 69, 75). At least some of these coactivators are acetyltransferases that can modify histones or other target proteins and may increase promoter access to DNA-binding proteins and the transcriptional machinery (1, 8, 56, 68, 77).
The identification of nonreceptor proteins that participate in hormone signaling provides insights not just into the mechanism of receptor-mediated transcription but also into ways in which the response to hormone can be regulated. Responses to steroid hormones are often tissue specific and sensitive to other signaling pathways. For example, the activation of protein kinase A modulates glucocorticoid responsiveness in a cell type-dependent manner (48, 55). Proteins that regulate the hormone response may confer tissue specificity to ubiquitously expressed receptors, if present in only some cell types, or integrate signaling information, if targeted by other signaling pathways. However, among the known nonreceptor proteins, few of them are expressed in a tissue-specific manner (e.g., ACTR) (8) or respond to other signals (e.g., CBP) (7, 41, 52).
Functional genetic strategies in easily manipulatable systems are powerful tools for identifying modulators that act at any step of a regulatory circuit. Although steroid receptors are not naturally present in the yeast Saccharomyces cerevisiae, the basic machinery of this organism is permissive to their function (46, 65). We have taken advantage of the ability of steroid receptors to mediate hormone-dependent transcription in yeast to identify proteins that can regulate hormone responsiveness. In previous genetic screens, we isolated yeast mutants and identified genes (LEM1 to LEM4, for ligand effect modulator) that negatively regulate responses to hormones (31; R. Sitcheran, R. Emter, A. Kralli, and K. R. Yamamoto, submitted for publication). In this study, we use a genetic scheme to identify directly mammalian proteins that enhance responses to glucocorticoid hormones. In principle, the screen can reveal both conserved proteins, whose yeast homologues are limiting or do not optimally interact with the mammalian receptor, and mammal-specific modulators, which may have evolved to confer specificity and regulation to steroid hormone responses.
MATERIALS AND METHODS
Yeast strains and cDNA library screening.
The yeast strain used in the cDNA library screen, YNK441, is a derivative of YPH499 (67) that has the endogenous HIS3 gene under the control of three copies of glucocorticoid response elements (GREs) from the tyrosine aminotransferase gene (26), a (GRE)×3-LACZ reporter cassette integrated at a disrupted pdr5::LEU2 locus (31), and a rat glucocorticoid receptor (GR) expression cassette integrated at the TRP1 locus. Integrations at the genomic loci were done sequentially, using the one-step replacement method (64) and DNA fragments from plasmids phis3::(GRE)×3-HIS3 (gift of J. Iniguez-Lluhi), pleu2::(GRE)×3-LACZ, and pBS/trp1::GR. Further information on these plasmids is available upon request. For the screen, YNK441 was transformed with a yeast expression HepG2 cDNA library (URA3, 2μm) (66) by a high-efficiency lithium acetate method (Technical Tips Online [http://tto.trends.com]) and plated on minimal medium lacking histidine and uracil and containing 15 mM 3-aminotriazole and either 4 μM dexamethasone or 0.5 μM corticosterone. Of 5.5 × 106 transformants, 719 grew and were analyzed for the β-galactosidase (β-gal) response to hormone. Plate and liquid β-gal assays were performed as described previously (25). Fifteen isolates showed an increased response to hormone that was lost when the library plasmid was selected against, on medium containing 5-fluoro-orotic acid. Plasmids were rescued, reintroduced to YNK441 to confirm their activity, and sequenced. The 15 plasmids carried three types of cDNA, encoding p23 (10×), ligand effect modulator 5 (LEM5) (1×), and the LEM6 HepG2 library isolate (LEM6H) (4×).
Isolation of LEM6 (hPGC-1) cDNA.
Sequencing of the LEM6H cDNAs isolated in the screen revealed two long open reading frames (ORFs) that were out of frame with each other and predicted the expression of a 272-amino-acid (aa) protein from the first ORF. Using oligonucleotides GCAGTGGTCTCAGTACCC and GTGGAGTTAGGCCTGCAG, which flank the junction where the two ORFs become discontinuous, and a proofreading DNA polymerase (Vent; New England Biolabs), we amplified by PCR the LEM6 cDNA from first-strand human liver cDNA (gift of R. Skoda). Sequencing of the amplified fragment showed the existence of one long ORF that had been split into two out-of-frame ORFs in the library isolate by a 32-bp insertion at nucleotide 804 (relative to the translation initiation site). The AatII-PstI 261-bp fragment of the liver LEM6 cDNA was used to replace the corresponding AatII-PstI 293-bp fragment of the library LEM6H isolate, thereby giving rise to the full-length clone that was used in subsequent experiments. The origin of the 32-bp insertion, which was present in all four LEM6 cDNAs isolated in the screen, has not been further explored. Sequencing also revealed high identity (95%) between LEM6 and the mouse PGC-1 (peroxisome proliferator-activated receptor gamma [PPARγ] coactivator 1) protein (62). Since LEM6 is the likely human homologue of PGC-1, it was renamed hPGC-1.
Plasmids.
The mammalian expression plasmids for rat GR (p6RGR, p6RN525, and p6R407C) (17, 18), rat mineralocorticoid receptor (MR) (p6RMR) (59), and the Gal4–ligand-binding domain (LBD) fusion [p6R.Gal(74/525)GR] are SP65-based vectors in which the Rous sarcoma virus promoter drives the expression of the respective genes. The simian virus 40 early promoter drives the expression of the human estrogen receptor (ER), androgen receptor (AR), and steroid receptor coactivator 1e (SRC-1e), the mouse NF-κB subunits p50 and p65, and the chimeric activator Gal4-VP16 in plasmids pSG5ER (72), pSVARo (gift of F. Hamy), pSG5-SRC-1e (gift of M. Parker) (27), pSG5-p50 and pSG5-RelA(p65) (gift of R. Nissen), and pSG424/Gal4-VP2, respectively. The luciferase reporter plasmids pTAT3Luc, pMMTVDLO, and pκB3DLO have three copies of the tyrosine aminotransferase GRE, the mouse mammary tumor virus (MMTV) long terminal repeat, and three copies of the interleukin 2α receptor NF-κB site, respectively, upstream of the minimal alcohol dehydrogenase promoter (25). The luciferase reporter plasmids pERE-TK-luc (two copies of the vitellogenin estrogen response element upstream of the thymidine kinase promoter) and pGK-1 (five Gal4 sites upstream of the E1b minimal promoter) have been described elsewhere (72).
All hPGC-1 expression vectors are derived from plasmid pBS/HA-hPGC-1, which carries the hPGC-1 cDNA (encoding aa 1 to 798) downstream of the hemagglutinin (HA) epitope-encoding sequence. To make pBS/HA-hPGC-1, the oligonucleotides 5′-GCCCGGATCCATGGCCTACCCATACGATGTCCCAGATTACGCCGGTCATATGGCGTGGGACATGTG-3′ (containing BamHI and NcoI sites, HA tag, NdeI site, and 17 nucleotides of hPGC-1 sequence; the underlined ATG is the translation initiation codon of hPGC-1) and 5′-GCCCGCGGCCGCG TCGAC TCAGTCAG TCACTCGAGT TACCTGCGCAAG - 3′ (containing NotI and SalI sites, three stop codons, XhoI site, natural stop codon of hPGC-1 [in bold], and last codons of hPGC-1) were used to amplify hPGC-1 DNA. The amplified fragment was digested with BamHI and NotI and subcloned into modified pBSKS (Stratagene). The hPGC-1 sequence was confirmed by sequencing.
Truncation and deletion variants of hPGC-1 were constructed in pBS/HA-hPGC-1. For N-terminal deletions, pBS/HA-hPGC-1 DNA was digested with NdeI and either NheI (for 91C), EcoRI (for 189C), or StuI (for 294C); for C-terminal deletions, pBS/HA-hPGC-1 DNA was digested with XhoI and either XbaI (for N408), StuI (for N293), EcoRI (for N186), or NheI (for N88). The ends were treated with Klenow polymerase and appropriate deoxynucleoside triphosphates and/or mung bean nuclease so as to preserve the correct reading frame and ligated. Similarly, internal deletions were made by digesting pBS/HA-hPGC-1 with NheI and EcoRI for hPGC-1Δ1 (Δ91-186), EcoRI and AgeI for hPGC-1Δ2L (Δ189-482), EcoRI and StuI for hPGC-1Δ2 (Δ189-293), and NheI and StuI for hPGC-1Δ3 (Δ91-293), treating the DNA ends with Klenow polymerase and/or mung bean nuclease, and ligating. All junctions were sequenced.
The full-length or variant HA–hPGC-1 constructs were then subcloned as BamHI-NotI fragments into the pcDNA3 vector (Invitrogen) downstream of the cytomegalovirus enhancer, for mammalian expression; as NcoI-SalI fragments into the pACT2 vector (Clontech) downstream of and in frame to the Gal4 activation domain (AD) for the expression of AD–hPGC-1 fusions in yeast; and as NcoI-SalI fragments into the pAS2-1 vector (Clontech) downstream of and in frame to the Gal4 DNA-binding domain (DBD), for the expression of Gal4–hPGC-1 fusions in yeast. For mammalian expression of the chimeric protein Gal4–hPGC-1, the NdeI-HindIII fragment of pBS/HA-hPGC-1 (lacking the HA epitope sequence and the last three codons of hPGC-1) was subcloned into vector pA4.7 (gift of Patrick Matthias) downstream of and in frame to the Gal4 DBD and upstream of and in frame to a C-terminal HA epitope sequence.
Northern analysis.
A human multiple-tissue Northern blot (Clontech) was hybridized with either a radioactively labeled 0.66-kb NdeI-PstI fragment of hPGC-1 cDNA or a β-actin-specific probe (Clontech), as recommended by the manufacturer, and exposed on X-ray film.
Two-hybrid interaction assay.
Plasmids expressing Gal4 DBD and Gal4 AD fusions (in vectors pAS2-1 and pACT2, respectively) were introduced by the lithium acetate transformation method (Technical Tips Online [http://tto.trends.com]) into yeast diploid cells (CG1945 × Y187; Clontech) that carry Gal4-driven LACZ reporters. Transformants carrying the plasmids were grown to stationary phase in 96-well plates, diluted 1:20 in selective medium (200 μl) containing either ethanol vehicle (0.25%) or 25 μM hormone (corticosterone or RU486), grown for an additional 16 to 18 h at 30°C in 96-well plates, and assayed for β-gal activity as described previously (25).
Cell culture, transient transfections, and reporter gene assays.
COS7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 9% charcoal-stripped fetal bovine serum. For experiments with the AR or the ER, media lacking phenol red were used. Cells from subconfluent 10-cm plates were diluted 8- to 10-fold and seeded in six-well plates 5 h prior to transfection by calcium phosphate precipitation (25). All transfections included 0.2 μg of p6RlacZ (59) to normalize for transfection efficiency. Expression vectors for receptors or other DNA-binding transcription factors were added at either 2 μg (AR and MR), 1 μg (GR, ER, p50, p65, and Gal4–hPGC-1), or 50 ng (Gal4-VP16) of DNA. Reporter plasmids were added at either 2 μg (for MR, p50, and p65) or 1 μg (for GR, ER, AR, Gal4-VP16, and Gal4–hPGC-1) of DNA. For the coactivators, 1 μg of hPGC-1 (or equivalent hPGC-1 variant) or 2 μg of SRC-1e expression plasmids was used, unless indicated otherwise (1 μg of SRC-1e DNA gave weaker enhancement of steroid receptor activity than 2 μg). After overnight exposure to the DNA-calcium phosphate precipitate, cells were washed with phosphate-buffered saline (PBS) lacking calcium and magnesium and incubated for an additional 24 h in fresh media containing either hormone or vehicle (0.1% ethanol). Cells were then washed with PBS and lysed in 100 μl of reporter lysis buffer (Promega). Lysates were cleared by centrifugation and assayed for luciferase activity with the Luciferase Assay System (Promega) and β-gal activity with the substrate chlorophenol red-β-d-galactopyranoside (25). Luciferase values normalized to the β-gal activity in the extracts are referred to as luciferase units.
GFP detection and immunofluorescence.
COS7 cells were seeded onto 18-mm coverslips and transiently transfected with hPGC-1 expression vectors by use of either the calcium phosphate precipitation method (for green fluorescent protein [GFP]–hPGC-1) or Effectene reagent (for HA–hPGC-1 constructs; used as recommended by the manufacturer, Qiagen). After 48 h, cells were washed with PBS, fixed for 15 min with 3% paraformaldehyde at room temperature, and permeabilized for 10 min with PBS containing 0.1% Triton X-100. Cells were then incubated for 10 min with 1% bovine serum albumin in PBS to block nonspecific binding. For immunodetection, a monoclonal antibody against the HA epitope (HA-11; BabCo) was used at a 1:1,000 dilution in PBS containing 0.5% bovine serum albumin, followed by a rhodamine-conjugated goat anti-mouse antibody (Jackson Immunoresearch Laboratories) at 1:100 in PBS.
Nucleotide sequence accession number.
The hPGC-1 sequence has been submitted to the GenBank database under accession no. AF186379.
RESULTS
Mammalian modulators of glucocorticoid signaling can be functionally isolated in yeast.
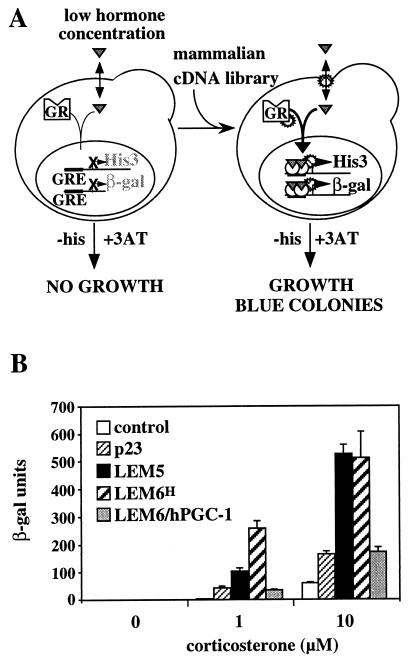
To identify proteins that enhance the cellular response to hormone, we exploited a yeast strain whose growth is dependent on glucocorticoid signaling. For this, the rat GR was expressed in yeast carrying two reporters under the control of GREs: the endogenous HIS3 and the bacterial β-gal-encoding gene (Fig. 1A). In the absence of hormone, the lack of HIS3 expression precludes growth in selective media. The addition of hormone induces HIS3 and the β-gal gene and restores growth. At suboptimal hormone concentrations insufficient to support growth, the expression of mammalian proteins that enhance the hormone response will activate HIS3, enable growth in selective media, and induce β-gal production (Fig. 1A).
FIG. 1.
A yeast screen can identify mammalian proteins that enhance steroid hormone signaling. (A) Schematic illustration of the yeast strain and strategy. The yeast strain YNK441 expresses GR constitutively and has two reporter genes, encoding His3 and β-gal, under the control of GREs. The expression of mammalian proteins that increase the response to hormone enables the activation of the pathway in the presence of low hormone concentrations, thereby allowing for growth and β-gal expression in selective media lacking histidine (−his) and containing the His3 inhibitor 3-aminotriazole (+3AT). The star-shaped forms indicate some of the possible interaction points for the mammalian modulators. (B) Mammalian p23, LEM5, and LEM6 (hPGC-1) enhance the response to corticosterone. Yeast strain YNK441 carrying a vector alone (control) or expression plasmids for the indicated cDNAs was incubated overnight with 0, 1, or 10 μM corticosterone and assayed for β-gal activity. Data represent the mean ± standard deviation of results from six independent yeast transformants. LEMGH, HepG2 library isolate; LEM6/hPGC-1, full-length cDNA.
Yeast was transformed with a human hepatoma-derived (HepG2) cDNA library and selected for growth in media containing either 4 μM dexamethasone or 0.5 μM corticosterone, i.e., hormone concentrations that did not allow growth of the parental strain. Fifteen yeast transformants grew under selective conditions and showed an increased β-gal response to hormone, in a manner dependent on the presence of the mammalian cDNA library plasmid (see Materials and Methods). Rescue of the plasmids and sequencing of the inserts revealed three types of cDNAs that enhanced the response to hormone in yeast (Fig. 1B). The first cDNA encoded the molecular chaperone p23, a known component of the glucocorticoid aporeceptor complex (61), thereby establishing that the screen can yield factors involved in mammalian glucocorticoid signaling. The other two cDNAs encoded novel proteins that we initially named LEM5 and LEM6. This study focuses on LEM6. The sequence of the LEM6 HepG2 library isolate (LEM6H in Fig. 1B) predicted the expression of a truncated protein. We therefore isolated additional LEM6 clones from human liver cDNA and constructed a full-length yeast expression plasmid (see Materials and Methods). As shown in Fig. 1B, full-length LEM6 also enhanced the receptor-mediated response to hormone, albeit to a lesser extent.
LEM6 is a tissue-specific nuclear protein and the human homologue of murine PGC-1.
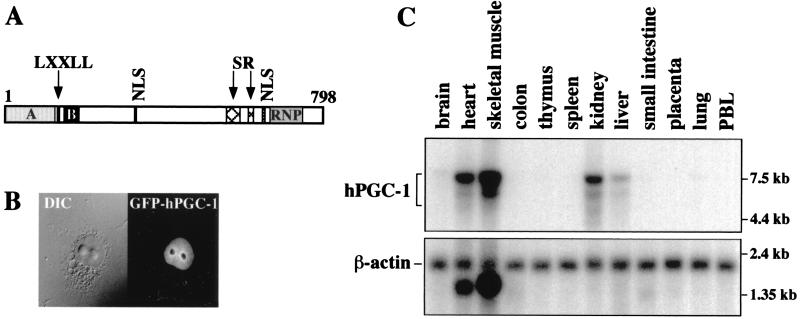
The full-length LEM6 cDNA sequence predicted a 798-aa protein that shares high identity (95%) with the mouse coactivator PGC-1 (62), suggesting that it is the human homologue of PGC-1. We therefore refer to LEM6 as hPGC-1. Like its mouse homologue, hPGC-1 is characterized by an N-terminal region rich in acidic amino acids (26.4% of aa 1 to 140), followed by a putative nuclear receptor interaction motif (LKKLL) at residues 144 to 148 and a predominantly basic stretch of amino acids at residues 168 to 207 (Fig. 2A). At the C terminus, there are two serine- and arginine (SR)-rich stretches (aa 566 to 599 and aa 621 to 631) and a putative RNA-binding domain (aa 677 to 753). Putative nuclear localization signals at residues 326 to 333, 627 to 633, and 651 to 667 predicted a nuclear protein. Indeed, fusion of hPGC-1 to GFP revealed exclusively nuclear fluorescence in transiently transfected mammalian cells (Fig. 2B).
FIG. 2.
(A) Schematic representation of the hPGC-1 protein. The N terminus is rich in charged residues (A, acidic; B, basic). The putative receptor interaction motif, LXXLL, is indicated (aa 144 to 148). The C terminus harbors two SR-rich stretches (aa 566 to 598 and aa 621 to 632) and a putative RNA-binding domain (RNP; aa 678 to 750). NLS, nuclear localization signals. (B) hPGC-1 is a nuclear protein. COS7 cells were transfected with a GFP–hPGC-1 expression vector, fixed, and analyzed for the localization of the fusion protein. (Left) Differential interference contrast (DIC) image. (Right) Fluorescence acquisition of the same field. (C) hPGC-1 mRNA is expressed in a tissue-specific manner. A human multiple-tissue northern blot (Clontech) was hybridized with an hPGC-1 probe (upper panel) or a β-actin-specific probe (lower panel). The positions of RNA markers (kilobases) are shown at the right of each panel. PBL, peripheral blood leukocytes.
Northern blot analysis of human tissues indicated that hPGC-1 is expressed in a tissue-specific manner. We detected at least two hPGC-1 mRNA transcripts (∼7.5 and 6 kb), predominantly in heart, skeletal muscle, kidney, and liver (Fig. 2C). Upon longer exposures, low levels of hPGC-1 expression could be seen first in brain and lung and then in small intestine, colon, and thymus. No expression was detectable in spleen, placenta, and peripheral blood leukocytes.
hPGC-1 is a potent activator of receptor-mediated transcription in mammalian cells.
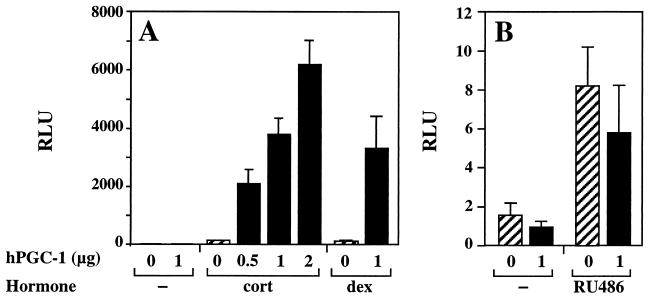
We isolated hPGC-1 based on its ability to enhance receptor-mediated transcription in yeast cells. To determine whether it displays similar activity in mammalian cells, we transiently transfected COS7 fibroblasts with hPGC-1 and GR expression plasmids and a GR-responsive luciferase reporter. In the absence of exogenous hPGC-1, the addition of hormones such as corticosterone and dexamethasone resulted in 50- to 60-fold induction of luciferase expression (Fig. 3A). Expression of hPGC-1 enhanced the hormone-induced transcription by another 20- to 60-fold, in a dose-dependent manner with respect to the amount of hPGC-1 plasmid transfected (Fig. 3A). Similar enhancement profiles were seen for cells treated with corticosterone, dexamethasone, or deoxycorticosterone (Fig. 3A and data not shown). Enhancement of transcription was strictly hormone dependent, as hPGC-1 expression did not increase luciferase expression in the absence of hormone (Fig. 3B).
FIG. 3.
hPGC-1 is a potent activator of hormone-dependent, receptor-mediated transcription in mammalian cells. COS7 cells were transfected with the receptor expression plasmid p6RGR, the indicated amounts of either the hPGC-1 expression vector pcDNA3/HA-hPGC-1 (black bars) or the empty vector pcDNA3 (hatched bars), and the GR-responsive luciferase reporter pTAT3Luc. The cells were treated for 24 h with either control vehicle (−) or 50 nM indicated ligand and assayed for luciferase activity. Luciferase activity in the presence of corticosterone (cort) and the absence of hPGC-1 (42,947 ± 17,676 luciferase units) was set equal to 100 within each experiment and used to normalize values from different experiments. Data represent the mean ± standard deviation of 8 to 10 transfections from four independent experiments. RLU, relative luciferase units; dex, dexamethasone.
The hormone requirement for the hPGC-1 effect on the glucocorticoid response could reflect the need for an agonist-induced conformation of the receptor or merely the translocation of the receptor to the nucleus and its binding to DNA. To distinguish between these possibilities, we tested whether hPGC-1 could enhance the transcriptional response to RU486. RU486 is an antagonist that displays partial agonist activity, depending on cell context, and induces a conformational change of GR distinct from that induced by pure agonists (19, 54). Treatment of transfected COS7 cells with RU486 led to the nuclear translocation of GR (data not shown) and a four- to fivefold activation of luciferase reporter expression (Fig. 3B). However, this modest induction was not further increased by hPGC-1, suggesting that hPGC-1 enhancement of the glucocorticoid response requires a specific, agonist-induced conformation of the receptor.
Specificity of hPGC-1 coactivator function.
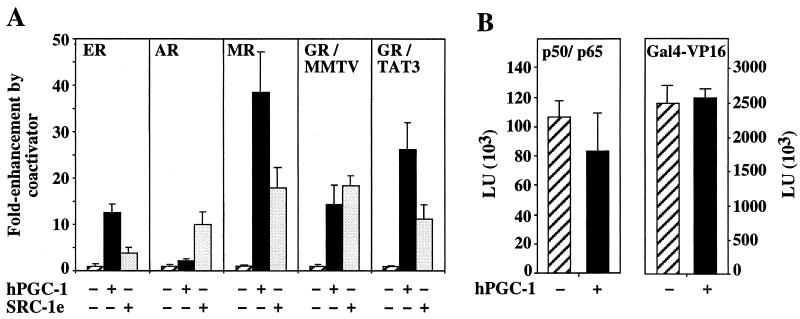
We next examined whether hPGC-1 could enhance hormone responsiveness for other steroid receptors. As shown in Fig. 4A, hPGC-1 expression enhanced the hormone-dependent transcription mediated by ER and MR strongly (by ∼13- and ∼38-fold, respectively) and that by AR only weakly (∼2-fold). The very weak effect of hPGC-1 on AR was confirmed in three different promoter contexts, the MMTV long terminal repeat response element (MMTV in Fig. 4A), the tyrosine aminotransferase HREs, and the probasin promoter (maximal enhancement, twofold; data not shown). AR activity was responsive to the effects of another coactivator, SRC-1e (27, 58), indicating that the lack of enhancement by hPGC-1 was not due to a general inability for increased transcription (Fig. 4A). Rather, a comparison of the coactivation conferred by SRC-1e and hPGC-1 on the different receptors and promoters tested suggests that the two coactivators act in a receptor- and promoter context-dependent manner. hPGC-1 was weaker than or equal to SRC-1e in certain contexts (AR on MMTV and TAT3; GR on MMTV) and considerably stronger in others (ER; MR and GR on TAT3) (Fig. 4A and data not shown).
FIG. 4.
(A) hPGC-1 enhances responses to steroids in a receptor-selective manner. Expression plasmids for ER, AR, MR, or GR were cotransfected into COS7 cells with control (hatched bars), hPGC-1 (black bars), or SRC-1e (gray bars) expression vectors and the following luciferase reporters: pERE-TK-luc for ER, pMMTVDLO (MMTV) for AR and GR, and pTAT3Luc (TAT3) for MR and GR. Cells were treated with 50 nM estradiol (ER), aldosterone (MR), or corticosterone (GR) or 100 nM dihydroxytestosterone (AR) for 24 h and assayed for luciferase activity. Results are expressed as fold enhancement by the coactivator in the presence of hormone; i.e., activity in the presence of hormone and absence of coactivator was set equal to 1 for all receptors. (B) hPGC-1 is not a general activator of transcription. COS7 cells were transfected with expression and reporter plasmids for either the two NF-κB subunits p50 and p60 (left panel) or the chimeric activator Gal4-VP16 (right panel) and assayed for luciferase activity. LU, luciferase units. Error bars show standard deviations.
To address whether hPGC-1 is a general activator of transcription, we tested its effect on two steroid-independent activators of transcription. The expression of the NF-κB subunits p50 and p65 (RelA) in COS7 cells resulted in a 40-fold induction of a luciferase reporter under the control of three NF-κB sites. hPGC-1 coexpression did not affect this induction (Fig. 4B). Similarly, hPGC-1 expression had no effect on the transcriptional activity of the chimeric activator Gal4-VP16 (Fig. 4B), suggesting that hPGC-1 is not a general coactivator of transcription.
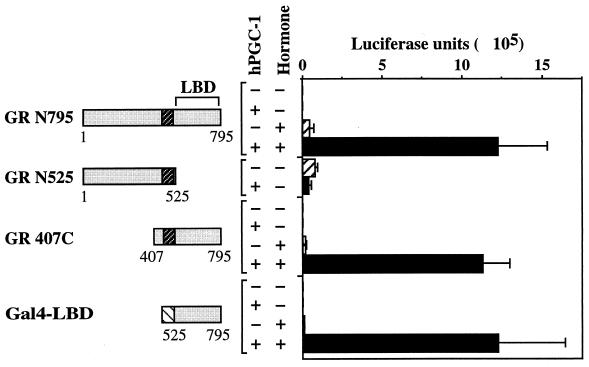
hPGC-1 acts via the LBD of GR.
Steroid receptors are modular proteins consisting of an N-terminal domain that carries a strong transcriptional activation function (AF1), a central domain that binds DNA, and a C-terminal domain (LBD) that binds hormone and carries a hormone-dependent transcriptional activation function (AF2). hPGC-1 could increase the response to hormone by interacting with a particular receptor domain and enhancing one or more of its functions, e.g., binding to DNA or activation of transcription. To delineate the affected receptor function, we measured the activity of truncated or chimeric variants of GR in the absence or presence of hPGC-1. As shown earlier, hPGC-1 stimulated efficiently the hormone-dependent induction mediated by full-length GR (N795 in Fig. 5). A C-terminal truncation of the receptor that removes the LBD and gives rise to a constitutively active transcription factor (18) eliminated the ability of hPGC-1 to enhance transcription (N525 in Fig. 5). In contrast, the receptor variant 407C, which lacks the N-terminal region but contains the DBD and the LBD, was still responsive to hPGC-1 (Fig. 5). Finally, the hormone-dependent activity of the chimeric Gal4-LBD protein, which contains the receptor LBD fused to the heterologous DBD of Gal4, was enhanced strongly by hPGC-1 (Fig. 5). The Gal4 DBD itself was unaffected (Fig. 4B). In conclusion, the LBD of the receptor is both essential and sufficient for a functional interaction with hPGC-1, suggesting that hPGC-1 is a coactivator of AF2, the hormone-dependent transcriptional activation function of the LBD.
FIG. 5.
hPGC-1 interacts functionally with the LBD of the receptor. COS7 cells were transfected with expression plasmids for either full-length GR (N795) (aa 1 to 795), the truncated GR variant N525 (aa 1 to 525) or 407C (aa 407 to 795), or the chimeric activator Gal4-LBD (aa 525 to 795 of GR); the GR- or Gal4-responsive reporter pTAT3Luc or pGK-1, respectively; and either vector alone (hatched bars) or the hPGC-1 expression vector (black bars). Cells were treated with 50 nM corticosterone (+) or carrier ethanol (−) for 24 h and assayed for luciferase activity. Data are the mean ± standard deviation luciferase units of eight transfections from four independent experiments.
The N terminus of hPGC-1 carries a potent transcriptional AD.
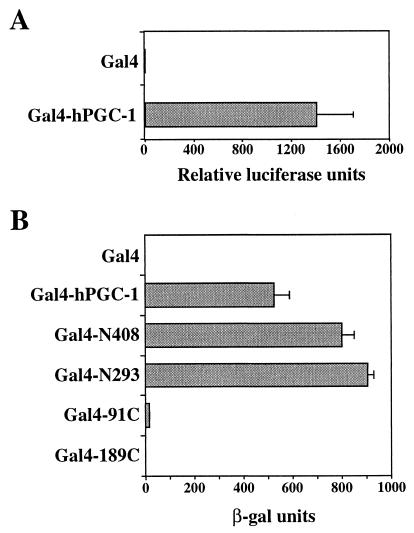
One of the hallmarks of transcriptional coactivators is that their recruitment to a specific promoter, either by protein-protein interactions with DNA-binding transcription factors such as GR or artificially by direct fusion to a DBD, activates transcription. This notion implies that coactivators contain transcriptional ADs. To determine whether hPGC-1 carries such a domain, we examined the ability of the chimeric Gal4–hPGC-1 protein, in which the coactivator is fused to the Gal4 DBD, to activate reporters under the control of Gal4-binding sites. As shown in Fig. 6, Gal4–hPGC-1 was a potent activator of transcription in both yeast and mammalian cells, indicating that hPGC-1 indeed harbors a transcriptional activation function.
FIG. 6.
hPGC-1 carries a transcriptional AD. (A) Gal4–hPGC-1 activates transcription in mammalian cells. COS7 cells transfected with the luciferase reporter plasmid pGK-1 and an expression vector for either the Gal4 DBD (aa 1 to 147 of Gal4) or the Gal4–hPGC-1 chimera were assayed for luciferase activity. Data are expressed relative to the activity seen with the Gal4 DBD alone (Gal4), which was set equal to 1, and are the mean ± standard deviation of six transfections from three independent experiments. (B) The hPGC-1 AD is in the N terminus. Yeast carrying a Gal4-responsive β-gal reporter and expression vectors for fusions of Gal4 (aa 1 to 147) to full-length or truncated hPGC-1 was assayed for β-gal activity. hPGC-1, aa 1 to 798; N408, aa 1 to 408; N293, aa 1 to 293; 91C, aa 91 to 798; 189C, aa 189 to 798. Data are the mean ± standard deviation β-gal activity of six or more independent yeast transformants.
To map the AD in hPGC-1, we tested the ability of truncated versions of hPGC-1 fused to Gal4 to activate transcription. Removal of the hPGC-1 C terminus did not reduce activity, suggesting that the AD is within the first 293 aa (N408 and N293 in Fig. 6B). Indeed, a deletion of the first 188 aa (189C) or 293 aa (294C) eliminated the ability to activate transcription (Fig. 6B and data not shown). Most of the activation function was in fact lost when just the first 90 aa were removed (91C). Since these constructs were expressed and functional in other assays (e.g., see Fig. 9 and data not shown), we concluded that the hPGC-1 AD lies in the N-terminal region, coinciding with the acidic amino acid stretch (Fig. 2A). We were unable to examine Gal4 fusions to smaller parts of the hPGC-1 N terminus, such as aa 1 to 90 or aa 1 to 186, because they were toxic. The toxicity of these constructs is consistent with a potent AD in this region, as similar toxicity has been observed with other strong transcriptional activators (15, 16).
FIG. 9.
The C terminus of hPGC-1 interacts with itself. Yeast carrying a Gal4-responsive β-gal reporter and expressing Gal4-294C was transformed with vectors for the Gal4 AD, either alone (AD) or fused to full-length and truncated hPGC-1 variants as indicated, and assayed for β-gal activity. hPGC-1 variants are named as in Fig. 6 and 7. Data are the mean ± standard deviation β-gal activity of six or more independent yeast transformants.
hPGC-1 and GR interact physically in a hormone-dependent manner.
The ability of hPGC-1 to enhance receptor-mediated transcription could be the result of its direct, hormone-dependent interaction with the receptor, leading to the recruitment of its strong transcriptional AD. To determine if the receptor and hPGC-1 interact physically, we used the yeast two-hybrid system, where the interaction between two proteins, one fused to a DBD and another fused to an AD, leads to the expression of β-gal (12). Yeast carrying a Gal4-responsive β-gal reporter was transformed with two vectors: one expressing the Gal4-LBD, shown above to be sufficient for the functional interaction with hPGC-1 (Fig. 5), and another expressing the Gal4 AD, either alone or fused to hPGC-1 (AD and AD–hPGC-1, respectively). As the transcriptional activation function of the receptor LBD is weak in yeast, the Gal4-LBD chimera was unable by itself to activate the β-gal reporter, even at high hormone concentrations (Fig. 7 and data not shown). In contrast, coexpression of Gal4-LBD and AD–hPGC-1 caused a strong induction of β-gal activity in the presence but not in the absence of hormone (Fig. 7), suggesting that the GR LBD and hPGC-1 interact physically in a hormone-dependent manner.
FIG. 7.
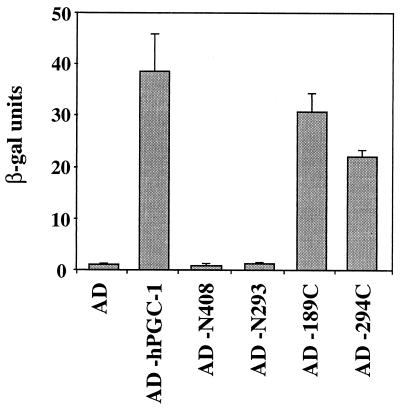
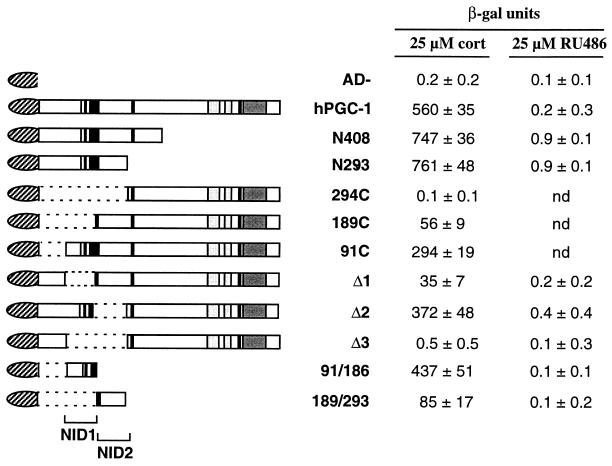
Two domains in hPGC-1 mediate a hormone-dependent interaction with the LBD of GR. hPGC-1 variants fused to the Gal4 AD were assayed for their ability to interact with a Gal4-LBD fusion (aa 525 to 795 of GR) in yeast carrying a Gal4-responsive β-gal reporter. Cells were treated with no hormone, 25 μM corticosterone (cort), or 25 μM RU486 for 20 h and assayed for β-gal activity. Values in the absence of hormone were <1 β-gal unit. Values shown are the mean ± standard deviation β-gal units of at least four independent yeast transformants. AD−, AD alone; nd, not determined. hPGC-1 variants are named as in Fig. 6; hPGC-1 Δ1, Δ2, and Δ3 carry deletions of aa 91 to 186, 189 to 293, and 91 to 293, respectively. 294C, aa 294 to 798; 91/186, aa 91 to 186; 189/293, aa 189 to 293.
To identify the domain(s) of hPGC-1 that mediates the interaction with the receptor, we tested fusions of the Gal4 AD to different parts of hPGC-1 in the two-hybrid assay. Deletion of large parts of the C terminus of hPGC-1 (as in N408 and N293) did not affect the interaction, suggesting that the interaction domain is within the first 293 aa (Fig. 7). Indeed, a deletion in this region (294C) gave rise to an otherwise functional protein (see Fig. 9) that did not interact with the receptor (Fig. 7). Further analysis of this region indicated the presence of two domains in hPGC-1 that can interact with the receptor; they were termed NID1 (aa 91 to 186) and NID2 (aa 189 to 293) (for nuclear receptor interaction domains [NIDs] 1 and 2, respectively). Constructs that have just one of the two NIDs, either NID1 (hPGC-1Δ2 and the minimal domain 91/186) or NID2 (189C, hPGC-1Δ1, and the minimal construct 189/293) showed a hormone-dependent interaction with the receptor (Fig. 7). In general, interactions mediated by NID1 were stronger (>250 β-gal units) than those mediated by NID2 (35 to 85 β-gal units). Deletion of both domains, as in hPGC-1Δ3, resulted in an hPGC-1 variant that was functional in other assays but could no longer interact with the receptor (Fig. 7 and data not shown).
The interactions between hPGC-1 and GR were dependent on the presence of corticosterone. To test whether the hormone dependence reflected a hormone-induced conformational change that enabled the interaction, we again tested the effect of the antagonist-partial agonist RU486. RU486 was unable to promote the interaction of GR with hPGC-1 (Fig. 7). Moreover, RU486 could compete with corticosterone for the hormone-induced interaction, i.e., acted as an antagonist (data not shown), demonstrating that both RU486 and corticosterone were able to bind the receptor but that only the full agonist corticosterone could promote a conformation competent to interact with hPGC-1.
The hPGC-1 AD and NIDs are essential for the coactivation of GR.
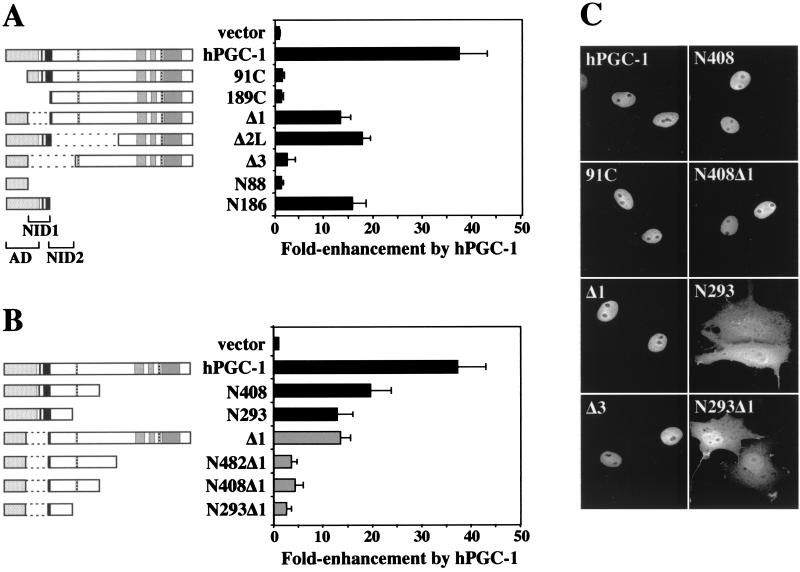
The functions encoded by hPGC-1 suggest that the mechanism by which it enhances the response to hormone involves first its recruitment by hormone-activated steroid receptors via the identified NIDs and second the enhancement of transcription via the identified AD. If this is the case, we would expect these domains to be essential for the hPGC-1 coactivator function. To test this notion, we assayed the ability of truncation and deletion variants of hPGC-1 to enhance hormone-dependent, receptor-mediated transcription in mammalian cells.
First, we addressed the importance of the AD. As shown before, full-length hPGC-1 enhanced strongly (35-fold) the transcriptional response to hormone (Fig. 8A). An hPGC-1 variant that lacked most of the AD (91C) could no longer enhance GR activity, indicating that the AD is essential for the coactivation function (Fig. 8A). The 91C protein was expressed at levels comparable to those of full-length hPGC-1, as determined by Western blot analysis (data not shown), and properly localized to the nucleus (Fig. 8C). A similar lack of activity was also seen with hPGC-1 constructs that lacked larger parts of the N terminus, such as 189C (Fig. 8A). Next, we determined the role of the two NIDs. Variants of hPGC-1 that lacked just NID1 (hPGC-1Δ1) or a region that includes NID2 (hPGC-1Δ2L) displayed reduced activity but were still able to enhance transcription by 14- to 18-fold (Fig. 8A). In contrast, deletion of both NIDs (hPGC-1Δ3) eliminated hPGC-1 activity (Fig. 8A). Again, constructs hPGC-1Δ1, hPGC-1Δ2L, and hPGC-1Δ3 expressed proteins that were properly localized to the nucleus, suggesting that the decrease in activity was due to the deletion of the interaction domains and not to changes in protein stability or localization (Fig. 8C and data not shown). In conclusion, our results indicate that both the AD and the ability to interact physically with the receptor are essential for the hPGC-1 coactivation function. While either NID in the context of full-length hPGC-1 is sufficient, the presence of both domains renders the coactivator more effective. Finally, the AD and the interaction with the receptor are sufficient for the coactivation of GR. As shown in Fig. 8A, an hPGC-1 construct that includes the AD and the stronger NID, NID1 (N186), enhanced transcription by 15-fold, suggesting that the first 186 aa form a core domain that, although compromised, can potentiate GR-mediated transcription.
FIG. 8.
Functional analysis of hPGC-1 domains. (A) The AD and NIDs are essential for hPGC-1 coactivation of GR. (B) The C-terminal domain contributes to hPGC-1 function. (A and B) COS7 cells were transfected with the GR expression vector p6RGR, the GR-responsive luciferase reporter plasmid pTAT3Luc, and either the control vector pcDNA3 or hPGC-1 variants as indicated, treated with 50 nM corticosterone for 24 h, and assayed for luciferase activity. Results are expressed as fold enhancement by hPGC-1, with activity in the presence of hormone and just the control vector pcDNA3 set equal to 1. Data are the mean ± standard deviation of 6 to 12 values from at least three independent experiments. hPGC-1 variants are named as in Fig. 6 and 7; hPGC-1 Δ2L carries a deletion of aa 189 to 482. N88, aa 1 to 88; N186, aa 1 to 186. (C) hPGC-1 variants are expressed and present in the nucleus. COS7 cells transfected with the indicated hPGC-1 constructs were analyzed for the expression and localization of the HA epitope-tagged protein by fluorescence microscopy. Note that cells transfected with hPGC-1 variants that lack the putative nuclear localization signals (N293 and N293Δ1) show some cytoplasmic staining but still have comparable levels of hPGC-1 in the nucleus.
The C terminus of hPGC-1 contributes to coactivator function.
The C terminus of hPGC-1 contains two recognizable features, an SR-rich region and a putative RNA-binding motif. To address a possible role of these domains in coactivation, we tested the ability of hPGC-1 variants that lack the C terminus to enhance receptor-mediated transcription. hPGC-1 constructs with different C-terminal truncations, such as N293 and N408, displayed reduced activity but were still good coactivators of transcription (Fig. 8B). Interestingly, deletion of the C terminus in hPGC-1 variants that already lacked NID1 (N482Δ1, N408Δ1, and N293Δ1) reduced hPGC-1 activity to almost the background level (Fig. 8B), suggesting that in the absence of the C terminus, the AD and the weaker NID, NID2, are not sufficient to enhance receptor activity. In conclusion, the hPGC-1 C terminus, although not essential, contributes to the coactivation function.
What might the role of the C terminus be? Motif prediction programs suggest that aa 635 to 670 may adopt a coiled-coil conformation (42). We therefore speculated that the C terminus could mediate homophilic interactions between hPGC-1 molecules. Oligomerization could stabilize hPGC-1 interactions with either steroid receptor dimers or other proteins. To test this notion, we used the two-hybrid system. Yeast carrying a Gal4-responsive β-gal reporter was transformed with vectors expressing the C terminus of hPGC-1 fused to the Gal4 DBD (Gal4-294C) as bait and hPGC-1 variants fused to the Gal4 AD as prey. As shown in Fig. 9, coexpression of Gal4-294C and AD–hPGC-1 induced β-gal activity, indicating that hPGC-1 can indeed interact with itself. This interaction was dependent on the C terminus, since it was detected with hPGC-1 proteins lacking the N terminus, such as 294C, but was lost in hPGC-1 variants N408 and N293, which lack the C-terminal region (Fig. 9).
DISCUSSION
In this work, we present a genetic scheme for the identification of mammalian proteins that regulate steroid hormone responses, based on their function in yeast. Using this approach, we have identified hPGC-1 as a protein that enhances glucocorticoid signaling. We show that hPGC-1 is a nuclear protein, expressed in a tissue-specific manner, and a potent coactivator of selective steroid hormone receptors.
The conservation of cellular pathways and molecular mechanisms from yeast to mammals has enabled genetic studies of higher eukaryotic processes in the simple, unicellular yeast. Hence, genes from several species have been cloned in yeasts by complementation of yeast mutants (33, 37, 39, 40, 66, 74) or in gain-of-function screens (44, 76). Since mammalian steroid receptors were first expressed in S. cerevisiae (46, 65), functional genetic screens have revealed yeast proteins participating in steroid signaling and led to the study of some of the conserved mammalian counterparts (6, 24, 29, 31, 32, 45). In parallel, the development and application of yeast two-hybrid screens has identified a number of proteins that interact physically with nuclear receptors (36). The approach that we present here, the functional screening of mammalian cDNA libraries in yeast engineered to reconstitute a mammalian pathway, should identify proteins regulating any step in the pathway, independent of a physical interaction or subcellular localization. We show that this approach can yield proteins of interest. Of the three proteins identified, the molecular chaperone p23 is a known component of steroid aporeceptor complexes. It interacts indirectly, via Hsp90, with the cytoplasmic, hormone-free GR and is thought to stabilize hormone-binding-competent receptors (11, 61). LEM5 is a novel, ubiquitous nuclear protein with a putative yeast homologue (A. Kaul and A. Kralli, unpublished data). In contrast, hPGC-1 (LEM6) is expressed in a tissue-specific manner and has no yeast or nematode homologue, suggesting that it could provide a regulatory function specific for vertebrates. Given our interest in proteins that confer specificity and sensitivity to lipophilic hormone signaling, we focused our studies on hPGC-1.
We show here that the nuclear protein hPGC-1 is a potent, bona fide coactivator of steroid receptors. It interacts with the hormone-activated form of GR, contains a transcriptional AD, and enhances strongly the transcriptional response mediated by GR, MR, and ER in mammalian cells. The high sequence identity of hPGC-1 with murine PGC-1 (95%) suggests that the two proteins are functional homologues. Murine PGC-1, isolated as a protein that interacts with the nuclear receptor PPARγ, has been shown to enhance transcription by PPARγ and TR and to be expressed in a tissue-specific manner (62). Taken together, these findings imply a wide role for hPGC-1 and mouse PGC-1 as tissue-specific regulators of nuclear receptor signaling.
Steroid receptors harbor two transcriptional ADs: the hormone-independent AF1 at the N terminus and the hormone-dependent AF2 at the C-terminal LBD. hPGC-1 enhanced the transcriptional activity of the LBD but not of the N-terminal part of the receptor, suggesting that it operates via AF2. In support of this notion, hPGC-1 enhanced the response to agonists that activate AF2 but not the response to partial agonists or antagonists, such as RU486, that do not induce proper LBD folding. Furthermore, one of the domains of hPGC-1 that interacts with the receptor LBD carries an LXXLL sequence, a motif that mediates the interaction of several proteins with the AF2 surfaces of receptors (20, 70). Altogether, these data suggest that hPGC-1 interacts with and enhances the activity of AF2.
Two adjacent domains in hPGC-1, NID1 and NID2, mediate the hormone-dependent interaction with the LBD of GR. NID1 (aa 91 to 186), as mentioned before, contains an LXXLL sequence. Secondary structure predictions for NID1 suggest that the LXXLL sequence and its flanking regions adopt a helical conformation, consistent with it being a nuclear receptor interaction surface. NID2 (aa 186 to 293) does not contain a consensus LXXLL motif but has a short sequence, LLKYLTTNDD, that resembles the third interaction box of the coactivator TIF2 (underlined amino acids are identical) and is also predicted to be part of an alpha helix. Future experiments will address the role of these motifs in the interactions mediated by NID1 and NID2. Finally, two lines of evidence suggest that NID1 is the major site of interaction with GR. First, the yeast two-hybrid assays show a stronger interaction of the receptor LBD with NID1 than with NID2 (Fig. 7). Second, hPGC-1 variants that lack NID1 display weaker GR coactivation than those that lack NID2, particularly when the C terminus of hPGC-1 is absent (compare N293Δ1 and N186 in Fig. 8).
Murine PGC-1 has also been reported to have two distinct interaction domains for PPARγ. One of them is in the N-terminal 180 aa and could be the same as NID1 of hPGC-1 (73). The second seems to be distinct from hPGC-1 NID2, since it is in a different region (aa 292 to 338 of murine PGC-1) and mediates a hormone-independent interaction with the DBD of PPARγ (62). These findings raise the interesting possibility that coactivators such as PGC-1 may have one interaction site (e.g., NID1) that directs them to a family of transcription factors, such as the nuclear receptors, and auxiliary sites (e.g., hPGC-1 NID2) that can discriminate among individual receptors within the large family. The first site could play a role in the strength of the interaction, and the second one could play a role in the specificity of the interaction. Interestingly, the coactivator GRIP1 was recently shown to have a C-terminal auxiliary domain that selectively enhances interactions with the LBDs of specific receptors (22).
Recruitment by the hormone-activated receptor would bring hPGC-1 to the vicinity of promoters under hormone regulation. What are the downstream effectors of hPGC-1? The presence of a strong transcriptional AD indicates that hPGC-1 makes direct contacts via its AD with either the basal transcription machinery or other transcription cofactors. The acidic nature of the hPGC-1 AD suggests contacts with targets of other acidic domains, e.g., TFIIA (30), histone acetyltransferase complexes (71), or the Mediator complex (also known as ARC and Srb) (5, 50). An LLXXLXXXL sequence at aa 88 to 96, similar to the CBP interaction motif in the p160 (NCoA) family of coactivators (70), implies possible contacts with CBP. Since Gal4–hPGC-1 is a potent transcriptional activator in both yeast and mammalian cells, it is possible that hPGC-1 can interact with more than one of these effectors, thereby activating multiple steps in transcription initiation and elongation.
Efficient coactivation of GR requires the C terminus of hPGC-1. A possible reason for this is the ability of this region to mediate homophilic hPGC-1 interactions. A “dimerization” surface could stabilize the binding of an hPGC-1 dimer to a receptor dimer, in a complex where each NID1 contacts one receptor AF2. Dimerization could also help NID2 to compensate for the loss of NID1. Deletion of the dimerization function might lead to two alternative complexes: (i) two hPGC-1 monomers and a receptor dimer, where the stronger NID (NID1) interacts with each AF2, or (ii) an hPGC-1 monomer and a receptor dimer, where NID1 and NID2 each interact with one receptor AF2. The latter case is similar to what has been observed for the crystal structure of the PPARγ LBD with an SRC-1 fragment that has two NIDs; a single SRC-1 molecule contacts via two LLXXLL motifs the two AF2 surfaces of the receptor dimer (53). In either case, a deletion of NID1 in the absence of the C terminus may leave the weaker NID (NID2) unable to mediate a stable interaction with the receptor dimer. This could explain why hPGC-1Δ1, which has both NID2 and the dimerization interface, but not N482Δ1, which has just NID2, can enhance GR activity (Fig. 8). A general role for dimerization is also consistent with the observations that receptor dimerization is important for other receptor-coactivator interactions (27) and that two molecules of the coactivator TIF2 bind cooperatively to a nuclear receptor heterodimer (38).
The C terminus of hPGC-1 also carries novel motifs for transcriptional coactivators, such as the SR-rich sequences and the putative RNA-binding domain. Although such features are often associated with posttranscriptional RNA-processing regulators, they have been found recently in other proteins that interact with transcription factors. The family of CTD-associated SR-like proteins (CASPs) interacts with the C-terminal domain of RNA polymerase II and has been suggested to couple transcription to pre-mRNA processing (10, 79). An RNA-binding protein, TLS, has been purified as a thyroid receptor-interacting protein (60). Although speculative at the moment, interesting roles for these domains can be proposed. First, they may bind an RNA cofactor (e.g., the steroid receptor RNA activator SRA [34]) that could play a structural role in interactions with other transcription factors or regulate hPGC-1 activity. More interestingly, they may enable coactivators such as hPGC-1 to regulate steps other than transcription initiation and elongation, e.g., pre-mRNA processing. Steroid receptors could recruit hPGC-1 and deliver it to the splicing machinery, possibly via RNA polymerase II, thereby providing hormone-regulated, gene-specific RNA splicing.
The identification of hPGC-1 establishes one more member of an already large group of coactivators for steroids and other nuclear receptors: the p160 family (represented by SRC-1, TIF2, and pCIP]), the histone acetyltransferases CBP and pCAF, and the multisubunit Mediator (also called SMCC, DRIP, TRAP, ARC, CRSP, and Srb) (reviewed in references 13, 28, 69, and 75). One of the challenges in transcriptional regulation by nuclear receptors is to understand how specificity (such as tissue, receptor, signal, or promoter specificity) and sensitivity to regulation (i.e., sensing and adapting to changes in cell state) are established. The existence of multiple coactivators could confer these properties, if the coactivators were expressed tissue specifically, showed selectivity for different receptors, affected different limiting steps in the control of gene expression, and were subject to regulation. hPGC-1 and murine PGC-1 display many of these properties. First, they are expressed tissue specifically. The predominant expression in heart, skeletal muscle, kidney, liver, and brown fat overlaps the expression of GR, MR, TR, and PPARγ and correlates with the sensitivity of these receptors to PGC-1. Second, a comparison of hPGC-1 and SRC-1e highlights receptor selectivity. The efficiency of the two coactivators is reversed on different receptors, SRC-1e being more potent with AR and hPGC-1 being stronger with ER, GR, and MR. Third, the effects of hPGC-1 and murine PGC-1 may depend on promoter context. Enhancement of GR activity by hPGC-1 but not by SRC1e was stronger at the TAT3 reporter than at the MMTV reporter. Similarly, Wu et al. reported that murine PGC-1 coactivation is promoter dependent, affecting the transcription of selective PPARγ-targeted genes in muscle (73). Finally, hPGC-1 and murine PGC-1 are good candidates for integrating other signaling pathways. Exposure of mice to cold and activation of β-adrenergic receptors lead to increased expression of murine PGC-1 (4, 62). The presence of putative phosphorylation sites suggests additional levels of regulation. We have observed that the ability of hPGC-1 to enhance GR activity is cell type dependent (data not shown). Understanding the regulatory mechanisms and the specificity of coactivator function will be the next challenge.
ACKNOWLEDGMENTS
We thank F. Hamy, J. Iniguez-Lluhi, P. Matthias, R. Nissen, M. Parker, D. Picard, and B. Starr for sharing plasmids; Chiron and Tony Brake for the HepG2 cDNA library; R. Skoda for the human liver cDNA; Exelgyn for mifepristone (RU486); and M. Hall, J. Iniguez-Lluhi, U. Müller, D. Picard, R. Sitcheran, M. Spiess, and B. Starr for discussions and helpful comments on the manuscript.
This work was supported by the Swiss National Science Foundation (A.K.), the Basel Chemical Industry (D.K.), and the Max Cloëtta Foundation (A.K.).
REFERENCES
- 1.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 2.Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 3.Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi M E, Taraseviciene L, Nordeen S K, Allegretto E A, Edwards D P. High-mobility-group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boss O, Bachman E, Vidal-Puig A, Zhang C Y, Peroni O, Lowell B B. Role of the beta(3)-adrenergic receptor and/or a putative beta(4)-adrenergic receptor on the expression of uncoupling proteins and peroxisome proliferator-activated receptor-gamma coactivator-1. Biochem Biophys Res Commun. 1999;261:870–876. doi: 10.1006/bbrc.1999.1145. [DOI] [PubMed] [Google Scholar]
- 5.Boyer T G, Martin M E, Lees E, Ricciardi R P, Berk A J. Mammalian Srb/mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 6.Cairns B R, Levinson R S, Yamamoto K R, Kornberg R D. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 1996;10:2131–2144. doi: 10.1101/gad.10.17.2131. [DOI] [PubMed] [Google Scholar]
- 7.Chawla S, Hardingham G E, Quinn D R, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 10.Corden J L, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 11.Dittmar K D, Demady D R, Stancato L F, Krishna P, Pratt W B. Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery. The role of p23 is to stabilize receptor.hsp90 heterocomplexes formed by hsp90.p60.hsp70. J Biol Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213. [DOI] [PubMed] [Google Scholar]
- 12.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 13.Freedman L P. Increasing the complexity of coactivation in nuclear receptor signaling. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 14.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert D M, Heery D M, Losson R, Chambon P, Lemoine Y. Estradiol-inducible squelching and cell growth arrest by a chimeric VP16-estrogen receptor expressed in Saccharomyces cerevisiae: suppression by an allele of PDR1. Mol Cell Biol. 1993;13:462–472. doi: 10.1128/mcb.13.1.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 17.Godowski P J, Picard D, Yamamoto K R. Signal transduction and transcriptional regulation by glucocorticoid receptor-LexA fusion proteins. Science. 1988;241:812–816. doi: 10.1126/science.3043662. [DOI] [PubMed] [Google Scholar]
- 18.Godowski P J, Rusconi S, Miesfeld R, Yamamoto K R. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature. 1987;325:365–368. doi: 10.1038/325365a0. . (Erratum 326:105.) [DOI] [PubMed] [Google Scholar]
- 19.Guido E C, Delorme E O, Clemm D L, Stein R B, Rosen J, Miner J N. Determinants of promoter-specific activity by glucocorticoid receptor. Mol Endocrinol. 1996;10:1178–1190. doi: 10.1210/mend.10.10.9121486. [DOI] [PubMed] [Google Scholar]
- 20.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 21.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 22.Hong H, Darimont B D, Ma H, Yang L, Yamamoto K R, Stallcup M R. An additional region of coactivator GRIP1 required for interaction with the hormone-binding domains of a subset of nuclear receptors. J Biol Chem. 1999;274:3496–3502. doi: 10.1074/jbc.274.6.3496. [DOI] [PubMed] [Google Scholar]
- 23.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 24.Imhof M O, McDonnell D P. Yeast RSP5 and its human homolog hRPF1 potentiate hormone-dependent activation of transcription by human progesterone and glucocorticoid receptors. Mol Cell Biol. 1996;16:2594–2605. doi: 10.1128/mcb.16.6.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iniguez-Lluhi J A, Lou D Y, Yamamoto K R. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- 26.Jantzen H M, Strahle U, Gloss B, Stewart F, Schmid W, Boshart M, Miksicek R, Schutz G. Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell. 1987;49:29–38. doi: 10.1016/0092-8674(87)90752-5. [DOI] [PubMed] [Google Scholar]
- 27.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingston R E. A shared but complex bridge. Nature. 1999;399:199–200. doi: 10.1038/20302. [DOI] [PubMed] [Google Scholar]
- 29.Knoblauch R, Garabedian M J. Role for Hsp90-associated cochaperone p23 in estrogen receptor signal transduction. Mol Cell Biol. 1999;19:3748–3759. doi: 10.1128/mcb.19.5.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi N, Horn P J, Sullivan S M, Triezenberg S J, Boyer T G, Berk A J. DA-complex assembly activity required for VP16C transcriptional activation. Mol Cell Biol. 1998;18:4023–4031. doi: 10.1128/mcb.18.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kralli A, Bohen S P, Yamamoto K R. LEM1, an ATP-binding-cassette transporter, selectively modulates the biological potency of steroid hormones. Proc Natl Acad Sci USA. 1995;92:4701–4705. doi: 10.1073/pnas.92.10.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kralli A, Yamamoto K R. An FK506-sensitive transporter selectively decreases intracellular levels and potency of steroid hormones. J Biol Chem. 1996;271:17152–17156. doi: 10.1074/jbc.271.29.17152. [DOI] [PubMed] [Google Scholar]
- 33.Lahue E E, Smith A V, Orr-Weaver T L. A novel cyclin gene from Drosophila complements CLN function in yeast. Genes Dev. 1991;5:2166–2175. doi: 10.1101/gad.5.12a.2166. [DOI] [PubMed] [Google Scholar]
- 34.Lanz R B, McKenna N J, Onate S A, Albrecht U, Wong J, Tsai S Y, Tsai M J, O'Malley B W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 35.Lavinsky R M, Jepsen K, Heinzel T, Torchia J, Mullen T M, Schiff R, Del-Rio A L, Ricote M, Ngo S, Gemsch J, Hilsenbeck S G, Osborne C K, Glass C K, Rosenfeld M G, Rose D W. Diverse signaling pathways modulate nuclear receptor recruitment of N- CoR and SMRT complexes. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J W, Choi H S, Gyuris J, Brent R, Moore D D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 37.Lee M G, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- 38.Leers J, Treuter E, Gustafsson J A. Mechanistic principles in NR box-dependent interaction between nuclear hormone receptors and the coactivator TIF2. Mol Cell Biol. 1998;18:6001–6013. doi: 10.1128/mcb.18.10.6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leopold P, O'Farrell P H. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell. 1991;66:1207–1216. doi: 10.1016/0092-8674(91)90043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lew D J, Dulic V, Reed S I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y Z, Chrivia J C, Latchman D S. Nerve growth factor upregulates the transcriptional activity of CBP through activation of the p42/p44(MAPK) cascade. J Biol Chem. 1998;273:32400–32407. doi: 10.1074/jbc.273.49.32400. [DOI] [PubMed] [Google Scholar]
- 42.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 43.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maser P, Sutterlin C, Kralli A, Kaminsky R. A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science. 1999;285:242–244. doi: 10.1126/science.285.5425.242. [DOI] [PubMed] [Google Scholar]
- 45.McDonnell D P, Vegeto E, O'Malley B W. Identification of a negative regulatory function for steroid receptors. Proc Natl Acad Sci USA. 1992;89:10563–10567. doi: 10.1073/pnas.89.22.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Metzger D, White J H, Chambon P. The human oestrogen receptor functions in yeast. Nature. 1988;334:31–36. doi: 10.1038/334031a0. [DOI] [PubMed] [Google Scholar]
- 47.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 48.Moyer M L, Borror K C, Bona B J, DeFranco D B, Nordeen S K. Modulation of cell signaling pathways can enhance or impair glucocorticoid-induced gene expression without altering the state of receptor phosphorylation. J Biol Chem. 1993;268:22933–22940. [PubMed] [Google Scholar]
- 49.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 51.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. The signal-dependent coactivator CBP is a nuclear target for pp90RSK. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 53.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 54.Nordeen S K, Bona B J, Moyer M L. Latent agonist activity of the steroid antagonist, RU486, is unmasked in cells treated with activators of protein kinase A. Mol Endocrinol. 1993;7:731–742. doi: 10.1210/mend.7.6.8395651. [DOI] [PubMed] [Google Scholar]
- 55.Nordeen S K, Moyer M L, Bona B J. The coupling of multiple signal transduction pathways with steroid response mechanisms. Endocrinology. 1994;134:1723–1732. doi: 10.1210/endo.134.4.8137736. [DOI] [PubMed] [Google Scholar]
- 56.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 57.Onate S A, Prendergast P, Wagner J P, Nissen M, Reeves R, Pettijohn D E, Edwards D P. The DNA-bending protein HMG-1 enhances progesterone receptor binding to its target DNA sequences. Mol Cell Biol. 1994;14:3376–3391. doi: 10.1128/mcb.14.5.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 59.Pearce D, Yamamoto K R. Mineralocorticoid and glucocorticoid receptor activities distinguished by nonreceptor factors at a composite response element. Science. 1993;259:1161–1165. doi: 10.1126/science.8382376. [DOI] [PubMed] [Google Scholar]
- 60.Powers C A, Mathur M, Raaka B M, Ron D, Samuels H H. TLS (translocated-in-liposarcoma) is a high-affinity interactor for steroid, thyroid hormone, and retinoid receptors. Mol Endocrinol. 1998;12:4–18. doi: 10.1210/mend.12.1.0043. [DOI] [PubMed] [Google Scholar]
- 61.Pratt W B, Toft D O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocrinol Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 62.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 63.Ribeiro R C J, Cavalieri R R, Lomri N, Rahmaoui C M, Baxter J D, Scharschmidt B F. Thyroid hormone export regulates cellular hormone content and response. J Biol Chem. 1996;271:17147–17151. doi: 10.1074/jbc.271.29.17147. [DOI] [PubMed] [Google Scholar]
- 64.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 65.Schena M, Yamamoto K R. Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science. 1988;241:965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- 66.Schild D, Brake A J, Kiefer M C, Young D, Barr P J. Cloning of three human multifunctional de novo purine biosynthetic genes by functional complementation of yeast mutations. Proc Natl Acad Sci USA. 1990;87:2916–2920. doi: 10.1073/pnas.87.8.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 69.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 70.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 71.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 72.Webb P, Lopez G N, Uht R M, Kushner P J. Tamoxifen activation of the estrogen receptor/AP-1 pathway: potential origin for the cell-specific estrogen-like effects of antiestrogens. Mol Endocrinol. 1995;9:443–456. doi: 10.1210/mend.9.4.7659088. [DOI] [PubMed] [Google Scholar]
- 73.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla R C, Spiegelman B M. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 74.Xiong Y, Connolly T, Futcher B, Beach D. Human D-type cyclin. Cell. 1991;65:691–699. doi: 10.1016/0092-8674(91)90100-d. [DOI] [PubMed] [Google Scholar]
- 75.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 76.Xu Q, Reed J C. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 77.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 78.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 79.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C C, Krieg S, Shapiro D J. HMG-1 stimulates estrogen response element binding by estrogen receptor from stably transfected HeLa cells. Mol Endocrinol. 1999;13:632–643. doi: 10.1210/mend.13.4.0264. [DOI] [PubMed] [Google Scholar]