ABSTRACT
In fission yeast, polarized cell growth stops during division and resumes after cytokinesis completes and cells separate. It is unclear how growth reactivation is timed to occur immediately after cell separation. We uncoupled these sequential events by delaying cytokinesis with a temporary Latrunculin A treatment. Mitotic cells recovering from treatment initiate end growth during septation, displaying a polar elongation simultaneous with septation (PrESS) phenotype. PrESS cell ends reactivate Cdc42, a major regulator of polarized growth, during septation, but at a fixed time after anaphase B. A candidate screen implicates Rga4, a negative regulator of Cdc42, in this process. We show that Rga4 appears punctate at the cell sides during G2, but is diffuse during mitosis, extending to the ends. Although the Morphogenesis Orb6 (MOR) pathway is known to promote cell separation and growth by activating protein synthesis, we find that, for polarized growth, removal of Rga4 from the ends is also necessary. Therefore, we propose that growth resumes after division once the MOR pathway is activated and the ends lose Rga4 in a cell-cycle-dependent manner.
KEY WORDS: Cdc42, Rga4, Cell cycle, Cell growth, Cytokinesis, Polarization
Summary: Resumption of polarized Cdc42 activation at growth sites depends on a cell-cycle cue. This is facilitated by the loss of a Cdc42 GAP, Rga4, from growth sites at the end of cell division.
INTRODUCTION
Most eukaryotic cells undergo polarized growth to achieve specific shapes necessary for function. Polarized growth requires polarization of growth-promoting signaling networks, actin organization and membrane trafficking (Etienne-Manneville, 2004; Nance and Zallen, 2011; Ridley, 2006). Polarization is also essential for cytokinesis, the final step in cell division (Albertson et al., 2005; Echard, 2008; Hercyk et al., 2019a; Wang et al., 2016). Because polarized growth and cytokinesis share the same polarization apparatus, they must be temporally distinct events. Accordingly, careful regulation ensures that cells alternate between growth and division. However, it is unclear how this is regulated at the molecular level. The major drivers of polarization are highly conserved among eukaryotes (Etienne-Manneville, 2004; Johnson, 1999). Therefore, we use fission yeast Schizosaccharomyces pombe to establish basic principles about how cells spatiotemporally regulate their polarization machinery.
In fission yeast, cytokinesis involves the formation of an actomyosin ring, which constricts along with an ingressing membrane barrier and septum to physically separate two daughter cells (Cheffings et al., 2016; Pollard, 2010, 2014). The septum matures and is eventually digested by glucanases, leading to cell separation (Cortés et al., 2016; García Cortés et al., 2016; Sipiczki, 2007). After cell separation, polarized growth initiates from the old ends, which existed in the previous generation (Mitchison and Nurse, 1985). Once the cells attain a certain size, the new ends, generated as a result of cell division, initiate growth by a process called new end take-off (NETO). The Rho GTPase Cdc42 is a major regulator of growth and polarity and is highly conserved among eukaryotes (Etienne-Manneville, 2004; Johnson, 1999). Cdc42 also plays a role in cytokinesis (Hercyk and Das, 2019a; Onwubiko et al., 2020; Wei et al., 2016). Cdc42 regulation determines when and where polarization occurs (Das et al., 2009, 2012; Etienne-Manneville, 2004; Howell et al., 2012). In S. pombe, Cdc42 is activated by two guanine-nucleotide-exchange factors (GEFs), Scd1 and Gef1 (Chang et al., 1994; Coll et al., 2003), and is inactivated by three GTPase-activating proteins (GAPs), Rga4, Rga6 and Rga3 (Das et al., 2007; Gallo Castro and Martin, 2018; Revilla-Guarinos et al., 2016; Tatebe et al., 2008). Of these, Rga4 is the primary GAP, while Rga6 and Rga3 play minor roles in polarity and cell pairing during sexual reproduction, respectively.
During mitosis, Cdc42 is inactivated at the ends, and polarized growth stops (Hercyk and Das, 2019b). Once two daughter cells have been generated via cytokinesis, Cdc42 is reactivated at the old ends and polarized growth resumes (Wei et al., 2016). The bio-probe CRIB-3xGFP specifically binds active Cdc42 and is used to identify sites of Cdc42 activity (Tatebe et al., 2008). CRIB-3xGFP localization indicates that Cdc42 is activated at growing cell ends during interphase (Das et al., 2012; Tatebe et al., 2005). During mitosis, active Cdc42 disappears from the ends and appears at the division site (Wei et al., 2016). After cell separation, Cdc42 is not immediately activated at a daughter cell's new end, but instead at its old end. It is unclear how Cdc42 activation transitions from the division site to the old ends after cell separation.
Although much is understood about how polarized growth occurs, little is known about the regulation of when polarized growth occurs. Cell-cycle-dependent signaling pathways temporally segregate cell division and growth phases (Ray et al., 2010; Simanis, 2015). During interphase, the Morphogenesis Orb6 (MOR) pathway promotes cell growth (Nunez et al., 2016; Ray et al., 2010). During cell division, the septation initiation network (SIN) is activated (Johnson et al., 2012; Simanis, 2015). SIN activation leads to MOR pathway inactivation, thus inhibiting cell growth during division (Ray et al., 2010). Subsequent SIN inactivation allows MOR pathway activation, leading to cell separation and growth (Gupta et al., 2013, 2014). This crosstalk between the SIN and MOR signaling pathways thus enforces temporal separation of cytokinesis and cell growth.
The MOR pathway promotes cell separation and polarized growth activation, which occur sequentially. It is unclear how the MOR pathway sequentially promotes these processes. The MOR pathway activates the Orb6/NDR kinase, which phosphorylates the exoribonuclease Sts5 (Nunez et al., 2016). Sts5 phosphorylation prevents its incorporation into processing bodies (P-bodies), where mRNA is stored or degraded (Nunez et al., 2016). Orb6 thus prevents Sts5-dependent mRNA degradation in the P-bodies, leading to increased protein synthesis. Among the mRNAs spared from degradation are those that encode the glucanases Eng1 and Agn1, and the Ras1 GEF Efc25 (Chen et al., 2019; Nunez et al., 2016). Whereas Eng1 and Agn1 promote septum digestion and cell separation, Efc25-mediated Ras1 activation promotes the localization of the Cdc42 GEF Scd1 to cell ends (García et al., 2005; Lamas et al., 2020a; Martin-Cuadrado et al., 2003; Papadaki et al., 2002). Thus, Orb6 activity promotes cell separation and polarized cell growth. However, constitutively activating the MOR pathway does not lead to constitutive cell growth, but rather to premature cell separation resulting in cell lysis (Gupta et al., 2014). This suggests that, once the MOR pathway is activated, it immediately promotes both cell separation and then growth initiation. Thus, the nature of the regulation that allows cell separation and growth initiation to occur sequentially is the focus of this study.
Here, we investigated how growth is reactivated at the ends after cell division. We observed that Cdc42 activity at the ends resumes at a fixed time after completion of anaphase B and is independent of cell separation. Our candidate screen for regulators involved in Cdc42 reactivation at cell ends identified the GAP Rga4. Rga4 localizes to cell sides, where it prevents Cdc42 activation (Das et al., 2007; Tatebe et al., 2008). We found that the localization pattern of Rga4 along the cortex changes in a cell-cycle-dependent manner. During interphase, Rga4 appears punctate along the cortex and is excluded from cell ends. During mitosis, Rga4 is diffuse and localizes all along the cortex, including the ends. We propose that Rga4 localization at ends during mitosis blocks Cdc42 activation at these ends. After mitosis, Rga4 is lost from the ends and Cdc42 activity resumes. This suggests that, after mitosis, growth activation at cell ends occurs once Rga4 is lost from these ends. In keeping with these findings, we show that, in mutants constitutively activating the MOR pathway, loss of rga4 leads to enhanced polarized cell growth. These data suggest that cell-cycle-dependent regulation of Rga4 paired with MOR pathway activation allows cell separation and polarized cell growth to occur sequentially.
RESULTS
Cytokinetic delay results in growth resumption without cell separation
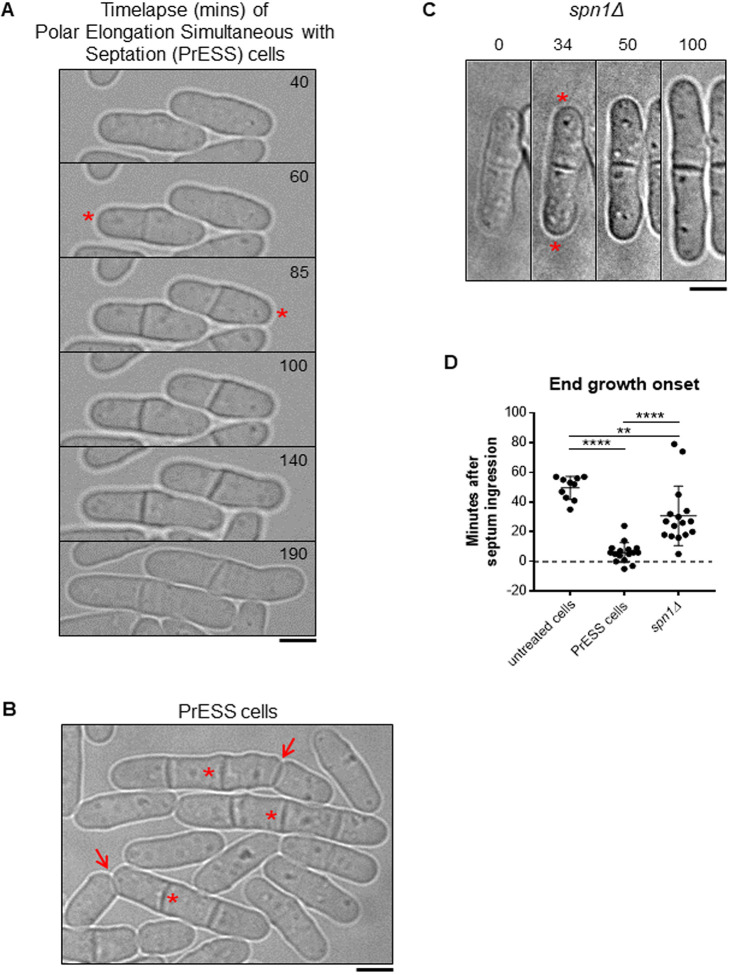
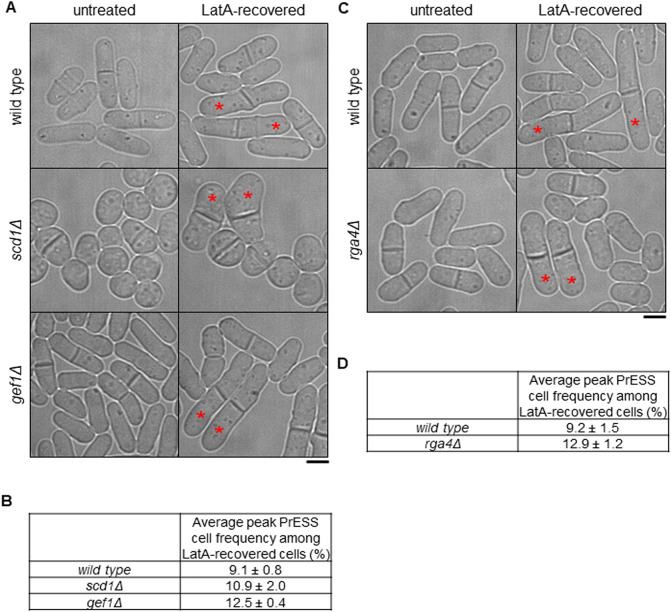
Because polarized growth occurs only after cell separation, we asked whether the timing of polarized growth initiation is impacted by cytokinesis. To test this, we used Latrunculin A (LatA) to induce a cytokinetic delay. LatA prevents actin polymerization, leading to loss of actin-based structures (Spector et al., 1983). Phalloidin staining shows loss of actomyosin rings upon LatA treatment (Fig. S1A). LatA treatment disrupts the actomyosin ring, and, upon washout, a new actomyosin ring assembles and cytokinesis resumes, albeit delayed (Fig. S1B) (Swulius et al., 2018). We treated an asynchronous population of wild-type cells with 10 µM LatA for 30 min and then washed it out. Cells typically take ∼45–60 min after LatA washout to recover before they display growth. After recovery, we observed that some cells (∼9%) show a unique phenotype in which they initiate growth at their ends as the cell is undergoing septation (Fig. 1A,B; Movie 1). We compared the timing of end-growth initiation with septum closure in these cells using bright-field imaging (Fig. S1C). In untreated wild-type cells, end growth initiates on average 50 min after septum closure. However, in cells recovering from LatA treatment, end growth initiates ∼6 min after septum closure while the septum is maturing (Fig. 1D). In some cells, growth initiates even before septum closure. We observed similar results when growth resumption was timed from the completion of ring closure, as depicted by Rlc1-tdTomato (Fig. S1D). Although these cells fail to separate, they enter the next cell cycle and undergo cell division. The septum formed in the second generation separates normally, suggesting that the LatA treatment only impacts the cell cycle in which it was treated (Fig. 1B, arrows). We call this phenotype polar elongation simultaneous with septation (PrESS). We also observed that the old ends of PrESS cells grow up to three times as fast as old ends in wild-type cells before NETO (Fig. S1E). The PrESS phenotype suggests that growth initiation after mitosis is not timed by cytokinetic events.
Fig. 1.
Cytokinetic delay uncouples end-growth resumption from cell separation. (A) Time lapse of cells displaying the PrESS phenotype after LatA washout. Growth initiation is indicated by asterisks. (B) Asynchronous wild-type cells were treated with 10 µM LatA for 30 min, then washed and allowed to recover. A subset of cells show the PrESS phenotype, in which they resume growth as the septum forms and eventually fail to separate (asterisks next to original septum). In the next cell cycle, these cells grow and separate normally (arrows). (C) Time lapse of growth after division in an spn1Δ cell. Asterisks denote growth initiation. (D) PrESS cells resume growth significantly earlier in relation to septum closure than untreated or spn1Δ cells (ordinary one-way ANOVA with Tukey’s multiple comparisons test; **P=0.004, ****P<0.0001; n≥10 cells). Timestamps in B and C refer to time since completion of septum closure. Scale bars: 5 μm.
PrESS cells visually resemble septin mutants, in which cell separation defects have been previously documented (Fig. 1C) (An et al., 2004; Berlin et al., 2003; Martín-Cuadrado et al., 2005; Tasto et al., 2003). However, although PrESS cells resume growth as the septum forms, it was unclear whether spn1Δ mutants behave similarly or whether they only resume growth after cell separation failure. We analyzed the timing of end-growth resumption in spn1Δ mutants compared to septum closure. We found that the ends resume growth on average ∼31 min after septum closure in spn1Δ cells, which fail to separate (Fig. 1D). This suggests that these cells first fail to separate, and then eventually growth resumes at the proper time. In contrast, in PrESS cells, cytokinesis and growth are uncoupled, such that growth resumes even before cell separation is possible.
Cdc42 activation at cell ends is cell-cycle dependent
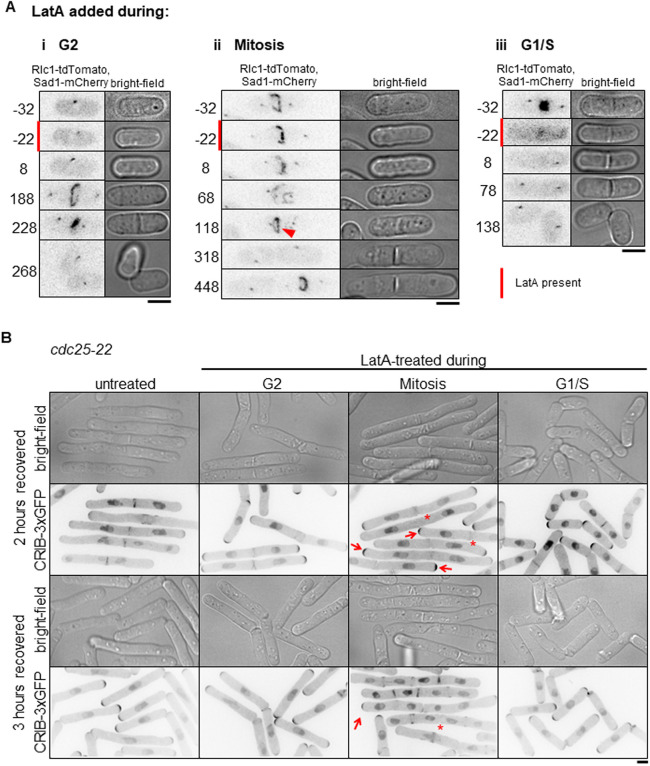
Only a subset of asynchronous cells exhibits the PrESS phenotype upon recovery from LatA treatment. We asked whether this is because the PrESS phenotype only occurs in cells in a certain cell-cycle stage. To address this, we imaged cells at various cell-cycle stages before, during and after LatA treatment by affixing the cells to the imaging dish using lectin (Tay et al., 2018), and performing LatA treatment and washout within the dish itself. Cells in lectin-coated dishes tend to grow slower than normal, but do not show any significant growth defects. We found that mitotic cells treated with LatA show the PrESS phenotype after recovery (Fig. 2Aii; Fig. S2A, Movie 2) and that cells treated during interphase or G1/S separate normally (Fig. 2Ai,iii; Fig. S2A). When treated with LatA, cells in anaphase proceed through mitosis, as detected with the spindle pole body marker Sad1-mCherry. However, the actomyosin ring disassembles, as marked by the type 2 myosin light chain Rlc1-tdTomato. After LatA washout, a new actomyosin ring assembles and then constricts along with septum ingression (Fig. 2Aii, arrowhead; Fig. S1B,C). However, instead of undergoing septum digestion and cell separation, these cells grow from the ends displaying the PrESS phenotype. Thus, the PrESS phenotype arises from cells that are in mitosis during LatA treatment.
Fig. 2.
The PrESS phenotype is cell-cycle dependent. (A) Time-lapse imaging of cells undergoing LatA treatment and recovery. Red bars denote cells undergoing LatA treatment. Time ‘0’ marks LatA washout. Rlc1-tdTomato and Sad1-mCherry mark the actomyosin ring and spindle pole bodies and indicate cell-cycle stage. Cells in G2 (i) or G1/S (iii) during LatA treatment do not show the PrESS phenotype. Cells in mitosis during LatA treatment (ii) show the PrESS phenotype. The arrowhead marks the recovered actomyosin ring undergoing constriction. (B) cdc25-22 cells containing CRIB-3xGFP were synchronized by cell-cycle block and release. During the indicated cell-cycle stages, occurring 0 (G2), 40 (Mitosis) and 80 (G1/S) min after release, cells were treated for 30 min with LatA and then washed. Cells recovering from LatA treatment in G2 or G1/S do not yield PrESS cells whereas those undergoing mitosis exclusively yield PrESS cells. Arrows mark PrESS cells with active Cdc42 at cell ends, asterisks indicate PrESS cells in which active Cdc42 is lost from the septum/division site. Scale bars: 5 μm.
To confirm this, we synchronized cdc25-22 cells by shifting them to restrictive temperature and arresting them in late G2 (Fantes and Nurse, 1978; Tormos-Pérez et al., 2016). Upon shifting back to permissive temperature, the synchronized cells enter mitosis. We took samples of cells released from arrest at regular intervals. The cell-cycle stage was determined by Phalloidin and 4′,6-diamidino-2-phenylindole (DAPI) staining after formaldehyde fixation of a fraction of the samples. Cells in G2 show a single nucleus with actin mainly at the ends. Cells in mitosis show a dividing nucleus with an actin ring at the cell middle (Fig. S2B). In G1/S, the cell middle shows a septum instead of an actin ring, with a single nucleus in each sister cell. The remaining fractions of the samples were treated with LatA like before and allowed to recover. We found that cells treated with LatA during late G2 went on to septate and divide normally (Fig. S2B). Cells released from restrictive temperature for 40 min were in mitosis at the time of LatA treatment. These cells almost exclusively displayed the PrESS phenotype during recovery. Cells released from restrictive temperature for 80 min were septated (in G1/S) at the time of LatA treatment, as evidenced by the divided nuclei and the lack of an actomyosin ring. These cells successfully separated after recovery and did not display the PrESS phenotype. To confirm that cells treated during mitosis were indeed able to grow from their ends, we looked for the presence of active Cdc42 at those ends. To do this, we treated cdc25-22 mutants expressing CRIB-3xGFP in the same way as above. Cells treated during mitosis displayed the PrESS phenotype, with CRIB3x-GFP at the ends and absent from the site of failed cell separation (Fig. 2B, arrows). Three hours after LatA washout, cells that were treated in G2 or G1/S had completed cell separation and had entered the next cell cycle. The cells treated in mitosis also entered the next cell cycle; however, the original division site still failed to separate and did not display CRIB-3xGFP (Fig. 2B, asterisks). These findings suggest that the PrESS phenotype is cell-cycle dependent and occurs in cells that are in mitosis during LatA treatment.
PrESS cells activate Cdc42 at the growing ends at the expense of the division site
Why do PrESS cells fail to separate? Cell separation requires the formation of a tri-layered septum, which is subsequently digested by glucanases (Cortés et al., 2016; García Cortés et al., 2016; Sipiczki, 2007). The septum is composed of a primary septum flanked by secondary septum. Glucanases are delivered to the cortex at the outer edge of the septum barrier to digest the primary septum and allow separation. Separation defects occur either due to inadequate primary septum formation or improper delivery of the glucanases. We asked whether separation failure in PrESS cells is due to improper primary septum formation. Using electron microscopy of cells recovering from LatA treatment, we found that the non-separating septum in PrESS cells is constructed properly, displaying a distinct tri-layer similar to untreated cells (Fig. S3A). These cells also showed proper recruitment of the primary septum-synthesizing enzyme Bgs1 during septation (Fig. S3B). Moreover, the original septum in a PrESS cell often does separate, albeit after a prolonged delay (Fig. S3C). The septum, therefore, appears competent for separation, suggesting that cell separation does not fail in PrESS cells due to a structural defect in the septum.
Cell separation failure can occur due to improper delivery of the septum-digesting glucanases to the division site (García Cortés et al., 2016). We asked whether cell separation failure in PrESS cells is due to improper glucanase delivery. We imaged the glucanases Eng1 and Agn1 (Dekker et al., 2004; Martin-Cuadrado et al., 2003) at the division site of PrESS cells. In untreated cells, Eng1 and Agn1 localize to the outer edge of the membrane barrier and appear as a ring (Fig. S3D) (Martín-Cuadrado et al., 2005). In PrESS cells, Eng1 and Agn1 appear as a patchy disc at the membrane barrier and do not localize to the outer edge (Fig. S3D). This altered localization pattern is similar to that observed in mutants that fail to deliver glucanases and thus fail to separate (Martín-Cuadrado et al., 2005; Perez et al., 2015; Santos et al., 2005; Wang et al., 2015). This suggests that cell separation failure in PrESS cells is due to improper delivery of the glucanases required for septum digestion.
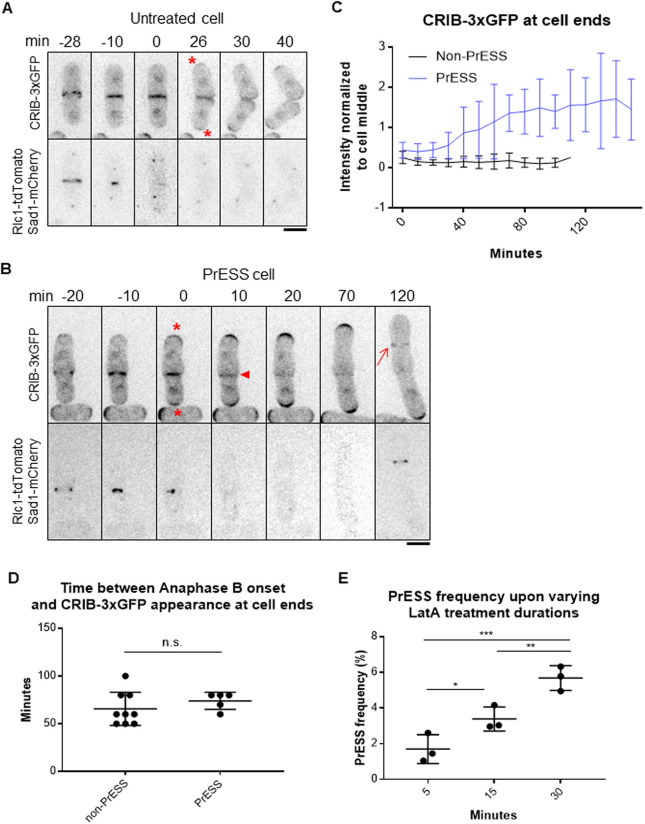
Glucanases do not localize properly to the outer edge of the membrane barrier in a hypomorphic mutant of the small GTPase Cdc42 (Onwubiko et al., 2020). Cdc42 activity stops at the ends during mitosis and transitions to the division site during cytokinesis (Merla and Johnson, 2000; Rincon et al., 2007; Wei et al., 2016). Once the cell separates, Cdc42 activity returns to the ends and growth resumes (Fig. 3A, upper panel, asterisks). The two growing ends compete for Cdc42 activity (Das et al., 2012). Normally, Cdc42 is not activated at the old ends of daughter cells until after cell separation has occurred (Fig. 3A). However, in PrESS cells, we found that Cdc42 activation occurs earlier at these ends (Fig. 3B, asterisks), while it gradually fades away from the septum (Fig. 3B, arrowhead). We quantified active Cdc42, by CRIB-3xGFP intensity, at the ends, normalized to the cell middle (Fig. 3C). In non-PrESS cells, Cdc42 activity was measured at the cell middle and the ends throughout cytokinesis through cell separation. We found that Cdc42 activity is higher at the cell middle than at the ends throughout cytokinesis. However, in PrESS cells, Cdc42 activation at the ends increases over time, soon overcoming the cell middle. We hypothesize that this is indicative of a competition for Cdc42 in PrESS cells between the ends and the septum. These sites ordinarily do not activate Cdc42 concurrently and thus do not compete with each other. The conjoined cells eventually enter the next cell cycle and show normal Cdc42 activation at the ends during interphase and at the new division site (Fig. 3B, arrow) during cytokinesis, as evident in the two sister cells at 120 min in Fig. 3B showing that subsequent cell cycles progress normally.
Fig. 3.
Cdc42 is activated at the ends prior to cell separation in PrESS cells. (A,B) Localization of the active Cdc42 marker CRIB-3xGFP in an untreated cell (A) and a PrESS cell (B). Asterisks denote growth onset. Arrowhead denotes loss of CRIB-3xGFP from the cell middle. Arrow shows new ring formation in the next cell cycle. Rlc1-tdTomato and Sad1-mCherry mark the actomyosin ring and spindle pole bodies. Time ‘0’ marks ring/septum closure. A single z-plane is shown for clarity and hence the spindle pole body is not visible in all the time frames. (C) Quantification of CRIB-3xGFP intensities at cell ends of non-PrESS and PrESS cells, normalized to the cell middle. n=6 cells. (D) The time between anaphase B completion and CRIB-3xGFP appearance at the ends is similar in non-PrESS and PrESS cells. (E) The PrESS frequency is lower in cells treated with LatA for a shorter duration than in cells treated for a longer duration. n>800 cells. An ordinary one-way ANOVA with two-stage step-up method of Benjamini, Krieger and Yekutieli multiple comparisons test was used for analysis (n.s., not significant; *P=0.0246; **P=0.0073; ***P=0.0004).
We observed that Cdc42 is activated at the ends prematurely with respect to cytokinesis in PrESS cells. Could an intrinsic timer, independent of cytokinesis, determine when Cdc42 is activated at the ends after division? To address this, we measured the timing of when CRIB-3xGFP returns to the ends after completion of anaphase B. In non-PrESS cells, CRIB-3xGFP appears at the ends ∼66 min after anaphase B. In PrESS cells, after accounting for the 30-min LatA treatment and 60-min recovery, we found that CRIB-3xGFP appears at the ends at about the same time (74 min, P=0.3364) as in non-PrESS cells (Fig. 3D). This indicates that an intrinsic timer within the cell determines when the ends activate Cdc42 after division. Cells undergoing this triggering event when treated with LatA will eventually show the PrESS phenotype. We thus speculated that fewer cells would undergo this Cdc42 activation trigger upon a shorter LatA treatment, resulting in fewer PrESS cells. Indeed, we found that the frequency of the PrESS phenotype decreases with decreasing duration of LatA treatment (Fig. 3E).
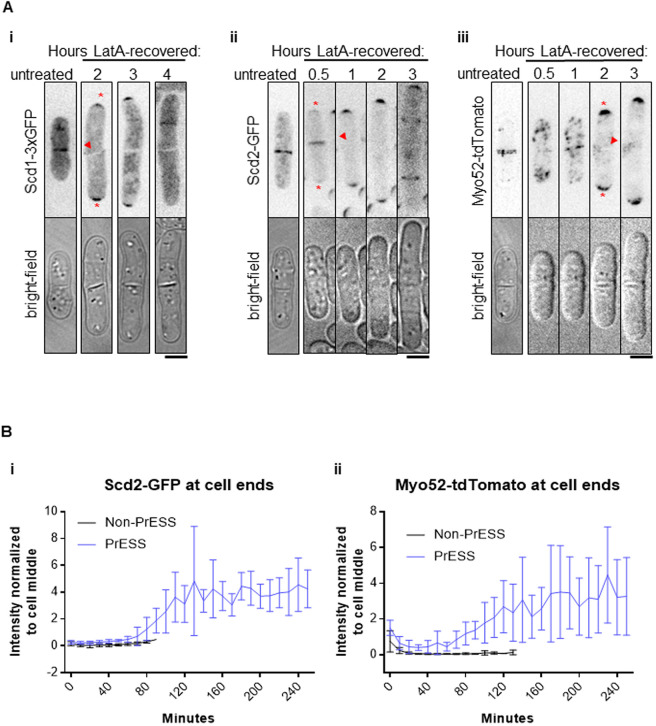
If Cdc42 activation resumes at the ends at the expense of the division site in PrESS cells, we should see this reflected in the levels of Cdc42 regulators in these cells. We analyzed the localization of the Cdc42 GEF Scd1 and its scaffold Scd2 (Chang et al., 1994). During cytokinesis, Scd1 and Scd2 localize to the division membrane barrier and remain there until cell separation (Hercyk et al., 2019b; Hirota et al., 2003). In PrESS cells, we found that Scd1-3xGFP and Scd2-GFP are lost from the division site (Fig. 4A,i,ii, arrowheads) and appear at the ends, where they become enriched over time (Fig. 4A,i,ii, asterisks). Although it is not easy to image Scd1-3xGFP over long periods of time due to bleaching, we were able to perform time-lapse imaging of Scd2-GFP. Quantification of Scd2-GFP over time revealed a localization pattern similar to CRIB-3xGFP, with PrESS cells showing an increase in signal intensity at the ends in relation to the cell middle (Fig. 4Bi).
Fig. 4.
Cdc42 effectors localize to the growing ends of PrESS cells. (A) In untreated cells, Scd1-3xGFP (i), Scd2-GFP (ii) and Myo52-tdTomato (iii) localize to the septum. When PrESS cells start to grow, all three disappear from the cell middle (arrowheads) and appear at the growing ends (asterisks). Maximum projections of Myo52-tdTomato and single planes of Scd1-3xGFP and Scd2-GFP are shown. For Scd1-3xGFP, images were taken every hour after LatA washout, and different cells are shown at each time point. Time-lapse imaging after LatA washout was performed for Scd2-GFP and Myo52-tdTomato. A different untreated Scd2-GFP or Myo52-tdTomato cell is shown while treated cells are the same cell over time. (B) Quantification of Scd2-GFP (i) and Myo52-tdTomato (ii) intensities at cell ends of non-PrESS and PrESS cells, normalized to the cell middle. n=6 cells. Scale bars: 5 μm.
Our data show that, in PrESS cells, glucanases required for cell separation are not trafficked properly during cytokinesis. Membrane trafficking occurs at sites of Cdc42 activation (Estravis et al., 2011, 2012; Harris and Tepass, 2010; Murray and Johnson, 2001). Thus, in PrESS cells, with Cdc42 activity decreasing at the cell middle and increasing at the ends, we expect a similar pattern for the membrane trafficking apparatus. We analyzed the recruitment of the type 5 myosin Myo52 to the division site (Win et al., 2001). Indeed, in PrESS cells, we observed that Myo52-tdTomato intensity gradually increases at the ends, whereas it decreases at the septum (Fig. 4Aiii,Bii). Myo52-tdTomato levels at the ends over time thus resemble those of CRIB-3xGFP and Scd2-GFP in PrESS cells (Fig. 3C, Fig. 4B). These findings together suggest that, in PrESS cells, Cdc42 activity, and consequently the membrane trafficking apparatus, transition to the ends from the division site. This leads to cell separation failure due to improper delivery of the glucanases required for primary septum digestion.
A candidate screen to identify cell-cycle-dependent regulators of Cdc42 activation
Next, we wanted to identify how Cdc42 is regulated in a cell-cycle-dependent manner at the ends. We hypothesized that a Cdc42 regulator is under cell-cycle control. Once cell division completes, this regulation allows Cdc42 activation at the ends. To identify the regulator, we performed a candidate screen of known mutants of Cdc42 regulators (Table S2). We measured the PrESS frequency in these mutants. We considered the different mutants’ cell sizes in computing the PrESS frequency, as described in the Materials and Methods. We observed the PrESS phenotype in all but one of the mutants analyzed. The PrESS phenotype did not occur in the cdc16-116 mutant (Table S2). Cdc16 is a GAP that inactivates the SIN pathway. Hypomorphic cdc16-116 mutant shows constitutive SIN activation leading to repeated septation and absence of cell growth (Schmidt et al., 1997). This suggests that the PrESS phenotype occurs as long as a cell is capable of growth. Cdc42 activation in LatA-treated cells is regulated by the MAP kinase Sty1 (Mutavchiev et al., 2016; Toda et al., 1996). We asked whether the PrESS phenotype is a Sty1-dependent stress response. The PrESS phenotype persisted in sty1Δ mutants, suggesting that this phenotype is not a stress response to LatA treatment (Fig. S4A). Cell polarity in fission yeast depends on the microtubule cytoskeleton. The microtubule-dependent Tea1 protein localizes polarity markers to the ends (Feierbach et al., 2004; Glynn et al., 2001). However, tea1Δ mutants show the PrESS phenotype after recovery from LatA treatment. This suggests that the PrESS phenotype is independent of the microtubule-associated polarity markers (Fig. S4B).
Polarized Cdc42 activation depends on Ras1 GTPase, which promotes polarized localization of the Cdc42 GEF Scd1 (Chang et al., 1994; Chen et al., 2019; Lamas et al., 2020a). However, ras1Δ mutants continue to display the PrESS phenotype after recovery from LatA treatment (Fig. S4C). Furthermore, loss of either of the partially redundant GEFs, Scd1 or Gef1, does not lead to reduced PrESS frequency (Fig. 5A,B). This suggests that cell-cycle-dependent activation of Cdc42 at the ends is not dependent on a specific GEF and that as long as a GEF is available, Cdc42 will be activated at the ends. We found that gef1Δ shows a higher PrESS frequency than wild type (Fig. 5B). We have previously shown that, in gef1Δ cells, the old end competes with the new end for Cdc42 activity more effectively than in wild-type cells (Hercyk et al., 2019b). Thus, in gef1Δ mutants, a robust old end can explain the higher PrESS frequency we observe.
Fig. 5.
Loss of negative regulation of Cdc42 increases the incidence of the PrESS phenotype. (A,B) PrESS phenotype in wild-type, scd1Δ (n.s.) and gef1Δ (P=0.015) cells (ordinary one-way ANOVA with Dunnett's multiple comparisons test; n.s., not significant). Asterisks denote PrESS cells. Repeated in triplicate, n≥350 cells per experiment. (C,D) PrESS phenotype in wild-type and rga4Δ cells (two-tailed unpaired Student's t-test; P=0.033). Repeated in triplicate, n≥350 cells per experiment. Scale bars: 5 μm.
Next, we analyzed the PrESS phenotype in deletion mutants of the Cdc42 GAPs Rga4 and Rga6. These GAPs inactivate Cdc42 at the cortex (Das et al., 2007; Revilla-Guarinos et al., 2016; Tatebe et al., 2008). We did not see any change in the PrESS frequency of rga6Δ cells. In contrast, rga4Δ shows a significantly higher incidence of the PrESS phenotype compared to wild type (P<0.05) (Fig. 5C,D). rga4Δ cells are wider and shorter than wild-type cells (Das et al., 2007). This observation is consistent among PrESS cells. One explanation for the higher PrESS frequency in gef1Δ and rga4Δ cells could be a longer mitotic phase or septation defect. However, mitosis is not prolonged in either gef1Δ mutants (Wei et al., 2016) or in rga4Δ mutants (Fig. S5C). We did observe a higher septation index in gef1Δ mutants but not in rga4Δ mutants (Fig. S5A,B). This indicates that the higher PrESS frequency in these mutants is not due to a mitotic delay or a septation defect.
Rga4 localization is regulated in a cell-cycle-dependent manner
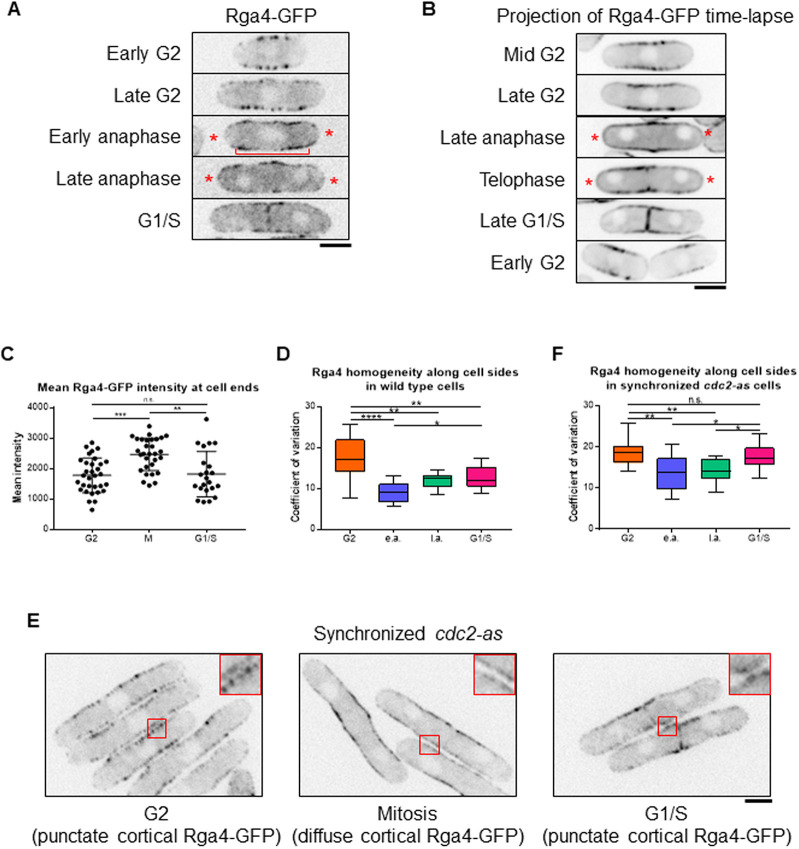
The enhanced Cdc42 activity at the ends of PrESS cells could be due to an increased activity of one of its GEFs or loss of one of its GAPs. Because the GEFs are non-essential for the PrESS phenotype, increased GEF activity is not responsible for this phenotype. As deletion of the Cdc42 GAP rga4 results in a higher PrESS frequency, we asked whether Rga4 has a role in regulating Cdc42 activity at the ends and thus growth during mitosis. Previous studies have shown that Rga4 localizes mainly to the cell sides, where it blocks Cdc42 activation (Das et al., 2007; Tatebe et al., 2008). In wild-type cells, we observed that Rga4-GFP localizes mostly to cell sides during interphase (Fig. 6A). As the cell enters division, Rga4-GFP can be found not only at the cell sides but also at the ends (Fig. 6A, asterisks). Time-lapse imaging of Rga4-GFP (10 s intervals for 1 min) indicated that Rga4-GFP at cell ends is dynamic (Fig. S6A). A time-lapse projection of Rga4-GFP (10 s intervals for 5 min) showed increased signal at the ends in mitotic cells compared to interphase cells (Fig. 6B, asterisks; Movies 3–8). We measured the mean intensity of Rga4-GFP at cell ends in these time-projected wild-type cells. Mitotic cells have significantly more Rga4-GFP at their cell ends than cells in G2 or G1/S (Fig. 6C). Also in time-projected cells, the intensity of Rga4-GFP at the cortex in relation to the cytoplasm is elevated in G2 and G1/S cells compared to mitotic cells (Fig. S6B).
Fig. 6.
Rga4 localization changes in a cell-cycle-dependent manner. (A) During G2 and septation (G1/S), Rga4-GFP localizes to the cortex along the cell sides as distinct puncta. During late and early anaphase, Rga4-GFP at the cortex appears diffuse (bracket) with localization extending to the ends (asterisks). (B) Time-lapse projection (10 s intervals over 5 min) of Rga4-GFP localization at different cell-cycle stages. Asterisks denote end localization in mitotic cells. (C) Quantification of Rga4-GFP intensities at cell ends during different cell-cycle stages. Significantly more Rga4-GFP localizes to cell ends during mitosis than during either G2 (P=0.0002) or G1/S (P=0.0027). (D) Quantification of Rga4-GFP localization pattern along cell sides (increased co-efficient of variance=decreased homogeneity=increased punctate appearance) in wild-type cells through different cell-cycle stages (n≥10 cells). e.a., early anaphase; l.a., late anaphase. (E) cdc2-as mutant cells blocked in G2 upon 1NM-PP1 treatment show distinct Rga4-GFP puncta along cell sides. After 1NM-PP1 washout, cdc2-as mutant enters mitosis and cortical Rga4-GFP appears more diffuse. In G1/S, Rga4-GFP again localizes as puncta. (F) Quantification of Rga4-GFP homogeneity along the sides, as done for D, in synchronized cdc2-as cells through different cell-cycle stages (n≥14 cells). Ordinary one-way ANOVA with Tukey's multiple comparisons was used for statistical analysis (n.s., not significant; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001). Scale bars: 5 µm.
We further observed that not only the localization, but also the distribution, of Rga4-GFP along the cortex changes throughout the cell cycle. In G2, Rga4-GFP appears as puncta dispersed along the cell sides (Fig. 6A). During mitosis, it spreads more homogeneously along the cortex (Fig. 6A, bracket). We quantified these changes by measuring the coefficient of variation of Rga4-GFP distribution along the cortex. The higher the coefficient of variation, the less homogeneous the distribution of Rga4-GFP. We found that the coefficient of variation is higher during G2 than during mitosis, reflecting our observation that Rga4-GFP appears more punctate during G2 and more diffuse during mitosis (Fig. 6D). The coefficient of variation drops in cells undergoing mitosis and appears to increase in G1/S cells. This suggests that, in mitotic cells, Rga4-GFP is distributed homogeneously along the cortex and, as the cells enter G1/S, gradually reverts back to a punctate appearance.
We next asked whether the Rga4 localization changes are indeed cell-cycle dependent. We analyzed the localization pattern of Rga4-GFP throughout the cell cycle via cell-cycle block and release. In our experience, Rga4-GFP localization shows artifacts at high temperature and hence we did not use the cdc25-22 mutants. Instead, we used an analog-sensitive mutant cdc2-as of the mitotic kinase Cdk1, which allows specific inhibition of its kinase activity upon addition of the inhibitor 1NM-PP1 (Aoi et al., 2014). In untreated and asynchronous cdc2-as cells, the Rga4-GFP distribution pattern is similar to that observed in control cells (Fig. S6C). We synchronized cdc2-as cells by adding the inhibitor 1NM-PP1. We distinguished between different cell-cycle stages by observing the nucleus/nuclei and Rga4 localization at the division site (Fig. S6D). After G2 arrest, we washed out the inhibitor to allow release into mitosis. Rga4-GFP distribution along the cortex was analyzed in cells at different cell-cycle stages. Cells arrested in G2 showed a high coefficient of variation, corresponding to a more punctate Rga4-GFP distribution (Fig. 6E,F). Approximately 20 min after removal of the inhibitor, the cells were in early anaphase. Under these conditions, the coefficient of variation of Rga4-GFP distribution is significantly reduced, corresponding to a more homogeneous distribution (Fig. 6E,F). Similarly, cells in late anaphase, ∼40 min after release, also showed a decreased coefficient of variation of Rga4-GFP distribution. The coefficient of variation in G1/S cdc2-as cells (∼70 min after release) is higher than in mitotic cells, suggesting that the punctate distribution reappears after mitosis. However, in asynchronous cells, the coefficient of variation in G1/S is lower than it is in G2 (Fig. S6C). During G1/S, Rga4-GFP distribution returns to a more punctate appearance, similar to G2. This change is likely better captured in synchronized cells. In cell-cycle-arrested cdc2-as cells, which are larger than wild-type cells, Rga4-GFP was not clearly visible at the ends. It is possible that the increased size of these cells leads to a decrease in the local concentration of Rga4-GFP at the ends.
Loss of Rga4 allows polarized growth activation after cell division
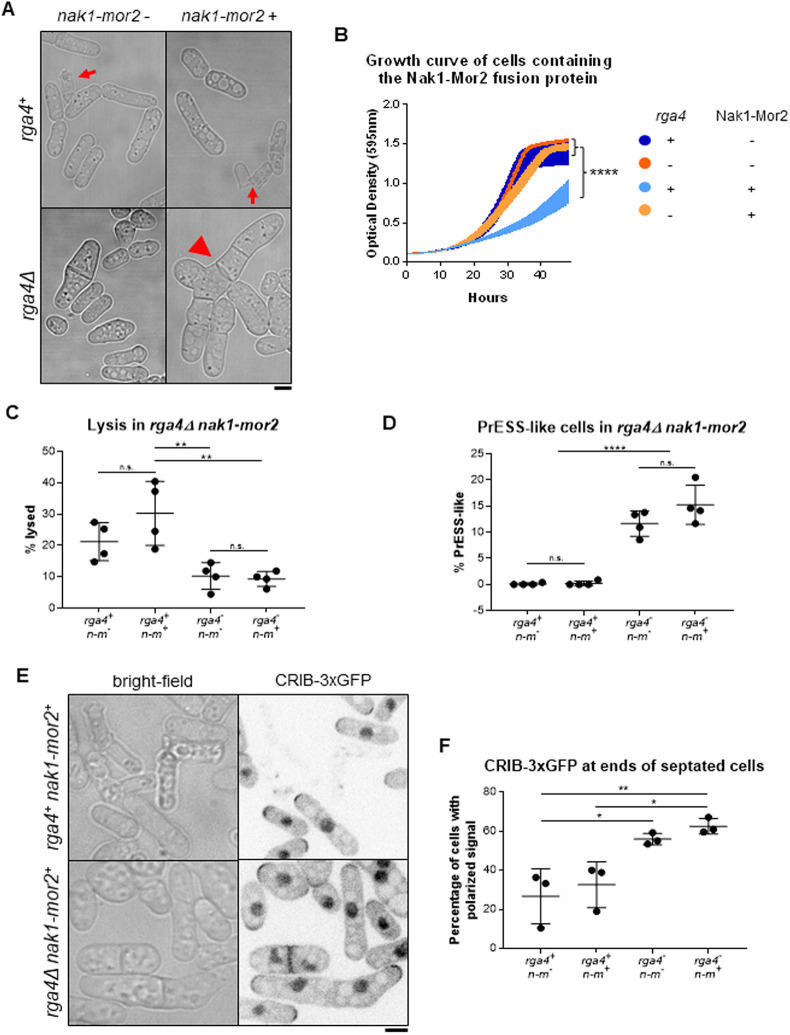
Our data suggest that Cdc42 regulation at the ends is cell-cycle dependent and that loss of rga4 enhances PrESS frequency. Moreover, we found that Rga4 localization to cell ends increases during mitosis, when Cdc42 activation at those ends is known to decline. We propose that two elements are necessary for polarized growth to occur: MOR pathway activation leading to protein synthesis and Cdc42 activation at the ends. During mitosis, the MOR pathway is inactive and Rga4 is present at the ends, and growth does not occur. As cell division completes, the ends lose Rga4 and the MOR pathway becomes active, allowing local Cdc42 activation. In PrESS cells, cytokinesis is delayed, but the MOR pathway and Cdc42 at the ends are activated as normal, resulting in polarized growth. We should thus be able to recapitulate the PrESS phenotype in cells both constitutively activating the MOR pathway and activating Cdc42 at the ends even without cytokinesis delay. Constitutive activation of the MOR pathway via expression of the nak1-mor2 fusion leads to premature protein synthesis during cytokinesis (Gupta et al., 2014). This includes the glucanases that are then prematurely delivered to the incomplete septum barrier, resulting in cell lysis (Fig. 7A, arrows; Fig. S7A). We hypothesized that if the ends were able to activate growth, the glucanases would be delivered to the ends rather than the division site, resulting in a PrESS-like phenotype. Based on our previous observations, we should accomplish this by deleting rga4 in a mutant constitutively activating the MOR pathway. Because nak1-mor2-expressing cells undergo cell lysis resulting in cell death, they display a low optical density (Gupta et al., 2014). We observed cell lysis in rga4+ cells with nmt1-nak1-mor2 under both expressing and repressing conditions. This is likely due to leaky expression of nak1-mor2 in these cells. However, cell lysis was enhanced under expressing conditions compared to repressing conditions, resulting in a significantly lower optical density in rga4+ cells expressing nak1-mor2 (Fig. 7B). Accordingly, the fraction of lysed cells in rga4Δ mutants expressing nak1-mor2 was reduced compared to that in rga4+ cells expressing nak1-mor2 (Fig. 7C). These cells failed to separate and continued to grow from their ends, similar to PrESS cells (Fig. 7A, arrowhead). Indeed, although rga4+ cells expressing nak1-mor2 did not show PrESS-like cells, rga4Δ cells expressing nak1-mor2 consistently showed this phenotype (Fig. 7D). Note that rga4Δ cells are capable of cell separation, and thus failure to separate in nak1-mor2-expressing conditions is due to premature growth at the ends, similar to PrESS cells.
Fig. 7.
Deletion of rga4 allows polarized growth in cells with constitutive Morphogenesis Orb6 (MOR) activity. (A) Cells expressing nak1-mor2 (thiamine) show cell lysis (arrows) at the division site during cytokinesis in rga4+ cells, while in rga4Δ mutants nak1-mor2 expression leads to a PrESS-like phenotype (arrowhead). (B) Growth curve of rga4+ or rga4Δ cells either expressing nak1-mor2 or repressing nak1-mor2. Optical density was measured every 15 min at 595 nm for 2 days (ordinary one-way ANOVA with Tukey's multiple comparisons; P<0.0001; five experiments averaged for each genotype). (C) rga4+ cells expressing nak1-mor2 (n-m) showed significantly more lysis than rga4Δ cells either repressing (P=0.0042) or expressing (P=0.0030) nak1-mor2. (D) Percentage of cells displaying a PrESS-like phenotype. rga4Δ cells containing nak1-mor2 (n-m) display a higher PrESS-like frequency than rga4+ cells containing nak1-mor2 (P<0.0001). (E) rga4+ and rga4Δ cells expressing nak1-mor2 and containing CRIB-3xGFP. Whereas only a few septated rga4+ cells show CRIB-3xGFP at their ends, a large fraction of septated rga4Δ cells do. (F) Quantification of CRIB-3xGFP at cell ends in rga4+ and rga4Δ cells expressing nak1-mor2 (n-m). Ordinary one-way ANOVA with Tukey's multiple comparisons was used for statistical analysis. Experiments repeated in triplicate. n.s., not significant; *P<0.05; **P<0.01; ****P<0.0001. Scale bars: 5 µm.
Next, we asked whether this PrESS-like phenotype is due to Cdc42 activation at the ends in dividing cells. PrESS cells have a septum and show active Cdc42 at their ends. We analyzed CRIB-3xGFP at the ends of septated cells in rga4+ and rga4Δ cells expressing nak1-mor2. In rga4Δ cells expressing nak1-mor2, a larger fraction of septated cells shows CRIB-3xGFP at the cells ends compared to the rga4+ control (Fig. 7E,F). We asked whether the PrESS-like phenotype was specific to Rga4. PrESS frequency also increases in gef1Δ mutants (Fig. 5), but we did not observe PrESS-like cells or rescue of cell lysis in gef1Δ cells expressing nak1-mor2 (Fig. S7B). It is possible that the PrESS-like phenotype could be due to loss of any Cdc42 GAP. However, in rga6Δ cells expressing nak1-mor2, we did not observe PrESS-like cells or rescue of cell lysis (Fig. S7B). This suggests that, under normal conditions, after MOR pathway activation, cell growth occurs once the ends lose Rga4.
DISCUSSION
It is unclear how polarized growth resumes at the ends after cell division completes. Specifically, it is unknown how Cdc42 activity transitions from the division site to the ends to drive polarized growth. Here, we show that the timing of Cdc42 activation at the old end is cell-cycle dependent.
The MOR pathway promotes cell separation and polarized growth after mitosis (Chen et al., 2019; Gupta et al., 2014; Nunez et al., 2016; Ray et al., 2010). Cytokinesis involves actomyosin ring formation and subsequent constriction in coordination with septum formation and membrane invagination (Cheffings et al., 2016; García Cortés et al., 2016; Pollard, 2010). After ring constriction, the septum matures to form a tri-layer – a primary septum flanked by secondary septum (Cortés et al., 2016; García Cortés et al., 2016). MOR pathway activation allows synthesis of glucanases, which are delivered to the outer edge of the membrane barrier to precisely digest the primary septum, resulting in cell separation (Martín-Cuadrado et al., 2005; Nunez et al., 2016; Santos et al., 2005). Once septum digestion starts, Cdc42 activity transitions to the old ends, and polarized growth initiates. It was unclear how cell separation and growth initiation occur sequentially because the MOR pathway promotes both processes. Constitutively activating the MOR pathway leads to cell lysis during division, which is caused by premature cell separation as a result of unregulated glucanase synthesis and delivery (Gupta et al., 2014). Here, we report the novel PrESS phenotype, in which polarized cell growth initiates during septation. In these cells, Cdc42 is activated at the ends at the expense of cell separation. Cdc42 activation at the ends of these cells coincides with a decrease in Cdc42 activity at the division site. Instead of delivering glucanases to the division site, resulting in premature cell lysis, the ends show polarized growth. This indicates that, under normal conditions, Cdc42 activity at the ends is inhibited while the division site successfully undergoes cell separation. The ends are able to activate Cdc42 only after cell separation completes, even though the MOR pathway is active throughout this stage. Indeed, we found that constitutively activating the MOR pathway does not lead to Cdc42 activation at the ends during division. These observations together suggest that, in addition to MOR activation, another pathway regulates Cdc42 at the ends after division.
Our results indicate that Rga4 is regulated at the ends during mitosis. We show that Rga4 localizes all the way to the ends during mitosis, instead of being restricted to the cell sides (Das et al., 2007). Rga4 at the ends during mitosis may block Cdc42 activation. Accordingly, we found that, in the absence of rga4, cells initiate growth at their ends during division when protein synthesis via the MOR pathway is active, thus recapitulating the PrESS phenotype.
How does Rga4 localize to the ends during mitosis? Rga4 typically localizes to the cell sides as distinct puncta during G2. However, during mitosis, Rga4 appears more diffuse along the cortex and extends all the way to the ends. It is unclear how Rga4 localization and distribution are regulated during mitosis. Rga4 could be a direct target of the mitotic kinase Cdc2 or an unknown cell-cycle-dependent regulatory module. In another study, Rga4 has been identified as a target of Cdk1 kinase activity; however, the physiological significance of this interaction is unknown (Swaffer et al., 2016). Rga4 still lingers at the ends after activation of the anaphase-promoting complex, which promotes Cdk1 inactivation. Rga4 may undergo some modification that gradually leads to its loss from the ends. This could be mediated by a phosphatase, by turnover of Rga4 or by an unknown mechanism. Although it is unknown how changes in the localization pattern of Rga4 affect Cdc42 regulation, a similar concept has been shown in another study (Gerganova et al., 2021 preprint). There, the authors show that artificially localizing a Cdc42 GAP to the cortex completely blocks Cdc42 activity, while conditions leading to its oligomerization result in its loss from the ends, enabling local Cdc42 activation.
Similar to our findings, a report in budding yeast shows that the start of polarization after cell division is independent of cytokinesis (Moran et al., 2019). Moreover, the start of polarization is earlier in mutants lacking the Cdc42 GAPs BEM3 and RGA2. However, another study shows that Cdc42 inactivation at the onset of mitosis is mediated by BEM3 (Gihana et al., 2021). Thus, the role of GAPs in the regulation of polarization after cell division may be conserved. Moreover, in budding yeast, CDKs Pho85 and Cdc28 phosphorylate the GAP Rga2 to restrain its activity and promote polarized growth (Sopko et al., 2007). Also, in budding yeast, the G1-cyclin-bound CDKs promote Cdc42 activation via the GEF Cdc24, thus allowing bud growth (McCusker et al., 2007). Our findings suggest a mitosis-dependent negative regulation of Cdc42 activity. It is unclear whether the G1-cyclin-CDK complex also regulates Cdc42 in fission yeast.
Although we find that Rga4 localizes to the ends during mitosis, loss of rga4 alone does not allow Cdc42 activation at the ends at this stage. This is probably because, even in the rga4 mutants, the MOR pathway is inhibited during mitosis. Cdc42 activation at the ends is not possible while the MOR pathway is inactive. Consequently, after division, Cdc42 activation at the ends requires both loss of Rga4 from the ends and MOR pathway activation. It is possible that, in an rga4Δ mutant, because cytokinesis is not delayed, the ends do not have a sufficient advantage in activating growth because the MOR pathway is only activated during septation. In rga4Δ cells constitutively activating the MOR pathway, the ends do have an advantage because the MOR pathway has been active even before septation, thus leading to growth. A recent report shows that presence of the GAPs at the cortex prevents local Cdc42 activation even upon localization of the scaffold Scd2, which recruits Scd1 to the cortex (Lamas et al., 2020b). Another report suggests that Cdc42 GAP levels in daughter cells determine the pattern of polarized growth in those cells (Pino et al., 2021). These reports and our findings together indicate that loss of Rga4 from the ends after mitosis is necessary to allow Cdc42 activation at these ends, so long as the MOR pathway is active.
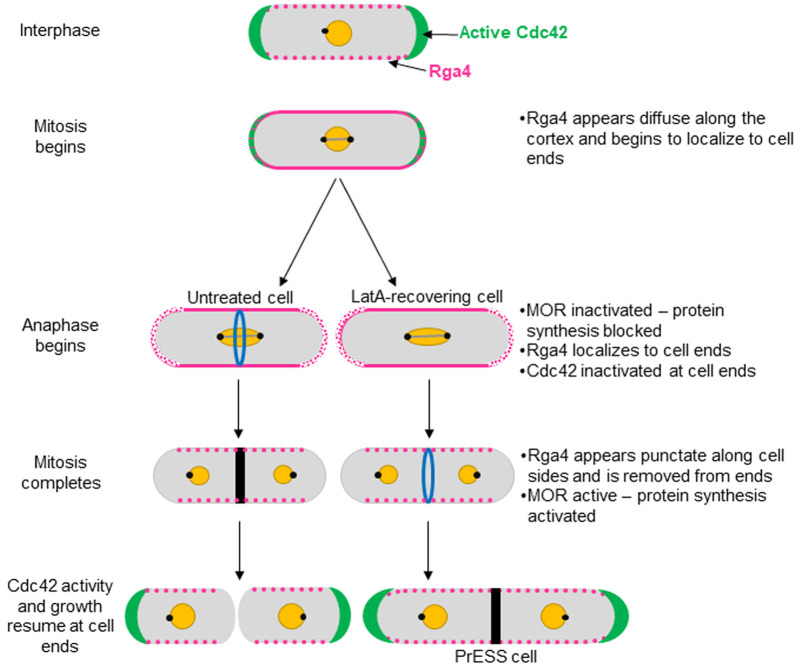
Once the MOR pathway is activated, cells first undergo separation and then initiate polarized growth at their ends. Based on our findings, we propose a model to describe how polarized growth is initiated after cell separation (Fig. 8). Cdc42 activity at the ends declines once the cell enters mitosis and is completely lost from these sites as mitosis progresses. At this stage, Rga4 no longer appears as distinct puncta along the cell sides. Rather, it displays a diffuse distribution along the cortex, which extends to the ends. When the cell enters anaphase, the SIN is activated whereas the MOR pathway is inactivated (Simanis, 2015). Polarized cell growth at the ends stops at this stage. Once mitosis is complete, the SIN is inactivated and the MOR pathway is activated, allowing synthesis of proteins necessary for cell separation and polarized growth. As the cells separate, Rga4 is gradually lost from the ends, its punctate distribution is restored and growth initiates. In LatA-treated cells, cytokinesis is delayed and mitosis progresses. In such cells, Rga4 is removed from the ends on time, allowing Cdc42 activation and growth at these sites because the MOR pathway is active, thus resulting in PrESS cells.
Fig. 8.
Model for growth resumption after cell division. Rga4 localizes to cell sides during G2 (magenta). Entering mitosis, Rga4 extends all the way to the ends (magenta). Upon anaphase-promoting complex activation, the MOR pathway is inactivated and Cdc42 activation and growth (green) stop (Ray et al., 2010; Nunez et al., 2016). After mitosis, MOR is activated and Rga4 is lost from the ends, which now resume growth (Ray et al., 2010; Nunez et al., 2016). In PrESS cells, LatA treatment delays cytokinesis while mitosis proceeds. In these cells, Rga4 is removed from cell ends before cytokinesis completes, resulting in local Cdc42 activation and growth at the expense of cytokinetic events at the cell middle. Thus, we propose that growth resumption upon completion of cell division is independent of cytokinesis and dependent on cell-cycle cues.
Through an artificial cytokinetic delay, we have uncovered a basic cellular principle that ensures that growth occurs at an appropriate time. Cdc42 activity is thus regulated based on a mitotic timer without regard to completion of cytokinesis. This especially makes sense in the context of pseudohyphal growth, which fission yeast typically undergo in nature. Our findings suggest that growth initiation is cell-cycle dependent and is timed to occur after cell separation. A recent report suggests that, in animal cells, growth stops as cells approach the metaphase-to-anaphase transition and resume in late cytokinesis (Miettinen et al., 2019). The Cdc42 activation pattern at cell ends in fission yeast is similar to these observations. The nature of cell-cycle-dependent regulation of polarized growth may be conserved throughout all eukaryotes. Our work provides a better understanding of how growth initiates after mitosis in fission yeast. An elegant cell-cycle-dependent system causes a subtle but effective localization change, controlling the timing of Cdc42 activation at the ends.
MATERIALS AND METHODS
Strains and cell culture
The S. pombe strains used in this study are listed in Table S1. Cells were cultured in yeast extract (YE) medium and grown exponentially at 25°C, unless otherwise specified (Moreno et al., 1991). Cells were grown for at least 3 days before imaging.
Latrunculin A treatments
Cells were treated with 10 µM LatA (Sigma-Aldrich #428021) for 30 min. The cells were washed twice with the appropriate medium, switched to a fresh tube and washed a third time.
PrESS frequency measurements
Wild type, scd1Δ and gef1Δ were all imaged on the same 3 days, and wild type and rga4Δ were both imaged on the same 3 days. Cells were treated with LatA and washed as described above, and then imaged immediately after washing as well as every hour after washing for 5 h. Untreated cells were imaged as a control for each strain. The length of each septated cell was measured, and the average and standard deviation were calculated. Any cell measured to be longer than two standard deviations more than the average for the corresponding untreated sample was counted as a PrESS cell. The reported peaks are the actual PrESS percentages minus the control PrESS percentage, which was always under 1%.
cdc25-22 synchronization
Strains synchronized using the cdc25-22 temperature-sensitive mutation were shifted to restrictive temperature, 36°C, for 4 h and then shifted back to permissive temperature, 25°C.
Cdc2-as inhibition and synchronization
Synchronization of cells containing cdc2-as was achieved via addition of 1NM-PP1 (Toronto Research Chemicals #A603003) to a concentration of 500 nM for 4 h. Cells were then washed three times with YE medium to remove the inhibitor.
Phalloidin staining
Phalloidin staining of actin structures was performed as described previously (Das et al., 2009; Pelham and Chang, 2002). Cells were fixed with 3.5% formaldehyde for 10 min at room temperature. After fixation, the cells were permeabilized with PM buffer (35 mM potassium phosphate pH 6.8, 0.5 mM MgSO4) with 1% Triton X-100, then washed with PM buffer and stained with Alexa-Fluor-Phalloidin (Molecular Probes) for 30 min.
Growth curve
Cells were grown at 25°C. To achieve plasmid expression, thiamine was washed out immediately before the experiment began. Cells were diluted to an optical density of 0.01 and loaded into a 96-well plate, which was read by a BioTek Cytation 5 plate reader. Optical density measurements at 595 nm were collected every 15 min for 48 h. During the experiment, the cells were maintained at 29°C, with shaking.
Analysis of Rga4 distribution and localization
To quantify Rga4 distribution along the cell sides, a line was drawn along the cell side in ImageJ for both sides of each cell in a single plane. Then, a plot profile was generated, and the intensity values were averaged to give an average coefficient of variation for Rga4-GFP distribution along the sides.
To quantify Rga4 localization at cell ends, background subtraction was first performed on each field of cells in ImageJ. A cap was drawn around each cell end, and the intensities of the two ends of each cell were averaged together to provide one value per cell.
Constitutive MOR activation
Cells were grown at 25°C in minimal medium with thiamine. Thiamine was washed out 16 h prior to imaging to allow plasmid expression.
Statistical tests
GraphPad Prism was used to perform statistical tests. A Student's t-test (two-tailed, unpaired, unequal variance) was used to determine significance when comparing two samples. When comparing three or more samples, one-way ANOVA was used alongside Tukey's multiple comparisons, unless otherwise mentioned.
Microscopy
Imaging was performed at room temperature (23–25°C). We used an Olympus IX83 microscope equipped with a VTHawk two-dimensional array laser scanning confocal microscopy system (Visitech International, Sunderland, UK), electron-multiplying charge-coupled device digital camera (Hamamatsu, Hamamatsu City, Japan) and a 100×/1.49 NA UAPO lens (Olympus, Tokyo, Japan). We also used a spinning disk confocal microscope system with a Nikon Eclipse inverted microscope with a 100×/1.49 NA lens, a CSU-22 spinning disk system (Yokogawa Electric Corporation) and a Photometrics EM-CCD camera. Images were acquired with MetaMorph (Molecular Devices, Sunnyvale, CA) and analyzed with ImageJ [National Institutes of Health, Bethesda, MD (Schneider et al., 2012)]. For still and z-series imaging, the cells were mounted directly on glass slides with a #1.5 coverslip (Thermo Fisher Scientific, Waltham, MA) and imaged immediately, and with fresh slides prepared every 10 min. Z-series images were acquired with a depth interval of 0.4 μm. For time-lapse images, cells were placed in 3.5 mm glass-bottom culture dishes (MatTek, Ashland, MA) and overlaid with YE medium containing 0.6% agarose and 100 μM ascorbic acid as an antioxidant to minimize toxicity to the cell, as reported previously.
For cells attached to MatTek dishes with lectin, we first let 10 μl of 1 mg/1 ml lectin (Sigma-Aldrich #L1395) incubate in the center of the dish at room temperature for 30 min. Then, we washed with 1 ml YE medium three times to remove excess lectin not adhered to the dish. We spun down 1 ml of cells at an optical density of ∼0.5 and removed all 950 μl of the supernatant, then resuspended and pipetted the remaining 50 μl into the center of the dish. We then rinsed the dish with YE medium three times to remove excess cells not adhered to the lectin. YE medium was subsequently kept in the dish at all times to prevent cell starvation, except during short time periods during washes and drug treatments.
Electron microscopy
Wild-type cells in late G2 were selected from a sucrose gradient. These cells were treated with 10 μM LatA for 30 min and then washed three times with YE medium. The untreated wild-type control cells were not selected from a sucrose gradient. Both samples were washed in sterile water three times and fixed in 2% potassium permanganate for 1 h at room temperature. They were again washed three times in sterile water and then resuspended in 70% ethanol and incubated overnight at room temperature. Dehydration was achieved via an ethanol series at room temperature: in order, two 15-min incubations in 70% ethanol, two 15-min incubations in 95% ethanol and finally three 20-min incubations in 100% ethanol. The cells were then incubated for 30 min in propylene oxide, followed by a 1-h incubation in a 1:1 mixture of propylene oxide and Spurr's resin, and lastly two 1-h incubations in neat Spurr's resin. The cells were pelleted via centrifugation and the supernatant carefully removed in between each incubation. The cells were transferred into small plastic tubes, and fresh Spurr's resin was layered above to the top of the tube. The two resulting tubes were incubated at 60°C overnight. Sectioning yielded ∼100 nm sections. Sections were post-stained with lead citrate for 8 min. Sections were washed by dunking in three sequential beakers of sterile water 30 times each and allowed to dry on filter paper. Samples were imaged with a Zeiss Dual Beam FIB/SEM microscope at the Joint Institute for Advanced Materials (Knoxville, TN). All images were collected at 15 kV.
Supplementary Material
Acknowledgements
We thank Das laboratory members for discussions and feedback; Kathy Gould, Sophie Martin, Jonathan Millar, Pilar Perez and Fulvia Verde for strains; Dannel McCollum for plasmids; and John Dunlap for electron microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.D., J.R.-R.; Methodology: J.R.-R., M.D.; Validation: J.R.-R.; Formal analysis: J.R.-R., M.D.; Investigation: J.R.-R., A.R., E.M.; Resources: M.D.; Data curation: J.R.-R., A.R., E.M.; Writing - original draft: J.R.-R.; Writing - review & editing: M.D.; Visualization: J.R.-R.; Supervision: M.D.; Project administration: M.D.; Funding acquisition: M.D.
Funding
This work was funded by a National Institute of General Medical Sciences R01 grant [1R01GM136847-01] awarded to M.D. J.R.-R. was awarded a National Science Foundation Graduate Research Fellowship [1452154] and a National Institutes of Health Initiative for Maximizing Student Development [R25GM086761]. Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.259291
References
- Albertson, R., Riggs, B. and Sullivan, W. (2005). Membrane traffic: a driving force in cytokinesis. Trends Cell Biol. 15, 92-101. 10.1016/j.tcb.2004.12.008 [DOI] [PubMed] [Google Scholar]
- An, H., Morrell, J. L., Jennings, J. L., Link, A. J. and Gould, K. L. (2004). Requirements of fission yeast septins for complex formation, localization, and function. Mol. Biol. Cell 15, 5551-5564. 10.1091/mbc.e04-07-0640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi, Y., Kawashima, S. A., Simanis, V., Yamamoto, M. and Sato, M. (2014). Optimization of the analogue-sensitive Cdc2/Cdk1 mutant by in vivo selection eliminates physiological limitations to its use in cell cycle analysis. Open Biol 4, 140063. 10.1098/rsob.140063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin, A., Paoletti, A. and Chang, F. (2003). Mid2p stabilizes septin rings during cytokinesis in fission yeast. J. Cell Biol. 160, 1083-1092. 10.1083/jcb.200212016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, E. C., Barr, M., Wang, Y., Jung, V., Xu, H. P. and Wigler, M. H. (1994). Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell 79, 131-141. [DOI] [PubMed] [Google Scholar]
- Cheffings, T. H., Burroughs, N. J. and Balasubramanian, M. K. (2016). Actomyosin ring formation and tension generation in eukaryotic cytokinesis. Curr. Biol. 26, R719-R737. 10.1016/j.cub.2016.06.071 [DOI] [PubMed] [Google Scholar]
- Chen, C., Rodriguez Pino, M., Haller, P. R. and Verde, F. (2019). Conserved NDR/LATS kinase controls RAS GTPase activity to regulate cell growth and chronological lifespan. Mol. Biol. Cell 30, 2598-2616. 10.1091/mbc.E19-03-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, P. M., Trillo, Y., Ametzazurra, A. and Perez, P. (2003). Gef1p, a new guanine nucleotide exchange factor for Cdc42p, regulates polarity in Schizosaccharomyces pombe. Mol. Biol. Cell 14, 313-323. 10.1091/mbc.e02-07-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés, J. C., Ramos, M., Osumi, M., Pérez, P. and Ribas, J. C. (2016). Fission yeast septation. Commun Integr Biol 9, e1189045. 10.1080/19420889.2016.1189045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M., Wiley, D. J., Medina, S., Vincent, H. A., Larrea, M., Oriolo, A. and Verde, F. (2007). Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol. Biol. Cell 18, 2090-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M., Wiley, D. J., Chen, X., Shah, K. and Verde, F. (2009). The conserved NDR kinase Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42. Curr. Biol. 19, 1314-1319. 10.1016/j.cub.2009.06.057 [DOI] [PubMed] [Google Scholar]
- Das, M., Drake, T., Wiley, D. J., Buchwald, P., Vavylonis, D. and Verde, F. (2012). Oscillatory dynamics of Cdc42 GTPase in the control of polarized growth. Science 337, 239-243. 10.1126/science.1218377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker, N., Speijer, D., Grun, C. H., van den Berg, M., de Haan, A. and Hochstenbach, F. (2004). Role of the alpha-glucanase Agn1p in fission-yeast cell separation. Mol. Biol. Cell 15, 3903-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard, A. (2008). Membrane traffic and polarization of lipid domains during cytokinesis. Biochem. Soc. Trans. 36, 395-399. 10.1042/BST0360395 [DOI] [PubMed] [Google Scholar]
- Estravís, M., Rincón, S. A., Santos, B. and Pérez, P. (2011). Cdc42 regulates multiple membrane traffic events in fission yeast. Traffic 12, 1744-1758. 10.1111/j.1600-0854.2011.01275.x [DOI] [PubMed] [Google Scholar]
- Estravis, M., Rincon, S. and Pérez, P. (2012). Cdc42 regulation of polarized traffic in fission yeast. Commun Integr Biol 5, 370-373. 10.4161/cib.19977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville, S. (2004). Cdc42 – the centre of polarity. J. Cell Sci. 117, 1291-1300. 10.1242/jcs.01115 [DOI] [PubMed] [Google Scholar]
- Fantes, P. A. and Nurse, P. (1978). Control of the timing of cell division in fission yeast. Cell size mutants reveal a second control pathway. Exp. Cell Res. 115, 317-329. [DOI] [PubMed] [Google Scholar]
- Feierbach, B., Verde, F. and Chang, F. (2004). Regulation of a formin complex by the microtubule plus end protein tea1p. J. Cell Biol. 165, 697-707. 10.1083/jcb.200403090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo Castro, D. and Martin, S. G. (2018). Differential GAP requirement for Cdc42-GTP polarization during proliferation and sexual reproduction. J. Cell Biol. 217, 4215-4229. 10.1083/jcb.201806016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García, I., Jiménez, D., Martín, V., Durán, A. and Sánchez, Y. (2005). The α-glucanase Agn1p is required for cell separation in Schizosaccharomyces pombe. Biol. Cell 97, 569-576. 10.1042/BC20040096 [DOI] [PubMed] [Google Scholar]
- García Cortés, J. C., Ramos, M., Osumi, M., Pérez, P. and Ribas, J. C. (2016). The cell biology of fission yeast septation. Microbiol. Mol. Biol. Rev. 80, 779-791. 10.1128/MMBR.00013-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerganova, V., Lamas, I., Rutkowski, D. M., Vještica, A., Castro, D. G., Vincenzetti, V., Vavylonis, D. and Martin, S. G. (2021). Cell patterning by secretion-induced plasma membrane flows. Sci. Adv. 7, eabg6718. 10.1126/sciadv.abg6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gihana, G. M., Cross-Najafi, A. A. and Lacefield, S. (2021). The mitotic exit network regulates the spatiotemporal activity of Cdc42 to maintain cell size. J. Cell Biol. 220, e202001016. 10.1083/jcb.202001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn, J. M., Lustig, R. J., Berlin, A. and Chang, F. (2001). Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11, 836-845. 10.1016/S0960-9822(01)00235-4 [DOI] [PubMed] [Google Scholar]
- Gupta, S., Mana-Capelli, S., McLean, J. R., Chen, C.-T., Ray, S., Gould, K. L. and McCollum, D. (2013). Identification of SIN pathway targets reveals mechanisms of crosstalk between NDR kinase pathways. Curr. Biol. 23, 333-338. 10.1016/j.cub.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Govindaraghavan, M. and McCollum, D. (2014). Cross talk between NDR kinase pathways coordinates cytokinesis with cell separation in Schizosaccharomyces pombe. Eukaryot. Cell 13, 1104-1112. 10.1128/EC.00129-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K. P. and Tepass, U. (2010). Cdc42 and vesicle trafficking in polarized cells. Traffic 11, 1272-1279. 10.1111/j.1600-0854.2010.01102.x [DOI] [PubMed] [Google Scholar]
- Hercyk, B. and Das, M. (2019a). Rho family GTPases in fission yeast cytokinesis. Commun Integr Biol 12, 171-180. 10.1080/19420889.2019.1678453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercyk, B. S. and Das, M. E. (2019b). F-BAR Cdc15 promotes Gef1-mediated Cdc42 activation during cytokinesis and cell polarization in S. pombe. Genetics 213, 1341-1356. 10.1534/genetics.119.302649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercyk, B. S., Onwubiko, U. N. and Das, M. E. (2019a). Coordinating septum formation and the actomyosin ring during cytokinesis in Schizosaccharomyces pombe. Mol. Microbiol. 112, 1645-1657. 10.1111/mmi.14387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercyk, B. S., Rich-Robinson, J., Mitoubsi, A. S., Harrell, M. A. and Das, M. E. (2019b). A novel interplay between GEFs orchestrates Cdc42 activity during cell polarity and cytokinesis in fission yeast. J. Cell Sci. 132, jcs236018. 10.1242/jcs.236018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, K., Tanaka, K., Ohta, K. and Yamamoto, M. (2003). Gef1p and Scd1p, the Two GDP-GTP exchange factors for Cdc42p, form a ring structure that shrinks during cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell 14, 3617-3627. 10.1091/mbc.e02-10-0665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, A. S., Jin, M., Wu, C.-F., Zyla, T. R., Elston, T. C. and Lew, D. J. (2012). Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell 149, 322-333. 10.1016/j.cell.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D. I. (1999). Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63, 54-105. 10.1128/MMBR.63.1.54-105.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A. E., McCollum, D. and Gould, K. L. (2012). Polar opposites: fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton 69, 686-699. 10.1002/cm.21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas, I., Merlini, L., Vještica, A., Vincenzetti, V. and Martin, S. G. (2020a). Optogenetics reveals Cdc42 local activation by scaffold-mediated positive feedback and Ras GTPase. PLoS Biol. 18, e3000600. 10.1371/journal.pbio.3000600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas, I., Weber, N. and Martin, S. G. (2020b). Activation of Cdc42 GTPase upon CRY2-induced cortical recruitment is antagonized by GAPs in fission yeast. Cells 9, 2089. 10.3390/cells9092089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado, A. B., Dueñas, E., Sipiczki, M., Vazquez de Aldana, C. R. and del Rey, F. (2003). The endo-β-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J. Cell Sci. 116, 1689-1698. 10.1242/jcs.00377 [DOI] [PubMed] [Google Scholar]
- Martín-Cuadrado, A. B., Morrell, J. L., Konomi, M., An, H., Petit, C., Osumi, M., Balasubramanian, M., Gould, K. L., Del Rey, F. and de Aldana, C. R. (2005). Role of septins and the exocyst complex in the function of hydrolytic enzymes responsible for fission yeast cell separation. Mol. Biol. Cell 16, 4867-4881. 10.1091/mbc.e04-12-1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker, D., Denison, C., Anderson, S., Egelhofer, T. A., Yates, J. R., 3rd, Gygi, S. P. and Kellogg, D. R. (2007). Cdk1 coordinates cell-surface growth with the cell cycle. Nat. Cell Biol. 9, 506-515. 10.1038/ncb1568 [DOI] [PubMed] [Google Scholar]
- Merla, A. and Johnson, D. I. (2000). The Cdc42p GTPase is targeted to the site of cell division in the fission yeast Schizosaccharomyces pombe. Eur. J. Cell Biol. 79, 469-477. 10.1078/0171-9335-00073 [DOI] [PubMed] [Google Scholar]
- Miettinen, T. P., Kang, J. H., Yang, L. F. and Manalis, S. R. (2019). Mammalian cell growth dynamics in mitosis. eLife 8, e447700. 10.7554/eLife.44700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, J. M. and Nurse, P. (1985). Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 75, 357-376. 10.1242/jcs.75.1.357 [DOI] [PubMed] [Google Scholar]
- Moran, K. D., Kang, H., Araujo, A. V., Zyla, T. R., Saito, K., Tsygankov, D. and Lew, D. J. (2019). Cell-cycle control of cell polarity in yeast. J. Cell Biol. 218, 171-189. 10.1083/jcb.201806196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., Klar, A. and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Murray, J. M. and Johnson, D. I. (2001). The Cdc42p GTPase and its regulators Nrf1p and Scd1p are involved in endocytic trafficking in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276, 3004-3009. 10.1074/jbc.M007389200 [DOI] [PubMed] [Google Scholar]
- Mutavchiev, D. R., Leda, M. and Sawin, K. E. (2016). Remodeling of the fission yeast Cdc42 cell-polarity module via the Sty1 p38 stress-activated protein kinase pathway. Curr. Biol. 26, 2921-2928. 10.1016/j.cub.2016.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance, J. and Zallen, J. A. (2011). Elaborating polarity: PAR proteins and the cytoskeleton. Development 138, 799-809. 10.1242/dev.053538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez, I., Rodriguez Pino, M., Wiley, D. J., Das, M. E., Chen, C., Goshima, T., Kume, K., Hirata, D., Toda, T. and Verde, F. (2016). Spatial control of translation repression and polarized growth by conserved NDR kinase Orb6 and RNA-binding protein Sts5. eLife 5, e14216 10.7554/eLife.14216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwubiko, U. N., Rich-Robinson, J., Mustaf, R. A. and Das, M. E. (2020). Cdc42 promotes Bgs1 recruitment for septum synthesis and glucanase localization for cell separation during cytokinesis in fission yeast. Small GTPases 12, 257-264. 10.1080/21541248.2020.1743926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki, P., Pizon, V., Onken, B. and Chang, E. C. (2002). Two ras pathways in fission yeast are differentially regulated by two ras guanine nucleotide exchange factors. Mol. Cell. Biol. 22, 4598-4606. 10.1128/MCB.22.13.4598-4606.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham, R. J. and Chang, F. (2002). Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature 419, 82-86. 10.1038/nature00999 [DOI] [PubMed] [Google Scholar]
- Pérez, P., Portales, E. and Santos, B. (2015). Rho4 interaction with exocyst and septins regulates cell separation in fission yeast. Microbiology 161, 948-959. 10.1099/mic.0.000062 [DOI] [PubMed] [Google Scholar]
- Pino, M. R., Nunez, I., Chen, C., Das, M. E., Wiley, D. J., D'Urso, G., Buchwald, P., Vavylonis, D. and Verde, F. (2021). Cdc42 GTPase activating proteins (GAPs) regulate generational inheritance of cell polarity and cell shape in fission yeast. Mol. Biol. Cell, mbcE20100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D. (2010). Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 22, 50-56. 10.1016/j.ceb.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D. (2014). The value of mechanistic biophysical information for systems-level understanding of complex biological processes such as cytokinesis. Biophys. J. 107, 2499-2507. 10.1016/j.bpj.2014.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, S., Kume, K., Gupta, S., Ge, W., Balasubramanian, M., Hirata, D. and McCollum, D. (2010). The mitosis-to-interphase transition is coordinated by cross talk between the SIN and MOR pathways in Schizosaccharomyces pombe. J. Cell Biol. 190, 793-805. 10.1083/jcb.201002055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-Guarinos, M. T., Martin-Garcia, R., Villar-Tajadura, M. A., Estravis, M., Coll, P. M. and Perez, P. (2016). Rga6 is a fission yeast Rho GAP involved in Cdc42 regulation of polarized growth. Mol. Biol. Cell 27, 1524-1535. 10.1091/mbc.E15-12-0818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, A. J. (2006). Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522-529. 10.1016/j.tcb.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Rincon, S., Coll, P. M. and Perez, P. (2007). Spatial regulation of Cdc42 during cytokinesis. Cell cycle 6, 1687-1691. 10.4161/cc.6.14.4481 [DOI] [PubMed] [Google Scholar]
- Santos, B., Martin-Cuadrado, A. B., Vazquez de Aldana, C. R., del Rey, F. and Perez, P. (2005). Rho4 GTPase is involved in secretion of glucanases during fission yeast cytokinesis. Eukaryot. Cell 4, 1639-1645. 10.1128/EC.4.10.1639-1645.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, S., Sohrmann, M., Hofmann, K., Woollard, A. and Simanis, V. (1997). The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 11, 1519-1534. 10.1101/gad.11.12.1519 [DOI] [PubMed] [Google Scholar]
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis, V. (2015). Pombe's thirteen – control of fission yeast cell division by the septation initiation network. J. Cell Sci. 128, 1465-1474. [DOI] [PubMed] [Google Scholar]
- Sipiczki, M. (2007). Splitting of the fission yeast septum. FEMS Yeast Res. 7, 761-770. 10.1111/j.1567-1364.2007.00266.x [DOI] [PubMed] [Google Scholar]
- Sopko, R., Huang, D., Smith, J. C., Figeys, D. and Andrews, B. J. (2007). Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 26, 4487-4500. 10.1038/sj.emboj.7601847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, I., Shochet, N. R., Kashman, Y. and Groweiss, A. (1983). Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219, 493-495. 10.1126/science.6681676 [DOI] [PubMed] [Google Scholar]
- Swaffer, M. P., Jones, A. W., Flynn, H. R., Snijders, A. P. and Nurse, P. (2016). CDK substrate phosphorylation and ordering the cell cycle. Cell 167, 1750-1761e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius, M. T., Nguyen, L. T., Ladinsky, M. S., Ortega, D. R., Aich, S., Mishra, M. and Jensen, G. J. (2018). Structure of the fission yeast actomyosin ring during constriction. Proc. Natl. Acad. Sci. USA 115, E1455-E1464. 10.1073/pnas.1711218115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasto, J. J., Morrell, J. L. and Gould, K. L. (2003). An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J. Cell Biol. 160, 1093-1103. 10.1083/jcb.200211126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe, H., Shimada, K., Uzawa, S., Morigasaki, S. and Shiozaki, K. (2005). Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr. Biol. 15, 1006-1015. 10.1016/j.cub.2005.04.061 [DOI] [PubMed] [Google Scholar]
- Tatebe, H., Nakano, K., Maximo, R. and Shiozaki, K. (2008). Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 18, 322-330. 10.1016/j.cub.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, Y. D., Leda, M., Goryachev, A. B. and Sawin, K. E. (2018). Local and global Cdc42 guanine nucleotide exchange factors for fission yeast cell polarity are coordinated by microtubules and the Tea1-Tea4-Pom1 axis. J. Cell Sci. 131, jcs216580. 10.1242/jcs.216580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, T., Niwa, H., Nemoto, T., Dhut, S., Eddison, M., Matsusaka, T., Yanagida, M. and Hirata, D. (1996). The fission yeast sts5+ gene is required for maintenance of growth polarity and functionally interacts with protein kinase C and an osmosensing MAP-kinase pathway. J. Cell Sci. 109, 2331-2342. 10.1242/jcs.109.9.2331 [DOI] [PubMed] [Google Scholar]
- Tormos-Pérez, M., Pérez-Hidalgo, L. and Moreno, S. (2016). Fission yeast cell cycle synchronization methods. Methods Mol. Biol. 1369, 293-308. 10.1007/978-1-4939-3145-3_20 [DOI] [PubMed] [Google Scholar]
- Wang, N., Wang, M., Zhu, Y.-H., Grosel, T. W., Sun, D., Kudryashov, D. S. and Wu, J.-Q. (2015). The Rho-GEF Gef3 interacts with the septin complex and activates the GTPase Rho4 during fission yeast cytokinesis. Mol. Biol. Cell 26, 238-255. 10.1091/mbc.E14-07-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N., Lee, I.-J., Rask, G. and Wu, J.-Q. (2016). Roles of the TRAPP-II complex and the exocyst in membrane deposition during fission yeast cytokinesis. PLoS Biol. 14, e1002437. 10.1371/journal.pbio.1002437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, B., Hercyk, B. S., Mattson, N., Mohammadi, A., Rich, J., DeBruyne, E., Clark, M. M. and Das, M. (2016). Unique spatiotemporal activation pattern of Cdc42 by Gef1 and Scd1 promotes different Events during cytokinesis. Mol. Biol. Cell 27, 1235-1245. 10.1091/mbc.E15-10-0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win, T. Z., Gachet, Y., Mulvihill, D. P., May, K. M. and Hyams, J. S. (2001). Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J. Cell Sci. 114, 69-79. 10.1242/jcs.114.1.69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.