Abstract
Coronavirus disease 2019 (COVID-19), caused by the highly contagious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become the worst pandemic disease of the current millennium. To address this crisis, therapeutic nanoparticles, including inorganic nanoparticles, lipid nanoparticles, polymeric nanoparticles, virus-like nanoparticles, and cell membrane-coated nanoparticles, have all offered compelling antiviral strategies. This article reviews these strategies in three categories: (1) nanoparticle-enabled detection of SARS-CoV-2, (2) nanoparticle-based treatment for COVID-19, and (3) nanoparticle vaccines against SARS-CoV-2. We discuss how nanoparticles are tailor-made to biointerface with the host and the virus in each category. For each nanoparticle design, we highlight its structure–function relationship that enables effective antiviral activity. Overall, nanoparticles bring numerous new opportunities to improve our response to the current COVID-19 pandemic and enhance our preparedness for future viral outbreaks.
Keywords: Viral pandemic, COVID-19, SARS-CoV-2, Nanotechnology, Nanoparticle
1. Introduction
The continued emergence of novel viruses poses a significant challenge and threat to global health [1]. While modern medicine has eradicated some viral diseases, the existence of others remains a fact of life, despite their deadly potential. The current coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has incited much panic in human beings [2]. Since the emergence of the COVID-19 pandemic, physicians and researchers have been actively seeking effective therapeutics against SARS-CoV-2 infection. Among these, nanotechnology-based approaches, especially therapeutic nanoparticles, are of great promise as novel antiviral solutions [3].
In the past few decades, nanoparticles have been developed to mitigate the limitations of free drug molecules and overcome biological barriers from systemic to cellular levels, leading to new therapeutics for various diseases [4], [5]. For drug delivery, nanoparticles can improve the solubility of poorly water-soluble drugs, prolong the circulation half-life through immune evasion, target and release drugs in a controlled fashion, and deliver drug combinations for synergy [6]. For disease detection, nanoparticle surfaces can be readily modified with multivalent ligands or other biomolecules for signal enhancement and readout [7]. In addition, their large surface-to-volume ratio offers unique electrochemical reactivity, catalytic ability, or optical property for biosensor design. For disease prevention, nanoparticle-based vaccines can either encapsulate antigens inside or carry antigens on their surface, thereby protecting antigens from premature degradation [8]. These vaccines can also target antigen-presenting cells (APCs), prolong antigen release, or mimic pathogens for multivalent antigen presentation, which together modulate immune activation for adequate protection. These established advantages have inspired researchers to design new therapeutic nanoparticles and tailor their performance to detect, treat, and prevent SARS-CoV-2 infection.
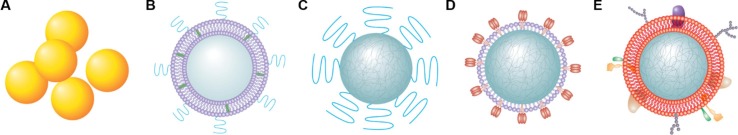
Since the beginning of the COVID-19 pandemic, researchers have tapped numerous nanoparticle platforms for antiviral solutions, including inorganic nanoparticles such as gold or silica nanoparticles, lipid nanoparticles (LNPs) containing a lipid bilayer with cholesterol and surface polyethylene glycol (PEG) for stabilization, polymeric nanoparticles made from self-assembled co-polymers, virus-like nanoparticles (VLPs) containing protein structures that resemble wild type viruses but do not have a viral genome or infectious ability, and cell membrane-coated nanoparticles containing natural cell membranes wrapping around a solid nanoparticle core (Fig. 1 ). Herein, we review antiviral strategies based on these nanoparticle platforms and discuss their progress against SARS-CoV-2 in three aspects. First, we discuss nanoparticle designs toward rapid, accurate, and convenient detection of SARS-CoV-2 virus. Second, we review nanoparticle designs aimed at boosting the efficacy of existing antiviral compounds or creating entirely new antiviral mechanisms through host mimicry. Third, we discuss nanoparticle-based vaccine platforms, especially LNPs and self-assembled VLPs, and their mechanisms to modulate the host immune system for protection. Progress made in these areas demonstrates the potential of nanoparticle approaches as effective antiviral strategies, which help to contain the COVID-19 pandemic and prepare for future viral outbreaks.
Fig. 1.
Schematic illustrations of major nanoparticle platforms explored for SARS-CoV-2 detection, treatment, and prevention. These platforms include (A) inorganic nanoparticles such as gold or silica nanoparticles, (B) lipid nanoparticles (LNPs) containing a lipid bilayer with cholesterol and surface polyethylene glycol (PEG) for stabilization, (C) polymeric nanoparticles made from self-assembled co-polymers, (D) virus-like nanoparticles (VLPs) containing protein structures that resemble wild type viruses but do not have a viral genome or infectious ability, and (E) cell membrane-coated nanoparticles containing natural cell membranes wrapping around a solid nanoparticle core.
2. Nanoparticle-enabled detection of SARS-CoV-2
Through surface functionalization, nanoparticles acquire the ability to interact with specific biomarkers associated with SARS-CoV-2 infection, such as viral proteins, viral RNA, and virus-specific antibodies [9], [10]. The unique physicochemical properties of nanoparticles, such as their optical, reactivity, or fluorescent properties, enable them to transduce biomarker-capturing events into measurable signals for detection [11], [12]. Various nanoparticle-enabled diagnostic platforms have been developed toward rapid and accurate SARS-CoV-2 detection.
Nanoparticles have been designed to detect SARS-CoV-2 viral proteins. In one study, gold nanoparticles (AuNPs) were functionalized with polyclonal antibodies targeting the spike protein (S protein), membrane protein, and envelope protein of SARS-CoV-2 [13]. These AuNPs had an average diameter of 20 nm, and multiple AuNPs bound to the surface of each viral particle. The binding interactions caused AuNP agglomeration and induced a redshift in the absorption peak of the colloidal solution from around 520–560 nm. Besides, the absorbance at 560 nm correlated with the viral load, allowing for quantitative measurements. This strategy was tested on nasal and throat swab specimens collected from 45 SARS-CoV-2-positive patients and 49 SARS-CoV-2-negative patients, confirmed by tests with real-time polymerase chain reaction (rtPCR). The results showed 96% sensitivity and 98% specificity. In another example, AuNPs were conjugated with α,N-acetyl neuraminic acid, a glycan moiety that binds specifically to SARS-CoV-2 S protein (glycan-AuNPs) [14]. The study used SARS-CoV-2 virus-like particles (VLPs) presenting S protein on the surface to mimic the viruses. When the VLPs were incubated with glycan-AuNPs, they formed the virus-glycan-AuNP complex. The sample was then loaded onto a lateral flow immunoassay (LFIA) strip. As the sample traveled along the strip, the virus-glycan-AuNP complex was captured by glycan molecules immobilized at the test line through interactions between the glycan and the S protein on VLPs in the complex. Accumulation of AuNPs at the test line generated a colorimetric signal for visual detection. This device detected SARS-CoV-2 VLPs with spike protein concentrations as low as 5 g ml−1.
Besides AuNPs, other nanoparticles have been designed to detect SARS-CoV-2 proteins. In one study, poly(lactic-co-glycolic) (PLGA) nanoparticles were encapsulated with 3,3′,5,5′-tetramethylbenzidine (TMB). The nanoparticles (denoted ‘TMB-PLGA NPs’) were then conjugated with antibodies against the SARS-CoV-2 S protein [15]. They were used in a microplate-based assay for S protein detection. In this assay, the S proteins were first captured by antibodies immobilized in the plate. Then anti-S protein antibody-functionalized TMB-PLGA NPs were added, which bound to immobilized S proteins. After removing unbound TMB-PLGA NPs, dimethyl sulfoxide was added to dissolve bound nanoparticles and release TMB. Subsequently, hydrogen peroxide and copper nanoparticles with peroxidase-like activities were added to oxidize TMB. The absorbance of the oxidized product correlated with S protein concentration. This assay detected S protein at concentrations in the femtogram ml−1 range.
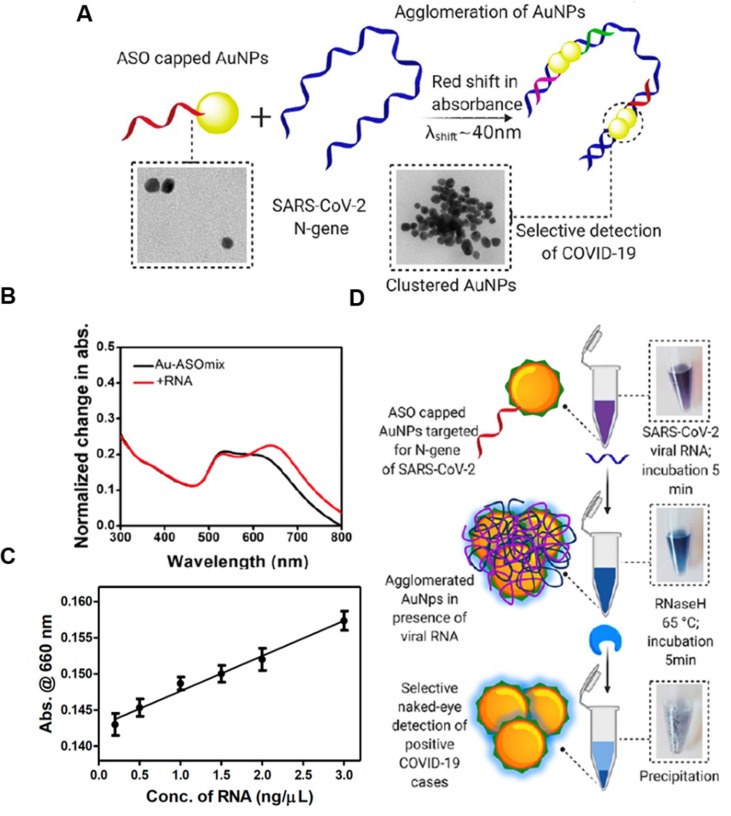
Nanoparticles have also been designed to detect SARS-CoV-2 RNA. For example, AuNPs were capped with thiol-modified antisense oligonucleotides (ASOs), which targeted SARS-CoV-2 RNA through complementary binding (Fig. 2 ) [16]. The targeted gene encoded the nucleocapsid protein (N-gene). Four ASO sequences targeting different regions of the N-gene were designed. Each was capped onto AuNPs separately, forming four types of ASO-functionalized AuNPs (ASO-AuNPs). These ASO-AuNPs had an average diameter of around 55 nm. Their simultaneous binding to the N-gene resulted in particle agglomeration and induced a redshift in the absorption peak from about 620–660 nm. When ASO-AuNPs were mixed with total RNA extracted from SARS-CoV-2 infected Vero cells, the increase in absorbance at 660 nm correlated with the RNA concentration, with a detection limit of 0.18 g ml−1. As a control, when ASO-AuNPs were mixed with total RNA extracted from non-infected cells, no increase in absorbance at 660 nm was observed. For visual detection, ribonuclease H was used to cleave the viral RNA strands bound to ASO-AuNPs. The cleavage destabilized ASO-AuNPs and induced particle precipitation out of the solution. The dramatic color change resulting from the AuNP precipitation was easily detected by naked eyes. This method was easy to operate and fast; it detected SARS-CoV-2 RNA within 10 min.
Fig. 2.
Design of a colorimetric assay based on antisense oligonucleotide-capped gold nanoparticles (ASO-AuNPs) to detect SARS-CoV-2 RNA. (A) Schematic illustration of the proposed mechanism for ASO-AuNP agglomeration in the presence of SARS-CoV-2 N-gene. (B) The absorbance spectra of ASO-AuNPs before and after addition of 1 g ml−1 of total RNA extracted from SARS-CoV-2 infected Vero cells. (C) Change in the absorbance at 660 nm when ASO-AuNPs were mixed with varying amounts of total RNA extracted from SARS-CoV-2 infected Vero cells. (D) Schematic illustration of the procedures for naked-eye detection of SARS-CoV-2 RNA. Ribonuclease H (RNase H) was used to cleave the viral RNA bound to ASO-AuNPs and induce particle precipitation for visual detection. Reproduced with permission from ref. [16]. Copyright 2020, the American Chemical Society.
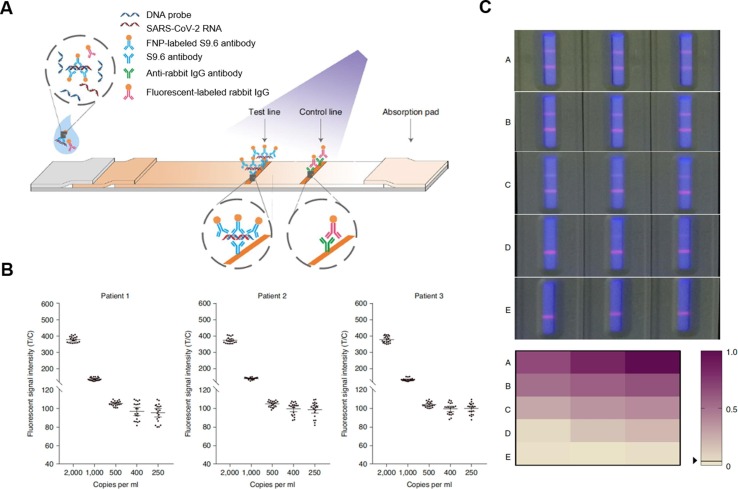
Other nanoparticles have also been designed to detect SARS-CoV-2 RNA. For example, fluorescent europium-chelate nanoparticles (FNPs) were conjugated with S9.6 antibodies, which bind specifically to RNA-DNA hybrid strands (S9.6-FNPs, Fig. 3 ) [17]. In this study, SARS-CoV-2 pseudovirus carrying viral RNA was incubated with S9.6-FNPs and DNA probes that specifically targeted SARS-CoV-2 RNA. During the incubation, the viral RNA bound to the DNA probe, forming RNA-DNA hybrid strands. The RNA-DNA hybrids then bound to S9.6-FNPs, forming RNA-DNA-S9.6-FNP complex. After the incubation, the sample was loaded onto an LFIA strip. On the strip, RNA-DNA-S9.6-FNP complex was captured by S9.6 antibodies immobilized at the test line. The accumulation of FNPs at the test line generated a fluorescent signal as a quantitative measurement of the viral RNA load. Due to the specificity of DNA probes, this method showed high specificity towards SARS-CoV-2 RNA with no apparent cross-reactivity with RNA of other viruses. When tested on 734 patient samples consisting of 593 throat swab specimens and 141 sputum specimens collected from 671 individuals, this method showed 100% sensitivity and 99% specificity when referenced to rtPCR tests.
Fig. 3.
Design of a lateral flow immunoassay (LFIA) based on S9.6 antibody-labelled europium-chelate fluorescent nanoparticles (S9.6-FNPs) to detect SARS-CoV-2 RNA. (A) Schematic illustration of the working principles of the S9.6-FNPs-based LFIA. (B) The fluorescent readouts of the assay when testing serially diluted throat swab samples from three SARS-CoV-2-positive patients. The fluorescent readouts are presented as the ratio of fluorescent intensities at the test line to those at the control line (T/C). The viral RNA loads in the samples were measured using polymerase chain reaction (PCR) and expressed in copies ml−1. The detection limit of the assay for viral RNA was determined to be 500 copies ml−1 with a positive rate higher than 95% (n = 20, means ± the standard deviation). (C) Photographs of typical assay results obtained from clinical samples under a fluorescent light source. The results were grouped into A, B, C, D, and E based on their fluorescent readouts. The fluorescent readouts decreased from group A to E. Group E consisted of SARS-CoV-2 negative results entirely. The fluorescent readouts were normalized to the maximum readout on a scale from 0 to 1. The normalized fluorescent readouts were presented in a color-gradient matrix. Reproduced with permission from ref [17]. Copyright 2020, Spring Nature.
Nanoparticles have also been designed to detect SARS-CoV-2-specific antibodies such as immunoglobulin M (IgM) and immunoglobulin G (IgG). For example, AuNPs were conjugated with anti-human IgM (anti-human IgM-AuNPs) and used to detect SARS-CoV-2 specific IgM in an LFIA strip [18]. The patient serum sample containing SARS-CoV-2 specific IgM was loaded onto the sample pad of the strip. The sample traveled along the strip to the conjugation pad, where anti-human IgM-AuNPs were stored. At the conjugation pad, SARS-CoV-2 specific IgM in the sample bound to anti-human IgM-AuNPs, forming (SARS-CoV-2-IgM)-(anti-human IgM)-AuNP complex. The complex traveled further along the strip and was captured by SARS-CoV-2 nucleoproteins immobilized at the test line through interactions between SARS-CoV-2 specific IgM in the complex and the nucleoproteins. Accumulation of AuNPs at the test line generated colorimetric signals for visual detection. This method was sensitive and fast; it only required 10-20 l of serum sample and was completed in 15 min. When tested on 19 patient serum samples collected from 5 SARS-CoV-2-positive (confirmed by rtPCR) patients and 14 healthy subjects, this method achieved 100% sensitivity and 93% specificity when referenced to rtPCR tests. AuNPs have also been used to detect multiple types of SARS-CoV-2 specific antibodies concurrently. For example, AuNPs were functionalized with SARS-CoV-2 S proteins (S-protein-AuNPs) and used to detect virus-specific IgM and IgG in an LFIA strip [19]. On the strip, S-protein-AuNPs bound to virus-specific IgM or IgG to form IgM-(S-protein)-AuNP or IgG-(S-protein)-AuNP complexes. These complexes were captured separately at two test lines, where anti-human IgM or anti-human IgG were immobilized, respectively. Color development at either test line was considered a positive result. When tested on venous blood specimens collected from 397 SARS-CoV-2-positive (confirmed by rtPCR) patients and 128 SARS-CoV-2-negative patients, this assay provided higher sensitivity than the single-antibody detection assay.
In addition to AuNPs, other nanoparticles have also been designed to detect SARS-CoV-2 specific antibodies. For example, selenium nanoparticles (SeNPs) were conjugated with SARS-CoV-2 nucleoprotein and used to detect virus-specific IgM and IgG in LFIA [20]. IgM or IgG-bound SeNPs generated colorimetric signals at the test line for visual detection. Compared with commercial AuNP-based LFIA kits, SeNP-based LFIA showed similar sensitivity for virus-specific IgG but achieved higher sensitivity for virus-specific IgM, possibly due to the higher affinity of IgM with SeNPs than AuNPs. In another design, lanthanide-doped polystyrene nanoparticles (LNPs) were conjugated with anti-human IgG and used to detect SARS-CoV-2 specific IgG in LFIA [21]. In this design, LNPs provided highly sensitive fluorescent signals for detection. This method detected virus-specific IgG in serum samples with 1:1000 dilution. In another example, SiO2 nanoparticles were coated with an Ag shell, and 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), a Raman-active compound, was loaded into the Ag shell. These nanoparticles exhibited a surface-enhanced Raman scattering (SERS) signal at 1328 cm−1 [22]. They were conjugated with SARS-CoV-2 S proteins and used to detect virus-specific IgM and IgG in LFIA. IgM or IgG-bound nanoparticles accumulated at the test line and provided SERS signals correlated with the amounts of virus-specific antibodies. This method was around 800 times more sensitive than AuNP-based LFIA.
Nanoparticle-based treatment for COVID-19 is summarized in Table 1 . Overall, these platforms have shown great potential in providing accurate and efficient detection of SARS-CoV-2. So far, most platforms are based on AuNPs, mainly for their compatibility for bioconjugation and unique optical properties. Research on other types of nanoparticles as described above for SARS-CoV-2 detection has also shown encouraging results. To further improve the performance, the physicochemical properties of the nanoparticles, such as their size and shape, need to be fine-tuned. The methods for surface functionalization of nanoparticles with bioactive moieties also need to be optimized so that these moieties can better retain their abilities to interact with the targets. In addition, many of the diagnostic platforms developed so far have not yet been tested on clinical patient samples or only on a relatively small number of samples. To validate the accuracy and reliability of these platforms, they need to be rigorously tested on a large and diverse pool of clinical patient samples. Lastly, it is necessary to develop large-scale production of these nanoparticle-based platforms with consistent quality and reproducible performance for successful clinical translation. With continuous development, nanoparticle-enabled platforms will make a more significant impact on the rapid and accurate detection of SARS-CoV-2 and other viral species.
Table 1.
Summary of nanoparticle-enabled detection of SARS-CoV-2.
| Platform | Selective example | Representative feature |
|---|---|---|
| Detection of viral protein |
|
|
| Detection of viral RNA |
|
|
| Detection of virus-specific antibody |
|
|
3. Nanoparticle-based treatment for COVID-19
Nanoparticles have been used as drug carriers to treat COVID-19. They improve existing antiviral drugs through their altered pharmacokinetics and biodistribution profiles [23], [24], [25]. Some nanoparticles do not carry drug payloads but exploit mechanisms such as disrupting the viral integrity, producing reactive oxygen species (ROS), or generating heat to kill the viruses. Recently, biomimetic nanoparticles were developed to leverage host cell membranes for viral inhibition, leading to a broad-spectrum antiviral mechanism [26], [27]. Overall, the development along these directions makes nanoparticles attractive to treat COVID-19.
3.1. Nanoparticles as antiviral drug carriers
Polymeric nanoparticles can encapsulate existing antiviral drugs for prolonged drug release and viral targeting, which together enhance antiviral potency. For example, polymeric nanoparticles made from poly(lactide-co-glycolide)-b-poly(ethylene glycol)-maleimide (PLGA-b-PEG-Mal) were used to deliver Ivermectin (IVM), a hydrophobic compound recently found effective against SARS-CoV-2 [28], [29], [30]. The nanoparticle IVM formulation crossed the blood-gut barrier when functionalized with an Fc immunoglobulin fragment that actively targets the neonatal Fc receptors expressed on intestinal epithelial cells of the host. Once in the circulation, the nanoparticles could eventually accumulate at the lung epithelia, making IVM more easily targeting SARS-CoV-2. The nanoparticles enhanced the antiviral potency by maintaining IVM levels around the minimum effective therapeutic dose but below the maximum tolerated dose.
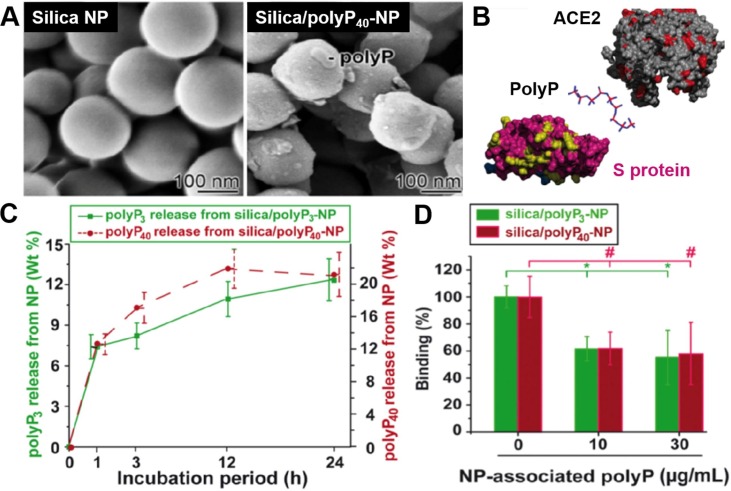
Besides polymeric nanoparticles, inorganic nanoparticles can also encapsulate and deliver anti-SARS-CoV-2 drug compounds. For example, silica nanoparticles encapsulated polyphosphate (polyP), a physiological polymer typically released from platelets (Fig. 4 )[31]. PolyP binds to the receptor-binding domain (RBD) of the viral S proteins via electrostatic interactions and prevents S protein from further interacting with the host receptor. The encapsulation with silica nanoparticles stabilized polyP against the degrading alkaline phosphatase. The study showed that the silica nanoparticles released polyP in vitro over 24 h. The nanoparticle polyP formulation inhibited S-protein binding with ACE2 at a polyP concentration equivalent to its blood concentration.
Fig. 4.
Nanoparticle-encapsulated polyphosphate blocks binding of SARS-CoV-2 S protein to ACE2. (A) Scanning electron microscopy (SEM) image of silica nanoparticle (silica-NP) and polyphosphate-loaded silica-NP (silica/polyP40-NP). (B) Schematic mechanism of polyphosphate binding to the viral S protein at the interface of S protein and ACE2 receptor. (C) In vitro release profile of polyP3 (3 phosphate units) or polyP40 (40 phosphate units) from silica nanoparticles. (D) Inhibition of S protein-ACE2 binding by polyP3 or polyP40 released from the silica nanoparticles. Data are presented as mean ± SEM (* p < 0.05). Reproduced with permission from Ref [31]. Copyright 2020 Elsevier.
Besides small molecules, nanoparticles were also used to deliver macromolecules such as messenger RNA (mRNA) to inhibit SARS-CoV-2. For example, LNPs were used to deliver in-vitro-transcribed mRNA (IVT mRNA) for rapid expression of soluble human ACE2 (hsACE2) in vivo. [32] Soluble ACE2 competes with endogenous ACE2 and binds with viruses for neutralization. The mRNA approach overcomes the issue of a relatively short half-life of the recombinant ACE2 in the bloodstream that would otherwise require repeated administrations to ensure long-term circulation of the protein for days after viral exposure. In this study, β-sitosterol, instead of more commonly used cholesterol, was used to formulate the LNPs. The replacement of cholesterol with β-sitosterol had little impact on nanoparticle physicochemical characteristics such as hydrodynamic size, polydispersity, and RNA encapsulation capacity. However, β-sitosterol dramatically enhanced mRNA transfection efficiency attributable to the multilamellar and polymorphic structures of the β-sitosterol-doped LNPs [33]. Using LNPs for mRNA delivery generated a transient but high expression of functional ACE2 without a risk of insertional mutagenesis associated with viral-based gene delivery. The synthesis of LNPs and mRNAs are economical and scalable, making this approach attractive.
Toward future development, nanocarriers can adopt emerging strategies, such as metabolic engineering and genetic engineering, to add functionalities for better delivery of antiviral drugs. Moreover, nanoparticles can be integrated into macroscopic matrices such as hydrogels, nanofibers, or autonomous nanorobots for intelligent drug release. In addition, nanoparticles can encapsulate multiple drugs to inhibit different stages of viral infection synergistically. As the nanoparticle designs are continuously refined, they are anticipated to play significant roles in antiviral drug delivery.
3.2. Nanoparticles for direct viral killing
Without carrying additional drug payloads, nanoparticles can exploit unique mechanisms for direct viral killing. One such mechanism is the disruption of viral proteins. For example, silver nanoparticles prevented VeroE6 cells from SARS-CoV-2 infection in a dose-dependent manner, with an effective virucidal concentration 10-times lower than the cytotoxic concentration [34]. Mechanistically, silver nanoparticles bind with SARS-CoV-2 surface proteins rich in sulfhydryl groups. Following the binding, they then cleave the disulfide bonds and disrupt the protein structure for killing. The virucidal effect of silver nanoparticles depended strongly on their sizes, with 2–15 nm nanoparticles demonstrating the highest protein affinity and virucidal activity.
Some nanoparticles generate oxidative radicals to kill viruses. For example, titanium dioxide (TiO2) nanoparticles generated free electrons upon UV excitation, which then produced ROS [35]. The ROS then turned into highly virucidal hydrogen peroxide (H2O2) and killed viruses effectively. One minute of UV radiation eradicated 100% of coronaviruses on a TiO2 nanoparticle-coated surface in the study. In contrast, approximately 10% of the viruses remained alive on the uncoated control surface. The results demonstrated that the TiO2 nanoparticles are useful to fabricate self-disinfecting surfaces in public and hospital settings.
Some nanoparticles harness the photothermal effect to converge heat and kill SARS-CoV-2 directly. For example, a semiconducting nanoparticle core made from poly[2,6-(4,4-bis-(2-ethylhexyl)-4H-cyclopenta [2,1-b;3,4-b′]dithiophene)-alt-4,7(2,1,3-benzothiadiazole)] (PCPDTBT) was coated with a lipid-PEG shell and further functionalized with anti-SARS-CoV-2 antibodies to capture the viruses [36]. When excited with light of 650 nm wavelength, the nanoparticles generated localized heat that killed SARS-CoV-2 captured by the nanoparticle. Without viral inactivation by heat, nanoparticle binding alone could not inhibit the viruses due to antibody-dependent enhancement (ADE), where the antibody-virus complex enters the host cells via Fc receptor-mediated phagocytic pathway [37].
With remarkable design flexibility, nanoparticles can be further optimized for higher potency in killing viruses. For example, silver nanoparticles' size and surface capping agents can be finely tailored to better interact with viral proteins. Such optimization may enhance the binding specificity toward viruses while reducing the toxicity toward the host cells. Moreover, nanoparticles can be immobilized on surfaces or formulated as spray disinfectants to inactivate surface-bound or airborne viruses before the viruses reach the host. Additionally, with the resurgence of variant strains of SARS-CoV-2, nanoparticles can be functionalized with antibodies against variant strains to improve viral binding and, subsequently, antiviral potency. These are exciting approaches that may allow for more functional and potent nanoparticles in killing SARS-CoV-2 and other viruses.
3.3. Host-mimicking nanoparticles for neutralizing SARS-CoV-2
Using cell membrane to coat nanoparticles is a top-down strategy to mimic host cells for biomedical application, including their recent use for viral neutralization [38], [39], [40], [41], [42], [43]. The resulting cell membrane-coated nanoparticles (denoted 'cellular nanosponges') inherit the critical surface antigens of their parent cells for viral binding. Thus, they bind to the viruses by acting as host cell decoys and diverting them away from the intended host targets. This working mechanism is different from conventional antiviral compounds, which require the identification of viral antigens for targeting. The knowledge in this respect is often limited. Besides, each antiviral compound selectively targets proteins encoded by a single virus. Therefore, the conventional “one drug-one virus” solution is ineffective. Notably, the infectivity of any virus relies on its binding with the protein receptors of the host cells, regardless of whether they are known or unknown. Therefore, the cellular nanosponge approach shifts the focus from the causative viruses to the hosts and overcomes the diversity of the viruses for neutralization. Some cells, such as macrophages, are the target of multiple viruses. Therefore, cellular nanosponges made from membranes of these cells unlock a broad-spectrum neutralization strategy against viral infection [43].
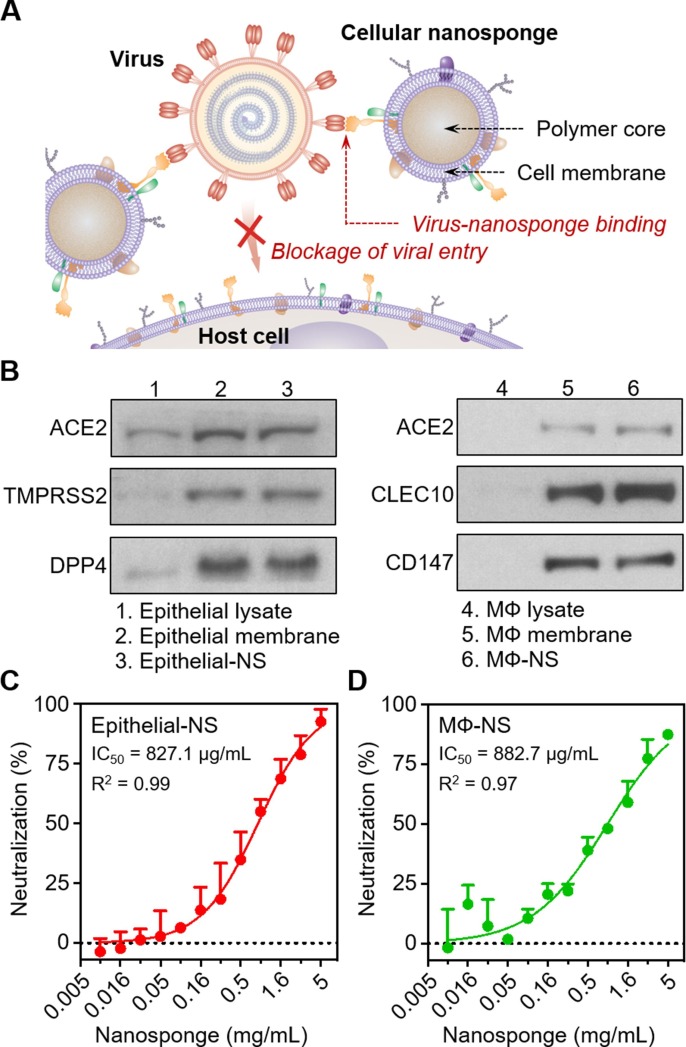
Recently, cellular nanosponges were demonstrated to inhibit the infectivity of SARS-CoV-2 (Fig. 5 )[44]. In the study, the nanosponges were made by coating PLGA cores with membranes of lung epithelial cells or macrophages, two cellular targets of the virus. The nanosponges displayed key cellular receptors for viral entry. When tested on Vero E6 cells, these nanosponges effectively blocked SARS-CoV-2 infection in a dose-dependent manner. When administered into the lungs of mice, the cellular nanosponges showed no apparent toxicity. Nanosponges are agnostic to viral mutations and potentially to different species of viruses and thus can protect the current and future emerging coronaviruses.
Fig. 5.
Cellular nanosponges neutralize SARS-CoV-2 infectivity. (A) Schematic mechanism of cellular nanosponges inhibiting SARS-CoV-2 infectivity. The nanosponges were constructed by wrapping polymeric nanoparticle (NP) cores with natural cell membranes derived from target cells such as human lung epithelial cells and macrophages (MФs). The resulting nanosponges (denoted “Epithelial-NS” and “MΦ-NS”, respectively) inherit the surface antigen profiles of the source cells and serve as decoys to bind with SARS-CoV-2. Such binding interaction blocks viral entry and inhibits viral infectivity. (B) Selective protein bands of cell lysate, cell membrane vesicles, and cellular nanosponges resolved with Western blotting analysis. (C-D) The neutralization against SARS-CoV-2 infection by Epithelial-NS (C), and MΦ-NS (D) was tested using live SARS-CoV-2 viruses on Vero E6 cells. The IC50 values for Epithelial-NS and MΦ-NS were found to be 827.1 and 882.7 μg/mL (membrane protein concentration), respectively. Data are presented as mean + standard deviation (n = 3). The horizontal dashed line marks the zero levels. IC50 values were derived from the variable slope model using Graphpad Prism 8. Reproduced with permission from Ref [44]. Copyright 2020 American Chemical Society.
Nanoparticles can also be made from membranes of genetically engineered cells expressing desired viral receptors for viral neutralization. For example, human embryonic kidney (HEK)-293 T cells were transiently transfected to highly express human ACE2 (hACE2) on the cell membranes[45]. The membranes of HEK-293 T-hACE2 cells were then isolated and formed cell membrane-based nanoparticles (CMBNPs) via sonication and extrusion. The CMBNPs bound with the S proteins of original SARS-CoV-2 and SARS-CoV-2 D614G, a more infectious variant strain[46], [47]. The study demonstrated the potential of host-mimicking nanoparticles in resisting mutations of the coronavirus, as long as ACE2 is required for viral entry into host cells.
Upon initial infection by SARS-CoV-2, the host immune system initiates an inflammatory response that upregulates abundant inflammatory cytokines for host protection [48]. However, sustained viral infection results in overexuberant inflammation characterized by systemic cytokine storm leading to immune dysfunction and organ damage [49]. Cellular nanosponges were previously demonstrated to neutralize multiple inflammatory cytokines in infectious diseases [39]. They were tested recently for concurrent virus and cytokine neutralization for COVID-19 treatment in animal models [50]. In this study, nanoparticles were fabricated by first fusing the membranes from HEK-293 T-hACE2 cells and human macrophage cells, followed by repeated sonication and extrusion through 100 nm nanopores. This method allows viral receptors and cytokine receptors packaged onto the surface of the same resulting nanoparticles. The study showed that the nanoparticles bound with patient-derived SARS-CoV-2 viruses and inhibited the viral binding and infection of Vero E6 cells in vitro. Additionally, in a mouse model of lipopolysaccharide-induced lung inflammation, intratracheally administered nanoparticles absorbed inflammatory cytokines including interleukin-6 (IL-6) and granulocyte–macrophage colony-stimulating factor (GM-CSF) in bronchoalveolar lavage fluid (BALF) in a dose-dependent manner.
Several challenges remain to be addressed for the translation of cell membrane-coated nanoparticles against SARS-CoV-2. For example, cell membrane-coated nanoparticles need to be manufactured at a scale to meet the demand in therapeutic doses. In this regard, large-scale cell farming and a high yield of cell membrane isolation are essential. T storage stability of cell membrane-coated nanoparticle formulations also needs to be thoroughly validated. The clinically standardized process of lyophilization, cold storage, and reconstitution can be employed for this purpose. Furthermore, the toxicity of cell membrane-coated nanoparticles remains to be characterized in more comprehensive studies. By addressing these challenges, we believe that cell membrane-coated nanoparticles will become a robust platform against SARS-CoV-2.
In summary, this section reviewed nanoparticles as antiviral drug carriers, nanoparticles for direct viral killing, and host-mimicking nanoparticles for viral neutralization. As summarized in Table 2 , the representative platforms in each category have demonstrated promise in combating SARS-CoV-2. By addressing the potential challenges of these platforms, we expect that their potency, safety, and functionality can be further improved.
Table 2.
Summary of nanoparticle-based treatment for COVID-19
| Platform | Selective example | Representative feature |
|---|---|---|
| Nanoparticles as antiviral drug carrier |
|
|
| Nanoparticles for direct viral killing |
|
|
| Host-mimicking nanoparticles for viral neutralization |
|
4. Nanoparticle vaccines against SARS-CoV-2
Vaccination is currently the most effective approach to control the COVID-19 pandemic. It activates the host's immune system, which generates neutralizing antibodies and T cell responses for protection [51]. So far, there are over 200 vaccine candidates in various stages of development. Among them, more than 50 are in clinical trials [52]. To date, three vaccines have been approved by the United States Food and Drug Administration (FDA), developed by Pfizer/BioNTech, Moderna, and Janssen (Johnson & Johnson)[53]. SARS-CoV-2 is an enveloped ssRNA virus with spike-like glycoproteins (S proteins) protruding from its surface. S proteins are responsible for the viral entry into the host cells by binding to host ACE2[54]. Therefore, the full-length S proteins or their receptor-binding domains (RBDs) are the primary targets for vaccine designs.
SARS-CoV-2 vaccine platforms can be divided into five major types: live attenuated virus, inactivated virus, engineered viral vectors, DNA/mRNA, and subunit-based vaccines, each with its advantages and limitations. While live attenuated and inactivated viruses are mature platforms of vaccines with a rapid manufacturing process against the outbreak of a pandemic, they are limited by the low scalability and safety concerns of potential reversion to the virulent form [55]. In contrast, subunit and nucleic acid-based vaccines can be manufactured fast with high scalability and safety. However, these vaccines suffer from instability and poor immunogenicity without the help of additional adjuvants [51]. Engineered viral vectors are non-replicable and highly immunogenic due to their inherent adjuvant qualities. Their production is also highly scalable. However, these vaccines are limited by possible pre-existing immunity in the vaccinated population against the engineered viral vectors, which is especially common in adenovirus-derived vectors [56].
Nanotechnology approaches have been combined with the vaccine development against SARS-CoV-2. Major nanoparticle platforms include LNPs and VLPs. LNPs consisting of ionizable lipids have been applied extensively to deliver genetic contents due to their high loading capacity and high transfection efficiency [57]. VLPs are non-infectious virus-mimicking particles formed by the self-assembly of protein monomers conjugated with viral capsid proteins [58]. So far, nanoparticle designs have been focused on modulating antigen transport to the lymph nodes, promoting antigen-adjuvant co-delivery, and protecting the vaccines from in vivo degradation [59].
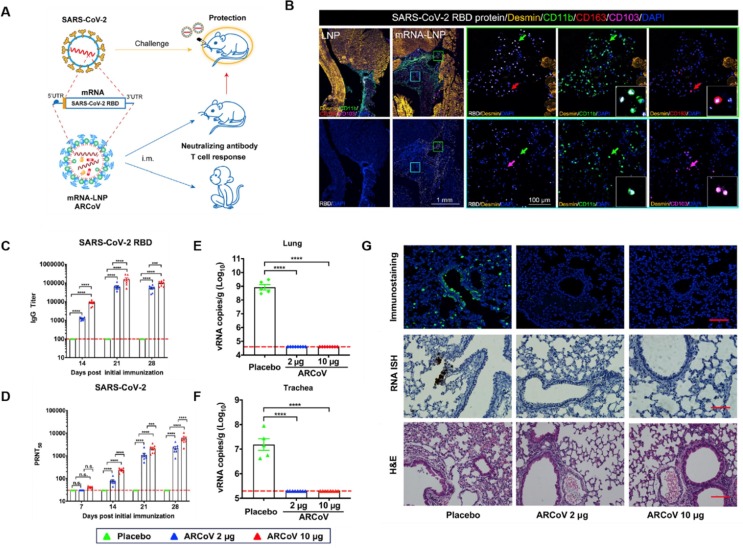
LNPs are neutrally charged at physiological pH but become positively charged at low pH. Their composition usually includes helper lipids for cell binding and cholesterols for structural stability [57]. These LNPs load negatively charged genetic materials with a high capacity. LNPs also facilitate nanoparticle endosomal escaping after the cell uptake. These properties make LNPs ideal to deliver nucleic acid-based vaccines [60]. For example, LNPs were first used to deliver mRNA encoding the RBD of SARS-CoV-2 (denoted “ARCoV”) (Fig. 6 A)[61]. In the study, these LNPs were injected intramuscularly into a mouse model of SARS-CoV-2 mouse-adapted strain, which was generated by intranasal inoculation of a mouse-adapted strain at passage 6 (MASCp6) in BALB/c mice [62]. The muscle tissue from the injection site showed high recombinant RBD expression, accompanied by APC recruitment. The vaccination led to a rapid elevation in the level of immunoglobulin G (IgG) neutralizing antibodies and virus-specific CD4+ and CD8+ effector memory T cells (Fig. 6B-D). Immunization of two doses of ARCoV in the SARS-CoV-2 mouse-adapted strain model showed complete prevention of viral replication in lungs and trachea after the viral challenge (Fig. 6E and F). In another study, BALB/c mice were immunized with LNPs containing mRNA encoding the full-length S protein [63]. Protective immune responses were also observed. These studies demonstrate a robust protection efficacy of the LNP-based mRNA vaccine against SARS-CoV-2 infection.
Fig. 6.
LNP-encapsulated mRNAs encoding the receptor-binding domains of SARS-CoV-2 (ARCoV) and their in vivo protective efficacy against SARS-CoV-2 challenge in mice. (A) Design features and proposed mechanism of ARCoV. (B) Multiplex immunostaining analysis for expression of ARCoV mRNA-LNP in mouse muscle tissues. Female BALB/c mice (n = 3) were immunized with 10 mg of ARCoV mRNA-LNP or empty LNP (n = 3). Muscle tissue at the injection site was collected 6 h after injection for multiplex immunofluorescent staining. Cell markers include SARS-CoV-2 (white), Desmin (gold), CD11b (green), CD163 (red), and CD103 (magenta). (C-G) Analysis of the protective immune responses elicited in SARS-CoV-2 challenged mice after ARCoV immunization. Female BALB/C mice were immunized with 2 ug ARCoV mRNA-LNP (n = 5), 10 ug ARCoV mRNA-LNP (n = 5), or placebo (n = 5) intramuscularly. Forty days after the initial immunization, mice were inoculated with the mouse-adapted SARS-CoV-2 (MASCp6), and the indicated tissues were collected 5 days after challenge for detection of viral loads and lung pathology. (C) The SARS-CoV-2-specific IgG antibody titer after immunization. (D) PRNT50 titer against SARS-CoV-2 after injection. (E-F) Viral RNA loads in the lungs (E) and trachea (F) of SARS-CoV-2 challenged mice after immunization. (G) Immunostaining with SARS-CoV-2-specific mAb, ISH assay for SARS-CoV-2 RNA, and H&E staining of lung tissues. Reproduced with permission from ref [61]. Copyright 2020 Elsevier.
LNP-based mRNA vaccines are considered safer and can be manufactured rapidly. As a result, they are now the leading vaccine against the COVID-19 pandemic [64]. Currently, BNT162b2 by Pfizer/BioNTech and mRNA-1273 by Moderna, the two mRNA-based LNP COVID-19 vaccines approved by FDA, showed an efficacy of 95% and 94.5%, respectively [65], [66], [67]. BNT162b2 contains a synthetic chemically modified RNA (modRNA) of 4284 nucleotides long, encoding a mutated form of the full-length S protein found on the surface of the SARS-CoV-2[68]. The modRNA and lipids, including ((4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate) (ALC-0315), 2-[(polyethylene glycol)-2000]-N,N-ditetradecylacetamide (ALC-0159), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and cholesterol, form LNPs, acting as both carriers and adjuvants. Similarly, the active ingredient in mRNA-1273 is an mRNA sequence containing 4101 nucleotides that encodes the full-length SARS-CoV-2 spike (S) glycoprotein with two mutations (K986P and V987P) designed to stabilize the pre-fusion conformation[69], [70]. The mRNA is encapsulated into LNPs consisting of DSPC, cholesterol, PEG 2000-dimyristoyl glycerol, and SM-102 (a synthetic amino lipid). To further enhance the vaccine efficacy, researchers added alphaviral replicase to the RNA sequences, enabling self-amplification of the RNA vaccine after cell uptake. They also replaced the untranslated region (UTR) of the mRNA with a synthetic UTR designed through de novo methods to enhance protein translation. Using LNPs to deliver these modified mRNAs led to effective anti-SARS-CoV-2 vaccination [71], [72].
Another distinct advantage of using nanoparticle-based vaccine platforms, typically the VLPs, is their capability of modulating host immune responses. Through the multivalent antigen presentation, VLPs usually generates more significant immune responses than recombinant antigens. This property is beneficial in delivering subunit-based vaccines with low immunogenicities, such as the full-length S protein or RBDs [73]. In some cases, VLP-based vaccines do not require boost immunization. For example, ferritin is a popular protein particle to make VLP vaccines [74], [75], [76]. Each ferritin particle can be conjugated with up to 24 viral antigens for simultaneous delivery and presentation to APCs [74]. Besides, ferritin-based VLPs can be manufactured rapidly in vitro through genetic fusion. Recently, full-length S protein/ferritin fusion protein was generated using Expi293F cells as a potential vaccine against SARS-CoV-2 (denoted “S-Fer”)[77]. Mice immunized with a single dose of S-Fer showed significantly higher neutralizing antibody titers than those immunized with RBD monomers or spike ectodomain trimers, implying the importance of multivalent presentation. Ferritin nanoparticles conjugated with RBDs, rather than full-length S proteins, also showed a better protective immune response when compared with unconjugated RBDs [78].
Recently, researchers used the SpyTag/SpyCatcher system to link ferritin nanoparticles with full-length S proteins or RBDs. The conjugation system enhanced ferritin expression otherwise restricted by the traditional direct genetic fusion approach [78], [79]. With the SpyTag/SpyCatcher system, multiple different antigens can also be conjugated onto the same ferritin nanoparticles for simultaneous presentation. For example, SARS-CoV-2 RBDs and heptad repeat (HR), which is part of the highly conserved S2 subunit of S protein, were conjugated onto the same ferritin nanoparticles [80]. The incorporation of HR onto VLPs provides the vaccine with broadened neutralization ability against neutralization-escape mutations. To develop more effective VLP vaccines, researchers also used other protein particles such as aldolase, which allow more antigens to be conjugated. For example, VLP formed by self-assembled RBD-conjugated aldolases showed higher efficacy in eliciting neutralizing antibody responses in mice or pigs than purified RBD or purified S protein alone [81], [82]. A higher capacity of antigen presentation makes these particles useful to develop more effective SARS-CoV-2 vaccines.
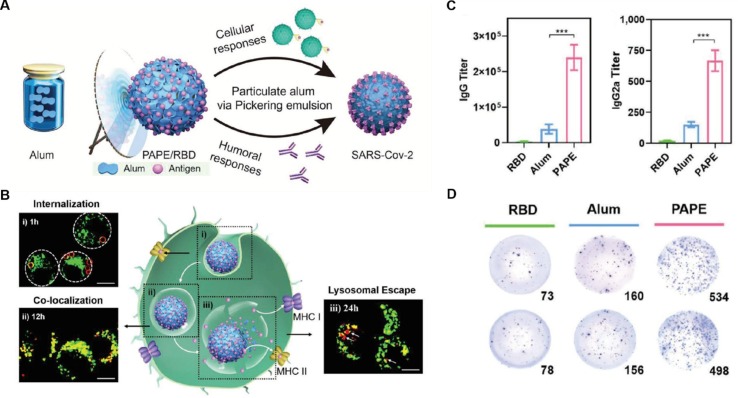
VLPs can also be made with synthetic nanoparticle substrates with adjuvant properties. Recently, a particulate alum‐stabilized Pickering emulsion (PAPE) was formulated by adding alum (aluminum hydroxide) to the squalene-water emulsion, where alum accumulated at the squalene-water interphase and stabilized the emulsion (Fig. 7 A)[83]. The cationic surface of the PAPE allows a dense layer of viral antigens to be absorbed. The PAPE also mimics the surface contour of the virus for antigen presentation. Furthermore, PAPE is more hydrophobic than traditional alum microgels. A high hydrophobicity favors its interaction with the macrophages and the phagocytosis along with co‐loaded antigens. The cationic PAPE interacts with anionic membranes inside the lysosomes, promoting lysosome disruption to release the antigens into the cytoplasm (Fig. 7B). In mice, PAPE loaded with RBDs elicited six times higher antigen‐specific antibody titer and three‐fold more IFN‐γ‐secreting T cells than mice immunized with a simple mixture of RBDs and alum (Fig. 7C and D). In another study, adjuvant-encapsulated liposomes were decorated with RBDs [84]. These VLPs induced prominent antibody titers in mice. When tested in vitro, these antibodies effectively neutralized pseudovirus cell entry, blocked RBD interaction with ACE2, and inhibited live virus replication.
Fig. 7.
Particulate alum via Pickering emulsion (PAPE) as an enhanced COVID-19 vaccine adjuvant. (A) A schematic of the PAPE synthesis process. (B) Uptake of PAPE for enhanced antigen-presentation to DCs in vitro. Confocal images showing the intracellular distribution of RBD (red) with lysosomes (green) of PAPE and alum. (C-D) Humoral responses to PAPE vaccination in BALB/c mice. 6–8-weeks-old BALB/c mice were vaccinated with 100 µl suspension of various formulations along with 5 µg of RBD on day 0 and day 14 through intramuscular injections (n = 6). Serum samples were collected 28 days after the initial immunization for IgG titer analysis, and splenocytes were collected 30 days after the initial immunization for cytokine analysis. (C) Serum RBD-specific IgG and IgG2a titers. (D) ELISpot analysis of IFN-γ spot-forming cells among splenocytes. Reproduced with permission from ref [83]. Copyright 2020 WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim.
Overall, nanoparticles can provide stability to the antigens, present antigens multivalently, and co-deliver adjuvants with the antigens. These advantages are the hallmarks of nanoparticle vaccines, including those developed against SARS-CoV-2 (Table 3 ). Nanoparticles delivering vaccines have also followed some distinct design strategies when compared to those for delivering antiviral drugs. For example, regarding the route of administration, nanoparticle vaccines are mainly designed for local and topical administration. In contrast, antiviral drug nanoparticle carriers likely prefer intravenous injection to maximize exposure. Regarding the surface charge, those for systemic drug delivery prefer a negative charge to minimize non-specific binding with the host [85]. In contrast, positively charged nanoparticles have shown higher immune stimulations that are favorable for vaccine delivery. Regarding PEGylation, nanoparticles designed for systemic circulation may favor a higher PEG density to achieve a longer circulation half-life. However, those intended for vaccine delivery may prefer a lower PEG density to minimize the barrier [86].
Table 3.
Summary of nanoparticle vaccines against SARS-CoV-2
| Platform | Selective example | Representative feature |
|---|---|---|
| Lipid nanoparticle (LNP) |
|
|
| Virus-like nanoparticle(VLP) |
|
The recent development of mRNA COVID-19 vaccines, such as BNT162b2 and mRNA-1273, also shows the advantages of scalability and cost-effectiveness. Unlike traditional vaccines, these mRNA vaccines can be produced in large quantities rapidly, with standard laboratory techniques. The first COVID-19 vaccine candidate started its clinical trials only 40 days after releasing the SARS-CoV-2 genomic report [55]. Such rapid development means nanoparticle vaccines can be easily produced in response to the outbreak of new viral infections. Nevertheless, to improve future nanoparticle vaccines, detailed studies on the in vivo host-nanoparticle interactions to optimize toxicity-efficacy are desirable. Knowledge in this perspective may further enhance the transfection efficiency of mRNA-based nanoparticle vaccines. In addition, improving the specificity of targeting nanoparticle vaccines to immune cells is a crucial challenge for optimizing vaccine dosage and efficacy. Furthermore, the stability of current mRNA vaccine formulations during storage and transportation requires further improvement. Both BNT162b2 and mRNA-1273 need to be stored at −20 and −70 °C, respectively. Such cold condition significantly increases the difficulties in vaccine distribution. As researchers continually address these challenges, nanoparticle vaccines can become safer and more effective in combating future pandemics.
5. Conclusion
Since the first case reported at the end of 2019, COVID-19 spread quickly, causing the current global pandemic. The virus also mutates rapidly, posing a significant challenge to the development of therapeutics and preventive medicine. The reality demands agile anti-viral responses. In this perspective, nanotechnology, particularly nanoparticle therapeutics, has provided new solutions and made significant contributions. Inorganic nanoparticles, LNPs, polymeric nanoparticles, VLPs, and cell membrane-coated nanoparticles have been developed against SARS-CoV-2, including new approaches for viral detection, treatment, and prevention. Continued development will lead to more sensitive, fast, and cost-efficient SARS-CoV-2 detection as well as more efficacious treatment and vaccines against viral infection. Notably, many nanoparticle strategies can be readily tailored against new variants or new viral species. Overall, nanoparticle-based technologies are expected to play a significant role in the management of viral pandemics.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by the Defense Threat Reduction Agency Joint Science and Technology Office for Chemical and Biological Defense under Grant Number HDTRA1‐18‐1‐0014.
References
- 1.Bekerman E., Einav S. Combating emerging viral threats. Science. 2015;348(6232):282–283. doi: 10.1126/science:aaa3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N.a., Zhang D., Wang W., Li X., Yang B.o., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in china. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou J., Krishnan N., Jiang Y., Fang R.H., Zhang L. Nanotechnology for virus treatment. Nano Today. 2021;36:101031. doi: 10.1016/j.nantod.2020.101031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell M.J., Billingsley M.M., Haley R.M., Wechsler M.E., Peppas N.A., Langer R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021;20(2):101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sindhwani S., Chan W.C.W. Nanotechnology for modern medicine: Next step towards clinical translation. J. Intern. Med. 2021;290(3):486–498. doi: 10.1111/joim.v290.310.1111/joim.13254. [DOI] [PubMed] [Google Scholar]
- 6.Ragelle H., Danhier F., Préat V., Langer R., Anderson D.G. Nanoparticle-based drug delivery systems: a commercial and regulatory outlook as the field matures. Expert Opin. Drug Deliv. 2017;14(7):851–864. doi: 10.1080/17425247.2016.1244187. [DOI] [PubMed] [Google Scholar]
- 7.Bellan L.M., Wu D., Langer R.S. Current trends in nanobiosensor technology. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2011;3(3):229–246. doi: 10.1002/wnan.v3.310.1002/wnan.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riley R.S., June C.H., Langer R., Mitchell M.J. Delivery technologies for cancer immunotherapy. Nat. Rev. Drug Discov. 2019;18(3):175–196. doi: 10.1038/s41573-018-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talebian S., Wallace G.G., Schroeder A., Stellacci F., Conde J. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 2020;15(8):618–621. doi: 10.1038/s41565-020-0751-0. [DOI] [PubMed] [Google Scholar]
- 10.Saxena A., Khare D., Agrawal S., Singh A., Dubey A.K. Recent advances in materials science: a reinforced approach toward challenges against COVID-19. Emergent Mater. 2021;4(1):57–73. doi: 10.1007/s42247-021-00179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A.D. Chintagunta, S.K. M, S. Nalluru, S.S. N, Nanotechnology: An emerging approach to combat COVID-19, Emergent. Mater. (2021) 1–12 10.1007/s42247-021-00178-6. [DOI] [PMC free article] [PubMed]
- 12.Hassanzadeh P. Nanotheranostics against COVID-19: From multivalent to immune-targeted materials. J. Control. Release. 2020;328:112–126. doi: 10.1016/j.jconrel.2020.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ventura B.D., Cennamo M., Minopoli A., Campanile R., Censi S.B., Terracciano D., Portella G., Velotta R. Colorimetric test for fast detection of SARS-CoV-2 in nasal and throat swabs. ACS Sensors 5. 2020;5(10):3043–3048. doi: 10.1021/acssensors.0c0174210.1021/acssensors.0c01742.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker A., Richards S.J., Congdon T., Hasan M., Guy C., Zwetsloot A., Gallo A., Lewandowski J., Stansfeld P., Straube A., et al. The SARS-CoV-2 spike protein binds sialic acids, and enables rapid detection in a lateral flow point of care diagnostic device. ACS Central. Sci. 2021;7:379–380. doi: 10.1021/acscentsci.1c00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khoris I.M., Ganganboina A.B., Suzuki T., Park E.Y. Self-assembled chromogen-loaded polymeric cocoon for respiratory virus detection. Nanoscale. 2021;13(1):388–396. doi: 10.1039/D0NR06893D. [DOI] [PubMed] [Google Scholar]
- 16.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by n gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D., He S., Wang X., Yan Y., Liu J., Wu S., Liu S., Lei Y., Chen M., Li L.i., Zhang J., Zhang L., Hu X., Zheng X., Bai J., Zhang Y., Zhang Y., Song M., Tang Y. Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4(12):1150–1158. doi: 10.1038/s41551-020-00655-z. [DOI] [PubMed] [Google Scholar]
- 18.Huang C., Wen T., Shi F.-J., Zeng X.-Y., Jiao Y.-J. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5(21):12550–12556. doi: 10.1021/acsomega.0c0155410.1021/acsomega.0c01554.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y.e., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.v92.910.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Zheng Z., Hu H., Zhou Q., Liu W., Li X., Liu Z., Wang Y., Ma Y. A point-of-care selenium nanoparticle-based test for the combined detection of anti-SARS-CoV-2 IgM and IgG in human serum and blood. Lab Chip. 2020;20(22):4255–4261. doi: 10.1039/D0LC00828A. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z., Zhang Z., Zhai X., Li Y., Lin L.i., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92(10):7226–7231. doi: 10.1021/acs.analchem.0c0078410.1021/acs.analchem.0c00784.s001. [DOI] [PubMed] [Google Scholar]
- 22.Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C., Wang S. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens. Actuators B Chem. 2021;329:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wankar J.N., Chaturvedi V.K., Bohara C., Singh M.P., Bohara R.A. Role of nanomedicine in management and prevention of COVID-19. Front. Nanotechnol. 2020;2 doi: 10.3389/fnano.2020.589541. [DOI] [Google Scholar]
- 24.Abd Ellah N.H., Gad S.F., Muhammad K., Batiha G.E., Hetta H.F. Nanomedicine as a promising approach for diagnosis, treatment and prophylaxis against COVID-19. Nanomedicine. 2020;15:2085–2102. doi: 10.2217/nnm-2020-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirtane A.R., Verma M., Karandikar P., Furin J., Langer R., Traverso G. Nanotechnology approaches for global infectious diseases. Nat. Nanotechnol. 2021;16(4):369–384. doi: 10.1038/s41565-021-00866-8. [DOI] [PubMed] [Google Scholar]
- 26.Singh L., Kruger H.G., Maguire G.E.M., Govender T., Parboosing R. The role of nanotechnology in the treatment of viral infections. Ther. Adv. Infect. Dis. 2017;4(4):105–131. doi: 10.1177/2049936117713593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang G., Chen S., Zhang J. Bioinspired and biomimetic nanotherapies for the treatment of infectious diseases. Front. Pharmacol. 2019;10:751. doi: 10.3389/fphar.2019.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surnar B., Kamran M.Z., Shah A.S., Dhar S. Clinically approved antiviral drug in an orally administrable nanoparticle for COVID-19. ACS Pharmacol. Transl. Sci. 2020;3(6):1371–1380. doi: 10.1021/acsptsci.0c0017910.1021/acsptsci.0c00179.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Surnar B., Kamran M.Z., Shah A.S., Basu U., Kolishetti N., Deo S., Jayaweera D.T., Daunert S., Dhar S. Orally administrable therapeutic synthetic nanoparticle for zika virus. ACS Nano. 2019;13(10):11034–11048. doi: 10.1021/acsnano.9b0280710.1021/acsnano.9b02807.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neufurth M., Wang X., Tolba E., Lieberwirth I., Wang S., Schröder H.C., Müller W.E.G. The inorganic polymer, polyphosphate, blocks binding of SARS-CoV-2 spike protein to ACE2 receptor at physiological concentrations. Biochem. Pharmacol. 2020;182:114215. doi: 10.1016/j.bcp.2020.114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J., Mukherjee A., Nelson D., Jozic A., Sahay G. Rapid generation of circulating and mucosal decoy ACE2 using mRNA nanotherapeutics for the potential treatment of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.07.24.205583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eygeris Y., Patel S., Jozic A., Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020;20(6):4543–4549. doi: 10.1021/acs.nanolett.0c0138610.1021/acs.nanolett.0c01386.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeremiah S.S., Miyakawa K., Morita T., Yamaoka Y., Ryo A. Potent antiviral effect of silver nanoparticles on SARS-CoV-2. Biochem. Biophys. Res. Commun. 2020;533(1):195–200. doi: 10.1016/j.bbrc.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khaiboullina S., Uppal T., Dhabarde N., Subramanian V.R., Verma S.C. Inactivation of human coronavirus by titania nanoparticle coatings and uvc radiation: throwing light on SARS-CoV-2. Viruses-Basel. 2021;13:19. doi: 10.3390/v13010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai X., Prominski A., Lin Y., Ankenbruck N., Rosenberg J., Chen M., Shi J., Chang E.B., Penaloza-MacMaster P., Tian B., et al. A neutralizing antibody-conjugated photothermal nanoparticle captures and inactivates SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.11.30.404624. [DOI] [Google Scholar]
- 37.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5(10):1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- 38.Hu C.-M.- J., Zhang L., Aryal S., Cheung C., Fang R.H., Zhang L. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc. Natl. Acad. Sci. U.S.A. 2011;108(27):10980–10985. doi: 10.1073/pnas.1106634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thamphiwatana S., Angsantikul P., Escajadillo T., Zhang Q., Olson J., Luk B.T., Zhang S., Fang R.H., Gao W., Nizet V., Zhang L. Macrophage-like nanoparticles concurrently absorbing endotoxins and proinflammatory cytokines for sepsis management. Proc. Natl. Acad. Sci. U.S.A. 2017;114(43):11488–11493. doi: 10.1073/pnas.1714267114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu C.-M., Fang R.H., Copp J., Luk B.T., Zhang L. A biomimetic nanosponge that absorbs pore-forming toxins. Nat. Nanotechnol. 2013;8(5):336–340. doi: 10.1038/nnano.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei X., Ran D., Campeau A., Xiao C., Zhou J., Dehaini D., Jiang Y., Kroll A.V., Zhang Q., Gao W., Gonzalez D.J., Fang R.H., Zhang L. Multiantigenic nanotoxoids for antivirulence vaccination against antibiotic-resistant gram-negative bacteria. Nano Lett. 2019;19(7):4760–4769. doi: 10.1021/acs.nanolett.9b0184410.1021/acs.nanolett.9b01844.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Y., Krishnan N., Zhou J., Chekuri S., Wei X., Kroll A.V., Yu C.L., Duan Y., Gao W., Fang R.H., Zhang L. Engineered cell-membrane-coated nanoparticles directly present tumor antigens to promote anticancer immunity. Adv. Mater. 2020;32(30):2001808. doi: 10.1002/adma.v32.3010.1002/adma.202001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei X., Zhang G., Ran D., Krishnan N., Fang R.H., Gao W., Spector S.A., Zhang L. T-cell-mimicking nanoparticles can neutralize HIV infectivity. Adv. Mater. 2018;30(45):1802233. doi: 10.1002/adma.v30.4510.1002/adma.201802233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q., Honko A., Zhou J., Gong H., Downs S.N., Vasquez J.H., Fang R.H., Gao W., Griffiths A., Zhang L. Cellular nanosponges inhibit SARS-CoV-2 infectivity. Nano Lett. 2020;20(7):5570–5574. doi: 10.1021/acs.nanolett.0c0227810.1021/acs.nanolett.0c02278.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang C., Wang S., Chen Y., Zhao J., Han S., Zhao G., Kang J., Liu Y., Wang L., Wang X., Xu Y., Wang S., Huang Y.i., Wang J., Zhao J. Membrane nanoparticles derived from ACE2-rich cells block SARS-CoV-2 infection. ACS Nano. 2021;15(4):6340–6351. doi: 10.1021/acsnano.0c0683610.1021/acsnano.0c06836.s00110.1021/acsnano.0c06836.s002. [DOI] [PubMed] [Google Scholar]
- 46.Hou Y.X.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., Leist S.R., Schafer A., Nakajima N., Takahashi K., et al. Sars-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Steiner S., Kelly J.N., Portmann J., Halwe N.J., Ulrich L., Trüeb B.S., Fan X., Hoffmann B., Wang L.i., Thomann L., Lin X., Stalder H., Pozzi B., de Brot S., Jiang N., Cui D., Hossain J., Wilson M.M., Keller M.W., Stark T.J., Barnes J.R., Dijkman R., Jores J., Benarafa C., Wentworth D.E., Thiel V., Beer M. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592(7852):122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- 48.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20(6):355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368(6490):473–474. doi: 10.1126/science:abb8925. [DOI] [PubMed] [Google Scholar]
- 50.Rao L., Xia S., Xu W., Tian R., Yu G., Gu C., Pan P., Meng Q.-F., Cai X., Qu D.i., Lu L.u., Xie Y., Jiang S., Chen X. Decoy nanoparticles protect against COVID-19 by concurrently adsorbing viruses and inflammatory cytokines. Proc. Natl. Acad. Sci. U.S.A. 2020;117(44):27141–27147. doi: 10.1073/pnas.2014352117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz‐Hitzky E., Darder M., Wicklein B., Ruiz‐Garcia C., Martín‐Sampedro R., del Real G., Aranda P. Nanotechnology responses to COVID-19. Adv. Healthc. Mater. 2020;9(19):2000979. doi: 10.1002/adhm.v9.1910.1002/adhm.202000979. [DOI] [PubMed] [Google Scholar]
- 52.Parker E.P.K., Shrotri M., Kampmann B. Keeping track of the SARS-CoV-2 vaccine pipeline. Nat. Rev. Immunol. 2020;20:650. doi: 10.1038/s41577-020-00455-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J.H., Marks F., Clemens J.D. Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med. 2021;27(2):205–211. doi: 10.1038/s41591-021-01230-y. [DOI] [PubMed] [Google Scholar]
- 54.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin M.D., Shukla S., Chung Y.H., Beiss V., Chan S.K., Ortega-Rivera O.A., Wirth D.M., Chen A., Sack M., Pokorski J.K., Steinmetz N.F. Covid-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020;15(8):646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- 56.Fausther-Bovendo H., Kobinger G.P. Pre-existing immunity against Ad vectors humoral, cellular, and innate response, what's important? Hum. Vacc. Immunother. 2014;10(10):2875–2884. doi: 10.4161/hv.29594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Let's talk about lipid nanoparticles, Nat. Rev. Mater. 6 (2021) 99–99 10.1038/s41578-021-00281-4. [DOI]
- 58.Zeltins A. Construction and characterization of virus-like particles: a review. Mol. Biotechnol. 2013;53(1):92–107. doi: 10.1007/s12033-012-9598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Medhi R., Srinoi P., Ngo N., Tran H.-V., Lee T.R. Nanoparticle-based strategies to combat COVID-19. ACS Appl. Nano Mater. 2020;3(9):8557–8580. doi: 10.1021/acsanm.0c01978. [DOI] [PubMed] [Google Scholar]
- 60.Ickenstein L.M., Garidel P. Lipid-based nanoparticle formulations for small molecules and RNA drugs. Expert Opin. Drug Deliv. 2019;16(11):1205–1226. doi: 10.1080/17425247.2019.1669558. [DOI] [PubMed] [Google Scholar]
- 61.Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G., Huang W.-J., Gao P., Zhou C., Zhang R.-R., Guo Y., Sun S.-H., Fan H., Zu S.-L., Chen Q.i., He Q.i., Cao T.-S., Huang X.-Y., Qiu H.-Y., Nie J.-H., Jiang Y., Yan H.-Y., Ye Q., Zhong X., Xue X.-L., Zha Z.-Y., Zhou D., Yang X., Wang Y.-C., Ying B.o., Qin C.-F. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182(5):1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu H., Chen Q.i., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X.-F., Li J., Zhang N.-N., Yang X., Chen S., Guo Y., Zhao G., Wang X., Luo D.-Y., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.-F., Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603–1607. doi: 10.1126/science:abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., Ojha A., Zhang L., Mu Z., Parks R., Manzoni T.B., Roper B., Strohmeier S., Tombácz I., Arwood L., Nachbagauer R., Karikó K., Greenhouse J., Pessaint L., Porto M., Putman-Taylor T., Strasbaugh A., Campbell T.-A., Lin P.J.C., Tam Y.K., Sempowski G.D., Farzan M., Choe H., Saunders K.O., Haynes B.F., Andersen H., Eisenlohr L.C., Weissman D., Krammer F., Bates P., Allman D., Locci M., Pardi N. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53(4):724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang F.Z., Kream R.M., Stefano G.B. An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med. Sci. Monitor. 2020;26 doi: 10.12659/MSM.924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nanomedicine and the COVID-19 vaccines, Nat. Nanotechnol. 15 (2020) 963–963 10.1038/s41565-020-00820-0. [DOI] [PMC free article] [PubMed]
- 66.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162B2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walsh E.E., Frenck R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Şahin U., Gruber W.C. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. N. Engl. J. Med. 2020;383(25):2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vogel A.B., Kanevsky I., Che Y.e., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., Loschko J., Maddur M.S., Ota-Setlik A., Tompkins K., Cole J., Lui B.G., Ziegenhals T., Plaschke A., Eisel D., Dany S.C., Fesser S., Erbar S., Bates F., Schneider D., Jesionek B., Sänger B., Wallisch A.-K., Feuchter Y., Junginger H., Krumm S.A., Heinen A.P., Adams-Quack P., Schlereth J., Schille S., Kröner C., de la Caridad Güimil Garcia R., Hiller T., Fischer L., Sellers R.S., Choudhary S., Gonzalez O., Vascotto F., Gutman M.R., Fontenot J.A., Hall-Ursone S., Brasky K., Griffor M.C., Han S., Su A.A.H., Lees J.A., Nedoma N.L., Mashalidis E.H., Sahasrabudhe P.V., Tan C.Y., Pavliakova D., Singh G., Fontes-Garfias C., Pride M., Scully I.L., Ciolino T., Obregon J., Gazi M., Carrion R., Alfson K.J., Kalina W.V., Kaushal D., Shi P.-Y., Klamp T., Rosenbaum C., Kuhn A.N., Türeci Ö., Dormitzer P.R., Jansen K.U., Sahin U. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592(7853):283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 69.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2-preliminary report. N. Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y.K., Barclay W.S., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeng C., Hou X., Yan J., Zhang C., Li W., Zhao W., Du S., Dong Y. Leveraging mRNA sequences and nanoparticles to deliver SARS-CoV-2 antigens in vivo. Adv. Mater. 2020;32(40):2004452. doi: 10.1002/adma.v32.4010.1002/adma.202004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang N., Shang J., Jiang S.B., Du L.Y. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kanekiyo M., Wei C.-J., Yassine H.M., McTamney P.M., Boyington J.C., Whittle J.R.R., Rao S.S., Kong W.-P., Wang L., Nabel G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499(7456):102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tokatlian T., Read B.J., Jones C.A., Kulp D.W., Menis S., Chang J.Y.H., Steichen J.M., Kumari S., Allen J.D., Dane E.L., Liguori A., Sangesland M., Lingwood D., Crispin M., Schief W.R., Irvine D.J. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science. 2019;363(6427):649–654. doi: 10.1126/science:aat9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanekiyo M., Bu W., Joyce M.G., Meng G., Whittle J.R., Baxa U., Yamamoto T., Narpala S., Todd J.-P., Rao S., McDermott A., Koup R., Rossmann M., Mascola J., Graham B., Cohen J., Nabel G. Rational design of an Epstein-Barr virus vaccine targeting the receptor-binding site. Cell. 2015;162(5):1090–1100. doi: 10.1016/j.cell.2015.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Powell A.E., Zhang K., Sanyal M., Tang S., Weidenbacher P.A., Li S., Pham T.D., Pak J.E., Chiu W., Kim P.S. A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Central. Sci. 2021;7(1):183–199. doi: 10.1021/acscentsci.0c01405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang W., Huang B., Zhu Y., Tan W., Zhu M. Ferritin nanoparticle-based SARS-CoV-2 RBD vaccine induces a persistent antibody response and long-term memory in mice. Cell Mol. Immunol. 2021;18(3):749–751. doi: 10.1038/s41423-021-00643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang B., Chao C.W., Tsybovsky Y., Abiona O.M., Hutchinson G.B., Moliva J.I., Olia A.S., Pegu A., Phung E., Stewart-Jones G.B.E., Verardi R., Wang L., Wang S., Werner A., Yang E.S., Yap C., Zhou T., Mascola J.R., Sullivan N.J., Graham B.S., Corbett K.S., Kwong P.D. A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-74949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma X., Zou F., Yu F., Li R., Yuan Y., Zhang Y., Zhang X., Deng J., Chen T., Song Z., Qiao Y., Zhan Y., Liu J., Zhang J., Zhang X.u., Peng Z., Li Y., Lin Y., Liang L., Wang G., Chen Y., Chen Q., Pan T., He X., Zhang H. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity. 2020;53(6):1315–1330.e9. doi: 10.1016/j.immuni.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tan T.K., Rijal P., Rahikainen R., Keeble A.H., Schimanski L., Hussain S., Harvey R., Hayes J.W.P., Edwards J.C., McLean R.K., Martini V., Pedrera M., Thakur N., Conceicao C., Dietrich I., Shelton H., Ludi A., Wilsden G., Browning C., Zagrajek A.K., Bialy D., Bhat S., Stevenson-Leggett P., Hollinghurst P., Tully M., Moffat K., Chiu C., Waters R., Gray A., Azhar M., Mioulet V., Newman J., Asfor A.S., Burman A., Crossley S., Hammond J.A., Tchilian E., Charleston B., Bailey D., Tuthill T.J., Graham S.P., Duyvesteyn H.M.E., Malinauskas T., Huo J., Tree J.A., Buttigieg K.R., Owens R.J., Carroll M.W., Daniels R.S., McCauley J.W., Stuart D.I., Huang K.-Y., Howarth M., Townsend A.R. A COVID-19 vaccine candidate using spycatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-020-20654-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Walls A.C., Fiala B., Schafer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O'Connor M.A., Chen C.B., et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382. doi: 10.1016/j.cell.2020.https://doi.org/10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peng S., Cao F., Xia Y., Gao X.-D., Dai L., Yan J., Ma G. Particulate alum via pickering emulsion for an enhanced COVID-19 vaccine adjuvant. Adv. Mater. 2020;32(40):2004210. doi: 10.1002/adma.v32.4010.1002/adma.202004210. [DOI] [PubMed] [Google Scholar]
- 84.Huang W.-C., Zhou S., He X., Chiem K., Mabrouk M.T., Nissly R.H., Bird I.M., Strauss M., Sambhara S., Ortega J., Wohlfert E.A., Martinez‐Sobrido L., Kuchipudi S.V., Davidson B.A., Lovell J.F. SARS-CoV-2 RBD neutralizing antibody induction is enhanced by particulate vaccination. Adv. Mater. 2020;32(50):2005637. doi: 10.1002/adma.v32.5010.1002/adma.202005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seyfoori A., Shokrollahi Barough M., Mokarram P., Ahmadi M., Mehrbod P., Sheidary A., Madrakian T., Kiumarsi M., Walsh T., McAlinden K.D., Ghosh C.C., Sharma P., Zeki A.A., Ghavami S., Akbari M. Emerging advances of nanotechnology in drug and vaccine delivery against viral associated respiratory infectious diseases (VARID) Int. J. Mol. Sci. 2021;22(13):6937. doi: 10.3390/ijms22136937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. MRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7(5):319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]