Summary
Deep learning (DL) models typically require large-scale, balanced training data to be robust, generalizable, and effective in the context of healthcare. This has been a major issue for developing DL models for the coronavirus disease 2019 (COVID-19) pandemic, where data are highly class imbalanced. Conventional approaches in DL use cross-entropy loss (CEL), which often suffers from poor margin classification. We show that contrastive loss (CL) improves the performance of CEL, especially in imbalanced electronic health records (EHR) data for COVID-19 analyses. We use a diverse EHR dataset to predict three outcomes: mortality, intubation, and intensive care unit (ICU) transfer in hospitalized COVID-19 patients over multiple time windows. To compare the performance of CEL and CL, models are tested on the full dataset and a restricted dataset. CL models consistently outperform CEL models, with differences ranging from 0.04 to 0.15 for area under the precision and recall curve (AUPRC) and 0.05 to 0.1 for area under the receiver-operating characteristic curve (AUROC).
Keywords: deep learning, machine learning, COVID-19, contrastive loss, electronic health records, recurrent neural network
Highlights
-
•
Contrastive loss consistently improves performance compared with cross-entropy loss
-
•
For imbalanced outcome data, only the contrastive loss model maintains proper clustering
-
•
Contrastive loss better identifies relevant features
The bigger picture
The abundance of health data provides exceptional opportunities for machine learning analyses to improve care in terms of enhanced screening, diagnosis, and prognosis. One such data type is electronic health records, which generally consist of demographics, diagnoses, laboratory tests, vital signs, medications, and clinical notes. While deep learning has emerged as a powerful analysis tool to process large-scale data by extracting useful patterns, creating robust and generalizable models with such data is notoriously challenging due to scale, complexity, and outcome imbalance. In this work, we develop and refine a new model architecture based on the recently proposed contrastive deep learning. As a relevant use case, we demonstrate the power of this framework for predicting critical events in coronavirus disease 2019 (COVID-19) patients as well as an enhanced ability to identify important features. Our work shows promise for datasets with high missingness and outcome imbalance that normally hinders performance.
Deep learning models applied on EHR data often utilize cross-entropy loss (CEL) as the primary optimization function, but CEL may not be suitable for real-world scenarios with imbalanced data. We develop a learning framework that incorporates both CEL and contrastive loss (CL) to tackle this issue. Our framework achieves better predictive performance and feature interpretability, particularly for imbalanced data.
Introduction
As of May 2021, coronavirus disease 2019 (COVID-19) has resulted in over 3.4 million reported deaths, with over 580,000 occurring in the United States and over 53,000 in New York State.1 Hospital resources such as medication supply and hospital beds quickly became constrained.2,3 Due to the novelty of the COVID-19 pandemic, there is a dearth of relevant data available for research purposes, so electronic health records (EHRs) became a valuable tool to study and characterize COVID-19 patients. EHRs have already been used in biomedical research for disease and patient clustering, patient trajectory modeling, disease prediction, and clinical decision support, among other things.4 Recent studies discovered important findings to better understand COVID-19 through EHR-based analyses.5, 6, 7, 8, 9, 10, 11
Machine learning (ML) is useful to examine resource allocation and risk stratification of patients, and ML models were successfully used to identify high-risk COVID-19 patients.12, 13, 14, 15, 16, 17, 18, 19, 20, 21
For temporal data modeling and prediction of patient outcomes in particular, deep learning (DL)22 holds promise over traditional ML techniques that require manual engineering of informative patient features.23, 24, 25 Heterogeneous graph networks are a powerful DL-based graph representation technique that have successfully been utilized in EHR-related studies.26,27 These models have a graphical structure that captures the underlying relationships between disparate medical concepts such as diagnoses and laboratory tests.28 Further, these graph convolutional models can be endowed with an attention mechanism29 to automatically identify how important local neighbors in the graph are to a given medical concept.30 Attention models such as reverse time attention (RETAIN) model provide a detailed interpretation of results and maintain high prediction accuracy by using reverse-time attention mechanisms to consolidate past visits.31
There are, however, substantial concerns about the limited generalizability of these models in COVID-19 (and in general) as they often underperform in external validation.32,33 Poor generalization of the models is normally due to underspecification.34 The underlying aspects of EHR data may also limit model effectiveness.35 Healthcare datasets often have inadequate sample sizes in terms of both small hospitals and rare disease populations and exhibit high class imbalance for key outcomes of interest, such as in rare events, as in the case with COVID-19. Several strategies have been utilized to mitigate these data and modeling challenges including up- and down-sampling, pre-training, transfer learning, and federated learning, but each has its limitations for use in EHR research.36 Other than these methods, the role of loss function has yet to be thoroughly investigated in the context of COVID-19 EHR work, which is the focus of this work. DL models often use cross-entropy loss (CEL) function, but CEL has been shown to potentially have poor classification margins, making the model less generalizable.37 Recently, supervised contrastive loss (CL) has been proposed to improve the classification performance of CEL.37 This original CL algorithm used sets of one anchor, many positives, and many negatives to maximize the similarities within the same class. Khosla et al.37 showed that CL is more general than triplet and N-pairs losses because, for any anchor, all positive pairs, including the augmentation of the anchor, are used for the loss. In addition, CL has a temperature parameter (τ) in loss that has been shown to improve learning. Triplet and N-pairs losses are special cases of supervised CL loss. When one positive and one negative pair are used, CL simplifies to triplet loss.38,39 When positive cases differ from anchor (i.e., excluding augmentations), more than one negative, and no τ are used, CL becomes equivalent to N-pairs loss.40,41
Although CL has already been applied to learn visual representations of medical images from paired images and text,42,43 it has not yet been used for EHR data and COVID-19 in particular. Here, we modify a CL-based architecture from its original formulation for EHR tasks. In the original work, CL was used for representation learning and CEL was used for the downstream classification. While we still utilize CL for patient representation learning, we explicitly incorporate a classification objective into our CL loss function. This additional term behaves similar to CEL and maximizes the similarities between patient representation (obtained from sequential models) and outcome representation (obtained from heterogeneous relational models).44
In this study, we compare different sequential models (RETAIN and recurrent neural network [RNN]) utilizing CEL and our developed CL in predicting critical outcomes of COVID-19 mortality, ICU transfers, and intubation for patients admitted to a large and diverse patient population from five hospitals within the Mount Sinai Health System in New York. Models are tested on a full dataset and a restricted dataset with severe class imbalance to better elucidate the impacts of these loss functions on model performance. Results are evaluated within the framework of three dimensions: predictive power, patient clustering, and feature importance.
Our main contributions can be summarized as:
-
1
We propose a novel deep learning framework for EHR data by adding a contrastive regularizer to CEL to improve the performance of prediction tasks.
-
2
We quantitatively examine the performance of CL for predicting critical events for COVID-19 patients.
-
3
We show the superior performance of CL framework compared with CEL and traditional ML algorithms, especially when data outcomes are more imbalanced.
-
4
We provide interpretability of these models that shows how feature importance becomes more clinically relevant using CL on datasets with imbalanced outcomes.
Results
Model comparison and evaluation of predictive performance
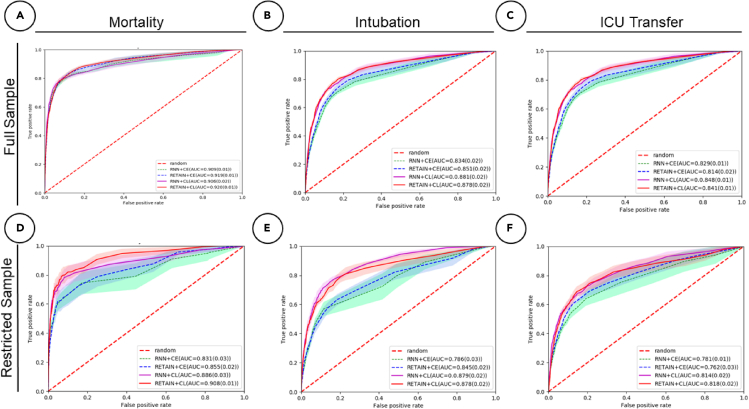
We first evaluate the results between loss functions using all data (i.e., full sample) for all three tasks, namely mortality, ICU transfer, and intubation prediction. The receiver operating characteristic (ROC) and precision-recall (PR) curves for these cross-validated results are shown in Figures 2A–2C and 3A–3C) and metrics are tabulated in Table 1. For mortality prediction (23% positive label percentage), we observe that the AUROC and AUPRC scores are similar between CEL and CL. For the RETAIN model, under 24-h prediction time frame, the AUROC and AUPRC scores are 0.92 ± 0.01, 0.82 ± 0.02 for CEL and 0.92 ± 0.01, 0.84 ± 0.02 for CL. These results indicate that two loss models achieve similar performance when we have relatively more balanced label ratios. For intubation prediction, the positive label percentage is 10.7%, which is less than half of the mortality label’s. We observe larger performance increases of 0.03 ± 0.02 for AUROC score and 0.09 ± 0.02 for AUPRC in CL compared with CEL for the RETAIN model. For ICU transfer prediction (17% positive label percentage) using RETAIN, we observe that the AUROC and AUPRC performances of CL are around 0.84 ± 0.01 and 0.60 ± 0.02, which are slightly higher than the CEL performances of 0.81 ± 0.01 and 0.57 ± 0.02.
Figure 2.
Receiver operating characteristic curves for all predictive tasks in a 24-h time frame
Performance is assessed for both contrastive loss (CL) and cross-entropy loss (CEL) for both RNN and RETAIN modeling strategies.
(A) Mortality with full dataset (23% positive labels).
(B) Intubation with full dataset (11% positive labels).
(C) ICU transfer with full dataset (17% positive labels).
(D) Mortality with restricted dataset (7% positive labels).
(E) Intubation with restricted dataset (5% positive labels).
(F) ICU transfer with restricted dataset (7% positive labels).
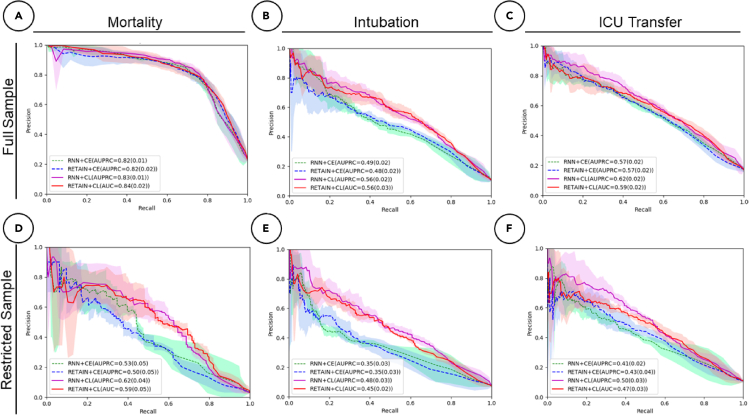
Figure 3.
PR curves for all event predictions in a 24-h time frame
Performance is assessed for both CL and CEL for both RNN and RETAIN modeling strategies.
(A) Mortality with full dataset (23% positive labels).
(B) Intubation with full dataset (11% positive labels).
(C) ICU transfer with full dataset (17% positive labels).
(D) Mortality with restricted dataset (7% positive labels).
(E) Intubation with restricted dataset (5% positive labels).
(F) ICU transfer with restricted dataset (7% positive labels).
Table 1.
Binary outcome prediction performance on different models using full sample data (positive label percentage: 23% for mortality, 10% for intubation, 17% for ICU transfer)
| Time window | Event | Model type | AUROC | AUPRC |
|---|---|---|---|---|
| 24 h | Mortality | LG | 0.85 (0.02) | 0.65 (0.01) |
| RF | 0.82 (0.02) | 0.63 (0.01) | ||
| SVM | 0.79 (0.02) | 0.61 (0.02) | ||
| XGB | 0.83 (0.02) | 0.65 (0.02) | ||
| RNN + CEL | 0.91 (0.01) | 0.82 (0.01) | ||
| RETAIN + CEL | 0.92 (0.01) | 0.82 (0.02) | ||
| RNN + CL | 0.91 (0.02) | 0.83 (0.01) | ||
| RETAIN + CL | 0.92∗ (0.01) | 0.84∗ (0.02) | ||
| Intubation | LG | 0.82 (0.01) | 0.47 (0.02) | |
| RF | 0.81 (0.01) | 0.46 (0.01) | ||
| SVM | 0.76 (0.02) | 0.42 (0.01) | ||
| XGB | 0.78 (0.02) | 0.46 (0.01) | ||
| RNN + CEL | 0.83 (0.02) | 0.49 (0.02) | ||
| RETAIN + CEL | 0.85 (0.02) | 0.48 (0.02) | ||
| RNN + CL | 0.91∗ (0.02) | 0.56∗ (0.02) | ||
| RETAIN + CL | 0.91 (0.02) | 0.56 (0.03) | ||
| ICU transfer | LG | 0.81 (0.01) | 0.52 (0.01) | |
| RF | 0.82 (0.02) | 0.55 (0.01) | ||
| SVM | 0.79 (0.01) | 0.52 (0.01) | ||
| XGB | 0.78 (0.02) | 0.55 (0.01) | ||
| RNN + CEL | 0.83 (0.01) | 0.57 (0.02) | ||
| RETAIN + CEL | 0.81 (0.02) | 0.57 (0.02) | ||
| RNN + CL | 0.86∗ (0.01) | 0.62∗ (0.02) | ||
| RETAIN + CL | 0.85 (0.01) | 0.59 (0.02) | ||
| 48 h | Mortality | LG | 0.85 (0.01) | 0.64 (0.02) |
| RF | 0.81 (0.01) | 0.61 (0.02) | ||
| SVM | 0.81 (0.01) | 0.63 (0.01) | ||
| XGB | 0.88 (0.01) | 0.69 (0.01) | ||
| RNN + CEL | 0.90 (0.02) | 0.82 (0.02) | ||
| RETAIN + CEL | 0.92 (0.01) | 0.83 (0.01) | ||
| RNN + CL | 0.92 (0.02) | 0.82 (0.01) | ||
| RETAIN + CL | 0.93∗ (0.01) | 0.84∗ (0.01) | ||
| Intubation | LG | 0.79 (0.01) | 0.45 (0.02) | |
| RF | 0.77 (0.02) | 0.44 (0.01) | ||
| SVM | 0.73 (0.01) | 0.39 (0.01) | ||
| XGB | 0.82 (0.01) | 0.49 (0.02) | ||
| RNN + CEL | 0.69 (0.04) | 0.40 (0.03) | ||
| RETAIN + CEL | 0.78 (0.03) | 0.39 (0.03) | ||
| RNN + CL | 0.86 (0.03) | 0.54 (0.02) | ||
| RETAIN + CL | 0.93∗ (0.01) | 0.51∗ (0.03) | ||
| ICU transfer | LG | 0.79 (0.02) | 0.50 (0.01) | |
| RF | 0.81 (0.01) | 0.54 (0.01) | ||
| SVM | 0.77 (0.02) | 0.49 (0.02) | ||
| XGB | 0.81 (0.01) | 0.57 (0.02) | ||
| RNN + CEL | 0.80 (0.01) | 0.54 (0.02) | ||
| RETAIN + CEL | 0.81 (0.02) | 0.52 (0.02) | ||
| RNN + CL | 0.83 (0.02) | 0.60 (0.02) | ||
| RETAIN + CL | 0.83∗ (0.01) | 0.59∗ (0.02) |
All predictions are calculated from 10-fold cross-validation, for which we record the mean value and standard deviation as confident intervals across folds. Asterisks (∗) indicate best model performance per event.
To further evaluate the above trends, we conducted an additional experiment to assess how CEL and CL functions perform on the same tasks with more imbalanced outcome ratios. This scenario may be the case in smaller hospital cohorts and for other outcomes. We perform random down-sampling on positive labels (i.e., restricted sample): for mortality, intubation, and ICU transfer prediction tasks, we randomly down-sampled the positive labels to 7%, 5%, and 7%, respectively. The ROC and PR curves are shown in Figures 2E, 2F, 3E, and 3F and the performance metrics are recorded in Table 2. The results of the experiment consistently show lower performance using CEL compared with CL. For the RETAIN model under a 24-h time frame, AUROC and AUPRC values are higher for CL than CEL for all outcomes. For instance, the AUROC increases from 0.80 ± 0.02 (CEL) to 0.88 ± 0.02 (CL) and AUPRC increases from 0.35 ± 0.03 (CEL) to 0.45 ± 0.02 (CL) for intubation prediction These finding show that, under cases with extremely unbalanced label data, models using CL perform better than using CEL.
Table 2.
Binary outcome prediction performance on different models using restricted sample data (positive label percentage: 7% for mortality, 5% for intubation, 7% for ICU transfer)
| Time window | Event | Model type | AUROC | AUPRC |
|---|---|---|---|---|
| 24 h | mortality | LG | 0.78 (0.02) | 0.51 (0.02) |
| RF | 0.61 (0.02) | 0.24 (0.03) | ||
| SVM | 0.60 (0.02) | 0.22 (0.02) | ||
| XGB | 0.65 (0.02) | 0.25 (0.01) | ||
| RNN + CEL | 0.83 (0.03) | 0.53 (0.05) | ||
| RETAIN + CEL | 0.86 (0.02) | 0.50 (0.05) | ||
| RNN + CL | 0.91∗ (0.03) | 0.62∗ (0.04) | ||
| RETAIN + CL | 0.91 (0.01) | 0.59 (0.05) | ||
| intubation | LG | 0.76 (0.02) | 0.33 (0.02) | |
| RF | 0.75 (0.01) | 0.34 (0.01) | ||
| SVM | 0.75 (0.02) | 0.32 (0.02) | ||
| XGB | 0.77 (0.02) | 0.39 (0.02) | ||
| RNN + CEL | 0.79 (0.03) | 0.35 (0.03) | ||
| RETAIN + CEL | 0.80 (0.02) | 0.35 (0.03) | ||
| RNN + CL | 0.88∗ (0.02) | 0.48∗ (0.03) | ||
| RETAIN + CL | 0.88 (0.02) | 0.45 (0.02) | ||
| ICU transfer | LG | 0.77 (0.01) | 0.39 (0.02) | |
| RF | 0.74 (0.01) | 0.36 (0.02) | ||
| SVM | 0.76 (0.02) | 0.38 (0.01) | ||
| XGB | 0.78 (0.02) | 0.36 (0.01) | ||
| RNN + CEL | 0.78 (0.01) | 0.41 (0.02) | ||
| RETAIN + CEL | 0.76 (0.03) | 0.43 (0.04) | ||
| RNN + CL | 0.86 (0.02) | 0.53∗ (0.03) | ||
| RETAIN + CL | 0.85∗ (0.02) | 0.51 (0.03) | ||
| 48 h | mortality | LG | 0.77 (0.02) | 0.33 (0.02) |
| RF | 0.57 (0.02) | 0.12 (0.02) | ||
| SVM | 0.62 (0.02) | 0.19 (0.03) | ||
| XGB | 0.69 (0.02) | 0.29 (0.02) | ||
| RNN + CEL | 0.85 (0.03) | 0.55 (0.04) | ||
| RETAIN + CEL | 0.90 (0.03) | 0.53 (0.03) | ||
| RNN + CL | 0.92∗ (0.03) | 0.63 (0.04) | ||
| RETAIN + CL | 0.91 (0.02) | 0.64∗ (0.04) | ||
| intubation | LG | 0.79 (0.01) | 0.36 (0.02) | |
| RF | 0.78 (0.02) | 0.35 (0.01) | ||
| SVM | 0.79 (0.01) | 0.34 (0.01) | ||
| XGB | 0.82 (0.02) | 0.40 (0.01) | ||
| RNN + CEL | 0.70 (0.03) | 0.34 (0.04) | ||
| RETAIN + CEL | 0.74 (0.02) | 0.31 (0.03) | ||
| RNN + CL | 0.83 (0.02) | 0.44 (0.02) | ||
| RETAIN + CL | 0.85∗ (0.02) | 0.44∗ (0.03) | ||
| ICU transfer | LG | 0.79 (0.02) | 0.41 (0.01) | |
| RF | 0.75 (0.01) | 0.35 (0.01) | ||
| SVM | 0.77 (0.01) | 0.37 (0.02) | ||
| XGB | 0.79 (0.01) | 0.41 (0.02) | ||
| RNN + CEL | 0.72 (0.04) | 0.38 (0.03) | ||
| RETAIN + CEL | 0.75 (0.04) | 0.43 (0.04) | ||
| RNN + CL | 0.82∗ (0.01) | 0.51∗ (0.02) | ||
| RETAIN + CL | 0.82 (0.02) | 0.50 (0.03) |
All predictions are calculated from 10-fold cross-validation, for which we record the mean value and standard deviation as confident intervals across folds. Asterisks (∗) indicate best model performance per event.
Our results show that CEL and CL have competitive performance in the full dataset but we generally find that, for all tasks in restricted datasets, models with CEL have lower performance compared with CL (Table 2). Other than the 24-h time frame, we also perform the same exact analysis for the 48-h time frame, which shows consistent trends. Finally, we use the RNN as the baseline model rather than the RETAIN model for all the predictions described earlier and our conclusions hold true (see Table 2).
We have also provided the results of using traditional ML algorithms in both tables. Our results generally show the superiority of DL with respect to these baselines.23
Identification of important clinical features
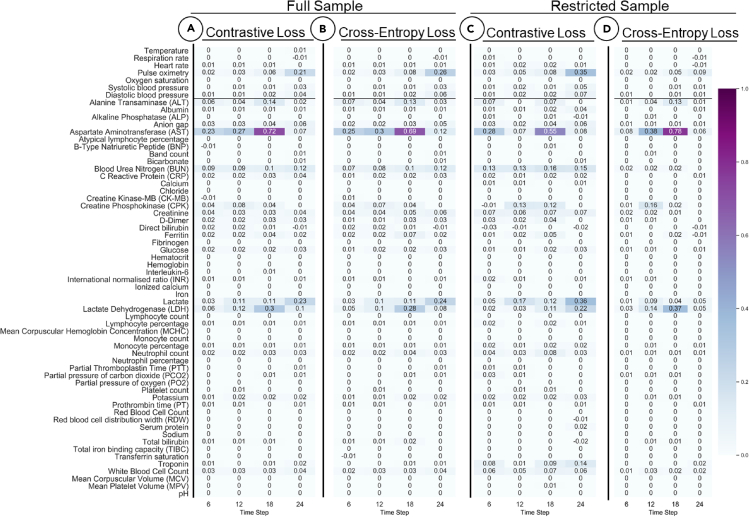
We assess the ability of CEL and CL functions in our models to identify relevant features of interest in full and restricted datasets. Specifically, we performed feature importance score calculations for the RETAIN model on predicting mortality as a representative case.
First, we generated the feature importance scores comparing two models (RETAIN-CL against RETAIN-CEL) over the 24-h time frame for the full dataset for mortality prediction. Figures 5A and 5B show the normalized feature importance heatmaps for CL and CEL with the columns representing four 6-h windows. These heatmaps display similar importance scores in terms of key features and their magnitudes. RETAIN identified laboratory tests and vital sign features that are considered important by both loss models: pulse oximetry (0.26 for CEL, 0.21 for CL); aspartate aminotransferase (0.69 for CEL, 0.72 for CL); blood urea nitrogen (0.12 for CEL, 0.12 for CL); lactate (0.24 for CEL, 0.23 for CL); lactate dehydrogenase (0.28 for CEL, 0.3 for CL). All these parameters indicate important aspects of an ill COVID-19 patient.
Figure 5.
Feature importance is predicted over four 6-h windows
(A) full sample withcontrastive loss (CL); (B) full sample with cross-entropy loss (CEL); (C) restrcited sample with CL; and (D) restrcited sample with CEL. The heat maps display similar importance scores in terms of key features and their magnitudes.
We then generated feature importance scores using two loss functions for mortality prediction in the restricted dataset (i.e., down-sampling the positive label to 7%) for the RETAIN model. We assessed how the variable importance score changes under these different conditions for different loss functions. The corresponding heatmap is plotted in Figures 5C and 5D. We observe that using CL can still capture the highly scored features identified in the full dataset (i.e., weigh similar key features). On the other hand, CEL fails to capture some important features. Of particular interest, the importance of pulse oximetry is no longer prioritized in the restricted sample using CL (importance value is 0.09 for CEL compared with 0.35 for CL). Also, blood urea nitrogen and lactate have lower importance values of 0.02 and 0.09 for CEL compared with 0.15 and 0.36 for CL. These findings reaffirm our hypothesis that CL is more robust when the outcome labels are highly imbalanced.
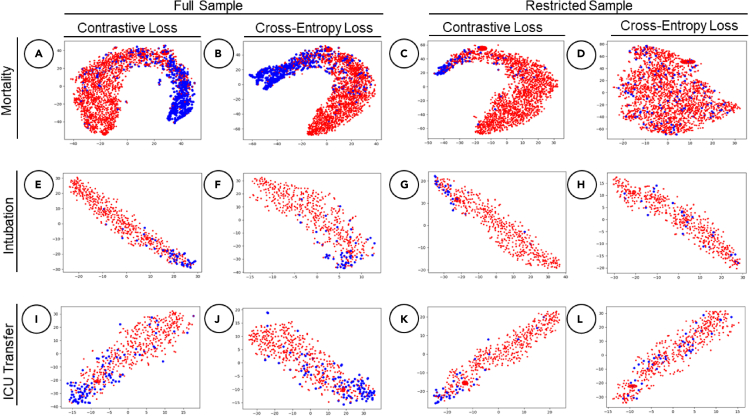
Visualizing patient embeddings
Finally, we generated two-dimensional t-distributed stochastic neighbor embedding (t-SNE) projections to compare patient embedding representations for RETAIN models between the CL and CEL in predicting all three medical events within 24-h intervals and the results are shown in Figure 4. The first two columns show patient embeddings using the full sample dataset, and the last two columns show embeddings for the restricted dataset. Blue dots represent positive labels and red dots represent negative labels. Models with both loss functions show clear clustering of positive and negative labels in the full datasets. However, when the dataset is restricted (i.e., lowered positive labels), the model with CEL consistently shows less clear patterns and poorer clustering of patients for all outcomes. In contrast, the RETAIN model with CL maintains its ability to group patients by their clinical outcomes.
Figure 4.
t-SNE latent embedding comparisons for all event predictions within a 24-h time frame using RETAIN
Blue dots represent positive labels and red dots represent negative labels. The plot is organized by outcome per row, namely first, mortality; second, intubation; and third, ICU transfer. The first and third columns represent CL plots and the second and fourth represent CEL.
(A) Mortality prediction with CL for the full dataset (23% positive labels).
(B) Mortality prediction with CL for the full dataset.
(C) Mortality prediction with CL for the restricted dataset (7% positive labels).
(D) Mortality prediction with CEL for the restricted dataset.
(E) Intubation prediction with CL for the full dataset (10% positive labels).
(F) Intubation prediction with CEL for the full dataset.
(G) Intubation prediction with CL for the restricted dataset (5% positive labels).
(H) Intubation prediction with CEL for the restricted dataset.
(I) ICU transfer prediction with CL for the full dataset (17% positive labels).
(J) ICU transfer prediction with CEL for the full dataset.
(K) ICU transfer prediction with CL for the restricted dataset (7% positive labels).
(L) ICU transfer prediction with CEL for the restricted dataset.
Discussion and conclusion
In this work, we develop a new DL model based on the CL for predicting three critical events (mortality, intubation, and ICU transfer) for COVID-19 patients using EHR data. Most DL-based EHR analyses utilize CEL as part of modeling. To the best of our knowledge, this is one of the first studies to demonstrate the utility of CL in EHR predictive analyses. We demonstrate the benefit of CL in multiple tasks with imbalanced outcome labels, which is particularly pertinent in the context of COVID-19. We compare the performance of different sequential architectures (RNN and RETAIN) for both CL and the conventional CEL model under two time window horizons. We also compare the performance of our developed framework with respect to traditional ML baselines. We conduct further experiments for each outcome in a restricted dataset of even more imbalanced outcomes and show the benefit of CL is even more pronounced via three separate experimental tasks, namely predictive performance, feature importance, and clustering.
The observed improvements in predictions come from the specific form of CL, which not only maximizes the similarity between patient and outcome embedding representations but also maximizes the similarities between patient representations related to a specific outcome. However, CEL mainly focuses on maximizing the similarity between patient and outcome one-hot representations. Therefore, CL tends to maximize the margins between classes better than CEL. The better margin classification ability of CL leads to higher performance for imbalanced data, for which the classification is difficult due to poor margin classification of CEL in addition to differences in data distributions for each class.
Our study contains several limitations that need to be addressed. First, we only compare CL with one other loss function, although it is widely used. Second, we only assess two sequential modeling techniques. Another main limitation of our study is excluding several laboratory values due to high levels of missingness, which may affect some of our interpretations. Furthermore, we utilize a specific time sequence modeling representation. Last, our work focuses only on one disease use case. For future studies, other modeling architecture can be assessed to evaluate the generalizability of our CL approach. Also, additional analysis is required to understand feature importance differences between the loss functions and to determine if our CL methodology is applicable to other healthcare datasets. We also plan to assess performance of this strategy for predictive tasks in other diseases, such as acute kidney injury. Another important investigation will be long-term predictions other than the 24- and 48-h windows studied in this work for longer-term critical events. We believe this work represents an effective demonstration of the power of using CL for ML work for predictive tasks using EHRs.
Experimental procedures
Resource availability
Lead contact
Any further information, questions, or requests should be sent to Benjamin S. Glicksberg (benjamin.glicksberg@mssm.edu)
Materials availability
Our study did not involve any physical materials.
Materials and methods
Clinical data and cohort
We obtained EHRs of COVID-19 patients from five hospitals within the Mount Sinai Healthcare System (MSHS). The collected EHR data contain the following information: COVID-19 status, demographics (age, gender, and race), 55 relevant laboratory test results (listed in Table S2), and vital signs, specifically heart rate, respiration rate, pulse oximetry, blood pressure (diastolic and systolic), temperature, height, weight, and 12 comorbidities (atrial fibrillation, asthma, coronary artery disease, cancer, chronic kidney disease, chronic obstructive pulmonary disease, diabetes mellitus, heart failure, hypertension, stroke, alcoholism, and liver disease). In addition, we collected information on clinically relevant outcomes of COVID-19: mortality, discharge, ICU transfer, and intubation. Laboratory tests and vital signs were measured at multiple time points along the hospital course. In our models, demographics features are used as static features.
Data pre-processing
We pre-process the vital signs, laboratory test, and static features by considering the values between 0.5 and 99.5 percentile to remove any inaccurate measurement. For any numerical data, we normalize the data by calculating the standard score (Z score). For categorical data, we use one-hot encoding representation. Numerical data with missing values are included with zeros.45
The initial feature input for vital signs at every time step is the vector representing the eight features (Figure 1A). Blood pressure is represented as two features: systolic and diastolic. Similar to vital signs, the initial feature input for laboratory tests is the vector representing the 55 features at each time step. If there is more than one vital sign or laboratory test for each patient, we average the values of the corresponding time step. We concatenate the vital sign vector and laboratory test vector to form the vector as the input at every time step; the subscripts v and l are dropped for simplicity.
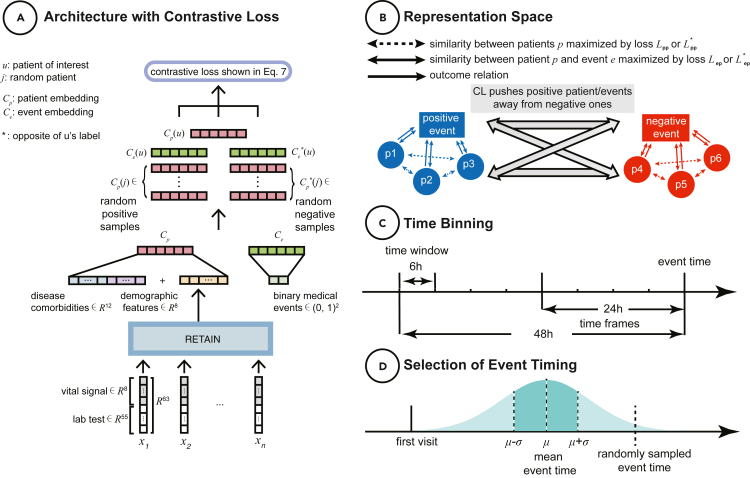
Figure 1.
Data and modeling schemas
(A) Architecture with CL. EHR data are modeled to create patient and event embedding representations, which are fed into our CL equation.
(B) Representation space. CL simultaneously pushes positive patients and event embeddings (i.e., concordant with respect to the outcome of the patient of interest, respectively) away from negative ones.
(C) Time binning. Schematic to visualize how we model time sequence. We have two outcome windows (i.e., 24and 48 h prior to event) and bin data by 6-h chunks.
(D) Selection of event timing for null outcomes. For patients that do not experience the outcome of interest, we generate a data-driven event time to align against as in (C). We compute the mean and standard deviation for the length of time that elapsed from admission for all patients with the affiliated outcomes independently. For patients without an event, we randomly pick a time to use as a reference end point using a Gaussian distribution with the mean and standard deviation obtained from the positive training data.
Static features consist of demographics (age, gender, and race) and disease comorbidity information, which are detailed in Table S2. For age, we record the normalized numerical value. We represent the gender feature as a two-dimensional (male and female) one-hot vector. We use a five-dimensional one-hot vector to represent the different groups for race (African American, white, Asian, other, and unknown). We represent disease comorbidity as a 12-dimensional one-hot vector. We concatenate all the demographic and disease comorbidity features into the vector .
Time sequence modeling
To model EHR data as a time sequence, we use a previously developed interpretable predictive model named RETAIN.31 This model is designed specifically to add feature explainability in terms of feature importance score on time sequence data.
For the RETAIN model, we present each patient for n time steps as in Choi et al.31:
| (Equation 1) |
where i denotes the time step, n is total number of time steps, is the attention vector of weights, is the attention weight for each feature, is a linear projection of the m dimensional input feature, and using projection matrix .
As another baseline architecture, we utilize a RNN model's Cp,seq with the original input feature vector :
| (Equation 2) |
We concatenate the static features vector Cp,static with the output of sequential models Cp,seq as illustrated in Figure 1A to get the final patient embedding
Our RNN architecture is based on long short-term memory (LSTM).
Heterogeneous relation modeling
Instead of treating the outcomes as labels, we model them as a directed heterogeneous bipartite graph as illustrated in Figure 1B. We create a triple relationship between patient, outcome, and event, where the outcome is the relation (or edge) between patient node p and event node e. Patient nodes with the same outcome relations are connected to the same event node. Since we are predicting binary outcomes, we have two event nodes representing positive and negative labels. Modeling the data as a bipartite graph provides both label information as well as event and clinical characteristic similarities.
Since the patient and event are two different node types, we use the heterogeneous relational model TransE44 to project patient and event node types and their outcome relationships into a shared latent space. The TransE model aims to relate different type of nodes by their relationship type and represents the relationship type (outcome) as a translation vector between the two node types. This relation is expressed as:
| (Equation 3) |
where Xe is the binary outcome representation, are learnable projection parameters for latent dimension l, are bias parameters, and δ is a non-linear activation function. We use a two-dimensional vector to represent positive or negative outcomes. is the latent representation of outcomes, Ce is translated representation from in the projection space by the learnable translation relational vector Ro, which is the relation vectors representing outcome relation that connects patients to positive event and negative event nodes, respectively.
After the projection, we apply similarity comparisons between these two representations (Cp and Ce) in the shared latent space.
Loss function delineation
After the assembly of the bipartite relational graph, we aim to predict the binary outcome of a patient by maximizing the similarity between the binary outcome latent representation and patient representation. The bipartite relational graph also considers the similarities within patient latent representations that connect to the same outcome. Therefore, the objective function is expressed as:
| (Equation 4) |
where u is the patient node of interest in the training sample, and Cp(u) is the latent representation of patient node u. Nc(u) is the patient node's neighboring nodes, which consist of binary outcome node representation Ce and similar patient nodes representations Cp. In Equation 4, we optimize the proximity between the representations of center patient node and its neighboring nodes.
The similarity between these latent representations is represented as an inner product, and we directly apply noise contrastive estimation (NCE) loss to capture the condition probability in Equation 4.46
| (Equation 5) |
where V is the training patients dataset, is the latent representation vector of j-th context node in , is the center node latent representation. K is the number of negative samples, and is the negative sampling distribution. are the co-occurrence positive representation pairs, and are the negative sampling pairs. The non-linear function captures the similarity score between the representation pairs.
We rewrite the objective function in Equation 5 in our own notations as follows:
| (Equation 6) |
where Cp is the projected latent representation of the binary outcome node that connects to a given patient representation Cp, and the inner product between Ce and Cp measures the similarity between these two representations. Superscript asterisks (∗) show the opposite outcome node of Cp and do not connect to the patient of interest u. Cp(j) is the similar context patient node representations that connect to the same outcome node as the patient node u. are the context patient node representations that connect to the opposite outcome node to the patient node.
The first two terms in Equation 6 capture the label information between outcome with the patient node, so they function as CEL. In the supplementary materials, we prove the lemma that the first two terms in Equation 6 are equivalent to the cross-entropy loss in general and therefore the improvements are primarily due to CL inter-patient terms. The last two terms provide additional information of similar patients that connects to the same outcome, and dissimilar patient information that connects to the other outcome.
We set a weight factor α to weigh the importance of the last two parts of Equation 6 that captures similar patient information, which is the main improvement due to CL. Our final objective function is as follows:
| (Equation 7) |
In this work, we use the optimal α = 0.8, which achieved the best performance for all models and tasks. We describe the experiments for exploring different values of α in Table S1.
After minimizing the CL from Equation 7, we obtain learned latent representation for events Ce, which are used to predict the probability of the events as follows:
| (Equation 8) |
where Ye represents the logit prediction for positive outcomes (mortality, intubation, and ICU transfer).
Feature importance scoring
The linear projection matrix Wp from the RETAIN model allows us to interpret variable importance at each time step. Our goal is to predict the probability of the outcome given a center patient representation. We can write this probability the same as Equation 8.
We can combine Equations 1, 3, and 8 to derive the similarity score as follows:
The contribution score for a specific feature k at time step i for input sample is derived as:
| (Equation 9) |
This is the similarity score between the positive outcome latent representation and patient latent representation for a RETAIN model with CL loss. The larger values of ω indicate that the feature k has a large contribution toward the prediction result.
For interpretability of a RETAIN model with CEL, we directly compute the importance score in a similar manner as in Choi et al.31:
| (Equation 10) |
where ym is the label for m-th sample.
Baselines
In this work, we are evaluating the performance of CL using two time-sequence models (RETAIN + CL and RNN + CL). As baseline models, we use the CEL with the same time-sequence models (RETAIN + CEL and RNN + CEL) to evaluate the potential improvements of CL.
As reference and comparison with the CL, the objective function for CEL is as follows:
| (Equation 11) |
where the logit output for the m-th sample is:
| (Equation 12) |
where Cp is the latent patient representation, and are the binary projection and bias parameters.We also compare the performance of the sequential models with respect to four traditional ML algorithms: logistic regression (LG), random forest (RF), support vector machine (SVM), and XGBoost (XGB).47
Experiment design
We perform three prediction tasks: mortality, ICU transfer, and intubation. We train our models on predicting events for two time frames: 24 and 48 h before the occurrence of a binary outcome. Longitudinal data (laboratory tests and vital signs) are binned within windows of 6 h and averaged if there is more than one measurement per window. The time binning representation is illustrated in Figure 1C. In the training phase, for each positive outcome, we utilize the exact time of an event to generate time frames for the experiments. For patients that did not have these outcome events, we needed a representative frame of reference to align against. Therefore, we compute the mean and standard deviation for the length of time that elapsed from admission for all the affiliated outcomes independently. For patients without an event, we randomly pick a time to use as a reference end point using a Gaussian distribution with the mean and standard deviation obtained from the positive training data. This procedure is shown in Figure 1D.
We also perform the comparisons of outcomes for both full sample and in artificial scenarios of more extreme imbalance (i.e., restricted sample) to determine the extent of performance differences between the two loss functions. In terms of generating the samples for any prediction using CEL, we find that a minimum of 5% positive labels is required to detect both negatives and positives. Therefore, we choose any positive label’s percentage to be greater than 5%. For the experiments with full sample, we use all available data where the percentages of positive labels is 23% mortality, 17% ICU transfer, and 10% intubation. For the experiments with restricted samples, we perform down-sampling to reduce the percentage of positive labels by randomly removing a percentage of positive labels. The percentages of positive labels in the down-sampled dataset are 7% mortality, 7% ICU transfer, and 5% intubation. In the restricted sample, the percentage of each positive label is less than half of the original positive label percentage.
Training details
In the training stage, after picking a patient node u, we select the binary outcome node that connects to u. Then we uniformly pick m similar patient nodes that also connect to this outcome node, and we use these samples as positive training pairs. For negative sampling, we first pick the binary outcome node that does not connect to the patient node u, and we then uniformly pick q similar patient nodes that connect to this outcome node. We utilize these samples as the negative training pairs. In this work, we use m = 2 and q = 2 to prioritize the positive samples.
For validation purposes, we perform 10-fold cross-validation for the patient of interest and record the mean evaluation values across 10 folds to determine the performance of the CL model against the CEL model.
All models and datasets are evaluated using the following metrics: area under the receiver-operating characteristic (AUROC) and area under the precision and recall curve (AUPRC). It is important to note that AUPRC is a more reliable metric for imbalanced samples because that takes into account negative predictive value (NPV) and positive predictive value (PPV).48
Acknowledgments
We thank the Clinical Data Science and Mount Sinai Data Warehouse teams for providing data. We appreciate all of the providers who contributed to the care of these patients. This work was supported by U54 TR001433-05, National Center for Advancing Translational Sciences, National Institutes of Health. Y.D. acknowledges support from Amazon Machine Learning Research Award. F.W. and C.Z. acknowledge the support from NSF 1750326 and 2027970, as well as the AWS Machine Learning for Research Award and Google Faculty Research Award.
Author contributions
T.W., F.W., A.Z., Y.D., and B.S.G. designed the study. T.W., H.H., and C.Z. performed the analyses. T.W., S.K.J., N.N., S.S., J.K.D.F., I.P., A.V., R.M., and G.N.N. processed the data. T.W., H.H., R.M., M.Z., Z.W., A.A., F.W., Y.D., and B.S.G. interpreted the results. A.A., F.W., Y.D., and B.S.G. supervised the work. All authors edited and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2021.100389.
Supplemental information
Data and code availability
Our data are not available due to institutional review board (IRB) rules for privacy protection. For reproducibility, our code is available at https://github.com/Tingyiwanyan/CL_covid and runs with TensorFlow 1.15.
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson C.N., Baumgartner J., Pichardo C., Toro B., Li L., Arciuolo R., Chan P.Y., Chen J., Culp G., Davidson A., et al. COVID-19 outbreak – New York City, February 29–June 1, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1725–1729. doi: 10.15585/mmwr.mm6946a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMahon D.E., Peters G.A., Ivers L.C., Freeman E.E. Global resource shortages during COVID-19: bad news for low-income countries. PLoS Negl. Trop. Dis. 2020;14:e0008412. doi: 10.1371/journal.pntd.0008412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glicksberg B.S., Johnson K.W., Dudley J.T. The next generation of precision medicine: observational studies, electronic health records, biobanks and continuous monitoring. Hum. Mol. Genet. 2018;27:R56–R62. doi: 10.1093/hmg/ddy114. [DOI] [PubMed] [Google Scholar]
- 5.Clifford C.T., Pour T.R., Freeman R., Reich D.L., Glicksberg B.S., Levin M.A., Klang E. Association between COVID-19 diagnosis and presenting chief complaint from New York City triage data. Am. J. Emerg. Med. 2020;46:520–524. doi: 10.1016/j.ajem.2020.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reeves J.J., Hollandsworth H.M., Torriani F.J., Taplitz R., Abeles S., Tai-Seale M., Millen M., Clay B.J., Longhurst C.A. Rapid response to COVID-19: health informatics support for outbreak management in an academic health system. J. Am. Med. Inform Assoc. 2020;27:853–859. doi: 10.1093/jamia/ocaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Somani S.S., Richter F., Fuster V., De Freitas J.K., Naik N., Sigel K., Mount Sinai C.I.C., Bottinger E.P., Levin M.A., Fayad Z., et al. Characterization of patients who return to hospital following discharge from hospitalization for COVID-19. J. Gen. Intern. Med. 2020;35:2838–2844. doi: 10.1007/s11606-020-06120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner T., Shweta F., Murugadoss K., Awasthi S., Venkatakrishnan A.J., Bade S., Puranik A., Kang M., Pickering B.W., O'Horo J.C., et al. Augmented curation of clinical notes from a massive EHR system reveals symptoms of impending COVID-19 diagnosis. eLife. 2020;9:e58227. doi: 10.7554/eLife.58227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z., Zheutlin A., Kao Y.H., Ayers K., Gross S., Kovatch P., Nirenberg S., Charney A., Nadkarni G., De Freitas J.K., et al. Hospitalised COVID-19 patients of the Mount Sinai Health System: a retrospective observational study using the electronic medical records. BMJ Open. 2020;10:e040441. doi: 10.1136/bmjopen-2020-040441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E., Curtis H.J., Mehrkar A., Evans D., Inglesby P., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 12.Yang H.S., Hou Y., Vasovic L.V., Steel P.A., Chadburn A., Racine-Brzostek S.E., Velu P., Cushing M.M., Loda M., Kaushal R. Routine laboratory blood tests predict SARS-CoV-2 infection using machine learning. Clin. Chem. 2020;66:1396–1404. doi: 10.1093/clinchem/hvaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., Bonten M.M., Dahly D.L., Damen J.A., Debray T.P. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;2020:369. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang W., Liang H., Ou L., Chen B., Chen A., Li C., Li Y., Guan W., Sang L., Lu J. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern. Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gysi D.M., Valle Í.D., Zitnik M., Ameli A., Gan X., Varol O., Sanchez H., Baron R.M., Ghiassian D., Loscalzo J. 2020. Network Medicine Framework for Identifying Drug Repurposing Opportunities for Covid-19.http://arxiv.org/abs/200407229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., Sun C., Tang X., Jing L., Zhang M. An interpretable mortality prediction model for COVID-19 patients. Nat. Machine Intelligence. 2020;2:1–6. [Google Scholar]
- 17.Vaishya R., Javaid M., Khan I.H., Haleem A. Artificial intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:337–339. doi: 10.1016/j.dsx.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei X., Lee H.-C., Diao K.-y, Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020:1–5. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaid A., Somani S., Russak A.J., De Freitas J.K., Chaudhry F.F., Paranjpe I., Johnson K.W., Lee S.J., Miotto R., Richter F. Machine learning to predict mortality and critical events in a cohort of patients with COVID-19 in New York City: model development and validation. J. Med. Internet Res. 2020;22:e24018. doi: 10.2196/24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alimadadi A., Aryal S., Manandhar I., Munroe P.B., Joe B., Cheng X. American Physiological Society; 2020. Artificial Intelligence and Machine Learning to Fight COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., Wang F., Tang J., Nussinov R., Cheng F. Artificial intelligence in COVID-19 drug repurposing. The Lancet Digital Health. 2020;2:e667. doi: 10.1016/S2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landi I., Glicksberg B.S., Lee H.C., Cherng S., Landi G., Danieletto M., Dudley J.T., Furlanello C., Miotto R. Deep representation learning of electronic health records to unlock patient stratification at scale. NPJ Digit Med. 2020;3:96. doi: 10.1038/s41746-020-0301-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi E., Bahadori M.T., Schuetz A., Stewart W.F., Sun J. Machine Learning for Healthcare Conference. 2016. Doctor ai: predicting clinical events via recurrent neural networks; pp. 301–318. [PMC free article] [PubMed] [Google Scholar]
- 24.Miotto R., Li L., Kidd B.A., Dudley J.T. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Scientific Rep. 2016;6:1–10. doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shickel B., Tighe P.J., Bihorac A., Rashidi P. Deep EHR: a survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J. Biomed. Health Inform. 2017;22:1589–1604. doi: 10.1109/JBHI.2017.2767063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi E., Bahadori M.T., Song L., Stewart W.F., Sun J. Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2017. GRAM: graph-based attention model for healthcare representation learning; pp. 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi E., Xu Z., Li Y., Dusenberry M., Flores G., Xue E., Dai A. Proceedings of the AAAI Conference on Artificial Intelligence. 2020. Learning the graphical structure of electronic health records with graph convolutional transformer; pp. 606–613. [Google Scholar]
- 28.Wanyan T., Kang M., Badgeley M.A., Johnson K.W., De Freitas J.K., Chaudhry F.F., Vaid A., Zhao S., Miotto R., Nadkarni G.N. International Conference on Artificial Intelligence in Medicine. Springer; 2020. Heterogeneous graph embeddings of electronic health records improve critical care disease predictions; pp. 14–25. [Google Scholar]
- 29.Veličković P., Cucurull G., Casanova A., Romero A., Lio P., Bengio Y. 2017. Graph Attention Networks.http://arxiv.org/abs/171010903 [Google Scholar]
- 30.Wang X., Ji H., Shi C., Wang B., Ye Y., Cui P., Yu P.S. The World Wide Web Conference. 2019. Heterogeneous graph attention network; pp. 2022–2032. [Google Scholar]
- 31.Choi E., Bahadori M.T., Sun J., Kulas J., Schuetz A., Stewart W., Retain An interpretable predictive model for healthcare using reverse time attention mechanism. Adv. Neural Inf. Process. Syst. 2016;29:3504–3512. [Google Scholar]
- 32.Barish M., Bolourani S., Lau L.F., Shah S., Zanos T.P. External validation demonstrates limited clinical utility of the interpretable mortality prediction model for patients with COVID-19. Nat. Machine Intelligence. 2020;1:1–3. [Google Scholar]
- 33.Gupta R.K., Marks M., Samuels T.H., Luintel A., Rampling T., Chowdhury H., Quartagno M., Nair A., Lipman M., Abubakar I. Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: an observational cohort study. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.03498-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Amour A., Heller K., Moldovan D., Adlam B., Alipanahi B., Beutel A., Chen C., Deaton J., Eisenstein J., Hoffman M.D. 2020. Underspecification Presents Challenges for Credibility in Modern Machine Learning.http://arxiv.org/abs/201103395 [Google Scholar]
- 35.Cyganek B., Graña M., Krawczyk B., Kasprzak A., Porwik P., Walkowiak K., Woźniak M. A survey of big data issues in electronic health record analysis. Appl. Artif. Intelligence. 2016;30:497–520. [Google Scholar]
- 36.Xu J., Glicksberg B.S., Su C., Walker P., Bian J., Wang F. Federated learning for healthcare informatics. J. Healthc. Inform Res. 2020:1–19. doi: 10.1007/s41666-020-00082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khosla P., Teterwak P., Wang C., Sarna A., Tian Y., Isola P., Maschinot A., Liu C., Krishnan D. 2020. Supervised Contrastive Learning.http://arxiv.org/abs/2004.11362 [Google Scholar]
- 38.Xu J., Xu Z., Walker P., Wang F. Proceedings of the AAAI Conference on Artificial Intelligence. 2020. Federated patient hashing; pp. 6486–6493. [Google Scholar]
- 39.Xu J., Xu Z., Yu B., Wang F. 2020 IEEE International Conference on Data Mining (ICDM) IEEE; 2020. Order-preserving metric learning for mining multivariate time series; pp. 711–720. [Google Scholar]
- 40.Zhang X., He L., Chen K., Luo Y., Zhou J., Wang F. AMIA Annual Symposium Proceedings. American Medical Informatics Association; 2018. Multi-view graph convolutional network and its applications on neuroimage analysis for Parkinson’s disease; p. 1147. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X., Chou J., Wang F. 2018 IEEE International Conference on Data Mining (ICDM) IEEE; 2018. Integrative analysis of patient health records and neuroimages via memory-based graph convolutional network; pp. 767–776. [Google Scholar]
- 42.Li Z., Roberts K., Jiang X., Long Q. Distributed learning from multiple EHR databases: contextual embedding models for medical events. J. Biomed. Inform. 2019;92:103138. doi: 10.1016/j.jbi.2019.103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Jiang H., Miura Y., Manning C.D., Langlotz C.P. 2020. Contrastive Learning of Medical Visual Representations from Paired Images and Text.http://arxiv.org/abs/2010.00747 [Google Scholar]
- 44.Bordes A., Usunier N., Garcia-Durán A., Weston J., Yakhnenko O. Proceedings of the 26th International Conference on Neural Information Processing Systems - Volume 2. Curran Associates Inc.; 2013. Translating embeddings for modeling multi-relational data; pp. 2787–2795. [Google Scholar]
- 45.Lipton Z.C., Kale D.C., Wetzel R. Modeling missing data in clinical time series with RNNs. Machine Learn. Healthc. 2016;56 https://arxiv.org/abs/1606.04130 [Google Scholar]
- 46.Levy O., Goldberg Y. Neural word embedding as implicit matrix factorization. Adv. Neural Inf. Process. Syst. 2014;27:2177–2185. [Google Scholar]
- 47.Rajkomar A., Dean J., Kohane I. Machine learning in medicine. New Engl. J. Med. 2019;380:1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 48.Saito T., Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015;10:e0118432. doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our data are not available due to institutional review board (IRB) rules for privacy protection. For reproducibility, our code is available at https://github.com/Tingyiwanyan/CL_covid and runs with TensorFlow 1.15.